Abstract
Epidermal growth factor receptor (EGFR) exon 19 deletion (E19del) is the most common activating mutation in advanced non–small cell lung cancer (NSCLC) and associates with the sensitivity of EGFR tyrosine kinase inhibitors (TKIs) treatment. However, not all mutant patterns of E19del have been well studied for the limited coverage of regular EGFR mutation testing. Here, we performed a retrospective cohort study of the C-helix E19del in advanced NSCLC patients based on the screening data by the next-generation sequencing (NGS) platform. From May 2012 to December 2019, clinical information and specimen from 7544 consecutive advanced (IIIB/IV) NSCLC patients were collected and screened for EGFR gene mutations by NGS from multicenters in China. The molecular characteristics and responsiveness to first-line EGFR TKIs therapy in NSCLC patients with C-helix E19del were analyzed. The clinical characteristics were also compared between patients with classical E19del and C-helix E19del. Thirty-eight (2.6%) patients with C-helix E19del and 1400 (97.4%) patients with classical E19dels were identified from 1438 patients with E19del. No significant difference in clinical characteristics was observed between the C-helix E19del and classical E19del groups (P > .05), except for histology (P < .001). All 22 patients with C-helix E19del as p.S752_I759del, p.A750_E758del, p.A750_E758delinsP, p.T751_A755delinsNY, p.T751_I759delinsG, p.T751_I759delinsLD, p.T751_I759delinsN, p.T751_L760delinsNL, and p.T751_D761delinsLY reached the best response as partial response rate (72.7%), and the progression-free survival (PFS) was 12.0 months. The PFS after EGFR TKIs in patients with C-helix E19del tended to be longer than patients with classical E19del but has no statistical significance (12.0 months vs 8.5 months, P = .06). The C-helix E19del could be a positive biomarker for predicting response to EGFR TKIs in advanced NSCLC patients. NGS should be the appropriate platform to identify this rare population, especially when patients harbor no actionable driver mutation initially and are reluctant to accept chemotherapy as first-line therapy.
Advanced lung cancer has been the leading life-threatening malignant carcinoma worldwide for decades [1]. About 85% of advanced lung cancers are non–small cell lung cancer (NSCLC) [2], and the efficacy of conventional chemotherapy for this population has reached a ceiling level around 30%-40% [3]. Fortunately, the turning point was the discovery of impressive sensitivity of tyrosine kinase inhibitors (TKIs) in advanced NSCLC patients with epidermal growth factor receptor (EGFR) active mutations [4]. EGFR targeting therapy has not only doubled the response rate of conventional chemotherapy but also prolonged the overall survival of the advanced NSCLC patients [5].
The activating EGFR gene mutants mainly occur in the 18-21 exon which encodes the intracellular tyrosine kinase (TK) domain [6]. The classical mutations refer to EGFR exon 19 deletion (E19del) and exon 21 point mutations which take about 85% of all EGFR mutations [7]. E19del was the most prevalent approximately 45% of all EGFR mutations and complex for many different mutant positions and patterns [7]. The mutant patterns of E19del are mainly deletion, while the point and insertion mutations are not common, respectively [8]. About 2.5% E19del would occur in the C-helix part of exon 19 [9] which could constructively impact the sensitivity of TKI treatment by activation of TK region [10]. However, the C-helix E19del could be undetected by routine genetic mutant testing which often does not cover the whole spectrum of exon 19. So far, the prevalence and effectiveness of EGFR TKIs therapy in this rare population have not been well understood.
To better address the clinical implication of the C-helix E19del in advanced NSCLC patients, we performed a large cohort study by next-generation sequencing (NGS) screening of EGFR mutations and analyzed the characteristics and responsiveness to TKIs in this population. The comparison of clinical characteristics between patients with classical E19del and C-helix E19del mutations was also discussed.
