Abstract
Severe Acute Respiratory Syndrome coronavirus 2 (SARS‐CoV‐2) is rapidly spreading around the world. There is no existing vaccine or proven drug to prevent infections and stop virus proliferation. Although this virus is similar to human and animal SARS‐CoVs and Middle East Respiratory Syndrome coronavirus (MERS‐CoVs), the detailed information about SARS‐CoV‐2 proteins structures and functions is urgently needed to rapidly develop effective vaccines, antibodies, and antivirals. We applied high‐throughput protein production and structure determination pipeline at the Center for Structural Genomics of Infectious Diseases to produce SARS‐CoV‐2 proteins and structures. Here we report two high‐resolution crystal structures of endoribonuclease Nsp15/NendoU. We compare these structures with previously reported homologs from SARS and MERS coronaviruses.
Keywords: COVID‐19, crystal structure, endoribonuclease, EndoU family, NendoU, Nsp15, SARS‐CoV‐2
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is an etiologic agent responsible for the current outbreak of coronavirus disease 2019 (COVID‐19). Over the past 3 months, the pathogen has infected over 1,600,000 people and caused at least 100,000 deaths. As we were preparing this report the numbers have nearly doubled. Although originally concentrated in China, the virus is spreading worldwide rapidly and is found in 183 countries and all continents (www.trackcorona.live). Millions of people are being quarantined and the epidemics impacts the world economy. There is no existing vaccine or proven drug for this disease, but various treatment options, for example, utilizing medicines effective in other ailments, are being attempted.
Coronaviruses are enveloped, nonsegmented positive‐sense RNA viruses from the order nidoviruses that have the largest genome among RNA viruses. The genome contains a large replicase gene encompassing nonstructural proteins (Nsps), followed by structural and accessory genes. Due to ribosomal frameshifting, the replicase gene encodes two open reading frames (ORFs), rep1a, and rep1b, that are translated into two large polyproteins, pp1a and pp1ab. 1 These polypeptides are processed by two viral proteases: 3C‐like protease (3CLpro, encoded by Nsp5), and papain‐like protease (PLP, encoded within Nsp3). The cleavage yields 15 or 16 viral Nsps 2 that assemble into a large membrane‐bound replicase complex and exhibits multiple enzymatic activities. While several functions of Nsps have been linked to RNA replication and processing of subgenomic RNAs, the roles of some proteins are poorly understood or remain unknown.
One of such enigmatic enzymes, corresponding to Nsp15, is a nidoviral RNA uridylate‐specific endoribonuclease (NendoU) carrying C‐terminal catalytic domain belonging to the EndoU family. EndoU enzymes are present in all kingdoms of life, where they play various biological functions associated with RNA processing. All characterized family members display an RNA endonuclease activity producing 2′‐3′ cyclic phosphodiester and 5′‐hydroxyl termini. 3 The viral and eukaryotic enzymes act on both, single‐stranded RNA and double‐stranded (ds) RNA and are specific for uridine. The prototypic member of the family was discovered in Xenopus laevis, and thus named XendoU, where it is associated with maturation of intron‐encoded small nucleolar RNAs (snoRNA).4, 5, 6, 7 The human homolog, placental protein 11 (PP11, HendoU) plays yet an unknown role in the placental tissue, 8 but is also expressed in cancer cells. In Drosophila, DendoU has been found to be relevant for nervous system physiology and pathology. 9 The EndoU representatives are also present in prokaryotes, with the best characterized enzyme lacking uridylate specificity of the family and functioning as a tRNase toxin in bacterial communication. 10
In viruses, the NendoU protein is conserved among coronaviruses, arteriviruses, and toroviruses, but is absent in nonvertebrate‐infecting representatives of the nidoviruses order: mesoniviruses and roniviruses. While initially Nsp15 was thought to directly participate in viral replication, it was later shown that Nsp15‐deficient coronaviruses were viable and replicating, rising doubts about the enzyme role in that process. More recently, it was proposed that NendoU activity of Nsp15 is responsible for the protein interference with the innate immune response, 11 though other studies indicate that the process is independent of the endonuclease activity. 12 There are also suggestions that Nsp15 degrades viral RNA to hide it from the host defenses. Nevertheless, Nsp15 is essential in coronavirus biology.
Nsp15s from coronaviruses share some sequence similarity with their arteriviruses counterpart Nsp11: The SARS‐CoV Nsp15 has only 22% identical residues with porcine reproductive and respiratory syndrome virus (PRRSV) Nsp11. Within arteriviruses alone sequence conservation of Nsp11 is generally low, while Nsp15 of coronaviruses are more conserved, with the current SARS‐CoV‐2 sharing 88% sequence identity and 95% similarity with its known closest homolog from SARS‐CoV. Nsp15 from Middle East Respiratory Syndrome coronavirus (MERS‐CoV) is more distant, with ~50% sequence identity and ~65% similarity and human coronavirus (H‐CoV‐229E) being even more distant with 43% sequence identity and 56% similarity.
After the 2002–2003 SARS outbreak there was an increased interest in virus‐encoded endoribonucleases. The first two structures of Nsp15 were determined from mouse hepatitis virus (MHV; Protein Data Bank [PDB] ids 2GTH, 2GTI 13 ) and SARS coronavirus (PDB id 2H85 14 ). Currently, there are in total eight structures available for Nsp15, including additional two from SARS‐CoV (PDB ids 2OZK, 2RHB 15 ), one from MERS‐CoV (PDB id 5YVD 16 ) and two from H‐CoV‐229E (PDB ids 4S1T, 4RS4 17 ). Among Nsp11 endoribonucleases, there are two structures determined for PRRSV homolog (PDB ids 5EYI, 5DA1 18 ), and two for equine arteritis virus (EAV, PDB ids 5HC1, 5HBZ 18 ). The structural studies of SARS‐ and MERS‐CoV Nsp15s showed that the protein forms hexamers made of dimers of trimers. The 39 kDa monomeric unit, composed of ~345 residues, folds into three domains: N‐terminal, middle domain, and C‐terminal catalytic NendoU domain.
