Abstract
The brain transforms nociceptive input into a complex pain experience comprised of sensory, affective, motivational, and cognitive components. However, it is still unclear how pain arises from nociceptive input and which brain networks coordinate to generate pain experiences. We introduce a new high-dimensional mediation analysis technique to estimate distributed, network-level patterns that formally mediate the relationship between stimulus intensity and pain. We applied the model to a large-scale analysis of functional magnetic resonance imaging data (N = 284), focusing on brain mediators of the relationship between noxious stimulus intensity and trial-to-trial variation in pain reports. We identify mediators in both traditional nociceptive pathways and in prefrontal, midbrain, striatal, and default-mode regions unrelated to nociception in standard analyses. The whole-brain mediators are specific for pain versus aversive sounds and are organized into five functional networks. Brain mediators predicted pain ratings better than previous brain measures, including the neurologic pain signature (Wager et al. 2013). Our results provide a broader view of the networks underlying pain experience, as well as novel brain targets for interventions.
Keywords: brain networks, fMRI, mediation analysis, pain, pattern analysis
Introduction
The brain is central to the generation of pain; it transforms sensory input into a complex set of pain-related responses, including subjective experience, autonomic responses, avoidance behavior, and activation of linked memories and concepts. However, the boundaries of the brain systems that mediate this series of transformations have been inconsistent across studies. Traditionally, pain processing has been associated with a discrete set of brain regions targeted by spinal nociceptive afferents, including primary (S1) and secondary (S2) somatosensory, and anterior midcingulate cortices (aMCC), medial and lateral thalamus, and posterior and midinsular cortices (Apkarian et al. 2005; Dum et al. 2009; Jensen et al. 2016). These have been referred to as the “pain matrix” and often treated as a unitary system, though this concept has been largely abandoned as its specificity to pain has been called into question. Other studies have found that additional regions are also involved in encoding the intensity of noxious stimuli and/or correlate with pain experience under some circumstances (Bingel et al. 2002; Büchel et al. 2002; Becerra et al. 2013), making the boundaries of the “pain matrix” elusive and context-dependent. And, though this set of regions has been grouped into subsystems (Craig et al. 2000)—for example, lateral and medial subsystems more closely related to sensory–discriminative and affective–motivational aspects of pain, respectively (Villemure et al. 2003)—empirical studies have shown that the division between sensory encoding and pain affect is not straightforward (Baliki et al. 2009; Atlas et al. 2010, 2014).
One source of complexity lies in the fact that “pain matrix” or “pain-processing” regions have been defined in multiple ways. Some studies identify regions based on stimulus intensity encoding, the tendency for a region to be activated by more versus less intense noxious stimuli (Peyron et al. 2002; Wager et al. 2004). Others identify regions based on correlations with pain reports (Coghill et al. 1999; Lindquist et al. 2017). Both are relevant: A region involved in pain generation should both encode stimulus intensity and correlate with reported subjective experience, even when intensity is matched. Accordingly, one step forward lies in characterizing formal brain “mediators” of the relationship between stimulus intensity and pain report (Atlas et al. 2010, 2014). Mediation is a statistical test that links experimentally manipulated variables, brain measures, and behavioral outcome variables in a single path model. It requires both an effect of an initial (often experimentally manipulated) variable and association with an outcome (e.g., pain) controlling for stimulus intensity. Applied to pain, it can be used to identify brain regions that both encode experimental manipulations in noxious stimulus intensity and correlate with pain experience controlling for intensity, with sufficient effect sizes in both tests to pass the more stringent test of mediation.
Previous studies have identified brain mediators of pain (Atlas et al. 2010, 2014), testing voxels one at a time. However, this kind of univariate approach can miss brain regions whose contributions to pain perception are conditional on other regions. More broadly, it is increasingly clear that much of the functional information encoded in the brain is encoded in distributed patterns across neural ensembles and systems (Pouget et al. 2000; Haxby et al. 2014), which requires brain information to be treated in a multivariate fashion (Kriegeskorte 2011; Woo et al. 2017). Mediation models with “multivariate” brain mediators are required to characterize these patterns, but have not been available until now. Here, we provide the first analysis of multivariate brain mediators of pain, using a novel statistical method called “principal directions of mediation” (PDM) (Chén et al. 2017). PDM decomposes activity across the brain into multiple networks that independently mediate stimulation effects on outcomes (Fig. 1A,B). This can help identify which brain systems mediate stimulus intensity effects on pain, taking the distributed nature of information encoding in the brain into account.
Figure 1.
Mediation analysis. (A) Schematic of the mediation analysis framework. Brain activity is an intermediate, mediating variable (M) between a manipulated noxious stimulus intensity (X) and perceived pain (Y). In the high-dimensional PDM approach, a linear combination of all brain voxels acts as mediators. Multiple, orthogonal mediators can be estimated. The weight vectors  (or PDMs) represent the contribution of individual voxels to the kth mediation pathway. Voxel weights (
(or PDMs) represent the contribution of individual voxels to the kth mediation pathway. Voxel weights ( ) are fit so that the indirect, mediated effect is maximized. (B) Mediation path coefficients for all 30 PDMs are shown with signs of path
) are fit so that the indirect, mediated effect is maximized. (B) Mediation path coefficients for all 30 PDMs are shown with signs of path  coefficients set to be positive. Path
coefficients set to be positive. Path 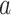 indicates the temperature-to-brain (PDM) relationship, path
indicates the temperature-to-brain (PDM) relationship, path 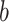 the PDM to pain rating relationship, and path
the PDM to pain rating relationship, and path 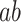 the indirect, mediated effect. Positive coefficients indicate that voxels with positive weights in a given PDM are positively related with temperature and/or rating. The first 10 PDMs explain more 99% of the total indirect effect. We focus on these PDMs in the following analyses (shaded area in right panel). (C) PDMs are estimated on the training data comprising a total of 209 participants from 7 pain studies. PDM validation is done on an independent eighth data set with 75 participants. (D) Individual PDMs can be combined into a single, cPDM by weighting and summing the individual, orthogonal PDMs. cPDM voxel weights are shown on the brain rendering and in key regions in the right panel.
the indirect, mediated effect. Positive coefficients indicate that voxels with positive weights in a given PDM are positively related with temperature and/or rating. The first 10 PDMs explain more 99% of the total indirect effect. We focus on these PDMs in the following analyses (shaded area in right panel). (C) PDMs are estimated on the training data comprising a total of 209 participants from 7 pain studies. PDM validation is done on an independent eighth data set with 75 participants. (D) Individual PDMs can be combined into a single, cPDM by weighting and summing the individual, orthogonal PDMs. cPDM voxel weights are shown on the brain rendering and in key regions in the right panel.
Another source of complexity lies in the fact that most pain studies have been necessarily limited in sample size (typical N < 50). Results from small studies are increasingly recognized as variable and prone to high levels of both false positive and false negative results (Button et al. 2013). We address this issue by analyzing within-person, single-trial pain data aggregated across eight individual studies (N = 284). In addition, the test study not only included heat pain stimuli but also two distinct types of sounds: physically aversive sounds (a “knife on plate”) and emotionally aversive sounds (gunshots, screams, etc.) This provides one of the first tests of specificity of multivariate pain-related patterns against sounds (cf. Horing et al. 2019; Liang et al. 2019) and the first test of functional specificity of brain mediators of pain.
Materials and Methods
Participants
The analysis included data from a total of 284 healthy participants from eight independent studies, with sample sizes ranging from N = 17 to N = 75 per study. Descriptive statistics on the age, sex, and other features of the subjects in each individual study are provided in Supplementary Tables S1–S3. Further details on studies 1–7, which were used to estimate the PDMs, are provided in Lindquist et al. (2017). Participants were recruited from New York City and Boulder/Denver Metro Areas. The institutional review board of Columbia University and the University of Colorado Boulder approved all the studies, and all participants provided written informed consent. Preliminary eligibility of participants was determined through an online questionnaire, a pain safety screening form, and a functional magnetic resonance imaging (fMRI) safety screening form.
We applied several exclusion criteria for analysis purposes. Participants with psychiatric, physiological or pain disorders, neurological conditions, and MRI contraindications were excluded prior to enrollment. In addition, participants were required to have at least 30 trials with low variance inflation factors (see Supplementary Materials and Methods), nonmissing rating, and stimulation intensity data. Based on these criteria, 18 participants from study 8 were excluded, resulting in a total of 209 participants for the primary PDM analysis and 75 participants for the validation sample.
Procedures
Overview
Participants in all studies underwent fMRI scanning while being exposed to varying levels of heat pain within-person and rating the perceived pain intensity (see Supplementary Tables S1–S3). For each participant, we recorded the temperature applied and the pain rating for each trial and estimated single-trial maps of brain activity. These three variables were used in the primary mediation analysis with temperature as the initial variable, brain activity as the mediator, and pain rating as the outcome variable (Fig. 1).
Using the high-dimensional mediation analysis model, we first estimated 30 whole-brain mediation patterns (PDMs). Each PDM specifies a linear combination of voxels across the brain maximizing the mediated effect from temperature to pain rating, while being orthogonal to other PDMs (Fig. 1A). Each PDM (or 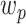 ) thus represents a formal, whole-brain mediator for pain. The voxel weights of each PDM inform about the contribution of individual brain regions to the generation of a painful experience following noxious stimulation.
) thus represents a formal, whole-brain mediator for pain. The voxel weights of each PDM inform about the contribution of individual brain regions to the generation of a painful experience following noxious stimulation.
Furthermore, as the PDM model is linear, independent PDMs can readily be fused into a single, combined PDM (cPDM) that can be prospectively applied to new data sets as a predictive model. We fused the individual brain mediator maps into a cPDM by estimating the weighted combination of individual PDMs that best predicted pain in the training sample (see section PDM and Fig. 1D). Prediction performance can then be evaluated against independent data test.
Thermal and Aversive Sound Stimulation
The number of noxious stimulation trials, stimulation sites, intertrial intervals, rating scales, and stimulus intensities and durations varied across studies, but were comparable; these variables are summarized in Supplementary Tables S2 and S3. Each study also comprised a specific psychological manipulation (except study 8), such as placebo treatment, which has been reported elsewhere (see Supplementary Table S1).
In studies 1–6, thermal stimulation was delivered to multiple skin sites using a TSA-II Neurosensory Analyzer (Medoc Ltd, Chapel Hill, NC) with a 16-mm Peltier thermode endplate. A pathway system (Medoc Ltd, Chapel Hill, NC) was used in studies 7 and 8. Participants rated the perceived magnitude of warmth or pain during or after each trial. Most studies applied heat to the left volar forearm (see Supplement Methods and Lindquist et al. [2017] for details).
Study 8, which was used for validation purposes (see below), also presented aversive sounds to participants. Trials with aversive sounds were used to test the specificity of the pain PDMs. Sounds included a physically aversive recording of nails on a chalkboard and a set of emotionally aversive sounds (attacks, screaming, and crying) from the International Affective Digital Sounds database (Bradley and Lang 2007). Aside from these sound trials, we focus on brain mediation of pain across all trials in the present paper, irrespective of the study-specific psychological and physical manipulations that influenced pain.
fMRI Data Processing
Preprocessing and Subject-Level Models
We chose to retain the original preprocessing used in each published paper for two reasons: (1) to establish, and test, robustness across minor variations in processing pipelines and (2) because study-specific analysis choices are appropriate in some cases, depending on the distribution of the data and study design (for details, see Supplementary Methods and Lindquist et al. [2017]). Briefly, structural, T1-weighted images were coregistered to the functional mean image and then normalized to MNI space using SPM. For each study, a single trial, or single epoch model, was estimated (Koyama et al. 2003; Rissman et al. 2010; Mumford et al. 2012). The single-trial brain activation estimates served as the basis for the subsequent analysis.
