Summary
In contrast to the current paradigm of using microbial mono‐cultures in most biotechnological applications, increasing efforts are being directed towards engineering mixed‐species consortia to perform functions that are difficult to programme into individual strains. In this work, we developed a synthetic microbial consortium composed of two genetically engineered microbes, a cyanobacterium (Synechococcus elongatus PCC 7942) and a heterotrophic bacterium (Pseudomonas putida EM173). These microbial species specialize in the co‐culture: cyanobacteria fix CO2 through photosynthetic metabolism and secrete sufficient carbohydrates to support the growth and active metabolism of P. putida, which has been engineered to consume sucrose and to degrade the environmental pollutant 2,4‐dinitrotoluene (2,4‐DNT). By encapsulating S. elongatus within a barium–alginate hydrogel, cyanobacterial cells were protected from the toxic effects of 2,4‐DNT, enhancing the performance of the co‐culture. The synthetic consortium was able to convert 2,4‐DNT with light and CO2 as key inputs, and its catalytic performance was stable over time. Furthermore, cycling this synthetic consortium through low nitrogen medium promoted the sucrose‐dependent accumulation of polyhydroxyalkanoate, an added‐value biopolymer, in the engineered P. putida strain. Altogether, the synthetic consortium displayed the capacity to remediate the industrial pollutant 2,4‐DNT while simultaneously synthesizing biopolymers using light and CO2 as the primary inputs.
This work describes the development of a synthetic microbial consortium composed of a cyanobacterium engineered to use light to fix inorganic carbon, and a heterotrophic bacterium engineered to degrade the environmental pollutant, 2,4‐dinitrotoluene. Together, the synthetic consortium displayed the capacity to remediate the industrial compound 2,4‐DNT while simultaneously synthesizing biopolymers using light and carbon dioxide as the primary inputs.
Introduction
In nature, bacteria typically coexist in communities with hundreds to thousands of species, creating a complex web of interspecies metabolic reactions (Little et al., 2008; Saxena, 2015; Dolinšek et al., 2016; Goldford et al., 2018). Most consortia exhibit a high degree of division of labour, where individual species display specialized metabolism and exchange small molecules and signals with neighbours (Tan et al., 2015). Interactions range from the cooperative degradation of toluene and other aromatic compounds (Nikel et al., 2014; Tecon and Or, 2017) to the consumption of metabolic waste products (Wilkinson et al., 1974). Compartmentalization of biochemical transformations across distinct species can confer metabolic capabilities on a consortium that may be difficult to engineer within any one individual species (Roell et al., 2019). Natural microbial consortia also exhibit other desirable features, such as robustness in the face of fluctuating environmental conditions and invasive microbes (Blasche et al., 2017). Against this background, there has been increasing interest in rationally engineering microbial consortia for desired outputs to capitalize upon some of these potential benefits (Ortíz‐Marquez et al., 2013; McCarty and Ledesma‐Amaro, 2019; Tsoi et al., 2019).
Recently, cyanobacteria have been engineered to efficiently produce and secrete simple sugars, making them attractive as alternative feedstock crops to produce carbohydrates for consumption by other industrially relevant microbes (Carrieri et al., 2012; Xu et al., 2013; Hays and Ducat, 2015; Song et al., 2016; Hays et al., 2017; Santos‐Merino et al., 2019; Vijay et al., 2019). In one such example, the heterologous expression of a sucrose permease gene (cscB) in Synechococcus elongatus PCC 7942 (hereafter, S. elongatus CscB) enabled light‐driven secretion of sucrose at rates estimated to exceed the productivities of traditional plant feedstock crops (Ducat et al., 2012). One approach to utilize the sugars secreted by such cyanobacteria is to cultivate them with a heterotrophic partner that can convert the carbohydrate into value‐added products. Recent reports have shown that S. elongatus CscB can be paired with a variety of heterotrophic microbes, including Escherichia coli and Bacillus subtilis (Hays et al., 2017), Azotobacter vinelandii (Smith and Francis, 2016, 2017), Halomonas boliviensis (Weiss et al., 2017), Pseudomonas putida (Löwe et al., 2017a), Saccharomyces cerevisiae (Ducat et al., 2012) and Cryptococcus curvatus and Rhodotorula glutinis (Li et al., 2017). Such co‐cultures are typically stable over long time periods (weeks to months) and the heterotrophic partner strain can be programmed to output value‐added biological products, such as α‐amylase (Hays et al., 2017), fatty acids (Li et al., 2017) and polyhydroxyalkanoates (PHAs) [e.g. poly(3‐hydroxybutyrate) (Smith and Francis, 2016; Smith and Francis, 2017; Weiss et al., 2017; Löwe et al., 2017a,b)].
The limited availability of potable water influences the feasibility of algal and cyanobacterial applications; cultivation using marginal water supplies may ultimately be more sustainable (Barry et al., 2016). Industrial wastewater streams are often unsuitable for plant‐based agriculture because of the toxic contaminants they contain. 2,4‐Dinitrotoluene (2,4‐DNT) is a nitroaromatic xenobiotic that is produced as a by‐product during the synthesis of polyurethane, pesticides and explosives (Spain, 1995; Ju and Parales, 2010). A single 2,4,6‐trinitrotoluene (TNT)‐manufacturing plant can contaminate up to 500 000 gallons of water with nitroaromatics in a single day (Yinon, 1990) – and 2,4‐DNT is a significant toxic contaminant found at dangerous levels in water and soils (Hooker and Skeen, 1999). 2,4‐DNT is highly stable within the environment and remediation costs via incineration have been estimated at 400 USD per cubic yard of soil (Griest et al., 1995, 1998).