Rapid upsurge and proliferation of SARS‐CoV‐2 raised questions about how this virus became so much more transmissible as compared to the SARS and MERS coronaviruses. The proteins of the coronavirus are mutating, although not very fast, but these changes may contribute to the virus virulence. Here, we report the first two crystal structures of SARS‐CoV‐2 Nsp15 determined at 1.90 Å and 2.20 Å resolution. The Nsp15 structure is very similar to the SARS‐CoV and MERS‐CoV homologs but it shows some differences that may contribute to altered SARS‐CoV‐2 virulence.
2. RESULTS AND DISCUSSION
2.1. Protein production and structure determination
We have used synthetic gene of SARS‐CoV‐2 coronavirus corresponding to sequence of Nsp15, which was codon optimized for expression in Escherichia coli, to produce soluble protein. Nsp15 was purified and crystallized exercising the well‐established structural genomics pipeline. The protein produced diffraction quality crystals under two conditions, yielding an apo form of Nsp15 (Nsp15/apo) and citrate‐bound form (Nsp15/cit), solved by molecular replacement and refined to 2.20 and 1.90 Å resolutions, as described in Section 4 and Table 1. The entire process from protein expression to structure deposition of the first Nsp15/apo model to the PDB took 5 days.
TABLE 1.
Data processing and refinement statistics
| Data processing | ||
|---|---|---|
| Structure | Nsp15/apo | Nsp15/cit |
| Wavelength (Å) | 0.9792 | 0.9792 |
| Resolution range (Å) a | 2.20–45.10 (2.20–2.24) | 1.90–44.90 (1.90–1.93) |
| Space group | P63 | P63 |
| Unit cell (Å) | a = 150.54, c = 111.31 | a = 150.78, c = 111.68 |
| Unique reflections, merged | 71,416 (3,176) | 111,894 (5,660) |
| Multiplicity | 5.1 (2.6) | 5.3 (5.1) |
| Completeness (%) | 98.5 (88.3) | 99.3 (99.7) |
| Mean I/sigma(I) | 10.1 (1.08) | 13.1 (1.43) |
| Wilson B‐factor (Å2) | 36.67 | 31.50 |
| R‐merge b | 0.174 (0.924) | 0.122 (1.525) |
| CC1/2 c | 0.967 (0.358) | 0.982 (0.586) |
| Refinement | ||
|---|---|---|
| Resolution range (Å) | 2.20–45.10 | 1.90–44.90 |
| Reflections work/test | 67,514/3,678 | 105,820/5,485 |
| R work/R free d | 0.158/0.178 | 0.161/0.185 |
| Number of nonhydrogen atoms | 6,009 | 6,261 |
| Macromolecules | 5,589 | 5,607 |
| Ligands/solvent | 74/346 | 135/519 |
| Protein residues | 348 × 2 | 348 × 2 |
| RMSD (bonds) (Å) | 0.002 | 0.011 |
| RMSD (angles) (°) | 0.433 | 0.982 |
| Ramachandran favored e (%) | 97.83 | 98.26 |
| Ramachandran allowed (%) | 2.17 | 1.74 |
| Ramachandran outliers (%) e | 0.0 | 0 |
| Rotamer outliers (%) e | 1.43 | 1.11 |
| Clashscore | 1.8 | 2.17 |
| Average B‐factor (Å2) | 44.0 | 41.0 |
| Macromolecules | 43.7 | 40.1 |
| Ligands | 69.4 | 62.9 |
| Solvent | 43.9 | 45.4 |
| Number of TLS groups | 9 | 11 |
| PDB ID | 6VWW | 6W01 |
Abbreviation: PDB, protein data bank; TLS, translation/libration/screw.
Values in parentheses correspond to the highest resolution shell.
Rmerge = ∑h∑j|Ihj–<Ih > |/∑h∑jIhj, where Ihj is the intensity of observation j of reflection h.
As defined by Karplus and Diederichs. 39
R = ∑h|Fo|–|Fc|/∑h|Fo| for all reflections, where Fo and Fc are observed and calculated structure factors, respectively. Rfree is calculated analogously for the test reflections, randomly selected and excluded from the refinement.
As defined by Molprobity. 40
Both structures of SARS‐CoV‐2 Nsp15 are of good quality and they refined to crystallographic R work of 15.8% and R free of 17.8% for the 2.20 Å structure and 16.1 and 18.5% for the 1.90 Å structure. The asymmetric unit contains two monomers of the Nsp15 protein. The electron density map is of high quality throughout the structure and the model covers the sequence from M1 to Q347. Currently, these are the most complete and the highest resolution structures of coronavirus Nsp15. In the final 2.20 Å structure 346 water molecules, eight glycerol molecules, three acetate ions, one magnesium, and one chloride ion were also identified in the electron density maps. For the 1.90 Å structure, 519 water molecules, 23 ethylene glycol molecules, three polyethylene glycol (PEG) molecules, and two citrate molecules were also found.