Methods
Patients and Procedures
Eligible patients were required to have pathologically confirmed NSCLC and sufficient tissue for analysis. EGFR mutations were assessed with NGS. Clinical and pathologic data retrospectively collected for analyses included age at diagnosis, gender, smoking status, stage, histology, and EGFR mutant status according to the regular guideline for practice. Twenty-two patients with C-helix E19del received EGFR TKIs treatment and had clinical data available on the outcome. Imaging data were independently reviewed by authors to evaluate their treatment responses according to the Response Evaluation Criteria in Solid Tumors version 1.1 PFS calculated from the date of initiating targeted drugs treatment to radiologic or clinical observation of disease progression. This study was approved by the ethics committee, and a written informed consent was obtained from each participant before the initiation of any study-related procedure.
Targeted NGS
DNA from formalin-fixed, paraffin-embedded tumor tissue and matched blood samples was extracted. Comprehensive genomic profiling was performed by NGS with a 381 cancer-related gene panel covering the whole exons of EGFR gene at a mean coverage depth of >800×. Genomic DNA sequencing libraries were prepared using the protocols recommended by the Illumina TruSeq DNA Library Preparation Kit. For samples close to the minimum input requirement, additional precapture PCR cycles were performed to generate sufficient PCR product for hybridization. The libraries were hybridized to custom-designed probes (Integrated DNA Technology) including all exons of 170 genes and selected intron of ALK, RET, and ROS1 for the detection of genomic rearrangements. DNA sequencing was performed on a HiSeq3000 sequencing system (Illumina, San Diego, CA) with 2×75-bp paired-end reads. The reads were aligned to the human genome build GRCh37 using BWA (a Burrows-Wheeler aligner). Somatic single nucleotide variant and indel calls were generated using MuTect and GATK, respectively. Somatic copy number alterations were identified with CONTRA. Genomic rearrangements were identified by the software developed in-house analyzing chimeric read pairs.
Statistical Analysis
Chi-square test or Fisher’s exact test was used to analyze correlations between the patterns of E19del and the characteristic of clinicopathologic factors in advanced NSCLC patients. Statistical analysis was performed using SPSS version 19.0 software (IBM, Armonk, NY). All P values were two-sided, and a P value of <.05 was considered statistically significant.
Results
Patients’ Characteristics and Treatments
From May 2012 to December 2019, a total of 7544 NSCLC patients were screened and 3099 (41.1%) were found to harbor EGFR mutations. Among them, 1438 (46.4%) patients included 97.4% (1400/1438) of the classical E19del and 2.6% (38/1438) of the C-helix E19del; the flowchart of the study design is shown in Figure 1. The baseline characteristics of the E19del harboring patients have been presented in Table 1, and no statistical difference was observed between patients with classical and C-helix E19del by the median age, gender, smoking status, and histology. A total of 1116 patients with classical E19del and 22 patients with C-helix E19del received EGFR TKIs treatment as first-line therapy, while others received conventional chemotherapy according to local practice. The EGFR TKIs drugs included gefitinib, erlotinib, and icotinib listed in Table 2.
Figure 1.
Flowchart of the study design.
Table 1.
Characteristics of NSCLC Patients with EGFR Exon19 Deletion
| Characteristics | N | No. of Patients (%) |
P | |
|---|---|---|---|---|
| Classical | C-Helix | |||
| Cases | 1438 | 1400 | 38 | |
| Median age | 58 (25-91) | 58 (25-91) | 57 (36-75) | .315 |
| Years (range) | ||||
| Gender | .282 | |||
| Male | 653 | 639 | 14 | |
| Female | 785 | 761 | 24 | |
| Smoking status | .924 | |||
| Prev/curr smoker | 350 | 341 | 9 | |
| Never smoker | 1088 | 1059 | 29 | |
| Histology | <.001 | |||
| Adenocarcinoma | 1412 | 1380 | 32 | |
| Squamous cell carcinoma | 9 | 9 | 0 | |
| Adenosquamous carcinoma | 7 | 3 | 4 | |
| Sarcomatoid carcinoma | 7 | 5 | 2 | |
| Adenoid cystic carcinoma | 3 | 3 | 0 | |
Table 2.
