Abstract
Nuclear receptor subfamily group H member 4 (NR1H4), also known as farnesoid X receptor, has been implicated in several cellular processes in the liver and intestine. Preclinical and clinical studies have suggested a role of NR1H4 in colon cancer development; however, how NR1H4 regulates colon cancer cell growth and survival remains unclear. We generated NR1H4 knockout (KO) colon cancer cells using clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein-9 nuclease (CAS9) technology and explored the effects of NR1H4 KO in colon cancer cell proliferation, survival, and apoptosis. Interestingly, NR1H4 KO cells showed impaired cell proliferation, reduced colony formation, and increased apoptotic cell death compared to control colon cancer cells. We identified MYC as an important mediator of the signaling pathway alterations induced by NR1H4 KO. NR1H4 silencing in colon cancer cells resulted in reduced MYC protein levels, while NR1H4 activation using an NR1H4 ligand, chenodeoxycholic acid, resulted in time- and dose-dependent MYC induction. Moreover, NR1H4 KO enhanced the anti-cancer effects of doxorubicin and cisplatin, supporting the role of MYC in the enhanced apoptosis observed in NR1H4 KO cells. Taken together, our findings suggest that modulating NR1H4 activity in colon cancer cells might be a promising alternative approach to treat cancer using MYC-targeting agents.
Keywords: colon cancer, Myc, NR1H4, signaling
INTRODUCTION
Colorectal cancer (CRC) is one of the most common cancers in the world; thus, the mechanisms underlying colorectal carcinogenesis have been extensively studied at the molecular level (Houlston, 2001). The recent advances in high-throughput screening, sequencing, and phenotyping have led to a better understanding of CRC based on integrated gene expression, cellular and molecular phenotype, and even clinical behavior data (Cancer Genome Atlas Network, 2012; Guinney et al., 2015). Four CRC consensus molecular subtypes (CMS1–4) have been suggested based on their gene expression profiles; CMS1, comprising approximately ~14% of all CRC cases, is characterized by hypermutation, microsatellite instability, and strong immune activation; CMS2 (~37%) is epithelial and has marked WNT and myelocytomatosis (MYC) signaling activation; CMS3 (~13%) is epithelial and characterized by profound metabolic dysregulation; and CMS4 (~23%) is characterized by transforming growth factor-β activation, stromal invasion, and angiogenesis (Guinney et al., 2015). Although studies in patient-derived cells, patient-derived xenografts, and cell lines have increased the understanding of CMSs (Okita et al., 2018; Sveen et al., 2018), further research is required for the development of more precise anti-cancer therapeutic strategies for CRC.
The role of oncogenes in metabolic reprogramming, one of the hallmarks of cancer, has also been investigated (DeBerardinis and Chandel, 2016; Nagarajan et al., 2016). Multiomics analyses in tissues from CRC patients and healthy individuals have suggested that metabolic reprogramming occurred at the adenoma stage of carcinogenesis (Satoh et al., 2017), and detailed integrated analyses supported a role of cellular Myc (c-Myc, hereafter Myc) in metabolic reprogramming observed in CRC, through the regulation of glucose and one-carbon metabolism (Satoh et al., 2017). Myc overexpression is a crucial oncogenic mechanism in several cancers, including colorectal carcinoma, since shown to be oncogenic in colorectal carcinoma when overexpressed (Smith et al., 1993); thus, interfering with the Myc-mediated metabolic reprogramming of cancer cells has been explored as a novel therapeutic approach (Altman et al., 2015; Hsieh et al., 2015; Luengo et al., 2017; Ortmayr et al., 2019). Myc functions are mainly mediated through transcriptional regulation. It has also been shown that Myc plays a role in RNA polymerase II pause release (Rahl et al., 2010). Okuyama and colleagues have shown that under conditions of limited energy sources, Myc protein is degraded (Okuyama et al., 2010), suggesting a relationship between energy metabolism and Myc protein levels.
Bile acids have been implicated in the regulation of lipid and glucose homeostasis in the liver and intestine, and have been considered to play an important role in the pathogenesis of several diseases, including liver and colon cancer. Increased levels of toxic bile acids are considered as one of the risk factors for CRC (Degirolamo et al., 2011; de Aguiar Vallim et al., 2013; Kuipers et al., 2015). Nuclear receptor subfamily 1 group H member 4 (NR1H4), also known as farnesoid X receptor (FXR), is involved in the synthesis and flux of bile acids in the liver and intestine (de Aguiar Vallim et al., 2013). It has been shown that NR1H4 expression is often downregulated in colon cancer, by DNA methylation and KRAS signaling (Bailey et al., 2014), implying a negative association between NR1H4 and tumor progression. Moreover, targeting NR1H4 using NR1H4 agonists has been shown to promote CRC progression, and that NR1H4 is involved in intestinal self-renewal (Fu et al., 2019). Maran and colleagues addressed the effect of NR1H4 deficiency on intestinal epithelial cell proliferation in vivo and suggested a role in tumor development (Maran et al., 2009). Another study in hepatocyte- and enterocyte-specific NR1H4 null mice revealed crosstalk between NR1H4 and Myc during carcinogenesis (Takahashi et al., 2018), suggesting a role of NR1H4 signaling in metabolic reprogramming in colon cancers.
However, the majority of studies have been focused on bile acid regulation, and its impact on metabolic alteration observed in the microenvironment. The role of NR1H4 in colon cancer development and progression at the molecular level remains elusive. Herein, we generated NR1H4 knockout (KO) colon cancer cell lines using CRISPR/CAS9 technology (Ran et al., 2013) and investigated the role of NR1H4 in colon cancer cell survival.