PDM Validation
The high-dimensional brain mediators (PDMs) were estimated on the training data comprised of studies 1–7 (Lindquist et al. 2017). Even though this data set is large (N = 209) and diverse, the possibility of overfitting to the training data might reduce the generalizability of the PDMs. To test for the generalizability of the PDMs, we validated the PDMs on independent data test (study 8, N = 75). Computing the inner product of each PDM with each single-trial beta image from study 8 resulted in 10 potential mediator variables. Each of these potential mediators was then subjected to a multilevel mediation analysis (Wager et al. 2009) with P values determined by a bootstrap procedure with 5000 iterations each. If the PDMs generalize to the new data set, paths 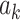 ,
, 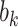 , and the indirect effect
, and the indirect effect 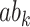 should be significant for all
should be significant for all 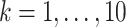 PDMs.
PDMs.
We also tested whether the PDMs specifically mediate the relationship between temperature and pain intensity. To this end, we also tested the original PDMs on the aversive sound trials from study 8. If the PDMs reflect specific patterns of brain activity involved in pain processing, they should not mediate the relationship between sound stimulation level and intensity ratings. We thus expect no significant indirect effect for the sound trials.
A further test to validate the stability of PDM estimation was conducted by switching training and data test. That is, pain PDMs were estimated on study 8 and tested on the original training data from studies 1 to 7 as described above.
Dimension Reduction
The training data set consisted of a total of 13 372 single-trial beta images (i.e., activation estimate images), each consisting of 229 519 voxels, from 209 participants. To reduce the dimensionality of the data to a computationally tractable size, a generalized version of population value decomposition (PVD) (Caffo et al. 2010; Crainiceanu et al. 2011; Chén et al. 2017) was applied (using PVD.m, included in the M3 mediation toolbox available at https://github.com/canlab/MediationToolbox). This procedure is similar to singular value decomposition (SVD) but decomposes the data matrix into both participant-specific and population-specific components. We chose a dimensionality of 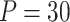 based on a tradeoff between variance explained and the number of trials available for each participant. The beta images were z-scored within each participant before PVD application. The reduced data matrix used for PDM estimation consisted of a matrix with dimensions 13 372 × 30.
based on a tradeoff between variance explained and the number of trials available for each participant. The beta images were z-scored within each participant before PVD application. The reduced data matrix used for PDM estimation consisted of a matrix with dimensions 13 372 × 30.
Principal Directions of Mediation Model
Let 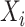 be the temperature,
be the temperature, 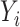 the reported pain, and
the reported pain, and 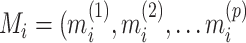 ) the brain activity over
) the brain activity over  voxels (i.e., the beta maps) measured between the application of the thermal stimuli and the pain report for observation (i.e., trial)
voxels (i.e., the beta maps) measured between the application of the thermal stimuli and the pain report for observation (i.e., trial)  . We are interested in determining how brain activation mediates the relationship between temperature and pain report. We can estimate the parameters of this model using the following set of equations:
. We are interested in determining how brain activation mediates the relationship between temperature and pain report. We can estimate the parameters of this model using the following set of equations:
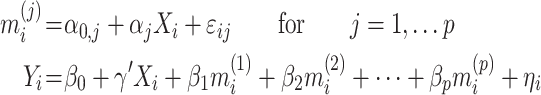 |
(1) |
Once the parameters have been estimated, we can express the total effect 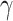 as the sum of the direct and indirect effects as follows:
as the sum of the direct and indirect effects as follows:
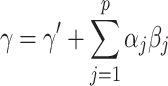 |
(2) |
If p is relatively small, the series of regressions described in equation (1) can be used to estimate the pertinent mediation effects. However, in our setting, there are too many mediators to allow reasonable interpretation (unless the model coefficients are highly structured), and there are many more mediators than subjects, precluding estimation using standard procedures. To overcome these problems, we introduce a transformation of the space of mediators, determined by finding linear combinations of the original mediators that (1) are orthogonal and (2) are chosen to maximize the indirect effect. The first constraint allows us to fit a separate linear model for each transformed variable. The second constraint allows us to limit our analysis to only those directions that contain the most information about the indirect effect. Here, we improve and extend the approach proposed by Chén et al. (2017) by choosing a different cost function, computing a cPDM, and analyzing an almost 10-times larger data set.
This new model, called the “PDM,” linearly combines activity in different voxels into a smaller number of orthogonal components, with components ranked based upon the proportion of the indirect effect that each accounts for. Ideally, the components form a small number of uncorrelated mediators that represent interpretable networks of voxels.
To illustrate, let 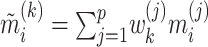 for
for 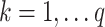 be a set of linear transformations of the mediators with
be a set of linear transformations of the mediators with 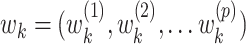 . Placing these new variables into our mediation model, we obtain:
. Placing these new variables into our mediation model, we obtain:
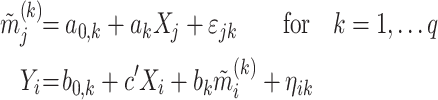 |
(3) |
Now, we can decompose the total effect into direct and indirect effects as follows:
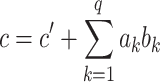 |
(4) |
The difference between this model and the standard mediation model described in equation (1) is that the  is unknown. In our approach
is unknown. In our approach  is chosen so that it maximizes the amount of the indirect effect that is explained (i.e.,
is chosen so that it maximizes the amount of the indirect effect that is explained (i.e., 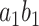 is maximized). We refer to
is maximized). We refer to 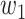 as the first “principal direction of mediation” (PDM). Note the first PDM corresponds to voxel-specific weights that can be mapped onto the brain and thus provides interpretable maps of brain networks in the same manner as independent component analysis (ICA) and principal component analysis (PCA). Subsequent directions
as the first “principal direction of mediation” (PDM). Note the first PDM corresponds to voxel-specific weights that can be mapped onto the brain and thus provides interpretable maps of brain networks in the same manner as independent component analysis (ICA) and principal component analysis (PCA). Subsequent directions 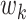 ,
, 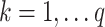 can be found that maximize the remaining indirect effect conditional on being orthogonal to previous PDMs. As the transformed mediators are ranked based upon the proportion of the indirect effect explained, one could potentially limit the number of PDMs computed to achieve dimension reduction. Hence, our approach is philosophically similar to PCA, but addresses a fundamentally different problem.
can be found that maximize the remaining indirect effect conditional on being orthogonal to previous PDMs. As the transformed mediators are ranked based upon the proportion of the indirect effect explained, one could potentially limit the number of PDMs computed to achieve dimension reduction. Hence, our approach is philosophically similar to PCA, but addresses a fundamentally different problem.
The individual, orthogonal PDMs can be fused into a cPDM by computing the following weighted sum:
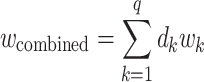 |
(5) |
The scalar weights 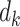 are estimated from the training sample using a linear model with the individual PDMs as regressors and reported pain as the response.
are estimated from the training sample using a linear model with the individual PDMs as regressors and reported pain as the response.
According to the model formulation, the signs of the PDMs are not identifiable, as any change in the sign of 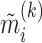 can be offset by a change in sign of both
can be offset by a change in sign of both 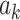 and
and 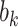 . We fix the signs of
. We fix the signs of 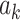 to be positive for easier interpretation, that is, positive voxel weights indicate higher brain activity for higher stimulus intensities. This is a similar constraint to the ICA approach often used in neuroimaging to detect networks. The orthogonality constraint does not reduce the total amount of variance explained by all PDMs.
to be positive for easier interpretation, that is, positive voxel weights indicate higher brain activity for higher stimulus intensities. This is a similar constraint to the ICA approach often used in neuroimaging to detect networks. The orthogonality constraint does not reduce the total amount of variance explained by all PDMs.
The problem of finding the 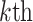 PDM involves finding the vector
PDM involves finding the vector 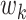 that maximizes
that maximizes 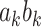 based on the constraint that
based on the constraint that 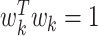 and
and 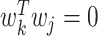 for all
for all 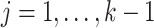 . This problem can be solved using a nonlinear programming solver such as the interior-point algorithm. Inference is performed using a bootstrap procedure with 5000 iterations, as described in Chén et al. (2017). PDM maps are thresholded at a false discovery rate (FDR) of
. This problem can be solved using a nonlinear programming solver such as the interior-point algorithm. Inference is performed using a bootstrap procedure with 5000 iterations, as described in Chén et al. (2017). PDM maps are thresholded at a false discovery rate (FDR) of  . The cPDM map in Figure 2 displays the top 5% of voxels based on their weight parameters, yielding a more conservative display than FDR. We present results of 10 PDMs accounting for more than 99% of the total indirect effect. A software implementation is available at (https://github.com/canlab/MediationToolbox) (multivariateMediation.m).
. The cPDM map in Figure 2 displays the top 5% of voxels based on their weight parameters, yielding a more conservative display than FDR. We present results of 10 PDMs accounting for more than 99% of the total indirect effect. A software implementation is available at (https://github.com/canlab/MediationToolbox) (multivariateMediation.m).
Figure 2.

PDM. Voxel maps for PDMs with individually significant voxels at FDR  . Tan backgrounds indicate PDMs with positive paths
. Tan backgrounds indicate PDMs with positive paths  and
and  . Blue backgrounds indicate PDMs with positive path
. Blue backgrounds indicate PDMs with positive path 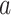 and negative path
and negative path 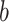 . Brain activity increases in voxels with positive weights (warm colors) with higher temperatures. Higher brain activity in these voxels is related to higher pain ratings in PDMs with positive path
. Brain activity increases in voxels with positive weights (warm colors) with higher temperatures. Higher brain activity in these voxels is related to higher pain ratings in PDMs with positive path 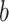 (tan panels) and negatively with negative path
(tan panels) and negatively with negative path 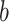 (blue panels). No voxels are individually significant in PDM 8. The bottom panel shows the cPDM, a weighted linear combination of the above 10 PDMs. The top 5% of voxels based on voxel weights are shown since almost all voxels survived the significance testing. All brain figures are displayed in neurological convention (left is left) and thresholded at FDR
(blue panels). No voxels are individually significant in PDM 8. The bottom panel shows the cPDM, a weighted linear combination of the above 10 PDMs. The top 5% of voxels based on voxel weights are shown since almost all voxels survived the significance testing. All brain figures are displayed in neurological convention (left is left) and thresholded at FDR 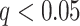 . MCC = midcingulate cortex, SMA = supplementary motor area, mPFC = medial prefrontal cortex, PAG = periaqueductal gray, midIns = mid-insula, dpIns = dorsal posterior insula, S2 = secondary somatosensory cortex, S1 = primary somatosensory cortex, M1 = primary motor cortex, mOFC = medial orbitofrontal cortex, RSC = retrosplenial cortex, SFG = superior frontal gyrus, vIns = ventral insula, dmPFC = dorsomedial prefrontal cortex, V1 = primary visual cortex, V2 = secondary visual cortex, vStriatum = ventral striatum, NAc = nucleus accumbens, mThal = medial thalamus, aIns = anterior insula, and SPL = superior parietal lobule.
. MCC = midcingulate cortex, SMA = supplementary motor area, mPFC = medial prefrontal cortex, PAG = periaqueductal gray, midIns = mid-insula, dpIns = dorsal posterior insula, S2 = secondary somatosensory cortex, S1 = primary somatosensory cortex, M1 = primary motor cortex, mOFC = medial orbitofrontal cortex, RSC = retrosplenial cortex, SFG = superior frontal gyrus, vIns = ventral insula, dmPFC = dorsomedial prefrontal cortex, V1 = primary visual cortex, V2 = secondary visual cortex, vStriatum = ventral striatum, NAc = nucleus accumbens, mThal = medial thalamus, aIns = anterior insula, and SPL = superior parietal lobule.
