Summary
Even when they no longer require the presence of iron, cells use zinc as a divalent cation, involved in a large variety of catalytic and regulatory functions. This metal is so important that it appears that ribosomes are instrumental in its ultimate storage. Here, we summarize a detailed analysis which investigates the way the global cell metabolism is integrated by zinc. This integration results from the zinc‐dependent way in which the one‐carbon metabolism is always coupled to the translation process, not only via methionine and S‐adenosylmethionine, but via the complex set‐up of the modification of the position 34 of the anticodon of tRNAs.
Global metabolic integration results from the zinc‐dependent way in which the one‐carbon metabolism is coupled to the translation process, via methionine, S‐adenosylmethionine, and the complex set up of the position 34 modification of the anticodon of tRNAs.
Much more than an anecdote: we keep behaving as the medieval alchemists who achieved the transmutation of metals. In contrast with physics, biology does not take great care about many details of its experimental implementations. When we grow microbes in ‘defined’ media – that is media supposed to contain all the ingredients required to support life [see reference recipes in (Miller, 1972; Lessard, 2013)] – many elements found in the cells come from nowhere. This is true even for metabolic engineering. To be sure, historically, biotechnology developed from industrial processes that were still plagued with vitalistic insights. Louis Pasteur changed the paradigm when he refuted the idea of spontaneous generation and decided to study the ‘diseases’ of beer and wine. However, while Pasteur took great care in the way he sterilized and kept the media he recommended, he did not consider the details of the growth media, except for their standard industrial set‐up. Alas, we know that the way we grow cells triggers considerable variability and inconsistencies in our observations [see, e.g. (Sridhar and Steele‐Mortimer, 2016)]. Here is a striking illustration that tells us that we must change the way we go. For decades, biochemists used supplementation of assay media with the fluoride ion as a way to explore energy metabolism (Elsbach and Schwartz, 1959; Manno and Schachter, 1970), PEP‐dependent transport (Cirillo and Razin, 1973; Saier and Feucht, 1980) or the activity of adenylate cyclase via interference with guanine nucleotide‐binding proteins [G‐proteins, (Tao and Lipmann, 1969; de Haën, 1974)]. The role of fluoride long remained ‘a frustrating mystery’ (Maguire et al., 1975). Having serendipitously found that changing glassware for plasticware abolished the fluoride effect, Sternweis and Gilman reported that the purified G‐protein regulation of adenylate cyclase depended on the presence of aluminium traces (Sternweis and Gilman, 1982). Subsequently, Chabre discovered that fluoride extracts aluminium from glass, forming the anion, isosteric with phosphate, and takes its place (Bigay et al., 1985). is since then used as an authentic mimic of phosphate (Jin et al., 2017).
Yet, despite cogent examples such as this one, we use costly and time‐consuming technology to study living processes still based on their unfolding in uncontrolled environments. It is therefore no surprise that reproducibility/replicability is not a strong marker of ‘scientific’ publications in the domain of biology (National Academies of Sciences, Engineering, and Medicine; Policy and Global Affairs; Committee on Science, Engineering, Medicine, and Public Policy; Board on Research Data and Information; Division on Engineering and Physical Sciences; Committee on Applied and Theoretical Statistics; Board on Mathematical Sciences and Analytics; Division on Earth and Life Studies; Nuclear and Radiation Studies Board; Division of Behavioral and Social Sciences and Education; Committee on National Statistics; Board on Behavioral, Cognitive, and Sensory Sciences; Committee on Reproducibility and Replicability in Science, 2019). What if one of the overlooked components played a critical role in major life processes? We have now hints that zinc, as the divalent cation Zn2+ and not monitored in the vast majority of growth media or biochemical assays, is integrating major metabolic features via the coordination of translation with one‐carbon metabolism [Danchin et al. 2020 and Fig. 1].
Fig. 1.
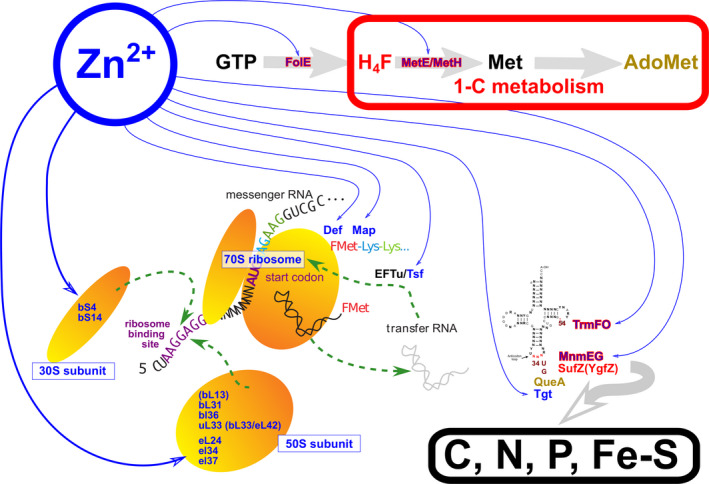
Zinc as an integrator of metabolism. Zinc‐binding proteins are in blue. When they also bind tetrahydrofolate, they are bordered by red. Methylations are not displayed, but the role of S‐adenosylmethionine (AdoMet) in the formation of queuine is indicated in brown. The ribosome acts as a Zn2+ store. The MnmEG/SufZ complex acts as a coordinator of carbon, nitrogen, phosphate and iron‐sulfur metabolism.
