Abstract
Medin, a 50-amino-acid cleavage product of the milk fat globule-EGF factor 8 protein, is one of the most common forms of localized amyloid found in the vasculature of individuals older than 50 years. Medin induces endothelial dysfunction and vascular inflammation, yet despite its prevalence in the human aorta and multiple arterial beds, little is known about the nature of its pathology. Medin oligomers have been implicated in the pathology of aortic aneurysm, aortic dissection, and more recently, vascular dementia. Recent in vitro biomechanical measurements found increased oligomer levels in aneurysm patients with altered aortic wall integrity. Our results suggest an oligomer-mediated toxicity mechanism for medin pathology. Using lipid bilayer electrophysiology, we show that medin oligomers induce ionic membrane permeability by pore formation. Pore activity was primarily observed for preaggregated medin species from the growth-phase and rarely for lag-phase species. Atomic force microscopy (AFM) imaging of medin aggregates at different stages of aggregation revealed the gradual formation of flat domains resembling the morphology of supported lipid bilayers. Transmission electron microscopy images showed the coexistence of compact oligomers, largely consistent with the AFM data, and larger protofibrillar structures. Circular dichroism spectroscopy revealed the presence of largely disordered species and suggested the presence of β-sheets. This observation and the significantly lower thioflavin T fluorescence emitted by medin aggregates compared to amyloid-β fibrils, along with the absence of amyloid fibers in the AFM and transmission electron microscopy images, suggest that medin aggregation into pores follows a nonamyloidogenic pathway. In silico modeling by molecular dynamics simulations provides atomic-level structural detail of medin pores with the CNpNC barrel topology and diameters comparable to values estimated from experimental pore conductances.
Significance
Medin is the main component of aortic medial amyloid, one of the most common forms of localized amyloid in the human body. Despite its relevance, little is known about the molecular mechanisms of medin toxicity. Here, we show that medin oligomers form membrane pores, which induce unregulated ionic currents through a lipid membrane and could alter cellular homeostasis. These results suggest a potential mechanism for medin involvement in vascular dysfunction.
Introduction
Amyloidogenic proteins (amyloids) are characterized by their spontaneous aggregation into β-sheet-containing fibers and display a cross-β diffraction pattern when observed by x-ray diffraction (1,2). These proteins exhibit physiological roles in microorganisms and humans (3, 4, 5) and have potential applications as nanostructured materials (6, 7, 8, 9, 10). However, the main interest in this class of proteins stems from their involvement in over 30 human diseases (2,11), including Alzheimer’s disease (linked to the amyloid-β [Aβ] and Tau proteins) (12, 13, 14, 15), Parkinson’s disease (linked to α-synuclein) (16,17), and light chain amyloidosis (18,19).
Aortic medial amyloid (AMA) is one of the most common forms of aging-related amyloid and is primarily located within the medial layer of the aorta, especially in individuals older than 50 years (20, 21, 22, 23, 24). Medin, the principal protein component of AMA (20), is a 50-amino-acid cleavage product of the milk fat globule-EGF factor 8 protein and shares 16% of its global sequence with the Aβ peptide (23,25), the putative agent underlying Alzheimer’s disease (26). Medin and Aβ have high local similarities within residues 23–29 (23), a section that incorporates the turn in NMR-based U-shaped motifs that is critical for Aβ fibril and pore formation (23,27).
Despite its prevalence in the human vasculature, little is known about medin pathology when compared with other amyloid proteins. Medin may have a role in thoracic aortic aneurysm and dissection (28,29), a leading cause of rupture of the aorta (29). Furthermore, AMA deposits are found closely associated with elastic structures such as the vascular extracellular matrix (30), which can influence arterial stiffness and development of arterial hypertension (31). We recently showed that medin is present in the cerebral arteries of elderly brain donors, with higher cerebrovascular medin in vascular dementia patients versus cognitively normal subjects (32). We also showed that physiologic doses of medin induce endothelial dysfunction, endothelial cell (EC) immune activation, and cytotoxicity (33), which appear to enhance neuroinflammation (32), pointing to medin’s potential role in the pathophysiology of cerebrovascular disease and vascular dementia. Importantly, both medin and Aβ induce profound endothelial dysfunction and oxidative stress in human arterioles (33,34), suggesting a potential for in vivo interactions between both amyloids. Our previous ex vivo experiments suggest that medin-induced EC oxidative stress is a potential mechanism of EC dysfunction (35); however, the underlying mechanisms of medin-induced pathology remain poorly understood and have rarely been addressed (35).
Although amyloid fibers are the defining characteristic of amyloid pathology and form the basis for diagnosis of amyloidogenic diseases (36), it is not fully understood whether fibers or intermediate, nonfibrillar species formed during the aggregation process are more important to disease mechanisms. Significant ex vivo and occasional in vivo evidence suggests that oligomeric species are toxic (37, 38, 39, 40) and induce membrane permeability that leads to cell dysfunction (41, 42, 43, 44, 45). One of the most plausible mechanisms leading to membrane permeability is the amyloid pore hypothesis (46, 47, 48, 49). According to this mechanism, oligomeric species self-assemble into membrane pores that permeabilize the plasma membrane, causing an abrupt change in cell ionic concentration that leads to loss of cellular homeostasis. The pore activity of multiple amyloidogenic proteins has been primarily characterized using suspended planar lipid bilayers (also known as black lipid membranes (BLMs)) (49, 50, 51, 52, 53, 54, 55, 56, 57) but also by optical patch clamping (58) and cell electrophysiology (50). Unlike ion channels, amyloid pores do not have a unitary conductance, are not gated (59,60), and have doughnut-like morphologies (27,48,49,61, 62, 63).
Here, we focus on an oligomer-mediated toxicity mechanism of medin proteins: membrane permeability induced by pore formation. Using biophysical techniques and in silico modeling, we demonstrate that medin induces pore activity in anionic membranes with lipid compositions previously validated as useful model systems of Aβ membrane pore formation (51,64,65). Our data indicate that the observed pore activity depends on medin aggregation, which is dependent on agitation. Pore activity was primarily observed for preaggregated species and seldom for unaggregated, lag-phase species. Medin aggregates emitted lower thioflavin T (ThT) fluorescence than Aβ, and their circular dichroism (CD) spectra revealed predominantly unfolded species and suggested the presence of β-sheets. After medin aggregation, atomic force microscopy (AFM) imaging revealed the formation of planar patches resembling the morphologies of supported lipid bilayers formed from liposome rupture and fusion (66). High-resolution transmission electron microscopy (TEM) images displayed two distinct morphologies resulting from the aggregation of medin oligomers. Amyloid fibers were observed neither by AFM nor by TEM imaging. In silico modeling of medin membrane pores yielded stable CNpNC barrel topology (where C and N are the C-terminal and N-terminal strands, respectively, and p denotes a central pore), with pore diameters in the range of the experimental values determined from BLM electrophysiology data. These results—the first, to our knowledge, to provide evidence for the formation of medin membrane pores in a lipid bilayer—provide a potential mechanism for medin oligomer-induced vascular pathology (29).
Materials and Methods
Protein preparation
Medin samples were prepared by recombinant methods, as described elsewhere (67). Briefly, medin was expressed using pOPINS-medin in Lemo 21 (DE3) cells, induced with isopropyl-β-D-thiogalacto-pyranoside (1 mM) for 16 h at 18°C. Cells were harvested by centrifugation and pellets resuspended in 6 M guanidine hydrochloride (GdmCl), 20 mM sodium phosphate, and 20 mM NaCl (pH 8.0). Cells were homogenized and cell debris removed by centrifugation. The supernatant was loaded onto a nickel-nitrilotriacetic acid (Ni2+-NTA) column, washed with 6 M GdmCl (pH 8 and pH 6), and eluted with 6 M GdmCl (pH 2). Fusion protein was buffer exchanged into 20 mM Tris-Cl, 0.5 M NaCl (pH 7.4) and the His6-SUMO tag removed with SUMO protease I. The protease was removed using a Ni2+-NTA column, and the flowthrough containing medin was buffer exchanged into 150 mM KCl, 10 mM HEPES, 1 mM MgCl2 solution, referred to hereinafter as “HEPES buffer,” and flash frozen. Medin was confirmed at >95% purity by sodium dodecyl sulfate polyacrylamide gel electrophoresis.
ThT protocol
The as-received medin samples (1.5 mL microcentrifuge tubes containing 500 μL of 50 μM medin in HEPES buffer) were stored at −80°C upon arrival and used as needed. Before use, the samples were defrosted in 37°C water baths for 15 min. Subsequently, 240 μL of the defrosted medin samples was mixed with 336 μL HEPES buffer and 24 μL ThT (to a concentration of 20 μM ThT and 20 μM medin), and then 100 μL was added to wells of 96-well plates (black walled, clear bottom; Corning, Corning, NY). Serial dilutions were performed to produce lower 10 and 5 μM medin concentrations by adding additional buffer and ThT. Three wells were prepared as replicates for each treatment. The plates were incubated for several hours at 37°C inside a Clariostar microplate reader (BMG Labtech, Cary, NC) undergoing 5 min cycles of 4 min vigorous shaking (300 RPM orbital shaking) and 1 min of ThT (450 nm excitation/490 nm emission) data acquisition. The resulting aggregation curves of ThT fluorescence versus time were fitted using a sigmoidal function to extract kinetic aggregation parameters (23,68,69).
| (1) |
where t is time, t1/2 is the time to reach the half maximum of ThT fluorescence, y0 is the initial fluorescence intensity, A is the amplitude of the maximal intensity, and k is the rate constant. The lag-phase time was calculated from the fitting parameters above, using
| (2) |
For Aβ1–42 experiments, 1 mg of Aβ1–42 in powder form (>90% purity; Anaspec, Fremont, CA) was dissolved in 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP; Sigma-Aldrich, St. Louis, MO) for 30 min at 25°C to a 250 μM concentration and aliquoted in 50 μL microcentrifuge tubes. After 2 h of HFIP evaporation from the aliquoted samples using a Thermo Scientific Savant DNA 120 Speedvac (Thermo Fisher Scientific, Waltham, MA), the samples were stored at −80°C and used as needed. For ThT experiments, the samples were resolubilized in 40 μL buffer (50 mM KCl and 10 mM HEPES) and 10 μL (1%) NH4OH, vortexed for 5 min, and sonicated for 15 min in a 1.9 L ultrasonic bath (Thermo Fisher Scientific). These samples were then mixed in another buffer (150 mM KCl, 10 mM HEPES) and ThT to a 20 μM Aβ1–42 (20 μM ThT) solution. Wells of a 96-well plate were prepared as described for medin. ThT fluorescence was read every 5 min on the Clariostar plate reader, with 5 s of 200 RPM orbital shaking between each read at 25°C.