An alternative strategy for remediation of sites contaminated with 2,4‐DNT is the use of bacterial strains capable of mineralizing this toxic nitroaromatic (Symons and Bruce, 2006; Aburto‐Medina et al., 2017). Generally, two biological pathways that transform 2,4‐DNT are found in nature. The non‐specific reductive pathway is thought to involve reduction of 2,4‐DNT by common electron carriers such as flavin‐ or iron center‐containing electron donors (e.g. nitroreductases; French et al., 2001; Shemer et al., 2018). Reduction of the nitro group of 2,4‐DNT creates highly reactive nitroso‐intermediates and hydroxylamino‐derivatives, which are also toxic due to their capacity to bind a range of biomolecules, frequently causing cellular damage by forming DNA and protein adducts (Spain, 1995; Achtnich et al., 1999; Padda et al., 2003). The toxic derivatives formed by the reductive pathway are not fully mineralized by most bacteria and also have long persistence times in natural environments (Achtnich et al., 1999; Shemer et al., 2018). A separate pathway that initiates with 2,4‐DNT oxidation has been identified in some prokaryotes, including Burkholderia sp. R34 (McCormick et al., 1978; Spanggord et al., 1991; Suen et al., 1996), where it is encoded in a single gene cluster (Suen and Spain, 1993; Haigler et al., 1994; Parales et al., 1996; Johnson et al., 2000; Nishino et al., 2000). Enzymes encoded by the dnt gene cluster catalyse the oxidation of 2,4‐DNT through a series of steps ultimately resulting in the release of nitrite, pyruvate and propionyl‐coenzyme A (CoA) (Johnson et al., 2002). Because the products of the oxidative pathway are non‐toxic metabolites common to most microorganisms, the oxidative pathway is preferable from the perspective of bioremediation.
In this study, we expanded upon prior efforts of engineering synthetic cyanobacteria/heterotroph consortia (Hays and Ducat, 2015; Smith and Francis, 2016, 2017; Hays et al., 2017; Weiss et al., 2017; Li et al., 2017; Löwe et al., 2017a) with the goal of generating co‐cultures that can utilize contaminated water supplies and perform a bioremediation function while also producing a valuable bioproduct from light and CO2 (Fig. 1). We desired a heterotrophic partner that was genetically tractable and which could tolerate the toxic effects of 2,4‐DNT – therefore, we turned our attention to the soil bacterium P. putida, an emerging microbial chassis (Calero and Nikel, 2019) recognized for its high tolerance to aromatic organic compounds, versatile central metabolism and genetic tractability (Park et al., 2006; Nikel et al., 2016; Nikel and de Lorenzo, 2018; Wirth et al., 2020). We explored the capacity of S. elongatus CscB to support engineered strains of P. putida in direct co‐culture without supplementation of other organic carbon sources. Furthermore, P. putida was engineered to express the oxidative 2,4‐DNT degradation pathway and the capacity of these synthetic co‐cultures to remove this common toxin from the stream was evaluated both in batch and repeated‐batch cultures. Finally, we explored the capability of these engineered co‐cultures to produce PHA as an accessory function.
Figure 1.
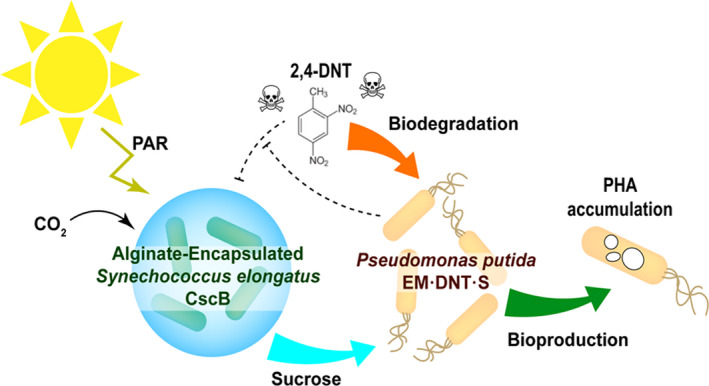
Photosynthetic co‐culture of engineered Synechococcus elongatus and Pseudomonas putida designed for simultaneous biodegradation and bioproduction. S. elongatus CscB, encapsulated within alginate beads, fix carbon dioxide via the Calvin‐Benson‐Bassham cycle driven by photosynthetically active radiation (PAR). The fixed carbon is converted into sucrose and exported through a heterologous sucrose permease (CscB) into the culture medium. The exported sucrose is consumed by P. putida EM·DNT·S for growth, supporting biotransformation of the toxic pollutant 2,4‐dinitrotoluene (2,4‐DNT), and concurrent production of polyhydroxyalkanoate (PHA).
Results
Alginate‐encapsulated S . elongatus CscB cells are protected from the toxic effects of 2,4‐DNT
2,4‐DNT is broadly toxic to many prokaryotes and eukaryotes (Yoon et al., 2006; Kuperman et al., 2009; Rocheleau et al., 2010); therefore, we first characterized the susceptibility of S. elongatus to this nitroaromatic compound at concentrations relevant for subsequent co‐culture applications. We observed that low concentrations of 2,4‐DNT (8 μM) started to cause a substantial (ca. 60%) growth impairment in planktonic cultures of S. elongatus, while growth was arrested at concentrations ≥ 15 μM (Fig. 2A). Likewise, the content of chlorophyll a [Chl(a)], the loss of which is an indicator of physiological stress and cell death (Sauer et al., 2001; Latifi et al., 2009; Korosh et al., 2018), declined rapidly in S. elongatus cells exposed to 2,4‐DNT concentrations above 30 μM (Fig. 2B and Fig. S1). In particular, we could detect a decrease of ca. 40% in the relative Chl(a) content over the first 6 h of incubation – and visible bleaching of cultures occurred by 48 h at 2,4‐DNT concentrations ≥ 30 μM. By comparison, soils and water from contaminated sites are routinely reported to have 2,4‐DNT levels near or above 100 μM (Griest et al., 1995). These preliminary experiments indicated that low 2,4‐DNT levels could significantly affect the growth and physiological status of planktonic S. elongatus cells, a complication that could preclude their use to support bioremediation of this compound in co‐cultures.
