Abstract
Circular RNAs (circRNAs) sequester microRNAs (miRNAs) and repress their endogenous activity. We hypothesized that artificial circRNA sponges (circmiRs) can be constructed to target miRNAs therapeutically, with a low dosage requirement and extended half-lives compared to current alternatives. This could present a new treatment approach for critical global pathologies, including cardiovascular disease. Here, we constructed a circmiR sponge to target known cardiac pro-hypertrophic miR-132 and -212. Expressed circmiRs competitively inhibited miR-132 and -212 activity in luciferase rescue assays and showed greater stability than linear sponges. A design containing 12 bulged binding sites with 12 nucleotides spacing was determined to be optimal. Adeno-associated viruses (AAVs) were used to deliver circmiRs to cardiomyocytes in vivo in a transverse aortic constriction (TAC) mouse model of cardiac disease. Hypertrophic disease characteristics were attenuated, and cardiac function was preserved in treated mice, demonstrating the potential of circmiRs as novel therapeutic tools. Subsequently, group I permutated intron-exon sequences were used to directly synthesize exogenous circmiRs, which showed greater in vitro efficacy than the current gold standard antagomiRs in inhibiting miRNA function. Engineered circRNAs thus offer exciting potential as future therapeutics.
Keywords: circRNA, microRNA interference, cardiac hypertrophyy, RNA therapy
Graphical Abstract
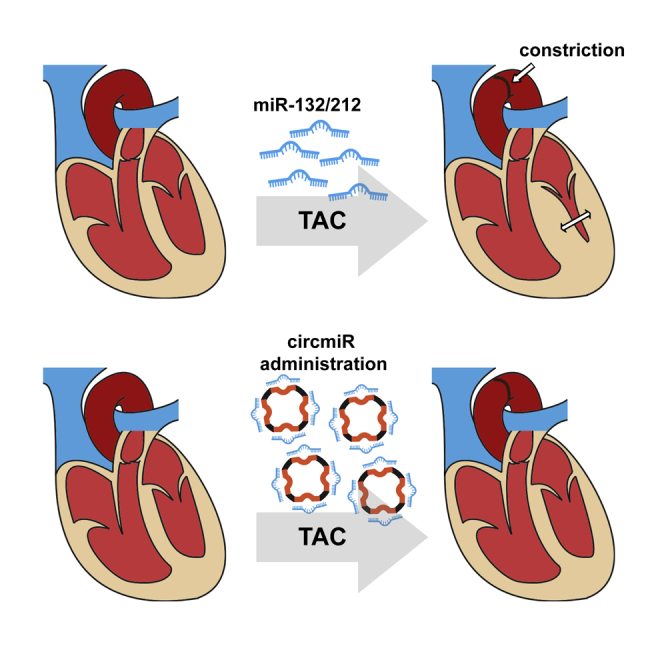
Lavenniah and colleagues detail considerations involved in custom-engineering circular miRNA sponges and demonstrate their greater stability and miRNA antagonism compared to linear miRNA inhibition technology. In vivo delivery of an engineered circular miRNA sponge improved cardiac function and inhibited hypertrophy in a mouse model of left ventricular pressure overload.
Introduction
Heart failure is the final common pathology for a myriad of cardiovascular diseases such as hypertension, metabolic syndrome, valve disease, and others.1 It is a major cause of mortality and morbidity worldwide,2 and poses a significant healthcare burden, with the pressing need for novel therapeutic approaches. Here, we present the development of a treatment strategy, using artificially designed circular RNA (circRNA) to target microRNAs (miRNA) in order to ameliorate pathological left ventricular hypertrophy.
As fine-tuners of gene expression, miRNAs play essential roles in normal development and homeostasis, dysregulation of which has been implicated in the pathogenesis of various diseases.3,4 Hence, targeting candidate miRNAs presents an exploitable therapeutic avenue. The first anti-miRNA therapeutic drug, Miravirsen, has shown efficacy in phase II clinical trials against hepatitis C virus infection with minimal side effects.5,6 While the continuous development and optimization of existing miRNA interference technology has conferred therapeutic benefits in clinical trials, many challenges remain. These include short half-lives, off-target effects, and potential accumulation of non-metabolizable molecules such as locked nucleic acid (LNA) nucleotides.5, 6, 7, 8
circRNAs belong to an emerging class of noncoding RNA that exist in circular instead of canonical linear form. The cellular splicing mechanism plays a central role in the biogenesis of circRNAs from pre-mRNAs. circRNA are generated through a back-splicing reaction where the 5′ splice donor site of a downstream exon is fused to the 3′ splice acceptor site of an upstream exon.9, 10, 11 Recently, there has been emerging evidence of endogenous circRNAs functioning as miRNA sponges. The first two circRNAs to be elucidated as miRNA sponges were ciRS-7 and circSRY.12 More than 70 seed sites for miR-7 were identified in ciRS-7, and 16 miR-138 sites were identified in circSRY.12 Of note, following miR-7 binding, ciRS-7 levels remained unaffected, whereas co-tested linear miR-7 sponge constructs saw an approximate 2-fold decrease in abundance, presumably the result of exonucleolytic degradation.12 Thus, the circularity of the circRNA may confer resistance to degradation upon miRNA binding.12 circRNAs are also protected against RNase mediated exonucleolytic decay due to non-requirement of a 5′ cap and a 3′ poly(A) tail.13 This is consistent with the typical observation of deadenylation and decapping when miRNA bind to their canonical linear mRNA targets.13
From a therapeutic viewpoint, an ideal miRNA interference technology should exhibit high miRNA inhibitory activity at a low dosage and enhanced stability. circRNAs carry the valuable trait of resistance against nuclease degradation without need for chemical modifications. Importantly, these stable circles thus likely enter a natural biological degradation pathway, which may not be the case for mixmer anti-miR drugs. With foresight, circRNAs have the potential to sustain miRNA suppression for prolonged time periods and would in turn require minimal dosage, or longer dosing intervals, as compared to linear miRNA inhibitor administration.
In this study, we constructed a circular miRNA sponge, termed “circmiR,” which was engineered as a custom sponge to sequester target miRNAs of interest. As proof of concept, we chose the mouse pressure-overload-induced cardiac hypertrophy disease model to test our circmiR. Pathological hypertrophy associates with cardiac dysfunction and fibrotic remodeling, leading to wall stiffness, which compromises systolic and diastolic function, and ultimately progresses to heart failure.14 Cardiac hypertrophy is one of the strongest prognostic factors in patients with heart disease,15 and reduction in pathological hypertrophy or adverse myocardial remodeling represents a therapeutic goal for cardiovascular pharmacotherapy.14,16 It was recently shown that miR-212/132 family expression is upregulated in cardiomyocytes following hypertrophic stimuli and miR-212/132 necessarily drive pathological hypertrophy.17 Pharmacological targeting of miR-132 by antagomiRs reduces cardiac hypertrophy progression, and abrogates heart failure in rodent models,17 and is currently entering phase I clinical trials.18,19 Hence, the circmiR was designed to target the miR-212/132 family to test its effectiveness against miRNA inhibition as a new development for pharmacotherapy.
Here, we address the importance of optimizing crucial parameters in engineered circmiRs such as the number and type of binding sites to be incorporated, as well as between-site spacer sizes. Cardiomyocyte-specific in vivo delivery of optimized circmiRs, targeting the miR-212/132 family, attenuated left ventricular hypertrophy. Furthermore, circmiRs exhibited improved efficacy compared to equimolar pharmacological antagomiRs in vitro and enhanced stability compared to linear counterparts. Our study introduces the first instance of therapeutic application of a targeted miRNA interference technology, circmiRs, in vivo.