Medin aggregation protocol
To accelerate medin aggregation into the growth phase, medin samples were prepared at high agitation rates. Agitation was performed on an orbital shaker (Labline, Melrose Park, IL) at typical rates of 300 RPM in 1.5 mL microcentrifuge tubes containing 20 μM medin in 600 μL HEPES buffer. To monitor medin aggregation, 96-well plates were placed alongside the microcentrifuge tube sample, with four of the wells containing 20 μM medin samples prepared using a similar protocol as the protocol followed for ThT experiments; 20 μM ThT was used, and the amount of HEPES buffer was adjusted accordingly. The plate was transferred to the plate reader at regular intervals (typically 15–30 min) for ThT data acquisition, and agitation of the microcentrifuge tube was stopped during this period of time.
CD spectroscopy
Far-ultraviolet CD spectra were recorded on a JASCO J-1100 spectropolarimeter (JASCO, Japan) at 25°C. Spectra were recorded over the 260–192 nm range at a scan rate of 50 nm/min with 1 s integration time. Spectra were recorded as the average of 10 scans using a 0.2 mm demountable cuvette containing a solution with 50 μM HEPES and the salt content described above.
Dynamic light scattering
A series of 16 measurements were taken at 25°C on a Zetasizer Nano dynamic light scattering (DLS) machine from Malvern Instruments (Malvern, UK) using back scatter. Averaged profiles were produced from three sets of measurements.
Lipid preparation
All phospholipids were purchased from Avanti Polar Lipids (Alabaster, AL). BLMs were prepared in a 1 mL glass vial using an equimolar mixture of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylethanolamine (POPE) and 1,2-dioleoyl-sn-glycero-3-phosphatidyl-L-serine (DOPS) dissolved in chloroform (Fisher Scientific, Fair Lawn, NJ). Chloroform from the vial was evaporated in vacuum for 1 h using a Rotary Evaporator (Heidolph North America, Elk Grove Village, IL). The lipid film deposited on the vial was re-dissolved in 50 μL n-Decane (from Fisher Scientific) to a final lipid concentration of 30 mg/mL.
BLM recording
All measurements were conducted using a Planar Lipid Bilayer Workstation (Warner Instruments, Hamden, CT), equipped with a BC-535 amplifier (from Warner) and an Axon Digidata 1550B digitizer (Molecular Devices, San Jose, CA) run by Clampex 10.6.2.2 data acquisition software (Molecular Devices). The analog signal acquired in gap free mode was first filtered by a 5 kHz eight-pole Bessel filter, followed by a second Bessel filter (four pole) set at 1 kHz (effective cutoff frequency of both filters ∼0.98 kHz), and then sampled at a 10 kHz sampling rate (5 kHz Nyquist frequency), which ensured the absence of aliasing. Typically, the flickering in our traces had frequencies ≪0.1 kHz, which could readily be resolved without significant distortion. A few events with frequencies >0.3 kHz, not considered in our analysis, had reduced amplitudes because of filtering. Another low-pass filter (100 Hz) was applied to all traces postrecording. We used Ag/AgCl2 electrodes, prepared by first polishing Ag wire electrodes with sandpaper, followed by washing of the electrodes with Milli-Q water and ethanol, and final chlorination by submersing the electrodes in bleach for 15 min. The Ag/AgCl2 electrodes were then placed in the BLM apparatus for measurements.
BLM formation
We used glass capillary tubes (whose end tip had been melted into a small knob by the flame of a Bunsen burner) to paint re-dissolved lipid onto the 200 μm diameter aperture of a 1 mL delrin “Classic Bilayer cup” (Warner Instruments). 1 mL of HEPES buffer was added on each side of the aperture. Further painting of lipid on the aperture was usually required for bilayer formation. Only lipid bilayers that were stable for at least 10 min with a capacitance over 80 pF and a baseline conductance below 20 pS were used for medin experiments. After the formation of a stable bilayer, medin sample was added to the delrin cup until the protein concentration was 12.5 μM. The resulting suspension was briefly mixed by repeated pipetting immediately thereafter. 100 mV was typically used as the bias voltage, but lower values were used occasionally if the current was close to the amplifier saturation current. Successful bilayers were recorded for several hours (typically up to 4 h, including reforming bilayers after breakages).
Data analysis
Clampfit (Molecular Devices) was used to analyze pore conductances, opening times, and mean ionic currents. The conductance of a step event was determined from the mean value of a step in the I vs. t trace. Conductances were also found from the peaks of all-point histograms (these plot the number of data points found in a given current interval of a trace). Because two adjacent data points in a trace are separated by 0.1 ms, the total number of data points in an interval gives the time that a membrane was “open” in a certain conductance range. From these conductance data, the pore diameters, d, were estimated using Eq. 3 (54,68,70,71):
| (3) |
with d0 = G/2σ, G the single-pore conductance, σ the solution conductivity (∼1.5 S/m for 150 mM KCl), and L the pore length (∼6 nm). This model assumes a hole with cylindrical geometry through the membrane for the pore, with the hole containing buffer solution. A similar model was used to estimate the diameters of Aβ1–42 pores in cellular membranes (50). Ionic permeability values were compared using repeated measures analysis of variance after natural log transformation to make the data normally distributed (Sigmastat 3.5; Systat Software, San Jose, CA). Post hoc pairwise comparison was done using the Holm-Sidak method. Significant value was set to p < 0.05 (two-sided).
AFM imaging
For AFM imaging, 100 μL was removed from the microcentrifuge tube taken at times corresponding to different stages of the ThT plate aggregation: before agitation, immediately after detecting an increase of fluorescence in the wells, and 2 h after the plateau of the aggregation curve was reached. 5 μL of each of these medin samples was added on a freshly peeled mica surface (muscovite mica V-1 quality from Electron Microscopy Sciences, Hatfield, PA) glued to a metal specimen disk (Ted Pella, Redding, CA) and dried for 3 h inside a vacuum desiccator. For AFM imaging, 60 μL of HEPES buffer was added on mica, and the sample was transferred to the AFM sample stage. AFM imaging was performed in PeakForce Tapping mode using the fluid cell of a Multimode 8 AFM (Bruker, Santa Barbara, CA) equipped with a NanoScope V controller (Bruker). PEAKFORCE-HIRS-F-B cantilevers (Bruker) were used. Values of 0.079 N/m were measured for the cantilever spring constant using the Thermal tune calibration method (72) implemented in the AFM used. The nominal tip radius is 1 nm, according to specifications by the manufacturer. All image analysis was performed using the NanoScope analysis software v 1.7 (Bruker). The particle analysis option included in this software was used to estimate the volume of medin species.
TEM imaging
Aggregated samples (5 μL) were loaded onto carbon-coated copper grids and negatively stained with 4% uranyl acetate before visualization on a Tecnai 10 electron microscope (FEI Company, Hillsboro, OR) at 120 kV. Image J (Analyze Particles) was used to perform size analysis of the compact oligomers. The distribution of oligomer areas was further fitted using a Gaussian function to find the oligomer diameter.
Molecular dynamics simulations
The U-shaped medin monomer structure was obtained from the homology model of medin fibril generated in Modeler using the Aβ1–40 protofibril (Protein Data Bank, PDB: 2LMN) as a template (23). The U-shaped motif of medin has a turn at Asp25-Lys30. To construct the β-barrel structure, the U-shaped peptide was inclined ∼37° relative to the pore axis, and then an 18-fold rotational symmetry operation was performed with respect to the pore axis, creating an 18-mer medin barrel (Fig. S1). Depending on the direction of the rotational symmetry, two medin barrels with different topologies can form: CNpNC and NCpCN, where N and C denote the N- and C-terminal strands, respectively, and p denotes the solvated central pore. The former has a central pore enclosed by the N-terminal strands, whereas the latter’s pore is enclosed by the C-terminal strands. The medin barrel is embedded in a zwitterionic lipid bilayer containing 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) phospholipid and an anionic lipid bilayer composed of POPE/DOPS (1:1 molar ratio) lipids. The unit cell containing the medin barrel and lipid bilayer was solvated with TIP3P water at both sides and neutralized by cations Mg2+, K+, Ca2+, and Zn2+. In addition to the counterions, the system has a total cation concentration near 100 mM. In the initial construction of the system, the updated CHARMM (73) all-atom additive force field for lipids (C36) (74) was used to generate the set of starting points and to relax the systems to a production ready stage. Our simulations closely followed the same protocol for the Aβ pore simulations as in our previous works (51,75,76). In the pre-equilibrium, we performed a series of minimization and dynamic cycles for the initial configurations to gradually relax the solvents around the harmonically restrained peptides with electrostatic cutoffs (12 Å). At the final pre-equilibrium stage, the harmonic restraints on the backbones of medin were gradually removed through dynamic cycles with the full Ewald electrostatics calculation. The production runs were performed with the Langevin temperature control, which maintains the constant temperature at 303 K, and the Nosé-Hoover Langevin piston pressure control, which sustains the pressure at 1 atm. For t < 50 ns, our simulation employed the constant number of atoms, pressure, surface area, and temperature ensemble with a constant normal pressure applied in the direction perpendicular to the membrane. After t = 50 ns, the simulations employed the NPT ensemble. Production runs of 1 μs for the starting points with the NAMD code (77) were performed on a Biowulf cluster at the National Institutes of Health. Averages were taken after 100 ns, discarding initial transients. Analysis was performed with the same CHARMM (73) programming package as used in the initial construction.