In summary, we obtain scalar coefficients for paths 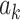 ,
, 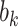 , and
, and 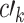 , as well as the indirect effect
, as well as the indirect effect 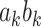 for each PDM as in a standard, univariate mediation analysis. In addition, we obtain the voxel weight vector
for each PDM as in a standard, univariate mediation analysis. In addition, we obtain the voxel weight vector 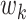 that maximizes the indirect effect
that maximizes the indirect effect 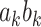 .
.
Cluster Analysis
The voxel weight maps for the mutually independent 10 PDMs span a high-dimensional space of brain mediators of pain perception. In order to reduce the dimensionality of that space and identify brain regions with similar activation profiles, we conducted a two-stage cluster analysis. The procedure is described in detail in Kober et al. (2008) and Atlas et al. (2014). Briefly, for significant voxels from the 10 PDMs, we extracted single-trial activity estimates, resulting in a 13 372 trials × 25 469 voxels matrix. We then used SVD to reduce the dimensionality of the voxel space. We kept 364 components that explained 95% of the variance. Next, we clustered voxels into 250 spatial parcels using hierarchical clustering. We then computed average single-trial activity within each parcel and used nonmetric multidimensional scaling (NMDS) and hierarchical clustering to further reduce the dimensionality of the data. Inspection of the Shepard plot suggested a NMDS dimensionality of 15 with stress indices below 0.05. Stress indices (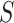 ) are computed according to Shepard (1980) with
) are computed according to Shepard (1980) with
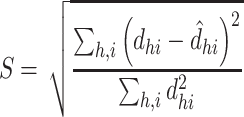 |
(6) |
Here, 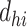 is the pairwise empirical dissimilarity and
is the pairwise empirical dissimilarity and 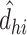 is the distance implied by the current solution between two brain regions
is the distance implied by the current solution between two brain regions 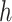 and
and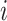 . Hierarchical clustering was then used to cluster the 250 parcels into 33 regions that coactivate across trials. These regions were not necessarily contiguous, and some spanned multiple anatomical regions, for example, covering right midinsula and dorsal insula plus operculum. Since we used voxel-wise FDR correction on the 10 PDMs, we expect some false positive values. Accordingly, some of the functional regions were located in the cerebrospinal fluid or outside the gray matter. We thus removed seven smaller functional clusters that were considered highly unlikely to be true gray matter region. We then averaged brain activity within the remaining 26 functional regions or nodes. NMDS was used to reduce the dimensionality again to 10 dimensions based on stress values. Applying hierarchical clustering again on the regions identified in the previous step identified large-scale functional brain networks. Permutation tests indicated that five networks provided the best clustering solution in terms of improvement over solutions on permuted data. Similarity of those five networks with the binarized PDM maps was assessed by Dice coefficients, which represents the true positive rate of the intersection between two maps.
. Hierarchical clustering was then used to cluster the 250 parcels into 33 regions that coactivate across trials. These regions were not necessarily contiguous, and some spanned multiple anatomical regions, for example, covering right midinsula and dorsal insula plus operculum. Since we used voxel-wise FDR correction on the 10 PDMs, we expect some false positive values. Accordingly, some of the functional regions were located in the cerebrospinal fluid or outside the gray matter. We thus removed seven smaller functional clusters that were considered highly unlikely to be true gray matter region. We then averaged brain activity within the remaining 26 functional regions or nodes. NMDS was used to reduce the dimensionality again to 10 dimensions based on stress values. Applying hierarchical clustering again on the regions identified in the previous step identified large-scale functional brain networks. Permutation tests indicated that five networks provided the best clustering solution in terms of improvement over solutions on permuted data. Similarity of those five networks with the binarized PDM maps was assessed by Dice coefficients, which represents the true positive rate of the intersection between two maps.
Local Pattern Expression Analyses
To summarize the cPDM pattern weights as a function of known regions or networks, we calculated the pattern energy within three sets of predefined regions. The first was a set of regions identified as contributing to nociceptive pain pathways based on prior work. We identified regions in the cortex using the atlas of Glasser et al. (2016), the thalamus using the atlas of Morel et al. (1997), and key brainstem regions based on previous papers: for parabrachial nucleus (PBN), Fairhurst et al. (2007), and for rostral ventral medulla (RVM), Brooks et al. (2017). For the PAG, we (T.D.W.) hand-drew a region on the 7T high-resolution group T1 of Keuken et al. (2014) and segmented out the cerebral aqueduct to exclude it. Nociceptive thalamic zones were included as defined below.
The second set of regions was based on Morel et al. (1997). We grouped the 80 or so thalamic/epithalamic regions into 17 functional zones likely to be detectable using fMRI (see Fig. 3 for a complete list). Nociceptive thalamic zones included the ventral–posterior–lateral (VPL) and ventral–posterior–medial (VPM) zones, the intralaminar group, and the mediodorsal “association nucleus” (MD).
Figure 3.
cPDM weights in nociceptive pathways. (A) Pattern weights in 24 cerebral targets of nociceptive pathways. The results show strong contributions from nociceptive subregions in the thalamus and brainstem, including both ventrolateral (sensory) and medial (affective) thalamic zones, and the parabrachial complex (PBN). These are critical pathways in animal models but have been seldom identified in human pain studies. (B) Pattern weights across thalamic regions as defined by the Morel atlas. Predictive weights are distributed unevenly across the thalamus, and positive weights (red) are concentrated in sensory (VPM/VPL) and medial nociceptive targets (MD and intralaminar) and anterior nuclei (VA, VM, VL). They are also positive in the habenula (Hb). By contrast, they are absent in visual/auditory regions (MGN, LGN, pulvinar). This demonstrates selectivity for expected pain-related nuclei.
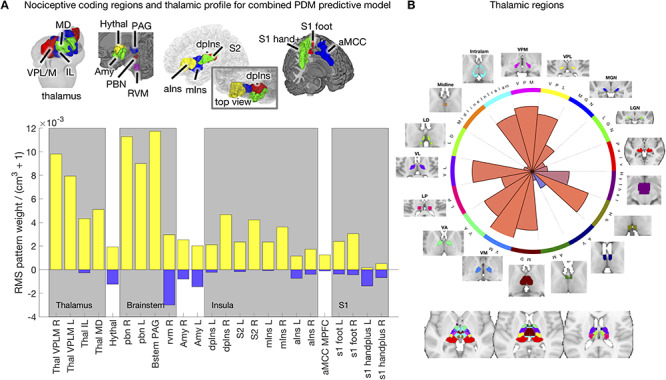
The third set of regions was a set of cortical networks defined based on resting-state connectivity, which we used to map cPDM weights onto established large-scale networks. We extracted loadings from 16 unique networks described in Schaefer et al. (2018) and manually separated them into left- and right-hemisphere components to examine lateralization (see Fig. 4 for a complete list).
Figure 4.
Cortical network profile for the cPDM. Pattern energy in resting-state cortical networks are distributed unevenly with strong, positive weights (red wedges) present in somatomotor B, ventral attention A, and control C networks. The first two match broadly on known nociceptive processing areas, while parts of control C (e.g., precuneus) are less known for pain processing.
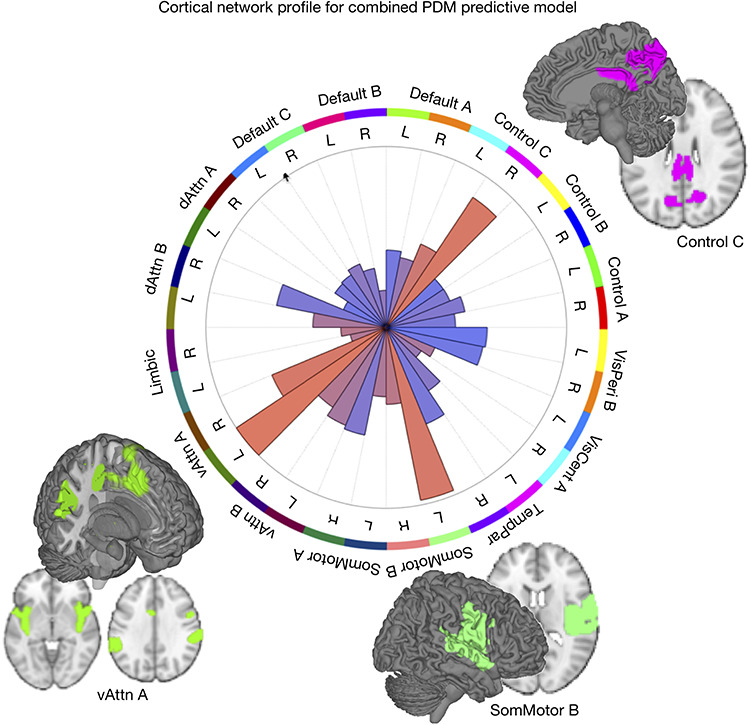
To summarize pattern weights in each local region or network, we calculated a measure of “pattern energy,” related to the absolute magnitude of predictive weights:
 |
(7) |
Er is the root-mean-square of weights in region (or network mask) r per cubic cm of brain tissue. w denotes the vector of weights for in-region voxels and V is the volume of the region in cm3. As the variance of Er varies inversely with region volume, the constant 1 is added to regularize the volume and thus avoid noise-driven, large-magnitude estimates for small regions. The area of the wedges in Figures 3 and 4 is proportional to Er.
We also defined a measure of “pattern valence” in predefined regions, which is the degree to which voxel weights are uniformly positive or negative. Pattern valence is defined as the cosine similarity of the pattern weights with the unit vector across in-region voxels. It is bounded at 1 and − 1, where 1 indicates uniform positive weights across voxels and − 1 indicates uniform negative weights. The color of the wedges in Figures 3 and 4 are proportional to the pattern valence; red colors indicate homogenous positive weights, blue homogenous negative weights, and purple mixed, variable weights across in-region voxels.
Comparison with Other Multivariate Models
To compare the PDM approach to other multivariate models of pain processing, we compared PDM1 and cPDM to the neurologic pain signature (NPS; Wager et al. 2013), to the stimulus intensity-independent pain signature 1 (SIIPS1; Woo et al. 2017), and the combination of NPS and SIIPS1. Furthermore, we compared the PDM approach against pain prediction by the Neurosynth reverse inference map for the term “pain” (https://neurosynth.org/analyses/terms/pain/). For each map, we correlated the predicted pain outcomes with the actual pain ratings on a single-trial level. In addition, we correlated the predictive brain maps with each other to evaluate similarities and differences in spatial pattern weights. Prediction–outcome correlations were compared using paired t-tests of Fisher-z–transformed correlation values.
Univariate Mediation Analysis
In univariate mediation analyses, a mediation model is estimated separately for every brain voxel (Wager et al. 2008; Atlas et al. 2010, 2014). Univariate mediation analysis produces three sets of brain maps—one for each path—in contrast to the PDM approach, which estimates only one set of paths for each PDM map. Previous studies also used smaller sample sizes available than the present study and had thus less statistical power than the present study. We ran a univariate mediation analyses on the training data set to directly compare the univariate results to the PDM approach. Univariate multilevel mediation analysis was conducted using the Multilevel Mediation and Moderation (M3) Toolbox for MATLAB (https://github.com/canlab/MediationToolbox). Voxel-wise significance was determined using a bootstrap procedure with 5000 iterations. An FDR of 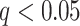 was used to control for multiple comparisons.
was used to control for multiple comparisons.
