Abstract
The majority of the Earth’s microbial biomass exists in the deep biosphere, in the deep ocean, and within the Earth’s crust. Although other physical parameters in these environments, such as temperature or pH, can differ substantially, they are all under high pressures. Beyond emerging genomic information, little is known about the molecular mechanisms underlying the ability of these organisms to survive and grow at pressures that can reach over 1000-fold the pressure on the Earth’s surface. The mechanisms of pressure adaptation are also important in food safety, with the increasing use of high-pressure food processing. Advanced imaging represents an important tool for exploring microbial adaptation and response to environmental changes. Here, we describe implementation of a high-pressure sample chamber with a two-photon scanning microscope system, allowing for the first time, to our knowledge, quantitative high-resolution two-photon imaging at 100 MPa of living microbes from all three kingdoms of life. We adapted this setup for fluorescence lifetime imaging microscopy with phasor analysis (FLIM/Phasor) and investigated metabolic responses to pressure of live cells from mesophilic yeast and bacterial strains, as well as the piezophilic archaeon Archaeoglobus fulgidus. We also monitored by fluorescence intensity fluctuation-based methods (scanning number and brightness and raster scanning imaging correlation spectroscopy) the effect of pressure on the chromosome-associated protein HU and on the ParB partition protein in Escherichia coli, revealing partially reversible dissociation of ParB foci and concomitant nucleoid condensation. These results provide a proof of principle that quantitative, high-resolution imaging of live microbial cells can be carried out at pressures equivalent to those in the deepest ocean trenches.
Significance
The majority of the Earth’s microbial biomass exists in high-pressure environments in which pressures can reach over 100 MPa. The molecular mechanisms that allow microbes to flourish under such extreme conditions remain to be discovered. The high-pressure, high-resolution imaging system presented here revealed pressure-dependent changes in metabolism and protein interactions in live microbial cells, demonstrating great promise for understanding deep life.
Introduction
Life on Earth exists in a wide range of environmental conditions, including extremes of pressure, temperature, pH, or salt concentrations (1). Indeed, over 90% of the Earth’s microbial biomass is thought to exist in high-pressure environments (2). Pressures encountered by living organisms on Earth can reach >100 MPa in the deepest ocean trench (the Challenger Deep in the Mariana trench) or within the Earth’s crust. Several dozen microbial isolates from the deep biosphere have been cultured, and a few of their genomes have been sequenced (3), but these represent only a small fraction of the diversity of species anticipated for this ecosystem. To date, the molecular mechanisms that allow these organisms to survive and proliferate at high pressure are not understood. Progress in this area would have implications for understanding the emergence of life on Earth, mechanisms that control carbon cycling, and the search for life elsewhere. High-pressure food processing is today a 14-billion-dollar industry and is expected to quadruple in the next decade (4). Acquired resistance to pressure treatment by foodborne pathogens represents a serious economic issue (5,6), the mitigation of which will require insight into the molecular mechanisms of microbial adaptation and response to pressure.
Direct observation at high pressure of organisms from the deep biosphere and their atmospheric-pressure-adapted counterparts using advanced microscopy would yield important information about these mechanisms. However, the poor pressure resistance of sample holder materials has required high pressure (HP) microscopy cells with thick windows, severely limiting optical resolution (7). Here, we present the adaptation of a fused silica capillary system, first used for in vitro fluorescence correlation spectroscopy (8,9), to high-pressure, high-resolution fluorescence imaging. We have implemented improvements in sample loading, adherence, and washing to apply such a system to the study of live microbial cells using quantitative high-resolution imaging techniques directly under pressure.
We report here the first high-resolution quantitative observations by two-photon microscopy of molecular processes taking place in live microbial cells under high hydrostatic pressure. We examined the pressure response of microbes from all three kingdoms of life; two adapted to atmospheric pressure, the Gram-positive bacterium Escherichia coli and the yeast Saccharomyces cerevisiae, as well as the anaerobic piezophilic thermophilic archaeon Archaeoglobus fulgidus, a sulfur-metabolizing organism found in hydrothermal vents. The natural autofluorescence from metabolic enzyme cofactors (NAD(P)H and flavin adenine dinucleotide (FAD)) (10) represents an alternative live-cell imaging contrast to fluorescent proteins. We coupled two-photon excitation of NAD(P)H (740 nm) with scanning fluorescence lifetime imaging microscopy (FLIM) and phasor analysis (11). FLIM with phasor (FLIM/Phasor) analysis of NAD(P)H fluorescence provides a metabolic footprint of live bacterial cells (12). In E. coli, we compared total NAD(P)H to the metabolic state before, during, and after pressurization. These studies revealed a strong and complex response of E. coli metabolism to pressure.
In cases for which fluorescent proteins can be genetically introduced into the sample strains, quantitative imaging modalities that rely on the fluorescence intensity fluctuations such as scanning number and brightness (sN&B) (13) and raster imaging correlation scanning (RICS) (14) analyses can be applied. These approaches yield protein stoichiometry, absolute concentration, and diffusion properties (13,14). Here, we applied fluorescence fluctuation-based methods, sN&B and RICS, to monitor the effect of pressure on the stoichiometry, localization, and dynamics of two fluorescent protein fusions implicated in nucleoid structure and plasmid partitioning and coexpressed in E. coli cells. Our results revealed pressure-induced dissociation of partition complexes and pressure-induced foci formation of the nucleoid-associated protein. These pressure effects were reversible for only a subset of cells.