Results
Medin aggregation in pore-forming environments
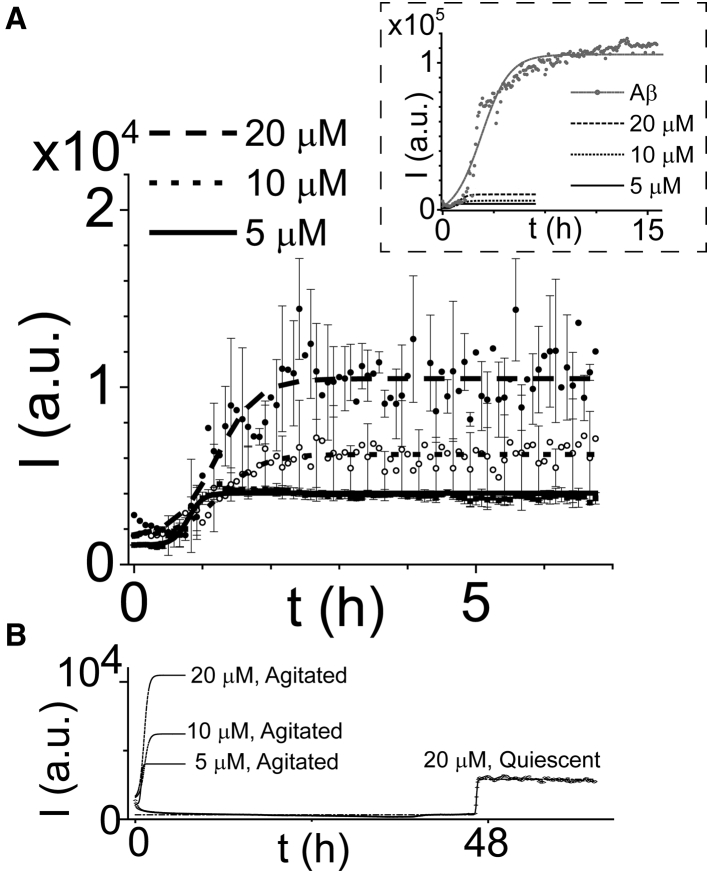
Amyloid aggregation is known to be dependent on experimental conditions, including the buffer used and sample agitation (78,79). Using ThT fluorescence, a qualitative measure of the number of β-sheets created during amyloid fiber formation (80, 81, 82), we characterized medin aggregation in the same buffer used for our electrophysiology experiments. For agitated samples, we found increasing ThT fluorescence for higher medin concentrations (Fig. 1 A) and a significantly decreased lag phase compared to quiescent samples (Fig. 1 B). The ThT fluorescence was more uniform for replicates of the lower concentration samples, as indicated by the smaller error bars. Under the agitation conditions used for most experiments, the lag-phase time obtained from fitting was 34 min and the aggregation rate constant was 9.3 h−1 for samples with 5 μM concentration. Comparable lag-phase times (37 and 35 min) and lower rate constants (3.2 and 3.3 h−1) were found for the 10 and 20 μM samples, respectively; however, these values are likely to have larger errors than the values for the 5 μM sample because of the decreasing fluorescence in the initial sections of the 10 and 20 μM curves. For quiescent samples, the lag-phase times found from fitting were 48.3 h, and the aggregation rate constants were 14.7 h−1 (Fig. 1 B). The rate constants found here for the 5 μM and quiescent samples are about an order of magnitude larger than the reported rates of medin (20 μM) aggregation in phosphate buffers (23). Notably, the ThT fluorescence in the plateau sections of the medin curves was minimal when compared with the plateau sections observed for Aβ1–42 aggregation in the same buffer (Fig. 1, inset). The rate constants found here for medin are also an order of magnitude larger than the rate constants of Aβ1–42 aggregation observed here and reported previously (68). To complement these ThT results, we performed CD spectroscopy measurements to determine the secondary structure of medin aggregates (Fig. S2 A). BeStSel analysis (83) of the spectra revealed a majority of disordered species and suggested the presence of β-sheet content.
Figure 1.
ThT aggregation curves for (A) agitated (300 RPM orbital shaking) medin samples with 5 μM (solid line), 10 μM (dotted line), and 20 μM (dashed line) concentrations. The data points and error bars represent the mean values and standard deviations for replicates in three wells. For clarity, only 50 (25) error bars are displayed for the 20 μM (10 μM) curve. Data fitting was performed using a sigmoidal function, as described in Materials and Methods. The inset shows the much higher ThT fluorescence observed during Aβ1–42 (20 μM) aggregation (gray). (B) The aggregation curves of quiescent medin samples displayed much longer lag-phase times than agitated samples. Data points and fitted curve are shown for 20 μM quiescent medin (only the fitted curves of the agitated samples shown in A are displayed here).
Morphology of medin aggregates
We used AFM to image the morphology of medin samples in three stages of their ThT aggregation curve (Fig. 2 A, inset): 1) t1, before agitation (start of lag phase); 2) t2, agitated until ThT fluorescence was detected (growth phase); and 3) t3, agitated for 2 h after the plateau was reached. At t1, the medin sample only had very few and scattered aggregates with heights above 1 nm (Fig. 2 A). At higher magnification (Fig. 2 B), it was possible to identify several structures with heights below 1 nm. The average height of these structures was 0.9 nm, they had a mean diameter (width at half height) of ∼3.5 nm, and their resulting volume was ∼9 nm3 (assuming a cylindrical shape). These volumes are slightly larger than the volume of monomers in U-shaped configurations (∼5.5 nm3) in Fig. S1, suggesting that the structures are primarily composed of medin monomers and low n oligomers (low n-mers). Assuming lag-phase monomers on mica have flexible random coil configurations with slightly lower volumes than U-shaped monomers and can be slightly compressed by the AFM tip during imaging, we think n ∼ 1–3 is a reasonable estimate.
Figure 2.
AFM images and corresponding cross sections displaying the morphology of medin aggregates transferred on a mica surface from solutions in three aggregation stages (indicated by the ThT curve in the inset A): (A and B) lag phase (t1), (C and D) growth phase (t2), and (E and F) plateau phase (t3). The low-magnification images in the upper row show the formation of large medin domains, resembling the morphology of supported lipid bilayers. The higher-magnification images in the lower row show the presence of monomeric and small oligomeric species in (B), larger oligomers in (D), and the large domains in (F). The domains have two phases separated by a height difference of less than 1 nm. Growth- and plateau-phase samples also contained smaller species of similar size to those from the lag phase (insets in D and F). For (A5/6N/AF) image sizes are provided at the bottom right. The scale bars in the insets of (D and F) represent 30 nm.
At t2, two types of aggregates were discerned: large flat domains (displayed in brighter contrast) with heights of 4.7 nm, occupying regions that extend over several micrometers, and much smaller structures, located principally in the left upper region of the image (Fig. 2 C). Higher-magnification imaging shows that the smaller structures have average heights of 5.7 nm and diameters of 21 nm (Fig. 2 D). We found each average-sized oligomer at t2 is composed of ∼200 monomers by performing particle size analysis to estimate the volumes of oligomer structures found at t2 and t1 (Fig. 2, B and D) and assuming that the structures at t1 are monomers.
At t3, almost all the structures observed were similar to the domains seen at t2. Two phases separated by a height difference of ∼1 nm could be identified in the domains, where the height of the lower-lying phase was similar to the height of the domains observed at t2. These domains resemble the morphology of lipid bilayer patches formed by rupture and fusion of liposomes on a solid surface. Some of these domains had globular-shaped structures (appearing in white contrast) on their surfaces (two are seen in Fig. 2 F). Their heights ranged from multiple values of the domains, consistent with multilayered structures, to several hundreds of nanometers. At both times, t2 and t3, growth- and plateau-phase samples still contained populations of small oligomers with comparable dimensions to those seen in Fig. 2 B (insets, Fig. 2, D and F). DLS measurements of the size distribution of medin aggregates in solution (Fig. S2 B) displayed peaks corresponding to oligomer dimensions below 10 nm and above 100 nm, consistent with the sizes of the species found by AFM imaging.
