Abstract
Coronavirus disease (COVID‐19), caused by a novel betacoronavirus, severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), has rapidly developed into a pandemic since it was first reported in December 2019. Nucleic acid testing is the standard method for the diagnosis of viral infections. However, this method reportedly has a low positivity rate. To increase the sensitivity of COVID‐19 diagnoses, we developed an IgM‐IgG combined assay and tested it in patients with suspected SARS‐CoV‐2 infection. In total, 56 patients were enrolled in this study and SARS‐CoV‐2 was detected by using both IgM‐IgG antibody and nucleic acid tests. Clinical and laboratory data were collected and analyzed. Our findings suggest that patients who develop severe illness might experience longer virus exposure times and develop a more severe inflammatory response. The IgM‐IgG test is an accurate and sensitive diagnostic method. A combination of nucleic acid and IgM‐IgG testing is a more sensitive and accurate approach for diagnosis and early treatment of COVID‐19.
Keywords: COVID‐19, diagnosis, IgG, IgM, SARS‐CoV‐2
Highlights
Combined detection of IgM and IgG is of great value to improve the clinical sensitivity of early diagnosis of COVID‐19.
Numerous differences were found between patients in severe group and those who were not.
IgM may be an indicator of inflammation severity in acute infection in COVID‐19 patients.
1. INTRODUCTION
Since severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) first emerged in Wuhan, China, on 12 December 2019, it has spread quickly across the world and developed into a pandemic. 1 , 2 , 3 The World Health Organization (WHO) announced a new name for the disease: coronavirus disease (COVID‐19). By 31 March 2020, more than 700 000 COVID‐19 cases were confirmed in over 100 countries and regions. To date, the rapid spread of SARS‐CoV‐2 has caused considerable harm to public health and the economy. 4 , 5 Clinical manifestations of COVID‐19 include fever, dry cough, and fatigue. Approximately half of the infected patients developed severe pneumonia, and nearly one‐third of the patients develop acute respiratory distress syndrome. 6 , 7 , 8 However, there is currently no specific treatment for COVID‐19. Due to the obstacle of collecting high‐quality throat swab samples in different stages of infection, nucleic acid test for SARS‐CoV‐2 showed high false negative rate. It is difficult to identify and quarantine the infected individuals to effectively break the spread chain and curb the infection. Therefore, it is of urgent need to develop a more sensitive diagnostic method that can rapidly identify SARS‐CoV‐2 infected patients with high accuracy.
Currently, the viral nucleic acid real‐time polymerase chain reaction (RT‐PCR) test based on patient nasopharyngeal and throat swabs is the standard method for clinical diagnosis of COVID‐19. Despite its crucial role in identifying SARS‐CoV‐2 infection in patients at the start of the epidemic, the limitations of this method soon became obvious. For example, one recent study demonstrated that RT‐PCR only showed a positive test rate of 38% in a total of 4880 specimens with a significant number of false negative cases. 9 It is accepted that IgM is the early immunoglobulin in response to the virus invasion and IgG has the highest opsonization and neutralization activities in humoral immune response. Previous studies have reported that IgM‐IgG seroconversion can start as early as 4 days after the onset of SARS infection. 10 Li et al 11 has developed a rapid point‐of care‐lateral flow immunoassay that can detect IgM and IgG levels within 15 minutes for the rapid screening of SARS‐CoV‐2 infection at different stages. Therefore, testing for antibodies specific to SARS‐CoV‐2 protein in patient serum samples might be an alternative method for rapid and highly sensitive laboratory diagnosis.
Presently, there are relatively few reports on COVID‐19 patient diagnoses through serological tests. Here, we retrospectively describe the clinical and laboratory characteristics of 56 patients with COVID‐19 diagnosed using IgM‐IgG antibody tests. All patients were admitted to Unit Z6 in the Cancer Center of Wuhan Union Hospital. This study may provide a reference for clinical profiles of patients with COVID‐19 confirmed using antibody detection.