How is this unfolding? Zinc is a fairly unobtrusive metal in life processes but it is referenced in many articles (almost 150 000 at PubMed in January 2020) despite its still elusive behaviour as a biochemical entity (Krezel and Maret, 2016). The most familiar role of zinc is related to its presence in ‘zinc‐finger’ domains of proteins, in particular regulatory proteins or protein–protein interfaces (Eom et al., 2016). As cases in point, zinc‐finger proteins have been used as molecular scissors for gene editing before the advent of the CRISPR‐Cas9 technology and they are still used (Broeders et al., 2019). Zinc is a critical cofactor in a great many enzymes – as much as 9% of the entire proteome in eukaryotes and from 5% to 6% in prokaryotes (Andreini et al., 2009). Yet, its singularity has been virtually overlooked [see however studies such as (Bütof et al., 2019)]. Unexpectedly, it was recently observed that the Aspergillus fumigatus mycotoxin, gliotoxin, had a function specifically involving zinc (Seo et al., 2019), and, because this metabolite could act as a specific zinc chelator, this opens up approaches to investigate in depth the role of zinc.
Indeed, the main reason for ignoring metal ions in growth media stems from the considerable difficulty we have to carefully control their concentration. Transition metal ions, in particular, lose their 4s2 electrons, and the corresponding Me2+ ions have more or less the same size, with fairly common hydration properties. Also, transition metals tend to favour binding to the same atoms (oxygen, nitrogen, also sulfur). The consequence is that it is difficult to distinguish between their individual transport systems or binding sites. A rule of thumb tells us that, when catalysis involve redox reactions, the nature of the cation has considerable influence on the outcome of the reaction, while non‐redox reactions – typical of reactions involving Zn2+ such as hydrolases (Coleman, 1998) – are much less sensitive to the ion (Valasatava et al., 2018). The consequence is that it is next to impossible to use standard chelators such as ethylenediaminetetraacetate (EDTA) to exert a tight control on their relative concentration (Chen et al., 2008). This same obstacle is likely to exist also for the cell, which has to scavenge many metals from its environment. This circumstance created a remarkable selective pressure that resulted in the design of highly specific chelators or storage proteins. However, while the process has been explored for the acquisition of iron – showing competition or cooperation via a large variety of siderophores with cells creating progressively stronger siderophore to outcompete those which make weaker ones (Kramer et al., 2019) – comparable studies have seldom been extended to other metals. The widespread interest in competition for iron as a major trigger of microbial virulence (Palmer and Skaar, 2016) made that investigators overlooked much details of how cells would acquire other metal divalent ions. Yet, in the case of zinc, it is well established that the cation is both essential and toxic if too concentrated [see a general discussion, centred on plants in (Cabot et al., 2019)]. As a consequence, its availability must be exquisitely regulated by a homeostatic process.
In a surprising turn of things, the intricate coupling between one‐carbon metabolism and translation – initially witnessed as the omnipresent requirement for a methionine residue at the start of translated polypeptides – revealed that the connection was established via the homeostasis of Zn2+ (Danchin et al. 2020, and Fig. 1), mediated by the ribosomes as a main Zn2+ store (Hensley et al., 2012). To be sure, in the three domains of life, several ribosomal proteins comprise a zinc‐binding domain. Furthermore, in Eukarya a number of ribosomal proteins also bind zinc‐finger regulators [see, e.g. (Dionne et al., 2019)]. This role must now be connected to the observation that in the model bacterium Bacillus subtilis a sophisticated regulatory circuit was recruited to optimize Zn2+ acquisition and cellular distribution when the ion becomes limiting (Nies, 2019). Miscellaneous studies suggest that this role of zinc is general. Homeostasis is based on zinc‐dependent enzymes – in particular enzymes involved in folic acid synthesis – that probe the pool of available Zn2+ ions and then amplify this signal to control the activity of Zn2+ chaperones, as well as modulation of the zinc content of ribosomes.