2.2. Overall structure
Except for the N‐ and C‐terminal residues, the difference between the two structures is very small as the root mean sqare deviation (RMSD) of Cα atoms is 0.44 Å for both chains. The structure of SARS‐CoV‐2 Nsp15 monomer is very similar to other Nsp15s from coronaviruses (Figures 1 and 2). It features three distinct domains. The N‐terminal domain is composed of an antiparallel β‐sheet (strands β1, β2, and β3) wrapped around two α‐helices (α1 and α2). The subsequent middle domain is formed by 10 β‐strands organized in three β‐hairpins (β5–β6, β7–β8, and β12–β13), a mixed β‐sheet (β4, β9, β10, β11, β14, and β15), and three short helices, two α, and one 310 (α3, η4, and α5). The C‐terminal catalytic NendoU domain contains two antiparallel β‐sheets (β16–β17–β18 and β19–β20–β21) with their edges hosting a catalytic site. β18, even though is broken in the secondary structure annotation, is treated here as a single element. The concave surface of the β‐sheets is flanked by five α‐helices (α6, α7, α8, α9, and α10).
FIGURE 1.
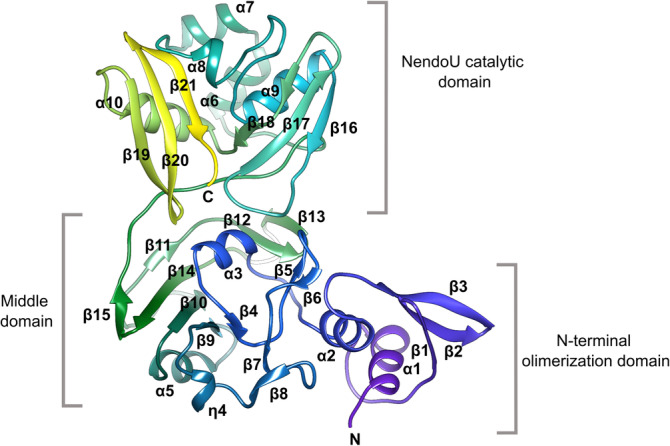
The structure of SARS‐CoV‐2 monomer
FIGURE 2.
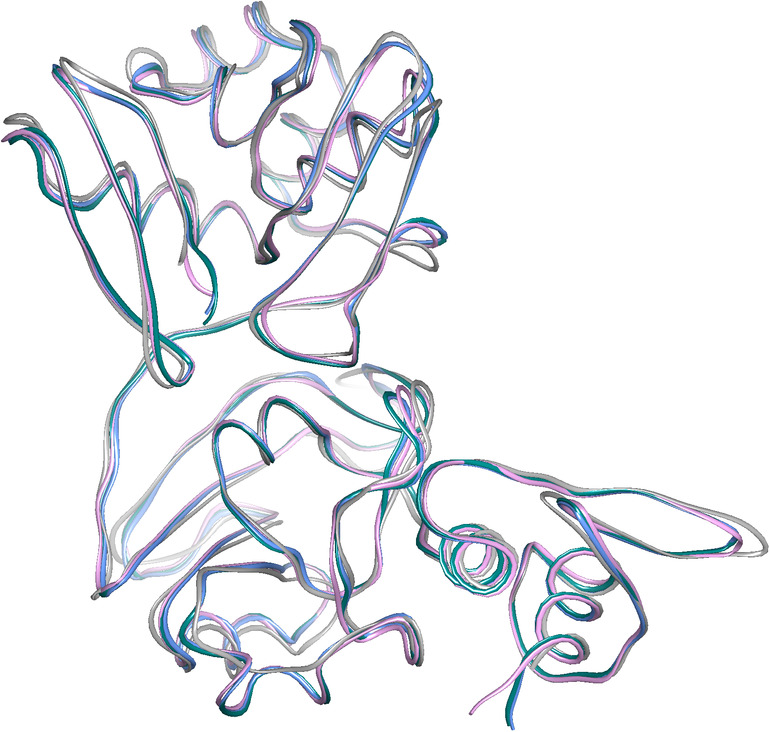
Superposition of Nsp15 monomers: SARS‐CoV‐2 (teal, Chain A; blue, Chain B), SARS‐CoV (pink), and MERS‐CoV (grey)
We use higher resolution structure Nsp15/cit form for comparisons with SARS‐ and MERS‐CoVs homologs. As expected from high sequence identity (Figure 3), the structure aligns best with SARS‐CoV Nsp15 (0.52 Å RMSD of Chain A with PDB id: 2H85, Chain A; sequence identity 88%), and also shows good agreement with Nsp15 from MERS‐CoV (1.16 Å RMSD of Chain A with PDB id 5YVD, Chain A; sequence identity 51%). The structural homology is not only observed in positions of α‐helices and β‐strands but surprisingly in several loop regions (e.g., the conformation of loop L1 [F16–P24] is virtually identical in all three proteins). But there are some interesting differences between SARS‐ and MERS‐CoVs proteins in loop regions (β8–β9 and β10–β11) with the largest differences (more than 5 Å) centering on the loop between strand β13 and β14 where MERS‐CoV Nsp15 has three‐residues insertion. These relatively small changes in the subunit structures translate into larger shifts in the hexamer (see below).
FIGURE 3.