Results
Principal Directions of Mediation
For each individual PDM, we estimated path 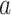 (stimulus intensity to brain), path
(stimulus intensity to brain), path 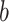 (brain to pain report), and mediation (
(brain to pain report), and mediation (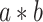 ) effects as in a standard mediation model (Figs 1 and 2). A positive path
) effects as in a standard mediation model (Figs 1 and 2). A positive path 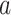 indicates that higher temperatures lead to more activity in voxels with positive PDM weights (yellow in brain figures) and less activity in voxels with negative PDM weights (blue in brain figures). A positive path
indicates that higher temperatures lead to more activity in voxels with positive PDM weights (yellow in brain figures) and less activity in voxels with negative PDM weights (blue in brain figures). A positive path 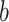 indicates that voxels with positive weights contribute positively to the pain rating after controlling for temperature. This pattern would be expected for regions that receive spinothalamic input, for example, the dorsal posterior insula or S2 (Willis and Westlund 1997; Dum et al. 2009) and possibly other mediating regions as well.
indicates that voxels with positive weights contribute positively to the pain rating after controlling for temperature. This pattern would be expected for regions that receive spinothalamic input, for example, the dorsal posterior insula or S2 (Willis and Westlund 1997; Dum et al. 2009) and possibly other mediating regions as well.
The absolute coefficient values for the indirect 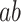 path assess how much of the effect of the manipulated temperature on pain ratings is explained by the brain mediator, that is, individual PDM pattern. Here, the first 10 PDMs accounted for 99.1% of the total mediation effect (Figs 1B and S1). We thus focus on the first 10 PDMs in all subsequent analyses with minimal loss of information. In order to analyze the contribution of individual brain regions to the mediation of pain, the signs of both paths
path assess how much of the effect of the manipulated temperature on pain ratings is explained by the brain mediator, that is, individual PDM pattern. Here, the first 10 PDMs accounted for 99.1% of the total mediation effect (Figs 1B and S1). We thus focus on the first 10 PDMs in all subsequent analyses with minimal loss of information. In order to analyze the contribution of individual brain regions to the mediation of pain, the signs of both paths 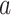 and
and 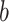 and the sign of the voxel weights have to be considered: Voxel weights are multiplied by the respective path coefficients to determine a region’s relationship to stimulation intensity and pain rating.
and the sign of the voxel weights have to be considered: Voxel weights are multiplied by the respective path coefficients to determine a region’s relationship to stimulation intensity and pain rating.
When considering the signs of the voxels weights, four different kinds of relationship are possible: (1) positive to temperature, positive to pain; (2) negative to temperature, negative to pain; (3) positive to temperature, negative to pain; and (4) negative to temperature, positive to pain. Here, type (1) is the standard, positive mediator case expected from nociceptive coding regions and type (2) represents a negative mediator, in which greater deactivation to the stimulus mediates increased pain (MacKinnon et al. 2000). Types (3) and (4) are suppressor effects (MacKinnon et al. 2000); for example, for type (3), brain activity increases with stimulus intensity that suppress pain and may thus be involved in stimulus-engaged regulatory processes and other negative feedback loops. Note that the values of path coefficients shown in Figure 1 depend on the scaling of the predictor (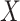 ), mediator (
), mediator (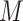 ), and outcome (
), and outcome (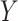 ). The fact that path
). The fact that path 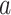 coefficients are an order of magnitude larger than path
coefficients are an order of magnitude larger than path 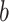 is solely related to differences in scaling and does not relate to their relevance. Please note that mixing of signals from distinct neural populations within fMRI voxels is common in similar types of analysis such as ICA and manifests itself in different weight patterns across PDMs, for example, for left S1.
is solely related to differences in scaling and does not relate to their relevance. Please note that mixing of signals from distinct neural populations within fMRI voxels is common in similar types of analysis such as ICA and manifests itself in different weight patterns across PDMs, for example, for left S1.
PDM 1 has both positive path 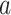 and
and 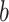 coefficients. Brain regions with positive weights (representing positive mediators, type 1 with positive paths
coefficients. Brain regions with positive weights (representing positive mediators, type 1 with positive paths 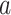 and
and 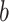 ) are shown in warm colors in Figure 2. These include brain regions commonly associated with pain processing, such as the dorsal posterior and midinsula, S1, S2, MCC, and the PAG (Fig. 2). Significant voxels in MCC stretch into the supplementary motor area (SMA), dorsal of the cingulate sulcus. In addition, PDM 1 contains negative, type (2), mediators, including the medial prefrontal cortex (mPFC) and left S1/M1. The negative weights indicate that these regions show less activation with increasing temperatures and lower regional activation is related to higher pain ratings. Such relationships are to be expected for brain regions whose function is inhibited by nociceptive input or that are deactivated with increased pain-related processing.
) are shown in warm colors in Figure 2. These include brain regions commonly associated with pain processing, such as the dorsal posterior and midinsula, S1, S2, MCC, and the PAG (Fig. 2). Significant voxels in MCC stretch into the supplementary motor area (SMA), dorsal of the cingulate sulcus. In addition, PDM 1 contains negative, type (2), mediators, including the medial prefrontal cortex (mPFC) and left S1/M1. The negative weights indicate that these regions show less activation with increasing temperatures and lower regional activation is related to higher pain ratings. Such relationships are to be expected for brain regions whose function is inhibited by nociceptive input or that are deactivated with increased pain-related processing.
Brain regions positively mediating the relationship between temperature and pain rating (type 1) in other PDMs are S1, M1, superior frontal gyrus (SFG), frontotemporal operculum, temporal poles, temporal operculum, ventral insula, pons, and the cerebellum (Fig. 2; yellow regions in, e.g., PDMs 1, 2, 4, and 6 in particular). These positive mediators include regions, like the temporal regions, that are traditionally not considered to be pain-processing regions. Brain regions acting as negative mediators (type 2) in other PDMs include medial orbitofrontal cortex (mOFC), dorsomedial prefrontal cortex (dmPFC), superior parietal lobule (SPL), retrosplenial cortex (RSC), precuneus, and cuneus (blue regions in PDMs 1, 2, 4, and 6). Those regions belong to systems that are deactivated consequent to pain or nociception (e.g., systems mediating competing functions).
A more complex function is indicated by positive path 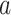 coefficients, but negative path
coefficients, but negative path 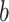 coefficients (types 3 and 4, PDMs 3, 5, 7, and 9). In type (3), regions with positive voxel weights show a positive relationship with temperature, that is, higher temperatures lead to more activity. However, the negative path
coefficients (types 3 and 4, PDMs 3, 5, 7, and 9). In type (3), regions with positive voxel weights show a positive relationship with temperature, that is, higher temperatures lead to more activity. However, the negative path 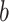 indicates that these regions are negatively related to pain ratings controlling for temperature, that is, more activity is related to lower pain ratings. Regions with such a profile fit a pain-inhibitory role, as they are activated by painful stimulation but serve to dampen pain—a negative feedback loop. Parts of the mOFC/vmPFC, the cerebellum, precuneus, S1, temporal–parietal junction, and the left dlPFC fit this pain-inhibitory profile (see yellow areas in PDMs 3, 5, and 7 in particular).
indicates that these regions are negatively related to pain ratings controlling for temperature, that is, more activity is related to lower pain ratings. Regions with such a profile fit a pain-inhibitory role, as they are activated by painful stimulation but serve to dampen pain—a negative feedback loop. Parts of the mOFC/vmPFC, the cerebellum, precuneus, S1, temporal–parietal junction, and the left dlPFC fit this pain-inhibitory profile (see yellow areas in PDMs 3, 5, and 7 in particular).
A final set of regions shows a negative relationship with temperature (positive path 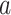 , but negative weights; blue in PDMs 3, 5, and 7) and a positive relationship with pain ratings, controlling for temperature (negative voxel weights and negative path
, but negative weights; blue in PDMs 3, 5, and 7) and a positive relationship with pain ratings, controlling for temperature (negative voxel weights and negative path 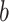 resulting in a net positive relationship; type 4). Such regions show stimulus intensity-dependent deactivation, with stronger deactivation mediating decreased pain, consistent with regulatory negative feedback mechanisms. Regions with this profile include parts of the mOFC, the parahippocampal gyrus, visual cortices, and the NAc. For example, NAc shows decreased activation for high temperatures, which may relate to punishment or negative reinforcement signals. At the same time, controlling for temperature, stronger NAc deactivation is related to lower pain ratings, potentially signaling reduced motivational relevance.
resulting in a net positive relationship; type 4). Such regions show stimulus intensity-dependent deactivation, with stronger deactivation mediating decreased pain, consistent with regulatory negative feedback mechanisms. Regions with this profile include parts of the mOFC, the parahippocampal gyrus, visual cortices, and the NAc. For example, NAc shows decreased activation for high temperatures, which may relate to punishment or negative reinforcement signals. At the same time, controlling for temperature, stronger NAc deactivation is related to lower pain ratings, potentially signaling reduced motivational relevance.
In the individual PDMs, each voxel is assigned a weight value—as in ICA, a voxel can thus participate in multiple components or “networks,” potentially revealing multiple functional roles of a voxel. Some regions participated in multiple PDMs in this fashion—most notably, mOFC appears to play roles as types (2–4) mediator in PDMs 2, 5, and 7. This may reveal a complex function of the mOFC in pain and a mixing together of signal from multiple distinguishable neural populations.
Combined PDM
The individual PDMs can be fused into a single, cPDM since the individual PDMs are orthogonal to each other. The weights are estimated from the training sample using a linear model with the individual PDMs as regressors and reported pain as the response. Summing the weighted PDMs results in a cPDM map (see Fig. 1D and section Methods). In doing so, we lose information about multiple functional roles played by each voxel or region, but we obtain a single overall characterization of each voxel and a map that can be applied as a predictive model. Voxel weights may be both positive and negative in different PDMs, because voxels may include neural ensembles participating in different distributed circuits related to either more or less pain. Thus, the individual PDMs represent a decomposition of voxels’ activity into different distributed components, while the cPDM reflects each voxel’s net contribution (controlling for other voxels). Computing and analyzing the cPDM can thus help to clarify overall relationships between regional activity and the predictor and outcome variables.
Within the cPDM, individually significant clusters of positive mediators included S2, MCC, SMA, PAG, insula (including anterior and dorsal–posterior parts), and the medial thalamus (Figs 1D and 2). Negative mediators (stimulus-induced deactivations mediating increased pain) included mPFC, SPL, S1, and M1.
To further characterize the weights of the cPDM in nociception- and pain-related regions of interest, we identified 24 distinct anatomical regions that encode pain in human and animal literature and examined the cPDM weights in each of these regions (Fig. 1D, right and Fig. 3). The regions were divided according to the thalamic atlas of Morel et al. (1997) and cortical atlas of Glasser et al. (2016). Positive weights were found in elements of the spinothalamic tract (bilateral VPL thalamus and dorsal posterior insula [dpINS] and also S1), spino-parabrachial tract (bilateral PBN and amygdala), spinoreticular tract (RVM and PAG), spinohypothalamic tract (posterior hypothalamus), and spinolimbic tract (mediodorsal [MD] and intralaminar [IL] nuclei of the thalamus and aMCC). Many of these subcortical regions have not been consistently identified in human studies, but are crucial mediators of pain in animal models. These results do not tell us that the activity in question is due to direct spinal input, but they robustly identify a set of targets in areas containing known pathways. Activity was bilateral in most regions, though the amygdala and spinothalamic regions (S1, S2, and dpINS) showed right-hemisphere dominance (most studies involved left-sided stimulation).