Materials and Methods
Cell preparation for imaging
E. coli K12 reference strain MG1655 and its mrr− derivative (15), devoid of the Mrr endonuclease, were used for FLIM experiments. For sN&B measurements, the E. coli strain DLT3053 was used for constitutive coexpression of the HU-mCherry fusion encoded by a chromosomal insertion and ParB-mVenus fusion encoded by the plasmid pJYB234 (16). The protocol was similar to that described previously (17). Overnight, cultures were diluted in LB medium and grown at 37°C until the OD600 reached 0.6. The cells were centrifuged and resuspended in a few microliters to a final OD600 ∼25 in minimal M9 medium supplemented with 0.4% glucose. S. cerevisiae BY4741 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ) was grown on full synthetic complete (SC) medium agar plate, and then a single colony was used to set an overnight culture in SC medium supplemented by 2% glucose and diluted the next morning. Similar to bacteria, a high density of cells was required for the injection in the capillary. A. fulgidus was grown in a liquid heterotrophic (lactate) sulfate-reduction medium using anoxic techniques ((18) and references therein). A cell pellet of an overnight culture was resuspended, under anoxic conditions, to obtain a highly concentrated cell suspension before injection. A few microliters of the highly concentrated cell preparation were injected into the coated capillary. The cells were left for a few minutes (∼10 min) to attach to the surface. Those that were not attached were rinsed with the fresh appropriate medium (M9 minimal medium for bacteria, SC medium for yeast, and media as described previously (18) for the archaea used to purge the entire system to prevent cell death.
Capillary preparation and coating
The middle of the 15-inch capillary was burned with a lighter for 2–3 s to remove the outer polyimide coating. The capillary was passed through the two glands (Fig. 1 A) and glued to the drilled pressure plug using epoxy glue. A solution of 100× chitosan for immobilization (19) was freshly prepared with ∼0.015 g of chitosan powder diluted in 900 μL of 2 M (= 10%) glacial acetic acid in an Eppendorf tube. Then, 60 μL of this 100× solution was diluted in 900 μL of distilled water. The chitosan solution was passed into the capillary using the peristatic pump until the solution was apparent in the output tubing and the connector. Incubation was allowed for 20–30 min at room temperature. The chitosan solution was rinsed by flushing the system with the minimal medium required by the cells during the experiment.
Figure 1.
Microscope setup used to perform high-resolution quantitative imaging of live cells under pressure. (A) shows a microscopy stage showing the capillary immobilized in a holder (attofluor) between a coverslip and the objective with glycerol as a coupling media. (B) shows a cross section and photo of the fused silica square capillary with an external polyimide coating inserted and glued in drilled plugs. (C) shows a schematic of the HP connections. Two valves (V) make it possible to switch with either the peristatic pump or the pressure pump to load or apply pressure, respectively (V3 and V4). (D) shows a photograph of the cart (with pressure pump, lines, and valves, as designated in the schematic, and the connection to the microscope with the mounted capillary). To see this figure in color, go online.
Sample loading
The capillary was moved into the modified attofluor holder, which was placed on the microscope stage so that the burned portion of the capillary was located at the objective. The attofluor is slitted at 180° such that the capillary is held in place. The 15-inch length of the capillary allows for easy adjustment of the capillary position on the microscope stage. The capillary was seated on a rubber gasket at the bottom of the attofluor to avoid breakage, and a second rubber gasket was placed on top of the capillary. Then, the capillary was immobilized in the stainless-steel holder with two coverslips (N2; VWR, Philadelphia, PA) on the top to prevent bending upon contact with the objective, and a stainless-steel gasket was placed within the ring of the attofluor. The weight of this piece maintains the capillary z-position during focusing. Glycerol was used as a coupling medium. By closing V3 and opening V4, the system was connected to the peristatic pump. A drop (less than 10 μL) of concentrated solution of cells (OD600 ∼25) was deposited on the plug located at the end of the capillary near V5, and the peristaltic pump was run backward until cells passing through the capillary were visible on the camera or at the objective. The pump was then turned off, and the capillary was tightly connected to the pressure output valve V5. The cells were allowed to attach to the surface for 10 min, and then with V4 closed and V3 and V5 open, unattached cells were rinsed away with minimal medium supplemented with 0.4% glucose for E. coli cells and the growth medium noted above for the yeast and archaea at a low flow rate from the pressure pump (0.05 mL/min). This method allowed a flat field of view (FOV) with a single layer of immobilized cells that remained attached to the surface with increasing pressure (Fig. S1). When there were no more unattached cells (Fig. S1), the pressure pump was switched off and V5 closed, leaving the system ready for high-pressure experiments.
All pressure connections (tubing, lines, glands) were 1/8 in diameter (High Pressure Equipment, Erie, PA) to minimize the footprint of the high-pressure apparatus. Moreover, we used an RF-1700 constant high-pressure pump from Pressure BioSciences (South Easton, MA) with a maximum pressure set to 15,000 psi (∼100 MPa). To match the fused silica capillary refraction index, oil was replaced by glycerol as the coupling medium. All tubing and pumps were purged with minimal media to minimize background fluorescence. Pressure was increased using a low flow rate (0.05 μL/min) such that reaching 100 MPa required 2–3 min. Pressure was released by slowly opening the output valve.