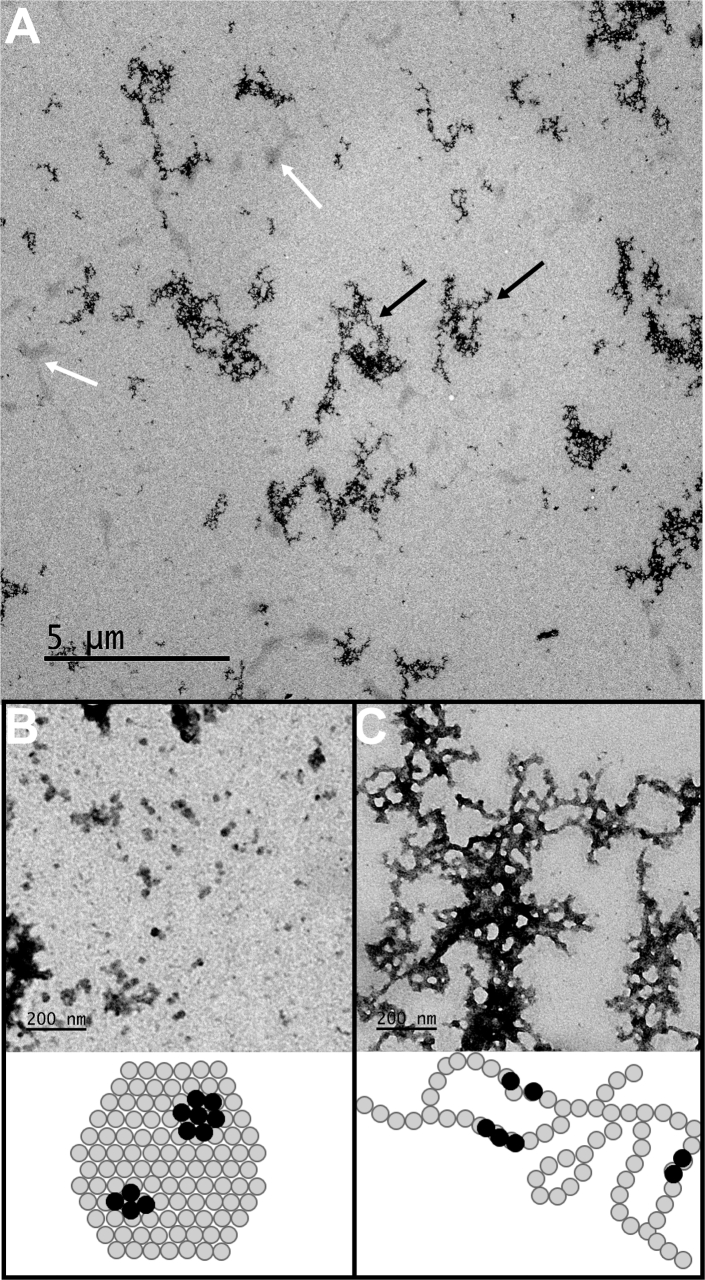
TEM imaging was used to gain further insight into the morphology and dimensions of medin aggregates (Fig. 3). Overview images (Fig. 3 A) revealed the coexistence of two distinct types of structures, which somewhat resembled the structures seen in Fig. 2 C. The small, compact structures (Fig. 3 B and white arrows in Fig. 3 A), typically ∼10–100 nm in size (average diameter: 19 nm), were in good agreement with the morphology and dimensions of the aggregates found by AFM (Fig. 2 D). The larger protofibrillar structures (Fig. 3 C and black arrows in Fig. 3 A) extended over hundreds of nanometers to a few micrometers and still somewhat resembled the elongated structures seen by AFM (Fig. 2 C). These structures suggest the aggregation mechanisms displayed in the schematics below the TEM images (Fig. 3, B and C).
Figure 3.
TEM images of medin aggregates. (A) Low-magnification overview images reveal two types of structures, indicated by white and black arrows. (B and C) High-magnification images of the structures indicated by arrows in (A) are given. (B) The compact aggregates shown by white arrows in (A) have a diameter of 19 ± 12 nm. The error is given as the HWHM of the Gaussian curve used to fit the diameter distribution. The schematic below the image (not drawn to scale) depicts oligomers (gray circles) aggregated into a compact structure. The black circles represent oligomers located on top of another oligomer seen in darker (brighter) contrast in the TEM (AFM) images. (C) Larger protofibrillar structures indicated by black arrows in (A) are shown. The schematic shows a potential mechanism of oligomer aggregation. The gray and black circles have the same meaning as in (B).
Pore activity of medin aggregates
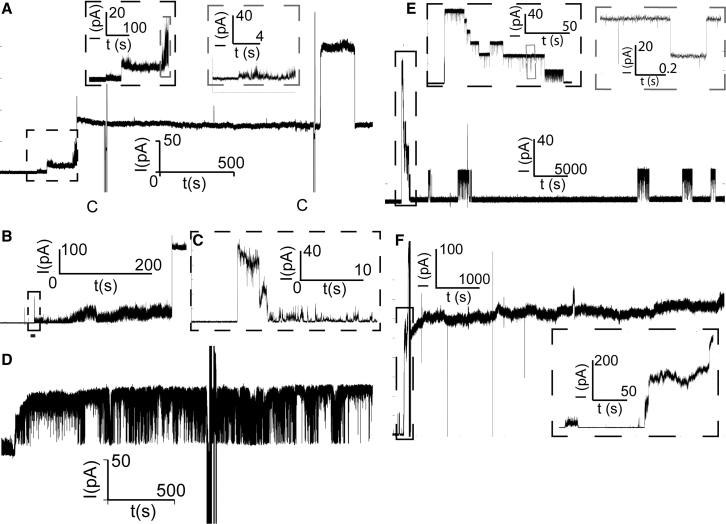
We used planar lipid bilayer electrophysiology measurements to determine the pore activity of medin proteins in the three different aggregation stages described above (Fig. 2 A, inset). Following the aggregation protocol described for the AFM experiments, medin samples were subsequently added to one of the compartments separated by a BLM, and the ionic current flowing through the membrane was monitored over time. After times that fluctuated between 45 and 120 min, we observed discrete events characteristic of membrane pore formation (Fig. 4 A). In addition to large step events (three are shown in Fig. 4 A), medin pores also displayed burst events characteristic of amyloid pores. Bursts were characterized by rapid fluctuations of current and had smaller conductance values than steps (Fig. 4 A, insets and Fig. 4, B and C). These bursts often preceded a step event (inset in gray, Fig. 4, A and B) and then continued superimposed on the step, likely indicating the rapid opening and closing of small pores (as well as possible membrane instabilities accompanying pore formation) before the formation of a larger pore with a slightly fluctuating pore diameter. Spike events, i.e., short-lived step events that only last a few milliseconds to seconds, were also observed as part of a longer burst (Fig. 4 C), in isolation, or superimposed on a step. In addition to steps, bursts, and spikes, medin pores also displayed (flickering) events in which the ionic current fluctuated between two values (Fig. 4 D) and that are compatible with the rapid opening and closing of a single pore. Current (I) versus time (t) traces acquired over several hours could display all these different behaviors.
Figure 4.
I vs. t traces displaying the different types of pore events observed for samples in the growth phase. Sections of the trace in which the capacitance was measured are indicated by a “C.” (A) The sequence of steps with different conductances is shown. The last step has a conductance above 1 nS. The burst activity preceding the first steps is shown in the insets. (B) Spike and burst activity before another step event is shown. The spike in (C) is a transient step event indicating the rapid opening and closing of a single pore, and the following burst is interpreted as either the rapid opening and closing of multiple small pores or membrane destabilization. (D) Long-lasting sequence of flickering events, indicating the opening and closing of a single pore or similar pores of ∼500 pS conductance, is shown. (E and F) I vs. t traces displaying pore events of medin proteins in the (E) lag phase and (F) plateau phase are given. The trace shown in (E) was the only trace of medin in the lag phase that displayed pore activity. The insets display a region of stepwise activity with step conductances in the range of 200–300 pS and transient step events with durations below 1 s. (F) A trace of proteins in the plateau phase displaying gigantic conductance levels (above 7 nS) and stable pore events is shown.
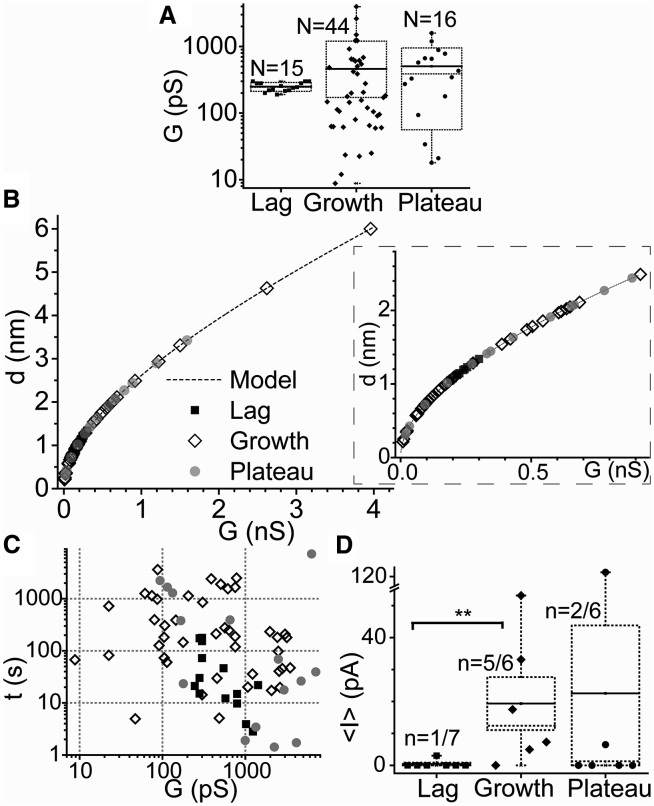
We compared the activity of pores formed in the growth phase with the pore activity of medin samples in the lag (Fig. 4 E) and plateau phases (Fig. 4 F). Only a minority (14 and 33%, respectively) of these samples displayed pore activity, and this behavior differed from the behavior of pores found in the growth phase. The large majority of pore events from samples in the lag phase had a narrow range of step conductances (200–300 pS) (Fig. 5 A), suggesting that these events originated from the opening and closing of either a single or very similar pores with estimated diameters of 1–1.5 nm (Fig. 5 B). In comparison, pores found in samples in the growth and plateau phases had a broader distribution of step conductances, with the majority of values between 10 and 1000 pS (Fig. 5 A) and pore diameters in the range of 0.1–2.5 nm (Fig. 5 B). Two pores from growth-phase samples had large diameters (4–6 nm). Only two out of six samples from the plateau phase induced pore activity; however, some of the pores from one of those samples had very large conductances and remained stable for hours with small relative fluctuations in current (Fig. 4 F). Although these events were rarely observed, they have the potential of significantly altering cellular homeostasis and inducing rapid cytotoxicity. Scatter plots showing the durations of step events spent at a given conductance level revealed that the majority of pores were stable over a range of tens to thousands of seconds (Fig. 5 C), and a significant number of them had conductance levels above 1000 pS because of the opening of multiple pores (Fig. 5, A and B). It should be noted that the average current of a step event was used for this analysis, whereas the difference of the upper and lower currents at a step event was used for Fig. 5, A and B. Short transient events with durations of less than 1 s were also observed (Fig. 4 C and inset in gray rectangle of Fig. 4 E); however, they were not included in this analysis.