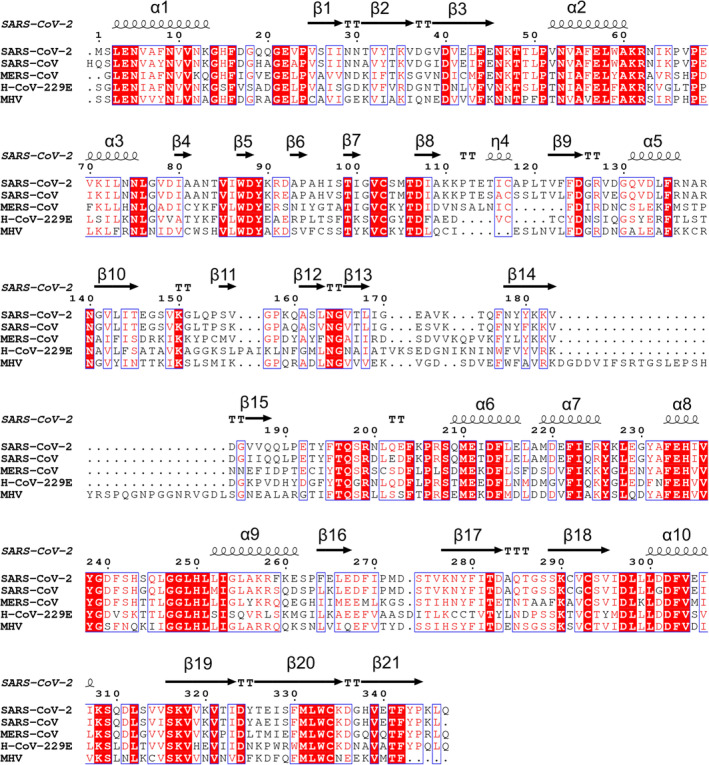
Sequence alignment of SARS‐CoV‐2 Nsp15 coronaviral homologs with structures available in the PDB: SARS‐CoV‐2 (6VWW), SARS‐CoV (2H85), MERS‐CoV (5YVD), and H‐CoV‐229E (4RS4) MHV (2GTH). The secondary structure elements labeled for SARS‐CoV‐2 Nsp15
The SARS‐CoV‐2 NendoU monomers assemble into a double‐ring hexamer, generated by a dimer of trimers (Figure 4). This is in agreement with protein interfaces, surfaces and assemblies calculations, 19 estimating trimers to be a stable form of the enzyme, and the previous experimental work capturing SARS‐CoV Nsp15 trimeric intermediates. 20 However, it was shown that hexamer is essential for the enzymatic activity. A ~100 Å long and narrow (10–15 Å diameter) channel runs down the threefold axis. This channel is accessible to solvent from the top, bottom, and also through three side openings in the middle of hexamer. The hexamer is stabilized by the interactions of N‐terminal oligomerization domains, but also each subunit domain contributes to oligomer interface. As a result, monomers interact extensively with all five other subunits of the hexamer, making the hexamer potentially very sensitive to mutations and small molecules that can disrupt oligomeric assembly (Figure 4). The middle domains are the most transposed out in the hexamer creating the concave surfaces that may serve as an interaction hubs with other proteins and RNA.
FIGURE 4.
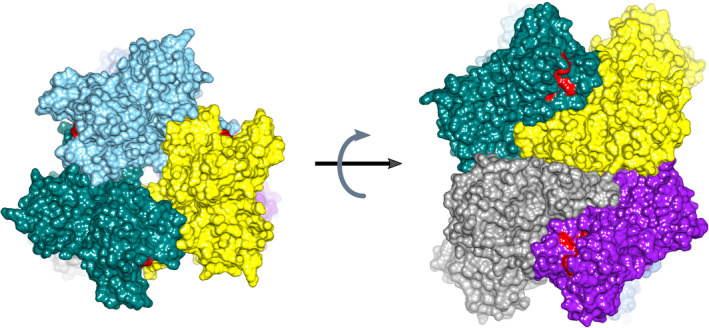
Structure of SARS‐CoV‐2 hexamer in surface representation with each subunit shown in different color. The active site residues are colored red
The SARS‐CoV‐2 NendoU oligomer resembles those of SARS‐CoV, H‐CoV‐229E, and MERS‐CoV enzymes. As with the monomeric folds, the SARS‐CoV‐2 hexamer shows higher similarity to the SARS‐CoV assembly than to H‐CoV‐229E and MERS‐CoV. The largest difference between SARS‐CoV‐2 and SARS‐CoV seems to occur in the position of middle domains. The differences with H‐CoV‐229E are still more significant and show shifts in positions of α‐helices, β‐sheets, and loops. Similarly, MERS‐CoV enzyme show significant structural changes, particularly in the loops of the middle domain. This suggests that the SARS‐CoV‐2 enzyme operates most likely in a manner very similar to SARS‐CoV, H‐CoV‐229E, and MERS‐CoV homologs, though it still may display different catalytic properties and potentially altered substrate specificity.
2.3. NendoU active site
Recombinant Nsp15s were shown to have Mn2+‐dependent endoribonuclease activity that cut dsRNA substrates with specificity towards uridylate in unpaired regions.21, 22, 23, 24 While the early work suggested cleavage upstream and downstream of U,21, 24 the more detailed 2006 study demonstrated the reaction occurs on the 3′ end of U. 23 The enzyme carries out transesterification reaction releasing 2′–3′ cyclic phosphate end, 24 as has been demonstrated for other members of EndoU family,25, 26 though some studies report subsequent hydrolysis of the cyclic compound. 25
The catalytic function of Nsp15 resides in the C‐terminal NendoU domain. The active site, located in a shallow groove between the two β‐sheets, carries six key residues conserved among SARS‐CoV‐2, SARS‐CoV, and MERS‐CoV proteins: His235, His250, Lys290, Thr341, Tyr343, and Ser294 (Figure 5a). The main chain architecture of this region as well as side chain conformations of the active site residues (with exception of Lys290) are conserved between all three proteins. The two histidine residues are contributed by the helical layer of the domain, while pairs lysine/serine and threonine/tyrosine originate from two β‐strands representing edges of the β‐sheets. His235, His250, and Lys290 have been proposed to constitute the catalytic triad, based on the similarity of their mutual arrangement to the active site of ribonuclease A. 14 In this scenario, His235 plays a role of the general acid while His250 acts a base. By the same reasoning, Ser294 together with Tyr343 are believed to govern U specificity, resembling the roles of Phe120 and Thr45 (the B1 subsite specific for pyrimidine base) in RNase A in base recognition. 27 Specifically, Ser294 analogs have been proposed to interact with the carbonyl oxygen atom O2 of uracil via the main chain nitrogen atom, while the hydroxyl group would bind to the nitrogen atom.21, 22, 23, 24 Notably though, not all mutational data support the hypothesis of the discriminatory role of Ser and, as an alternative, Thr341 has been also considered. 14
FIGURE 5.
