Abstract
A pandemic of coronavirus disease 2019 (COVID‐19) caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has been spreading throughout the world. Though molecular diagnostic tests are the gold standard for COVID‐19, serological testing is emerging as a potential surveillance tool, in addition to its complementary role in COVID‐19 diagnostics. Indubitably quantitative serological testing provides greater advantages than qualitative tests but today there is still little known about serological diagnostics and what the most appropriate role quantitative tests might play. Sixty‐one COVID‐19 patients and 64 patients from a control group were tested by iFlash1800 CLIA analyzer for anti‐SARS CoV‐2 antibodies IgM and IgG. All COVID‐19 patients were hospitalized in San Giovanni di Dio Hospital (Florence, Italy) and had a positive oro/nasopharyngeal swab reverse‐transcription polymerase chain reaction result. The highest sensitivity with a very good specificity performance was reached at a cutoff value of 10.0 AU/mL for IgM and of 7.1 for IgG antibodies, hence near to the manufacturer's cutoff values of 10 AU/mL for both isotypes. The receiver operating characteristic curves showed area under the curve values of 0.918 and 0.980 for anti‐SARS CoV‐2 antibodies IgM and IgG, respectively. iFlash1800 CLIA analyzer has shown highly accurate results for the anti‐SARS‐CoV‐2 antibodies profile and can be considered an excellent tool for COVID‐19 diagnostics.
Keywords: coronavirus, humoral immunity, immune responses, SARS coronavirus, virus classification
1. INTRODUCTION
Coronavirus disease 2019 (COVID‐19) is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), which first appeared in Wuhan, China, in December 2019 and is now spreading worldwide. COVID‐19 is currently diagnosed through detection of the responsible microorganism SARS‐CoV‐2 in upper and lower respiratory specimens by molecular tests, such as real‐time reverse‐transcription polymerase chain reaction (RT‐PCR). 1 , 2 , 3 However, these methods are dependent on the time‐window of viral replication, low viral titer, and subject to incorrect sample collection which is why they can all potentially cause low predictive rate results, thereby limiting the usefulness of RT‐PCR in the field.
During a pandemic, false negative results can produce grave consequences by facilitating the circulation of contagious individuals who spread the virus. Anti‐SARS‐CoV‐2 antibodies may represent a tool that can both help close the RT‐PCR negative gap as well as significantly increase diagnostic sensitivity for COVID‐19 patients, especially by detecting IgM antibodies which are swiftly formed in response to infection. 4 , 5 Even if testing specific SARS‐CoV‐2 antibodies has a faster turn‐around time and high‐throughput, and proves to be simpler and cheaper than molecular tests, it is important to underline that the detection of SARS‐CoV‐2 viral nucleic acid by RT‐PCR test is still the current standard diagnostic method for COVID‐19. Moreover, it becomes more and more evident that, notwithstanding the importance of the diagnostic role of SARS‐CoV‐2 antibodies testing, its epidemiologic potential to evaluate a population's immunization state is increasingly important. 6 This means then that it can determine, together with the swab negative test, which healthcare workers are immune and when they can return to work, as well as effectively establish which businesses outside the healthcare system including schools, public transportation services, and such, can resume operations. Vaccine research would also benefit. 7 Nevertheless, global supply challenges and huge demand for PCR primers and positive controls have sent diagnostic companies scrambling to produce antibody tests, as a key reaction to virus transmission and to assure timely treatment of patients. Because of the need to accelerate progress in diagnostics, serological tests have been developed. More than 200 different assays have been proposed so far but almost all have poor regulatory status and lack clinical and analytical performance review. 8 In fact the speed with which they are released in the market and the versatility of immunoassays such as source of antigen and secondary antibody conjugate, make them poorly evaluated tests. Given that during the outbreak test validation is not a priority and given that nonlaboratory specialists are allowed to handle these tests because of limited staff resources has meant that unregulated testing has spread widely. In particular, since rapid tests do not require any instruments or laboratory personnel they could be set up anywhere and at any time, especially in developing nations with limited healthcare resources and in remote settings. The more relaxed rules of the FDA's “Policy for Diagnostic Tests for Coronavirus Disease‐2019 during the Public Health Emergency” issued on 16 March 2020, 9 has allowed the market easier access to these tests as well as easier and faster diagnostics, but the lack of control in the production process is also dangerous making these tests potentially less reliable. Along with chromatographic rapid immunoassays as qualitative tests, 10 quantitative antibodies detection tests, such as enzyme‐linked immunosorbent assay and chemiluminescence immunoassay (CLIA), have spread often by fully automated analyzers.