Results
Design and Circularization of miRNA Sponge Sequences
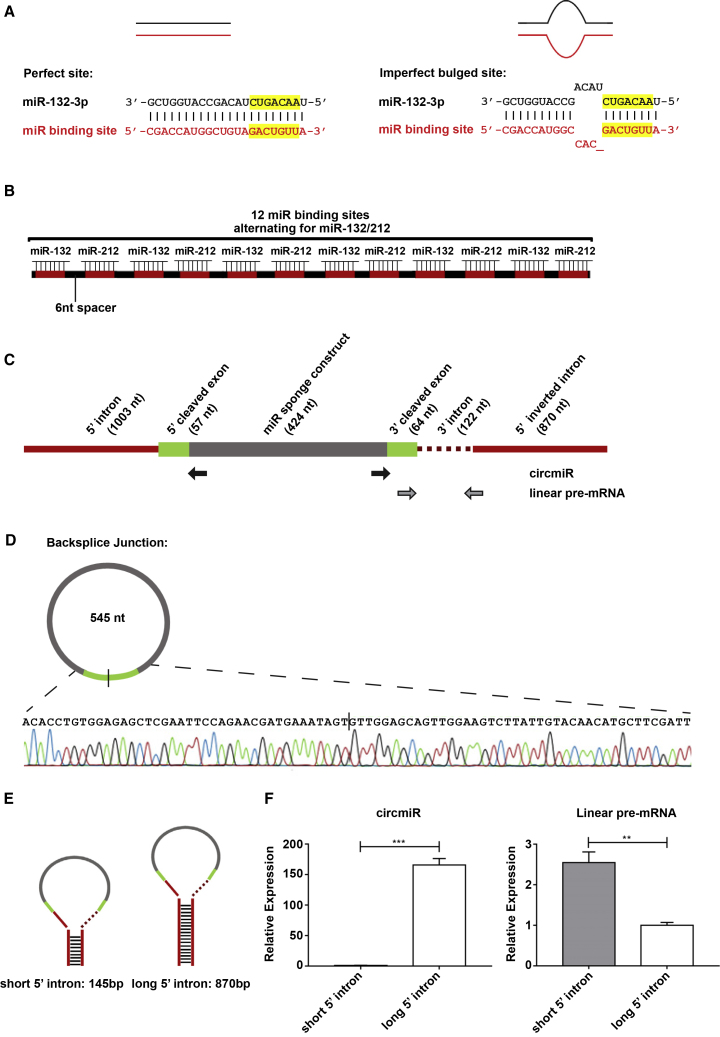
miRNA binding sites (MBSs) were engineered as illustrated in Figure 1A, and bulged binding sites were introduced by one deletion and three mismatches in the MBSs (Figure 1A). The miRNA sponge was designed initially to carry a total of 12 alternating bulged miRNA binding sites, 6 for each of miR-132 and miR-212, with a 6 nucleotide (nt) separation space between miRNA binding sites (Figure 1B). The values for these parameters were based on optimization studies previously carried out for linear miRNA sponge design.20,21
Figure 1.
Engineering of a Circular miRNA Sponge
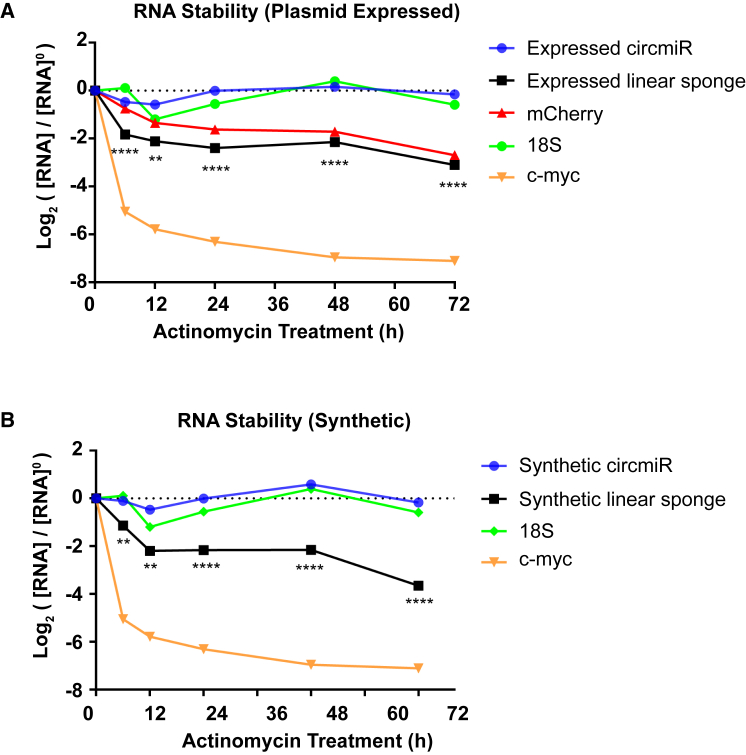
(A) Design of a perfect complementary or imperfect bulged miRNA binding site. The bulge is created by one base deletion and three base mismatches at positions 9–12 nt. Seed regions are highlighted in yellow. (B) Schematic illustration of miRNA sponge construct carrying 12 binding sites separated by 6 nt spacers. (C) Schematic illustration of circmiR expression construct, indicating positions of the convergent (gray arrows) and circmiR-specific divergent (black arrows) PCR primer binding sites. (D) Sanger sequencing of PCR product following amplification with divergent circmiR primers confirming back-splicing of the miRNA sponge construct. (E) Schematic of varying length of intronic sequences, short and long, flanking the miRNA sponge sequence. (F) Expression abundance of circRNA derived from constructs with short or long flanking intronic sequences in transfected HEK293T cells using qPCR. (n = 3); ∗∗p < 0.01, ∗∗∗p < 0.001. Student’s t test.
To circularize the miRNA sponge, we flanked the sequence with inverted complementary introns (Figure 1C). The head-to-tail junction of the consequent circmiR was detected by quantitative real-time PCR using divergent primers and confirmed by Sanger sequencing (Figure 1D). To test whether shorter flanking intronic sequences could also lead to efficient circularization, we trimmed the intronic sequences (Figure 1E). However, a significant decrease in circmiR and increase in linear pre-mRNA levels was observed, suggesting lower circularization efficiency with shorter flanking introns (Figure 1F). Hence, the construct with long inverted introns was used for the rest of the study.