Cases with EGFR Exon 19 C-Helix Deletion
| ID | Amino Acid Change | Gender | Age, Year | Histology | Smoking Status | TKI | Best Response | PFS (Months) |
|---|---|---|---|---|---|---|---|---|
| 1 | S752_I759del | Female | 48 | Adenocarcinoma | Never smoker | Gefitinib + erlotinib | PR | 12.0 |
| 2 | S752_I759del | Female | 64 | Adenocarcinoma | Never smoker | - | - | - |
| 3 | S752_I759del | Female | 67 | Adenocarcinoma | Never smoker | - | - | - |
| 4 | S752_I759del | Male | 52 | Adenocarcinoma | Prev/curr smoker | - | - | - |
| 5 | S752_I759del | Female | 67 | Adenocarcinoma | Never smoker | Icotinib | PR | 18.0 |
| 6 | S752_I759del | Male | 70 | Adenocarcinoma | Prev/curr smoker | Icotinib | SD | 6.5+ |
| 7 | S752_I759del | Female | 52 | Adenocarcinoma | Never smoker | Gefitinib | PR | 17.0+ |
| 8 | S752_I759del | Female | 44 | Adenosquamous carcinoma | Never smoker | Icotinib | PD | 2.0 |
| 9 | S752_I759del | Male | 71 | Adenocarcinoma | Prev/curr smoker | Icotinib | PR | 54.0+ |
| 10 | S752_I759del | Female | 47 | Adenocarcinoma | Never smoker | Icotinib | PR | 8.0 |
| 11 | S752_I759del | Female | 67 | Sarcomatoid carcinoma | Never smoker | |||
| 12 | S752_I759del | Male | 53 | Adenocarcinoma | Never smoker | Gefitinib | SD | 12.0 |
| 13 | S752_I759del | Female | 69 | Adenocarcinoma | Never smoker | Icotinib | PR | 34.5 |
| 14 | S752_I759del | Male | 62 | Adenocarcinoma | Prev/curr smoker | |||
| 15 | S752_I759del | Female | 59 | Adenosquamous carcinoma | Never smoker | Gefitinib | SD | 4.5+ |
| 16 | S752_I759del | Female | 49 | Adenocarcinoma | Never smoker | Erlotinib | PR | 6.5 |
| 17 | S752_I759del | Female | 47 | Sarcomatoid carcinoma | Never smoker | |||
| 18 | S752_I759del | Male | 63 | Adenocarcinoma | Prev/curr smoker | Gefitinib | PR | 9.5 |
| 19 | S752_I759del | Female | 66 | Adenocarcinoma | Never smoker | Icotinib | PD | 2.0 |
| 20 | A750_E758del | Male | 65 | Adenocarcinoma | Prev/curr smoker | - | - | - |
| 21 | A750_E758del | Male | 51 | Adenocarcinoma | Never smoker | - | - | - |
| 22 | A750_E758del | Female | 64 | Adenocarcinoma | Never smoker | Gefitinib | PR | 7.0 |
| 23 | A750_E758delinsP | Female | 62 | Adenocarcinoma | Never smoker | Gefitinib | PR | 9.0 |
| 24 | L747_A755delinsSKG | Female | 50 | Adenosquamous carcinoma | Never smoker | - | - | - |
| 25 | E749_K754del | Male | 75 | Adenocarcinoma | Never smoker | - | - | - |
| 26 | T751_A755delinsNY | Female | 64 | Adenocarcinoma | Never smoker | Icotinib | PR | 7.0 |
| 27 | T751_E758del | Female | 49 | Adenocarcinoma | Never smoker | - | - | - |
| 28 | T751_I759delinsG | Female | 36 | Adenocarcinoma | Never smoker | Gefitinib | PR | 18.0 |