Figure 3 shows the pattern energy (root-mean-square weights per cubic cm of tissue) for positive and negative weights in yellow and blue, respectively. It shows that weights are mixed in the hypothalamus, RVM, amygdala, and S1, suggesting that the local pattern weights in these regions are particularly important, as they make the pattern response different from the average across the anatomic region. Figure 3B shows the pattern energy for all subdivisions of the thalamus, not only those related to nociception. The pattern energy is high (large wedge area) and weights are fairly uniformly positive (red color) in thalamic nuclei known to have nociceptive inputs: VPL, VPM, IL, and MD nuclei and the habenula (Hb). Roles for all of these in pain are well documented (Willis and Westlund 1997; Shelton et al. 2012). Pattern energy is low and with mixed signs on weights (purple color) in other sensory nuclei, including lateral/medial geniculate and pulvinar nuclei, along with other nuclei. Though a perfect match between any anatomical atlas and functional imaging data cannot be guaranteed, these findings suggest that the distribution of weights even across the relatively small volume of the thalamus is meaningful. They also confirm specific nuclei known mainly from animal and invasive human studies as pain-related in human fMRI data and suggest new potential pain-related nuclei to be confirmed and further characterized, such as ventrolateral (VL), ventromedial (VM), and anterior medial (AM) nuclei.
Examining the cPDM weights in established resting-state cortical networks also yielded a selective profile across networks, with weights concentrated in a few networks (Fig. 4). cPDM weights were high and relatively uniformly positive (red wedges in Fig. 4) in “somatomotor B,” “ventral attention A,” and “control C” networks. (We adopt these names by convention only, and do not suggest that the networks’ functions map onto these labels.) The first two broadly match previous analyses of nociceptive activity, though they provide additional information on mapping to subnetworks. The latter is more surprising, as it includes regions that are not typically nociceptive such as precuneus and parts of posterior cingulate. “Somatomotor” and “ventral attention” weights were left-lateralized, and “somatomotor” most strongly so. “Control C” weights were right-lateralized.
Clustering PDMs into Functional Networks
While the previous analyses examined the relationship of cPDM weights with independently defined anatomical regions and functional brain networks, in the next step we analyzed the spatial clustering arising from the individual PDMs themselves. The PDMs provide a dimensional view of coherent, distributed processes, with each PDM a distinct dimension; clustering the PDMs can reveal the network structure of the interregional relationships. To do this, we used an iterative clustering procedure to group regions based on interregional correlations in stimulus-evoked responses across trials without considering stimulation temperatures or pain ratings (Kober et al. 2008; Atlas et al. 2014). The cluster analysis of single-trial activity from significant voxels within 10 PDMs revealed 26 functional regions organized into five different functional networks (Fig. 5A,B). Though somewhat limited, for exploratory purposes a functional description of these networks was determined by computing the similarity of each network with feature maps generated by the meta-analytic tools on neurosynth.org (Yarkoni et al. 2011). The top 10 features for each network are shown in Table 1. Network names were chosen based on the functional associations with neurosynth.org terms. For example, the top three feature associations for network 1 were somatosensory, motor, and stimulation. Based on these associations, we labeled network 1 as “sensorimotor network.”
Figure 5.
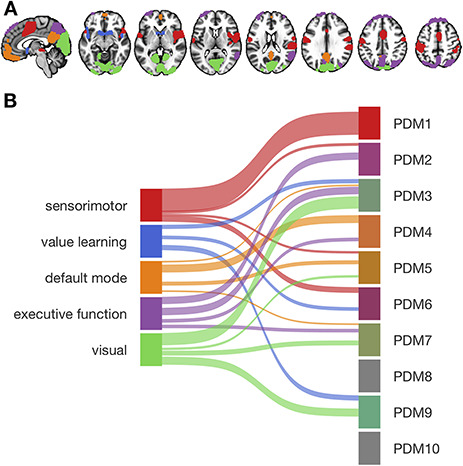
Functional networks mediating pain processing. (A) Five functional networks based on the clustering of brain activity in significant voxels from the PDM analysis. Labels for colors are shown in (B). (B) Associations between functional networks and PDMs. Ribbon width represents Dice coefficient similarity between networks and PDMs.
Table 1.
Neurosynth.org network associations
| Sensorimotor | Value learning | Default mode | Executive function | Visual | |||||
|---|---|---|---|---|---|---|---|---|---|
| r | Features | r | Features | r | Features | r | Features | r | Features |
| 0.369 | Somatosensory | 0.313 | Reward | 0.207 | Self-referential | 0.139 | Mental | 0.223 | Visual |
| 0.304 | Motor | 0.255 | Money | 0.202 | Person | 0.124 | Intention | 0.152 | Eye |
| 0.301 | Stimulation | 0.252 | Anticipation | 0.201 | Self | 0.117 | Stories | 0.141 | Eyes |
| 0.272 | Sensorimotor | 0.252 | Rewards | 0.197 | Default | 0.115 | Attention | 0.137 | Color |
| 0.266 | Muscle | 0.251 | Incentive | 0.176 | Autobiographical | 0.115 | Visuospatial | 0.126 | Shape |
| 0.257 | Sensory | 0.240 | Monetary | 0.157 | Resting state | 0.114 | Story | 0.108 | Shapes |
| 0.256 | Pain | 0.236 | Outcome | 0.149 | Social | 0.108 | Reasoning | 0.105 | Spatial |
| 0.245 | Movements | 0.196 | Outcomes | 0.149 | Mentalizing | 0.107 | Default | 0.102 | Development |
| 0.245 | Production | 0.185 | Dopamine | 0.148 | Personal | 0.106 | Calculation | 0.097 | Distractor |
| 0.240 | Painful | 0.179 | Reinforcement | 0.135 | Thought | 0.106 | Retrieval | 0.097 | Target |
Note: Top 10 features from neurosynth.org showing the highest Pearson’s correlation (r) with each network.
Network 1 (“sensorimotor”) included somatosensory regions like dpIns, midinsula, S2, and S1 but also the PAG, MCC, SMA, M1, and cerebellum. The second network (“value learning”) included the NAc, ventral anterior insula, frontal operculum, and temporal poles. Network 3 consisted of regions that are part of the default mode network (DMN), including mPFC, mOFC, and RSC. The fourth network (“executive function”) included precuneus, inferior parietal lobule (IPL), SPL, dorsal lateral occipital cortex (dLOC), temporal–parietal junction (TPJ), SFG, and dlPFC. Finally, network 5 (“visual”) included mostly occipital, visual areas and parts of the parahippocampal gyrus. The variety of functions ascribed to the five networks mediating pain indicate that pain processing involves multiple, distinct brain networks in addition to somatosensory systems.
We next investigated associations between functional networks and individual PDMs by computing pairwise Dice similarity coefficients across voxels, estimating the spatial similarity of the PDMs and network maps (Fig. 5B). PDM 1 (type 1/2 mediators) had the greatest overall similarity with any single network, namely, with the sensorimotor network (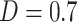 ). No other network was substantially associated with PDM 1 (all
). No other network was substantially associated with PDM 1 (all 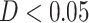 ). PDMs 2, 5, and 6 are also associated to the sensorimotor network (PDMs 2 and 6 are type 1/2 mediators). The value learning network was related to PDMs 3, 6, and 9, with the highest similarity to PDM 9 (
). PDMs 2, 5, and 6 are also associated to the sensorimotor network (PDMs 2 and 6 are type 1/2 mediators). The value learning network was related to PDMs 3, 6, and 9, with the highest similarity to PDM 9 (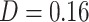 )—thus, mainly components with type (3)/(4) mediation (PDMs 3 and 9). Similarity between the DMN and PDM 4 was highest (
)—thus, mainly components with type (3)/(4) mediation (PDMs 3 and 9). Similarity between the DMN and PDM 4 was highest (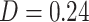 , mainly type 2 mediators). Parts of the DMN also overlapped with PDMs 3, 5, and 7. The executive function network was associated with PDM 2 (
, mainly type 2 mediators). Parts of the DMN also overlapped with PDMs 3, 5, and 7. The executive function network was associated with PDM 2 (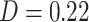 ) and PDM 3 (
) and PDM 3 (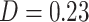 ) and, to a lesser degree, with PDMs 4 and 7. Finally, the visual network was related to PDM 3 (
) and, to a lesser degree, with PDMs 4 and 7. Finally, the visual network was related to PDM 3 (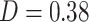 ) and to a lesser degree to PDMs 5, 7, and 9. The overall similarity pattern between functional networks and PDMs shows that in contrast to PDM1, few of the remaining PDMs are dominated by a single network. More often PDMs were comprised of a mix of two or three networks that together act as a pain mediator, reflecting the complexity of the transformation from nociception into pain experience.
) and to a lesser degree to PDMs 5, 7, and 9. The overall similarity pattern between functional networks and PDMs shows that in contrast to PDM1, few of the remaining PDMs are dominated by a single network. More often PDMs were comprised of a mix of two or three networks that together act as a pain mediator, reflecting the complexity of the transformation from nociception into pain experience.
Validation on an Independent Cohort
Although we estimated PDMs on a large and diverse data set, there is a risk that the PDMs may overfit noise inherent in the training data, potentially preventing generalization to other data sets. We thus applied the PDMs to an independent test data set, without re-estimating any model parameters. The resulting vectors of potential mediators (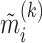 ) were then entered into standard multilevel mediation models. If the PDMs generalize to the new data, the indirect
) were then entered into standard multilevel mediation models. If the PDMs generalize to the new data, the indirect 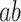 effects should be significant on the data test.
effects should be significant on the data test.
Applying the PDMs to independent pain data test (N = 75, an independent community sample cohort of mixed races and sex) revealed significant paths 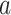 and
and 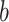 for all 10 PDMs and the cPDM (Fig. 6A). The indirect path
for all 10 PDMs and the cPDM (Fig. 6A). The indirect path 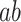 was also significant for the cPDM and all individual PDMs, suggesting that all PDMs are reliably related to pain and generalize across cohorts. The magnitude of the indirect effects (path
was also significant for the cPDM and all individual PDMs, suggesting that all PDMs are reliably related to pain and generalize across cohorts. The magnitude of the indirect effects (path  ) is monotonically decreasing for the training data (Fig. 1B). On the data test, indirect path coefficients were not strictly monotonically decreasing from PDM 1 to PDM 10 (Figs 6A and S2), indicating some variability of the PDM order across data sets, as expected. The cPDM and the first two individual PDMs had the strongest effect in both data sets, suggesting that they capture the most important brain activity for pain across data sets. Figure 6C shows the predicted pain from the cPDM plotted against the empirical pain ratings for pain training and data test.
) is monotonically decreasing for the training data (Fig. 1B). On the data test, indirect path coefficients were not strictly monotonically decreasing from PDM 1 to PDM 10 (Figs 6A and S2), indicating some variability of the PDM order across data sets, as expected. The cPDM and the first two individual PDMs had the strongest effect in both data sets, suggesting that they capture the most important brain activity for pain across data sets. Figure 6C shows the predicted pain from the cPDM plotted against the empirical pain ratings for pain training and data test.
Figure 6.
