Summary
Filamentous fungi are important cell factories for large‐scale enzyme production. However, production levels are often low, and this limitation has stimulated research focusing on the manipulation of genes with predicted function in the protein secretory pathway. This pathway is the major route for the delivery of proteins to the cell exterior, and a positive relationship between the production of recombinant enzymes and the unfolded protein response (UPR) pathway has been observed. In this study, Aspergillus nidulans was exposed to UPR‐inducing chemicals and differentially expressed genes were identified by RNA‐seq. Twelve target genes were deleted in A. nidulans recombinant strains producing homologous and heterologous GH10 xylanases. The knockout of pbnA (glycosyltransferase), ydjA (Hsp40 co‐chaperone), trxA (thioredoxin) and cypA (cyclophilin) improved the production of the homologous xylanase by 78, 171, 105 and 125% respectively. Interestingly, these deletions decreased the overall protein secretion, suggesting that the production of the homologous xylanase was specifically altered. However, the production of the heterologous xylanase and the secretion of total proteins were not altered by deleting the same genes. Considering the results, this approach demonstrated the possibility of rationally increase the production of a homologous enzyme, indicating that trxA, cypA, ydjA and pbnA are involved in protein production by A. nidulans.
Filamentous fungi are important cell factories for large‐scale enzyme production. However, production levels are often low, and this limitation has stimulated research focusing on the manipulation of genes with predicted function in the protein secretory pathway. Single deletion of four genes with predicted function in different metabolic processes improved the production of an Aspergillus nidulans homologous xylanase.

Introduction
Microbial enzymes find a myriad of applications in many fields such as chemical, fermentation, agricultural, textile, pharmaceuticals and food production (Singh et al., 2019). Choosing a suitable expression system is critical for high‐yield enzyme production, and many bacteria, filamentous fungi and yeasts have been commonly used to express recombinant enzymes (Demain and Vaishnav, 2009). In the bioenergy field, techno‐economic analysis demonstrated that enzyme costs are much higher than generally considered. Therefore, the optimization of cell factories for large‐scale processes can contribute to reducing the costs of enzyme production (Klein‐Marcuschamer et al., 2012).
Aspergillus is one of the most studied genera of filamentous fungi due to the medical, food spoilage and industrial relevance of some species (Gabrielli et al., 2014; Frisvad et al., 2018; Taniwaki et al., 2018). Several Aspergilli have a long history as producers of plant polysaccharide modifying and degrading enzymes, which allows broad utilization of carbon sources from different substrates. Aspergillus nidulans is a genetic model that has been extensively studied regarding metabolism, cellular development and regulation, contributing to the understanding of eukaryotic cell biology and molecular processes (Brandl and Andersen, 2017). The main advantage of filamentous fungi, in relation to other microbial cell factories, is the potential to produce high quantities of extracellular enzymes (Brandl and Andersen, 2017; de Vries et al., 2017). Differently from prokaryotes, which have simpler pathways of protein secretion and is frequently essential for the organism fitness and pathogenicity (Maffei et al., 2017), the secretion of proteins in eukaryotes, such as filamentous fungi, involves a more complex machinery in which N‐glycosylation can influence considerably (Dai et al., 2013).
Random mutagenesis generally used for strain breeding may introduce many mutations into genomic DNA, resulting in higher productivity, though mutations are mostly unknown. On the other hand, rational genetic manipulation of target genes is time‐consuming (Nevalainen and Peterson, 2014). Strain engineering by deletion of extracellular protease genes (Zhang et al., 2014), deletion of autophagic genes (Yoon et al., 2013), overexpression or deletion of genes associated with the secretory pathway (Schalén et al., 2016) and manipulation of unfolded protein response (UPR) pathway (Yu et al., 2017) have been commonly explored as a rational approach to increase protein production in fungal systems. Furthermore, improvement of protein secretion has also been obtained by introducing multiple copies of heterologous genes (Liu et al., 2014), by using native or artificial strong regulators (Zhang et al., 2017), gene fusion to a well‐secreted homologous protein, using native signal sequences and others (Schalén et al., 2016).
An important drawback when using fungi as cell factory is that the expressed heterologous protein can be lost or stuck in the secretory pathway due to issues in processing, post‐translational modifications (PTMs) or misfolding, thus inducing the endoplasmic reticulum (ER) stress. ER stress activates UPR to alleviate stress and restore homeostasis, promoting cell survival and adaptation (Heimel, 2015; Sun and Su, 2019). Under low levels of unfolded proteins, Ire1 is bound to BiP that is an ER‐resident Hsp70 chaperone. However, in cases of high levels of unfolded proteins, BiP is dissociated from Ire1, resulting in the Ire1 oligomerization and trans‐autophosphorylation. Consequently, Ire1 conformation is altered, which enables the ability to bind in cis‐acting elements present in the promoters of UPR‐target genes, known as UPREs. Ire1 homodimer activates the RNase domain that has hac1u (uninduced) as its substrate, removing an unconventional intron resulting in the hac1i (induced) mRNA. This results in the translation of Hac1p, the bZIP transcription factor responsible for the restoration of ER homeostasis (Krishnan and Askew, 2014). Sims et al. (2005) showed that approximately 23% of genes upregulated in A. nidulans recombinant strain producing chymosin were also upregulated when UPR was chemically induced by dithiothreitol (DTT). In addition, transcriptional analysis of mutant yeast strains with improved secretion of a heterologous α‐amylase showed a series of altered cellular processes, and the balancing of amino acid biosynthesis seemed to be particularly important (Huang et al., 2017). Approximately 600 strains of Neurospora crassa carrying single deletion in genes with predicted functions in the secretory pathway were investigated for alterations in secretion, and seven strains hyperproducing cellulolytic enzymes were identified. Mutants implicated the loss of the Sterol Regulatory Element Binding Protein (SREBP) pathway (Reilly et al., 2015; Qin et al., 2017).
Here, a data bank of A. nidulans differentially expressed (DE) genes under chemically induced ER stress was generated by RNA‐seq using DTT and tunicamycin (Tm). Twelve target DE genes with predicted function in the secretory pathway were selected for single‐gene deletions in A. nidulans recombinant strains producing homologous and heterologous xylanases. After the single deletion of four genes with predicted function in different metabolic processes, the production of the homologous xylanase was increased, whereas the production of the heterologous xylanase remained unaltered. In addition, we investigated the presence of UPREs in the promoters of the deleted genes since those elements were described in Aspergillus niger (Mulder et al., 2006), but not in A. nidulans.