Functional Efficacy Testing and Optimization of circmiR Design In Vitro
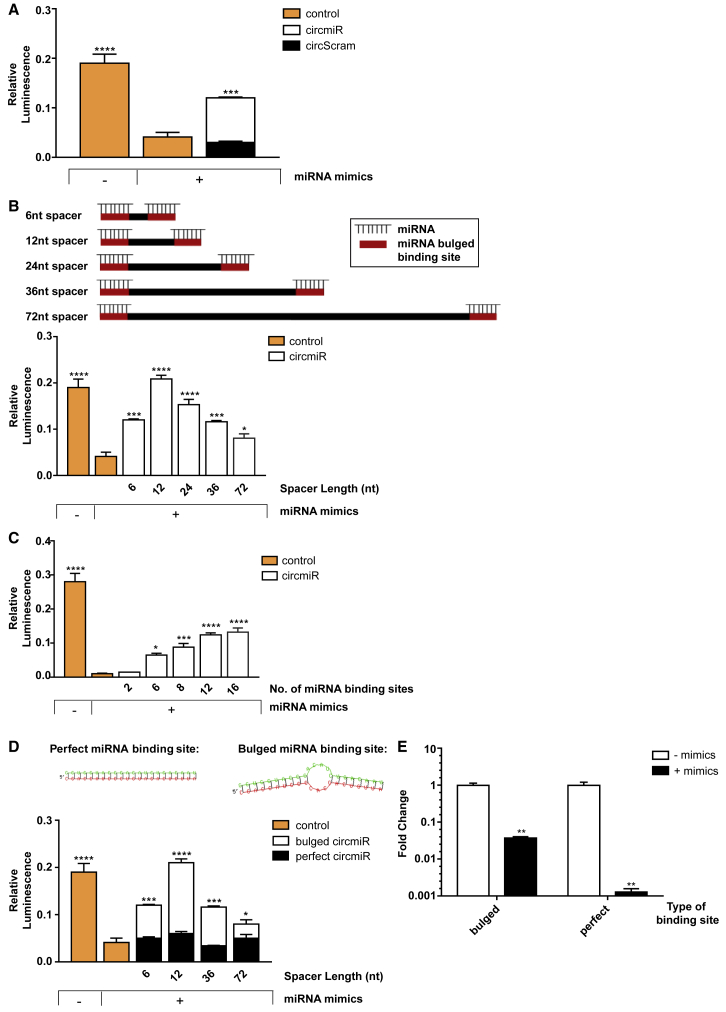
First, to validate circmiR function, miR-132 and -212 binding sites were inserted into the 3′ untranslated region (UTR) of a Renilla luciferase construct from the dual-luciferase reporter system. Co-transfecting this construct with miR-132 and miR-212 mimics into HEK293T cells caused a significant reduction in Renilla activity, as expected (Figure 2A). Upon introduction of circmiR carrying bulged miRNA binding sites, Renilla activity was significantly rescued, compared to introduction of a negative control sponge (circScram), in which miR binding sites were scrambled (Figure 2A).
Figure 2.
Engineered circmiRs Are Efficient Sponges of miR-132 and -212
Luciferase rescue reporter assays using dual reporter constructs with miR-132 and -212 binding sites inserted into the 3′ UTR of the Renilla luciferase gene. HEK293T cells were co-transfected with dual reporter plasmid psiCheck2, miR-132 and -212 mimics, and respective circRNA expression constructs for 48 h, to determine (A) the effect of circmiR versus circScram, (B) the effect of circmiR with different spacer lengths as follows: 6, 12, 24, 36, and 72 nucleotides, (C) the effect of circmiR with different numbers of miRNA binding sites as follows: 2, 6, 8, 12, 16, and (D) the effect of bulged versus perfect complementary miRNA binding sites. RNAhybrid prediction of structures are shown for each binding site type upon miRNA binding. (n = 3); ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 relative to control with mimics. One-way ANOVA with Benjamini-Hochberg adjustment. (E) Expression abundance 48 h post-transfection of circmiRs carrying either 12 bulged or perfect miRNA binding sites in the presence or absence of mimics in HEK293T cells using qPCR. (n = 3); ∗∗p < 0.01 relative to control without mimics. Student’s t test.
The circmiR structural design was next optimized by testing the effect of different spacer lengths, type of binding sites, and total number of binding sites. Previously, linear miRNA sponges showed effective miRNA inhibition with short spacers between miRNA binding sites.21 However, short spacer sequences in a circular structure may conceivably exert tension on neighboring binding sites, affecting miRNA binding. To examine whether longer spacer lengths are preferable, we constructed bulged circmiRs with different spacer lengths as follows: 6, 12, 24, 36, and 72 nt. The 12-nt spacer construct produced the greatest rescue effect (Figure 2B). Spacer sizes greater than 12 nt showed reduced rescue of Renilla activity (Figure 2B). Notably, the 6-nt and 36-nt spacer constructs showed similar rescue effects (Figure 2B).
Bulged circmiR constructs were also generated containing 2, 6, 8, 12, and 16 binding sites, separated by 12-nt spacers, per sponge. The rescue effect of circmiR increased with increasing number of binding sites, but no significant difference was seen between circmiRs containing 12 and 16 binding sites (Figure 2C). Bulged miRNA binding sites are preferred over perfect binding sites in linear sponge sequences since the latter result in degradation via RISC-mediated endonucleolytic cleavage upon miRNA binding.21,22 To determine whether circmiRs suffer the same fate, we compared circmiRs with either bulged or perfect miRNA binding sites. Regardless of the spacer size, expression of perfect circmiRs failed to rescue Renilla activity (Figure 2D). However, bulged circmiRs rescued Renilla expression consistently, and this effect was influenced by their spacer sizes as before (Figure 2D). Interestingly, upon miRNA mimic treatment, both bulged and perfect circmiRs carrying 12 miRNA binding sites separated by 12-nt spacers were degraded (Figure 2E). However, the perfect circmiRs were degraded to a much greater extent than bulged circmiRs (785-fold and 27-fold respectively; Figure 2E). These luciferase experiments were replicated in H9C2 rat cardiomyocytes where luciferase activity was similarly rescued only by bulged but not perfect circmiRs, each carrying 12 miRNA binding sites separated by 12-nt spacers (Figure S1).
Altogether, we concluded an optimal circmiR design, comprising of 12-nt spacers with 12 bulged miRNA binding sites. This design was used for all further experiments.
In Vivo circmiR Administration Attenuates Disease in a Mouse TAC Model
Hypertrophic stimuli reportedly upregulate cardiomyocyte expression of miR-132 and miR-212, which are necessary to drive pathological hypertrophy.17 We confirmed the upregulation of both miR-132 and miR-212 in our in vivo transverse aortic constriction (TAC) cardiac pressure overload mouse model, although at a later time point than previously reported (Figure S2).
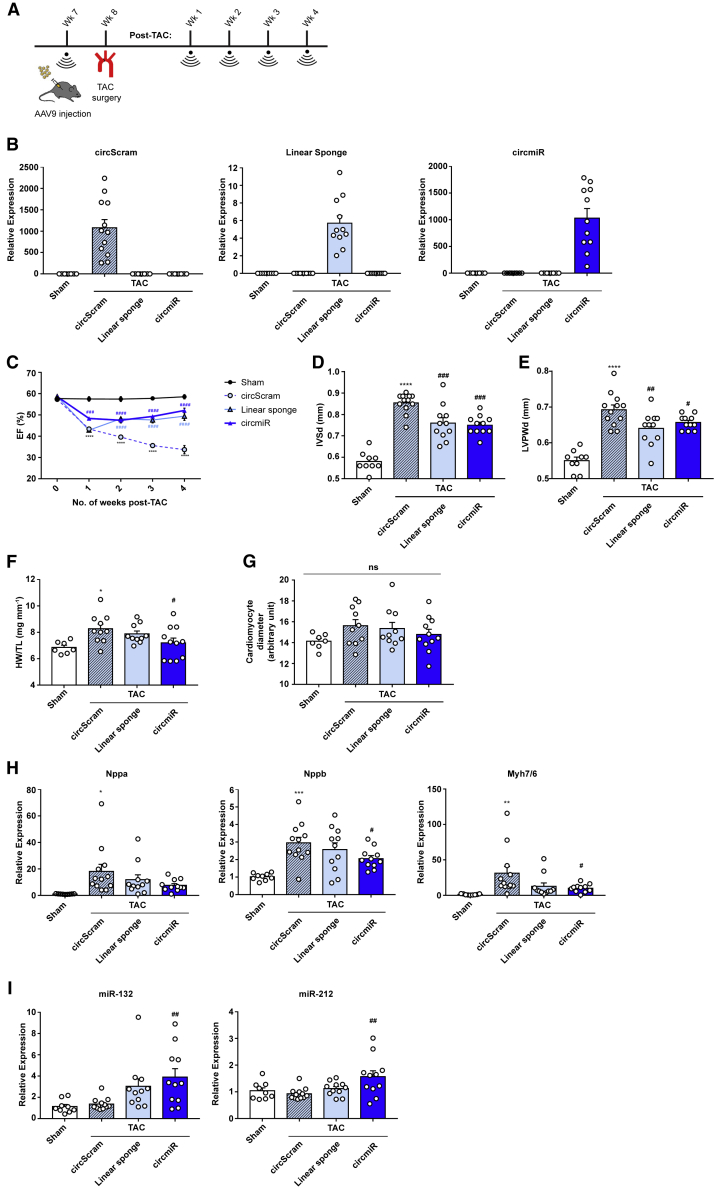
Next, adeno-associated virus (AAV) serotype 9 vectors were employed to deliver and express constructs in vivo, specifically in cardiomyocytes (CMs), using the cardiac troponin T (cTnT) promoter.23 AAV vectors expressing either circScram, circmiR, or a linear miR-212/132 sponge (linsp) were injected intra-peritoneally 1 week before TAC surgery (Figure 3A). Successful targeted expression was validated by quantitative PCR (qPCR)analysis of isolated CMs (Figure 3B). Specific divergent primers detected circScram and circmiR constructs, respectively, whereas convergent primers detected linsp constructs, as expected (Figures 3B and S3).