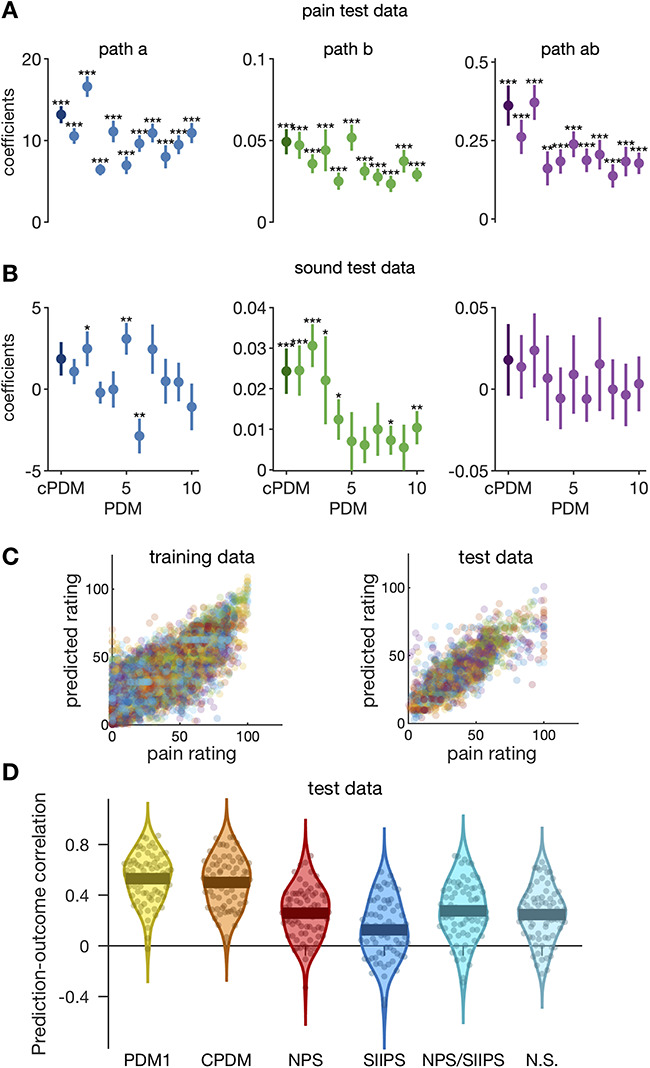
Validation on independent data (N = 75). (A) The cPDM (dark circle) and all 10 individual PDMs (lighter circles) are significant mediators for independent pain data test. (B) PDMs show specificity with respect to aversive sounds because no indirect effect is significant here. (C) Scatterplots of pain predicted from the cPDM against empirical pain ratings for training (left) and test (right) pain data. Individual trials from all subjects are shown. Colors indicate different subjects. (D) The prediction–outcome correlations between reported pain and pattern responses for the first PDM (PDM1), the cPDM, the NPS, the stimulus intensity-independent pain signature 1 (SIIPS1), the combination of NPS and SIIPS1, and the Neurosynth reverse inference map for the term “pain.”
To further corroborate the generalizability and robustness of the PDMs, we also estimated 10 PDMs on the original test data set (study 8) and cross-validated the new PDMs on the original training data set (studies 1–7). The results were similar to the main results presented here. Six out of 10 indirect paths were significant when PDM estimation was done on the smaller sample. The indirect  path coefficients for the first four PDMs were highest when applying the new PDMs to the original training data (Fig. S3). Generalization thus does not depend strongly on the choice of the training data.
path coefficients for the first four PDMs were highest when applying the new PDMs to the original training data (Fig. S3). Generalization thus does not depend strongly on the choice of the training data.
In order to test whether PDMs are mediators specifically for somatic pain, we also applied the original PDMs to other, nonpainful aversive stimuli in study 8—physically (knife on plate) and emotionally (screaming, crying, etc.) aversive sounds with three predefined intensity levels of each stimulus type. Study 8 was designed to test specificity versus generalizability to aversive sounds and match duration and approximate aversiveness ratings based on pilot studies; trials were randomly intermixed with heat pain trials. Application of the original PDMs on the sound data revealed no significant indirect effects (Figs 6B and S4) and only nine significant paths  or
or  in total. Thus, pain-derived PDMs do not mediate the relationship between sound intensity and intensity ratings for either type of sound. However, they are not perfectly selective as the expression of some PDMs correlates positively with ratings of sound unpleasantness (path
in total. Thus, pain-derived PDMs do not mediate the relationship between sound intensity and intensity ratings for either type of sound. However, they are not perfectly selective as the expression of some PDMs correlates positively with ratings of sound unpleasantness (path 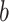 ). In summary, these results nevertheless indicate some degree of specificity to somatic pain versus sound.
). In summary, these results nevertheless indicate some degree of specificity to somatic pain versus sound.
Comparison with Other Multivariate Models
Previous studies have investigated the direct relationship between brain responses and pain reports, both using univariate (Coghill et al. 1999; Bornhövd et al. 2002; Ploner et al. 2010) and multivariate approaches (Marquand et al. 2010; Brodersen et al. 2012; Wager et al. 2013; Geuter et al. 2014; Woo et al. 2017). One study trained a multivariate pattern, termed the NPS, which predicts pain reports from brain activity that can be easily applied to new data sets (Wager et al. 2013). In contrast to the present approach, the estimation of the NPS did not account for temperature–brain relationships; its goal was rather to predict pain intensity without demonstrating mediation. In addition, we compared it to the stimulus intensity-independent pain signature (SIIPS1), which was trained to predict pain ratings after removing linear effects of stimulus intensity on brain activity and ratings (Woo et al. 2017). Additionally, the combination of NPS and SIIPS1 as well as the Neurosynth reverse inference map for the term “pain” was compared.
To examine relationships among the PDM models and other established models, we compared prediction–outcome correlations (see Fig. 6D) Correlations were calculated across individual differences in response values for each model. The first PDM performed best (mean r = 0.53), followed by the cPDM (mean r = 0.50). The NPS, the combination of NPS and SIIPS1, and the Neurosynth reverse inference map performed roughly equivalently (mean r = 0.26, 0.28, and 0.25, respectively). Finally, SIIPS1 performed the worst (mean r = 0.13). Prediction performance between PDM1 and cPDM did not differ significantly, while both PDM-based models significantly outperformed the remaining models (qFDR < 0.05, all P < 0.2e−11, all t(74) > 8.4).
The cPDM was highly correlated with PDM1 (r = 0.95), but only moderately correlated with NPS (r = 0.64) and Neurosynth (r = 0.66). The NPS, by contrast, was more strongly correlated with Neurosynth (r = 0.82) than the PDM models. The SIIPS1 pattern was distinct, and essentially uncorrelated with either the PDM models, NPS, or the Neurosynth (r ≤ 0.15). This is expected, as the SIIPS was designed to be independent of stimulus intensity.
Comparison with Univariate Mediation Analysis
In contrast to the present multivariate PDM approach, mass-univariate mediation analyses of fMRI data estimate independent mediation models for each voxel (Wager et al. 2008; Atlas et al. 2014). The intersection of voxels with significant paths 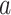 ,
,  , and
, and 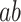 is then interpreted as a set of mediating brain regions. In order to compare the novel high-dimensional PDM approach to the univariate mediation analysis, we first computed a mass-univariate mediation analysis on the training data set (studies 1–7).
is then interpreted as a set of mediating brain regions. In order to compare the novel high-dimensional PDM approach to the univariate mediation analysis, we first computed a mass-univariate mediation analysis on the training data set (studies 1–7).
The univariate analysis identified the MCC, cerebellum, posterior and midinsula, S2, and S1 as brain mediators defined as the intersection of the coefficient maps for paths 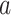 ,
, 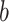 , and
, and 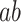 at FDR
at FDR 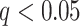 (Fig. 7). Comparing these results to the cPDM revealed both similarities and some notable differences (Figs 2 and 7). Both maps include somatosensory regions in aMCC, insula, and S2, as well as the cerebellum. Additional regions with positive weights in the cPDM included the thalamus, PAG, and other midbrain regions like the PBN not included in the univariate model. Furthermore, negative contributions in SPL and S1 were not identified in the univariate model. Such results are expected if some brain regions make detectable contributions only after controlling for the influences of other brain regions; this is an advantage of multivariate predictive approaches to neuroimaging analysis.
(Fig. 7). Comparing these results to the cPDM revealed both similarities and some notable differences (Figs 2 and 7). Both maps include somatosensory regions in aMCC, insula, and S2, as well as the cerebellum. Additional regions with positive weights in the cPDM included the thalamus, PAG, and other midbrain regions like the PBN not included in the univariate model. Furthermore, negative contributions in SPL and S1 were not identified in the univariate model. Such results are expected if some brain regions make detectable contributions only after controlling for the influences of other brain regions; this is an advantage of multivariate predictive approaches to neuroimaging analysis.
Figure 7.
Comparison with univariate mediation analysis. Top three panels show individually significant voxels for paths  (blue),
(blue),  (green), and
(green), and 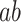 (purple) from a univariate mediation analysis at FDR
(purple) from a univariate mediation analysis at FDR 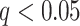 . Panel 4 shows voxels mediating the relationship between temperature and pain, that is, the overlap between the three paths (red).
. Panel 4 shows voxels mediating the relationship between temperature and pain, that is, the overlap between the three paths (red).
Discussion
In brief, our analyses identified brain mediators of pain that extend substantially beyond the boundaries of traditional “pain matrix” regions, including prefrontal and midbrain regions previously thought to play an “extranociceptive” or modulatory role (Kucyi et al. 2012; Geuter et al. 2013; Seminowicz and Moayedi 2017). Integrated activity across these pathways, as reflected in the cPDM, predict human pain intensity more accurately than previous pain predictive models.
Brain regions primarily associated with motivational, learning, or executive functions have been considered to modulate activity in a pain-processing brain system. The involvement of those brain regions in pain processing has been shown in a multitude of studies (e.g., Bushnell et al. 2013; Seminowicz and Moayedi 2017). Here, we show that many of these regions, including NAc, dlPFC, mPFC, and mOFC, are formal mediators of the stimulus–pain relationship. Their activity is directly related to stimulus intensity and at the same time to pain when controlling for stimulus intensity. The broad range of psychological functions associated with the regions serving as formal pain mediators is in line with the idea that brain regions related to motivational, learning, and executive functions are much more directly involved in the generation of pain. Their mediating role suggests that they are not necessarily external modulators to a distinct pain system but that primarily non–pain-processing brain regions play more a direct role in generating the pain experience, further blurring the boundaries of a so-called pain matrix.
One of the most prominent functions of pain is its motivational drive since it is associated with tissue damage (Navratilova and Porreca 2014; Geuter et al. 2016). Learning about painful stimuli is important to learn to minimize future harm. Pain stimulation relates to activity in the NAc, a brain region associated with motivational learning (Becerra et al. 2013; Woo et al. 2015). In line with the NAc’s role in pain in humans (Baliki et al. 2010, 2012) and animal models (Chang et al. 2014; Navratilova and Porreca 2014; Schwartz et al. 2014; Ren et al. 2016), NAc also acts as a formal mediator between nociceptive stimuli and pain. Furthermore, we show that NAc function for pain is based on opposing relationships of NAc activity with stimulus intensity (negative) and pain (positive)—NAc shows stimulus intensity-dependent deactivation, with stronger deactivation mediating decreased pain, consistent with regulatory negative feedback mechanisms. The NAc might exert its control in this feedback loop indirectly via its connections with the hypothalamus or mPFC as indicated by studies in humans and animals (Baliki et al. 2012; Schwartz et al. 2014; Lee et al. 2015; Woo et al. 2015). However, the exact contribution of the NAc to pain perception might rely on more complex temporal dynamics that cannot be resolved in the current data set and are still a matter of debate (Baliki et al. 2010; Becerra et al. 2013) as is its role in aversive learning more generally (Roy et al. 2014; Matsumoto et al. 2016).
Notably, another novel feature of the present cPDM map is that it contains positive weights in the bilateral PBN and specific parts of the amygdala, as well as RVM and PAG. Though definitive localization to these nuclei is difficult with any human method, activation is consistent with their locations in atlases and previous studies, and identifying them robustly in humans could provide an important step forward in the ability to study both bottom-up and top-down effects on crucial nociceptive and pain-modulatory pathways.
The PAG and RVM receive nociceptive afferents and form a major descending bulbospinal tract that controls the balance of descending pain-inhibitory and facilitatory projections to the spinal dorsal horn (Fields 2004; De Felice et al. 2011; Wager and Atlas 2015; Geuter et al. 2017b). Imaging studies have identified PAG and RVM activation during both evoked pain and pain-modulatory conditions like placebo analgesia (Tracey et al. 2002; Eippert et al. 2009a; Tinnermann et al. 2017). But these regions have not, to our knowledge, been identified as mediators of human stimulus–pain relationships.