Figure 5.
Summary of the pore characteristics obtained from all traces. Events in the lag, growth, and plateau phases are represented by squares, diamonds, and circles, respectively, throughout the figure. (A) A box plot of step conductances (in log scale) obtained by subtracting the ionic currents on the top and bottom of a step is given. The dotted lines on top and bottom of the box plots show the standard deviations, and the dotted (solid) lines inside the box indicate the median (mean) of the values inside the box. (B) Estimated pore diameters for the step conductances in (A) using the models mentioned in the main text are given. The inset shows an amplified section for the events below 1 nS conductance (89.7% of all events). (C) Scatter plots of step durations at a given conductance level obtained directly from the ionic current of the step are given. The values are larger than the step conductances shown in (A) and (B). Only events that lasted more than 1 s are plotted. (D) A box plot displaying the ionic permeabilities (average ionic charge per unit time allowed through medin pores during an I vs. t trace) is given. The box is similarly constructed as in (A), except that the standard errors instead of the standard deviations are displayed here. The events in the lag and growth phases were statistically different (∗∗p < 0.010) according to a post hoc pairwise comparison performed using the Holm-Sidak method.
We calculated the mean ionic current, <I>, (the integral of an I vs. t trace over the total time of the trace, T, ) through the BLM as a measure of the membrane ionic permeability induced by medin proteins over the course of a measurement, and we averaged these values for all samples in an aggregation phase (Fig. 5 D). The ionic permeability induced by medin proteins in the lag phase was significantly lower than for medin in the growth and plateau phases (Fig. 5 D). Although medin proteins in the growth and plateau phases induced comparable permeability on average, the majority of samples in the plateau phase did not induce any permeability, and only one of these samples (Fig. 4 F) induced very large and stable pores, which skewed the average value toward higher values. The ionic permeability induced by the pore events from this sample was over five times larger than the average permeability induced by all samples in the growth and lag phases. On the contrary, growth-phase samples consistently induced pore activity, and some of the pore events had very large conductances (Fig. 5, A–C). We further calculated the average rates of pore formation (opening and closing events of the same pore were not counted) for samples in the three different phases. The results were 0.5 h−1 for lag-phase, 1.8 h−1 for growth-phase, and 0.7 h−1 for plateau-phase species (Fig. 5, A and D). These values represent the average number of different pores opened per membrane per hour during the time of a typical measurement (∼4 h).
Molecular dynamics simulations
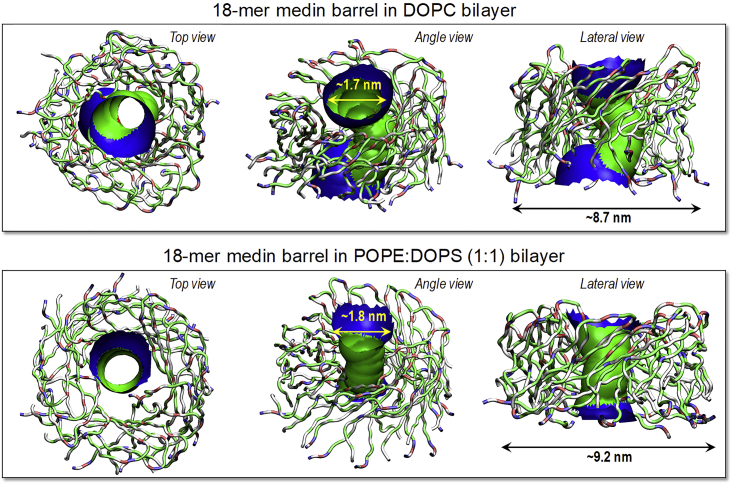
We performed 1-μs all-atom molecular dynamics simulations on 18-mer medin barrels with the CNpNC and NCpCN topologies embedded in the zwitterionic DOPC and anionic POPE/DOPS (1:1 molar ratio) lipid bilayers. The U-shaped medin monomers were initially preassembled as an annular shape, forming a β-barrel morphology. During the simulation, we observed that the medin barrel with the NCpCN topology was unstable. The β-barrel dismissed the annular shape and induced a collapsed pore (Fig. S3). Thus, hereafter we only consider the medin barrel with the CNpNC topology, referred to as just the medin barrel. In our previous simulations, we observed that Aβ barrels and channels also preferred the CNpNC topology in the lipid bilayers (51,52,75,76,84). In our simulations, the medin barrel also presents heterogeneity in barrel conformations, as observed in the Aβ barrels and channels (Fig. 6). Although gradual relaxation of the lipid bilayers induces discontinuities in the β-sheet network between the relaxed peptides in the barrel, the overall barrel conformation with the solvated central pore is preserved. The averaged pore and outer diameters are ∼1.7 and ∼8.7 and ∼1.8 and ∼9.2 nm for the 18-mer medin barrels in the zwitterionic and anionic lipid bilayers, respectively, which are highly comparable to the sizes of Aβ barrels (27).
Figure 6.
Averaged pore structures calculated with HOLE program embedded in the average barrel conformations during the simulations for the 18-mer medin barrels in the DOPC and POPE:DOPS (1:1 molar ratio) bilayers. In the barrel structures with the ribbon representations, hydrophobic, polar/Gly, positively charged, and negatively charged residues are colored white, green, blue, and red, respectively. For the pore structures, green denotes pore diameter in the range, 1.4 ≤ d ≤ 2.0 nm, and blue denotes pore diameter of d > 2.0 nm.
To observe ion activity in the solvated pore, we calculated the probability distribution for ions across the bilayer (Fig. 7, left panels). In our simulations, no ions were initially placed in the solvated pore. However, during the course of simulations, these ions rapidly migrated into the pore, and some cations even penetrated to the pore, as indicated in the probability distribution. High probabilities at z = ±2.0 nm represent cations interacting with lipid headgroups. Thus, probability distribution at −2.0 < z < 2.0 nm represents those cations located in the solvated pore. We observed that the medin barrel in the anionic bilayer has slightly higher probabilities of pore cations than that in the zwitterionic bilayer. Pore migrating cations can reflect ion fluctuations across the pore. To observe the fluctuations, we calculated the change in total charge in the pore as a function of the simulation time. In the calculations, we selected two pore lengths along the pore axis, −1.0 < z < 1.0 and −1.8 < z < 1.8 nm, ensuring that the charge fluctuations exclude a contribution of ion interactions with the lipid headgroups. For |z| < 1.0 nm, charge fluctuation in the total charge for the medin barrel in the zwitterionic bilayer is similar to that in the anionic bilayer. However, for |z| < 1.8 nm, the medin barrel in the anionic bilayer exhibits large charge fluctuation, suggesting that anionic lipids can attract more cations and induce these ions to localize near the gate of the solvated pore.
Figure 7.
Probability distribution functions for Mg2+ (green line), K+ (red line), Ca2+ (blue line), Zn2+ (cyan line), and Cl− (gray line) as a function of the distance along the pore center axis for the 18-mer medin barrels in the DOPC and POPE:DOPS (1:1 molar ratio) bilayers (left panels). Change in total charge in the pore is shown as a function of the simulation time for the 18-mer medin barrels in the DOPC and POPE:DOPS (1:1 molar ratio) bilayers (right panels). In the calculation, two pore height cutoffs, −1.0 < z < 1.0 and −1.8 < z < 1.8 nm, along the pore axis were used. C∗ denotes an effective Coulomb with the relationship, C∗ = C/n, where n is 1010.
Discussion
Medin is the main constituent of AMA, the most common form of localized amyloid (20). Little is known about its exact in vivo pathological role; however, ex vivo evidence suggests medin is implicated in multiple cardiovascular disorders (28, 29, 30, 31), including vascular contributions to neurodegeneration (33,34) and vascular dementia (32). An in vivo study found higher abundance of nonamyloidogenic medin species in aortas from patients with either thoracic aortic aneurysms or aortic dissection than in control individuals with normal aortas (29). The trend was reversed for amyloid particles (i.e., lower abundances of amyloidogenic medin deposits were found in diseased than in control aortas), thus suggesting that the nonamyloidogenic oligomers could be the toxic species in the aneurysm and dissection patients (29). Also, we recently found increased levels of oligomeric species in a subgroup of aneurysm patients with altered aortic wall integrity; however, we could not determine with certainty if these species were medin (28). Here, we have investigated an oligomer-mediated toxicity mechanism: do medin oligomers induce ionic permeability in lipid membranes? Using BLM electrophysiology, we found sudden, stepwise increases of ionic current through lipid bilayers upon exposure to oligomeric medin species (Fig. 4), indicative of the formation of medin membrane pores. These results open the possibility to the presence of medin-induced unregulated ionic currents through the plasma membrane, which could alter cellular ionic homeostasis and lead to the endothelial dysfunction and medin-induced EC cytotoxicity observed in vitro (33).