These technologies characterized by high‐throughput and low complexity have helped us to use serological testing more accurately during both antibody development and monitoring the different phases of the disease. Indeed, being able to receive information about the antibody concentration and time kinetics of humoral response is very important for diagnostic, prognostic, and therapeutic applications. 11
The aim of the this study was to assess the diagnostic performance of a novel fully automated CLIA for the quantitative detection of anti‐SARS‐CoV‐2 IgM and IgG antibodies.
2. MATERIAL AND METHODS
2.1. Methods
SARS‐CoV‐2 antibodies IgM and IgG CLIA kits were from Shenzhen YHLO Biotech Co, Ltd (China), with two antigens of SARS‐CoV‐2 coated on the magnetic beads of the CLIA (nucleocapsid protein or N protein and spike protein or S protein). All antibody tests were performed by iFlash1800 fully automatic CLIA analyzer from YHLO biotechnology Co (LTD, Shenzhen, China). The amount of anti‐SARS‐CoV‐2 antibodies IgM and IgG is positively correlated with the relative light units (RLU) measured by the chemiluminescence analyzer. iFlash1800 CLIA analyzer automatically calculates the concentration (AU/mL) based on the calibration curve. Cut off value proposed by manufacturer is 10 AU/mL both for IgM and IgG antibodies: hence samples with IgM and IgG concentration more than equal to 10 AU/mL are considered positive (reactive).
2.2. Patients
This study enrolled a total of 61 patients (59 ± 23 years; 35 women and 26 men) hospitalized in San Giovanni di Dio Hospital (Florence, Italy) for COVID‐19 and a pre‐COVID‐19 (2018‐2019) disease control group of 44 patients (49 ± 17 years; 35 women and 9 men) who had rheumatic diseases (n = 31) and infectious diseases (n = 13). Twenty blood donors from the COVID‐19 era (winter 2019) (44 ± 11 years; 8 women and 12 men) also participated in the study.
All COVID‐19 patients were confirmed to be infected with SARS‐CoV‐2 detected in oropharyngeal and nasopharyngeal swabs by use of RT‐PCR (confirmed by two SARS‐CoV‐2 nucleic acid tests). Thirty out of the 61 patients had mild to moderate symptoms, while 31 with severe pneumonia required admission to the intensive care unit (ICU). Blood samples had a mean duration of 12 days (range 8‐17 days) from the onset of symptoms.
3. RESULTS
The Reciever Operating Characteristics (ROC) performance curves showed Area Under the Curve (AUC) values of 0.918 and 0.980 for anti‐SARS‐CoV‐2 antibodies IgM and IgG, respectively (Figure 1).
Figure 1.
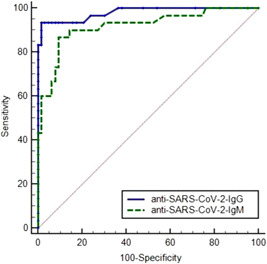
Reciever operating characterstic analysis for anti‐SARS‐CoV‐2 antibodies detection
At the manufacturer's cutoff value of 10 AU/mL, sensitivity was 73.3% and 76.7% and specificity was 92.2% and 100% for IgM and IgG antibodies, respectively (Figure 2). We reported four IgM positive results in the control group: two cases of cytomegalovirus infection, one scleroderma, and one lupus erythematosus systemic patients. Diagnostic performances of the anti‐SARS‐CoV‐2 antibodies at different cutoff values are described in Table 1. The highest sensitivity with a good specificity performance was reached at a cutoff of 10.0 AU/mL for IgM (positive negative value [PPV] 81.5% and negative predictive value [NPV] 88.1%) and of 7.1 for IgG (PPV 100%, NPV 92.8).