Figure 3.
circmiR Therapy Attenuates Pressure Overload-Induced Hypertrophy
(A) Experimental strategy to test circmiR therapeutic or circScram/linsp control constructs in vivo by AAV9 injection to 7-week-old mice. TAC surgery was performed one week later. Weekly echocardiography was conducted up to 4 weeks post TAC surgery before sacrifice. (B) Expression abundance of products from AAV construct circScram, linsp, and circmiR in isolated cardiomyocytes using qPCR. (C–E) Echocardiographic analysis of (C) ejection fraction, (D) interventricular septal thickness, and (E) left ventricular posterior wall thickness in circScram, linsp, and circmiR injected mice and sham-operated controls. (F) Heart weight to tibia length ratios and (G) cardiomyocyte diameter measured by immunofluorescence analysis of WGA and cTnI staining to visualize cell membrane and cardiomyocytes respectively in circScram, linsp, and circmiR injected mice and sham-operated controls. (H) Expression levels of cardiac stress response genes Nppa, Nppb, and Myh7/Myh6 ratio in isolated cardiomyocytes using qPCR. (I) miR-132 and miR-212 expression levels in isolated cardiomyocytes using qPCR. For all data, circScram (n = 12), circmiR (n = 11), linsp (n = 11), and sham (n = 9) except (F) and (G) where circScram (n = 10), circmiR (n = 11), linsp (n = 10) and sham (n = 7). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 (sham versus circScram). #p < 0.05, ##p < 0.01, and ###p < 0.001 (circScram versus circmiR/linsp). One-way ANOVA with Benjamini-Hochberg adjustment.
Cardiac function was evaluated by weekly echocardiographic measurements. Ejection fraction, a parameter of systolic cardiac function, was significantly compromised in TAC-operated circScram mice, whereas both circmiR and linsp mouse groups showed improved preservation of cardiac function up to 4 weeks post-TAC (Figure 3C). Pressure overload induces pathological cardiac wall thickening (hypertrophy).24 4 weeks post-TAC, interventricular septal (IVS) and left ventricular posterior wall (LVPW) thickness of both circmiR and linear sponge groups were significantly reduced, compared to circScram (Figures 3D and 3E). Thus, pathological hypertrophy was significantly attenuated in the circmiR and linear sponge treated groups, compared to circScram controls.
The heart weight to tibia length ratio, another measure of cardiac hypertrophy, increased post-TAC in both circScram and linsp mice, whereas heart weight in circmiR mice remained similar to sham levels (Figure 3F). Morphologically, CM diameter trended to increase in both circScram and linsp mice more so than circmiR mice, in which CM diameter was again similar to those of sham mice, although these changes did not reach statistical significance (Figure 3G).
The expression levels of canonical CM stress markers Nppa, Nppb, and Myh7 were significantly increased post-TAC in circScram-treated hearts compared to sham levels (Figure 3H). These stress markers were attenuated in circmiR-treated hearts, for which decreases in Nppb and Myh7 were statistically significant (Figure 3H). Attenuation of stress markers was also observed in linsp-treated hearts but did not reach statistical significance (Figure 3H). Interestingly, the detected abundance of miR-132 and -212 were markedly higher in both circmiR and linsp groups compared to circScram controls (Figure 3I). Conceivably, this may be caused by retention of sequestered miRNAs without degradation, due in part to bulged binding sites (Figure 2E). Both circmiR and linsp did conversely appear to reduce miR-212/132 abundance in HEK293T cells in vitro (Figure S4), possibly reflecting the considerable differences between cell types, and in vitro versus in vivo environments.
Generation of Synthetic circmiRs
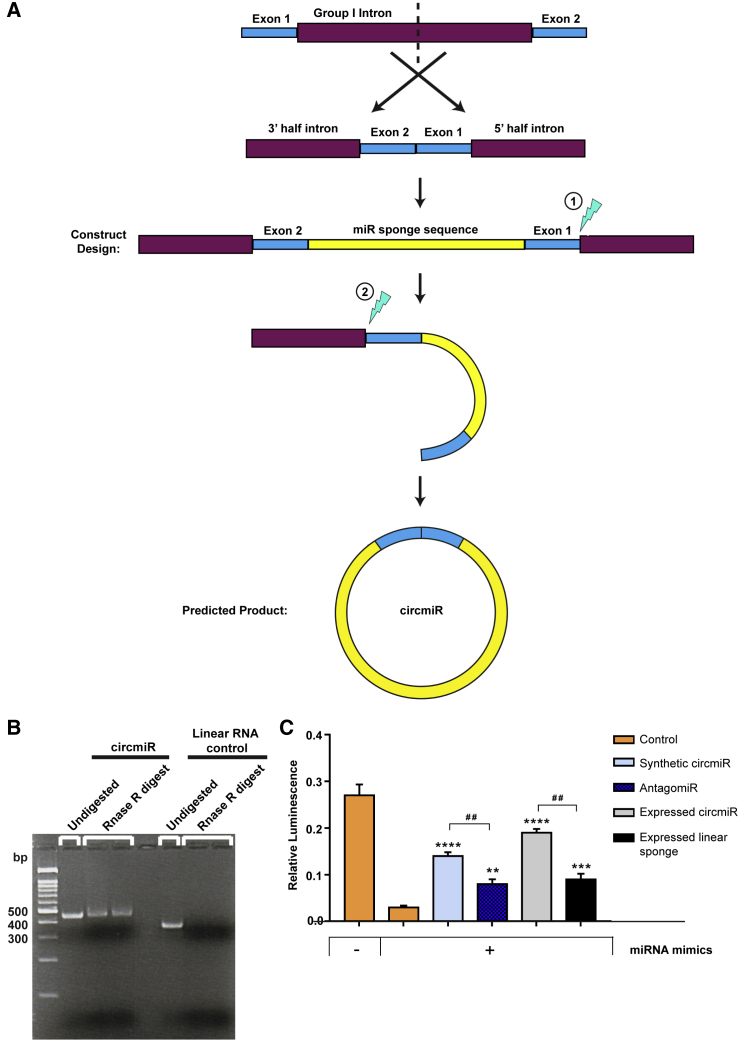
Next, we sought to compare the efficacy of synthesized circmiRs against pre-existing antagomiR technology. Synthetic circmiRs, in contrast to vector driven expression, allow for a direct route of delivery and controlled, temporal dosing. Dosage comparisons can also be made directly between the two technologies, unlike potentially saturating expression through AAV, which may be long-term when transduced in non-dividing adult cardiomyocytes. We leveraged the permutated intron-exon (PIE) method to generate in vitro synthesized circmiRs (Figure 4A).25, 26, 27 Group I introns, a class of autocatalytic ribozymes, carry out efficient splicing between reversed splice sites.25, 26, 27 Constructs therefore consisted of fused exons flanked by half-intron sequences and were made to undergo two successive transesterifications, resulting in the excision of a circular miRNA sponge product (Figure 4A).