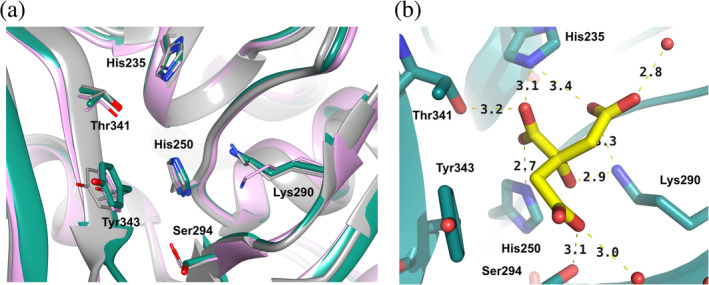
Active site of SARS‐CoV‐2 Nsp15 NendoU. (a) Superposition of the SARS‐CoV‐2 Nsp15 active site with its homologs: SARS‐CoV‐2 (teal, Chain A), SARS‐CoV (pink), and MERS‐CoV (grey) and (b) Binding of citrate to the Nsp15 active site
In the structure of Nsp15/cit the citrate ion is found bound in the active site (Figure 5b). It forms hydrogen bonds with several key active site residues, including His235, His250, Lys290, and Thr341, as well as with two water molecules and PEG. It also makes van der Waals contact with Tyr343. These interactions stabilize the active site residues and contribute to order of the EndoU domain.
The Mn2+ dependence has been observed for most EndoU members, and appears to be a common feature of NendoU subfamily, but for example Nsp11 does not show such behavior. 25 However, the metal binding site was never located, though it is important to note that there is no structure of the protein/RNA complex. In our Nsp15/apo structure, there are two subunits in the asymmetric unit. One of them has an electron density peak near the active site that may correspond to metal ion and we tentatively modeled it as a magnesium ion, present in the crystallization solution, although the coordination sphere is poor. The metal ion is coordinated by oxygen atom of carboxylate Asp283 (2.61 Å), hydroxyl group of Ser262 (2.08 Å), and main chain carbonyl oxygen atom of Pro263 (2.28 Å). Nearby, there is a side chain of Arg258. If this would be a manganese ion, this arginine could coordinate the metal ion using nitrogen of guanidinium moiety (distance to magnesium ion is 4.1 Å). These residues are conserved in SARS‐CoVs but not in MERS‐CoVs enzymes. We propose that this is the metal binding site required for maintaining conformation of the active site and substrate during catalysis.
In the hexameric context, the active sites are located on the top and bottom of the assembly. NendoU binds single and ds RNA and large substrate can access these six sites from a site of the hexamer. All six sites can be occupied simultaneously. Due to extensive interactions between protein subunits, there is possibility for cooperativity or anti‐cooperativity between binding sites. By analogy to SARS‐CoV Nsp15 and SARS‐CoV‐2 Nsp15, the hexamer most likely represents an active form of the enzyme.
2.4. NendoU properties
We have tested Nsp15 endoribonuclease activity using EndoU fluorescent probe 5′‐6‐FAM‐dArUdAdA‐6‐TAMRA‐3′ and used competition to test binding of several RNA and DNA oligonucleotides. The protein cleaves the single‐stranded probe, presumably at uridine position, as shown in Figure 6 with the apparent reaction rate k = 0.0071 s−1. The cleavage is consistent with previously reported values for EndoU enzymes. 7 We have also tested stability of the protein and we have observed two transition temperatures, one at ~45°C and second at ~59°C. These transitions may correspond to hexamer dissociation and protomer unfolding, respectively. Presence of metal ions at 0.1 mM concentrations (Mg2+, Mn2+, and Fe3+) have very small effect on the Nsp15 thermal unfolding.
FIGURE 6.
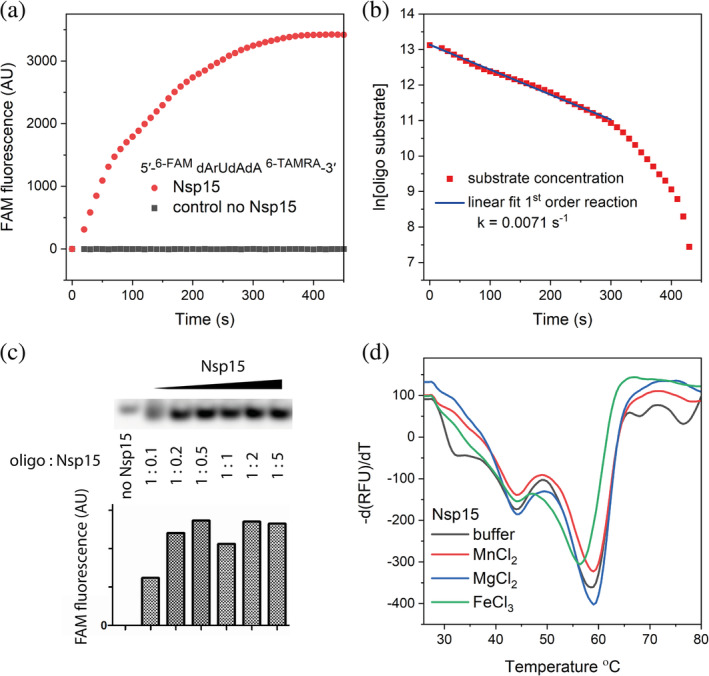
Nsp15 endoribonuclease assay and differential scanning fluorimetry. (a) Time‐dependent cleavage of the oligonucleotide 5′‐6‐FAM‐dArUdAdA‐6‐TAMRA‐3′, (b) Kinetics data are plotted against substrate concentration, (c) The cleavage of the oligonucleotide is monitored on NuPAGE, and (d) DSF of the Nsp15 in the absence and presence of metal ions
3. CONCLUSIONS
We have determined the high‐resolution crystal structures of endoribonuclease NendoU from SARS‐CoV‐2. These structures are homologous to SARS‐ and MERS‐CoVs Nsp15s and show a hexamer, the functionally active form of the endoribonuclease. The active site residues are conserved both in terms of sequence and conformation and the enzyme cleaves single‐stranded RNA. The structural comparisons suggest that inhibitors of SARS‐CoV Nsp15 have good chance to inhibit also the SARS‐CoV‐2 homolog but inhibitors of MERS‐CoV NendoU are unlikely to inhibit the enzyme.