Figure 2.
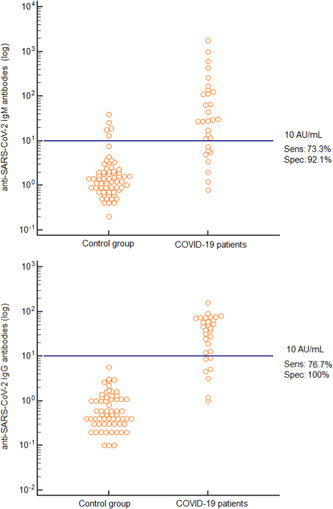
Distribution of anti‐SARS‐CoV‐2 IgM and IgG antibodies levels in COVID‐19 patients and in the control group at the manufacturer's cutoff
Table 1.
Performance characteristics (with 95% confidence intervals) of anti‐SARS‐CoV‐2 antibodies IgM and IgG at different cutoff values as determined by CLIA method
| Anti‐SARS‐CoV‐2 IgM antibodies | |||||||
|---|---|---|---|---|---|---|---|
| Cutoff value | 6.7 AU/mL | 7.5 AU/mL | 9.4 AU/mL | 10.0 AU/mL | 11.3 AU/mL | 12.2 AU/mL | 13.4 AU/mL |
| Sensitivity | 76.7% (59.7‐89.2) | 73.3% (56.0‐86.8) | 73.3% (56.0‐86.8) | 73.3% (56.0‐86.8) | 70.0% (52.4 ‐ 84,3) | 66.7% (48.9‐81.7) | 66.7% (48.9‐81.7) |
| Specificity | 90.6% (81.9‐96.2) | 90.6% (81.9‐96.2) | 92.2% (84.0‐97.1) | 92.2% (84.0‐97.1) | 92.2% (84.0 ‐ 97,1) | 92.2% (84.0‐97.1) | 93.7% (86.1‐98.0) |
| PPV | 79.3% (62.5‐91.2) | 78.6% (61.3‐90.9) | 81.5% (64.3‐92.9) | 81.5% (64.3‐92.9) | 80.8% (63.1‐92.6) | 80.0% (61.8‐92.3) | 83.3% (65.4‐94.5) |
| NPV | 89.2% (80.2‐95.2) | 87.9% (78.6‐94.3) | 88.1% (78.9‐94.4) | 88.1% (78.9‐94.4) | 86.8% (77.4‐93.4) | 85.5% (76.0‐92.5) | 85.7% (76.3‐92.6) |
| LR+ | 8.18 | 7.82 | 9.39 | 9.39 | 8.96 | 8.53 | 10.7 |
| LR− | 0.26 | 0.29 | 0.29 | 0.29 | 0.33 | 0.36 | 0.36 |
| OR | 31.8 | 26.6 | 32.5 | 32.5 | 27.5 | 23.6 | 30.0 |
| Anti‐SARS‐CoV‐2 IgG antibodies | |||||||
|---|---|---|---|---|---|---|---|
| Cutoff value | 5.4 AU/mL | 7.1 AU/mL | 8.9 AU/mL | 10.0 AU/mL | 10.7 AU/mL | 12.6 AU/mL | 15.9 AU/mL |
| Sensitivity | 83.3% (67.5‐93.7) | 83.3% (67.5‐93.7) | 80.0% (63.6‐91.5) | 76.7% (59.7‐89.2) | 76.7% (59.7‐89.2) | 73.3% (56.0‐86.8) | 70.0% (52.4‐84.3) |
| Specificity | 98.45 (93.3‐99.9) | 100% (94.3‐100) | 100% (94.3‐100) | 100% (94.3‐100) | 100% (94.3‐100) | 100% (94.3‐100) | 100% (94.3‐100) |
| PPV | 96.2% (84.1‐99.8) | … | … | … | … | … | … |
| NPV | 92.6% (84.9‐97.3) | 92.8% (85.1‐97.3) | 91.4% (83.4‐96.5) | 90.1% (81.8‐95.6) | 90.1% (81.8‐95.6) | 88.9% (80.3‐94.8) | 87.7% (78.9‐93.9) |
| LR+ | 53.3 | … | … | … | … | … | … |
| LR− | 0.17 | 0.17 | 0.20 | 0.23 | 0.23 | 0.27 | 0.30 |
| OR | 315 | … | … | … | … | … | … |
Abbreviations: CLIA, chemiluminescence immunoassay; LR + , positive likelihood ratio; LR−, negative likelihood ratio; NPV, negative predictive value; OR, odds ratio; PPV, positive predictive value.
