Abstract
As an emerging infectious disease, the clinical course and virological course of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection remain to be further investigated. In this case report, we described a case of SARS‐CoV‐2 infection with the clinical course for more than 2 months. This patient had recovered from pneumonia after treatment. The viral RNA of throat swabs became negative and the viral‐specific antibodies were produced during the recovery period. However, the viral RNA reappeared and additionally persisted in throat swabs for more than 40 days. In addition, the viral RNA was detected in multiple types of specimens with extremely high titers in the saliva. In conclusion, these findings indicate that SARS‐CoV‐2 can cause a long clinical course. The coexistence of viral RNA and viral‐specific antibodies may imply an immune evasion of SARS‐CoV‐2 from the host's immune system.
Keywords: coronavirus, saliva, SARS‐CoV‐2, virus shedding
Highlights
This case of SARS‐CoV‐2 infection exhibited several remarkable characteristics, including long clinical course, viral RNA relapse, and coexistence of viral RNA and viral specific antibodies during recovery period of the disease.
These findings may imply an immune evasion and chronic infection of SARAS‐CoV‐2 in some patients.
1. INTRODUCTION
The emergence and spread of the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) have become a global health concern. As the name of the virus, SARS‐CoV‐2 is presumed to cause an acute infection, the clinical spectrum encompasses asymptomatic infection, mild upper respiratory tract illness, and severe viral pneumonia with respiratory failure and even death. 1 , 2 , 3 However, as an emerging infectious disease, the clinical course and virological course of SARS‐CoV‐2 infection remain to be further investigated. In this study, we are reporting a case of SARS‐CoV‐2 infection with the clinical course lasted for more than 2 months.
1.1. Clinical and laboratory data of the subject
On 1 February 2020, a 44‐year‐old man was admitted to a hospital in Wuhan, with the presentation of fever, malaise, and fatigue for 11 days. The patient did not have a past medical history of immunodeficiency‐related diseases. His temperature was 37.1°C, oxygen saturation was 92% on room air, and respiratory rate was 26 breaths per minute—rough breathing sounds but without any signs of dry or wet rales. Laboratory examinations showed decreased lymphocytes (1.03 × 109/L), increased monocyte (1.03 × 109/L), and C‐reactive protein (61.5 mg/L) without any abnormalities in the liver, heart, and renal functions. As shown in Figure 1, the computed tomography (CT) scan showed that the infection infiltrated in the lower left lobe on 23 January and diffused throughout the lungs on 1 February. Assays for influenza viruses and a respiratory panel were negative, but a throat swab was positive for SARS‐CoV‐2 RNA (cycle threshold, C t = 33.2) on reverse‐transcription quantitative polymerase chain reaction (two pairs of primers targeting at regions of ORF1 and N) assays on 3 February.
Figure 1.
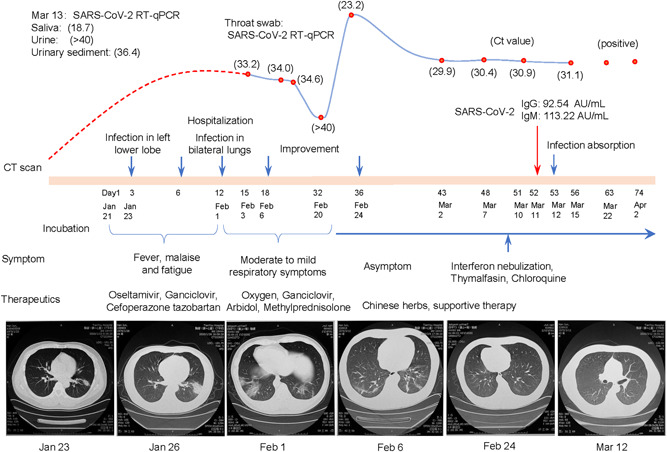
The virological course of SARS‐CoV‐2 infection in the patient. The curve presents the changes in throat swab viral RNA. To visually reflect the relative quantification of viral RNA, the cycle threshold (C t, red points) values showed on the curve were converted according to the equation of log2 (40 − x), x = C t value. The C t of 40 is the lower limit of detection on the RT‐qPCR assay. The red‐dash line describes putative changes in viral RNA before detection. The last two dictions of throat swab were reported as qualitative analyses (positive) in another hospital. The symptoms and main therapies are listed under the timeline. The time points of CT scan are labeled as blue arrows. CT, computed tomography; IgG, immunoglobulin G; IgM, immunoglobulin M; RT‐qPCR, reverse‐transcription quantitative polymerase chain reaction; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
After hospitalization, the patient received a low dose of methylprednisolone and oxygen therapies, as well as anti‐infective therapy including ganciclovir, arbidol, and moxifloxacin. His body temperature returned to normal and his symptoms resolved within 1 week. The CT scan revealed a significant improvement in pulmonary infection and the throat swab became negative (C t > 40) for SARS‐CoV‐2 RNA on 20 February (Figure 1). Then, he continued to receive consolidated treatment including traditional Chinese medicine, while methylprednisolone and oxygen therapies were removed. As preparation for discharge, the patient was retested for viral RNA.