2. MATERIALS AND METHODS
2.1. Patients
In this study, 56 patients were enrolled from Unit Z6 at the Cancer Center of Wuhan Union Hospital between 15 and 25 February 2020. Most patients were admitted to the hospital due to fever or respiratory symptoms. Nasopharyngeal and throat swabs were used for respiratory pathogen testing. Serum levels of IgM‐IgG antibodies targeting SARS‐CoV‐2 were tested upon patient admission. Medical history about when the clinical symptoms appeared was asked and the time interval between clinical symptoms and antibody testing was recorded in detail. Physical findings and hematological and biochemical results were also collected. All patients enrolled in this study were diagnosed according to the 5th edition of the Guideline on diagnosis and treatment of COVID‐19 established by China's National Health Commission, including patient's epidemic history, clinical characteristics, chest computed tomographic (CT) scan, and laboratory findings. Patients with COVID‐19 having severe illness were defined having one of the following criteria: (a) respiratory distress with respiratory frequency (RP) more than or equal to 30/min, (b) pulse oximeter oxygen saturation less than or equal to 93% at rest, or (c) oxygenation index (arterial partial pressure of oxygen/inspired oxygen fraction [PaO2/FiO2]) less than or equal to 300 mm Hg. Clinical characteristics were compared between severe and nonsevere cases. This study was approved by the Ethics Committee of the First Affiliated Hospital of USTC. This is a retrospective and observational study and the informed consent was obtained.
2.2. Real‐time reverse transcription polymerase chain reaction assay
The presence of SARS‐CoV‐2 was detected using RT‐PCR. Viral RNA was extracted from nasopharyngeal and throat swabs using the QIAamp RNA virus kit (Qiagen, Heiden, Germany). The open reading frame 1ab (ORF1ab) and nucleocapsid protein (N) were simultaneously amplified and tested using RT‐PCR. The following primers were used: Target 1 (ORF1ab) forward primer: 5′‐CCCTGTGGGTTTTACACTTAA‐3′; reverse primer: 5′‐ACGATTGTGCATCAGCTGA‐3′; probe: 5′‐VIC‐CCGTCTGCGGTATGTGGAAAGGTTATGG‐BHQ1‐3′; Target 2 (N) forward primer: 5′‐GGGGAACTTCTCCTGCTAGAAT‐3′; reverse primer: 5′‐CAG ACATTTTGCTCTCAAGCTG‐3′; probe: 5′‐FAM‐TTGCTGCTGCTTGACAGATT‐TAMRA‐3′. The conditions for the amplification were 50°C for 20 minute, 95°C for 10 minute, followed by 40 cycles of denaturation at 95°C for 15 second, and extension and fluorescence collection at 60°C.
2.3. IgM‐IgG test for SARS‐CoV‐2
Anti‐human IgG and IgM assays were purchased from YHLO Biological Technology Co, Ltd, Shenzhen, China. In all patients, IgG and IgM antibodies against the SARS‐CoV‐2 envelope (E) protein and nucleocapsid (N) protein in serum samples were measured using chemiluminescence immunoassay. The cutoff value for a positive result was 10, and samples with values greater than or equal to 10 AU/mL were considered positive for SARS‐CoV‐2 infection.
2.4. Statistical analysis
Categorical variables are presented as numbers (%) and continuous measurements as medians (interquartile range [IQR]). Antibody concentration was reported as the geometric mean (SD). Continuous variables were analyzed using the Mann‐Whitney test or unpaired t test. The correlation of IgM and IgG quantitative detection with hematological profiles was analyzed using Pearson correlation. Graphpad Prism 8.3 was used for all statistical analyses. A two‐sided α value less than .05 was considered statistically significant.
3. RESULTS
3.1. Comparison of IgM‐IgG antibodies with nucleic acid test
IgM‐IgG antibody levels and nucleic acid test results are summarized in Table 1. Of the 56 patients, 40 (71.43%) showed negative nucleic acid tests and 16 (28.57%) were positive. Among the 40 negative patients, 34 (85%) tested positive for the presence of IgM antibodies. Among the 16 patients who tested positive with nucleic acid tests, one patient showed a negative IgM level. The IgG antibody test was positive in all 56 patients.