Results and discussion
Initially, two A. nidulans strains producing recombinant endo‐xylanases (EC 3.2.1.8) were constructed to evaluate the expression and production of recombinant enzymes classified in the same glycoside hydrolase (GH) family but from different microbial sources. The gene encoding for the heterologous xylanase (tpet_0854) was isolated from the hyperthermophilic bacterium Thermotoga petrophila, while AN7401/xlnE was selected as a homologous model. These xylanases share 53% of amino acid sequence similarity and belong to the family GH10 (Fig. 1A), and both enzymes have two predicted N‐glycosylation sites (Fig. S1).
Figure 1.
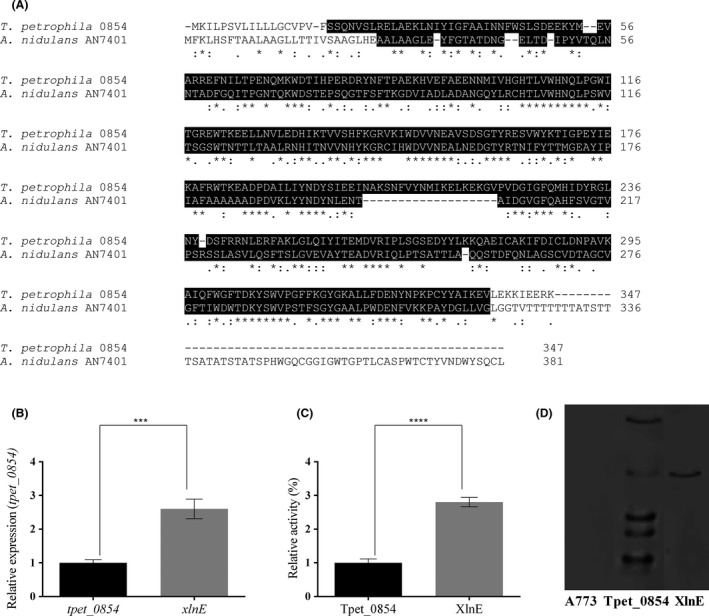
Overview of homologous and heterologous xylanase sequence, expression and production in A. nidulans. The A. nidulans Tpet_0854 and XlnE strains were grown in liquid MM supplemented with 2% maltose for 36 h at 37°C.
A. In silico protein alignment of xylanase sequences. GH10 domains are highlighted in black.
B. Gene expression calculated by qPCR using the Relative Standard Curve Method. The tubC (tubulin) and tpet_0854 genes were used as reference and calibrators respectively. (***) P‐values between 0.001 and 0.0001.
C. Xylanase activity in the crude extracellular filtrate produced by the A. nidulans Tpet_0854 and XlnE strains. (***) P‐values between 0.001 and 0.0001.
D. Southern analysis. Probes were obtained by PCR amplification of the xylanase gene from Thermotoga petrophila and the region between AN7401 and the pyrGAf gene for the A. nidulans Tpet_0854 and XlnE strain, respectively. A773: wild‐type.
Both genes were cloned into the pEXPYR expression vector and transformed into A. nidulans (Segato et al., 2012). xlnE was more expressed (Fig. 1B), and XlnE was more secreted than Tpet_0854 (Fig. 1C and Fig. S2), despite the higher copy numbers of tpet_0854 (5 copies) compared with the single copy of xlnE in the genomes of the recombinant strains (Fig. 1D) (Appendix S1). Although the native gene was present in the genome and the reinsertion of the xlnE in random locus was carried out, the transformed xlnE was not inserted into the native site (Fig. S3). Moreover, no native xylanase production was detected in all the enzyme production experiments carried out in minimal media supplemented with maltose. Only the recombinant XlnE was produced since it is under the control of the glucoamylase promoter.
To generate a database of genes induced by chemical stress, the expression of canonical UPR markers, such as bipA and hacA (hacAi and hacAu), was analysed by quantitative PCR (qPCR) (Appendix S1). The expression levels of these genes were higher at early stages (2 h) than at later periods (8 h) after chemical stress induction (Fig. S4), and thus short exposition periods were selected to perform RNA‐seq analysis (Appendix S1). Cellular stress was induced with Tm and DTT, which prevent N‐glycosylation of newly synthesized proteins and the formation of disulphide bonds, respectively (Fan et al., 2018; Yoo et al., 2018). A total of 294 million reads were generated, more than 94% of the filtered reads were mapped to the A. nidulans FGSC A4 reference genome, and a total of 10,774 genes were analysed (Table S1). For DTT, 1905 and 1172 genes were DE at 2 and 8 h, respectively, while 312 and 1862 genes were DE at 2 and 8 h of exposition to Tm respectively (Fig. S5A and B, Table S2).
Gene ontology (GO) enrichment of upregulated genes was performed for functional analysis (Appendix S1). The GO term oxidoreductase activity was enriched in the 2 h DTT treatment. Catalytic activity was found highly enriched, and lipid metabolic process, carboxylic acid metabolic process, organic acid metabolic process, cellular amino acid metabolic process and oxoacid metabolic process were moderately enriched in the 8 h DTT treatment. Regarding downregulated genes, the GO terms organonitrogen compound metabolic process, lipid metabolic process, catalytic activity and cytoplasm were enriched in 2 h DTT, whereas oxidoreductase activity and extracellular region were enriched in 8 h DTT. Additionally, the GO term secondary metabolic process was enriched in the upregulated genes in the 2 h Tm treatment, whereas intracellular was enriched in 8 h Tm (Fig. S5C).
The RNA‐seq data demonstrated to be consistent with previous works (Travers et al., 2000; Sims et al., 2005; Guillemette et al., 2007; Carvalho et al., 2012). The presence of transcripts related to oxidoreductase activity, mainly at the 8 h chemical treatment, could be partially explained by the role of DTT as a reducing agent. In addition, sustained ER stress was reported to cause oxidative stress in yeast cells by the UPR‐regulated oxidative folding machinery in the ER and mitochondria (Haynes et al., 2004). This seems to be a ‘side effect’ of cellular response to ER stress, that is the oxidative protein folding intensified by loading of client proteins into the ER somehow results in ROS production. Lipid and cellular amino acid metabolic processes were also enriched, which could be explained by the ER expansion to accommodate higher influx of newly synthesized proteins (Pineau and Ferreira, 2010) and by the supply of cells with amino acids to synthesize proteins involved in the protection against reactive species respectively (Harding et al., 2003; Herzog et al., 2013). Moreover, downregulation of extracellular proteins could be associated with the so‐called process repression under secretion stress (RESS), which is a feedback mechanism activated in response to impairment in protein folding or transport (Pakula et al., 2003).