Figure 2.
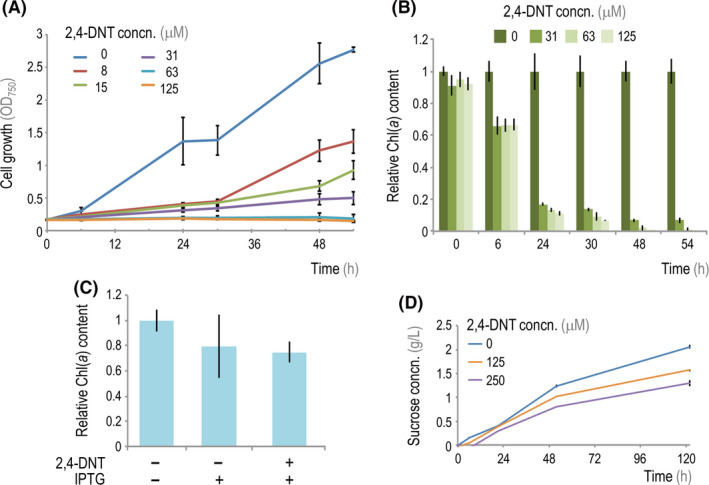
Toxic effects of 2,4‐DNT on planktonic and alginate‐encapsulated S. elongatus CscB.
A. Effect of increasing 2,4‐DNT concentrations (concn.) on the growth of S. elongatus CscB in planktonic cultures, assessed as the optical density measured at 750 nm (OD750).
B. The chlorophyll a [Chl(a)] content in cells of the cultures shown in panel (A) was measured and normalized to that in control cultures with no added 2,4‐DNT.
C. Chl(a) content of alginate‐encapsulated S. elongatus CscB cells after 5 days of exposure to 2,4‐DNT (250 μM) in M3 minimal medium with or without IPTG (1 mM) as indicated. Chl(a) content values were normalized to that in control cultures without any additives.
D. Total sucrose concentration in the culture supernatant of induced (via addition of 1 mM IPTG) cultures of alginate‐encapsulated S. elongatus CscB grown in the presence of different 2,4‐DNT concentrations. In panels (A–C), the mean values for n = 3 are displayed, and error bars represent standard deviations; in panel (D), the mean values for n = 3 with three technical replicates are shown, and error bars represent standard deviations.
Previously, we have shown that S. elongatus cells encapsulated within an alginate hydrogel remain metabolically active for months under prolonged nitrogen‐depletion stress (Weiss et al., 2017). Similarly, immobilization of a variety of cell types in hydrogels has been reported to increase stress tolerance and cell longevity (Bailliez et al., 1985; Gilleta et al., 2000; Leino et al., 2012; Therien et al., 2014; Ruiz‐Güereca and Sánchez‐Saavedra, 2016). We therefore examined the viability of alginate‐encapsulated S. elongatus CscB cells and found that they maintained relative Chl(a) contents at a level similar to that of planktonic cells, even when exposed to 2,4‐DNT concentrations near the solubility limits (250 μM) (Fig. 2C and Fig. S2 and S3). As indicated in Fig. S3B, the Chl(a) content per cell in alginate‐encapsulated S. elongatus CscB was ca. 1 × 10–8 μg cell−1, irrespective of the addition of 250 μM 2,4‐DNT and 1 mM IPTG (the inducer needed to stimulate sucrose excretion by means of the CscB exporter). Furthermore, these encapsulated S. elongatus CscB cells also retained metabolic activity for at least 5 days when exposed to 250 μM 2,4‐DNT – as assayed by the sucrose secreted to the supernatants of IPTG‐induced cultures (Fig. 2D). In particular, the sucrose concentration in supernatants of cultures added with 250 μM 2,4‐DNT reached ca. 1.2 g l−1 by the end of the experiment. Therefore, these experiments indicate that alginate encapsulation of S. elongatus CscB sufficiently protects cells from the acute toxicity brought about by 2,4‐DNT addition without affecting the capacity of the cells to produce and secrete sugars, which led us to explore co‐cultivation strategies as indicated below.
Co‐cultivation of engineered derivatives of P . putida EM173 with alginate‐encapsulated S . elongatus CscB
We next constructed a strain of P. putida that is capable of (i) utilizing sucrose as a carbon source and (ii) transforming 2,4‐DNT through the oxidative pathway from Burkholderia sp. R34. P. putida does not naturally utilize sucrose as a carbon source, but has been previously engineered to grow on this substrate (Löwe et al., 2017a,b). We first transformed the genetically tractable, prophage‐less strain EM173 of P. putida (Martínez‐García et al., 2015) with the low‐copy‐number plasmid pSEVA221·cscRABY (Löwe et al., 2019) that bears the sucrose‐utilization genes from P. protegens Pf‐5 (Fig. 3A). The resulting strain was named P. putida EM·S. Plasmid pSEVA221·cscRABY, a derivative of vector pSEVA221 (Silva‐Rocha et al., 2013), constitutively expresses the genes encoding a sucrose hydrolase (CscA, PFL_3237) and a sucrose permease (CscB, PFL_3238). These functions enable the uptake and catabolism of the disaccharide, together with a cognate transcriptional regulator (CscR, PFL_3236). Separately, a synthetic mini‐Tn7 transposon, carrying the functions required for 2,4‐DNT biotransformation from Burkholderia sp. R34 (Fig. 3A), was constructed as indicated in Experimental Procedures. The dnt gene cluster in this transposon was delivered into the unique att·Tn7 site within the chromosome of P. putida EM·S, resulting in a stable, engineered strain designed for both oxidative 2,4‐DNT transformation and sucrose consumption (hereafter referred to as P. putida EM·DNT·S).
Figure 3.