Table 1.
Comparison of IgM‐IgG antibodies with nucleic acid test
| No. (%) | IgM | IgG | ||
|---|---|---|---|---|
| Nucleic acid detection | + | − | + | − |
| Negative 40 (71.43%) | 34 (85%) | 6 (15%) | 40 (100%) | 0 |
| Positive 16 (28.57%) | 15 (93.75%) | 1 (6.25%) | 16 (100%) | 0 |
| Total | 49 (87.5%) | 7 (12.5%) | 56 (100%) | 0 |
Note: The data are presented as no. (%). No. is the number of patients with available data.
3.2. Clinical characteristics of patients with COVID‐19
Among the 56 hospitalized patients with clinically confirmed COVID‐19, the median age was 56.5 years (IQR, 49.25‐64.75), and 32 (57.14%) of the patients were women. In addition, 34 (60%) patients had severe symptoms because of organ dysfunction. Less than half of the patients had underlying diseases (14 [25.00%]), including hypertension (3 [5.36%]), diabetes (7 [12.5%]), hyperlipidemia (1 [1.79%]), hyperuricemia (1 [1.79%]), multiple myeloma (1 [1.79%]), and chronic hepatitis B virus infection (1 [1.79%]). The most common symptoms at onset were fever (42 [75%]), cough (23 [41.07%]), and chest tightness (20 [35.71%]). Less common symptoms were fatigue (16 [28.57%]), abdominal distension and diarrhea (9 [16.07%]), sore throat (7 [12.5%]), muscle soreness (4 [7.14%]), and chills (3 [5.36%]) (Table 2).
Table 2.
Baseline characteristics of 56 COVID‐19 patients
| No. (%) | |||
|---|---|---|---|
| Total (n = 56) | Severe (n = 34) | Nonsevere (n = 22) | |
| Age, y, median (IQR) | 56.5 (49.25‐64.75) | 60 (50.75‐67.0) | 54.0 (46.5‐58.75) |
| Sex | |||
| Female | 32 (57.14%) | 16 (47.06%) | 16 (72.73%) |
| Male | 24 (42.86%) | 18 (52.94%) | 6 (27.27%) |
| Chronic medical illness | 14 (25.00%) | ||
| High blood pressure | 7 (12.50%) | 6 (17.65%) | 1 (4.55%) |
| Diabetes | 3 (5.36%) | 2 (5.88%) | 1 (4.55%) |
| Hyperlipidemia | 1 (1.79%) | 1 (2.94%) | 0 |
| Hyperuricemia | 1 (1.79%) | 1 (2.94%) | 0 |
| Multiple myeloma | 1 (1.79%) | 1 (2.94%) | 0 |
| Chronic hepatitis B virus infection | 1 (1.79%) | 0 | 1 (4.55%) |
| Signs and symptoms | |||
| Fever (℃) | |||
| <37.3 | 14 (25.00%) | 9 (26.47%) | 5 (22.73%) |
| 37.3‐38.0 | 28 (50.00%) | 16 (47.06%) | 12 (54.55%) |
| 38.1‐39.0 | 12 (21.43%) | 7 (20.59%) | 5 (22.73%) |
| >39.0 | 2 (3.57%) | 2 (5.88%) | 0 |
| Cough | 23 (41.07%) | 15 (44.12%) | 12 (54.55%) |
| Chest tightness | 20 (35.71%) | 12 (35.29%) | 8 (36.36%) |
| Fatigue | 16 (28.57%) | 7 (20.59%) | 9 (40.91%) |
| Abdominal distension and diarrhea | 9 (16.07%) | 3 (8.82%) | 5 (22.73%) |
| Sore throat | 7 (12.50%) | 6 (17.65%) | 1 (4.55%) |
| Muscle soreness | 4 (7.14%) | 3 (8.82%) | 1 (4.55%) |
| Chills | 3 (5.36%) | 2 (5.88%) | 1 (4.55%) |
Note: Data are presented as median (IQR) or No. (%). No. is the number of patients with available data.
Abbreviations: COVID‐19, coronavirus disease; IQR, interquartile range.