Among the COVID‐19 patients 64.1% (41/64) had both IgM and IgG positive test results, while 4.7% (3/64) and 7.8% (5/64) had only IgM, and only IgG positive results, respectively. The average concentration among COVID‐19 positive sera was 69.8 AU/mL for IgM and 48.95 AU/mL for IgG antibodies.
4. DISCUSSION
The most widely used biomarkers for COVID‐19 are IgM and IgG antibodies produced from the second week of viral infection. IgM can be detected in the patient samples from 10 to 30 days after SARS‐CoV‐2 infection, while IgG appears at day 20 onwards. 11 IgM manifests earlier than IgG, but it then weakens and disappears. IgG however can persist for a long time following infection and may potentially have a protective role. For the purpose of monitoring kinetics of the antibodies, quantitative assays are preferable to qualitative tests, even if available assays have not yet been widely validated. 12 , 13 , 14 , 15
However, it is unclear which antibodies are optimally effective in the scenario of COVID‐19 and which of them are neutralizing. There is also uncertainty as to which antibody isotype (IgM, IgG or IgA) (single or combined) is the best choice in these different contexts. 16
As with most existing studies on the diagnostic performance of the SARS‐CoV‐2 antibodies, our preliminary data showed that most COVID‐19 patients have both IgM and IgG, and only few of them have isolated IgG or IgM antibodies. On the one hand, in reference to IgM and IgG combination, the overall sensitivity of 75% may reflect that some patients may not yet develop antibodies or will never develop (the length of time from the symptoms onset to serological test ranged from 8 to 17 days); on the other hand, the 100% specificity performance of IgG antibodies makes them an appropriate test for the different immunization protocols. With regard to IgM false positive results, it's important to underline that we designed a disease control group made up of (a) donors from last winter when other coronaviruses were active who had all negative results; (b) autoimmune and infectious diseases dating back at least 1 year in which we found four reactive sera. This means that we had no cross reaction with other coronaviruses but two CMV infections and two rheumatic diseases interfered with the test, even if with a low titer. This data can be added to the known issues concerning IgM by rapid tests such as the lack of specificity together with the low sensitivity due to low antibody concentrations or to their short duration. We speculate that some patients have not been produced yet, turned already to negative, might not develop IgM or produce any response at all.
Considering that the best cutoff value is related to the specific use of the test, our internal ROC curves showed very similar values to the ones proposed by the manufacturer, if we hypothesize a screening application. In fact further refinement of the manufacture's cutoff is always advisable to calibrate the kit to a specific population.
To the best of our knowledge this is one of the first studies on anti‐SARS‐CoV‐2 IgM and IgG antibodies by CLIA method on an Italian population. In fact, previous studies are few and mainly involving small Chinese cohorts. Our study results have some limitations: the time between the onset of symptoms and serum sample vary among patients; COVID‐19 patients were not enrolled at the early stage of disease, and neither was a COVID‐19 group who had provided a negative nasopharyngeal/oropharyngeal swab.