MATERIALS AND METHODS
Materials
McCoy’s 5A medium (for HT29 cells) and defined fetal bovine serum (FBS) were obtained from Gibco (USA). Mouse monoclonal antibodies against β-actin, c-Myc, and CyclinD1 were obtained from Santa Cruz Biotechnology (USA). Rabbit monoclonal antibodies against p-p38/p-38, p-ERK/ERK, EGFR, B-cell lymphoma 2 (Bcl-2), Bcl-extra large (Bcl-xL), poly (ADP-ribose) polymerase (PARP), and cleaved PARP were obtained from Cell Signaling Technology (USA). Antibodies against glycogen synthase kinase-3beta (GSK3β) and p-AktSer473 were also purchased from Cell Signaling Technology. Anti-Myc was obtained from Invitrogen (Thermo Fisher Scientific, USA), while antibodies against p-MycSer62 and p-MycThr58 were obtained from Abcam (UK). The rabbit monoclonal antibody against NR1H4 was obtained from Novus Biologicals (USA). Horseradish peroxidase-conjugated anti-mouse and anti-rabbit antibodies were purchased from Cell Signaling Technology. Small interfering RNAs (siRNAs) targeting human NR1H4 were obtained from Santa Cruz Biotechnology. Gefitinib (Iressa, Iretinib; Chong Kun Dang Holdings, Korea), doxorubicin-HCl (A.D. Mycin, Boryung, Korea), and cisplatin (Cisplatin INJ.; Dong-A St., Korea) were obtained from the Medication Department of the National Cancer Center (Korea). Human NR1H4 cDNA (GenBank accession No. NM_001206979) was obtained from the DNASU Plasmid Repository (Arizona State University, USA) and cloned in an expression vector (pFLAG-CMV2-hNR1H4). The primer sequences used for polymerase chain reaction (PCR) and cloning are available on request. Fragments obtained by PCR and subcloning were confirmed by DNA sequencing. pSpCas9(BB)-2A-GFP (px458) was a gift from Feng Zhang (Addgene plasmid #48138; http://n2t.net/addgene:48138; RRID: Addgene_48138). Lipofectamine 2000 was purchased from Invitrogen (USA).
Cell culture
The human colon carcinoma cell lines HT29 were obtained from the American Type Culture Collection (USA). All cells were authenticated by short-tandem repeat PCR method in 2017 by National Cancer Center Omics Core facility. HT29 cells were maintained in McCoy’s 5A, supplemented with 10% heat-inactivated FBS. All cells were grown at 37°C in a humidified 5% CO2 atmosphere.
Downregulating NR1H4 expression in colon cancer cells
Generation of NR1H4 KO cell lines
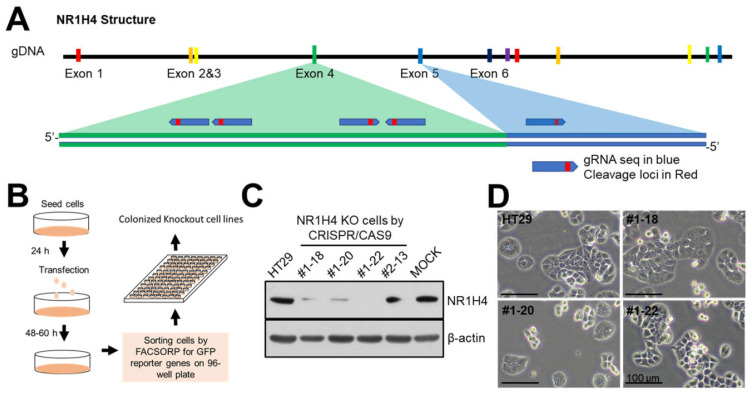
Clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein-9 nuclease (CAS9) system was applied to generate NR1H4 KO cells following the protocol with pSpCas9(BB)-2A-Green fluorescent protein (GFP, px458) (Ran et al., 2013). First guide RNAs specific for NR1H4 were designed using ChopChop site (http://chopchop.cbu.uib.no) and inserted to pSpCas9(BB) -2A-GFP, followed by sequencing for confirmation. Cells (5 × 105) were grown for 24 h and transfected with cloned CRISPR/CAS9 systems targeting NR1H4 or CRISPR/CAS9 vector (MOCK). After 48 h, cells with GFP reporter proteins were sorted by FACSORP and seeded onto 96-well plates and grown until each clone formed visible colony. Obtained colonies were evaluated by immunoblotting for NR1H4 expression. Four cell lines (#1-18, #1-20, #1-22, and #2-13) and one MOCK were established and three NR1H4 KO cell lines (#1-18, #1-20, and #1-22) and one MOCK cell lines were applied for this study.
Small interfering (si) RNA-mediated silencing of NR1H4
Cells were grown for 24 h and transfected with siRNAs against NR1H4 (Santa Cruz Biotechnology) by using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instruction. After 48 h, cells were harvested for immunoblotting.
Cell proliferation assay
Cells (5 × 104) were grown in 6-well plates upto 168 h and counted every 24 h after trypsinization for comparison. Some cells were grown on 6-well plates for 72 h and counted for comparison. At the same time, digital images of cells were obtained with 10× or 20× magnification by using light microscope, Olympus CKX53 (Olympus, Japan).
Clonogenic assay
Two hundreds of cells were plated on 6-well plates and grown for 10 days. Formed colonies were fixed and stained using a Diff-Quik stain kit (Sysmex, Japan). Then, stained colonies were air-dried and digital images of colonies were obtained with 10× or 20× magnification by using light microscope, Olympus CKX53 microscope. Colonies were counted by software Image J (width 12-pixel, threshold 1.5) as well as direct counting for statistical analyses.
Flowcytometry for analyzing cell death
Cells (5 × 105) were grown on 6-well plates for during 48 h and harvested for apoptosis assay according to manufacturer’s instruction (FITC Annexin V Apoptosis Detection Kit, 556547; BD Pharmingen, USA). The supernatants and cells were collected, centrifuged at 1,500 rpm for 5 min and washed with ice-cold phosphate buffered saline (PBS). Then, floating and adherent cells were suspended with Annexin V (ANX) binding buffer and treated with FITC ANX V and propidium iodide (PI) staining solution for 15 min in dark at room temperature for analyses with LSR Fortessa SORP (BD Biosciences, USA).
RNA extraction, reverse transcription (RT)-PCR, and PCR array analysis
RNA extraction and RT-PCR
Cells (5 × 105) were grown in 6-well plates for 24 h and harvested. Total RNA was extracted with the RNeasy kit (Qiagen, USA). RNA was quantified, and samples (5 µg) were reverse-transcribed at 42°C for 60 min in 20 µl buffer (10 mM Tris, pH 8.3, 50 mM KCl, 5 mM MgCl2, and 1 mM dNTP) in the presence of a random hexamer primer. Hot-start PCR was performed to increase the specificity of amplification. The PCR products were subjected to electrophoresis on 1.5% (w/v) agarose gels, and the resulting bands were visualized with ethidium bromide and photographed using the GelDoc program (Bio-Rad, USA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA was used as an endogenous control.