While preparing this manuscript we have determined and deposited the structure of Nsp15 in complex with uridine‐5'‐monophosphate bound to the active site. The structure was determined at 1.82 Å resolution. This structure shows how the enzyme discriminates between uracil and cytosine, adenine and guanine bases with Ser294 serving as the key residue. The details of this structure will be described in a separate manuscript. The coordinates of this complex are available in the PDB under id 6WLC.
4. MATERIALS AND METHODS
4.1. Gene cloning, protein expression, and purification
The gene cloning, protein expression, and purification were performed as reported previously. 28 Briefly, the gene for Nsp15 SARS‐CoV‐2 was optimized for E. coli expression using the optimumgene codon optimization algorithm followed by manual editing and then synthesized cloned directly into pMCSG53 vector (Twist Bioscience). The plasmid was transformed into the E. coli BL21(DE3)‐Gold strain (Stratagene). For large‐scale purification of the protein, a 4 L culture of LB Lennox medium was grown at 37°C (190 rpm) in presence of ampicillin 150 μg/ml. Once the culture reached OD600 ~ 1.0, the temperature setting was changed to 4°C. When bacterial suspension cooled down to 18°C it was supplemented with the following components to indicated concentration: 0.2 mM IPTG, 0.1% glucose and 40 mM K2HPO4. The temperature was set to 18°C for 20 hr incubation. Bacterial cells were harvested by centrifugation at 7,000g and cell pellets were resuspended in a 12.5 ml lysis buffer (500 mM NaCl, 5% [v/v] glycerol, 50 mM 2‐[4‐(2‐hydroxyethyl)piperazin‐1‐yl]ethanesulfonic acid (HEPES) pH 8.0, 20 mM imidazole, and 10 mM β‐mercaptoethanol) per liter culture and sonicated at 120 W for 5 min (4 s ON, 20 s OFF). The cellular debris was removed by centrifugation at 30,000g for 1 hr at 4°C. Supernatant was mixed with 4 ml of Ni2+ Sepharose (GE Healthcare Life Sciences) equilibrated with lysis buffer supplemented to 50 mM imidazole pH 8.0 and suspension was applied on Flex‐Column (420400‐2510) connected to Vac‐Man vacuum manifold (Promega). Unbound proteins were washed out via controlled suction with 160 ml of lysis buffer (50 mM imidazole). Bound proteins were eluted with 20 ml of lysis buffer supplemented to 500 mM imidazole pH 8.0. 2 mM DTT was added followed by tobacco etch virus (TEV) protease treatment at 1:20 protease:protein ratio. The solution was left at 4°C overnight. Unfortunately, for this particular construct, TEV protease was not able to cleave off the His tag. Nsp15 was successfully separated from TEV protease on Superdex 200 column equilibrated in lysis buffer where 10 mM β‐mercaptoethanol was replaced by 1 mM TCEP. Fractions containing Nsp15 were collected. Lysis buffer was replaced on 30 kDa MWCO filter (Amicon‐Millipore) via 10X concentration/dilution repeated three times to crystallization buffer (150 mM NaCl, 20 mM HEPES pH 7.5, 1 mM TCEP). Final concentration of Nsp15 was 36 mg/ml.
4.2. Crystallization
Crystallization experiments also were conducted as described previously, with slight modifications. 29 The sitting‐drop vapor‐diffusion method was used with the help of the Mosquito liquid dispenser (TTP LabTech) in 96‐well CrystalQuick plates (Greiner Bio‐One). Crystallizations were performed with the protein‐to‐matrix ratio of 1:1. MCSG4 (Anatrace), SaltRX (Hampton), and INDEX (Hampton) screens were used for protein crystallization at 16°C. The crystallization conditions were MCSG4 H11 (0.2 M calcium acetate, 0.1 M HEPES/NaOH pH 7.5, 10% PEG 8000). These crystals belong to hexagonal space group P63 diffract X‐rays to 2.20 Å resolution. Diffraction‐quality crystals of Nsp15 suitable for data collection appeared after 12 hr. The second hexagonal crystal form, also in P63 space group, was obtained from 0.1 M trisodium citrate pH 5.6, 10%(w/v) PEG4000, and 10%(v/v) isopropanol. These crystals diffracted X‐rays to 1.90 Å.