However, a positive SARS‐CoV‐2 RNA reappeared in the throat swab on 23 February and remained high (C t, ~30) in the following four tests before 15 March. During this period, the patient remained asymptomatic and had no abnormal signs. His body temperature kept below 36.5°C and SpO2 remained above 97%. The chest CT scan showed the infection lesion had been absorbed and the lungs returned to normal on 12 March. In addition, the serological SARS‐CoV‐2 antibodies (immunoglobulin M and immunoglobulin G, targeting at S and N proteins) were detected on 11 March. To actively inhibit the virus replication, interferon nebulization, thymalfasin, and chloroquine diphosphate were sequentially initiated since 8 March, according to the Chinese guideline on the novel coronavirus pneumonia diagnosis and treatment (trial version 7). However, the viral RNA remained positive in throat swab, saliva, and urine sediment (it was negative in urine) on 13 March. Particularly, an extremely high level of viral RNA (C t = 18.7) was detected in saliva. The patient was transferred to another hospital for centralized management and treatment since 15 March. Positive throat swabs (without C t value) were reported on 22 March and 2 April.
2. DISCUSSION
Up to the end of data collection, the clinical course of this patient had lasted for more than 70 days. To the best of our knowledge, this is the longest reported duration of clinical course in the SARS‐CoV‐2 infection. Although we presented a solo case in this study, there are still about 3000 patients hospitalized in Wuhan city by 1 April, and many of them were infected at the early stage of the outbreak in December 2019. In addition, many cases of recrudescence after discharge have been reported in the latest news. These indicate that the infection can last for several months in a considerable number of patients.
During the early course of the infection, the viral RNA declined gradually and became undetectable. Although the undetectable viral RNA might be caused by the low viral load or bias in sample collection of throat swab, the dynamic changes in viral titers showed that the host immunity suppressed viral replication during this stage. However, the viral RNA reappeared in throat swab 3 days later. Logically, the 3 days of virus vanishment is too short to be an incubation period for a new infection, thus we deduced that the reappearance of the viral RNA was a recrudescence, rather than a new infection. The recrudescence persisted for more than 40 days, this is the longest persistence of recrudescence ever reported to our knowledge. 5 , 6 In addition, the virus was detected in the multiple types of specimens including throat swab, salivary, and urine sediment; even though the antibodies targeting at the S protein had been produced. All of these results showed that the immune system had responded to the infection but it was incompetent to clear the viruses.
The viral persistence is possibly caused by the immune evasion, viral mutation, and/or replication at immune‐privileged sites. 7 Previous studies demonstrated that the salivary gland was a target of coronavirus infection and was an immune‐privileged site of cytomegalovirus immune evasion and persistence. 8 , 9 It is notable that an extremely high load of viral RNA was detected in saliva. Therefore, we hypothesize that the salivary gland is likely an immune‐privileged site of SARS‐CoV‐2 persistence. The high‐load salivary virus may contaminate and spread to the respiratory tract and digestive tract. This may explain the positive throat swab and feces in the asymptomatic carriers and the patients recovered from pneumonia. 10 , 11 However, it is noteworthy that the infectious virus can be readily isolated from respiratory tract‐derived samples only early in the disease process, but not from stool, blood, and urine samples. 12
In conclusion, this case of SARS‐CoV‐2 infection exhibited several remarkable characteristics, including long clinical course, persistent viral RNA positivity during the recovery period, and the inability of viral antibody to clear the virus. These findings may provide some useful information about this disease. First, the high load of the salivary virus during the recovery period indicates that this patient could be potent but hidden infectious sources of droplets and close contact transmission. Hence, we propose that saliva might be included in the etiological examination for this population. Second, the production of antibodies is not linked to clearance of the virus in this patient, suggesting a potential immune evasion of the virus. This may challenge the efficacy of the vaccine in developing. Third, the long clinical course may indicate that some patients likely develop chronic SARS‐CoV‐2 infection. However, it should be noted that the use of methylprednisolone may prolong the clinical course in this case.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China under Grant 81670530; the Start‐up Foundation of Hangzhou Normal University under Grant 2018QDL035; and the Basic Science and Advanced Technology Foundation of Chongqing Science and Technology Commission under Grant cstc2016jcyjA0158.
Yang J‐R, Deng D‐T, Wu N, Yang B, Li H‐J, Pan X‐B. Persistent viral RNA positivity during the recovery period of a patient with SARS‐CoV‐2 infection. J Med Virol. 2020;92:1681–1683. 10.1002/jmv.25940
Jian‐Rong Yang and Dao‐Ting Deng contributed equally to this study.
REFERENCES
- 1. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus‐infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507‐513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID‐19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054‐1062. 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen Dabiao, Xu Wenxiong, Lei Ziying, et al. Recurrence of positive SARS‐CoV‐2 RNA in COVID‐19: a case report. Int J Infect Dis. 2020;93:297‐299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang T, Cui X, Zhao X, et al. Detectable SARS‐CoV‐2 viral RNA in feces of three children during recovery period of COVID‐19 pneumonia. J Med Virol. 2020. 10.1002/jmv.25795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138:30‐50. [DOI] [PubMed] [Google Scholar]
- 8. Liu L, Wei Q, Alvarez X, et al. Epithelial cells lining salivary gland ducts are early target cells of severe acute respiratory syndrome coronavirus infection in the upper respiratory tracts of rhesus macaques. J Virol. 2011;85:4025‐4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Campbell AE, Cavanaugh VJ, Slater JS. The salivary glands as a privileged site of cytomegalovirus immune evasion and persistence. Med Microbiol Immunol. 2008;197:205‐213. [DOI] [PubMed] [Google Scholar]
- 10. Zhang W, Du RH, Li B, et al. Molecular and serological investigation of 2019‐nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9:386‐389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang W, Xu Y, Gao R, et al. Detection of SARS‐CoV‐2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. 10.1001/jama.2020.3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID‐2019. Nature. 2020. 10.1038/s41586-020-2196-x. [published online ahead of print, April 1, 2020]. [DOI] [PubMed] [Google Scholar]