Profiling signaling pathway with RT2 Profiler PCR array
To profile altered signaling pathway in NR1H4 KO in HT29 colon cancer cells, we conducted PCR array with RT2 Profiler PCR Array Human Signal Transduction Pathway Finder kit (PAHS-014ZF-12, 330231; Qiagen), according to manufacturer’s instruction. Growing cells were harvested for RNA extraction, which was qualified by bioanalyzer before cDNA synthesis. Qualified RNAs were used for cDNA synthesis, which were subjected to PCR array and analyzed using a Roche LightCycler 480 (Roche Diagnostics, USA).
Cell viability by MTT assay
Cells (2 × 103 or 5 × 103) were grown in 96-well plates for 24 h and treated with anticancer drugs such as 0.01-200 µM gefitinib, doxorubicin, cisplatin or vehicle for 96 h. Then, cells were harvested for viability assay using 3-(4,5-dimethylthiazol-2yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT). Before harvesting, cells were exposed to MTT agent (final concentration of 0.5 mg/ml) and aspirated. Formed formazan was melted with DMSO for 5 min, whose optical density (OD) at 540 nm and at 650 nm were measured. OD values (Absorbance at 540 nm - Absorbance at 640 nm -blank) were used for viability comparison. Treated cells were observed after treated 96 h. Cell viability was analyzed by MTT.
Immunoblotting
Protein samples were heated at 95°C for 7-10 min and separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis using 8% to 15% acrylamide gels, followed by transfer to polyvinylidene difluoride membranes. The membranes were blocked for 1 h in Tris-buffered saline with 0.01% Tween-20 (TBST) with 5% non-fat dried milk, after which they were incubated overnight with primary antibody in TBST with 2% non-fat dried milk or 3% bovine serum albumin (BSA), followed by incubation with horseradish peroxidase-conjugated anti-mouse or -rabbit antibodies. The blots were developed with an enhanced chemiluminescence kit (West-ZOL plus, Western Blot Detection System; iNtRON Biotechnology, Korea), and quantification of band intensity on XAR-5 film (Eastman Kodak, USA) was measured with Quantity One software (Bio-Rad).
Statistical analysis
All data are expressed as percentages of the control and shown as mean ± SE. Statistical comparisons between groups were performed using Student’s t-tests. Values of P < 0.05 were considered significant. All analyses were performed using GraphPad Prism 5 software (GraphPad Software, USA).
RESULTS
Generation of NR1H4 KO cell lines
To assess the oncogenic role of NR1H4 in colon cancer, we generated NR1H4 KO colon cancer cells. Guide RNAs (gRNAs) against NR1H4 were cloned into pSpCas9(BB)-2A-GFP, which is an all-in-one CRISPR/CAS9 system encoding CAS9 and gRNA in a single vector (Ran et al., 2013). Cells were transfected with NR1H4-pSpCas9(BB)-2A-GFP and flow-sorted for GFP after incubation for 48-60 h. GFP-positive single cells were seeded into a 96-well plate and incubated until visible colonies were formed. Colonies were subjected to immunoblotting for NR1H4 expression (Fig. 1). Four KO cell line clones (#1-18, #1-20, #1-22, and #2-13) were confirmed as NR1H4 KO. Three of the KO cell line clones (#1-18, #1-20, and #1-22), as well as one MOCK cell line clone, were used for further analysis.
Fig. 1. Generation of NR1H4 KO cell lines using CRISPR/CAS9 technology.
(A) Simplified gene structure of NR1H4. (B) Workflow of the methodology used to generate NR1H4 KO cell lines. (C) Cells were grown for 24 h in 6-well plates and harvested for immunoblotting to evaluate NR1H4 expression. (D) Representative light microscopy images of sub-confluent cells. Results shown are representative of at least three independent experiments. Scale bars = 100 µm.
NR1H4 KO affects cell viability in HT29 colon cancer cells
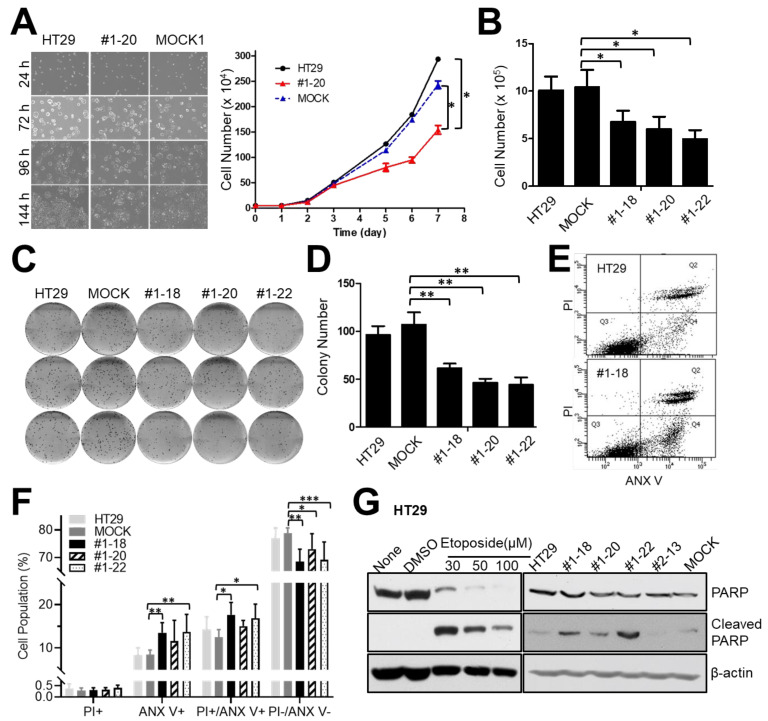
Interestingly, we observed that NR1H4 KO cells showed impaired cell growth and proliferation compared with MOCK and HT29 parental cells. To quantify the impact of NR1H4 KO on colon cancer cell growth and proliferation, we seeded 10,000 cells into 35 mm plates and incubated for 8 days; cells were counted every day. NR1H4 KO clone #1-20 showed a drastic reduction in cell proliferation compared with parental and MOCK HT29 cells 3 days after incubation (Figs. 2A and 2B). NR1H4 KO clones #1-18 and #1-22 showed a reduction in growth rate, similar to clone #1-20. Clonogenic assays revealed that all three NR1H4 KO clones showed reduced survival compared with MOCK and parental HT29 cells (Figs. 2C and 2D), suggesting a role of NR1H4 in colon cancer cell growth and survival. We also assessed the impact of NR1H4 KO on the cell cycle by flow cytometry. Cells were grown for 24, 48, and 72 h followed by cell cycle analysis. Clone #1-20 showed a higher percentage of cells in sub-G1 phase compared with parental or MOCK HT29 cells, suggesting a role of NR1H4 in cell cycle progression and cell death (data not shown). To investigate whether NR1H4 expression affects cell death in colon cancer cells, all clones were grown in regular media supplemented with 10% FBS for 3 days and then harvested for cell death analysis by flow cytometry. At 48 h, the percentage of living cells (ANX V-negative and PI-negative) was significantly lower in NR1H4 KO clones #1-18, #1-20, and #1-22 compared with parental and MOCK HT29 cells (Figs. 2E and 2F), while a higher percentage of early-apoptotic (ANX V-positive) cells was observed in NR1H4 KO cells. The percentage of non-apoptotic (PI-positive) cells did not differ significantly between the groups. Furthermore, the levels of cleaved PARP were higher in all NR1H4 KO clones (Fig. 2G), supporting a role of NR1H4 in the evasion of apoptotic cell death.