We characterized the activity of medin pores in the three characteristic states of medin aggregation: the lag, growth, and plateau phases. Medin aggregates emitted significantly lower ThT fluorescence than Aβ1–42 under comparable experimental conditions (Fig. 1); however, the activity of medin pores (Fig. 5) was at least comparable to the activity reported for Aβ1–40/42 pores (46,49,51,85). The low percentage (14%) of lag-phase samples that induced pore activity indicates that pore aggregation is a highly unlikely pathway for lag-phase species. Although the pore formation rates were generally not very high, a significant number of single and cumulative pore events were extremely large (G > 1 nS) and stable (Figure 4, Figure 5 C), providing a significant risk of homeostasis loss by a single event (46,51,59). We primarily used membranes with a high anionic lipid content (50% PS and 50% PE) for consistency with previous work on Aβ; however, we also observed pore activity when we employed membranes containing a majority of zwitterionic lipids (60% phosphatidylcholine, PC, and 30% PE), and a lower content of anionic lipids (10% PS) (Fig. S4). We did not perform a detailed comparison of pore activity as a function of lipid composition (because it is beyond the scope of this study); however, the in silico models (Fig. 6) of medin pores in PC zwitterionic membranes predict similar architectures and diameters as pores in anionic membranes.
AFM and TEM imaging of medin aggregates revealed the presence of small, compact aggregates (Figure 2, Figure 3 B) and larger protofibrillar structures (Figure 2, Figure 3 C). The reasonable agreement of the aggregate sizes seen by both imaging techniques and the DLS results (Fig. S2 B) suggests that both structures were present in solution. The flat domains observed by AFM closely resemble the morphology of patches formed by supported lipid bilayers after rupture and fusion of liposomes (Fig. 2; (66)). By analogy, these morphologies could suggest that compact domains originate from medin micelles in solution; however, this model would have difficulty explaining the presence of protofibrils (Fig. 3 C). Therefore, we believe that compact structures and protofibrils originate from two different aggregation patterns of smaller oligomers (schematics in Fig. 3, B and C). Although the AFM and TEM data are largely consistent with one another, it is possible that the planar morphology of the aggregates found by AFM was induced by their interaction with the mica substrate. The low β-sheet content of the aggregates (Fig. S2 A) when compared with the β-sheet content of fibrillar medin (65%) (23,78) is consistent with the absence of amyloid fibers in the AFM and TEM images (Figs. 2 and 3), and the low ThT fluorescence emitted by medin aggregates (Fig. 1).
Our previous studies indicated that agitation reduces the time of medin aggregation into amyloid fibers by a factor of ∼18 (78). Here, we found that heavy agitation reduces the (lag-phase) time of medin aggregation into pore-forming species by a factor of ∼30 (Fig. 1 B). The influence of agitation in aggregation may have implications in the formation of medin toxic species and deposition in atherosclerosis-prone vascular regions subjected to abnormal shear stress hemodynamics, a subject requiring further empiric study. Possibly related to this, we recently found that 89% (eight out of nine) of identified oligomer-rich thoracic aortic aneurysm patients had diagnosed hypertension, compared with 54% (seven out of 13) of patients with syndromic thoracic aortic aneurysm due to bicuspid valve disorder (28). We routinely observe medin aggregation into amyloid fibers when phosphate buffers are used for medin incubation (23,78); however, the protofibrillar structures formed by medin aggregates (Figs. 2 and 3), their low ThT fluorescence (Fig. 1), and the lack of amyloid fibers observed by AFM and TEM imaging suggest a nonamyloidogenic pathway to pore formation. We attribute this seeming discrepancy to the use of a different buffer in these experiments. HEPES was used for medin aggregation and BLM electrophysiology for consistency with previous BLM work (51,52,64,65). The differential effects of phosphate and HEPES buffers on Aβ fibrillation and oligomerization have been previously documented (79).
The significantly lower membrane permeability induced by lag-phase species compared with growth-phase aggregates (Fig. 5 D) indicates that medin solution preaggregation into β-sheet species (Fig. S2 A) is necessary for pore formation. The rate-limiting step in the formation of pore-forming medin oligomers is likely to be a conformational change from lag-phase species into β-sheet-containing growth-phase oligomers, which is also influenced by interactions with membrane lipids (41,42). This influence is possibly reflected in the activity induced by one of our lag-phase samples during the time of data acquisition (∼4 h). After this conformational change, pore-forming species could insert in the membrane and self-assemble into pores or insert as complete pores upon self-assembly on the membrane surface. We hypothesize that low n-mers get inserted in the membrane as β-sheet species, and then self-assemble in the membrane. Very small species (i.e., monomers and dimers), however, can lose their β-sheet structure in the lipid bilayer more easily than slightly larger oligomers (e.g., pentamers), as suggested by our previous simulations of Aβ17–42 membrane insertion (86).
Previous planar lipid bilayer results showed that full-length Aβ40–42 peptides (59,85,87), as well as fragments (52) and post-translational modifications (51) thereof, induced membrane pores with multiple conductances and selectivity to divalent cations, most notably Ca2+ and Mg2+. Here, similarly to these Aβ pores, we found a wide range of single-pore conductances for medin pores (Fig. 5 B). Medin pores also preserved a large pore (Fig. 6), wide enough for conducting water and ions. However, our in silico models suggest that medin pores have low ionic selectivity. Unlike Aβ pores (27,52,60,75,76,84), the central water channel of medin pores lacks a cationic ring able to selectively trap divalent cations (Fig. 6; Fig. S1). Even without potential gradients across the bilayer, the simulations verify that medin pores induce nonselective ion translocation in the central water channel. Cation transport across the solvated pore can yield a high transmembrane conductance when a local potential gradient is established across the membrane-embedded pore.
Medin and Aβ sequences have high local similarities within amino acid residues critical for Aβ fibril and pore formation (23,27). We constructed in silico models of membrane pores using the U-shaped β-strand-turn-β-strand motif of Aβ monomers (27,52,88), with the turn section in residues Asp25-Lys30 for medin (Fig. S1). The in silico models showed stable CNpNC barrel topology in both zwitterionic and anionic bilayers (Fig. 6). The pore diameters, ∼1.8 nm for the anionic membranes, corresponded to pores with ∼0.5 nS conductances (Fig. 5 B). The multiple conductances of amyloid pores suggest that pores have different diameters and consist of varying numbers of monomers. Although beyond the scope of this work, stable pores with similar architectures to the 18-mer barrel and diameters spanning the range of experimental diameters (Fig. 5) could be constructed for different n-mer configurations. Previous simulations performed for 12-, 16-, 20-, 24-, and 36-mers of the Aβ9–42 fragment suggest amyloid pores have varying diameters and are composed of dynamic β-sheet subunits (60). A similar mechanism involving mobile subunits may also explain the multiple conductances observed experimentally with medin pores. The large sizes of growth-phase oligomers seen in AFM images (Fig. 2, C and D) suggests that the majority of these oligomers are unlikely to form pores. Membrane insertion of large species is sterically hindered, and even when inserted, they could adopt non-pore-forming configurations. The smaller species still seen in growth-phase and plateau-phase samples are more likely to contain pore-forming oligomers (insets in Fig. 2, D and F); however, non-pore-forming configurations are also possible. Although we think 16–32 monomers represent the essential n-mers in a barrel (Fig. S1), we could also speculate of pore configurations containing the barrel and additional monomers around the barrel providing further stability to the structure.
In conclusion, we have used BLM electrophysiology to show that medin (the main component of AMA) induces abrupt, stepwise ionic currents in lipid membranes, consistent with the formation of membrane pores. Pore activity was primarily observed for preaggregated medin and seldom for monomeric and low n-mer species from the lag phase. Our results show medin aggregates did not form amyloid fibers, emitted significantly lower ThT fluorescence than Aβ, and had lower β-sheet content than previously studied medin fibers, thus strongly suggesting a nonamyloidogenic pathway to pore formation. Atomistic in silico modeling produced stable CNpNC barrel topology, with pore diameters of ∼1.8 nm corresponding to experimental pore conductances of ∼0.5 nS. Membrane permeability induced by pore formation is a novel, to our knowledge, potential mechanism and therapeutic target of medin pathology.
Author Contributions
F.T.A., S.Y., J.M., and R.Q.M. conceived and designed research. H.A.D. and J.M. prepared protein samples. S.Y. prepared samples for BLM and ThT experiments. S.Y., M.J.N., H.A.D., J.M., and F.T.A. performed experiments. H.J. performed MD simulations. F.T.A., J.G.N.G., and R.N. oversaw research. S.Y., H.A.D., J.M., F.T.A., H.J., and R.N. analyzed and interpreted data. F.T.A., H.J., J.M., and R.Q.M. wrote the manuscript with input from all authors. All authors edited and approved the manuscript.
Acknowledgments
F.T.A. thanks Ratnesh Lal (University of California, San Diego) for sharing his insight during valuable discussions. J.M. thanks Heike Arnolds for assistance with DLS measurements.
F.T.A. acknowledges funding from the Arizona Alzheimer’s Consortium. J.M. acknowledges funding from the British Heart Foundation (FS/12/61/29877). This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN26120080001E. This research was supported (in part) by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. R.Q.M. acknowledges VA Merit BX007080 and Department of Defense W81XWH-17-1-0473 funding. All Multimode AFM images and data were collected in the W.M. Keck Center for Nano-Scale Imaging in the Department of Chemistry and Biochemistry at the University of Arizona. This instrument purchase was supported by Arizona Technology and Research Initiative Fund (A.R.S.§15-1648). All simulations were performed using the high-performance computational facilities of the Biowulf PC/Linux cluster at the National Institutes of Health, Bethesda, MD (https://hpc.nih.gov/). The content of this publication does not necessarily reflect the views or policies of the U.S. Department of Health and Human Services, Veterans Affairs, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Editor: Charles Deber.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.04.026.
