Main Text
In vivo genome editing with CRISPR-Cas9 systems is garnering widespread attention and enthusiasm for possible treatment of genetic disorders. However, the consequences of potential immunogenicity of the bacterial Cas9 protein have been the subject of considerable speculation. In fact, a fraction of the human population is positive for pre-existing immunity to the most commonly used Cas9 variants, presumably a result of exposure to these microbes.1, 2, 3, 4 In vivo delivery of CRISPR-Cas9 is typically achieved with adeno-associated viral (AAV) vectors, which persist long-term in post-mitotic cells, further elevating concerns regarding sustained exposure to the foreign Cas9 antigen. Adaptive immune responses to Cas9 following AAV-CRISPR treatment have been observed in various mouse models with no pre-existing Cas9 immunity, but, interestingly, this does not seem to be an obstacle to long-term persistence of edited cells in these systems.5, 6, 7, 8 However, early studies in mouse models have indicated that pre-existing immunity to Cas9 could limit gene editing efficacy.8 In this issue of Molecular Therapy, Li et al.9 provide valuable insights to this topic, although it is also important to acknowledge the caveats and limitations of this study in the broader context of therapeutic genome editing.
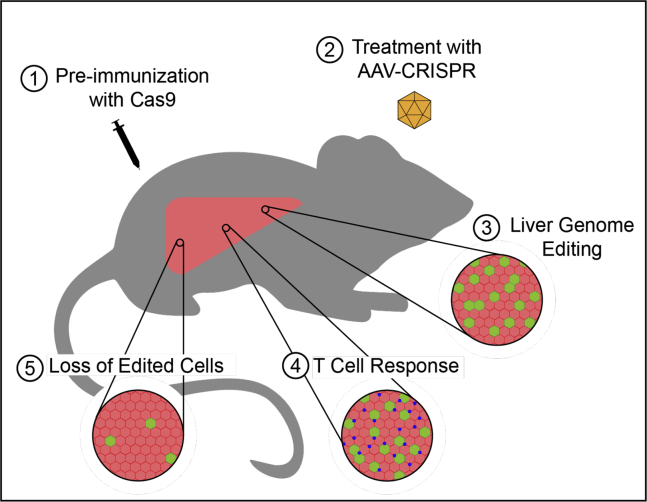
Li et al.9 made an important step toward understanding how pre-existing immunity to Cas9 may affect genome editing outcomes. They accomplished this by pre-immunizing mice with subcutaneous injections of Staphylococcus aureus Cas9 (SaCas9) protein before subsequent genome editing in the liver by intraperitoneal injection of AAV-CRISPR carrying SaCas9 and gRNA 1 week later (Figure 1). While gene editing was readily detected in the livers of mice with or without SaCas9 immunization, in the mice immunized with SaCas9 there was also strong a cytotoxic T cell response and the percentage of edited cells diminished after 12 weeks. This indicates that exposure to a controlled amount of Cas9 protein, in a format that essentially serves as a vaccine, is an obstacle to achieving therapeutic levels of Cas9 genome editing in the liver of this preclinical model. However, the mechanisms by which humans are exposed to and develop immunity to Cas9 are likely diverse, and the exact frequency of anti-Cas9 immunity in the human population is still unclear. Consequently, this study underscores that further investigation is needed to fully characterize this cellular response in the diverse applications for which AAV-CRISPR is being developed.
Figure 1.
A Model System of Pre-existing Immunity to Cas9
Systemic AAV delivery of CRISPR-Cas9 and gRNA is a well-established method to achieve stable genome editing in the livers of mice. However, the stability of that editing is unknown in the context of pre-existing immunity to Cas9. Li et al.9 found that when mice are immunized with Cas9 protein prior to AAV-CRISPR treatment, the initial genome editing is followed by a T cell response that results in loss of the edited cells.