4.3. Endoribonuclease assay
The substrate used in the endoribonuclease assay possesses carboxyfluorescein (FAM) at 5′ terminus of the oligonucleotide and quencher molecule tetramethylrhodamine (TAMRA) at 3′ end. The oligonucleotide 5′‐6‐FAM‐dArUdAdA‐6‐TAMRA‐3′ was synthetized by Genscript. We used 0.5 μM substrate with 2 μM Nsp15 in a buffer containing 20 mM HEPES pH 7.8, 100 mM NaCl, 5 mM MnCl2. Detection of the emission of FAM fluorescence signal was done on CFX Connect Real‐Time System (BioRad).
We tested 5′‐6‐FAM‐dArUdAdA‐6‐TAMRA‐3′ substrate cleavage using polyacrylamide gel electrophoresis. The concentration of labeled substrate was 10 μM and Nsp15 concentration varies to achieve different molar ratios of substrate to Nsp15. Samples were mixed in a buffer with 20 mM HEPES pH 7.8, 100 mM NaCl, 5 mM MnCl2. After 10 min of incubation samples were loaded on NuPAGE Tris‐Acetate 3–8% gel and the electrophoresis was done on 80 V for 1 hr. The detection of FAM emission on gel was done using Gel Doc EZ system (BioRad).
4.4. Protein differential scanning fluorimetry
For protein differential scanning fluorimetry (DSF) measurements, we used 10 μM Nsp15 in a buffer composed of 20 mM Tris/HCl pH 7.5, 100 mM NaCl, 1 mM TCEP with the addition of five times concentrated SYPRO Orange dye (Invitrogen). To investigate how metal ions affect Nsp15 stability, the buffer was supplemented with 100 μM MgCl2, MnCl2, or FeCl3. Then, 50 μl samples were loaded on 96‐well PCR plates (BioRad), DSF assay was done on CFX Connect Real‐Time System (BioRad). Samples were equilibrated at 20°C for 30 min. The acquisition of fluorescence signal was done with the temperature ramp rate of 1°C per 60 s. Values of first derivatives of fluorescence signal were depicted on graphs.
4.5. Data collection, structure determination, and refinement
Prior to data collection at 100 K, all cryoprotected crystals of Nsp15 were flash‐cooled in liquid nitrogen. The X‐ray diffraction experiments were carried out at the Structural Biology Center 19‐ID beamline at the Advanced Photon Source, Argonne National Laboratory. The diffraction images were recorded from both crystal forms on the PILATUS3 X 6M detector using 0.5° rotation and 0.5 s exposure for 100°. The data set was processed and scaled with the HKL3000 suite. 30 Intensities were converted to structure factor amplitudes in the Ctruncate program31, 32 from the CCP4 package. 33 The structure was determined using molrep 34 implemented in the HKL3000 software package and SARS‐CoV Nsp15 structure (PDB id 2H85) as a search model. The initial solution was manually adjusted using COOT 35 and then iteratively refined using COOT, PHENIX, 36 and REFMAC.33, 37 Initially REFMAC refinement worked better for the structure which was subsequently moved to PHENIX to finalize the refinement. Throughout the refinement, the same 5% of reflections were kept out throughout from the refinement (in both REFMAC and PHENIX refinement). The final structure converged to a good R work = 0.158 and R free = 0.178 with regards to data quality. The stereochemistry of the structure was checked with PROCHECK 38 and the Ramachandran plot and validated with the PDB validation server. The data collection and processing statistics are given in Table 1. The atomic coordinates and structure factors have been deposited in the PDB under accession code 6VWW and 6W01 for Nsp15/apo and Nsp15/cit, respectively.
AUTHOR CONTRIBUTIONS
Youngchang Kim: Conceptualization; formal analysis; investigation; methodology; validation. Robert Jedrzejczak: Conceptualization; formal analysis; investigation; methodology. Natalia Maltseva: Investigation; methodology. Mateusz Wilamowski: Formal analysis; investigation; methodology. Michael Endres: Formal analysis; investigation; methodology. Adam Godzik: Conceptualization; data curation. Karolina Michalska: Formal analysis; investigation; writing‐original draft. Andrzej Joachimiak: Conceptualization; formal analysis; funding acquisition; investigation; project administration; resources; supervision; writing‐review and editing.
ACKNOWLEDGEMENTS
We truthfully thank the members of the SBC at Argonne National Laboratory, especially Darren Sherrell and Alex Lavens for their help with setting beamline and data collection at beamline 19‐ID. We thank Lukasz Jaroszewski and Monica Rosas Lemus for construct design. Funding for this project was provided in part by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract HHSN272201700060C. The use of SBC beamlines at the Advanced Photon Source is supported by the U.S. Department of Energy (DOE) Office of Science and operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE‐AC02‐06CH11357.
Kim Y, Jedrzejczak R, Maltseva NI, et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARS‐CoV‐2. Protein Science. 2020;29:1596–1605. 10.1002/pro.3873
Youngchang Kim and Robert Jedrzejczak provided equal contribution.