| 29 | T751_I759delinsLD | Female | 40 | Adenocarcinoma | Never smoker | Icotinib | PR | 15.5 |
| 30 | T751_I759delinsN | Female | 65 | Adenocarcinoma | Never smoker | Icotinib | PR | 22.0 |
| 31 | T751_I759del | Female | 58 | Adenocarcinoma | Never smoker | - | - | - |
| 32 | T751_L760delinsNL | Male | 53 | Adenosquamous carcinoma | Never smoker | Icotinib | SD | 5.0 |
| 33 | T751_D761delinsLY | Female | 39 | Adenocarcinoma | Never smoker | Icotinib | PR | 9.5+ |
| 34 | P753_I759delinsG | Male | 60 | Adenocarcinoma | Prev/curr smoker | - | - | - |
| 35 | A750_E758delinsP | Female | 46 | Adenocarcinoma | Never smoker | - | - | - |
| 36 | A750_I759delinsG | Male | 56 | Adenocarcinoma | Prev/curr smoker | |||
| 37 | A750_E758del | Male | 65 | Adenocarcinoma | Prev/curr smoker | Gefitinib | PR | 8.0+ |
| 38 | A750_E758del | Male | 51 | Adenocarcinoma | Never smoker | - | - | - |
Distribution of C-Helix E19del Screening by NGS
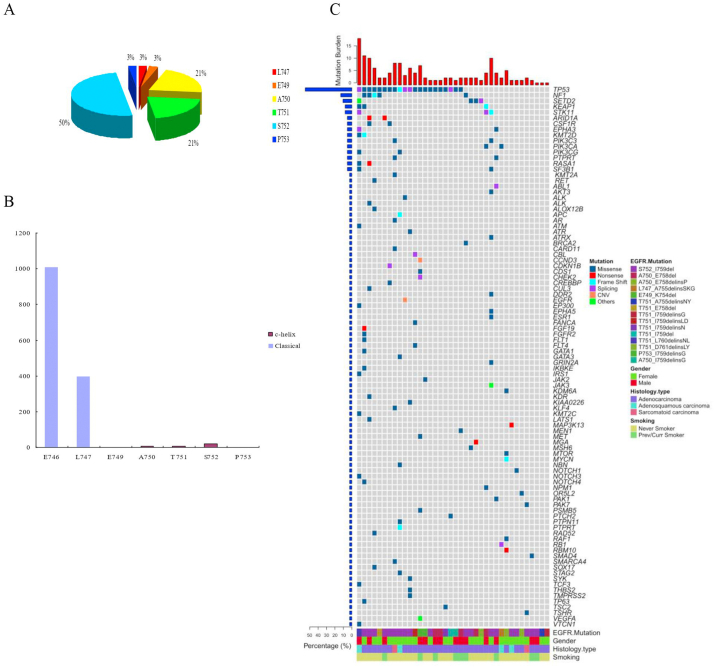
Of the 38 patients with EGFR exon 19 C-helix deletion, 50.0% (19/38) of locus occurred on S752; 21.1% (8/38) of locus occurred on A750 and T751; and 2.6% (1/38) occurred on L747, E749, and P753 (Table 2, Figure 2A). The locus of classical E19del was mainly distributed from E746 to P747 and C-helix E19del was distributed from P747 to P753 (Figure 2B). Furthermore, TP53 mutant (55.3%, 21/38) was the most frequent coexisting genetic alteration in all patients with EGFR exon 19 C-helix deletion (Figure 2C).
Figure 2.
EGFR exon 19 c-helix deletion mutation distribution. (A) Distribution of EGFR exon 19 c-helix patients categorized according to locus. (B) Comparison of classical and c-helix loci from E746 to P753. (C) Heatmap showed the next-generation sequencing results for co-mutation spectrum of EGFR exon 19 c-helix deletion mutations in all patients.
Clinical Outcome of EGFR Exon 19 C-Helix Deletion Patients
After median follow-up duration of 94.0 months, all 22 patients with C-helix E19del as p.S752_I759del, p.A750_E758del, p.A750_E758delinsP, p.T751_A755delinsNY, p.T751_I759delinsG, p.T751_I759delinsLD, p.T751_I759delinsN, p.T751_L760delinsNL, and p.T751_D761delinsLY reached the best response as partial response (PR) rate (72.7%), and the progression-free survival (PFS) was 12.0 months. The response rate to TKIs as first-line palliative therapy in patients with C-helix E19del in our study (Table 2). Kaplan-Meier survival curves showed no statistical significance of PFS after EGFR TKIs between classical (n=1400) and c-helix (n=38) with E19del patients (8.5 months vs 12.0 months, P = .06) (Figure 3).