FLIM/Phasor analysis
The advantage of this phasor approach for FLIM analysis lies in the easy interpretation of the raw data without any fitting of lifetime decay curves (11). Each pixel of the FLIM image was transformed into a pixel on the phasor plot, with g and s coordinates calculated from the fluorescence intensity decay using the following transformation, with i and j corresponding to a pixel of the image and ω the frequency determined by the laser repetition rate (80 MHz):
| (1) |
| (2) |
Pixels corresponding to a single species with a single-exponential fluorescence intensity decay are located on the universal semicircle limiting the phasor plot, and the position on the semicircle depends on the lifetime. Short lifetimes are located on the right with high g-values and small s-values, whereas long lifetimes are near the origin of the semicircle. Multiexponential decays are located inside the semicircle, at a position defined by the lifetime values and their relative fractional intensities. For a mixture of two single-exponential decays, the phasor will be located on a line between the two single-decay positions on the universal circle, the position being weighted by the fractional contribution of each single-exponential component to the decay. Indeed, the coordinates g and s are described following Eqs. 3 and 4, with hk the intensity and τk the lifetime of component, k:
| (3) |
| (4) |
Thus, the global phasor at each pixel of an image is the sum of the independent phasors of each fluorescent decay. To highlight the spatial localization of lifetime components, we selected a cluster of pixels within the phasor plot and visualized the localization of these pixels on the fluorescent image using the VistaVision software (ISS, Champaign, IL). FLIM experiments were performed at 740 nm with a two-photon excitation and a frequency of 80 MHz. The number of frames, the pixel dwell time, and the laser power were adjusted to collect 100 counts in the brightness pixel of the FOV. We have generally acquired a single frame with 1 ms of pixel dwell time for an FOV of 13 × 13 μm and 256 × 256 pixels.
Number and brightness
Number and brightness (N&B) is an image-based implementation of fluctuation spectroscopy (13). Here, we imaged fluorescent protein fusions expressed in bacteria. A series of raster scans (25 frames, in our case) was acquired using a pixel dwell time (40 μs) faster than the diffusion time. This provides fluorescence intensity values over time for each pixel, from which fluorescence fluctuations (variance) and average intensity (F) can be calculated and used to deconvolve the fluorescence intensity into the brightness (B) and the number of molecules (N) in the excitation volume (F = N × B). The shot-noise-corrected brightness (e) values and the number (n) of diffusing molecules were calculated at each pixel:
| (5) |
| (6) |
The brightness is directly proportional to the number of fluorophore-containing molecules diffusing together, so that the brightness of a monomeric fluorescent protein can be used to calculate the stoichiometry of an oligomeric complex. However, the autofluorescence of the background contribution tends to decrease the values of brightness (esample) and fluorescence intensity (Fsample) measured, which is why we have removed its contribution using N&B measurements on a background strain that does not express FP-tagged fluorescence molecules (brightness ebg and fluorescence Fbg) following Eq. 7:
| (7) |
These analyses were carried out using the Patrack software (Patrick Dosset, CBS, Montpellier, France) (20). The average values were calculated for all pixels within the central region of all cells in the image, as previously described for N&B studies on bacteria (21).
Scanning N&B was performed as previously described by Bourges et al. (17) except that an FOV was scanned 25 times instead of 50 times to control the time of exposure to pressure. The size of an FOV was 13 × 13 μm. All experiments on fluorescent protein fusions were performed with a two-photon excitation at 930 nm. For the simultaneous excitation of mVenus and mCherry, a 580LP mirror was used to separate the emission light on two channels with a 530/43 nm (mVenus) or 650/50 nm (mCherry) filter (Chroma Technologies, Bellows Falls, VT). Merged images of the average fluorescence intensity images were obtained using the software package, Fiji (available online at http://fiji.sc/). The background-corrected brightness values presented were calculated using the background strain E. coli MG1655 (17).
RICS
From the same raster scans obtained in N&B, the diffusion coefficient of the fluorescent molecules can be extracted by fitting the pixel pair spatiotemporal correlation function (22),
where ξ and ψ are the spatial increments of the scanning laser in x and y, respectively. The S(ξ, ψ) and G(ξ, ψ) functions correspond to the correlation due to scanning and that due to diffusion, respectively:
and
where τp and τl are the pixel dwell time and line time, respectively, and δr is the distance between pixels in a line.
The pixel dwell time used was 40 μs, which means that one frame in sN&B took 2–3 s. Thus, fluorescence fluctuations were observed even for particles diffusing very slowly on the millisecond timescale. The vertical (line to line) spatiotemporal correlation function is on the millisecond timescale and is thus the most useful. It reflects the fact that the probability of seeing a molecule in the next line is higher if the molecule diffuses slowly. Conversely, if the molecule diffuses quickly, such as free diffusing green fluorescent protein (GFP), the fluorescence intensity correlation decreases rapidly from line to line and is represented by a rapid drop in the vertical autocorrelation curve. Raster scans were converted to RICS curves and analyzed using SimFCS (E. Gratton, Laboratory for Fluorescence Dynamics, Irvine, CA).
Results and Discussion
Live-cell imaging compatible high-pressure sample chamber
The major modifications presented here with respect to prior high-pressure single-molecule in vitro setups involved implementing a sample-loading system that was compatible with immobilizing and washing microbial cells and scaling down the size of the high-pressure components, as described in the Materials and Methods. We employed square capillary tubing, as previously described (23), which, although presenting lower pressure limits (∼150 MPa), has significantly better optical properties and allows for efficient immobilization of microbial cells. The thickness of the capillary walls (150 μm) matches the working distance of high numerical aperture objectives. Rather than sealing one end of the capillary with a blow torch after sample loading (8,23) (a process that would be highly detrimental to live cells), a drilled plug system sealed around the capillary with epoxy glue was used for both ends (Materials and Methods and Fig. 1 B). Thus, both ends of the capillary were glued into high-pressure plugs that were drilled with a 400-mm bore. Because both ends were connected to the pressure system via the drilled plugs, a peristaltic pump could be used to easily load the microbial cells into the capillary (Fig. 1 B; Fig. S1). After loading the cells and washing out nonadhered cells with medium, the capillary was connected to the pressure lines by switching between two valves, V3 and V4 (Fig. 1 B). Pressure experiments were performed by closing a valve located at the output of the capillary (V5 on Fig. 1 B). The two-photon scanning microscope (Alba; ISS) was the same as previously described in Bourges et al. (17), with the exception of the objective that has been replaced by an 60× 1.4 NA oil immersion objective (APO, VC; Nikon Instruments, Melville, NY) (Fig. 1).