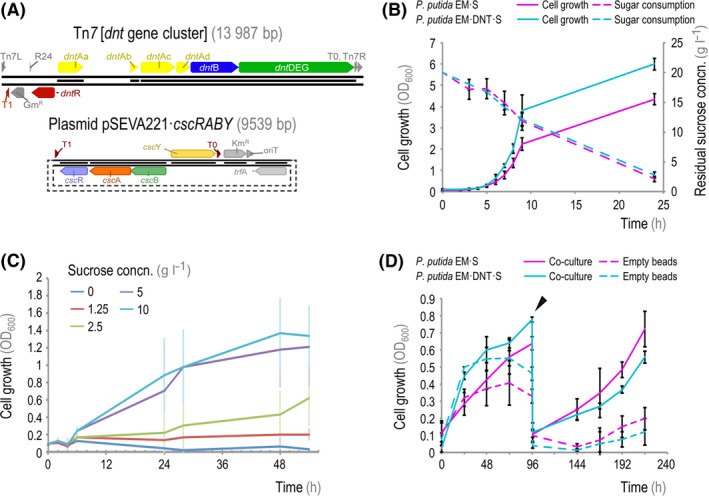
Growth characterization of P. putida strains engineered for sucrose consumption and 2,4‐DNT transformation.
A. Schematic representation of the gene cluster encoding the enzymes involved in 2,4‐DNT degradation from Burkholderia sp. R34, transferred into the P. putida chromosome via Tn7 transposition (top panel), and plasmid pSEVA221·cscRABY (Löwe et al., 2019), encoding the functions needed for sucrose utilization from P. protegens Pf‐5 (bottom panel).
B. Growth and sucrose utilization by engineered P. putida strains in M9 minimal medium containing 20 g l−1 sucrose. Bacterial growth was assessed as the optical density measured at 600 nm (OD600); residual sucrose concentration (concn.) in culture supernatants was determined with an enzymatic assay. (C) Growth of P. putida EM·DNT·S in M3 minimal medium containing different concentrations of sucrose (0, 1.25, 2.5, 5 and 10 g l−1).
D. The growth of P. putida EM·S and P. putida EM·DNT·S, inoculated from overnight cultures with 20 g l−1 sucrose, was monitored with or without encapsulated S. elongatus CscB (indicated as co‐culture). OD600 measurements track only the growth of the P. putida cells (i.e. planktonic cells). At 96 h post‐inoculation (slanted arrowhead), all of the M3 minimal medium was removed and replaced with fresh medium, allowing the residual P. putida cells on the surface of the alginate beads to repopulate the culture. In panels (B) and (C), the mean values for n = 4 are displayed, and error bars represent standard deviations. In panel (D), the mean values for n > 3 are displayed from experiments carried in different days, and error bars represent standard deviations.
We then examined the sucrose catabolism capacity of these modified P. putida strains under a range of physiologically relevant growth conditions. We observed that the pSEVA221·cscRABY plasmid was sufficient to confer sucrose utilization in both P. putida EM·S and P. putida EM·DNT·S when grown in M9 minimal medium supplemented with 20 g l−1 sucrose (Fig. 3B). Both P. putida EM·S and P. putida EM·DNT·S grew exponentially over the course of the first 9 h post‐inoculation, and by 24 h had reached final cell densities of OD600 (optical density measured at 600 nm) of 4.3 and 6 respectively. The specific growth rates of the two engineered strains were very similar, and the pattern of sucrose consumption was almost identical in either case. Indeed, during this incubation period, sucrose concentration declined in a nearly linear fashion from 20 to < 3 g l−1 at 24 h (Fig. 3B). We similarly evaluated the growth profile of engineered P. putida at lower sucrose concentrations within M3 minimal medium, designed for cyanobacterial growth (derived from BG‐11 minimal medium; see Table S1). To this end, P. putida EM·DNT·S was first inoculated in M3 minimal medium without supplementing a carbon source and incubated overnight in order to deplete internal carbon stores, then washed and re‐inoculated into fresh M3 minimal medium supplemented with a range of sucrose concentrations (from 0 to 10 g l−1). Bacterial growth was evident at sucrose concentrations ranging from 1.25 to 10 g l−1, after a lag phase longer than that observed in M9 minimal medium (Fig. 3C). In particular, we observed that at sugar concentrations > 5 g l−1 the bacterial cultures attained OD600 values well over 1 unit after 48 h of incubation. Taken together, these results indicate that this cyanobacterial culture medium supports growth of engineered P. putida strains when sucrose is present as the sole carbon source – thereby enabling its use for designing co‐cultures.
Next, we implemented and evaluated co‐cultures by inoculating P. putida EM·S and P. putida EM·DNT·S into flasks containing either empty alginate beads or alginate‐encapsulated S. elongatus CscB (Fig. 3D). Within the first 48 h, P. putida cells grew in all flasks – even in those containing empty alginate beads. Residual carbon stores within P. putida and/or trace amounts of sugars present in the alginate beads likely drove the residual bacterial growth observed in flasks containing empty beads, and the growth of these cultures was minor compared to that of cells co‐cultivated with alginate‐encapsulated (and sucrose‐producing) S. elongatus CscB (Fig. 3D). Furthermore, when the alginate beads were re‐suspended in fresh M3 minimal medium at 96 h post‐inoculation, growth of engineered P. putida strains was only observed in flasks with alginate‐encapsulated S. elongatus CscB cells, where they grew exponentially. Additionally, we did not observe degradation of the alginate beads in these (nor in subsequent) experiments, indicating that P. putida cannot utilize the hydrogel itself as a carbon source, in agreement with previous work (Löwe et al., 2017a). With these results at hand, we next evaluated the use of this co‐culture setup for 2,4‐DNT transformation.