Genes with predicted function in the secretory pathway were selected using a previously reported secretory model for Aspergillus oryzae (Liu et al., 2014). A total of 375 homologous genes were found in A. nidulans by using the Aspergillus Genome Database (AspGD; Table S3). Among them, 114 were upregulated and 53 genes were downregulated under DTT treatment, whereas 27 genes were upregulated and 13 genes downregulated under Tm treatment (Fig. S6A). Target genes for single deletion in A. nidulans were selected based on three criteria: (i) genes up‐ or downregulated in both treatment periods (2 and 8 h); (ii) top 25 genes with the highest or lowest fold‐change; and (iii) genes with no redundant function in the secretory pathway, covering a wide range of biological processes. These rankings resulted in the selection of 12 genes (Fig. S6B and Table S4).
The following step in our strategy was the deletion of the target genes in the A. nidulans strains producing XlnE and Tpet_0854 in order to evaluate their influence on the production of distinct recombinant xylanases. Initially, auxotrophy for uridine and uracil was regenerated by selecting XlnE and Tpet_0854 strains growing in MM with 5‐fluorotic acid (5‐FOA) (Appendix S1). A total of 20‐30 colonies presenting 5‐FOA‐resistance were obtained after three days of cultivation. After monosporic purification, more than 95% of the isolates showed uridine and uracil auxotrophy (pyrG‐) while maintaining equivalent levels of xylanase secretion compared with their parental strains (data not shown). Gene deletions in XlnE and Tpet_0854 pyrG‐ strains were confirmed by diagnostic PCR and Southern blot (Appendix S1) (Fig. S7).
XlnE, ΔpbnA, ΔydjA, ΔtrxA and ΔcypA strains showed 78, 171, 105 and 125% higher xylanase activity than the control strain at 36 h of induction (Fig. 2A), respectively, and even higher after 48 h (Fig. S8) (Appendix S1).
Figure 2.
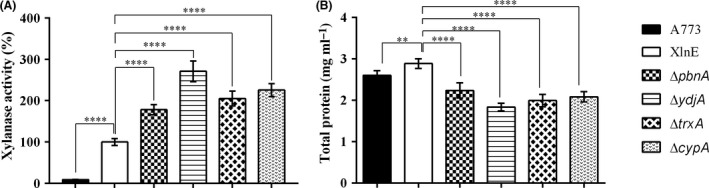
Extracellular xylanase and total proteins secretion by the A. nidulans XlnE mutant strains. The A. nidulans XlnE strain and mutants were grown in liquid MM supplemented with 2% maltose for 36 h at 37°C.
A. Xylanase activity was assayed with Azo‐Xylan as substrate, pH 5.5 at 50°C, and data regarding the XlnE strain are shown.
B. Protein quantification was assayed by the BCA method. (**) P‐values between 0.01 and 0.001; (****) P‐values <0.0001.
Additionally, we investigated whether the improvement observed in xylanolytic activity resulted because of a specific increase in XlnE production or an increase in total protein secretion. Interestingly, lower protein secretion was observed in the mutant strains, suggesting that single deletions specifically increased the XlnE production (Fig. 2B). On the other hand, no changes in xylanase production, nor in the levels of secreted proteins were observed in the Tpet_0854 mutant strains (Fig. S9).
In an attempt to address how the single deletions increased secretion of the homologous xylanase, we investigated the presence of UPREs in the promoters of pbnA, cypA, ydjA and trxA genes using the JASPAR database (Khan et al., 2018). Our analysis of A. nidulans promoters using the UPREs motifs found in Aspergillus niger was inconclusive. Thus, we used the hac1 upstream consensus sequence from S. cerevisiae (profile MA0310.1) as a model and the highest relative score was found for the following motifs: GACACGTC (0.969), AACACGTC (0.937), GCCACGTA (0.955), AACACATA (0.802), ACCACGTT (0.900), CCCACGTT (0.854) and GACACGTA (0.984) in the promoters of bipA, pdiA, hacA, pbnA, ydjA, trxA and cypA, respectively, suggesting that these genes may be regulated by UPR. The CACGT motif was not found in the pbnA promoter, suggesting that this gene is probably independent of hacA‐mediated UPR (Fig. 3A).
Figure 3.
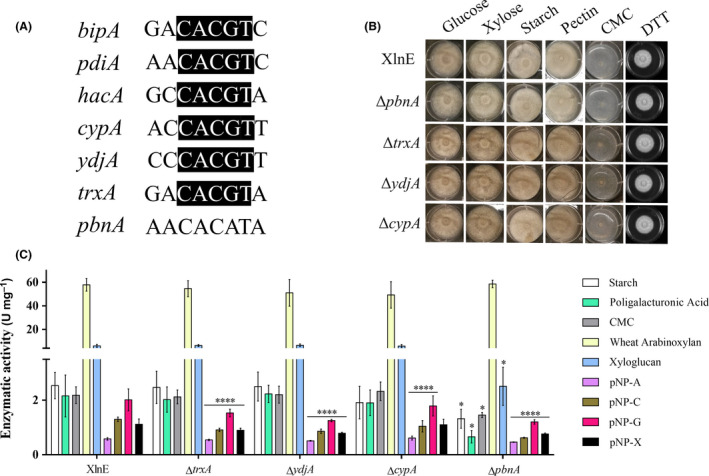
Carbohydrate‐active enzymes production in A. nidulans XlnE mutant strains.
A. Alignment of the putative promoter consensus sequences found in bipA, pdiA, hacA, cypA, ydjA, trxA and pbnA genes. Black box: identical sequence.
B. Growth of mutant strains on agar plates with different substrates and DTT for 3 days at 37°C.
C. Enzymatic activities in the supernatants of A. nidulans strains grown on 1% hydrothermal‐pretreated sugarcane bagasse, pH 6.5 for 3 days at 37°C. Enzymatic activity assays were performed with different substrates, pH 5.5 and at 50°C. (*) P‐values between 0.05 and 0.01 and (****) P‐values <0.0001.