Fig. 2. NR1H4 KO affects cell proliferation, survival, and apoptosis of HT29 colon cancer cells.
(A) Cells (5 × 104) were seeded into 35 mm plates and incubated for 144 h. Cells were counted every 24 h and taken pictures at 10× magnification. (B) Cells (1 × 105) were plated into 6-well plates and incubated for 72 h. The cell number was quantified. (C and D) Cells (200/well) were plated into 6-well plates and incubated for 10 days. Colonies were stained using the Diff-Quik system. Digital images were obtained by light microscopy (C) and quantified using ImageJ software (D). (E and F) Cells were grown for 48 h and harvested for flow cytometry to measure apoptotic cell death after staining with ANX V and PI. The percentage of non-apoptotic (PI+/ANX V–), early apoptotic (PI–/ANX V+), dying (PI+/ANX V+), and living (PI–/ANX V–) parental, MOCK, and NR1H4 KO HT29 cells were compared. (G) Cells were incubated for 72 h and harvested for immunoblotting. Some cells were grown for 24 h and exposed to Etoposide (30-100 µM) or DMSO (vehicle) for an additional 48 h, followed by immunoblotting. Results shown are representative of at least three independent experiments. Data are expressed as mean ± SE of at least three independent experiments. Statistical significance was assessed using unpaired Student’s t-test or one-way ANOVA. *P < 0.005, **P < 0.001, ***P < 0.0001.
NR1H4 KO affects MYC expression in HT29 colon cancer cells
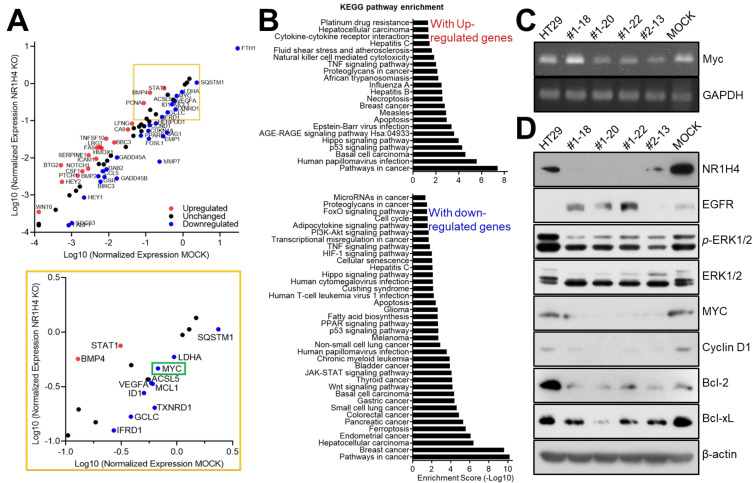
We performed a PCR array using the RT2 Profiler PCR Array (Signal Transduction Pathway Finder, 330231; Qiagen) to identify alterations in cell signaling in NR1H4 KO colon cancer cells. Parental, MOCK, and #1-20 HT29 cells were grown in 60 mm dishes for 24 h and harvested for RNA extraction. After RT, cDNA from each cell line was subjected to a PCR array. A total of 80 genes important for cancer cell signaling were analyzed (Fig. 3A). The expression of 18 genes, including NOTCH1, HEY2, CSF1, BTG2, STAT1, and WNT6, was elevated in NR1H4 KO cells; the signaling pathways involved in platinum drug resistance and cytokine-cytokine receptor interaction were found to be enriched by KEGG pathway analysis (Fig. 3B, upper panel). The expression of more than 20 genes, including MYC, was decreased in NR1H4 KO cells; these genes were involved in cell cycle, fatty acid synthesis, PPAR signaling, p53 signaling, ferroptosis, and apoptosis (Fig. 3B, lower panel). Importantly, MYC was downregulated in all NR1H4 KO clones, both at the mRNA (Fig. 3C) and protein level (Fig. 3D), suggesting that NR1H4 regulates Myc expression. All NR1H4 KO clones showed impaired activation of extracellular signal-regulated kinases (ERKs) and a lower expression of CyclinD1 compared with MOCK and parental HT29 cells. The levels of anti-apoptotic proteins, such as Bcl-2 and Bcl-xL, were also decreased in NR1H4 KO cells. These findings further supported our results that NR1H4 KO cells showed cell cycle progression impairment and subsequent apoptotic cell death, possibly through regulating Myc expression (Chen et al., 2018; Conacci-Sorrell et al., 2014; Dang, 2012; Garcia-Gutierrez et al., 2019).
Fig. 3. NR1H4 KO affects MYC expression in HT29 colon cancer cells.
(A and B) Cells (1 × 106) were incubated for 24 h and harvested for RNA extraction and reverse-transcription. RT2 Profiler PCR Array for Human Signal Transduction Pathway was performed. Gene expression alterations were analyzed by scatter plot (A) and DAVID analyses, followed by KEGG pathway enrichment analysis (B). (C) Subconfluent cells were harvested for RT-PCR to validate MYC expression at the RNA level. (D) Cells were incubated for 24 h and harvested for immunoblotting to examine the expression of several cellular proteins. Results shown are representative of at least three independent experiments.