Figure 4.
Engineering of Synthetic circmiRs In Vitro Using Group I Permutated Intron-Exon Splicing
(A) Schematic illustration of permutated intron-exon synthetic circmiR construct design and predicted splice product. Partial sections of the group I intron of T4 bacteriophage Td gene are reversed to allow the intron to fold into a stabilized structure required for splicing. Downstream exon 2 fragment is ligated to upstream exon 1 fragment. The miRNA sponge sequence is inserted between the exon 2-exon 1 junction. During autocatalytic splicing, a two-step transesterification reaction (lightning symbol) occurs that results in ligation of both ends of the miRNA sponge sequence. (B) RNase R digests separated on agarose gel to confirm circmiR circularization. (C) Luciferase reporter assays using dual reporter constructs with miR-132 and -212 binding sites inserted into the 3′ UTR of Renilla. HEK293T cells were co-transfected with dual reporter plasmid psiCheck2, miR-132 and -212 mimics, and respective constructs to determine the effect of antagomiRs versus circmiRs. (n = 3); ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 relative to control with mimics. One-way ANOVA with Benjamini-Hochberg adjustment. ##p < 0.01 as indicated. Student’s t test.
Using this strategy, we inserted our miRNA sponge sequence between two exon regions (exon 1 and 2) of the PIE construct flanked by 3′ and 5′ intron halves of the permutated group I intron of the T4 phage thymidylate synthase (Td) gene (Figure 4A). circmiRs synthesized by in vitro transcription were confirmed to be resistant to RNase R degradation (Figure 4B). Interestingly, however, subsequent Sanger sequencing revealed that splicing had not occurred exactly at the predicted regions. Instead, self-ligation occurred between a region within the 3′ half intron and at the end of the 3′ miRNA sponge sequence (Figure S5). Regardless, the resulting circmiRs were purified at high yields, and significantly rescued Renilla activity by 4-fold when co-transfected with miR-212/132 mimics in our in vitro luciferase reporter model (Figure 4C). Individual datapoints are displayed in Figure S6. Moreover, this effect was significantly greater than the 2-fold rescue effect of Renilla activity by equimolar antagomiRs (Figure 4C). Synthetic circmiR was more efficacious than 2-, 4-, and 6-fold higher molar concentrations of antagomiRs (Figure S7; see Materials and Methods for calculation of molar concentrations). Hence, in this system, our synthetic circmiR product appeared more efficacious than antagomiRs in vitro. However, the rescue effect was lower than that of plasmid driven expression of circmiRs, potentially due to plasmid transfection producing higher copy numbers of circRNA products in cells (Figure 4C).
circmiRs Are More Stable Than Linear miRNA Sponges
Finally, the stability of circmiRs in comparison to linear miRNA sponge constructs was investigated. HEK293T cells were transfected with plasmids driving either circmiR or linear sponge expression or with synthetically generated circmiR or linear sponge constructs. Transcription was inhibited with actinomycin D and total RNA was harvested at indicated time points.
As anticipated,28,29 measured by qPCR, positive control 18S abundance was highly stable across the 72 h time period, whereas a decrease in abundance of the less stable c-Myc transcript (MYC) was observed, exhibiting a half-life of 6 h. Plasmid-expressed circmiRs were resistant to nuclease degradation and were stable for up to 72 h (Figure 5A). In contrast, the turnover rate of the linear sponge RNA was comparable to that of an mCherry reporter (half-lives < 24 h), also expressed from the same construct. Similar to plasmid-expressed circmiR, the synthetic circmiR was stable for up to 72 h, compared to synthetic linsp RNA (half-life < 24 h) (Figure 5B). Thus, these results strongly demonstrate that regardless of the source of circmiR production, plasmid-driven, or synthetically produced, circular constructs are more stable than their linear counterparts.
Figure 5.
circRNA Offers Superior Protection against Endogenous Nuclease Degradation
RNA stability assay of sponge constructs in HEK293T cells following actinomycin D treatment. 18S and MYC act as endogenous control transcripts while mCherry is a plasmid driven control transcript. RNA levels at each time point were measured by qPCR, to enable comparison of RNA stability between (A) plasmid-driven circmiRs versus linear sponges, and (B) in vitro synthesized circmiRs versus linear sponges. ∗∗p < 0.01 and ∗∗∗∗p < 0.0001 (circmiR versus linear sponge). One-way ANOVA with Benjamini-Hochberg adjustment.
Discussion
circRNAs have been propelled to recent attention, highlighted by their abundant endogenous expression, and function as miRNA sponges.12,28 Compared to other anti-miR technology implemented to date, circmiRs, without the requirement for chemical modifications, are likely to be well tolerated in biological systems. Our present study exploits the natural potential of the circular structure to engineer a therapeutic tool for miRNA antagonism. Here we demonstrate the successful construction and in vivo testing of artificial circmiRs as miR-132 and miR-212 antagonists.
circRNAs are more stable than linear RNAs due to the covalent linkage of their ends and our in vitro experimental evidence concurs with this greater stability, compared to linear sponge constructs. This is an advantageous feature of circRNAs as a therapeutic tool. Longer flanking introns promote more efficient back-splicing than shorter flanking introns and this has been well reported.28,30,31 In our study, we used the longer flanking introns to maximize circularization efficiency. However, if the circmiR design needs to be more compact due to viral vector space constraints, shorter flanking introns could be incorporated.
Importantly, we found that circmiRs constructed with 12 perfect sites showed poor miRNA inhibitory effect compared to bulged circmiRs. This could be due to the susceptibility of circmiRs containing perfect binding sites to degradation upon miRNA binding by Ago2-mediated cleavage, which is not the case for bulged sites.22,32,33 Furthermore, it has been reported that the presence of a near-perfect miR-671 site abolished functionality of an endogenous circular miRNA sponge, ciRS-7, through RISC mediated endonucleolytic cleavage.12,34 Altogether, the differences in spatial and structural distribution have varying effects on circmiR functionality. Hence, circmiR design plays utmost importance to its resulting structure that in turn determines its effectiveness as a miRNA inhibitor.
In a recent study describing the synthesis of a circular sponge comprising 8 miRNA binding sites, both bulged or perfect binding sites were equally effective for miRNA inhibition.35 This discrepancy could be accounted for by the shorter post-transfection (4 h) luciferase activity measurement during which susceptibility of the bulged or perfect binding sites to degradation may not be detected.