The XlnE mutant strains were cultivated on MM agar plates supplemented with glucose, CMC, xylose, starch or pectin to investigate growth phenotypes as well as in the presence of DTT to evaluate chemical cellular stress resistance. Deleted genes were not essential to carbohydrate metabolism nor to metabolize DTT since all four mutant strains seem to equally grow in the different conditions (Fig. 3B). In addition, mutant strains were grown on hydrothermal‐pretreated sugarcane bagasse in order to quantify the secretion of different carbohydrate‐active enzymes (CAZymes) in the crude supernatants. Arabinofuranosidase, cellobiohydrolase, β‐glucosidase and β‐xylosidase activities were slightly lower in the XlnE mutant strains (except for β‐xylosidase activity in the ΔcypA strain), whereas amylase, pectinase, endoglucanase, xylanase and xyloglucanase activities were lower only in the ΔpbnA mutant (Fig. 3C) (Appendix S1). Furthermore, no morphological differences were detected in the mutant strains hyphae (Fig. S10).
For further understanding of pbnA, ydjA, trxA and cypA functions, a weighted gene co‐expression network analysis (WGCNA) was performed, resulting in the identification of 21 modules (Fig. S11). The pbnA, ydjA, trxA and cypA genes were, respectively, found in the following modules: lightyellow, brown, darkmagenta and lightcyan (Table S5). GO enrichment analysis of the modules demonstrated that these genes have potential functional links with several GO terms involved in a wide range of processes related to protein secretion (Fig. S12).
To date, pbnA was not characterized in A. nidulans, whereas the orthologous in S. cerevisiae (pbn1) encodes a chaperone‐like protein, an essential component of the glycosylphosphatidylinositol‐mannosyltransferase I complex. The lightyellow module was especially enriched for proteasome complex activity (Fig. S12A and Table S6). pbn1 physically interacts with prb1 (vacuolar proteinase B) and ecm27 (Na+/Ca2+ exchanger) in S. cerevisiae. The expression of pdiA and bipA was measured to evaluate whether pbnA deletion induced UPR activation or not. The genes pdiA and bipA were not overexpressed in the knockout strain, suggesting that UPR activation is not involved in the higher production of XlnE at least in 36 h (Fig. S13). This result, in addition to the reduction in total protein secretion (Fig. 2B) and the higher XlnE secretion (Fig. 2A), suggests that the cell converged for the XlnE production by reducing the traffic of other proteins in the ER. Such a compensation mechanism could then avoid UPR activation.
A group of chaperones that includes ydj1 is induced in strains overexpressing hac1 (Graf et al., 2008). ydjA was detected in the brown module (Fig. S12B and Table S6). This result indicates that this gene might have functions on intracellular protein transmembrane transport, protein refolding and ERAD pathway. In S. cerevisiae, ydj1 physically interacts with prd1, which encodes an intracellular proteinase involved in protein degradation (Buchler et al., 1994). In addition, ydj1 genetically interacts with pbn1 (pbnA in A. nidulans), tsa1 and cne1 (calnexin). Calnexin is a key ER chaperone that assists in folding and subunit assembly of most Asn‐linked glycoproteins passing through ER (Leach and Williams, 2013). Our results suggest ydjA is a chaperone that contributes to the folding of different A. nidulans proteins since ydjA knockout strain showed lower total protein secretion (Fig. 2B).
trxA encodes thioredoxin (trx1 orthologous in S. cerevisiae) that contributes to protection against reactive oxygen species (ROS; Thön et al., 2007). Detoxification mechanisms include the production of superoxide dismutases, catalases, peroxiredoxins, glutathione and the thioredoxin system, a dual system composed of thioredoxin and thioredoxin reductase (TrxR; Thön et al., 2007). trxA was co‐expressed with genes predicted to have function in cell redox homeostasis, fatty acid transport and ubiquitin‐dependent protein catabolism in the darkmagenta module (Fig. S12C and Table S6). trx1 has predicted physical interactions with trx2 (thioredoxin), ero1 (protein disulphide isomerase), met16 (3′‐phosphoadenylsulphate reductase), ahp1 (thiol‐specific peroxiredoxin), tsa1 (thioredoxin peroxidase) and mxr1 (methionine‐S‐sulfoxide reductase) in S. cerevisiae. Analysis of the genetic interactions showed crosstalk between trx1 and pbn1 networks through tsa1 and prb1 in S. cerevisiae. tsa1 genetically interacts with prb1, which, in turn, is physically linked to pbn1. It is likely to suggest that, although TrxA is very important for the folding of several proteins in A. nidulans, it did not affect the XlnE production.
Finally, cypA is a homologue to cpr1 in S. cerevisiae, which encodes a peptidyl‐prolyl cis‐trans isomerase (PPiases) catalysing the cis‐trans isomerization of peptide bonds in N‐terminal proline residues. cpr1 showed physical and genetic interactions with set3, hos4, hos2, sif2 and snt1 in S. cerevisiae. These genes, including cpr1, form the Set3p complex (SET3C), in which the subunits Hos2, Set3, Sif2 and Snt1 form the core complex, whereas Hst1p, Sum1p and Cpr1p are peripherally associated (Pijnappel et al., 2001). Many of these proteins are histone deacetylases (HDACs), and deacetylation has been identified as a major regulator of eukaryotic gene transcription due to a repressive chromatin structure (Kurdistani and Grunstein, 2003). Then, the SET3C complex is responsible for transcriptional repression of some genes related to the early/middle class of sporulation‐specific genes, including the key meiotic regulator in yeasts (Pijnappel et al., 2001). cypA was co‐expressed with genes with predicted functions in the hyphal tip, vacuole organization and response to heat (Fig. S12D and Table S6). How the deletion of pbnA, ydjA, trxA and cypA exactly impact XlnE production, but not Tpet_0854, remains to be determined.
Cellular stress was chemically induced in A. nidulans using DTT and Tm. Based on the RNA‐seq database, twelve genes with predicted function in the secretory pathway were deleted in A. nidulans recombinant strains producing homologous and heterologous GH10 endo‐xylanase. Higher enzyme production occurred only in the mutant strains (ΔpbnA, ΔydjA, ∆trxA and ΔcypA) producing the homologous xylanase. These four deleted genes have predicted functions in processes such as proteasome activity, protein refolding, ERAD pathway, hyphal tip, vacuole organization, autophagy, cell redox homeostasis, fatty acid transport and ubiquitin‐dependent degradation. The continuing characterization of pbnA, ydjA, trxA, cypA, as well as other mutants, will enhance our understanding of protein secretion in filamentous fungi, including industrially relevant species.