Interestingly, there were differences in laboratory findings between the groups with severe and nonsevere COVID‐19 symptoms. This included higher neutrophil counts, neutrophil percentage (NEU%), and fibrinogen level, and lower lymphocyte counts and lymphocyte percentages (LYM%) (P < .05) (Figure 1). The median D‐dimer level was increased in the group with severe symptoms, but the difference was not significant. Procalcitonin and hypersensitive C‐reactive protein levels were in the normal range in the majority of patients (Table 3).
Figure 1.
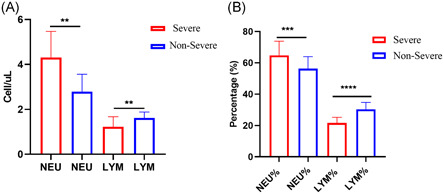
Differences in laboratory findings between severe and nonsevere groups (A) NEU and LYM cell counts, (B) percentage of NEU and LYM. Data are presented as the median (interquartile range [IQR]) and were analyzed using Mann‐Whitney test. All statistical analyses were performed using GraphPad Prism 8.3 (**P < .005, ***P < .001, ****P < .0001). P < .05 was considered statistically significant. LYM%, lymphocyte percentage; NEU%, neutrophil percentage
Table 3.
Laboratory findings of COVID‐19 patients on admission to hospital
| Median (IQR) | |||||
|---|---|---|---|---|---|
| Blood routine | Normal range | Total (n = 56) | Severe (n = 34) | Nonsevere (n = 22) | P |
| Leukocytes, ×109/L | 3.5‐9.5 | 5.61 (4.54‐7.16) | 6.46 (4.67‐7.51) | 5.13 (4.46‐5.98) | .0685 |
| Neutrophils, ×109/L | 1.8‐6.3 | 3.33 (2.63‐4.68) | 4.31 (2.88‐5.47) | 2.78 (2.35‐3.56) | .001** |
| Percentage of neutrophils, % | 40‐75.0 | 63.26 (56.26‐69.23) | 64.74 (61.6‐73.82) | 56.31 (50.07‐63.96) | .0001*** |
| Lymphocytes, ×109/L | 1.1‐3.2 | 1.44 (1.06‐1.75) | 1.22 (0.93‐1.68) | 1.62 (1.31‐1.88) | .008** |
| Percentage of lymphocytes, % | 20‐50.0 | 24.65 (19.08‐31.15) | 21.65 (15.88‐25.25) | 30.35 (26‐34.78) | <.0001**** |
| Platelets, ×109/L | 125‐350.0 | 204.5 (157.5‐262.8) | 210 (154.5‐308.8) | 204.5 (161‐243) | .5856 |
| Hemoglobin, g/L | 115‐150.0 | 120 (116.0‐131.0) | 125 (117.0‐135.0) | 117.5 (113.3‐120.5) | .0185* |
| CD3, % | 58.17‐84.22 | 75 (70.57‐79.61) | 74.68 (70.81‐78.92) | 77.18 (68.35‐82.03) | .2459 |
| CD4, % | 25.34‐51.37 | 45.49 (40.59‐52.52) | 44.92 (40.46‐53.49) | 46.29 (40.7‐52.36) | .9303 |
| CD8, % | 14.23‐38.95 | 24.56 (17.37‐30.52) | 23.52 (17.8‐30.23) | 25.16 (16.15‐31.41) | .9967 |
| CD4/CD8 | 0.41‐2.72 | 1.94 (1.42‐2.88) | 1.88 (1.39‐2.85) | 1.99 (1.52‐3.19) | .8581 |
| Coagulation function | |||||
| Activated partial thromboplastin time, s | 28‐43.5 | 35.6 (32.90‐38.45) | 36 (34.05‐39.85) | 34.15 (32.48‐36.95) | .0802 |
| Prothrombin time, s | 11‐16.0 | 13.25 (12.80‐13.78) | 13.4 (12.68‐13.9) | 13.1 (12.8‐13.6) | .3388 |