In our in vivo study, both circmiR and linsp, bearing binding sites for miR-132 and miR-212, attenuated cardiac hypertrophy and heart failure progression to a similar extent. This effect concurs with a previous study in which the pharmacologic inhibition of miR-132 by antagomiR injection suppressed pressure-overload induced hypertrophy.17 While ideal as proof-of-concept, cardiac AAV vector systems drive high levels of constant expression,36,37 which therefore likely preclude the benefit of the higher stability circmiR compared to its linear counterpart. However, therapeutically, a pharmacological agent that is more precisely and temporally dose controlled is typically more desirable. It is in this context that we propose that the markedly improved stability of a directly administered synthetic circmiR may have the advantage against linear therapeutics. This will require further rigorous in vivo testing.
Accordingly, we also synthesized and purified our circmiRs to enable future testing of route of delivery and dosage and conducted direct comparisons to antagomiRs as the current technology leaders in this field. Remarkably, circmiRs showed favorable functional effectiveness when compared to equimolar antagomiRs in vitro. Thus far, few publications have described the in vitro synthesis of circular miRNA sponges. A circular sponge that inhibits miR-21 in gastric carcinoma cells, and another that inhibits miR-122 from hepatitis C virus, have been reported to function in vitro.35,38 Notably, both studies employed enzymatic ligation of linear RNA generated from in vitro transcription as the method of RNA circularization. We and others have recently shown efficacy of expressing endogenous circRNAs in mammalian models of disease.39,40 In the present study, we utilized endogenous splice machinery to generate custom-designed circular miRNA sponges and delivered them into an in vivo mouse model using AAVs.
It will be of great importance to determine the optimal delivery platform for circmiRs. Currently, in addition to aforementioned limitations, the AAV system applied here as proof of concept risks undesired immunological and long-term off-target effects. Future studies to package in vitro synthesized circmiRs into targeted nanoparticles could allow for a more refined route of delivery in vivo. In conclusion, our findings demonstrate the promising potential of circmiRs as therapeutic miRNA antagonists. We have applied this treatment and demonstrate benefits in an in vivo model of cardiovascular disease, which represents the primary cause of death globally, and anticipate that future development can expand this scope significantly.
Materials and Methods
Cell Culture and Transfections
HEK293T and H9C2 cells were maintained in DMEM (GE Healthcare Life Science) supplemented with 10% FBS (Capricorn Scientific), 100 U/mL penicillin, and 100 μg/mL streptomycin (Nacalai Tesque) in a humidified atmosphere at 37°C, 5% CO2. Transient transfection of plasmids and/or miRNA mimics (QIAGEN) into both cell lines was carried out with Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s protocol.
circmiR Construct Design
The miRNA sponge sequence was constructed using Ultramer DNA oligos (Integrated DNA Technologies, IDT) designed to contain 2, 4, or 6 miRNA binding sites. Binding sites were designed as the reverse complement of the mature sequences of mmu-miR-132-3p or mmu-miR-212-3p (miRBase). Bulged sites carried one deletion and three base mismatches outside the seed regions as described,22 while perfect sites had complete complementarity to the mature miRNA sequences (Table S1). Spacer regions of different sizes and a scrambled sequence were created using a random oligo generator (http://mkwak.org/oligorand/). The online RNAhybrid tool41,42 was used to confirm binding of designed miRNA sponge sequences to target miR-212/132 sequences.
1.1 kb of the 5′ intronic sequence upstream and 1.1 kb of the 3′ intronic sequence downstream of exon 2 of the mouse Slc8a1 locus (encoding a known Slc8a1 circRNA39,43,44) was PCR amplified (Table S1). Both fragments were cloned into an AAV9-cTnT-eGFP plasmid backbone (gift from Dr. Jiang Jianming, Cardiovascular Research Institute, National University of Singapore). The miRNA sponge oligos were cloned between these fragments to generate circmiRs carrying 2 or 6 binding sites. For circmiRs carrying 8, 12, or 16 binding sites, additional miRNA sponge oligos were sequentially cloned45 between the inverted intronic repeats. The linear sponge sequence comprised of 12 bulged miRNA binding sites, constructed by sequential cloning45 of miRNA sponge oligos carrying 6 binding sites into the AAV9-cTnT-eGFP plasmid, without inverted intronic repeats. circScram was constructed by inserting a scrambled sequence, ordered as a gBlock gene fragment (IDT), between the inverted intronic repeats in the AAV9-cTnT-eGFP plasmid. The entire circmiR/linear/circScram sponge sequences were cloned into pCAG-mCherry plasmid backbones for in vitro applications.
Luciferase Reporter Assays
miRNA-212/132 binding sites were cloned downstream of the Renilla luciferase gene of the psiCHECK-2 vector (Promega). 5 × 104 HEK293T cells were co-transfected with 50 ng of the psiCHECK-2-miR-212/132 or empty psiCHECK-2 vector, 50 ng of circmiR/circScram vectors, and 10 pmol of equal molar mix of miR-212/132 mirVana miRNA mimics (QIAGEN), using Lipofectamine 2000 reagent (Invitrogen) according to manufacturer’s instructions. T7 synthesized circmiRs or miRvana miRNA inhibitors (QIAGEN) were also co-transfected with 10 pmol of equal molar mix of miR-212/132 mirVana miRNA mimics (QIAGEN). H9C2 cells (a kind gift from Dr. Zhou Yue, Cardiovascular Research Institute, National University of Singapore) were co-transfected with 50 ng of psiCHECK-2-miR-212/132 or empty psiCHECK-2 vector, 200 ng of circmiR/circScram vectors and 0.2 pmol of equal molar mix of miR-212/132 mirVana miRNA mimics (QIAGEN). Luciferase activity was measured 48 h after transfection using the Dual-Glo Luciferase Assay System (Promega) according to manufacturer’s protocol and read on a GloMax multi-plate reader (Promega). Results are expressed as Renilla luciferase normalized against Firefly luciferase.
RNA Isolation, cDNA Synthesis, and Quantitative Real-Time PCR
Total RNA was extracted from either isolated adult mouse cardiomyocytes or HEK293T cells using TRIzol Reagent (Thermo Fisher Scientific) according to standard procedures. Complementary DNA (cDNA) was synthesized with random primers using the qScript cDNA Synthesis Kit (Quantabio). qPCR was performed with Perfecta SYBR Green FastMix (Quantabio) on a LightCycler 480 (Roche) according to the manufacturers’ instructions using primers listed in Table S2. Primer design to distinguish between circmiR and linear sponge has been detailed in Figure S3. All qPCR data were normalized to expression of the housekeeping genes GAPDH (for human genes) or 18S (for mouse genes). miRNA reverse transcription reactions were carried out using the miRCURY LNA Universal RT Kit (QIAGEN) according to manufacturer’s protocol. qPCR was performed with Perfecta SYBR Green FastMix (Quantabio) on a Rotorgene Q cycler (QIAGEN) using miRCURY LNA miRNA PCR assay (QIAGEN) primer sets for mmu-miR-132-3p and mmu-miR-212-3p. Results were normalized to expression of 18S. All qPCR reactions were carried out in duplicates.
TAC Model and Cardiomyocyte Isolation
All animal procedures were approved by the National University of Singapore Institutional Animal Care and Use Committee and were undertaken in strict accordance with Singapore National Advisory Committee for Laboratory Animal Research guidelines. Adult mice were housed in individually ventilated cages, with sex-matched littermates, under standard conditions. Food and water were available ad libitum. TAC or sham surgery was performed on 8-week-old male C57BL/6 mice as previously described.46 Left ventricular cardiomyocytes were isolated as described47 from AAV treated mice 4 weeks after surgery. Transthoracic echocardiography was performed according to the manufacturer’s guide for small animal echocardiography (Vevo 2100 Imaging System, Visualsonics). Doppler velocity measurements of right and left carotid arteries across the aortic constriction was performed at weeks 1 and 4 post-TAC mice to confirm the consistency of the surgical procedure (Vevo 2100 Imaging System, Visual Sonics).