Conflict of interest
The authors declare no conflicts of interest.
Supporting information
Fig. S1. Structure and N‐glycosylation analysis of XlnE and Tpet_0854. The 3D models were constructed using Swiss Model based on Aspergillus aculeatus endo‐xylanase PDB http://www.rcsb.org/pdb/search/structidSearch.do?structureId=6Q8M (100% identity) for AN7401 XlnE, and T. petrophila xylanase 10B PDB http://www.rcsb.org/pdb/search/structidSearch.do?structureId=3NIY (100% identity) for Tpet_0854. In addition, the accessible surface area (ASA) and N‐glycosylation prediction were calculated for each site.
Fig. S2. Production of recombinant GH10 xylanases in A. nidulans. Secreted proteins were resolved by SDS‐PAGE. The A. nidulans strains XlnE and Tpet_0854 were grown in minimum media containing 2% maltose for 72 h at 37°C. The arrow indicates the position of the homologous and heterologous xylanase.
Fig. S3. Southern blot analysis. The probe was obtained by PCR amplification of the xlnE gene from A. nidulans. A773: reference strain; pEXPYR: A773 transformed with empty pEXPYR plasmid; AN7401 pyrG ‐: A773 transformed with the AN7401 gene (xlnE) cloned into the pEXPYR plasmid followed by pyrG recycling; AN7401 pyrG +: A773 transformed with the AN7401 gene (xlnE) cloned into the pEXPYR plasmid.
Fig. S4. Expression of UPR genes in A. nidulans A773 during exposure to DTT (20 mM) by qPCR analysis. (A) hacA total; (B) hacAi; C) bipA.
Fig. S5. Analysis of differentially expressed genes in A. nidulans exposed to cellular stress‐inducing chemicals. The A. nidulans A773 strain was grown in MM for 24 h, exposed to DTT and Tm for 2 and 8 h, and then analyzed by RNA‐seq. (A) Number of up‐ and down‐regulated genes. (B) Venn diagram showing the DE genes. (C) Enrichment analysis of GO terms over‐represented in the DE genes using the Blast2Go software. For the differential expression analysis, an adjusted P‐value ≤ 0.01 was used as threshold, and a cutoff of log2 fold change ≥ 1 or ≤ −1 for Tm treatment and log2 fold change ≥2 or ≤ −2 for DTT treatment.
Fig. S6. Expression profile of genes with predicted function in the A. nidulans secretory pathway under stress conditions. Genes identified in A. nidulans after DTT and Tm treatment for 2 and 8 h at 37°C. (A) Number of genes up‐ and down‐regulated. (B) Expression profile of the twelve genes chose to be deleted in A. nidulans recombinant strains.
Fig. S7. Strategy used for construction and confirmation of A. nidulans knocked out strains. (A) ΔtrxA, (B) Δcyp1, (C) ΔydjA and (D) ΔpbnA. The pyrG gene from Aspergillus fumigatus (pyrGAf) was used as a selectable marker. The transformants were purified by five rounds of monosporic purification and colonies containing genetic identical nuclei were submitted to two independent PCRs, one using primers anchored outside the flanking targeting sequence (F2/R1) and a second PCR using primers anchored within the cassette (F1/R1). After PCR, Southern Blot analysis was performed to confirm the deletion. Mutants gDNA were digested with NcoI and BamHI and 5′ flanking regions of each gene was used as a probe. Differences among the size of the detected fragment allowed differentiation of mutant and wild‐type strain. This strategy was used for all deleted genes.
Fig. S8. Extracellular xylanase activity. The A. nidulans XlnE strain and mutants were grown in liquid MM supplemented with 2% maltose for 48 h at 37°C. Xylanase activity was assayed with Azo‐Xylan as substrate, pH 5.5 at 50°C, and the data are shown relative to the XlnE strain. (****) adjusted P‐values < 0.0001.
Fig. S9. Xylanase activity and protein secretion by the A. nidulans Tpet_0854 mutant strains. (A) Enzymatic activity measured with Azo‐xylan. The assay was performed using 1 ug of protein at pH 5.5 and 50°C and expressed in relation to the activity of the control strain. (B) Evaluation of protein secretion yield.
Fig. S10. Mycelia morphology of A. nidulans strains at 400× (right column) and 1000× (left column) magnifications under an optical microscope. Strains were grown at 37°C in MM for 96 h. Wild‐type hyphae observed on control strain (XlnE). Mutant strains hyphae showed no morphological differences.
Fig. S11. Hierarchical cluster tree showing 21 modules of co‐expressed genes. Each of the 10 586 genes is represented by a leaf in the tree, and each of the 21 modules by a major tree branch. The lower panel shows modules in designated colors after merged, such as Black, Darkred, Pink and others.
Fig. S12. GO Biological Process enrichment analysis of the modules lightyellow/pbnA (A), brown/ydjA (B), darkmagenta/trxA (C) and lightcyan1/cypA (D).
Fig. S13. Expression of UPR targets. The A. nidulans XlnE and mutant strains were grown in liquid MM supplemented with 2% maltose for 36 h at 37°C. (B) Gene expression calculated by qPCR using the Relative Standard Curve Method. The gene tubC (tubulin) and was used as reference.
Methodology S1. R script of the WGCNA analysis.
Table S1. Expression data of A. nidulans genes after DTT and TM treatment.
Table S2. Differentially expressed (DE) genes in Aspergillus nidulans mycelia after DTT and TM treatment. For the differential expression analysis, an adjusted P‐value ≤ 0.01 was used as threshold, and a cutoff of log2 fold change ≥1 or ≤ −1 for TM treatment and log2 fold change ≥2 or ≤−2 for DTT treatment.
Table S3. Genes encoding predicted secretory proteins based on a previously reported secretory model from Aspergillus oryzae. A total of 375 homologous genes were found in A. nidulans by performing a search in the Aspergillus Genome Database (AspGD).
Table S4. List of selected genes deleted in A. nidulans.
Table S5. Modules of co‐expressed genes in the A. nidulans network.