The PBN has rarely been reported in neuroimaging studies but is an emerging target of great importance in representing danger signals related to pain and other bodily inputs. The PBN is a major center for pain and other forms of interoception and chemical sensation, including taste, itch, dyspnea, and vagally mediated immune surveillance and sickness behavior (Goehler et al. 2000; Kelley et al. 2003). A major pathway composed of CGRP neurons projects from the PBN to the central nucleus of the amygdala. This pathway is activated in response to danger signals across multiple sensory modalities, including visceral and cutaneous pain (chemical, mechanical, thermal, and electrical) and itch, across ascending trigeminal, spinal, and vagal sensory pathways (Han et al. 2015; Campos et al. 2018). It is crucial for representing danger signals that produce avoidance behavior, including joint control of learned pain avoidance and food intake, which is strongly inhibited by its activation (Han et al. 2015; Sato et al. 2015; Campos et al. 2018). PBN also plays a role in pain regulation by sending non-CGRP projections to the RVM (Roeder et al. 2016). PBN stimulation in humans can reduce chronic pain (Katayama et al. 1985). FMRI activation consistent with human PBN has been found to respond to noxious stimulation, correlate with human parasympathetic activity (Napadow et al. 2008), and respond to vagal stimulation (Frangos et al. 2015) and acupuncture (Napadow et al. 2009). Our results suggest it may be possible to measure activity in human PBN–central amygdala and other rubrospinal pathways.
Among regions commonly associated with pain in neuroimaging studies, including the medial thalamus, PAG, S2, insula, MCC, SMA, and S1 (Dum et al. 2009), activity increased due to increasing temperatures, and higher activity was related to stronger pain, controlling for temperature. This set of pain-associated regions (Apkarian et al. 2005; Bushnell et al. 2013; Duerden and Albanese 2013; Jensen et al. 2016) was complemented by anterior temporal regions and the cerebellum, which share the same functional response profile. A positive relationship with both temperature and pain rating is in-line with a traditional, feed-forward encoding view of nociception (Bushnell et al. 2013; Atlas et al. 2014; cf. Geuter et al. 2017a).
By contrast, the mPFC, SPL, RSC, precuneus, and parts of S1 and M1 were negatively related to both temperature and pain. The mPFC, RSC, and precuneus are part of the DMN, which has been associated with mind-wandering and internal thoughts (Andrews-Hanna et al. 2010; Kucyi and Davis 2015). The negative mediating role of the DMN regions could be related to the disruption of ongoing thought processes by the painful stimulation or attentional refocusing from internal to external sensations. The observation that some regions, like S1, participate in both, positive and negative, mediation relationships may indicate the mixing of signals from distinct neuronal populations within single fMRI voxels. Such mixing of activity patterns is also observed across components from ICA.
In addition, there are multiple sources of endogenous variation in drivers of pain beyond stimulus intensity, including attention, arousal, and endogenous variation in ascending spinal afferents due to processing within the spinal cord (Eippert et al. 2009b; Geuter and Büchel 2013; Tinnermann et al. 2017). Because each study included a different psychological manipulation to modulate pain, our approach will only identify brain mediators common to all studies. Differences across studies thus increase the robustness of the identified brain mediators but also reduce the amount of variance that will be explained in each single study. Additional factors introducing variance across studies include context effects (Leknes et al. 2013), temporal effects within and across trials (Jepma et al. 2014), and prestimulus fluctuations in brain activity (Ploner et al. 2010). Variation in pain related to variability of afferent input, for example, endogenous trial-to-trial variation in spinal cord processing, will be captured in path b in the mediation model. Other endogenous sources of variation unrelated to afferent input may be captured in the direct effect (path 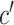 ) and will not be reflected in a brain mediation model that seeks to connect noxious stimulus intensity with pain. Similarly, potential nonlinear relationships between stimulus intensity and pain or differences across participants might not be adequately represented by linear brain mediators. However, using linear models instead of nonlinear models offers better interpretability.
) and will not be reflected in a brain mediation model that seeks to connect noxious stimulus intensity with pain. Similarly, potential nonlinear relationships between stimulus intensity and pain or differences across participants might not be adequately represented by linear brain mediators. However, using linear models instead of nonlinear models offers better interpretability.
In summary, the new high-dimensional mediation analysis revealed a comprehensive picture of brain responses underlying the complex, multifaceted pain experience. Several brain regions, such as the mPFC, thalamus, NAc, and PBN, are shown to directly and formally mediate stimulus–pain relationships. The functional diversity of the brain mediators observed here offers a better understanding of the brain responses underlying the complexity of the pain experience.
Funding
German Research Foundation (DFG) (GE 2774/1-1 to S.G.) and the National Institutes of Health (grant R01DA035484 to T.D.W., grant 2R01MH076136 to T.D.W., grant R01DA027794 to T.D.W., grant R01 EB016061 to M.A.L., grant R01 EB026549 to M.A.L., and grant P41 EB015909 to M.A.L.). Conflict of Interest: None declared.
Supplementary Material
References
- Andrews-Hanna JR, Reidler JS, Huang C, Buckner RL. 2010. Evidence for the default network’s role in spontaneous cognition. J Neurophysiol. 104:322–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apkarian AV, Bushnell MC, Treede R-D, Zubieta J-K. 2005. Human brain mechanisms of pain perception and regulation in health and disease. Eur J Pain. 9:463–484. [DOI] [PubMed] [Google Scholar]
- Atlas LY, Bolger N, Lindquist MA, Wager TD. 2010. Brain mediators of predictive cue effects on perceived pain. J Neurosci. 30:12964–12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlas LY, Lindquist MA, Bolger N, Wager TD. 2014. Brain mediators of the effects of noxious heat on pain. Pain. 155:1632–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliki MN, Geha PY, Apkarian AV. 2009. Parsing pain perception between nociceptive representation and magnitude estimation. J Neurophysiol. 101:875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliki MN, Geha PY, Fields HL, Apkarian AV. 2010. Predicting value of pain and analgesia: nucleus accumbens response to noxious stimuli changes in the presence of chronic pain. Neuron. 66:149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliki MN, Petre B, Torbey S, Herrmann KM, Huang L, Schnitzer TJ, Fields HL, Apkarian AV. 2012. Corticostriatal functional connectivity predicts transition to chronic back pain. Nat Neurosci. 15:1117–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra L, Navratilova E, Porreca F, Borsook D. 2013. Analogous responses in the nucleus accumbens and cingulate cortex to pain onset (aversion) and offset (relief) in rats and humans. J Neurophysiol. 110:1221–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingel U, Quante M, Knab R, Bromm B, Weiller C, Büchel C. 2002. Subcortical structures involved in pain processing: evidence from single-trial fMRI. Pain. 99:313–321. [DOI] [PubMed] [Google Scholar]
- Bornhövd K, Quante M, Glauche V, Bromm B, Weiller C, Büchel C. 2002. Painful stimuli evoke different stimulus–response functions in the amygdala, prefrontal, insula and somatosensory cortex: a single-trial fMRI study. Brain. 125:1326–1336. [DOI] [PubMed] [Google Scholar]
- Bradley MM, Lang PJ. 2007. The international affective digitized sounds (; IADS-2): affective ratings of sounds and instruction manual. Gainesville, FL: University of Florida, Tech Rep B-3. [Google Scholar]
- Brodersen KH, Wiech K, Lomakina EI, Lin C, Buhmann JM, Bingel U, Ploner M, Stephan KE, Tracey I. 2012. Decoding the perception of pain from fMRI using multivariate pattern analysis. Neuroimage. 63:1162–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks JCW, Davies W-E, Pickering AE. 2017. Resolving the brainstem contributions to attentional analgesia. J Neurosci. 37:2279–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büchel C, Bornhövd K, Quante M, Glauche V, Bromm B, Weiller C. 2002. Dissociable neural responses related to pain intensity, stimulus intensity, and stimulus awareness within the anterior cingulate cortex: a parametric single-trial laser functional magnetic resonance imaging study. J Neurosci. 22:970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell MC, Čeko M, Low LA. 2013. Cognitive and emotional control of pain and its disruption in chronic pain. Nat Rev Neurosci. 14:502–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Button KS, Ioannidis JPA, Mokrysz C, Nosek BA, Flint J, Robinson ESJ, Munafò MR. 2013. Power failure: why small sample size undermines the reliability of neuroscience. Nat Rev Neurosci. 14:365–376. [DOI] [PubMed] [Google Scholar]
- Caffo BS, Crainiceanu CM, Verduzco G, Joel S, Mostofsky SH, Bassett SS, Pekar JJ. 2010. Two-stage decompositions for the analysis of functional connectivity for fMRI with application to Alzheimer’s disease risk. Neuroimage. 51:1140–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos CA, Bowen AJ, Roman CW, Palmiter RD. 2018. Encoding of danger by parabrachial CGRP neurons. Nature. 555:617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P-C, Pollema-Mays SL, Centeno MV, Procissi D, Contini M, Baria AT, Martina M, Apkarian AV. 2014. Role of nucleus accumbens in neuropathic pain: linked multi-scale evidence in the rat transitioning to neuropathic pain. Pain. 155:1128–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chén OY, Crainiceanu C, Ogburn EL, Caffo BS, Wager TD, Lindquist MA. 2017. High-dimensional multivariate mediation with application to neuroimaging data. Biostatistics. 19:121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghill RC, Sang CN, Maisog JM, Iadarola MJ. 1999. Pain intensity processing within the human brain: a bilateral, distributed mechanism. J Neurophysiol. 82:1934–1943. [DOI] [PubMed] [Google Scholar]
- Craig AD, Chen K, Bandy D, Reiman EM. 2000. Thermosensory activation of insular cortex. Nat Neurosci. 3:184–190. [DOI] [PubMed] [Google Scholar]
- Crainiceanu CM, Caffo BS, Luo S, Zipunnikov VM, Punjabi NM. 2011. Population value decomposition, a framework for the analysis of image populations. J Am Stat Assoc. 106:775–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Felice M, Sanoja R, Wang R, Vera-Portocarrero L, Oyarzo J, King T, Ossipov MH, Vanderah TW, Lai J, Dussor GO et al. 2011. Engagement of descending inhibition from the rostral ventromedial medulla protects against chronic neuropathic pain. Pain. 152:2701–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerden EG, Albanese M-C. 2013. Localization of pain-related brain activation: a meta-analysis of neuroimaging data. Hum Brain Mapp. 34:109–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dum RP, Levinthal DJ, Strick PL. 2009. The spinothalamic system targets motor and sensory areas in the cerebral cortex of monkeys. J Neurosci. 29:14223–14235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eippert F, Bingel U, Schoell ED, Yacubian J, Klinger R, Lorenz J, Büchel C. 2009a. Activation of the opioidergic descending pain control system underlies placebo analgesia. Neuron. 63:533–543. [DOI] [PubMed] [Google Scholar]
- Eippert F, Finsterbusch J, Bingel U, Büchel C. 2009b. Direct evidence for spinal cord involvement in placebo analgesia. Science. 326:404. [DOI] [PubMed] [Google Scholar]
- Fairhurst M, Wiech K, Dunckley P, Tracey I. 2007. Anticipatory brainstem activity predicts neural processing of pain in humans. Pain. 128:101–110. [DOI] [PubMed] [Google Scholar]
- Fields HL. 2004. State-dependent opioid control of pain. Nat Rev Neurosci. 5:565–575. [DOI] [PubMed] [Google Scholar]