AAV9 Viral Production and Purification
circScram, circmiRs, and linear sponge constructs were cloned into AAV9-cTnT-eGFP vectors as described above. The target AAV9 vectors were packaged by a triple transfection method with helper plasmids pAdΔF6 and pAAV2/9 (Penn Vector Core) as previously described.48 All constructs were administered at a titer of 5 × 1010 virus genome (vg)/kg via thoracic cavity injection to 7-week-old mice.
Heart Weight Measurement and Immunofluorescence
Mice were anesthetized by isoflurane inhalation. The heart was arrested in diastole by injecting 500 μL of a 15% potassium chloride solution into the inferior vena cava. Hearts were excised and flushed with saline solution via retrograde perfusion. Aorta and auricles were trimmed, and hearts were dried by removing excess fluid with forceps. Heart weight was measured, after which hearts were immersed in 4% buffered formalin and embedded in paraffin blocks according to standard procedures.
5 μM sections were co-stained with cardiac troponin I (cTnI; Abcam, ab56357) and biotinylated wheat germ agglutinin (WGA; Vectorlabs, B-1025) with streptavidin linked Alexa Fluor 488 conjugate (Life Technologies). Images were analyzed using the NIKON-NIS-Elements Viewer software and ImageJ.49 For each heart, the diameter of 200 cardiomyocytes were counted and a mean value was obtained, with experimenters blinded to experimental condition until data collection was finalized.
Synthetic circmiR Generation
Based on established methods, the 3′ and 5′ group I permutated intron-exon (PIE) sequences27 from the T4 phage were synthesized as gBlocks (IDT). These were inserted downstream of a T7 promoter in the pcDNA3.1 plasmid vector (Addgene). A miRNA sponge sequence carrying 12 bulged binding sites was cloned between these intron-exon sequences. To create a linear RNA control for comparison of exonuclease degradation susceptibility, we removed the exon and 5′ half intron of the PIE sequence downstream of the miRNA sponge sequence by restriction enzyme digestion during plasmid linearization prior to in vitro transcription. Both circular and linear constructs were then synthesized by in vitro transcription from linearized plasmid DNA template using TranscriptAid T7 High Yield Transcription Kit (Thermo Fisher Scientific). RNA was purified from the reactions using TRIzol-LS reagent (Thermo Fisher Scientific). To confirm circularization, we digested 1 μg of RNA with 2 U RNase R (Epicenter) at 37°C for 30 min. Digests were separated on 2% agarose gels and bands were visualized using the ChemiDoc Imaging System (BioRad). Molar concentrations of synthetic circmiR were calculated using the NEBioCalculator tool (https://nebiocalculator.neb.com/#!/ssrnaamt).
RNA Stability Assay
1.2 × 105 HEK293T cells per well were seeded on 24-well culture plates. 0.25 μg of plasmids driving either circmiR or linear sponge expression were transfected using jetPRIME transfection reagent (Polyplus Transfection). Alternatively, 0.5 μg of T7 synthesized circmiRs and linear sponge RNA constructs were transfected using Lipofectamine 2000 (Invitrogen). 48 h post-transfection, cells were treated with 10 μg/mL actinomycin D (Sigma Aldrich) in fresh media. Cells were harvested at 0-, 6-, 12-, 24-, 48-, and 72-h time points post-actinomycin treatment. RNA was isolated and equal RNA quantities were subjected to reverse-transcriptase-PCR and qPCR as described above. Each RNA level was normalized against the 0 h time point to calculate log2 fold change.
Statistical Analysis
All results are presented as mean + standard error of the mean (SEM). Two-tailed, unpaired Student’s t test was performed for comparison between two groups. One-way ANOVA followed by the Benjamini-Hochberg adjustment was used to compare more than two groups. All tests were performed using GraphPad Prism 7. p < 0.05 was considered significant.
Author Contributions
A.L., T.D.A.L., Y.P.L., and M.A.-J. conducted the experiments. A.L., M.A.-J., and R.S.-Y.F. designed the experiments and analyzed the data. T.B.L. and J.J. provided material support. A.L., M.A.-J., and R.S.-Y.F. drafted and edited the manuscript. All authors approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
A.L. was funded by a YLL School of Medicine, National University Singapore PhD scholarship, and R.S.-Y.F. is funded by National Medical Research Grants (CSA SI and CS-IRG).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2020.04.006.
Supplemental Information
References
- 1.Creemers E.E., Wilde A.A., Pinto Y.M. Heart failure: advances through genomics. Nat. Rev. Genet. 2011;12:357–362. doi: 10.1038/nrg2983. [DOI] [PubMed] [Google Scholar]
- 2.Benjamin E.J., Muntner P., Alonso A., Bittencourt M.S., Callaway C.W., Carson A.P., Chamberlain A.M., Chang A.R., Cheng S., Das S.R., American Heart Association Council on Epidemiology and Prevention Statistics Committee and Stroke Statistics Subcommittee Heart Disease and Stroke Statistics-2019 Update: A Report From the American Heart Association. Circulation. 2019;139:e56–e528. doi: 10.1161/CIR.0000000000000659. [DOI] [PubMed] [Google Scholar]
- 3.McDermott A.M., Heneghan H.M., Miller N., Kerin M.J. The therapeutic potential of microRNAs: disease modulators and drug targets. Pharm. Res. 2011;28:3016–3029. doi: 10.1007/s11095-011-0550-2. [DOI] [PubMed] [Google Scholar]
- 4.van Rooij E. The art of microRNA research. Circ. Res. 2011;108:219–234. doi: 10.1161/CIRCRESAHA.110.227496. [DOI] [PubMed] [Google Scholar]
- 5.Gebert L.F.R., Rebhan M.A.E., Crivelli S.E.M., Denzler R., Stoffel M., Hall J. Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic Acids Res. 2014;42:609–621. doi: 10.1093/nar/gkt852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janssen H.L.A., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-Torres M., Patel K., van der Meer A.J., Patick A.K., Chen A., Zhou Y. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 7.Swayze E.E., Siwkowski A.M., Wancewicz E.V., Migawa M.T., Wyrzykiewicz T.K., Hung G., Monia B.P., Bennett C.F. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 2007;35:687–700. doi: 10.1093/nar/gkl1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakraborty C., Sharma A.R., Sharma G., Doss C.G.P., Lee S.S. Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine. Mol. Ther. Nucleic Acids. 2017;8:132–143. doi: 10.1016/j.omtn.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 10.Chen I., Chen C.Y., Chuang T.J. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip. Rev. RNA. 2015;6:563–579. doi: 10.1002/wrna.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 13.Ebbesen K.K., Kjems J., Hansen T.B. Circular RNAs: Identification, biogenesis and function. Biochim. Biophys. Acta. 2016;1859:163–168. doi: 10.1016/j.bbagrm.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 14.McKinsey T.A., Kass D.A. Small-molecule therapies for cardiac hypertrophy: moving beneath the cell surface. Nat. Rev. Drug Discov. 2007;6:617–635. doi: 10.1038/nrd2193. [DOI] [PubMed] [Google Scholar]
- 15.Ruilope L.M., Schmieder R.E. Left ventricular hypertrophy and clinical outcomes in hypertensive patients. Am. J. Hypertens. 2008;21:500–508. doi: 10.1038/ajh.2008.16. [DOI] [PubMed] [Google Scholar]