| D‐dimer, mg/L | <0.5 | 0.43 (0.22‐0.80) | 0.59 (0.22‐1.0) | 0.34 (0.22‐0.54) | .1796 |
| Fibrinogen, g/L | 2.0‐4.0 | 4.06 (3.13‐5.25) | 4.51 (3.39‐5.45) | 3.51 (2.90‐4.39) | .0154* |
| Blood biochemistry | |||||
| Alanine aminotransferase, IU/L | 8‐40.0 | 30.5 (20‐52.75) | 33.5 (21‐62.75) | 26 (17.5‐43.5) | .3974 |
| Aspartate aminotransferase, IU/L | 5‐35.0 | 26.50 (21‐39.25) | 28 (22‐40.75) | 23.5 (18.75‐36.75) | .1794 |
| Serum creatinine, μmol/L | 44‐106.0 | 69.5 (62‐79.75) | 70.95 (60.75‐84) | 67.95 (62‐77.5) | .4407 |
| Creatine kinase, IU/L | 22.0‐269.0 | 52 (33.25‐70.75) | 50.5 (33‐73.25) | 52 (37‐62.75) | .9635 |
| Creatine kinase‐MB, ng/mL | <6.6 | 0.5 (0.3‐0.7) | 0.45 (0.3‐0.7) | 0.5 (0.3‐0.93) | .4376 |
| Lactate dehydrogenase, U/L | 109‐245.0 | 194 (155‐232.5) | 200 (168‐244) | 188 (145.8‐206.8) | .1223 |
| Hypersensitive troponin I, ng/L | <26.2 | 2.15 (1.2‐4.38) | 3 (1.2‐5.9) | 1.6 (1.08‐3.43) | .0994 |
| Infection‐related biomarkers | |||||
| C‐reactive protein, mg/L | 0‐8.0 | 33.1 (11.83‐67.45) | 33.1 (15.40‐80.20) | 30.08 (8.26‐51.9) | 0.8235 |
| Hypersensitive C‐reactive protein, mg/L | <4.0 | 1.9 (0.98‐3.31) | 2.61 (1.56‐3.69) | 1.25 (0.08‐2.5) | .147 |
| Procalcitonin, ng/mL | 0‐0.5 | 0.2 (0.19‐0.21) | 0.2 (0.19‐0.21) | <0.13 | / |
Note: Data are presented as the median (interquartile range [IQR]). Statistical analysis was performed using the Mann‐Whitney test. P values indicate differences between severe and nonsevere patients. *P < .05 was considered statistically significant.
Abbreviation: COVID‐19, coronavirus disease.
*P < .05. **P < .005. ***P < .001. ****P < .0001.
IgM‐IgG antibody levels in patients in the groups with severe and nonsevere symptoms during the same time period are shown in Figure 2A. IgG maintains high levels after SARS‐CoV‐2 infection. IgM increased in 1 week and then started to decline 4 to 5 weeks after the onset of illness. In contrast to the profiles of immune responses against acute virus infections, a simultaneous or earlier IgG response against SARS‐CoV‐2, compared with IgM, was observed. IgM‐IgG levels between the two groups were further analyzed, and significant differences were noted (Figure 2B,C). Pearson correlation was used to explore the relationship between immune activation and SARS‐CoV‐2 infection. IgG levels showed no correlation with NEU% and LYM% in either group, while IgM levels showed a weak correlation with NEU% in patients in the severe group (R = .34, P = .0468), suggesting that IgM could be considered an indicator of severe inflammation during acute infection (Table 4; Figure 2D).
Figure 2.