Biotransformation of 2,4‐DNT by P . putida EM·DNT·S
As indicated previously, the dnt gene cluster of Burkhoderia sp. R34 encodes the enzymes needed for oxidative breakdown of 2,4‐DNT through oxygenation reactions (Spain, 1995; Symons and Bruce, 2006). The enzymes within the 2,4‐DNT degradation pathway catalyse two successive oxygenation reactions mediated by DntA and DntB (Fig. 4A). 2,4‐DNT is firstly dioxygenated by DntA to form 4‐methyl‐5‐nitrocatechol (4M5NC), a compound with a strong absorption peak at 420 nm (de las Heras et al., 2011). 4M5NC is the substrate of DntB, which yields 2‐hydroxy‐5‐methylquinone (2H5MQ), an intermediate with an absorption peak at 485 nm (de las Heras et al., 2011). In the native context of this catabolic route, 2H5MQ is then processed by the products of a number of additional genes encoded in the pathway to finally yield pyruvate and propionyl‐CoA (Fig. 4A).
Figure 4.
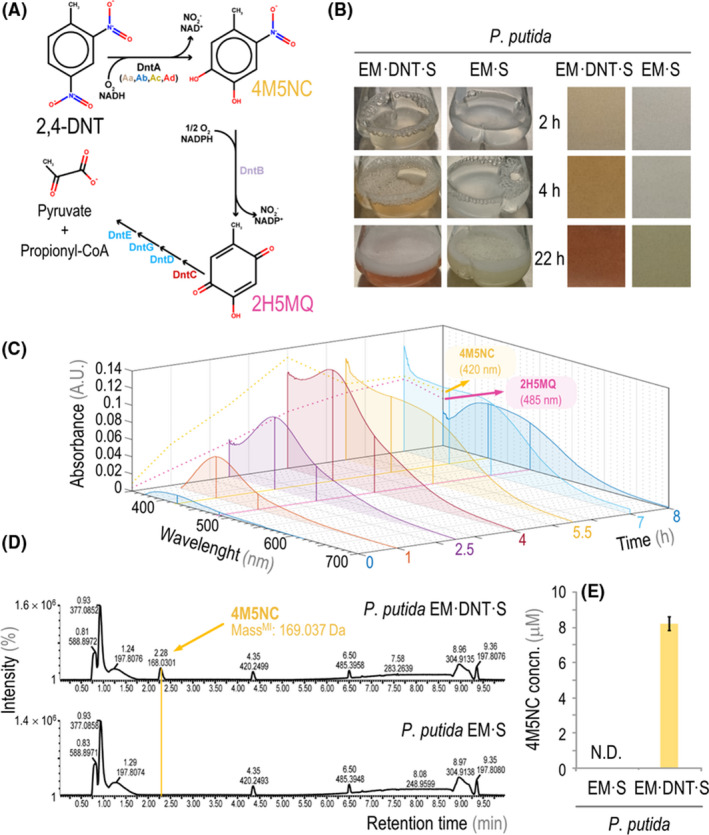
2,4‐DNT biotransformation in mono‐cultures of engineered P. putida.
A. Pathway for the oxidative degradation of 2,4‐DNT in Burkholderia sp. R34. The first step, executed by DntA, is a di‐oxygenation reaction that leads to the formation of 4‐methyl‐5‐nitrocatechol (4M5NC). This intermediate is the substrate for the DntB mono‐oxygenase, yielding 2‐hydroxy‐5‐methylquinone (2H5MQ). In the native context, the pathway leads to the conversion of 2,4‐DNT into pyruvate and propionyl‐coenzyme A (CoA).
B. Representative images of axenic cultures of P. putida EM·DNT·S and P. putida EM·S grown in the presence of 250 μM 2,4‐DNT over time (left column); the zoom‐in of supernatants from these cultures shows the characteristic changes in pigmentation (right column).
C. Time‐course of averaged spectral signatures (n = 3) in supernatants from P. putida EM·DNT·S cultures grown in the presence of 250 μM 2,4‐DNT, as measured via scanning spectrophotometry. The signature absorbance peaks of 4M5NC and 2H5MQ are indicated. A.U., arbitrary units.
D. LC‐MS analysis of culture supernatants of P. putida EM·DNT·S (top panel) and P. putida EM·S (bottom panel) after 4 h of incubation with 250 μM 2,4‐DNT. The peak corresponding to the monoisotopic mass of 4M5NC (massMI) is indicated by a yellow line.
E. LC‐MS quantification of the 4M5NC concentration (concn.) in supernatants of P. putida EM·S and P. putida EM·DNT·S cultures after 4 h incubation with 250 μM 2,4‐DNT. All cultures were grown in M3 minimal medium with sucrose at either 20 g l−1 (B) or 2 g l−1 (C–E). N.D., not detected.