Microbial metabolic responses to pressure
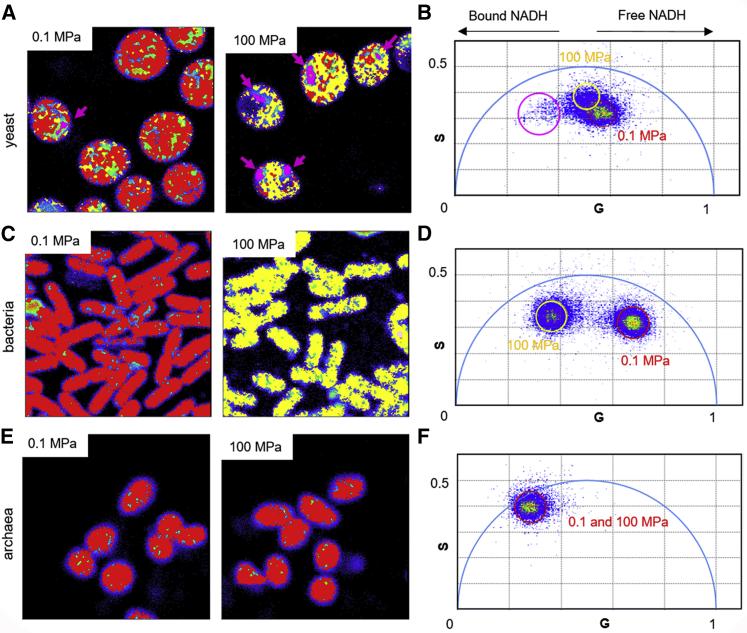
Measurement of fluorescence lifetime with phasor analysis of the enzyme cofactors NADH or NADPH provides a fingerprint of microbial metabolic states (12). The bound cofactors exhibit a significantly longer fluorescence lifetime than the free forms, although NADH and NAD(P)H cannot be distinguished from each other. The difference in fluorescence lifetimes for bound and free NAD(P)H results in distinct positioning on the phasor plot. We demonstrate here the proof of principle that such measurements can be made on live microbial cells at high pressure. As described in the Materials and Methods, the g and s coordinates in the phasor plot (Fig. 2, B, D, and F) represent, respectively, the cosine and sine Fourier transforms of the intensity decay function. Single-exponential decays will have s- and g-phasor values such that they lie on the universal half circle (Fig. S2), with long lifetimes toward the left and short lifetimes toward the right. For samples with emitting species exhibiting different lifetimes, the phasor position will be defined by the fractional contribution of each of the species. In an image, the phasor position of each pixel within a FOV is uniquely defined by the fluorescence decay at that pixel and the acquisition frequency (here, 80 MHz). Pixels with a mixture of bound and free NAD(P)H (excited here by two-photon excitation at 740 nm with a pulsed infrared laser) will be situated below the universal circle at a position between the bound and free forms that depends upon their ratio. Thus, metabolic changes will affect the position of the phasor position of pixels for images of NAD(P)H autofluorescence (10). For example, bacterial response to antibiotic stress is associated with higher ratios of free/bound NAD(P)H (12). In Bacillus subtilis, the switch from growth on a glycolytic to a gluconeogenic carbon source led to a shift toward a higher free/bound NAD(P)H ratio (Fig. S2), reflecting a change from catabolic to anabolic activity.
Figure 2.
Effect of pressure on FLIM/Phasor of NADH for yeast (S. cerevisiae), bacteria (E. coli MG1655), and archaea (A. fulgidus). (A), (C), and (E) are fluorescent images with pixels colored according to their position on the phasor plots (B), (D), and (F), respectively. Pixels are colored according to their phasor positions. Red signifies 0.1 MPa, and yellow and pink signify 100 MPa, corresponding to the circles in the phasor plots. On the top row are images of S. cerevisiae, pink arrows corresponding to foci with high bound/free NAD(P)H ratios. In the middle row are images of E. coli. On the bottom row are images of A. fulgidus. For A. fulgidus, at 100 MPa, most pixels remained within the red circle at atmospheric pressure. Images correspond to a field of view of 13 × 13 μm for yeast and bacteria and 10 × 10 μm for archaea and 256 × 256 pixels. Excitation was 740 nm, and phasor frequency was 80 MHz. At least three fields of view were acquired at each pressure point, and the experiment was repeated three times with the same observations. To see this figure in color, go online.
To demonstrate that such measurements can be made at high pressure, the pressure response of the autofluorescence lifetimes exciting at 740 nm for three different cell types, E. coli, A. fulgidus (type strain VC-16), and S. cerevisiae strain BY1741 (bacteria, archaea, and yeast) was tested in the high-pressure capillary microscope system (Fig. 2). A. fulgidus is ubiquitous in subsurface, high-pressure environments and has been reported to grow up to 60 MPa (18). It is also an obligate anaerobic thermophile (∼83°C maximum growth temperature) (24), whereas the yeast and bacterium are both mesophilic organisms that grow aerobically at atmospheric pressure. Although the high-pressure experiments on yeast and E. coli were carried out aerobically, we maintained anaerobic conditions during the experiment with A. fulgidus by purging the entire system with oxygen-depleted minimal medium before loading the cells.
At atmospheric pressure, most pixels for the yeast cells were positioned at g-values >0.5, indicating a significant fraction of free NAD(P)H (Fig. 2, A and B, red). At 100 MPa, most pixels in the yeast cells shifted slightly to the left, generally toward bound NAD(P)H, although the trajectory of the shift was not linear, but rather described an arc, indicative of heterogeneity beyond that of free and bound NAD(P)H. The pink pixels (Fig. 2 A, right and Fig. 2 B) exhibited phasors with lower g-values, yet similar s-values to that at atmospheric pressure, and were localized in small foci, indicating association of metabolic enzymes into discrete foci upon high-pressure stress. Most other pixels (yellow) shifted left and slightly upward (lower g-values, higher s-values), which, as noted above, indicates heterogeneity of the contributions to yeast autofluorescence. Indeed, yeast autofluorescence is known to have a strong contribution from riboflavin and its derivatives (25) in addition to NAD(P)H. The absorption spectrum of riboflavin (two maxima at 350 and 450 nm), along with its peak emission at 520 nm, would be detected under our imaging conditions (740 nm excitation, GFP filter). We note that the phasor position of FAD, a riboflavin derivative, and that of bound NADH are similar (10,26,27).