Table S7. Strains used in this work.
Table S8. Primers used in this study.
Appendix S1. Experimental procedure.
Acknowledgements
MPZ, MVR, CRFT, EPA, FJC and AD were supported by FAPESP (São Paulo Research Foundation), grants no. 2014/15403‐6, 2013/24988‐5, 2016/16306‐0, 2017/26315‐9, 2017/10083‐1, 2012/20549‐4 and 2015/50612‐8. AD and CRFT were also supported by CNPq (Brazilian Council for Scientific and Technological Development), grants no. 404654/2018‐5 and 304816/2017‐5 and 420392/2018‐1 respectively. The authors acknowledge Espaço da Escrita – Pró‐Reitoria de Pesquisa – UNICAMP – for the language services provided. Research supported by LNBR – Brazilian Biorenewables National Laboratory (CNPEM/MCTIC) during the use of the NGS ‐ Next Generation Sequencing open access facility.
Microbial Biotechnology (2020) 13(4), 1245–1253
Funding information
MPZ, MVR, CRFT, EPA, FJC and AD were supported by FAPESP (São Paulo Research Foundation), grants no. 2014/15403‐6, 2013/24988‐5, 2016/16306‐0, 2017/26315‐9, 2017/10083‐1 and 2012/20549‐4. AD and CRFT were also supported by CNPq (Brazilian Council for Scientific and Technological Development), grants no. 404654/2018‐5 and 304816/2017‐5 and 420392/2018‐1 respectively.
References
- Brandl, J. , and Andersen, M.R. (2017) Aspergilli: models for systems biology in filamentous fungi. Curr Opin Syst Biol 6: 67–73. [Google Scholar]
- Buchler, M. , Tisljar, U. , and Wolf, D.H. (1994) Proteinase yscD (oligopeptidase yscD). Structure, function and relationship of the yeast enzyme with mammalian thimet oligopeptidase (metalloendopeptidase, EP 24.15). Eur J Biochem 219: 627–639. [DOI] [PubMed] [Google Scholar]
- Carvalho, N.D. , Jørgensen, T.R. , Arentshorst, M. , Nitsche, B.M. , van den Hondel, C.A. , Archer, D.B. , and Ram, A.F. (2012) Genome‐wide expression analysis upon constitutive activation of the HacA bZIP transcription factor in Aspergillus niger reveals a coordinated cellular response to counteract ER stress. BMC Genomics 13: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, Z. , Aryal, U.K. , Shukla, A. , Qian, W.J. , Smith, R.D. , Magnuson, J.K. , et al (2013) Impact of alg3 gene deletion on growth, development, pigment production, protein secretion, and functions of recombinant Trichoderma reesei cellobiohydrolases in Aspergillus niger . Fungal Genet Biol 61: 120–132. [DOI] [PubMed] [Google Scholar]
- Demain, A.L. , and Vaishnav, P. (2009) Production of recombinant proteins by microbes and higher organisms. Biotechnol Adv 27: 297–306. [DOI] [PubMed] [Google Scholar]
- Fan, F. , Zhang, Y. , Wang, S. , Han, Y. , Wang, L. , and Lu, D. (2018) Characterization of the oxidative protein folding activity of a unique plant oxidoreductase, Arabidopsis protein disulfide isomerase‐11. Biochem Biophys Res Commun 495: 1041–1047. [DOI] [PubMed] [Google Scholar]
- Frisvad, J.C. , Møller, L.L.H. , Larsen, T.O. , Kumar, R. , and Arnau, J. (2018) Safety of the fungal workhorses of industrial biotechnology: update on the mycotoxin and secondary metabolite potential of Aspergillus niger, Aspergillus oryzae, and Trichoderma reesei . Appl Microbiol Biotechnol 102: 9481–9515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielli, E. , Fothergill, A.W. , Brescini, L. , Sutton, D.A. , Marchionni, E. , Orsetti, E. , et al (2014) Osteomyelitis caused by Aspergillus species: a review of 310 reported cases. Clin Microbiol Infect 20: 559–565. [DOI] [PubMed] [Google Scholar]
- Graf, A. , Gasser, B. , Dragosits, M. , Sauer, M. , Leparc, G.G. , Tüchler, T. , et al (2008) Novel insights into the unfolded protein response using Pichia pastoris specific DNA microarrays. BMC Genomics 390: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette, T. , Van Peij, N.N.M.E. , Goosen, T. , Lanthaler, K. , Robson, G.D. , Van Hondel, C.A. , et al (2007) Genomic analysis of the secretion stress response in the enzyme‐producing cell factory Aspergillus niger . BMC Genomics 8: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding, H.P. , Zhang, Y. , Zeng, H. , Novoa, I. , Lu, P.D. , Calfon, M. , et al (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11: 619–633. [DOI] [PubMed] [Google Scholar]
- Haynes, C.M. , Titus, E.A. , and Cooper, A.A. (2004) Degradation of misfolded proteins prevents ER‐derived oxidative stress and cell death. Mol Cell 15: 767–776. [DOI] [PubMed] [Google Scholar]
- Heimel, K. (2015) Unfolded protein response in filamentous fungi – implications in biotechnology. Appl Microbiol Biotechnol 99: 121–132. [DOI] [PubMed] [Google Scholar]
- Herzog, B. , Popova, B. , Jakobshagen, A. , Shahpasandzadeh, H. , and Braus, G.H. (2013) Mutual cross talk between the regulators Hac1 of the unfolded protein response and Gcn4 of the general amino acid control of Saccharomyces cerevisiae . Eukaryot Cell 12: 1142–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, M. , Bao, J. , Hallström, B.M. , Petranovic, D. , and Nielsen, J. (2017) Efficient protein production by yeast requires global tuning of metabolism. Nat Commun 8: 1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, A. , Fornes, O. , Stigliani, A. , Gheorghe, M. , Castro‐Mondragon, J.A. , van der Lee, R. , et al (2018) JASPAR 2018: update of the open‐access database of transcription factor binding profiles and its web framework. Nucleic Acids Res 46: 260–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein‐Marcuschamer, D. , Oleskowicz‐Popiel, P. , Simmons, B.A. , and Blanch, H.W. (2012) The challenge of enzyme cost in the production of lignocellulosic biofuels. Biotechnol Bioeng 109: 1083–1087. [DOI] [PubMed] [Google Scholar]
- Krishnan, K. , and Askew, D.S. (2014) The fungal UPR: A regulatory hub for virulence traits in the mold pathogen Aspergillus fumigatus . Virulence 5: 334–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdistani, S.K. , and Grunstein, M. (2003) Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol 4: 276–284. [DOI] [PubMed] [Google Scholar]
- Leach, M.R. , and Williams, D.B . (2013) Calnexin and Calreticulin. Molecular Chaperones of the Endoplasmic Reticulum In Madame Curie Bioscience Database. Austin, TX, USA: Landes Bioscience, pp. 2000–2013. [Google Scholar]