We next examined the functional capacity of the dnt gene cluster introduced into P. putida EM·DNT·S by following the kinetics of 2,4‐DNT transformation in axenic cultures. M3 minimal medium, supplemented with 250 μM 2,4‐DNT and 2 g l−1 sucrose, was inoculated with P. putida EM·S or P. putida EM·DNT·S at an initial OD600 = 0.1. Consistent with the sequential formation of two expected oxidative intermediates, the supernatant of the P. putida EM·DNT·S cultures visibly changed pigmentation over the course of the growth experiment (Fig. 4B), whereas cultures of P. putida EM·S did not show any visible changes in coloration over the entire incubation period. Within 2–4 h following inoculation of P. putida EM·DNT·S, the supernatant turned yellow, consistent with the accumulation of the first intermediate, 4M5NC. Subsequently, the supernatant transitioned from yellow to orange (around 4 h) and then to red (22 h after inoculation), which reveals the presence of the second intermediate, 2H5MQ, and the breakdown of 4M5NC. These results are indicative of the biotransformation of 2,4‐DNT and, as reported previously, P. putida strains carrying the dnt gene cluster were not able to grow on 2,4‐DNT as the only carbon source (Akkaya et al., 2018). A separate quantitative analysis of the absorbance spectra of P. putida EM·DNT·S supernatants focused upon the first 8 h following 2,4‐DNT addition, revealing a characteristic absorbance rise at 420 nm (between 1‐4 h post‐inoculation). A second absorption peak arose at 485 nm (4–8 h), during the time period where the characteristic peak of 4M5NC gradually decreased (Fig. 4C). These spectroscopic changes were neither observed in control experiments with the non‐degrading P. putida EM·S strain (Fig. S4) nor in the absence of 2,4‐DNT. We directly confirmed the accumulation of 4M5NC by liquid chromatography coupled to mass spectrometry (LC‐MS). P. putida EM·DNT·S supernatants exhibited a peak matching the 4M5NC standard (Fig. 4D), whereas no 4M5NC was detected in control (P. putida EM·S) cultures under any condition. Direct quantification of 4M5NC levels by LC‐MS indicated a peak consistent with 4M5NC accumulation (ca. 8 μM) at 4 h post‐inoculation (Fig. 4E). Again, we could not detect any 4M5NC formation in control cultures of the non‐degrading strain P. putida EM·S. As an additional control of the activity of the first enzymes within the pathway, we carried experiments in which the release of nitrite from the substrate was measured by a colorimetric method. Nitrite could be detected in culture supernatants when P. putida EM·DNT·S was incubated in the presence of 250 μM 2,4‐DNT and using sucrose at 2 g l−1 as the sole carbon source. After 4 h, the nitrite concentration reached 7.3 ± 0.2 μM, and it peaked at 59.8 ± 1.3 μM by 24 h, suggesting that the oxidative pathway for 2,4‐DNT degradation plays a significant role in the removal of the nitroaromatic. The decrease in the 4M5NC together with the release of nitrite are indicative of 2,4‐DNT biotransformation. As previously mentioned, no bacterial growth was detected when P. putida EM·DNT·S cells were incubated in the mineral medium containing 2,4‐DNT but no added carbon source – which indicates that this engineered strain can transform the substrate without net assimilation of carbon (or at least not to the extent that would support growth on 2,4‐DNT).
The disappearance of 2,4‐DNT from the supernatant of P. putida cultures was directly assessed by gas chromatography coupled to mass spectrometry (GC‐MS) in all the experiments. We observed that the soluble fraction of 2,4‐DNT declined significantly within the first 4 h, and dropped to undetectable levels by 22 h. This kinetic pattern of 2,4‐DNT transformation is similar to what has been reported for Burkholderia sp. R34 (Pérez‐Pantoja et al., 2013) and other engineered P. putida strains (Akkaya et al., 2018, 2019). Soluble levels of 2,4‐DNT declined in cultures of P. putida EM·S as well (Fig. S5A), whereas the concentration of the nitroaromatic remained constant in control flasks that were not inoculated with P. putida cells (Fig. S5B). These observations can be accounted for by the fact that a small fraction of 2,4‐DNT became most likely adsorbed (i.e. physically associated) to cell surfaces, a known property of nitroaromatic compounds (Achtnich et al., 1999). Additionally, the reductive pathway (catalysed by non‐specific nitroreductases, which have been reported to be present in many bacterial species) could result in the conversion of 2,4‐DNT into other intermediates (Fig. S5C). To investigate this possibility, we assessed culture supernatants for the presence of 2‐amino‐4‐nitrotoluene/4‐amino‐2‐nitrotoluene (2A4NT/4A2NT), two key isomer intermediates of the reductive pathway, by LC‐MS. These two compounds were detected at low levels (< 2.5 μM) in the supernatant of cultures of both P. putida EM·DNT·S and P. putida EM·S, but the formation of 2A4NT/4A2NT was significantly lower in cultures of P. putida EM·DNT·S (Fig. S5D). This result suggests that the oxidative route competes with the non‐specific reductive pathway in removing 2,4‐DNT in engineered P. putida strains, with a predominant role of the former over the later in P. putida EM·DNT·S. With this information at hand, we set to establish co‐cultures of engineered P. putida and S. elongatus as indicated in the next section.
Biotransformation of 2,4‐DNT by a synthetic consortium of engineered P . putida and S . elongatus CscB
We next introduced 2,4‐DNT into co‐cultures of P. putida with alginate‐encapsulated S. elongatus CscB cells. Both P. putida EM·S and P. putida EM·DNT·S were able to grow with the beaded S. elongatus CscB cells in the presence of 250 μM 2,4‐DNT (Fig. 5A). Cultures containing only alginate‐encapsulated S. elongatus CscB or empty alginate beads served as controls for both optical density measurements as well as any potential 2,4‐DNT adsorption to abiotic surfaces. As previously observed, no noticeable bacterial growth was detected in the absence of S. elongatus CscB in these experiments. GC‐MS analysis of supernatants demonstrated that cultures containing engineered P. putida or S. elongatus cells removed 2,4‐DNT from the culture, while the 2,4‐DNT concentration did not change in empty‐bead control experiments (Fig. 5B). Similarly, the supernatant of co‐cultures containing P. putida EM·DNT·S exhibited visual changes in colour similar to those reported in Fig. 4, indicating activity of the oxidative dnt pathway. Accordingly, LC‐MS analysis validated that the oxidative pathway intermediate 4M5NC transiently accumulated in the supernatants of P. putida EM·DNT·S–containing co‐cultures (Fig. 5C), reaching a concentration of ca. 3 μM within the first hour of incubation and completely disappearing after 24 h. Conversely, 4M5NC was not detected in P. putida EM·S–containing co‐cultures. These results indicate that the synthetic consortium can efficiently remove 2,4‐DNT in a single batch culture.
Figure 5.