Of the two most frequently used Cas9 orthologs, Streptococcus pyogenes (SpCas9) and SaCas9, SaCas9 is often used for in vivo applications because its smaller size is more compatible with the limits of AAV packaging.10 Pre-clincial studies with SaCas9 have been perfomed in heart and skeletal muscle for Duchenne muscular dystrophy, the liver for a variety of metabolic and cardiovascular diseases, the CNS for neurodegenerative conditions, and the retina for forms of congenital blindness.6,11, 12, 13, 14 An SaCas9-based therapy delivered to the retina with AAV5 is currently in clinical trials for treatment of Leber congenital amaurosis type 10.15 These various in vivo applications have demonstrated AAV-CRISPR to be a relatively efficient method to provide long-term correction of disease-causing mutations.
Despite the rapid pace of development for AAV-CRISPR therapies, some concerns remain regarding their general safety and efficacy. At the top of the list are how the immune system will interact with the components of such a therapy and what effect it will have on both safety and efficacy. Our current understanding of this process is informed by decades of research in gene therapy. As the use of AAV for gene replacement is a step ahead of its use for genome editing, we have an expanding understanding of how the immune system responds to the influx of diverse viral capsids.16 Pre-existing adaptive immunity to AAV in the form of neutralizing antibodies can prevent viral transduction, and their presence is currently a disqualifying factor for participation in many gene therapy clinical trials. Cellular responses to the AAV capsid are being managed clinically with transient immunosuppression. However, it has remained unclear whether the stable expression of the bacterial Cas9 protein will be tolerated.
Streptococcus pyogenes is a common human pathogen and Staphylococcus aureus is a commensal species and opportunistic pathogen, so recent reports of pre-existing immunity to SpCas9 and SaCas9 in human populations were not entirely unexpected.1, 2, 3, 4 Charlesworth et al.1 reported antibodies against SpCas9 and SaCas9 in 58% and 78% of blood donors, respectively, and anti-SpCas9 and anti-SaCas9 T cells in 67% and 78% of donors, respectively. Wagner et al.2 and Ferdosi et al.3 identified anti-SpCas9 antibodies in 85% and 5% of donors, respectively, as well as anti-SpCas9 T cells. Simhadri et al.4 found the prevalence of anti-SaCas9 and anti-SpCas9 antibodies to be 10% and 2.5%, respectively. These disparate studies demonstrate the need for further research that takes into account the characteristics of populations tested, the assay design and sensitivity, and the possibility of cross-reactivity between Cas9 orthologs and other antigens to fully understand the prevalence of pre-existing anti-Cas9 immunity. While these studies investigate the frequency of potential patients with previous exposure to Cas9, we must learn how this pre-existing immunity affects the ability to safely deliver Cas9 and stably edit the genome in order to fully understand the scale and scope of this challenge.
To this point, Li et al.9 use a simple experimental setup in which mice are exposed to SaCas9 protein before AAV-CRISPR treatment. The authors determine the effect of previous exposure on subsequent genome editing and the cellular immune response. As mice raised in pathogen-free facilities should not have been previously exposed to SaCas9, mice in the study were immunized against 25 μg SaCas9 protein or 100 μg ovalbumin control. One week later, the mice were treated with an AAV8 vector encoding SaCas9 driven by a liver-specific HLP promoter and a U6 promoter driving a gRNA targeting the low-density lipoprotein receptor (Ldlr) gene, a non-essential gene in the liver. Intrahepatic T cells were analyzed at time points up to 12 weeks post-AAV administration. The proportion of CD8+ T cells to total T cells increased in the SaCas9-immunized mice compared to the ovalbumin control at weeks 1 through 4 post-administration, with a peak at week 2. This CD8+ T cell infiltration was accompanied by an increase in serum alanine transaminase (ALT) activity, a marker of liver injury, and apoptotic cells in the liver, followed by a period of hepatocyte proliferation.