- 16.Frey N., Katus H.A., Olson E.N., Hill J.A. Hypertrophy of the heart: a new therapeutic target? Circulation. 2004;109:1580–1589. doi: 10.1161/01.CIR.0000120390.68287.BB. [DOI] [PubMed] [Google Scholar]
- 17.Ucar A., Gupta S.K., Fiedler J., Erikci E., Kardasinski M., Batkai S., Dangwal S., Kumarswamy R., Bang C., Holzmann A. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat. Commun. 2012;3:1078. doi: 10.1038/ncomms2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu D., Thum T. RNA-based diagnostic and therapeutic strategies for cardiovascular disease. Nat. Rev. Cardiol. 2019;16:661–674. doi: 10.1038/s41569-019-0218-x. [DOI] [PubMed] [Google Scholar]
- 19.Foinquinos A., Batkai S., Genschel C., Viereck J., Rump S., Gyöngyösi M., Traxler D., Riesenhuber M., Spannbauer A., Lukovic D. Preclinical development of a miR-132 inhibitor for heart failure treatment. Nat. Commun. 2020;11:633. doi: 10.1038/s41467-020-14349-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebert M.S., Sharp P.A. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otaegi G., Pollock A., Sun T. An optimized sponge for microRNA miR-9 affects spinal motor neuron development in vivo. Front. Neurosci. 2012;5:146. doi: 10.3389/fnins.2011.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gentner B., Schira G., Giustacchini A., Amendola M., Brown B.D., Ponzoni M., Naldini L. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat. Methods. 2009;6:63–66. doi: 10.1038/nmeth.1277. [DOI] [PubMed] [Google Scholar]
- 23.Jiang J., Wakimoto H., Seidman J.G., Seidman C.E. Allele-specific silencing of mutant Myh6 transcripts in mice suppresses hypertrophic cardiomyopathy. Science. 2013;342:111–114. doi: 10.1126/science.1236921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anversa P., Ricci R., Olivetti G. Quantitative structural analysis of the myocardium during physiologic growth and induced cardiac hypertrophy: a review. J. Am. Coll. Cardiol. 1986;7:1140–1149. doi: 10.1016/s0735-1097(86)80236-4. [DOI] [PubMed] [Google Scholar]
- 25.Ford E., Ares M., Jr. Synthesis of circular RNA in bacteria and yeast using RNA cyclase ribozymes derived from a group I intron of phage T4. Proc. Natl. Acad. Sci. USA. 1994;91:3117–3121. doi: 10.1073/pnas.91.8.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puttaraju M., Been M.D. Group I permuted intron-exon (PIE) sequences self-splice to produce circular exons. Nucleic Acids Res. 1992;20:5357–5364. doi: 10.1093/nar/20.20.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Umekage S., Kikuchi Y. In vitro and in vivo production and purification of circular RNA aptamer. J. Biotechnol. 2009;139:265–272. doi: 10.1016/j.jbiotec.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 28.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meganck R.M., Borchardt E.K., Castellanos Rivera R.M., Scalabrino M.L., Wilusz J.E., Marzluff W.F., Asokan A. Tissue-Dependent Expression and Translation of Circular RNAs with Recombinant AAV Vectors In Vivo. Mol. Ther. Nucleic Acids. 2018;13:89–98. doi: 10.1016/j.omtn.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L., Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Ebert M.S., Neilson J.R., Sharp P.A. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tay F.C., Lim J.K., Zhu H., Hin L.C., Wang S. Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells. Adv. Drug Deliv. Rev. 2015;81:117–127. doi: 10.1016/j.addr.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 34.Hansen T.B., Wiklund E.D., Bramsen J.B., Villadsen S.B., Statham A.L., Clark S.J., Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jost I., Shalamova L.A., Gerresheim G.K., Niepmann M., Bindereif A., Rossbach O. Functional sequestration of microRNA-122 from Hepatitis C Virus by circular RNA sponges. RNA Biol. 2018;15:1032–1039. doi: 10.1080/15476286.2018.1435248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonçalves M.A. Adeno-associated virus: from defective virus to effective vector. Virol. J. 2005;2:43. doi: 10.1186/1743-422X-2-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCarty D.M., Young S.M., Jr., Samulski R.J. Integration of adeno-associated virus (AAV) and recombinant AAV vectors. Annu. Rev. Genet. 2004;38:819–845. doi: 10.1146/annurev.genet.37.110801.143717. [DOI] [PubMed] [Google Scholar]
- 38.Liu X., Abraham J.M., Cheng Y., Wang Z., Wang Z., Zhang G., Ashktorab H., Smoot D.T., Cole R.N., Boronina T.N. Synthetic Circular RNA Functions as a miR-21 Sponge to Suppress Gastric Carcinoma Cell Proliferation. Mol. Ther. Nucleic Acids. 2018;13:312–321. doi: 10.1016/j.omtn.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim T.B., Aliwarga E., Luu T.D.A., Li Y.P., Ng S.L., Annadoray L., Sian S., Ackers-Johnson M.A., Foo R.S.Y. Targeting the highly abundant circular RNA circSlc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovasc. Res. 2019;115:1998–2007. doi: 10.1093/cvr/cvz130. [DOI] [PubMed] [Google Scholar]
- 40.Shen S., Wu Y., Chen J., Xie Z., Huang K., Wang G., Yang Y., Ni W., Chen Z., Shi P. CircSERPINE2 protects against osteoarthritis by targeting miR-1271 and ETS-related gene. Ann. Rheum. Dis. 2019;78:826–836. doi: 10.1136/annrheumdis-2018-214786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krüger J., Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451-4. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan W.L.W., Lim B.T.S., Anene-Nzelu C.G.O., Ackers-Johnson M., Dashi A., See K., Tiang Z., Lee D.P., Chua W.W., Luu T.D.A. A landscape of circular RNA expression in the human heart. Cardiovasc. Res. 2017;113:298–309. doi: 10.1093/cvr/cvw250. [DOI] [PubMed] [Google Scholar]
- 44.Lim T.B., Lavenniah A., Foo R.S.Y. Circles in the heart and cardiovascular system. Cardiovasc. Res. 2020;116:269–278. doi: 10.1093/cvr/cvz227. [DOI] [PubMed] [Google Scholar]
- 45.Blachinsky E., Marbach I., Cohen R., Grably M.R., Engelberg D. Procedure for controlling number of repeats, orientation, and order during cloning of oligonucleotides. Biotechniques. 2004;36:933–936. doi: 10.2144/04366BM02. [DOI] [PubMed] [Google Scholar]
- 46.Rockman H.A., Ross R.S., Harris A.N., Knowlton K.U., Steinhelper M.E., Field L.J., Ross J., Jr., Chien K.R. Segregation of atrial-specific and inducible expression of an atrial natriuretic factor transgene in an in vivo murine model of cardiac hypertrophy. Proc. Natl. Acad. Sci. USA. 1991;88:8277–8281. doi: 10.1073/pnas.88.18.8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ackers-Johnson M., Li P.Y., Holmes A.P., O’Brien S.M., Pavlovic D., Foo R.S. A Simplified, Langendorff-Free Method for Concomitant Isolation of Viable Cardiac Myocytes and Nonmyocytes From the Adult Mouse Heart. Circ. Res. 2016;119:909–920. doi: 10.1161/CIRCRESAHA.116.309202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wakimoto H., Seidman J.G., Foo R.S.Y., Jiang J. AAV9 Delivery of shRNA to the Mouse Heart. Curr. Protoc. Mol. Biol. 2016;115:1–23. doi: 10.1002/cpmb.9. [DOI] [PubMed] [Google Scholar]
- 49.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.