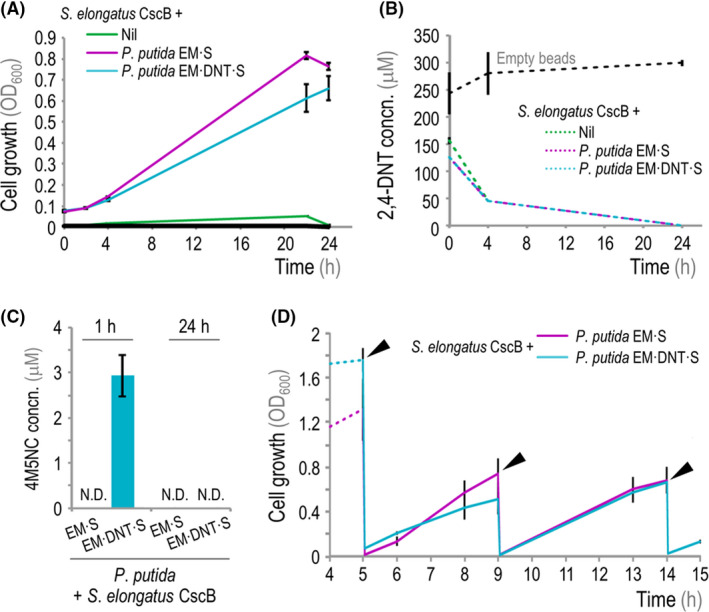
Biotransformation of 2,4‐DNT in co‐cultures of engineered S. elongatus and P. putida.
A. Growth of engineered P. putida strains [assessed as the optical density measured at 600 nm (OD600)] in co‐culture with alginate‐encapsulated S. elongatus CscB in the presence of 250 μM 2,4‐DNT. The black line indicates control experiments with empty beads.
B. GC‐MS analysis of 2,4‐DNT concentration (concn.) in supernatants of co‐cultures during 24 h. 2,4‐DNT concentrations dropped below detectable levels by 24 h post‐inoculation.
C. LC‐MS quantification of 4M5NC concentration (concn.) in supernatants of co‐cultures after addition of 2,4‐DNT at 250 μM at 1 and 24 h post‐inoculation. N.D., not detected.
D. Growth of engineered P. putida strains in long‐term co‐cultures with encapsulated S. elongatus CscB. The culture medium was exchanged at each indicated time‐point (slanted arroheads), and IPTG and 2,4‐DNT were added at 1 and 250 μM respectively. In panels (A–C), the mean values for n = 3 are displayed, and error bars represent standard deviations. In panel (D), the mean values for n = 14 are indicated, and error bars represent standard deviations.
Based on these results, we next evaluated the capacity of the co‐culture to remediate multiple (i.e. repeated) batches of 2,4‐DNT–containing streams by extending the co‐culture experiment to 15 days and exchanging the culture supernatant every 4–5 days with fresh medium containing 250 μM 2,4‐DNT (Fig. 5D). In each case, P. putida was able to quickly re‐grow from residual planktonic cells after removal and replacement of the culture supernatant – a phenomenon likewise observed for P. putida EM·S and P. putida EM·DNT·S. Again, colorimetric changes in the supernatant were evident only in the P. putida EM·DNT·S–containing co‐cultures after each back‐dilution, with added 2,4‐DNT. Direct measurement of the residual nitroaromatic concentration indicates that the total mass of 2,4‐DNT cleared from solution by the P. putida EM·DNT·S strain over this period in repeated‐batch co‐cultures amounts to 136 mg l−1.
Simultaneous removal of 2,4‐DNT and accumulation of PHA by engineered strains of P. putida in a synthetic consortium
Wild‐type P. putida strains are known to accumulate PHA as a carbon storage (López et al., 2015; Prieto et al., 2016), particularly under nitrogen‐depleted conditions in the presence of an abundant carbon source (Hoffmann and Rehm, 2004), which prompted us to investigate the opportunity to both remove 2,4‐DNT and simultaneously produce a valuable by‐product. In these experiments, engineered P. putida cells were inoculated into the alginate‐encapsulated and IPTG‐induced S. elongatus CscB culture flasks at an OD600 ~ 0.5 in the presence of 250 μM 2,4‐DNT (Fig. 6A). These flasks contained either M3 or M3‐N minimal medium, the latter of which has a reduced nitrogen content to trigger polymer accumulation (2 mM NaNO3; Table S1). The pattern of cell growth was very similar for the two strains under evaluation, and bacterial growth was not significantly affected by the reduced nitrogen conditions in M3‐N minimal medium. Twenty‐four hours after cycling co‐cultures into either nitrogen‐rich or nitrogen‐depleted media (i.e. M3 and M3‐N minimal medium, respectively), planktonic P. putida cells were harvested and the intracellular PHA content was evaluated by means of LC‐MS measurements of crotonic acid as indicated in Experimental Procedures. Both engineered P. putida strains were able to accumulate the polymer under nitrogen‐limited conditions – but not in co‐cultures run in nitrogen‐rich M3 minimal medium (Fig. 6B). In particular, P. putida EM·S and P. putida EM·DNT·S co‐cultures grown under nitrogen‐depleted conditions in M3‐N minimal medium accumulated the polymer at a specific volumetric productivity of 4.9 and 5.1 mg PHA l−1 day−1 of culture, with a polymer content in biomass of 22 and 23.4 mg of PHA g cell dry weight‐1, respectively. These results indicate that the accumulation of PHA from sucrose as the only carbon source by engineered P. putida is not affected by the presence of 2,4‐DNT (or its transformation products) and that the polymer can be obtained in co‐cultures designed for 2,4‐DNT removal as an added‐value by‐product.
Figure 6.