What is more, many zinc‐dependent enzymes that are required for translation – with tRNA modifications as substantiating evidence in all domains of life – are directly dependent on 1‐C metabolism, not only via methylations but also directly involving tetrahydrofolate. This strongly argues for a functional dependency relating translation, 1‐C metabolism and zinc homeostasis. Analysis of the underlying enzyme activities revealed that this possibly happened via management of iron‐sulfur clusters (Danchin et al. 2020). The key metabolites of this network are formaldehyde, methionine, S‐adenosylmethionine (AdoMet) and tetrahydrofolate. Remarkably, a triad of tRNA anticodon modification enzymes, namely MnmE, MnmG and SufZ (YgfZ), are integrated together via a role of zinc coupling translation, folic acid metabolism, management of iron‐sulfur clusters and zinc availability. The MnmEG complex, previously identified as coupling carbon metabolism with replication (Shippy and Fadl, 2015) as a consequence of tRNA modifications, is further used for phosphate homeostasis by thiolation of the same U34 base (Gupta et al., 2019), making this position ideal to balance carbon, nitrogen, sulfur and phosphate metabolism via a Zn2+‐mediated translation control of Fe2+‐S cluster synthesis, input into polypeptides during translation and maintenance.
Finally, this puzzling role of zinc asked for an investigation of its origins, in particular by revisiting the available scenarios of the origins of life – essentially plagued by creeds, and fairly far from authentic science. If the metabolic origin of biological macromolecules was distributed within large populations of cells that kept exchanging metabolic pathways and primitive genetic set‐ups, we favoured the scenario proposed by Charles Kurland where primitive cells made populations of fairly large scavenger organisms (a last ancestral cell ensemble – LACE) in a predator/ prey dialog (see https://www.youtube.com/watch?v=3iD0RiNw9B4 and Danchin et al. 2020). In this scenario, H4F, formate and nucleotides were primitive compounds. They were linked to metabolic pathways involving iron‐sulfur clusters, followed by emergence of methionine and AdoMet, then RNAs and RNA metabolism. A crucial step at this early stage was the RNA‐dependent formation of polypeptides that ended up in the process of translation. A crucial family of associated functions was that of hydrolases, enzymes that often use Zn2+ at their catalytic centre. This ion would thus have gained its present role as an integrator of metabolism very early on. Highly specific chelators such as gliotoxin should help us investigate further in depth this scenario, which, if substantiated, will be critical for evolved metabolic engineering.
Conflict of interest
None declared.
Microbial Biotechnology (2020) 13(4), 895–898
Funding Information
No funding information provided.
References
- Andreini, C. , Bertini, I. , and Rosato, A. (2009) Metalloproteomes: a bioinformatic approach. Acc Chem Res 42: 1471–1479. [DOI] [PubMed] [Google Scholar]
- Bigay, J. , Deterre, P. , Pfister, C. , and Chabre, M. (1985) Fluoroaluminates activate transducin‐GDP by mimicking the gamma‐phosphate of GTP in its binding site. FEBS Lett 191: 181–185. [DOI] [PubMed] [Google Scholar]
- Broeders, M. , Herrero‐Hernandez, P. , Ernst, M.P.T. , van der Ploeg, A. T. , and Pijnappel, W.W.M.P. (2019) Sharpening the molecular scissors: Advances in gene‐editing technology. iScience 23: 100789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bütof, L. , Große, C. , Lilie, H. , Herzberg, M. , and Nies, D.H. (2019) Interplay between the Zur regulon components and metal resistance in Cupriavidus metallidurans . J Bacteriol 201: e00192–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabot, C. , Martos, S. , Llugany, M. , Gallego, B. , Tolrà, R. , and Poschenrieder, C. (2019) A Role for zinc in plant defense against pathogens and herbivores. Front Plant Sci 10: 1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z. , Sun, Q. , Xi, Y. , and Owens, G. (2008) Speciation of metal‐EDTA complexes by flow injection analysis with electrospray ionization mass spectrometry and ion chromatography with inductively coupled plasma mass spectrometry. J Sep Sci 31: 3796–3802. [DOI] [PubMed] [Google Scholar]
- Cirillo, V.P. , and Razin, S. (1973) Distribution of a phosphoenolypyruvate‐dependent sugar phosphotransferase system in mycoplasms. J Bacteriol 113: 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, J.E. (1998) Zinc enzymes. Curr Opin Chem Biol 2: 222–234. [DOI] [PubMed] [Google Scholar]