5. CONCLUSION
Our experience highlights the importance of a CLIA method, not only to overcome the problems of the subjective reading of the band (especially weak) in the rapid tests, but for the wide range of potentials inherent to a quantitative method, such as assisting with diagnosis and evaluating the disease through antibodies profiles. Furthermore, selection of IgG antibodies at high level concentrations may be helpful in developing vaccines and treating SARS‐CoV‐2 by convalescent plasma therapy.
Infantino M, Grossi V, Lari B, et al. Diagnostic accuracy of an automated chemiluminescent immunoassay for anti‐SARS‐CoV‐2 IgM and IgG antibodies: an Italian experience. J Med Virol. 2020;92:1671–1675. 10.1002/jmv.25932
REFERENCES
- 1. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus‐infected pneumonia. N Engl J Med. 2020;382:1199‐1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. World Health Organization. (2020) . Laboratory testing for coronavirus disease 2019 (COVID‐19) in suspected human cases: interim guidance, 2 March 2020. World Health Organization.
- 4. Infantino M, Damiani A, Gobbi FL, et al. Serological assays for SARS‐CoV‐2 infectious disease: benefits, limitations and perspectives. Isr Med Assoc J. 2020;22:203‐210. [PubMed] [Google Scholar]
- 5. Okba NMA, Müller MA, Li W. SARS‐CoV‐2 specific antibody responses in COVID‐19 patients. medRxiv. 2020. [Google Scholar]
- 6. Centers for Disease Control and Prevention . Coronavirus disease 2019 (COVID‐19)–Information for Laboratories. https://www.cdc.gov/coronavirus/2019‐ncov/lab/index.html. Accessed on April 11, 2020.
- 7. Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID‐19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020;38:1‐9. [DOI] [PubMed] [Google Scholar]
- 8. Cassaniti I, Novazzi F, Giardina F, et al. Performance of VivaDiag COVID‐19 IgM/IgG Rapid Test is inadequate for diagnosis of COVID‐19 in acute patients referring to emergency room department. J Med Virol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. U.S. Food & Drug Administration , Policy for Diagnostic Tests for Coronavirus Disease‐2019 during the Public Health Emergency. https://www.fda.gov/media/135659/download. Accessed on March 16, 2020.
- 10. Li Z, Yi Y, Luo X, et al. Development and clinical application of a rapid IgM‐IgG combined antibody test for SARS‐CoV‐2 infection diagnosis. J Med Virol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tan W, Lu Y, Zhang J, et al. Viral kinetics and antibody responses in patients with COVID‐19. medRxiv. 2020. [Google Scholar]
- 12. Zeng F, Dai C, Cai P, et al. A comparison study of SARS‐CoV‐2 IgG antibody between male and female COVID‐19 patients: a possible reason underlying different outcome between gender. medRxiv 2020. [DOI] [PMC free article] [PubMed]
- 13. Padoan A, Cosma C, Sciacovelli L, Faggian D, Plebani M. Analytical performances of a chemiluminescence immunoassay for 2019‐nCov IgM/IgG and antibody kinetics. Clin Chem Lab Med. 2020. [DOI] [PubMed] [Google Scholar]
- 14. Jin Y, Wang M, Zuo Z, et al. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int J Infect Dis. 2020;94:49‐52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lippi G, Salvagno GL, Pegoraro M, et al. Assessment of immune response to SARS‐CoV‐2 with fully‐automated MAGLUMI 2019‐nCoV IgG and IgM chemiluminescence immunoassays. Clin Chem Lab Med. 2020. [DOI] [PubMed] [Google Scholar]
- 16. Binnicker MJ. Emergence of a novel coronavirus disease (COVID‐19) and the importance of diagnostic testing: why partnership between clinical laboratories, public health agencies, and industry is essential to control the outbreak. Clin Chem. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]