NR1H4 affects MYC stability in HT29 colon cancer cells
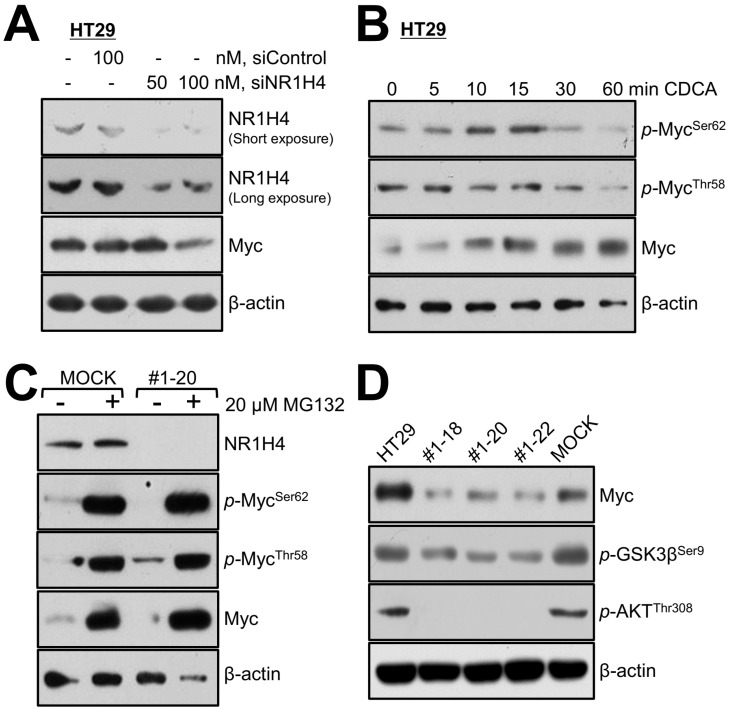
To investigate whether NR1H4 expression affects Myc expression and stability, we transiently silenced NR1H4 expression in HT29 parental cells using siRNA (Fig. 4A). NR1H4 silencing resulted in a profound decrease in MYC protein levels, which was more drastic at 48 h than 24 h, supporting the hypothesis that NR1H4 regulates Myc expression indirectly. In the presence of growth factors, ERK mediates Myc phosphorylation at Ser62, increasing its stability and activity; however, phosphorylation of Thr58 by GSK3β promotes ubiquitinylation-mediated degradation (Cao et al., 2011; Kazi et al., 2018; Sears et al., 2000). When cells were treated with the proteasome inhibitor MG132, Myc expression and phosphorylation levels were similar in MOCK and #1-20 cells, regardless of NR1H4 expression (Fig. 4C). Interestingly, the phosphorylation levels of Myc on Thr58 were higher in #1-20 compared with MOCK cells, suggesting phosphorylation-mediated protein degradation of Myc in NR1H4 KO cells. When parental HT29 cells were treated with chenodeoxycholic acid, a metabolic ligand for NR1H4, Myc protein levels increased within 1 h, while Thr58 phosphorylation levels decreased (Fig. 4B). As both GSK3β and AKT mediate phosphorylation of Thr58 of Myc, their protein levels were investigated by immunoblotting. We found that NR1H4 KO clones had lower levels of phosphorylated GSK3β (active) and AKT (inactive), suggesting that both inactivation of AKT and activation of GSK3β contribute to MYC phosphorylation at Thr58 in NR1H4 KO cells (Fig. 4D).
Fig. 4. NR1H4 activity is closely related to MYC expression and stability in HT29 colon cancer cells.
(A) Cells were grown in 6-well plates for 24 h and transfected with siRNAs targeting NR1H4 for 48 h. (B) Cells were grown in 6-well plates for 24 h and exposed to 30 µM CDCA for the indicated period of time (0-60 min), following which cells were harvested for immunoblotting. Cells were grown in 6-well plates for 24 h and treated with 20 µM MG143 for 6 h, followed by immunoblotting. Cells were grown in 6-well plates for 24 h and harvested for immunoblotting. Results shown are representative of at least three independent experiments.
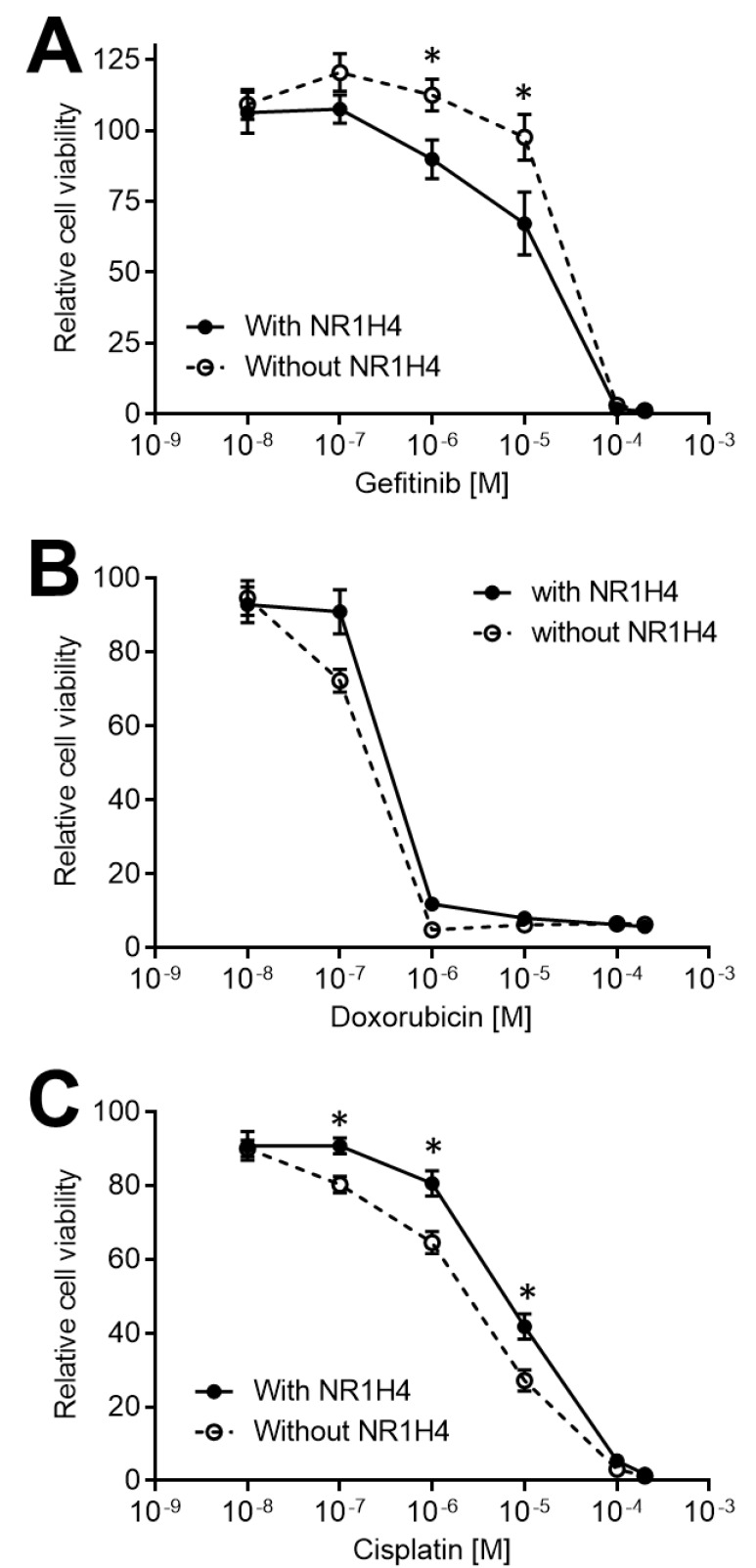
NR1H4 KO modulates drug sensitivity and apoptosis in HT29 colon cancer cells
Myc is a master regulator of several cellular processes, including cell growth, survival, and death, mediated by downstream signaling pathways in a cell context-dependent manner (Conacci-Sorrell et al., 2014; Dang, 2012). Myc has been implicated in several aspects of carcinogenesis, including metabolism (Satoh et al., 2017; Soucek et al., 2008; Stine et al., 2015) and cell cycle (Altman et al., 2015; Garcia-Gutierrez et al., 2019). Therefore, several emerging anti-cancer therapeutic strategies aim to inhibit Myc (Chen et al., 2018; Kazi et al., 2018; Sears et al., 2000; Soucek et al., 2008). Importantly, Myc has also been implicated in the efficacy of anti-cancer drugs (Frenzel et al., 2011; Jo et al., 2015; Leonetti et al., 1999). Leonetti and colleagues have shown that MYC silencing using anti-sense oligonucleotides increased cisplatin sensitivity in human metastatic melanoma (Leonetti et al., 1999). Another study has identified cytotoxic drugs that selectively target tumor cells that overexpress Myc, such as doxorubicin (Frenzel et al., 2011). To assess the impact of NR1H4-mediated Myc expression changes on the response to anti-cancer drugs, we compared the responsiveness of HT29 cells to doxorubicin, cisplatin, and gefitinib. Cells were treated with 0, 0.01, 0.1, 1, 10, 100, and 200 µM of cisplatin, doxorubicin, or gefitinib; as a control, cells were also treated with vehicles (DMSO or PBS). After incubation for 96 h, the relative cell survival for each group was assessed by MTT assay (Fig. 5). NR1H4 KO clones were more sensitive to cisplatin-induced cell death compared with MOCK or