In contrast to the strong pressure response of yeast, we found no pressure response of the autofluorescence of the piezophile A. fulgidus (Fig. 2, E and F). Even at atmospheric pressure, the phasor values for its autofluorescence were shifted strongly toward bound either bound NAD(P)H or FAD, and this did not change at 100 MPa, even though the pressure used was higher than its maximum survival pressure (80 MPa) (18). It should be noted that A. fulgidus analyses were conducted at room temperature, well below the optimal growth range for this thermophilic species. Thus, the absence of a response to elevated pressures might be explained by the cells undergoing temperature stress well before pressurization. We are currently developing protocols to implement temperature control in the high-pressure imaging system.
The response of E. coli to pressure was stronger than for the other two microbes (Fig. 2, C and D). At atmospheric pressure compared to the yeast sample, the average phasor position of all pixels was shifted toward free NAD(P)H (Fig. 2 C, left, Fig. 2 D, red), indicative of lower metabolic activity, perhaps because of the switch to minimal medium. In all experiments at 100 MPa, a significant fraction of pixels of NAD(P)H fluorescence in E. coli exhibited a shift of their phasor positions to lower g-values (e.g., Fig. 2 C, right, Fig. 2 D, yellow), indicating a pressure-induced increase in the fraction of bound NAD(P)H (10). Note that the exact position and distribution of the atmospheric phasor values varied somewhat between experiments. Also note that no time dependence of the changes was observed after the 10-min equilibration time before measurements.
The MG1655 reference strain of E. coli used in the experiments in Fig. 2 harbored a pressure-activated restriction endonuclease, Mrr, which leads to a pressure-induced SOS response (17,28,29). To ascertain whether response of E. coli metabolism to pressure was linked to this Mrr-induced SOS response, we carried out FLIM/Phasor analysis on the autofluorescence of an E. coli strain in which the mrr gene had been deleted (mrr−) (15). Comparison of the average intensity images with the phasor maps of an E. coli strain at atmospheric pressure shows that regardless of the overall NAD(P)H content (total intensity), most pixels exhibited similar fractions of bound/free cofactor (Fig. 3, A–C; Fig. S3 A). These were somewhat shifted to lower g-values as compared to Fig. 2, perhaps reflecting differences in the growth rate for this sample. At 100-MPa pressure (Fig. 3, D–F; Fig. S3 B), in contrast to the mrr+ strain, a bimodal pressure response of NAD(P)H lifetimes and intensity was observed. Comparison of the average intensity (which is correlated to the concentration of NAD(P)H) with the phasor patterns revealed that bacterial cells exhibiting a higher fraction of longer-lived, bound NAD(P)H (Fig. 3 E, green pixels, left-shifted) also exhibited a much lower overall autofluorescence intensity (Fig. 3 D, green arrow). In contrast, cells that exhibited higher autofluorescent intensity were those with NAD(P)H phasor values shifted to the right toward shorter-lived, free NAD(P)H (Fig. 3, D–F, yellow pixels and arrow). This shift to a higher fraction of free cofactor indicates decreased metabolic activity in response to pressurization. Thus, the bimodal response in the lifetimes was accompanied by a bimodal response in the intensity of individual cells (Fig. S3 B). Upon return to atmospheric pressure, cells exhibiting low autofluorescence intensity mostly retained the left-shifted phasor pixels (Fig. 3, G–I, green arrows and pixels) and did not exhibit a recovery of their intensity (Fig. 3 C), indicating that they had been irreversibly compromised by pressure.
Figure 3.
Comparison of the pressure response of total autofluorescence intensity with FLIM/Phasor analysis of NAD(P)H for E. coli MG1655mrr−. (A–C) Atmospheric pressure is shown. Fluorescence intensity, phasor map, and phasor plots, respectively, are also shown. The red circle in (C) corresponds to the phasor positions of the red pixels in (B). (D–F) 100 MPa fluorescence intensity, phasor map, and phasor plots, respectively, are shown. The red, green, and yellow circles in (F) correspond to the phasor positions of the red, green, and yellow pixels in (E), respectively. The green arrows indicate a cell with a low autofluorescence intensity and left-shifted phasor positions, whereas the yellow arrows indicate a cell with a high autofluorescence intensity and right-shifted phasor values. Red pixels in the phasor plot represent positions that are unchanged with respect to atmospheric pressure. (G–I) A return to 0.1 MPa (atmospheric pressure) is shown. Fluorescence intensity, phasor map, and phasor plots, respectively, are also shown. The red, green, and yellow circles in (I) correspond to the phasor positions of the red, green, and yellow pixels in (H), respectively. Green arrows indicate a cell with a low autofluorescence intensity and left-shifted phasor positions, whereas yellow arrows indicate a cell with a high autofluorescence intensity and right-shifted phasor values. Red arrows represent a cell with a high intensity and phasor positions that are equivalent to those at atmospheric pressure. Images correspond to a field of view of 13 × 13 μm and 256 × 256 pixels. Excitation was 740 nm, and phasor frequency was 80 MHz. At least three fields of view were acquired at each pressure point. Intensity scales for images in (A), (D), and (G) are arbitrary. To see this figure in color, go online.