Funding information National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, Grant/Award Number: HHSN272201700060C; U.S. Department of Energy (DOE) Office of Science, Grant/Award Number: DE‐AC02‐06CH11357
REFERENCES
- 1. Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baez‐Santos YM, St John SE, Mesecar AD. The SARS‐coronavirus papain‐like protease: Structure, function and inhibition by designed antiviral compounds. Antivir Res. 2015;115:21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ulferts R, Ziebuhr J. Nidovirus ribonucleases: Structures and functions in viral replication. RNA Biol. 2011;8:295–304. [DOI] [PubMed] [Google Scholar]
- 4. Caffarelli E, Arese M, Santoro B, Fragapane P, Bozzoni I. In vitro study of processing of the intron‐encoded U16 small nucleolar RNA in Xenopus laevis . Mol Cell Biol. 1994;14:2966–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Caffarelli E, Maggi L, Fatica A, Jiricny J, Bozzoni I. A novel Mn++‐dependent ribonuclease that functions in U16 SnoRNA processing in X. laevis . Biochem Bioph Res Co. 1997;233:514–517. [DOI] [PubMed] [Google Scholar]
- 6. Gioia U, Laneve P, Dlakic M, Arceci M, Bozzoni I, Caffarelli E. Functional characterization of XendoU, the endoribonuclease involved in small nucleolar RNA biosynthesis. J Biol Chem. 2005;280:18996–19002. [DOI] [PubMed] [Google Scholar]
- 7. Laneve P, Altieri F, Fiori ME, Scaloni A, Bozzoni I, Caffarelli E. Purification, cloning, and characterization of XendoU, a novel endoribonuclease involved in processing of intron‐encoded small nucleolar RNAs in Xenopus laevis . J Biol Chem. 2003;278:13026–13032. [DOI] [PubMed] [Google Scholar]
- 8. Laneve P, Gioia U, Ragno R, et al. The tumor marker human placental protein 11 is an endoribonuclease. J Biol Chem. 2008;283:34712–34719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Laneve P, Piacentini L, Casale AM, et al. Drosophila CG3303 is an essential endoribonuclease linked to TDP‐43‐mediated neurodegeneration. Sci Rep. 2017;7:41559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Michalska K, Quan Nhan D, Willett JLE, et al. Functional plasticity of antibacterial EndoU toxins. Mol Microbiol. 2018;109:509–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deng X, Hackbart M, Mettelman RC, et al. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci USA. 2017;114:E4251–E4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu X, Fang P, Fang L, et al. Porcine deltacoronavirus nsp15 antagonizes interferon‐beta production independently of its endoribonuclease activity. Mol Immunol. 2019;114:100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu X, Zhai Y, Sun F, et al. New antiviral target revealed by the hexameric structure of mouse hepatitis virus nonstructural protein nsp15. J Virol. 2006;80:7909–7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ricagno S, Egloff MP, Ulferts R, et al. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc Natl Acad Sci USA. 2006;103:11892–11897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Joseph JS, Saikatendu KS, Subramanian V, et al. Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch. J Virol. 2007;81:6700–6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang L, Li L, Yan L, et al. Structural and biochemical characterization of endoribonuclease Nsp15 encoded by Middle East respiratory syndrome coronavirus. J Virol. 2018;92:e00893‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huo T, Liu X. Crystallization and preliminary X‐ray crystallographic analysis of a nonstructural protein 15 mutant from human coronavirus 229E. Acta Cryst. 2015;F71:1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang M, Li X, Deng Z, et al. Structural biology of the arterivirus nsp11 endoribonucleases. J Virol. 2017;91:e01309‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. [DOI] [PubMed] [Google Scholar]
- 20. Guarino LA, Bhardwaj K, Dong W, Sun J, Holzenburg A, Kao C. Mutational analysis of the SARS virus Nsp15 endoribonuclease: Identification of residues affecting hexamer formation. J Mol Biol. 2005;353:1106–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bhardwaj K, Guarino L, Kao CC. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J Virol. 2004;78:12218–12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bhardwaj K, Palaninathan S, Alcantara JM, et al. Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. J Biol Chem. 2008;283:3655–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bhardwaj K, Sun J, Holzenburg A, Guarino LA, Kao CC. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J Mol Biol. 2006;361:243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ivanov KA, Hertzig T, Rozanov M, et al. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc Natl Acad Sci USA. 2004;101:12694–12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nedialkova DD, Ulferts R, van den Born E, et al. Biochemical characterization of arterivirus nonstructural protein 11 reveals the nidovirus‐wide conservation of a replicative endoribonuclease. J Virol. 2009;83:5671–5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Renzi F, Caffarelli E, Laneve P, Bozzoni I, Brunori M, Vallone B. The structure of the endoribonuclease XendoU: From small nucleolar RNA processing to severe acute respiratory syndrome coronavirus replication. Proc Natl Acad Sci USA. 2006;103:12365–12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. delCardayre SB, Raines RT. Structural determinants of enzymatic processivity. Biochemistry. 1994;33:6031–6037. [DOI] [PubMed] [Google Scholar]
- 28. Makowska‐Grzyska M, Kim Y, Maltseva N, et al. Protein production for structural genomics using E. coli expression. Methods Mol Biol. 2014;1140:89–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim Y, Babnigg G, Jedrzejczak R, Eschenfeldt WH, et al. High‐throughput protein purification and quality assessment for crystallization. Methods. 2011;55:12–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL‐3000: The integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr D. 2006;62:859–866. [DOI] [PubMed] [Google Scholar]
- 31. French S, Wilson K. Treatment of negative intensity observations. Acta Crystallogr A. 1978;A34:517–525. [Google Scholar]
- 32. Padilla JE, Yeates TO. A statistic for local intensity differences: Robustness to anisotropy and pseudo‐centering and utility for detecting twinning. Acta Crystallogr D. 2003;59:1124–1130. [DOI] [PubMed] [Google Scholar]
- 33. Winn MD, Ballard CC, Cowtan KD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D. 2011;D67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D. 2010;D66:22–25. [DOI] [PubMed] [Google Scholar]
- 35. Emsley P, Cowtan K. Coot: Model‐building tools for molecular graphics. Acta Crystallogr D. 2004;D60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 36. Adams PD, Afonine PV, Bunkoczi G, et al. PHENIX: A comprehensive python‐based system for macromolecular structure solution. Acta Crystallogr D. 2010;D66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum‐likelihood method. Acta Crystallogr D. 1997;D53:240–255. [DOI] [PubMed] [Google Scholar]
- 38. Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 39. Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: Structure validation and all‐atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. [DOI] [PMC free article] [PubMed] [Google Scholar]