Figure 3.
Comparison of classical and c-helix with EGFR-TKIs therapy (8.5 months vs 12.0 months, P = .06)
Discussion
Activating EGFR gene mutations are the well-known driver mutations in advanced NSCLC, occurring in 10%-15% and 50%-60% in white and Asian populations, respectively [11,12]. Mutations within exons 18, 19, and 21 are associated with the sensitivity to EGFR TKIs such as gefitinib, erlotinib, afatinib, and icotinib [13]; E19del are in-frame deletions and are found in about 48% of EGFR mutant lung cancers [14]. More than 50 patterns of E19del mutations have been reported, and p.E746_A750 is the most common deletion accounting for over 45% in all [15]. E19del mutations occur in a protein strand (called the β3 strand) adjacent to the C-helix which is within the N-lobe of EGFR TK domain [16]. It is postulated that reducing the length of this strand may also favor the activation of EGFR TK region [10]. However, the characteristics and responsiveness of EGFR TKIs in NSCLC patients with C-helix E19del are unclear partly because they are not hotspots and could be missed by the regular gene mutation testing, such as ARMS and direct sequencing [17].
In this study, we used massive parallel NGS platform to screen EGFR mutations and preliminarily found that about 2.4% of E19del occurred in the C-helix part consistent with result from previous study [9], and most of them combined with insertions at the same time. No significant different clinical characteristic of patients with C-helix E19del could be found compared with those that carried classical E19del, except for histology. All 22 patients with C-helix E19del as p.S752_I759del, p.A750_E758del, p.A750_E758delinsP, p.T751_A755delinsNY, p.T751_I759delinsG, p.T751_I759delinsLD, p.T751_I759delinsN, p.T751_L760delinsNL, and p.T751_D761delinsLY reached the best response as PR rate (72.7%), and the PFS was 12.0 months. As far as we know, the responsiveness to TKIs in EGFR p.T751_I759delinsG has never been reported before.
Schrock et al. [18] reported that 3.5% (14/390) of E19del in C-helix were within the 753-761 amino acid (AA) range and 96.5% (286/390) were classical E19del within the 743-754 AA range by genomic comprehensive profiling in previous EGFR-negative patients. The majority of C-helix E19del were within 752-759 AA [18]. We reported that the prevalence of C-helix E19del was 2.6% from large cohort, which is close to the result from this study. One patient with p.T751_I759delinsN received TKI treatment and achieved PR consistent according to their study, and the response was consistent with ours. In our study, p.S752_I759del, p.A750_E758del, p.A750_E758delinsP, p.T751_A755delinsNY, p.T751_I759delinsG, p.T751_I759delinsLD, p.T751_I759delinsN, p.T751_L760delinsNL, and p.T751_D761delinsLY have been proved to be sensitizing EGFR mutations in advanced NSCLC by longer than 1-year PFS after TKIs treatment. The unknown EGFR p.T751_I759delinsG was affected at the same position in C-helix like p.T751_I759delinsN but with different insertion of AA which could presumably be oncogenic either. However, p.T751_I759delinsS seemed to have intrinsic resistance to gefitinib, and PFS was only 2.0 months in another study [19]. In that case, we presumed that both of the deletion position and the insertion AA may influence the sensitivity of EGFR TKIs. Notably, those 22 patients would have no chance to accept TKIs therapy if the EGFR mutation was not detected by NGS platform. NGS has shown to have superior advantages in screening for uncommon EGFR mutations that allow detection of multiple genes and relevant mutant patterns simultaneously from tumor specimens [20]. There was no concurrent of well-known driver gene alterations in patients with C-helix E19del, such as ALK/RET/ROS1 rearrangement and KRAS mutation. It may reflect the pro-oncogenic nature of C-helix E19del in NSCLC patients similar to the mutual exclusiveness of classical EGFR mutations with other driver gene alterations [21]. Of note, most of the patients (35/38, 92.1%) carried concurrent gene alterations, and 55.3% (21/38) of them carried TP53 mutations. The mutation rate of TP53 was 56.1% in one study of 1441 metastatic NSCLC patients [22]. In their study, the TP53 mutation correlated with a negative prognosis regardless of EGFR mutation status. Furthermore, about half of TP53 mutations (11/21) have been detected in patients with S752_I749del, which has never been reported before. Another tumor suppressor gene NF1 mutation has also been detected in five patients, and four of them carried concomitant TP53 mutations. NF1 mutations have been reported to occur more often with oncogenic alterations and TP53 mutations [23], but the clinical significance is unknown.