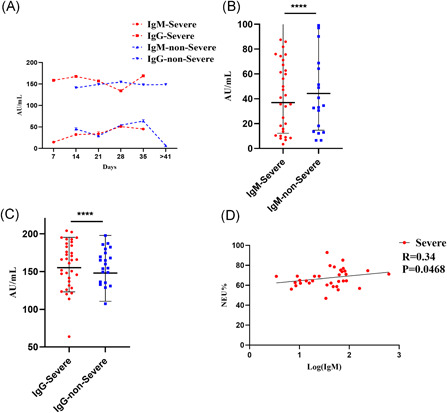
Analysis of IgM‐IgG findings in severe and nonsevere groups. A, Kinetic analysis of IgM‐IgG antibodies in severe and nonsevere groups; B, differences in total IgM levels between severe and nonsevere groups; C, differences in total IgG levels between severe and nonsevere groups; and D, correlation between IgM and NEU% in patients in the severe group. Antibody concentration was presented as the geometric mean (SD) and analyzed using an unpaired t test. The correlation of IgM‐IgG with hematological profiles was analyzed using Pearson correlation. All statistical analyses were performed using GraphPad Prism 8.3 (****P < .0001). P < .05 was considered statistically significant. NEU%, neutrophil percentage; SD, standard deviation
Table 4.
Pearson correlation analysis between IgM‐IgG antibody and laboratory profiles
| Total IgM | Total IgG | IgM‐severe | IgG‐severe | IgM‐nonsevere | IgG‐nonsevere | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker | R | P | R | P | R | P | R | P | R | P | R | P |
| NEU% | −.33 | .0333* | .21 | .19 | .34 | .0468* | −.17 | .33 | −.35 | .12 | .21 | .36 |
| LYM% | .27 | .08 | −.21 | .19 | −.25 | .15 | .17 | .35 | .30 | .18 | −.26 | .24 |
Note: Statistical analyses were performed using Pearson correlation.
Abbreviations: LYM%, lymphocyte percentage; NEU%, neutrophil percentage.
*P < .05 was considered statistically significant.
4. DISCUSSION
In this study, we analyzed the clinical features and immunological characteristics of 56 patients with COVID‐19. Despite negative nucleic acid test results, all patients showed high specific IgG concentrations, suggesting SARS‐CoV‐2 infection. Of the 56 patients, over 50% developed severe illness and required intensive care. Common symptoms were fever, cough, and chest tightness, which is consistent with previous studies. 4 , 6 , 12 Compared with patients with nonsevere symptoms, patients in the severe illness group had numerous laboratory abnormalities, such as higher neutrophil counts, NEU%, fibrinogen levels, lower lymphocyte counts, and lower LYM%. IgM was lower while IgG was higher in patients with severe symptoms. In addition, a weak correlation between IgM and NEU% was noted. These findings suggest that patients in the severe illness group might experience a longer virus exposure time and develop a more severe inflammatory response.
Currently, the nucleic acid test based on individual nasopharyngeal and throat swabs is the standard diagnostic method for COVID‐19. Although the RT‐PCR method is sensitive and effective, it still suffers from some limitations such as being labor‐intensive and time‐consuming. Furthermore, the respiratory tract might not be the only route for SARS‐CoV‐2 transmission. 13 While nucleic acid testing of pharyngeal swabs suggest negative results, whether subjects carry the virus in other organs such as the intestine remains unclear. Thus, rapid, low‐cost, and universal detection methods for SARS‐CoV‐2 are needed. Our study showed that the virus‐specific antibody test had a significantly higher positive result rate than the nucleic acid test. In addition to the self limitation of PCR test kits, the timing for specimens collection and PCR detection also does great impact on PCR results. In our study, due to the urgency condition and limitation of PCR kits supply in Wuhan, most patients might miss the best chance for early SARS‐CoV‐2 screening. Specimens collected in the late stage of illness while viral copies may have declined below detectable limits, which may lead to low positive rate for PCR and high titers of IgM and IgG. Furthermore, this may be one of the possible explanations for a large proportion of severe cases in our study.
Two consecutive negative results of the nucleic acid test in a 24‐hour interval are considered as clearance of COVID‐19. However, several reports showed that a small portion of such recovered patients tested positive for infection through the nucleic acid test again during a follow‐up visit. These results only indicate the presence of the SARS‐CoV‐2 virus in the patients, not whether reinfection or recurrence has occurred. One possible solution for this is to detect the titer of IgG in these patients, as the level of IgG usually increases with reinfection of the same virus.