- Danchin, A. , Fang, G. , Sekowska, A. , and You, C. (2020) One‐carbon metabolism, folate, zinc and translation. Microbial Biotech 13: 687–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne, K.L. , Bergeron, D. , Landry‐Voyer, A.‐M. , and Bachand, F. (2019) The 40S ribosomal protein uS5 (RPS2) assembles into an extraribosomal complex with human ZNF277 that competes with the PRMT3‐uS5 interaction. J Biol Chem 294: 1944–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsbach, P. , and Schwartz, I.L. (1959) Studies on the sodium and potassium transport in rabbit polymorphonuclear leukocytes. J Gen Physiol 42: 883–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom, K.S. , Cheong, J.S. , and Lee, S.J. (2016) Structural analyses of zinc finger domains for specific interactions with DNA. J Microbiol Biotechnol 26: 2019–2029. [DOI] [PubMed] [Google Scholar]
- Gupta, R. , Walvekar, A.S. , Liang, S. , Rashida, Z. , Shah, P. , and Laxman, S. (2019) A tRNA modification balances carbon and nitrogen metabolism by regulating phosphate homeostasis. Elife 8: e44795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haën, C. (1974) Adenylate cyclase. A new kinetic analysis of the effects of hormones and fluoride ion. J Biol Chem 249: 2756–2762. [PubMed] [Google Scholar]
- Hensley, M.P. , Gunasekera, T.S. , Easton, J.A. , Sigdel, T.K. , Sugarbaker, S.A. , Klingbeil, L. , et al (2012) Characterization of Zn(II)‐responsive ribosomal proteins YkgM and L31 in E. coli . J Inorg Biochem 111: 164–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Y. , Molt, R.W. , and Blackburn, G.M. (2017) Metal fluorides: tools for structural and computational analysis of phosphoryl transfer enzymes. Top Curr Chem (Cham) 375: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, J. , Özkaya, Ö. , and Kümmerli, R. (2019) Bacterial siderophores in community and host interactions. Nat Rev Microbiol 18: 152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krezel, A. , and Maret, W. (2016) The biological inorganic chemistry of zinc ions. Arch Biochem Biophys 611: 3–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard, J. C. (2013) Growth media for E. coli In, Methods in Enzymology. Amsterdam, Netherlands: Elsevier, pp. 181–189. [DOI] [PubMed] [Google Scholar]
- Maguire, M.E. , Sturgill, T.W. , and Gilman, A.G. (1975) Frustration and adenylate cyclase. Metab Clin Exp 24: 287–299. [DOI] [PubMed] [Google Scholar]
- Manno, J.A. , and Schachter, D. (1970) Energy‐coupled influx of thiomethylgalactoside into Escherichia coli . J Biol Chem 245: 1217–1223. [PubMed] [Google Scholar]
- Miller, J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory edition. [Google Scholar]
- National Academies of Sciences, Engineering, and Medicine; Policy and Global Affairs; Committee on Science, Engineering, Medicine, and Public Policy; Board on Research Data and Information; Division on Engineering and Physical Sciences; Committee on Applied and Theoretical Statistics; Board on Mathematical Sciences and Analytics; Division on Earth and Life Studies; Nuclear and Radiation Studies Board; Division of Behavioral and Social Sciences and Education; Committee on National Statistics; Board on Behavioral, Cognitive, and Sensory Sciences; Committee on Reproducibility and Replicability in Science (2019) Reproducibility and Replicability in Science. Washington, DC: National Academies Press (US). [Google Scholar]
- Nies, D.H. (2019) The ancient alarmone ZTP and zinc homeostasis in Bacillus subtilis . Mol Microbiol 112: 741–746. [DOI] [PubMed] [Google Scholar]
- Palmer, L.D. , and Skaar, E.P. (2016) Transition metals and virulence in bacteria. Annu Rev Genet 50: 67–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier, M.H. , and Feucht, B.U. (1980) Regulation of carbohydrate transport activities in Salmonella typhimurium: use of the phosphoglycerate transport system to energize solute uptake. J Bacteriol 141: 611–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H. , Kang, S. , Park, Y.‐S. , and Yun, C.‐W. (2019) The role of zinc in gliotoxin biosynthesis of Aspergillus fumigatus . Int J Mol Sci 20: E6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shippy, D.C. , and Fadl, A.A. (2015) RNA modification enzymes encoded by the gid operon: Implications in biology and virulence of bacteria. Microb Pathog 89: 100–107. [DOI] [PubMed] [Google Scholar]
- Sridhar, S. , and Steele‐Mortimer, O. (2016) Inherent variability of growth media impacts the ability of Salmonella typhimurium to interact with host cells. PLoS ONE 11: e0157043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternweis, P.C. , and Gilman, A.G. (1982) Aluminum: a requirement for activation of the regulatory component of adenylate cyclase by fluoride. Proc Natl Acad Sci USA 79: 4888–4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, M. , and Lipmann, F. (1969) Isolation of adenyl cyclase from Escherichia coli . Proc Natl Acad Sci USA 63: 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasatava, Y. , Rosato, A. , Furnham, N. , Thornton, J.M. , and Andreini, C. (2018) To what extent do structural changes in catalytic metal sites affect enzyme function? J Inorg Biochem 179: 40–53. [DOI] [PMC free article] [PubMed] [Google Scholar]