parental HT29 cells (Fig. 5C), supporting previous studies reporting a role of Myc in cisplatin sensitivity (Date and Ito, 2020; Leonetti et al., 1999). Moreover, NR1H4 KO cells showed increased cell death after doxorubicin treatment, compared with parental or MOCK HT29 cells (Fig. 5B), suggesting a role of Myc in resistance to doxorubicin. Sarosiek and colleagues have reported a role of Myc in apoptosis in a tissue-specific manner, through regulating mitochondrial function (Sarosiek et al., 2017). Protein profiling in MOCK and NR1H4 KO cells led to the observation that epidermal growth factor receptor (EGFR) was drastically upregulated in NR1H4 KO clones (except clone #2-13), suggesting that EGFR might have a role in NR1H4-mediated cell survival in colon cancer cells. NR1H4 KO cells showed higher viability after gefitinib treatment compared with MOCK or parental HT29 cells (Fig. 5A).
Fig. 5. NR1H4 KO modulates anti-cancer drug sensitivity and apoptosis in HT29 colon cancer cells.
NR1H4-expressing (MOCK) and NR1H4-deficient (#1-18, #1-20, and #1-22) HT29 cells were grown for 24 h and treated with gefitinib (A), doxorubicin (B), or cisplatin (C) for 96 h. Cells were harvested and cell viability was assessed using the MTT assay. Results shown are representative of at least three independent experiments. Data are expressed as mean ± SE of at least three independent experiments. Statistical significance was assessed using unpaired Student’s t-test or one-way ANOVA. *P < 0.05.
DISCUSSION
As a bile acid nuclear receptor, the role of NR1H4 in maintaining bile acid homeostasis in the liver-intestine axis is well established; however, the role of bile acid homeostasis dysregulation in liver and colon tumorigenesis is less clear. NR1H4 has been suggested to be an important regulator of colon and liver cancer development. Gomez-Ospina and colleagues have shown that loss-of-function variants of NR1H4 caused progressive familial intrahepatic cholestasis, implying a role of NR1H4 in liver protection (Gomez-Ospina et al., 2016), and highlighting its importance in the regulation of bile acid homeostasis. Some studies have also demonstrated that NR1H4 is involved in intestinal epithelial cell proliferation (Maran et al., 2009) and cholic acid-induced hepatocarcinogenesis (Kong et al., 2016). An inverse correlation between NR1H4 and MYC has also been shown. Interestingly, the comparison between whole-body, hepatocyte-specific, and enterocyte-specific NR1H4-null mice suggested that enterocyte-specific NR1H4 KO mice had a lower risk of hepatic tumor development (Takahashi et al., 2018). Herein, we addressed the role of NR1H4 in Myc expression in colon cancer cells. We are further investigating if the effect of NR1H4 on Myc expression might be altered by environmental stimuli from other types of cells in tissues. Our findings suggested that NR1H4 regulates the expression levels of MYC, as well as the stability of Myc protein, leading to enhanced cell death in response to anti-cancer agents, including doxorubicin (Fig. 5). We found that EGFR was expressed at higher levels in NR1H4 KO cells compared with MOCK cells; however, gefitinib, an EGFR targeting anti-cancer agent, did not show higher efficacy in NR1H4 KO cells (Fig. 5). Protein profiling showed that several proteins involved in cell cycle progression and cell growth, including ERK, Myc, and CyclinD1, were drastically downregulated in NR1H4 KO colon cancer cells, which suggests the EGFR signaling to cell survival might be not active in NR1H4 KO cells. Botany and colleagues demonstrated a role of the EGFR signaling pathways on human β-defensin-1 expression through Myc in gefitinib-treated colonic epithelial cells (Bonamy et al., 2018). Human β-defensin-1 has reported to be regulated by NR1H4 in liver and colonic epithelial cells (Klag et al., 2018; Lajczak et al., 2017), suggesting another crosstalk between EGFR-Myc-NR1H4 in colon cancer cells. The detailed mechanism between them is under being investigated.