- Liu, L. , Feizi, A. , Österlund, T. , Hjort, C. , and Nielsen, J. (2014) Genome‐scale analysis of the high‐efficient protein secretion system of Aspergillus oryzae . BMC Syst Biol 8: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffei, B. , Francetic, O. , and Subtil, A. (2017) Tracking proteins secreted by bacteria: what’s in the toolbox? Front Cell Infect Microbiol 7: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder, H.J. , Nikolaev, I. , and Madrid, S.M. (2006) HACA, the transcriptional activator of the unfolded protein response (UPR) in Aspergillus niger, binds to partly palindromic UPR elements of the consensus sequence 5′‐CAN(G/A)NTGT/GCCT‐3′. Fungal Genet Biol 43: 560–572. [DOI] [PubMed] [Google Scholar]
- Nevalainen, H. , and Peterson, R. (2014) Making recombinant proteins in filamentous fungi‐ are we expecting too much? Front Microbiol 5: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakula, T.M. , Laxell, M. , Huuskonen, A. , Uusitalo, J. , Saloheimo, M. , and Penttilä, M. (2003) The effects of drugs inhibiting protein secretion in the filamentous fungus Trichoderma reesei. Evidence for down‐regulation of genes that encode secreted proteins in the stressed cells. J Biol Chem 278: 45011–45020. [DOI] [PubMed] [Google Scholar]
- Pijnappel, W.W. , Schaft, D. , Roguev, A. , Shevchenko, A. , Tekotte, H. , Wilm, M. , et al (2001) The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic‐specific repressor of the sporulation gene program. Genes Dev 15: 2991–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau, L. , and Ferreira, T. (2010) Lipid‐induced ER stress in yeast and β cells: parallel trails to a common fate. FEMS Yeast Res 10: 1035–1045. [DOI] [PubMed] [Google Scholar]
- Qin, L. , Wu, V.W. , and Glass, N.L. (2017) Deciphering the regulatory network between the SREBP pathway and protein pecretion in Neurospora crassa . MBio 8: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly, M.C. , Qin, L. , Craig, J.P. , Starr, T.L. , and Glass, N.L. (2015) Deletion of homologs of the SREBP pathway results in hyper‐production of cellulases in Neurospora crassa and Trichoderma reesei . Biotechnol Biofuels 8: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalén, M. , Anyaogu, D.C. , Hoof, J.B. , and Workman, M. (2016) Effect of secretory pathway gene overexpression on secretion of a fluorescent reporter protein in Aspergillus nidulans . Fungal Biol Biotechnol 3: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segato, F. , Damásio, A.R.L. , Gonçalves, T.A. , de Lucas, R.C. , Squina, F.M. , Decker, S.R. , and Prade, R.A. (2012) High‐yield secretion of multiple client proteins in Aspergillus. Enzyme Microb Technol 51: 100–106. [DOI] [PubMed] [Google Scholar]
- Sims, A.H. , Gent, M.E. , Lanthaler, K. , Nigel, S. , Oliver, S.G. , Robson, G.D. , and Dunn‐coleman, N.S. (2005) Transcriptome analysis of recombinant protein secretion by Aspergillus nidulans and the unfolded‐protein response in vivo. Appl Environ Microbiol 71: 2737–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R.S. , Singh, T. , and Pandey, A . (2019) Microbial enzymes—an overview In Advances in Enzyme Technology Biomass, Biofuels, Biochemicals. Singh R.S., Singhania R.R., Pandey A., and Larroche C. (eds). Amsterdam, the Netherlands: Elsevier, pp. 1–40. [Google Scholar]
- Sun, X. , and Su, X. (2019) Harnessing the knowledge of protein secretion for enhanced protein production in filamentous fungi. World J Microbiol Biotechnol 35. [DOI] [PubMed] [Google Scholar]
- Taniwaki, M.H. , Pitt, J.I. , and Magan, N. (2018) Aspergillus species and mycotoxins: occurrence and importance in major food commodities. Curr Opin Food Sci 23: 38–43. [Google Scholar]
- Thön, M. , Al‐Abdallah, Q. , Hortschansky, P. , and Brakhage, A. (2007) The thioredoxin system of the filamentous fungus Aspergillus nidulans: impact on development and oxidative stress response. J Biol Chem 282: 27259–27269. [DOI] [PubMed] [Google Scholar]
- Travers, K.J. , Patil, C.K. , Wodicka, L. , Lockhart, D.J. , Weissman, J.S. , Walter, P. , and Francisco, S. (2000) Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER‐associated degradation. Cell 101: 249–258. [DOI] [PubMed] [Google Scholar]
- de Vries, R.P. , Riley, R. , Wiebenga, A. , Aguilar‐Osorio, G. , Amillis, S. , Uchima, C.A. , et al (2017) Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol 18: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, J. , Mashalidis, E.H. , Kuk, A.C.Y. , Yamamoto, K. , Kaeser, B. , Ichikawa, S. , and Lee, S.Y. (2018) GlcNAc‐1‐P‐transferase–tunicamycin complex structure reveals basis for inhibition of N‐glycosylation. Nat Struct Mol Biol 25: 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, J. , Kikuma, T. , Maruyama, J. , and Kitamoto, K. (2013) Enhanced production of bovine chymosin by autophagy deficiency in the filamentous fungus Aspergillus oryzae . PLoS One 8: e62512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X.W. , Sun, W.H. , Wang, Y.Z. , and Xu, Y. (2017) Identification of novel factors enhancing recombinant protein production in multi‐copy Komagataella phaffii based on transcriptomic analysis of overexpression effects. Sci Rep 7: 16249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, G. , Zhu, Y. , Wei, D. , and Wang, W. (2014) Enhanced production of heterologous proteins by the filamentous fungus Trichoderma reesei via disruption of the alkaline serine protease SPW combined with a pH control strategy. Plasmid 71: 16–22. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Li, Y. , Zhao, X. , and Bai, F. (2017) Constitutive cellulase production from glucose using the recombinant Trichoderma reesei strain overexpressing an artificial transcription activator. Bioresour Technol 223: 317–322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Structure and N‐glycosylation analysis of XlnE and Tpet_0854. The 3D models were constructed using Swiss Model based on Aspergillus aculeatus endo‐xylanase PDB http://www.rcsb.org/pdb/search/structidSearch.do?structureId=6Q8M (100% identity) for AN7401 XlnE, and T. petrophila xylanase 10B PDB http://www.rcsb.org/pdb/search/structidSearch.do?structureId=3NIY (100% identity) for Tpet_0854. In addition, the accessible surface area (ASA) and N‐glycosylation prediction were calculated for each site.