One possible reason for the decrease in intensity in these cells is pressure-induced oxidation of a large fraction of the NAD(P)H to NAD(P), which is nonfluorescent. Another possibility is that the viability of these bacteria were compromised by high pressure and that much of their free NAD(P)H had diffused out of the cells because of pressure-induced membrane alteration. Although absolute concentrations of NAD(P)H can be extracted from phasor data (30), this information would not allow us to distinguish between these two hypotheses, although such analyses would be useful in other circumstances. The limited reversibility in both lifetime and intensity for the cells with left-shifted phasor values supports the hypothesis that the loss of intensity is due to leakage through compromised membranes. Because the enzyme-bound fraction is less likely to diffuse out of these cells because of its larger molecular weight than the free cofactor, the fraction of bound NAD(P)H would be expected to increase in these cells.
Some of the cells that retained their high autofluorescence intensity upon return to atmospheric pressure also retained the right-shifted pixels corresponding to lower metabolic activity (Fig. 3, G–I, yellow pixels and arrows). In contrast, in other cells with high autofluorescence intensity upon pressure release, the pixels recovered their original phasor positions (Fig. 3, G–I, red pixels and arrows), indicating complete reversibility of the pressure effects on metabolic state for these cells. Hence, in this E. coli mrr− strain, a significant fraction of the cell population can sustain high-pressure treatment and rapidly resume normal metabolic activity after pressure release. These FLIM/Phasor results highlight the stochastic nature and complexity of the response of bacterial metabolism to pressure. They also establish that phenomena other than the SOS response triggered by the Mrr endonuclease participate in pressure-induced loss of cell viability.
Pressure effects on the bacterial partition machinery
Large-scale transposon mutagenesis on a moderate piezophile, Photobacterium profundum, revealed that the largest fraction of loci-conferring pressure sensitivity to this organism were implicated in chromosome structure and function (31). This underscores the importance of proteins involved in chromosome function in pressure adaptation. Hence, we were interested in ascertaining the effects of pressure on bacterial proteins involved in such processes. As an example of such a system, we characterized the effects of pressure on the ParB partition complex protein of E. coli using our high-pressure capillary microscopy system coupled with sN&B (13) and RICS (14). In E. coli, the type I partition systems are involved in segregation of the F and P1 low-copy number plasmids (32). The ParB protein recognizes parS sites on the plasmid and recruits the ParA ATPase. ParB is known to form dimers organized into foci (one per plasmid origin of replication) containing ∼200 ParB monomers (33,34). These foci are thought to correspond to liquid-liquid phase-separated droplets (16).
To monitor pressure effects on ParB in live E. coli cells, we employed a strain in which ParB, fused to monomeric mVenus fluorescent protein, was expressed from its natural locus on the F plasmid (16). This strain expressed, as well, a chromosomal mCherry fusion of the nucleoid-associated protein, HU, to allow visualization of the entire nucleoid. At atmospheric pressure, discrete ParB-mVenus foci were apparent in all cells (Fig. 4 A, green; Fig. S4), whereas the HU-mCherry protein (Fig. 4 A, red) was distributed homogeneously on the nucleoid. Application of 100-MPa pressure led to disruption of the ParB clusters in all cells (Fig. 4 B, green; Fig. S4). In contrast, pressure caused the HU protein to form foci in ∼60% of cells (Fig. 4 B, red; Fig. S4). Because this E. coli strain also expressed the pressure-activated Mrr endonuclease, the formation of HU foci probably corresponded to DNA condensation after double-strand breaks and induction of the SOS response by Mrr (29). After pressure release, some bacterial cells appeared to recover, as evidenced by the reappearance of ParB-mVenus foci in 24% of cells (Fig. 4 C, green; Fig. S4) and the dispersion of the HU-mCherry signal in 29% of cells (Fig. 4 C, red; Fig. S4), whereas others did not. This indicates for these cells irreversible condensation of the chromosome, likely due to the SOS response, was accompanied by a lack of reassembly of ParB at the plasmidic origin of replication.
Figure 4.
Effect of pressure on ParB partition protein clusters and on the chromosome in E. coli. (A–C) Images correspond to the merging of the fluorescence intensity images of ParB-mVenus (in green) and HU-mCherry (in red) at (A) atmospheric pressure (0.1 MPa), (B) under pressure (100 MPa), and (C) back to atmospheric pressure (0.1 MPa). Images are 13 × 13 μm and 256 × 256 pixels. (D) and (E) show a comparison of the vertical autocorrelation profile from RICS analyses at 0 MPa (black squares), 100 MPa (red triangles), and back to 0.1 MPa (gray diamonds) of (D) freely diffusing GFPmut2 in E. coli cytoplasm and (E) mVenus fusion with ParB proteins in E. coli. (F) Molecular brightness from the sN&B analyses of free diffusing GFPmut2 (green) and ParB-mVenus (yellow) at atmospheric pressure (0.1 MPa), high pressure (100 MPa), and after pressure is released (back to 0 MPa). GFPmut2 is expressed in the E. coli MG1655 chromosome from the inducible PBAD promotor with 0.4% arabinose, and HU-mCherry/ParB-mVenus protein fusions are constitutively expressed in E. coli DLT3053 with the plasmid pJYB234. The experiment was repeated two times with eight FOVs. Each RICS vertical autocorrelation curve is the average vertical autocorrelation profile of eight FOVs. The brightness values correspond to the average brightness values of the pixels inside bacteria of 16 FOVs per pressure point with an average of 20–25 bacteria per FOV. Brightness values from all FOVs from 2 days of experiments were averaged and corrected for background as described in Materials and Methods. Error bars represent the standard deviation of the mean for all 16 FOVs. To see this figure in color, go online.