The main limitation of our research was that we retrospectively collected data and some bias in our conclusion could not be avoided. Although all 22 patients with C-helix deletion showed good response to TKIs and their disease was well controlled, we could not confirm the predictive value of C-helix deletion since the outcome of those who harbored the same deletion but accepted chemotherapy was unknown. Longer follow-up and further structural and functional research for those uncommon deletions should be conducted in future to verify our findings.
In summary, our study increases the evidence supporting EGFR TKIs treatment as first-line therapy for advanced NSCLC patients with C-helix E19del. Reliable detection of drug-sensitive EGFR mutations is crucial in the care of advanced NSCLC patients. NGS platform should be more suitable for screening those uncommon mutants than regular testing, especially in those patients who are reluctant to receive chemotherapy.
Conflict of Interest
None has any conflict of interest.
Acknowledgements
This study was supported in part by grants from Zhejiang Public Welfare Technology Research Program (LGJ20H160001), Science and Technology Planning project of Zhejiang Province (LGF19H160002), Medical Scientific Research Foundation of Zhejiang Province of China (2019RC027), Zhejiang Traditional Chinese Medicine Science Fund Project (2020ZB037), Scientific Research Foundation of Zhejiang Medical Association (2019ZYC-A76), and Xisike-Hanson Cancer Research Foundation (Y-HS2019-20).
Contributor Information
Li-yun Miao, Email: liyunmiao462@163.com.
Li-ping Wang, Email: 215974914@qq.com.
Yong Song, Email: yong_song6310@yahoo.com.
References
- 1.Bray F., Ferlay J., Soerjomataram I. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Shepherd F.A. Chemotherapy for non–small cell lung cancer: have we reached a new plateau? Semin Oncol. 1999;26(1 Suppl 4):3–11. [PubMed] [Google Scholar]
- 4.Zhou C., Wu Y. L, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non–small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12(8):735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 5.Sebastian M., Yang J.C.-H., Sequist L. Analysis of overall survival (OS) in two large open-label phase III studies (LUX-Lung 3 [LL3] and LUX-Lung 6 [LL6]) comparing afatinib with chemotherapy (CT) in patients (pts) with advanced non–small cell lung cancer (NSCLC) harboring common (Del19/L858R) epidermal growth factor receptor mutations (EGFR mut) Eur Respir J. 2014;44(Suppl 58):1929. [Google Scholar]
- 6.Hodoglugil U., Carrillo M.W., Hebert J.M. PharmGKB summary: very important pharmacogene information for epidermal growth factor receptor (EGFR) Pharmacogenet Genom. 2013;23(11):636. doi: 10.1097/FPC.0b013e3283655091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shigematsu H., Lin L., Takahashi T. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer I. 2005;97(5):339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 8.Lee V.H., Tin V.P. Choy T-s, et al. Association of exon 19 and 21 EGFR mutation patterns with treatment outcome after first-line tyrosine kinase inhibitor in metastatic non–small-cell lung cancer. J Thorac Oncol. 2013;8(9):1148–1155. doi: 10.1097/JTO.0b013e31829f684a. [DOI] [PubMed] [Google Scholar]