Supporting Material
References
- 1.Eisenberg D., Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrison R.S., Sharpe P.C., Fairlie D.P. Amyloid peptides and proteins in review. Rev. Physiol. Biochem. Pharmacol. 2007;159:1–77. doi: 10.1007/112_2007_0701. [DOI] [PubMed] [Google Scholar]
- 3.Barnhart M.M., Chapman M.R. Curli biogenesis and function. Annu. Rev. Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linder M.B., Szilvay G.R., Penttilä M.E. Hydrophobins: the protein-amphiphiles of filamentous fungi. FEMS Microbiol. Rev. 2005;29:877–896. doi: 10.1016/j.femsre.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 6.Knowles T.P., Oppenheim T.W., Welland M.E. Nanostructured films from hierarchical self-assembly of amyloidogenic proteins. Nat. Nanotechnol. 2010;5:204–207. doi: 10.1038/nnano.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang S., Yokoi H., Horii A. Designer self-assembling peptide nanofiber scaffolds. In: Silva G.A., Parpura V., editors. Nanotechnology for Biology and Medicine: At the Building Block Level. Springer; 2012. pp. 123–147. [Google Scholar]
- 8.Cherny I., Gazit E. Amyloids: not only pathological agents but also ordered nanomaterials. Angew. Chem. Int.Engl. 2008;47:4062–4069. doi: 10.1002/anie.200703133. [DOI] [PubMed] [Google Scholar]
- 9.Gras S.L. Amyloid fibrils: from disease to design. New biomaterial applications for self-assembling cross-β fibrils. Aust. J. Chem. 2007;60:333–342. [Google Scholar]
- 10.Gras S.L., Tickler A.K., MacPhee C.E. Functionalised amyloid fibrils for roles in cell adhesion. Biomaterials. 2008;29:1553–1562. doi: 10.1016/j.biomaterials.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 11.Benson M.D., Buxbaum J.N., Westermark P. Amyloid nomenclature 2018: recommendations by the international society of amyloidosis (ISA) nomenclature committee. Amyloid. 2018;25:215–219. doi: 10.1080/13506129.2018.1549825. [DOI] [PubMed] [Google Scholar]
- 12.Blennow K., de Leon M.J., Zetterberg H. Alzheimer’s disease. Lancet. 2006;368:387–403. doi: 10.1016/S0140-6736(06)69113-7. [DOI] [PubMed] [Google Scholar]
- 13.Tanzi R.E., Bertram L. Twenty years of the Alzheimer’s disease amyloid hypothesis: a genetic perspective. Cell. 2005;120:545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Jakob-Roetne R., Jacobsen H. Alzheimer’s disease: from pathology to therapeutic approaches. Angew. Chem. Int.Engl. 2009;48:3030–3059. doi: 10.1002/anie.200802808. [DOI] [PubMed] [Google Scholar]
- 15.DeToma A.S., Salamekh S., Lim M.H. Misfolded proteins in Alzheimer’s disease and type II diabetes. Chem. Soc. Rev. 2012;41:608–621. doi: 10.1039/c1cs15112f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts R.F., Wade-Martins R., Alegre-Abarrategui J. Direct visualization of alpha-synuclein oligomers reveals previously undetected pathology in Parkinson’s disease brain. Brain. 2015;138:1642–1657. doi: 10.1093/brain/awv040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Recchia A., Debetto P., Giusti P. α-synuclein and Parkinson’s disease. FASEB J. 2004;18:617–626. doi: 10.1096/fj.03-0338rev. [DOI] [PubMed] [Google Scholar]
- 18.Migrino R.Q., Harmann L., Hari P. Clinical and imaging predictors of 1-year and long-term mortality in light chain (AL) amyloidosis: a 5-year follow-up study. Heart Vessels. 2014;29:793–800. doi: 10.1007/s00380-013-0419-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Migrino R.Q., Truran S., Hari P. Human microvascular dysfunction and apoptotic injury induced by AL amyloidosis light chain proteins. Am. J. Physiol. Heart Circ. Physiol. 2011;301:H2305–H2312. doi: 10.1152/ajpheart.00503.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Häggqvist B., Näslund J., Westermark P. Medin: an integral fragment of aortic smooth muscle cell-produced lactadherin forms the most common human amyloid. Proc. Natl. Acad. Sci. USA. 1999;96:8669–8674. doi: 10.1073/pnas.96.15.8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsson A., Söderberg L., Westermark P. Unwinding fibril formation of medin, the peptide of the most common form of human amyloid. Biochem. Biophys. Res. Commun. 2007;361:822–828. doi: 10.1016/j.bbrc.2007.06.187. [DOI] [PubMed] [Google Scholar]
- 22.Kholová I., Niessen H.W. Amyloid in the cardiovascular system: a review. J. Clin. Pathol. 2005;58:125–133. doi: 10.1136/jcp.2004.017293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies H.A., Madine J., Middleton D.A. Comparisons with amyloid-β reveal an aspartate residue that stabilizes fibrils of the aortic amyloid peptide medin. J. Biol. Chem. 2015;290:7791–7803. doi: 10.1074/jbc.M114.602177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mucchiano G., Cornwell G.G., III, Westermark P. Senile aortic amyloid. Evidence for two distinct forms of localized deposits. Am. J. Pathol. 1992;140:871–877. [PMC free article] [PubMed] [Google Scholar]
- 25.Davies H.A., Rigden D.J., Madine J. Probing medin monomer structure and its amyloid nucleation using 13C-direct detection NMR in combination with structural bioinformatics. Sci. Rep. 2017;7:45224. doi: 10.1038/srep45224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardy J.A., Higgins G.A. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 27.Jang H., Arce F.T., Nussinov R. Disordered amyloidogenic peptides may insert into the membrane and assemble into common cyclic structural motifs. Chem. Soc. Rev. 2014;43:6750–6764. doi: 10.1039/c3cs60459d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies H.A., Caamaño-Gutiérrez E., Madine J. Idiopathic degenerative thoracic aneurysms are associated with increased aortic medial amyloid. Amyloid. 2019;26:148–155. doi: 10.1080/13506129.2019.1625323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng S., Larsson A., Westermark P. Role of aggregated medin in the pathogenesis of thoracic aortic aneurysm and dissection. Lab. Invest. 2007;87:1195–1205. doi: 10.1038/labinvest.3700679. [DOI] [PubMed] [Google Scholar]
- 30.Peng S., Glennert J., Westermark P. Medin-amyloid: a recently characterized age-associated arterial amyloid form affects mainly arteries in the upper part of the body. Amyloid. 2005;12:96–102. doi: 10.1080/13506120500107006. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y., Feng X., Zhao W. Is vascular amyloidosis intertwined with arterial aging, hypertension and atherosclerosis? Front. Genet. 2017;8:126. doi: 10.3389/fgene.2017.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karamanova N., Truran S., Migrino R.Q. Endothelial immune activation by medin: potential role in cerebrovascular disease and reversal by monosialoganglioside-containing nanoliposomes. J. Am. Heart Assoc. 2020;9:e014810. doi: 10.1161/JAHA.119.014810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Migrino R.Q., Davies H.A., Madine J. Amyloidogenic medin induces endothelial dysfunction and vascular inflammation through the receptor for advanced glycation endproducts. Cardiovasc. Res. 2017;113:1389–1402. doi: 10.1093/cvr/cvx135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Migrino R.Q., Truran S., Beach T.G. Human cerebral collateral arteriole function in subjects with normal cognition, mild cognitive impairment, and dementia. Am. J. Physiol. Heart Circ. Physiol. 2018;315:H284–H290. doi: 10.1152/ajpheart.00206.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davies H.A., Phelan M.M., Madine J. Oxidative stress alters the morphology and toxicity of aortic medial amyloid. Biophys. J. 2015;109:2363–2370. doi: 10.1016/j.bpj.2015.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sipe J.D., Cohen A.S. Review: history of the amyloid fibril. J. Struct. Biol. 2000;130:88–98. doi: 10.1006/jsbi.2000.4221. [DOI] [PubMed] [Google Scholar]
- 37.Cline E.N., Bicca M.A., Klein W.L. The amyloid-β oligomer hypothesis: beginning of the third decade. J. Alzheimers Dis. 2018;64:S567–S610. doi: 10.3233/JAD-179941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glabe C., Kayed R., Hall J. Common structure and mechanism of soluble amyloid oligomer pathogenesis in degenerative diseases. Neurobiol. Aging. 2004;25:S75–S76. doi: 10.1016/j.neurobiolaging.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 39.Kayed R., Sokolov Y., Glabe C.G. Permeabilization of lipid bilayers is a common conformation-dependent activity of soluble amyloid oligomers in protein misfolding diseases. J. Biol. Chem. 2004;279:46363–46366. doi: 10.1074/jbc.C400260200. [DOI] [PubMed] [Google Scholar]
- 40.Winner B., Jappelli R., Riek R. In vivo demonstration that α-synuclein oligomers are toxic. Proc. Natl. Acad. Sci. USA. 2011;108:4194–4199. doi: 10.1073/pnas.1100976108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butterfield S.M., Lashuel H.A. Amyloidogenic protein-membrane interactions: mechanistic insight from model systems. Angew. Chem. Int.Engl. 2010;49:5628–5654. doi: 10.1002/anie.200906670. [DOI] [PubMed] [Google Scholar]
- 42.Relini A., Cavalleri O., Gliozzi A. The two-fold aspect of the interplay of amyloidogenic proteins with lipid membranes. Chem. Phys. Lipids. 2009;158:1–9. doi: 10.1016/j.chemphyslip.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 43.Di Paolo G., Kim T.W. Linking lipids to Alzheimer’s disease: cholesterol and beyond. Nat. Rev. Neurosci. 2011;12:284–296. doi: 10.1038/nrn3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams T.L., Serpell L.C. Membrane and surface interactions of Alzheimer’s Aβ peptide--insights into the mechanism of cytotoxicity. FEBS J. 2011;278:3905–3917. doi: 10.1111/j.1742-4658.2011.08228.x. [DOI] [PubMed] [Google Scholar]