A second AAV vector was also co-injected with the AAV-CRISPR vector, expressing a GFP reporter to track transduced cells. To further understand the impact of the CD8+ T cell response, the number of copies of both AAV genomes in the liver was measured by qPCR at each time point. While the copies of both genomes were sustained for the first 6 weeks, there was a >35-fold drop in both the AAV-CRISPR and AAV-GFP genomes between weeks 6 and 12 in the SaCas9-immunized mice compared to ovalbumin-immunized mice. The loss of AAV-CRISPR vector genomes between weeks 6 and 12 is proposed to be due to clearance of SaCas9-expressing cells and their replacement by proliferating non-transduced cells, but it could also be due to the proliferation of transduced cells and the dilution of episomes. To clarify this point, the editing frequency of the Ldlr gene was quantified at each time point. Consistent with the quantification of AAV-CRISPR vector genomes, the percent of edited alleles was stable or increasing for the first 6 weeks, followed by a sharp decline between weeks 6 and 12 from about 20% to 3%. The loss of both AAV-CRISPR genomes and edited cells supports the claim that cells transduced and edited by SaCas9 were cleared between weeks 6 and 12 and replaced by unedited cells.
As one of the first studies to begin characterizing the effects of previous exposure to SaCas9 on subsequent genome editing in the liver, these results indicate that anti-Cas9 immunity could pose a challenge to AAV-CRISPR therapies. But they also highlight the need for additional studies. Testing in this study was limited to a single gRNA that targets a single gene in a single organ with a single promoter and a single AAV capsid, and the immune response could manifest differently in other contexts, though the results are supported by previous observations.8 The major loss of edited cells in this study occurred between weeks 6 and 12, and further resolution of the changes during that time, and past 12 weeks, would help to further characterize this response. Additional assays, such as ELISpot to verify the specificity of the T cell response to SaCas9, and T cell receptor profiling to understand immunogenic epitopes, would be valuable to inform potential strategies to circumvent this response. A significant challenge for the field will be understanding the differences in the naturally occurring pre-existing immunity found in humans and the SaCas9 vaccination model used here. Notably, a recent study also showed that vaccination with AAV-CRISPR limited the level of gene editing when those mice were re-dosed with subsequent AAV-CRISPR via a distinct AAV capsid, presumably due to the anti-Cas9 immunity.8 Finally, an important and obvious next step is extending these studies to large animal models with immune systems that better recapitulate human biology.
Overcoming the responses of the immune system is well-trodden ground for the gene therapy field, as demonstrated by the emerging successes of AAV-based gene transfer. If additional research shows that pre-existing immunity is a major hurdle for AAV-CRISPR therapies, a variety of strategies for evading the immune response could be implemented. Minimizing the duration of Cas9 expression should diminish the cellular response and could be achieved with alternative transient delivery vehicles17 or self-cleaving systems.18 CRISPR-Cas9 systems from exotic species not typically seen by humans could provide a workaround. Alternatively, current Cas9 systems could be engineered to remove immunogenic epitopes.3 Interestingly, the human pre-existing immunity to Cas9 includes Cas9-reactive regulatory T cells,2 and these cells were also elevated in the livers treated with AAV-CRISPR in this study,9 indicating a possible mechanism for inducing tolerance to Cas9. As transient immunosuppression is also now a standard addition to AAV administration for current gene therapies to avoid anti-capsid responses, it would likely also help in ameliorating the response to Cas9.
The dream of stably altering the genome in vivo for therapeutic effect is not going to be achieved without challenges, and only by doing the difficult work to shine light on those challenges can the field overcome them.