RICS analysis of an E. coli MG1655 strain expressing free GFPmut2, a fast-maturing GFP variant (35), from a plasmid revealed that pressure did not affect the diffusion of free-monomeric GFPmut2 (Fig. 4 D). This is an interesting observation because it indicates that the viscosity of the cytosol does not change significantly at 100-MPa pressure. In contrast, the average ParB-mVenus diffusion was faster under pressure and after release (Fig. 4 E), consistent with dissociation of the foci at high pressure and limited recovery upon return to 0.1 MPa. In addition, the molecular brightness of mVenus fused to ParB calculated by sN&B decreased two-fold under pressure, indicating that pressure led to the dissociation of ParB-mVenus dimers to monomers (Fig. 4 F). Note that the molecular brightness of free GFPmut2 is not significantly affected by pressure in vivo. Thus, pressure appeared to dissociate both the ParB dimers and the foci themselves to form monomers that diffuse freely in the cytoplasm. As noted, it has been suggested that the ParB foci correspond to liquid-liquid phase-separated droplets (16), which, in this case, were disrupted by HP.
Conclusions
We have shown for the first time, to our knowledge, that quantitative, high-resolution two-photon imaging modalities can be applied to study the response of live bacterial, archaeal, and eukaryotic microbial cells to high hydrostatic pressure. Significant changes in metabolic state and protein interactions were observed for the mesophilic organisms in response to pressure, whereas the extremophile archaeon exhibited no pressure-induced change in metabolism. Future work will focus on the underlying molecular mechanisms for these changes and how they depend upon pressure. The fact that pathogens such as certain strains of E. coli evolve tolerance to and even the ability to grow at high pressure in the context of pressure treatment of food products (6,36, 37, 38, 39) underscores the importance of understanding the mechanism underlying these adaptations. Moreover, to the extent that extremophilic organisms can be genetically manipulated, as is the case with P. profundum SS9 (31), it will be possible to engineer fluorescent protein and promoter fusions in these organisms, opening up large avenues of investigation. Given the sheer scale of life in the deep biosphere and our limited knowledge of the molecular mechanisms at play, high-pressure, high-resolution quantitative imaging will be extremely useful for understanding molecular and cellular adaptations to the environments of the deep biosphere.
Author Contributions
A.C.B. performed research. A.L. contributed to system design. N.D. interpreted results and wrote the article. K.L.R. provided samples and assisted with sample preparation. C.A.R. designed research, interpreted results, wrote the article.
Acknowledgments
We thank Jean-Yves Bouet for providing the E. coli strain DLT3053 with the plasmid pJYB234.
This research was funded in part by the Alfred P. Sloan Foundation through the Deep Life Community of the Deep Carbon Observatory.
Editor: Jochen Mueller.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.04.017.
Supporting Material
References
- 1.Rampelotto P.H. Extremophiles and extreme environments. Life (Basel) 2013;3:482–485. doi: 10.3390/life3030482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bar-On Y.M., Phillips R., Milo R. The biomass distribution on earth. Proc. Natl. Acad. Sci. USA. 2018;115:6506–6511. doi: 10.1073/pnas.1711842115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.L’Haridon S., Corre E., Jebbar M. Complete genome sequence of the halophilic methylotrophic methanogen archaeon Methanohalophilus portucalensis strain FDF-1T. Genome Announc. 2018;6:e01482-17. doi: 10.1128/genomeA.01482-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duffy M. HPP keeps food safe, while extending shelf life. Food Safety Tech. 2018 [Google Scholar]
- 5.Vanlint D., Rutten N., Aertsen A. Emergence and stability of high-pressure resistance in different food-borne pathogens. Appl. Environ. Microbiol. 2012;78:3234–3241. doi: 10.1128/AEM.00030-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vanlint D., Rutten N., Aertsen A. Exposure to high hydrostatic pressure rapidly selects for increased RpoS activity and general stress-resistance in Escherichia coli O157:H7. Int. J. Food Microbiol. 2013;163:28–33. doi: 10.1016/j.ijfoodmicro.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Vass H., Black S.L., Allen R.J. A multipurpose modular system for high-resolution microscopy at high hydrostatic pressure. Rev. Sci. Instrum. 2010;81:053710. doi: 10.1063/1.3427224. [DOI] [PubMed] [Google Scholar]
- 8.Müller J.D., Gratton E. High-pressure fluorescence correlation spectroscopy. Biophys. J. 2003;85:2711–2719. doi: 10.1016/s0006-3495(03)74694-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicolini C., Celli A., Winter R. Pressure tuning of the morphology of heterogeneous lipid vesicles: a two-photon-excitation fluorescence microscopy study. Biophys. J. 2006;91:2936–2942. doi: 10.1529/biophysj.106.088716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stringari C., Nourse J.L., Gratton E. Phasor fluorescence lifetime microscopy of free and protein-bound NADH reveals neural stem cell differentiation potential. PLoS One. 2012;7:e48014. doi: 10.1371/journal.pone.0048014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Digman M.A., Caiolfa V.R., Gratton E. The phasor approach to fluorescence lifetime imaging analysis. Biophys. J. 2008;94:L14–L16. doi: 10.1529/biophysj.107.120154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhattacharjee A., Datta R., Hochbaum A.I. Metabolic fingerprinting of bacteria by fluorescence lifetime imaging microscopy. Sci. Rep. 2017;7:3743. doi: 10.1038/s41598-017-04032-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Digman M.A., Dalal R., Gratton E. Mapping the number of molecules and brightness in the laser scanning microscope. Biophys. J. 2008;94:2320–2332. doi: 10.1529/biophysj.107.114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown C.M., Dalal R.B., Gratton E. Raster image correlation spectroscopy (RICS) for measuring fast protein dynamics and concentrations with a commercial laser scanning confocal microscope. J. Microsc. 