Fig. S2. Production of recombinant GH10 xylanases in A. nidulans. Secreted proteins were resolved by SDS‐PAGE. The A. nidulans strains XlnE and Tpet_0854 were grown in minimum media containing 2% maltose for 72 h at 37°C. The arrow indicates the position of the homologous and heterologous xylanase.
Fig. S3. Southern blot analysis. The probe was obtained by PCR amplification of the xlnE gene from A. nidulans. A773: reference strain; pEXPYR: A773 transformed with empty pEXPYR plasmid; AN7401 pyrG ‐: A773 transformed with the AN7401 gene (xlnE) cloned into the pEXPYR plasmid followed by pyrG recycling; AN7401 pyrG +: A773 transformed with the AN7401 gene (xlnE) cloned into the pEXPYR plasmid.
Fig. S4. Expression of UPR genes in A. nidulans A773 during exposure to DTT (20 mM) by qPCR analysis. (A) hacA total; (B) hacAi; C) bipA.
Fig. S5. Analysis of differentially expressed genes in A. nidulans exposed to cellular stress‐inducing chemicals. The A. nidulans A773 strain was grown in MM for 24 h, exposed to DTT and Tm for 2 and 8 h, and then analyzed by RNA‐seq. (A) Number of up‐ and down‐regulated genes. (B) Venn diagram showing the DE genes. (C) Enrichment analysis of GO terms over‐represented in the DE genes using the Blast2Go software. For the differential expression analysis, an adjusted P‐value ≤ 0.01 was used as threshold, and a cutoff of log2 fold change ≥ 1 or ≤ −1 for Tm treatment and log2 fold change ≥2 or ≤ −2 for DTT treatment.
Fig. S6. Expression profile of genes with predicted function in the A. nidulans secretory pathway under stress conditions. Genes identified in A. nidulans after DTT and Tm treatment for 2 and 8 h at 37°C. (A) Number of genes up‐ and down‐regulated. (B) Expression profile of the twelve genes chose to be deleted in A. nidulans recombinant strains.
Fig. S7. Strategy used for construction and confirmation of A. nidulans knocked out strains. (A) ΔtrxA, (B) Δcyp1, (C) ΔydjA and (D) ΔpbnA. The pyrG gene from Aspergillus fumigatus (pyrGAf) was used as a selectable marker. The transformants were purified by five rounds of monosporic purification and colonies containing genetic identical nuclei were submitted to two independent PCRs, one using primers anchored outside the flanking targeting sequence (F2/R1) and a second PCR using primers anchored within the cassette (F1/R1). After PCR, Southern Blot analysis was performed to confirm the deletion. Mutants gDNA were digested with NcoI and BamHI and 5′ flanking regions of each gene was used as a probe. Differences among the size of the detected fragment allowed differentiation of mutant and wild‐type strain. This strategy was used for all deleted genes.
Fig. S8. Extracellular xylanase activity. The A. nidulans XlnE strain and mutants were grown in liquid MM supplemented with 2% maltose for 48 h at 37°C. Xylanase activity was assayed with Azo‐Xylan as substrate, pH 5.5 at 50°C, and the data are shown relative to the XlnE strain. (****) adjusted P‐values < 0.0001.
Fig. S9. Xylanase activity and protein secretion by the A. nidulans Tpet_0854 mutant strains. (A) Enzymatic activity measured with Azo‐xylan. The assay was performed using 1 ug of protein at pH 5.5 and 50°C and expressed in relation to the activity of the control strain. (B) Evaluation of protein secretion yield.
Fig. S10. Mycelia morphology of A. nidulans strains at 400× (right column) and 1000× (left column) magnifications under an optical microscope. Strains were grown at 37°C in MM for 96 h. Wild‐type hyphae observed on control strain (XlnE). Mutant strains hyphae showed no morphological differences.
Fig. S11. Hierarchical cluster tree showing 21 modules of co‐expressed genes. Each of the 10 586 genes is represented by a leaf in the tree, and each of the 21 modules by a major tree branch. The lower panel shows modules in designated colors after merged, such as Black, Darkred, Pink and others.
Fig. S12. GO Biological Process enrichment analysis of the modules lightyellow/pbnA (A), brown/ydjA (B), darkmagenta/trxA (C) and lightcyan1/cypA (D).
Fig. S13. Expression of UPR targets. The A. nidulans XlnE and mutant strains were grown in liquid MM supplemented with 2% maltose for 36 h at 37°C. (B) Gene expression calculated by qPCR using the Relative Standard Curve Method. The gene tubC (tubulin) and was used as reference.
Methodology S1. R script of the WGCNA analysis.
Table S1. Expression data of A. nidulans genes after DTT and TM treatment.
Table S2. Differentially expressed (DE) genes in Aspergillus nidulans mycelia after DTT and TM treatment. For the differential expression analysis, an adjusted P‐value ≤ 0.01 was used as threshold, and a cutoff of log2 fold change ≥1 or ≤ −1 for TM treatment and log2 fold change ≥2 or ≤−2 for DTT treatment.
Table S3. Genes encoding predicted secretory proteins based on a previously reported secretory model from Aspergillus oryzae. A total of 375 homologous genes were found in A. nidulans by performing a search in the Aspergillus Genome Database (AspGD).
Table S4. List of selected genes deleted in A. nidulans.
Table S5. Modules of co‐expressed genes in the A. nidulans network.
Table S7. Strains used in this work.
Table S8. Primers used in this study.
Appendix S1. Experimental procedure.