References
- 1.Charlesworth C.T., Deshpande P.S., Dever D.P., Camarena J., Lemgart V.T., Cromer M.K., Vakulskas C.A., Collingwood M.A., Zhang L., Bode N.M. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 2019;25:249–254. doi: 10.1038/s41591-018-0326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagner D.L., Amini L., Wendering D.J., Burkhardt L.M., Akyüz L., Reinke P., Volk H.D., Schmueck-Henneresse M. High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nat. Med. 2019;25:242–248. doi: 10.1038/s41591-018-0204-6. [DOI] [PubMed] [Google Scholar]
- 3.Ferdosi S.R., Ewaisha R., Moghadam F., Krishna S., Park J.G., Ebrahimkhani M.R., Kiani S., Anderson K.S. Multifunctional CRISPR-Cas9 with engineered immunosilenced human T cell epitopes. Nat. Commun. 2019;10:1842. doi: 10.1038/s41467-019-09693-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simhadri V.L., McGill J., McMahon S., Wang J., Jiang H., Sauna Z.E. Prevalence of Pre-existing Antibodies to CRISPR-Associated Nuclease Cas9 in the USA Population. Mol. Ther. Methods Clin. Dev. 2018;10:105–112. doi: 10.1016/j.omtm.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson C.E., Wu Y., Gemberling M.P., Oliver M.L., Waller M.A., Bohning J.D., Robinson-Hamm J.N., Bulaklak K., Castellanos Rivera R.M., Collier J.H. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat. Med. 2019;25:427–432. doi: 10.1038/s41591-019-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thakore P.I., Kwon J.B., Nelson C.E., Rouse D.C., Gemberling M.P., Oliver M.L., Gersbach C.A. RNA-guided transcriptional silencing in vivo with S. aureus CRISPR-Cas9 repressors. Nat. Commun. 2018;9:1674. doi: 10.1038/s41467-018-04048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chew W.L., Tabebordbar M., Cheng J.K., Mali P., Wu E.Y., Ng A.H., Zhu K., Wagers A.J., Church G.M. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat. Methods. 2016;13:868–874. doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreno A.M., Palmer N., Alemán F., Chen G., Pla A., Jiang N., Leong Chew W., Law M., Mali P. Immune-orthogonal orthologues of AAV capsids and of Cas9 circumvent the immune response to the administration of gene therapy. Nat. Biomed. Eng. 2019;3:806–816. doi: 10.1038/s41551-019-0431-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li A., Tanner M.R., Lee C.M., Hurley A.E., De Giorgi M., Jarrett K.E., Davis T.H., Doerfler A.M., Bao G., Beeton C., Lagor W.R. AAV-CRISPR Gene Editing Is Negated by Pre-existing Immunity to Cas9. Mol Ther. 2020;28:1432–1441. doi: 10.1016/j.ymthe.2020.04.017. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Castellanos Rivera R.M., Madhavan S., Pan X., Ran F.A., Yan W.X. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y., Wang L., Bell P., McMenamin D., He Z., White J., Yu H., Xu C., Morizono H., Musunuru K. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol. 2016;34:334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ekman F.K., Ojala D.S., Adil M.M., Lopez P.A., Schaffer D.V., Gaj T. CRISPR-Cas9-Mediated Genome Editing Increases Lifespan and Improves Motor Deficits in a Huntington’s Disease Mouse Model. Mol. Ther. Nucleic Acids. 2019;17:829–839. doi: 10.1016/j.omtn.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCullough K.T., Boye S.L., Fajardo D., Calabro K., Peterson J.J., Strang C.E., Chakraborty D., Gloskowski S., Haskett S., Samuelsson S. Somatic Gene Editing of GUCY2D by AAV-CRISPR/Cas9 Alters Retinal Structure and Function in Mouse and Macaque. Hum. Gene Ther. 2019;30:571–589. doi: 10.1089/hum.2018.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maeder M.L., Stefanidakis M., Wilson C.J., Baral R., Barrera L.A., Bounoutas G.S., Bumcrot D., Chao H., Ciulla D.M., DaSilva J.A. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 2019;25:229–233. doi: 10.1038/s41591-018-0327-9. [DOI] [PubMed] [Google Scholar]
- 16.Colella P., Ronzitti G., Mingozzi F. Emerging Issues in AAV-Mediated In Vivo Gene Therapy. Mol. Ther. Methods Clin. Dev. 2017;8:87–104. doi: 10.1016/j.omtm.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finn J.D., Smith A.R., Patel M.C., Shaw L., Youniss M.R., van Heteren J., Dirstine T., Ciullo C., Lescarbeau R., Seitzer J. A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing. Cell Rep. 2018;22:2227–2235. doi: 10.1016/j.celrep.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 18.Li A., Lee C.M., Hurley A.E., Jarrett K.E., De Giorgi M., Lu W., Balderrama K.S., Doerfler A.M., Deshmukh H., Ray A. A Self-Deleting AAV-CRISPR System for In Vivo Genome Editing. Mol. Ther. Methods Clin. Dev. 2018;12:111–122. doi: 10.1016/j.omtm.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]