- 9.O’Kane G.M., Bradbury P.A., Feld R. Uncommon EGFR mutations in advanced non–small cell lung cancer. Lung Cancer. 2017;109:137–144. doi: 10.1016/j.lungcan.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 10.Politi K., Lynch T.J. Two sides of the same coin: EGFR exon 19 deletions and insertions in lung cancer. Clin Cancer Res. 2012;18(6):1490–1492. doi: 10.1158/1078-0432.CCR-11-3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirsch F.R., Bunn P.A. EGFR testing in lung cancer is ready for prime time. Lancet Oncol. 2009;5(10):432–433. doi: 10.1016/S1470-2045(09)70110-X. [DOI] [PubMed] [Google Scholar]
- 12.Shi Y., Li J., Zhang S. Molecular epidemiology of EGFR mutations in Asian patients with advanced non–small-cell lung cancer of adenocarcinoma histology—Mainland China subset analysis of the PIONEER study. Plos One. 2015;10(11) doi: 10.1371/journal.pone.0143515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang W., Wu X., Fang W. Network meta-analysis of erlotinib, gefitinib, afatinib and icotinib in patients with advanced non–small-cell lung cancer harboring EGFR mutations. Plos One. 2014;9(2) doi: 10.1371/journal.pone.0085245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitsudomi T., Yatabe Y. Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. FEBS J. 2010;277(2):301–308. doi: 10.1111/j.1742-4658.2009.07448.x. [DOI] [PubMed] [Google Scholar]
- 15.Kato Y., Peled N., Wynes M.W. Novel EGFR mutation specific antibodies for NSCLC: Immunohistochemistry as a possible screening method for EGFR mutations. J Thorac Oncol. 2010;5(10):1551. doi: 10.1097/JTO.0b013e3181e9da60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nie W., Tang L., Zhang H. Structural analysis of the EGFR TK domain and potential implications for EGFR targeted therapy. Int J Oncol. 2012;40(6):1763–1769. doi: 10.3892/ijo.2012.1356. [DOI] [PubMed] [Google Scholar]
- 17.Khoo C., Rogers T.M., Fellowes A. Molecular methods for somatic mutation testing in lung adenocarcinoma: EGFR and beyond. Transl Lung Cancer Res. 2015;4(2):126. doi: 10.3978/j.issn.2218-6751.2015.01.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schrock A.B., Frampton G.M., Herndon D. Comprehensive genomic profiling identifies frequent drug-sensitive EGFR exon 19 deletions in NSCLC not identified by prior molecular testing. Clin Cancer Res. 2016;22(13):3281–3285. doi: 10.1158/1078-0432.CCR-15-1668. [DOI] [PubMed] [Google Scholar]
- 19.Improta G., Zupa A., Natalicchio M.I. Uncommon frame-shift exon 19 EGFR mutations are sensitive to EGFR tyrosine kinase inhibitors in non–small cell lung carcinoma. Med Oncol. 2018;35(3):28. doi: 10.1007/s12032-018-1078-7. [DOI] [PubMed] [Google Scholar]
- 20.Lin M.T., Mosier S.L., Thiess M. Clinical validation of KRAS, BRAF, and EGFR mutation detection using next-generation sequencing. Am J Clin Pathol. 2014;141(6):856–866. doi: 10.1309/AJCPMWGWGO34EGOD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gainor J.F., Varghese A.M., Ou S.-H.I. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1, 683 patients with non–small cell lung cancer. Clin Cancer Res. 2013;19(15):4273–4281. doi: 10.1158/1078-0432.CCR-13-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiao X.D., Qin B.D., You P. The prognostic value of TP53 and its correlation with EGFR mutation in advanced non–small cell lung cancer, an analysis based on cBioPortal data base. Lung Cancer. 2018;123:70–75. doi: 10.1016/j.lungcan.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Redig A.J., Capelletti M., Dahlberg S.E. Clinical and molecular characteristics of NF1-mutant lung cancer. Clin Cancer Res. 2016;22(13):3148–3156. doi: 10.1158/1078-0432.CCR-15-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]