In conclusion, our study showed that NR1H4 KO resulted in decreased cell proliferation and cell survival, as well as in increased apoptotic cell death in colon cancer cells. Moreover, NR1H4 KO resulted in decreased MYC, Bcl-xL, Bcl-2, and CyclinD protein levels. We found that NR1H4 levels positively correlated to MYC protein levels and that NR1H4 KO altered the efficacy of anti-cancer drugs as well as apoptotic cell death in colon cancer cells.
ACKNOWLEDGMENTS
We thank Tae Sik Kim of the FACS core (National Cancer Center), Jung A Hwang of the Genomics core (National Cancer Center), Ji Yea Kim and Dr. Heesun Chung (National Cancer Center) for their expert assistance and helpful suggestions.
This work was supported in part by National Cancer Center Grant NCC-1710180 (to H.J.Y.) and NCC-1710252 (to H.J.Y.) and National Research Foundation of Korea Grant funded by the Korea Government (MSIP, South Korea) (No. 2015R1A2A2A01003829 to H.J.Y., No. 2019R1H1A2039666).
Footnotes
AUTHOR CONTRIBUTIONS
Y.J.L. and E.Y.L. performed experiments mainly. H.J.Y. designed the research study. E.Y.L., B.H.C., and H.J. contributed essential reagents or tools. Y.J.L., E.Y.L., J.K.M., and H.J.Y. analyzed the data. Y.J.L. and H.J.Y. wrote the manuscript. H.J.Y. supervised the research.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Altman B.J., Hsieh A.L., Sengupta A., Krishnanaiah S.Y., Stine Z.E., Walton Z.E., Gouw A.M., Venkataraman A., Li B., Goraksha-Hicks P., et al. MYC disrupts the circadian clock and metabolism in cancer cells. Cell Metab. 2015;22:1009–1019. doi: 10.1016/j.cmet.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A.M., Zhan L., Maru D., Shureiqi I., Pickering C.R., Kiriakova G., Izzo J., He N., Wei C., Baladandayuthapani V., et al. FXR silencing in human colon cancer by DNA methylation and KRAS signaling. Am. J. Physiol. Gastrointest. Liver Physiol. 2014;306:G48–G58. doi: 10.1152/ajpgi.00234.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonamy C., Sechet E., Amiot A., Alam A., Mourez M., Fraisse L., Sansonetti P.J., Sperandio B. Expression of the human antimicrobial peptide beta-defensin-1 is repressed by the EGFR-ERK-MYC axis in colonic epithelial cells. Sci. Rep. 2018;8:18043. doi: 10.1038/s41598-018-36387-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z., Fan-Minogue H., Bellovin D.I., Yevtodiyenko A., Arzeno J., Yang Q., Gambhir S.S., Felsher D.W. MYC phosphorylation, activation, and tumorigenic potential in hepatocellular carcinoma are regulated by HMG-CoA reductase. Cancer Res. 2011;71:2286–2297. doi: 10.1158/0008-5472.CAN-10-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Liu H., Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct. Target. Ther. 2018;3:5. doi: 10.1038/s41392-018-0008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conacci-Sorrell M., McFerrin L., Eisenman R.N. An overview of MYC and its interactome. Cold Spring Harb. Perspect. Med. 2014;4:a014357. doi: 10.1101/cshperspect.a014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C.V. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date Y., Ito K. Oncogenic RUNX3: a link between p53 deficiency and MYC dysregulation. Mol. Cells. 2020;43:176–181. doi: 10.14348/molcells.2019.0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Aguiar Vallim T.Q., Tarling E.J., Edwards P.A. Pleiotropic roles of bile acids in metabolism. Cell Metab. 2013;17:657–669. doi: 10.1016/j.cmet.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis R.J., Chandel N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016;2:e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degirolamo C., Modica S., Palasciano G., Moschetta A. Bile acids and colon cancer: solving the puzzle with nuclear receptors. Trends Mol. Med. 2011;17:564–572. doi: 10.1016/j.molmed.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Frenzel A., Zirath H., Vita M., Albihn A., Henriksson M.A. Identification of cytotoxic drugs that selectively target tumor cells with MYC overexpression. PLoS One. 2011;6:e27988. doi: 10.1371/journal.pone.0027988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu T., Coulter S., Yoshihara E., Oh T.G., Fang S., Cayabyab F., Zhu Q., Zhang T., Leblanc M., Liu S., et al. FXR regulates intestinal cancer stem cell proliferation. Cell. 2019;176:1098–1112.:e18. doi: 10.1016/j.cell.2019.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gutierrez L., Delgado M.D., Leon J. MYC oncogene contributions to release of cell cycle brakes. Genes (Basel) 2019;10:244. doi: 10.3390/genes10030244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Ospina N., Potter C.J., Xiao R., Manickam K., Kim M.S., Kim K.H., Shneider B.L., Picarsic J.L., Jacobson T.A., Zhang J., et al. Mutations in the nuclear bile acid receptor FXR cause progressive familial intrahepatic cholestasis. Nat. Commun. 2016;7:10713. doi: 10.1038/ncomms10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinney J., Dienstmann R., Wang X., de Reynies A., Schlicker A., Soneson C., Marisa L., Roepman P., Nyamundanda G., Angelino P., et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlston R.S. What we could do now: molecular pathology of colorectal cancer. Mol. Pathol. 2001;54:206–214. doi: 10.1136/mp.54.4.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh A.L., Walton Z.E., Altman B.J., Stine Z.E., Dang C.V. MYC and metabolism on the path to cancer. Semin. Cell Dev. Biol. 2015;43:11–21. doi: 10.1016/j.semcdb.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo M.J., Paek A.R., Choi J.S., Ok C.Y., Jeong K.C., Lim J.H., Kim S.H., You H.J. Regulation of cancer cell death by a novel compound, C604, in a c-Myc-overexpressing cellular environment. Eur. J. Pharmacol. 2015;769:257–265. doi: 10.1016/j.ejphar.2015.11.027. [DOI] [PubMed] [Google Scholar]