- Frangos E, Ellrich J, Komisaruk BR. 2015. Non-invasive access to the vagus nerve central projections via electrical stimulation of the external ear: fMRI evidence in humans. Brain Stimul. 8:624–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuter S, Boll S, Eippert F, Büchel C. 2017a. Functional dissociation of stimulus intensity encoding and predictive coding of pain in the insula. Elife. 6:e24770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuter S, Büchel C. 2013. Facilitation of pain in the human spinal cord by Nocebo treatment. J Neurosci. 33:13784–13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuter S, Cunningham JT, Wager TD. 2016. Disentangling opposing effects of motivational states on pain perception. Pain Rep. 1:e574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuter S, Eippert F, Hindi Attar C, Büchel C. 2013. Cortical and subcortical responses to high and low effective placebo treatments. Neuroimage. 67:227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuter S, Gamer M, Onat S, Büchel C. 2014. Parametric trial-by-trial prediction of pain by easily available physiological measures. Pain. 155:994–1001. [DOI] [PubMed] [Google Scholar]
- Geuter S, Koban L, Wager TD. 2017b. The cognitive neuroscience of placebo effects: concepts, predictions, and physiology. Annu Rev Neurosci. 40:167–188. [DOI] [PubMed] [Google Scholar]
- Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann CF, Jenkinson M et al. 2016. A multi-modal parcellation of human cerebral cortex. Nature. 536:171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehler LE, Gaykema RP, Hansen MK, Anderson K, Maier SF, Watkins LR. 2000. Vagal immune-to-brain communication: a visceral chemosensory pathway. Auton Neurosci. 85:49–59. [DOI] [PubMed] [Google Scholar]
- Han S, Soleiman MT, Soden ME, Zweifel LS, Palmiter RD. 2015. Elucidating an affective pain circuit that creates a threat memory. Cell. 162:363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haxby JV, Connolly AC, Guntupalli JS. 2014. Decoding neural representational spaces using multivariate pattern analysis. Annu Rev Neurosci. 37:435–456. [DOI] [PubMed] [Google Scholar]
- Horing B, Sprenger C, Büchel C. 2019. The parietal operculum preferentially encodes heat pain and not salience. PLoS Biol. 17:e3000205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Regenbogen C, Ohse MC, Frasnelli J, Freiherr J, Lundström JN. 2016. Brain activations during pain: a neuroimaging meta-analysis of patients with pain and healthy controls. Pain. 157:1279–1286. [DOI] [PubMed] [Google Scholar]
- Jepma M, Jones M, Wager TD. 2014. The dynamics of pain: evidence for simultaneous site-specific habituation and site-nonspecific sensitization in thermal pain. J Pain. 15:734–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y, Tsubokawa T, Hirayama T, Yamamoto T. 1985. Pain relief following stimulation of the pontomesencephalic parabrachial region in humans: brain sites for nonopiate-mediated pain control. Appl Neurophysiol. 48:195–200. [DOI] [PubMed] [Google Scholar]
- Kelley KW, Bluthé R-M, Dantzer R, Zhou J-H, Shen W-H, Johnson RW, Broussard SR. 2003. Cytokine-induced sickness behavior. Brain Behavior Immun. 17:112–118. [DOI] [PubMed] [Google Scholar]
- Keuken MC, Bazin P-L, Crown L, Hootsmans J, Laufer A, Müller-Axt C, Sier R, van der Putten EJ, Schäfer A, Turner R et al. 2014. Quantifying inter-individual anatomical variability in the subcortex using 7 T structural MRI. Neuroimage. 94:40–46. [DOI] [PubMed] [Google Scholar]
- Kober H, Barrett LF, Joseph J, Bliss-Moreau E, Lindquist K, Wager TD. 2008. Functional grouping and cortical–subcortical interactions in emotion: a meta-analysis of neuroimaging studies. Neuroimage. 42:998–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, McHaffie JG, Laurienti PJ, Coghill RC. 2003. The single-epoch fMRI design: validation of a simplified paradigm for the collection of subjective ratings. Neuroimage. 19:976–987. [DOI] [PubMed] [Google Scholar]
- Kriegeskorte N. 2011. Pattern-information analysis: from stimulus decoding to computational-model testing. Neuroimage. 56:411–421. [DOI] [PubMed] [Google Scholar]
- Kucyi A, Davis KD. 2015. The dynamic pain connectome. Trends Neurosci. 38:86–95. [DOI] [PubMed] [Google Scholar]
- Kucyi A, Hodaie M, Davis KD. 2012. Lateralization in intrinsic functional connectivity of the temporoparietal junction with salience- and attention-related brain networks. J Neurophysiol. 108:3382–3392. [DOI] [PubMed] [Google Scholar]
- Lee M, Manders TR, Eberle SE, Su C, D’amour J, Yang R, Lin HY, Deisseroth K, Froemke RC, Wang J. 2015. Activation of corticostriatal circuitry relieves chronic neuropathic pain. J Neurosci. 35:5247–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leknes S, Berna C, Lee MC, Snyder GD, Biele G, Tracey I. 2013. The importance of context: when relative relief renders pain pleasant. Pain. 154:402–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M, Su Q, Mouraux A, Iannetti GD. 2019. Spatial patterns of brain activity preferentially reflecting transient pain and stimulus intensity. Cereb Cortex. 29:2211–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist MA, Krishnan A, López-Solà M, Jepma M, Woo C-W, Koban L, Roy M, Atlas LY, Schmidt L, Chang LJ et al. 2017. Group-regularized individual prediction: theory and application to pain. NeuroImage. 145(Part B):274–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKinnon DP, Krull JL, Lockwood CM. 2000. Equivalence of the mediation, confounding and suppression effect. Prev Sci. 1:173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquand A, Howard M, Brammer M, Chu C, Coen S, Mourão-Miranda J. 2010. Quantitative prediction of subjective pain intensity from whole-brain fMRI data using Gaussian processes. Neuroimage. 49:2178–2189. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Tian J, Uchida N, Watabe-Uchida M. 2016. Midbrain dopamine neurons signal aversion in a reward-context-dependent manner. Elife. 5:e17328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel A, Magnin M, Jeanmonod D. 1997. Multiarchitectonic and stereotactic atlas of the human thalamus. J Comp Neurol. 387:588–630. [DOI] [PubMed] [Google Scholar]
- Mumford JA, Turner BO, Ashby FG, Poldrack RA. 2012. Deconvolving BOLD activation in event-related designs for multivoxel pattern classification analyses. Neuroimage. 59:2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napadow V, Dhond R, Conti G, Makris N, Brown EN, Barbieri R. 2008. Brain correlates of autonomic modulation: combining heart rate variability with fMRI. Neuroimage. 42:169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napadow V, Dhond R, Park K, Kim J, Makris N, Kwong KK, Harris RE, Purdon PL, Kettner N, Hui KKS. 2009. Time-variant fMRI activity in the brainstem and higher structures in response to acupuncture. Neuroimage. 47:289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navratilova E, Porreca F. 2014. Reward and motivation in pain and pain relief. Nat Neurosci. 17:1304–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyron R, Frot M, Schneider F, Garcia-Larrea L, Mertens P, Barral FG, Sindou M, Laurent B, Mauguière F. 2002. Role of Operculoinsular cortices in human pain processing: converging evidence from PET, fMRI, dipole modeling, and intracerebral recordings of evoked potentials. Neuroimage. 17:1336–1346. [DOI] [PubMed] [Google Scholar]
- Ploner M, Lee MC, Wiech K, Bingel U, Tracey I. 2010. Prestimulus functional connectivity determines pain perception in humans. Proc Natl Acad Sci USA. 107:355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouget A, Dayan P, Zemel R. 2000. Information processing with population codes. Nat Rev Neurosci. 1:125–132. [DOI] [PubMed] [Google Scholar]
- Ren W, Centeno MV, Berger S, Wu Y, Na X, Liu X, Kondapalli J, Apkarian AV, Martina M, Surmeier DJ. 2016. The indirect pathway of the nucleus accumbens shell amplifies neuropathic pain. Nat Neurosci. 19:220–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissman J, Greely HT, Wagner AD. 2010. Detecting individual memories through the neural decoding of memory states and past experience. Proc Natl Acad Sci USA. 107:9849–9854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder Z, Chen Q, Davis S, Carlson JD, Tupone D, Heinricher MM. 2016. Parabrachial complex links pain transmission to descending pain modulation. Pain. 157:2697–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy M, Shohamy D, Daw N, Jepma M, Wimmer GE, Wager TD. 2014. Representation of aversive prediction errors in the human periaqueductal gray. Nat Neurosci. 17:1607–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Ito M, Nagase M, Sugimura YK, Takahashi Y, Watabe AM, Kato F. 2015. The lateral parabrachial nucleus is actively involved in the acquisition of fear memory in mice. Mol Brain. 8:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Kong R, Gordon EM, Laumann TO, Zuo X-N, Holmes AJ, Eickhoff SB, Yeo BTT. 2018. Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cereb Cortex. 28:3095–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz N, Temkin P, Jurado S, Lim BK, Heifets BD, Polepalli JS, Malenka RC. 2014. Decreased motivation during chronic pain requires long-term depression in the nucleus accumbens. Science. 345:535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seminowicz DA, Moayedi M. 2017. The dorsolateral prefrontal cortex in acute and chronic pain. J Pain. 18:1027–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton L, Becerra L, Borsook D. 2012. Unmasking the mysteries of the habenula in pain and analgesia. Prog Neurobiol. 96:208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard RN. 1980. Multidimensional scaling, tree-fitting, and clustering. Science. 210:390–398. [DOI] [PubMed] [Google Scholar]
- Tinnermann A, Geuter S, Sprenger C, Finsterbusch J, Büchel C. 2017. Interactions between brain and spinal cord mediate value effects in nocebo hyperalgesia. Science. 358:105–108. [DOI] [PubMed] [Google Scholar]
- Tracey I, Ploghaus A, Gati JS, Clare S, Smith S, Menon RS, Matthews PM. 2002. Imaging attentional modulation of pain in the periaqueductal gray in humans. J Neurosci. 22:2748–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemure C, Slotnick BM, Bushnell MC. 2003. Effects of odors on pain perception: deciphering the roles of emotion and attention. Pain. 106:101–108. [DOI] [PubMed] [Google Scholar]
- Wager TD, Atlas LY. 2015. The neuroscience of placebo effects: connecting context, learning and health. Nat Rev Neurosci. 16:403–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager TD, Atlas LY, Lindquist MA, Roy M, Woo C-W, Kross E. 2013. An fMRI-based neurologic signature of physical pain. N Engl J Med. 368:1388–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager TD, Davidson ML, Hughes BL, Lindquist MA, Ochsner KN. 2008. Prefrontal-subcortical pathways mediating successful emotion regulation. Neuron. 59:1037–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager TD, Rilling JK, Smith EE, Sokolik A, Casey KL, Davidson RJ, Kosslyn SM, Rose RM, Cohen JD. 2004. Placebo-induced changes in FMRI in the anticipation and experience of pain. Science. 303:1162–1167. [DOI] [PubMed] [Google Scholar]
- Wager TD, Waugh CE, Lindquist M, Noll DC, Fredrickson BL, Taylor SF. 2009. Brain mediators of cardiovascular responses to social threat: part I: reciprocal dorsal and ventral sub-regions of the medial prefrontal cortex and heart-rate reactivity. NeuroImage, Brain Body Med. 47:821–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis WD, Westlund KN. 1997. Neuroanatomy of the pain system and of the pathways that modulate pain. J Clin Neurophysiol Pain. 14:2–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo C-W, Roy M, Buhle JT, Wager TD. 2015. Distinct brain systems mediate the effects of nociceptive input and self-regulation on pain. PLoS Biol. 13:e1002036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo C-W, Schmidt L, Krishnan A, Jepma M, Roy M, Lindquist MA, Atlas LY, Wager TD. 2017. Quantifying cerebral contributions to pain beyond nociception. Nat Commun. 8:14211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC, Wager TD. 2011. Large-scale automated synthesis of human functional neuroimaging data. Nat Methods. 8:665–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