Although the IgM‐IgG test is an important index for the diagnosis of COVID‐19, limitations still exist. The variation of the methodology and antigens used in the IgM and IgG antibody detection kits are essential for the testing sensitivity and specificity. Li et al 11 reported the testing sensitivity as 88.66% and specificity as 90.63% in the IgM‐IgG combined antibody testing kits by using receptor binding domain of SARS‐CoV‐2 Spike Protein as the antigen. Another study suggested a very poor sensitivity of VivaDiag COVID‐19 IgM‐IgG rapid test which could not be recommended for diagnosis of COVID‐19. 14 Considering the kits used in our study, the E protein functions to assemble the virions and N protein is the most conserved and stable protein among the CoV structural proteins. However, false positives might appear if cross‐reactivity with other coronaviruses occurs. Hence, more studies with larger sample sizes and controls are needed to further verify the specificity and accuracy of this test. In summary, the combination of nucleic acid testing and the IgM‐IgG antibody test is the optimal method for diagnosing SARS‐CoV‐2 infection.
CONFLICT OF INTERESTS
All the authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
HH and YG secured the funding for this study, designed the study, participated in data analysis, and extensively reviewed the manuscript. JX and CD analyzed the data and drafted the manuscript. Other authors contributed to clinical and laboratory data acquisition and reviewed the manuscript.
ACKNOWLEDGMENTS
We thank all health‐care workers involved in the diagnosis and treatment of COVID‐19 patients in the First Affiliated Hospital of USTC and Wuhan Union Hospital. This study was funded by the Special Project for Emergency Scientific and Technological Research on New Coronavirus Infection (no. YD2070002017) and the Hefei Comprehensive National Science Center.
Xie J, Ding C, Li J, et al. Characteristics of patients with coronavirus disease (COVID‐19) confirmed using an IgM‐IgG antibody test. J Med Virol. 2020;92:2004–2010. 10.1002/jmv.25930
Jiajia Xie and Chengchao Ding contributed equally to this work.
Contributor Information
Yong Gao, Email: ygao387@ustc.edu.cn.
Hongliang He, Email: hhl725@ustc.edu.cn.
REFERENCES
- 1. Lu H, Stratton CW, Tang YW. Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J Med Virol. 2020;92(4):401‐402. 10.1002/jmv.25678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou P, Yang X, Wang X, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lai C, Shih T, Ko W, et al. Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) and coronavirus disease‐2019 (COVID‐19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55(3):105924. 10.1016/j.ijantimicag.2020.105924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu Y, Yang Y, Zhang C, et al. Clinical and biochemical indexes from 2019‐nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. 2020;63(3):364‐374. 10.1007/s11427-020-1643-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang J, Dong X, Cao Y, et al. Clinical characteristics of 140 patients infected with SARS‐CoV‐2 in Wuhan, China [published online ahead of print February 19, 2020]. Allergy. 2020. 10.1111/all.14238 [DOI] [PubMed] [Google Scholar]
- 9. Liu R, Han H, Liu F, et al. Positive rate of RT‐PCR detection of SARS‐CoV‐2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta. 2020;505:172‐175. 10.1016/j.cca.2020.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hsueh PR, Huang LM, Chen PJ, et al. Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS‐associated coronavirus. Clin Microbiol Infect. 2004;10(12):1062‐1066. 10.1111/j.1469-0691.2004.01009.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Z, Yi Y, Luo X, et al. Development and clinical application of a rapid IgM‐IgG combined antibody test for SARS‐CoV‐2 infection diagnosis [published online ahead of print February 27, 2020]. J Med Virol. 2020. 10.1002/jmv.25727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507‐513. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang W, Du RH, Li B, et al. Molecular and serological investigation of 2019‐nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9(1):386‐389. 10.1080/22221751.2020.1729071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cassaniti I, Novazzi F, Giardina F, et al. Performance of VivaDiag COVID‐19 IgM/IgG rapid test is inadequate for diagnosis of COVID‐19 in acute patients referring to emergency room department [published online ahead of print March 30, 2020]. J Med Virol. 2020. 10.1002/jmv.25800 [DOI] [PMC free article] [PubMed] [Google Scholar]