2008;229:78–91. doi: 10.1111/j.1365-2818.2007.01871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tesfazgi Mebrhatu M., Wywial E., Aertsen A. Evidence for an evolutionary antagonism between Mrr and Type III modification systems. Nucleic Acids Res. 2011;39:5991–6001. doi: 10.1093/nar/gkr219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Gall A., Cattoni D.I., Nollmann M. Bacterial partition complexes segregate within the volume of the nucleoid. Nat. Commun. 2016;7:12107. doi: 10.1038/ncomms12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bourges A.C., Torres Montaguth O.E., Royer C.A. High pressure activation of the Mrr restriction endonuclease in Escherichia coli involves tetramer dissociation. Nucleic Acids Res. 2017;45:5323–5332. doi: 10.1093/nar/gkx192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliver G. Rensselaer Polytechnic Institute; 2019. Exploring microbial growth of a model extremophile, Archaeoglobus fulgidus, at elevated pressures. PhD thesis. [Google Scholar]
- 19.Faure L.M., Fiche J.-B., Mignot T. The mechanism of force transmission at bacterial focal adhesion complexes. Nature. 2016;539:530–535. doi: 10.1038/nature20121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Espenel C., Margeat E., Milhiet P.E. Single-molecule analysis of CD9 dynamics and partitioning reveals multiple modes of interaction in the tetraspanin web. J. Cell Biol. 2008;182:765–776. doi: 10.1083/jcb.200803010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferguson M.L., Le Coq D., Royer C.A. Absolute quantification of gene expression in individual bacterial cells using two-photon fluctuation microscopy. Anal. Biochem. 2011;419:250–259. doi: 10.1016/j.ab.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 22.Digman M.A., Brown C.M., Gratton E. Measuring fast dynamics in solutions and cells with a laser scanning microscope. Biophys. J. 2005;89:1317–1327. doi: 10.1529/biophysj.105.062836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patra S., Anders C., Winter R. Osmolyte effects on the conformational dynamics of a DNA hairpin at ambient and extreme environmental conditions. Angew. Chem. Int. Ed. Engl. 2017;56:5045–5049. doi: 10.1002/anie.201701420. [DOI] [PubMed] [Google Scholar]
- 24.Beeder J., Nilsen R.K., Lien T. Archaeoglobus fulgidus isolated from hot North Sea oil field waters. Appl. Environ. Microbiol. 1994;60:1227–1231. doi: 10.1128/aem.60.4.1227-1231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maslanka R., Kwolek-Mirek M., Zadrag-Tecza R. Autofluorescence of yeast Saccharomyces cerevisiae cells caused by glucose metabolism products and its methodological implications. J. Microbiol. Methods. 2018;146:55–60. doi: 10.1016/j.mimet.2018.01.017. [DOI] [PubMed] [Google Scholar]
- 26.Wright B.K., Andrews L.M., Gratton E. NADH distribution in live progenitor stem cells by phasor-fluorescence lifetime image microscopy. Biophys. J. 2012;103:L7–L9. doi: 10.1016/j.bpj.2012.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stringari C., Cinquin A., Gratton E. Phasor approach to fluorescence lifetime microscopy distinguishes different metabolic states of germ cells in a live tissue. Proc. Natl. Acad. Sci. USA. 2011;108:13582–13587. doi: 10.1073/pnas.1108161108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aertsen A., Van Houdt R., Michiels C.W. An SOS response induced by high pressure in Escherichia coli. J. Bacteriol. 2004;186:6133–6141. doi: 10.1128/JB.186.18.6133-6141.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghosh A., Passaris I., Aertsen A. Cellular localization and dynamics of the Mrr type IV restriction endonuclease of Escherichia coli. Nucleic Acids Res. 2014;42:3908–3918. doi: 10.1093/nar/gkt1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma N., Digman M.A., Gratton E. Measurements of absolute concentrations of NADH in cells using the phasor FLIM method. Biomed. Opt. Express. 2016;7:2441–2452. doi: 10.1364/BOE.7.002441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lauro F.M., Tran K., Bartlett D.H. Large-scale transposon mutagenesis of Photobacterium profundum SS9 reveals new genetic loci important for growth at low temperature and high pressure. J. Bacteriol. 2008;190:1699–1709. doi: 10.1128/JB.01176-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinto U.M., Pappas K.M., Winans S.C. The ABCs of plasmid replication and segregation. Nat. Rev. Microbiol. 2012;10:755–765. doi: 10.1038/nrmicro2882. [DOI] [PubMed] [Google Scholar]
- 33.Funnell B.E. ParB partition proteins: complex formation and spreading at bacterial and plasmid centromeres. Front. Mol. Biosci. 2016;3:44. doi: 10.3389/fmolb.2016.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez A., Cattoni D.I., Bouet J.Y. Stochastic self-assembly of ParB proteins builds the bacterial DNA segregation apparatus. Cell Syst. 2015;1:163–173. doi: 10.1016/j.cels.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 35.Cormack B.P., Valdivia R.H., Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 36.Vanlint D., Mitchell R., Aertsen A. Rapid acquisition of Gigapascal-high-pressure resistance by Escherichia coli. MBio. 2011;2 doi: 10.1128/mBio.00130-10. e00130–e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hauben K.J., Bartlett D.H., Michiels C.W. Escherichia coli mutants resistant to inactivation by high hydrostatic pressure. Appl. Environ. Microbiol. 1997;63:945–950. doi: 10.1128/aem.63.3.945-950.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marietou A., Nguyen A.T.T., Bartlett D.H. Adaptive laboratory evolution of Escherichia coli K-12 MG1655 for growth at high hydrostatic pressure. Front. Microbiol. 2015;5:749. doi: 10.3389/fmicb.2014.00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aertsen A., Michiels C.W. Mrr instigates the SOS response after high pressure stress in Escherichia coli. Mol. Microbiol. 2005;58:1381–1391. doi: 10.1111/j.1365-2958.2005.04903.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.