- 45.Sciacca M.F., Kotler S.A., Ramamoorthy A. Two-step mechanism of membrane disruption by Aβ through membrane fragmentation and pore formation. Biophys. J. 2012;103:702–710. doi: 10.1016/j.bpj.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arispe N., Pollard H.B., Rojas E. Giant multilevel cation channels formed by Alzheimer disease amyloid beta-protein [A beta P-(1-40)] in bilayer membranes. Proc. Natl. Acad. Sci. USA. 1993;90:10573–10577. doi: 10.1073/pnas.90.22.10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arispe N., Pollard H.B., Rojas E. The ability of amyloid beta-protein [A beta P (1-40)] to form Ca2+ channels provides a mechanism for neuronal death in Alzheimer’s disease. Ann. N. Y. Acad. Sci. 1994;747:256–266. doi: 10.1111/j.1749-6632.1994.tb44414.x. [DOI] [PubMed] [Google Scholar]
- 48.Lin H., Bhatia R., Lal R. Amyloid β protein forms ion channels: implications for Alzheimer’s disease pathophysiology. FASEB J. 2001;15:2433–2444. doi: 10.1096/fj.01-0377com. [DOI] [PubMed] [Google Scholar]
- 49.Quist A., Doudevski I., Lal R. Amyloid ion channels: a common structural link for protein-misfolding disease. Proc. Natl. Acad. Sci. USA. 2005;102:10427–10432. doi: 10.1073/pnas.0502066102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bode D.C., Baker M.D., Viles J.H. Ion channel formation by amyloid-β42 oligomers but not amyloid-β40 in cellular membranes. J. Biol. Chem. 2017;292:1404–1413. doi: 10.1074/jbc.M116.762526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gillman A.L., Jang H., Teran Arce F. Activity and architecture of pyroglutamate-modified amyloid-β (AβpE3-42) pores. J. Phys. Chem. B. 2014;118:7335–7344. doi: 10.1021/jp5040954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jang H., Arce F.T., Lal R. Truncated beta-amyloid peptide channels provide an alternative mechanism for Alzheimer’s disease and down syndrome. Proc. Natl. Acad. Sci. USA. 2010;107:6538–6543. doi: 10.1073/pnas.0914251107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin M.C., Kagan B.L. Electrophysiologic properties of channels induced by Abeta25-35 in planar lipid bilayers. Peptides. 2002;23:1215–1228. doi: 10.1016/s0196-9781(02)00057-8. [DOI] [PubMed] [Google Scholar]
- 54.Tosatto L., Andrighetti A.O., Dalla Serra M. Alpha-synuclein pore forming activity upon membrane association. Biochim. Biophys. Acta. 2012;1818:2876–2883. doi: 10.1016/j.bbamem.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 55.Kim H.Y., Cho M.K., Zweckstetter M. Structural properties of pore-forming oligomers of alpha-synuclein. J. Am. Chem. Soc. 2009;131:17482–17489. doi: 10.1021/ja9077599. [DOI] [PubMed] [Google Scholar]
- 56.Last N.B., Rhoades E., Miranker A.D. Islet amyloid polypeptide demonstrates a persistent capacity to disrupt membrane integrity. Proc. Natl. Acad. Sci. USA. 2011;108:9460–9465. doi: 10.1073/pnas.1102356108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mustata M., Capone R., Nussinov R. K3 fragment of amyloidogenic beta(2)-microglobulin forms ion channels: implication for dialysis related amyloidosis. J. Am. Chem. Soc. 2009;131:14938–14945. doi: 10.1021/ja9049299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Demuro A., Smith M., Parker I. Single-channel Ca(2+) imaging implicates Aβ1-42 amyloid pores in Alzheimer’s disease pathology. J. Cell Biol. 2011;195:515–524. doi: 10.1083/jcb.201104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arispe N., Rojas E., Pollard H.B. Alzheimer disease amyloid β protein forms calcium channels in bilayer membranes: blockade by tromethamine and aluminum. Proc. Natl. Acad. Sci. USA. 1993;90:567–571. doi: 10.1073/pnas.90.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jang H., Arce F.T., Nussinov R. Misfolded amyloid ion channels present mobile β-sheet subunits in contrast to conventional ion channels. Biophys. J. 2009;97:3029–3037. doi: 10.1016/j.bpj.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhatia R., Lin H., Lal R. Fresh and globular amyloid beta protein (1-42) induces rapid cellular degeneration: evidence for AbetaP channel-mediated cellular toxicity. FASEB J. 2000;14:1233–1243. doi: 10.1096/fasebj.14.9.1233. [DOI] [PubMed] [Google Scholar]
- 62.Lashuel H.A., Hartley D., Lansbury P.T., Jr. Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature. 2002;418:291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- 63.Lashuel H.A., Lansbury P.T., Jr. Are amyloid diseases caused by protein aggregates that mimic bacterial pore-forming toxins? Q. Rev. Biophys. 2006;39:167–201. doi: 10.1017/S0033583506004422. [DOI] [PubMed] [Google Scholar]
- 64.Martinez Hernandez A., Urbanke H., Fischer A. The diphenylpyrazole compound anle138b blocks Aβ channels and rescues disease phenotypes in a mouse model for amyloid pathology. EMBO Mol. Med. 2018;10:32–47. doi: 10.15252/emmm.201707825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arispe N., Pollard H.B., Rojas E. Zn2+ interaction with Alzheimer amyloid beta protein calcium channels. Proc. Natl. Acad. Sci. USA. 1996;93:1710–1715. doi: 10.1073/pnas.93.4.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Richter R., Mukhopadhyay A., Brisson A. Pathways of lipid vesicle deposition on solid surfaces: a combined QCM-D and AFM study. Biophys. J. 2003;85:3035–3047. doi: 10.1016/S0006-3495(03)74722-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davies H.A., Wilkinson M.C., Middleton D.A. Expression and purification of the aortic amyloid polypeptide medin. Protein Expr. Purif. 2014;98:32–37. doi: 10.1016/j.pep.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 68.Lee J., Gillman A.L., Teran Arce F. Role of the fast kinetics of pyroglutamate-modified amyloid-β oligomers in membrane binding and membrane permeability. Biochemistry. 2014;53:4704–4714. doi: 10.1021/bi500587p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arosio P., Knowles T.P., Linse S. On the lag phase in amyloid fibril formation. Phys. Chem. Chem. Phys. 2015;17:7606–7618. doi: 10.1039/c4cp05563b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sansom M.S. Structure and function of channel-forming peptaibols. Q. Rev. Biophys. 1993;26:365–421. doi: 10.1017/s0033583500002833. [DOI] [PubMed] [Google Scholar]
- 71.Cruickshank C.C., Minchin R.F., Martinac B. Estimation of the pore size of the large-conductance mechanosensitive ion channel of Escherichia coli. Biophys. J. 1997;73:1925–1931. doi: 10.1016/S0006-3495(97)78223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hutter J.L., Bechhoefer J. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 1993;64:1868–1873. [Google Scholar]
- 73.Brooks B.R., Bruccoleri R.E., Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 74.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jang H., Arce F.T., Nussinov R. β-barrel topology of Alzheimer’s β-amyloid ion channels. J. Mol. Biol. 2010;404:917–934. doi: 10.1016/j.jmb.2010.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jang H., Arce F.T., Nussinov R. Familial Alzheimer’s disease osaka mutant (ΔE22) β-barrels suggest an explanation for the different Aβ1-40/42 preferred conformational states observed by experiment. J. Phys. Chem. B. 2013;117:11518–11529. doi: 10.1021/jp405389n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Davies H.A., Lee C.F., Madine J. Insights into the origin of distinct medin fibril morphologies induced by incubation conditions and seeding. Int. J. Mol. Sci. 2018;19:E1357. doi: 10.3390/ijms19051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Garvey M., Tepper K., Fändrich M. Phosphate and HEPES buffers potently affect the fibrillation and oligomerization mechanism of Alzheimer’s Aβ peptide. Biochem. Biophys. Res. Commun. 2011;409:385–388. doi: 10.1016/j.bbrc.2011.04.141. [DOI] [PubMed] [Google Scholar]
- 80.Nilsson M.R. Techniques to study amyloid fibril formation in vitro. Methods. 2004;34:151–160. doi: 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 81.Biancalana M., Koide S. Molecular mechanism of thioflavin-T binding to amyloid fibrils. Biochim. Biophys. Acta. 2010;1804:1405–1412. doi: 10.1016/j.bbapap.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Khurana R., Coleman C., Singh S. Mechanism of thioflavin T binding to amyloid fibrils. J. Struct. Biol. 2005;151:229–238. doi: 10.1016/j.jsb.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 83.Micsonai A., Wien F., Kardos J. BeStSel: a web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018;46:W315–W322. doi: 10.1093/nar/gky497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jang H., Teran Arce F., Nussinov R. Structural convergence among diverse, toxic β-sheet ion channels. J. Phys. Chem. B. 2010;114:9445–9451. doi: 10.1021/jp104073k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hirakura Y., Lin M.C., Kagan B.L. Alzheimer amyloid abeta1-42 channels: effects of solvent, pH, and Congo Red. J. Neurosci. Res. 1999;57:458–466. [PubMed] [Google Scholar]
- 86.Jang H., Connelly L., Nussinov R. Mechanisms for the insertion of toxic, fibril-like β-amyloid oligomers into the membrane. J. Chem. Theory Comput. 2013;9:822–833. doi: 10.1021/ct300916f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kagan B.L., Azimov R., Azimova R. Amyloid peptide channels. J. Membr. Biol. 2004;202:1–10. doi: 10.1007/s00232-004-0709-4. [DOI] [PubMed] [Google Scholar]
- 88.Arce F.T., Jang H., Lal R. Polymorphism of amyloid β peptide in different environments: implications for membrane insertion and pore formation. Soft Matter. 2011;7:5267–5273. doi: 10.1039/C1SM05162H. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.