- Kazi A., Xiang S., Yang H., Delitto D., Trevino J., Jiang R.H.Y., Ayaz M., Lawrence H.R., Kennedy P., Sebti S.M. GSK3 suppression upregulates beta-catenin and c-Myc to abrogate KRas-dependent tumors. Nat. Commun. 2018;9:5154. doi: 10.1038/s41467-018-07644-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klag T., Thomas M., Ehmann D., Courth L., Mailander-Sanchez D., Weiss T.S., Dayoub R., Abshagen K., Vollmar B., Thasler W.E., et al. Beta-defensin 1 is prominent in the liver and induced during cholestasis by bilirubin and bile acids via farnesoid X receptor and constitutive androstane receptor. Front. Immunol. 2018;9:1735. doi: 10.3389/fimmu.2018.01735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong B., Zhu Y., Li G., Williams J.A., Buckley K., Tawfik O., Luyendyk J.P., Guo G.L. Mice with hepatocyte-specific FXR deficiency are resistant to spontaneous but susceptible to cholic acid-induced hepatocarcinogenesis. Am. J. Physiol. Gastrointest. Liver Physiol. 2016;310:G295–G302. doi: 10.1152/ajpgi.00134.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers E.J., Grady W.M., Lieberman D., Seufferlein T., Sung J.J., Boelens P.G., van de Velde C.J., Watanabe T. Colorectal cancer. Nat. Rev. Dis. Primers. 2015;1:15065. doi: 10.1038/nrdp.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajczak N.K., Saint-Criq V., O'Dwyer A.M., Perino A., Adorini L., Schoonjans K., Keely S.J. Bile acids deoxycholic acid and ursodeoxycholic acid differentially regulate human beta-defensin-1 and -2 secretion by colonic epithelial cells. FASEB J. 2017;31:3848–3857. doi: 10.1096/fj.201601365R. [DOI] [PubMed] [Google Scholar]
- Leonetti C., Biroccio A., Candiloro A., Citro G., Fornari C., Mottolese M., Del Bufalo D., Zupi G. Increase of cisplatin sensitivity by c-myc antisense oligodeoxynucleotides in a human metastatic melanoma inherently resistant to cisplatin. Clin. Cancer Res. 1999;5:2588–2595. [PubMed] [Google Scholar]
- Luengo A., Gui D.Y., Vander Heiden M.G. Targeting metabolism for cancer therapy. Cell Chem. Biol. 2017;24:1161–1180. doi: 10.1016/j.chembiol.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maran R.R., Thomas A., Roth M., Sheng Z., Esterly N., Pinson D., Gao X., Zhang Y., Ganapathy V., Gonzalez F.J., et al. Farnesoid X receptor deficiency in mice leads to increased intestinal epithelial cell proliferation and tumor development. J. Pharmacol. Exp. Ther. 2009;328:469–477. doi: 10.1124/jpet.108.145409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan A., Malvi P., Wajapeyee N. Oncogene-directed alterations in cancer cell metabolism. Trends Cancer. 2016;2:365–377. doi: 10.1016/j.trecan.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita A., Takahashi S., Ouchi K., Inoue M., Watanabe M., Endo M., Honda H., Yamada Y., Ishioka C. Consensus molecular subtypes classification of colorectal cancer as a predictive factor for chemotherapeutic efficacy against metastatic colorectal cancer. Oncotarget. 2018;9:18698–18711. doi: 10.18632/oncotarget.24617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuyama H., Endo H., Akashika T., Kato K., Inoue M. Downregulation of c-MYC protein levels contributes to cancer cell survival under dual deficiency of oxygen and glucose. Cancer Res. 2010;70:10213–10223. doi: 10.1158/0008-5472.CAN-10-2720. [DOI] [PubMed] [Google Scholar]
- Ortmayr K., Dubuis S., Zampieri M. Metabolic profiling of cancer cells reveals genome-wide crosstalk between transcriptional regulators and metabolism. Nat. Commun. 2019;10:1841. doi: 10.1038/s41467-019-09695-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl P.B., Lin C.Y., Seila A.C., Flynn R.A., McCuine S., Burge C.B., Sharp P.A., Young R.A. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarosiek K.A., Fraser C., Muthalagu N., Bhola P.D., Chang W., McBrayer S.K., Cantlon A., Fisch S., Golomb-Mello G., Ryan J.A., et al. Developmental regulation of mitochondrial apoptosis by c-Myc governs age- and tissue-specific sensitivity to cancer therapeutics. Cancer Cell. 2017;31:142–156. doi: 10.1016/j.ccell.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh K., Yachida S., Sugimoto M., Oshima M., Nakagawa T., Akamoto S., Tabata S., Saitoh K., Kato K., Sato S., et al. Global metabolic reprogramming of colorectal cancer occurs at adenoma stage and is induced by MYC. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E7697–E7706. doi: 10.1073/pnas.1710366114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears R., Nuckolls F., Haura E., Taya Y., Tamai K., Nevins J.R. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.R., Myint T., Goh H.S. Over-expression of the c-myc proto-oncogene in colorectal carcinoma. Br. J. Cancer. 1993;68:407–413. doi: 10.1038/bjc.1993.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L., Whitfield J., Martins C.P., Finch A.J., Murphy D.J., Sodir N.M., Karnezis A.N., Swigart L.B., Nasi S., Evan G.I. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stine Z.E., Walton Z.E., Altman B.J., Hsieh A.L., Dang C.V. MYC, metabolism, and cancer. Cancer Discov. 2015;5:1024–1039. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sveen A., Bruun J., Eide P.W., Eilertsen I.A., Ramirez L., Murumagi A., Arjama M., Danielsen S.A., Kryeziu K., Elez E., et al. Colorectal cancer consensus molecular subtypes translated to preclinical models uncover potentially targetable cancer cell dependencies. Clin. Cancer Res. 2018;24:794–806. doi: 10.1158/1078-0432.CCR-17-1234. [DOI] [PubMed] [Google Scholar]
- Takahashi S., Tanaka N., Fukami T., Xie C., Yagai T., Kim D., Velenosi T.J., Yan T., Krausz K.W., Levi M., et al. Role of farnesoid X receptor and bile acids in hepatic tumor development. Hepatol. Commun. 2018;2:1567–1582. doi: 10.1002/hep4.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]