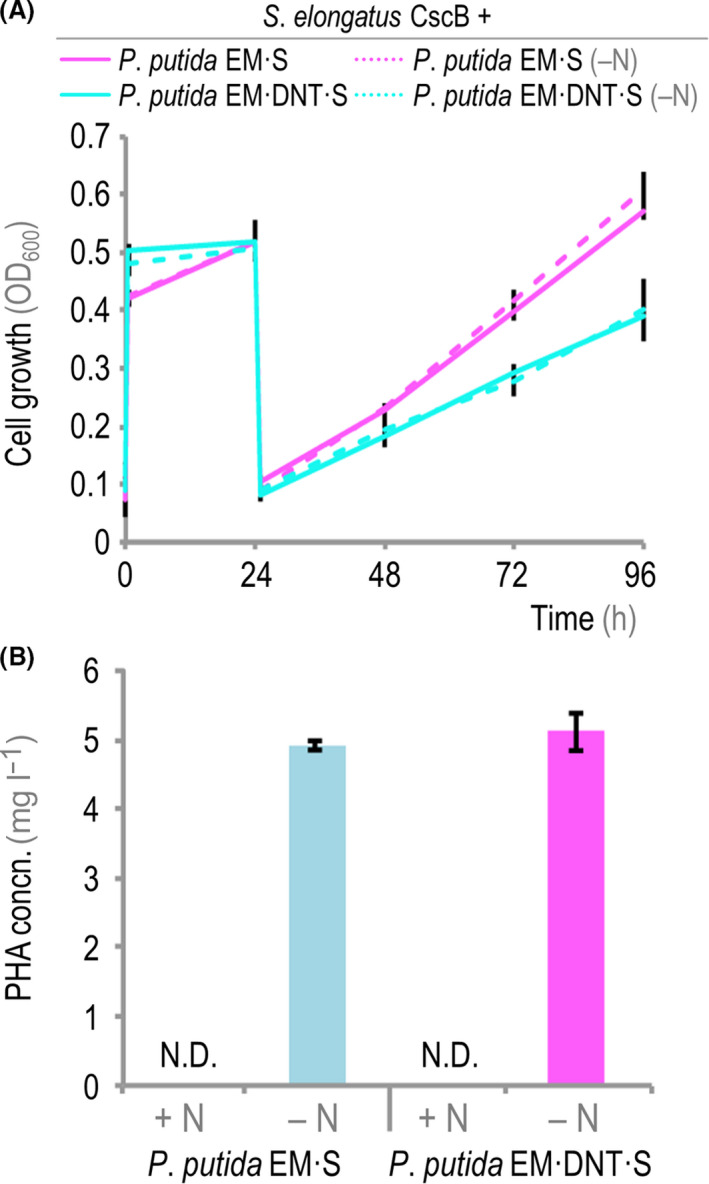
PHA accumulation by engineered P. putida in phototrophic co‐cultures.
A. Growth of engineered P. putida strains in co‐cultures of alginate‐encapsulated S. elongatus CscB in the presence of 250 μM 2,4‐DNT with or without nitrogen deprivation. Bacterial growth was assessed as the optical density measured at 600 nm (OD600). Separate experiments were run in M3 minimal medium with nitrate at the standard concentration (17.6 mM, indicated as + N) or at a reduced concentration (2 mM, indicated as –N; minimal M3‐N medium).
B. Quantification of the PHA concentration (concn.) in co‐cultures of engineered P. putida strains and alginate‐encapsulated S. elongatus CscB in the presence of 250 μM 2,4‐DNT after 24 h of incubation into nitrogen‐replete or nitrogen‐reduced minimal M3 medium. In all cases, average values from n = 3 are presented, and error bars represent standard deviations. N.D., not detected.
Discussion
Rational engineering of microbial communities is an emerging area of study that has implications for both applied and fundamental microbiology. Most natural microbial biochemistry occurs in the context of complex microbial consortia that are unlike the mono‐cultures routinely cultivated under controlled laboratory conditions (Jessup et al., 2004). Desirable system‐level features (e.g. robustness) are thought to emerge from simple interactions between individual species [e.g. by the division of metabolic labour (McCann, 2000; Ives and Carpenter, 2007; Volke and Nikel, 2018)], although these phenomena remain poorly understood. Gaining a deeper understanding of the design principles underlying natural microbial ecologies will likely require the application of top‐down, systems‐level analyses (e.g. meta‐omic approaches) and the development of flexible bottom‐up platforms that permit the assembly of simple, tractable microbial mixed cultures (Jessup et al., 2005; Momeni et al., 2011; Goers et al., 2014).
In this work, we constructed microbial co‐cultures capable of transforming the industrial toxin 2,4‐DNT by utilizing light and CO2 as the primary inputs. Our present contribution builds upon a flexible co‐cultivation system by demonstrating that (engineered) heterotrophs can be incorporated into designer communities in order to programme them for removal of environmental contaminants – thus extending the utility of synthetic consortia beyond the mere coexistence of microbial strains into performing complex biodegradation tasks. We chose 2,4‐DNT as a model toxicant, since this nitroaromatic compound is a chemical contaminant that poses a significant bioremediation challenge due to toxicity and persistence in natural environments. This synthetic consortium described herein consists of engineered strains of S. elongatus and P. putida designed to divide metabolic labour between the partner species – with S. elongatus CscB specializing in photosynthetic processes, generating and secreting soluble sugars sufficient to promote the growth and distinct metabolic activities of co‐cultivated engineered P. putida. We demonstrated that co‐cultures were reproducibly stable over a series of back‐dilutions (repeated‐batch cultivation), permitting the removal of soluble 2,4‐DNT in successive batch cultures for an extended period of time.
The selection of the partner heterotroph in our system is not arbitrary. P. putida has gained significant attention as a chassis for industrially relevant synthetic biology due to its metabolic diversity, high tolerance of oxidative stress, and capacity to conduct complex biochemical reactions involving aromatic substrates (Poblete‐Castro et al., 2017). In this context, the still‐evolving 2,4‐DNT degradation pathway from Burkholderia sp. R34 has been reported to lead to accumulation of oxidative stress an increased rate of mutation in its native context, whereas P. putida heterologously expressing the dnt gene cluster (P. putida EM·DNT and derivatives thereof) does not exhibit the same rate of DNA damage and mutation (Akkaya et al., 2019). P. putida EM·DNT·S co‐cultures removed 2,4‐DNT from the soluble phase partly via this oxidative degradation pathway. However, under the conditions of our co‐cultures, a portion of 2,4‐DNT is also transformed via reductive processes (Fig. S5D) and/or gets adsorbed to surfaces. Because multiple products and nitroamino‐adducts are formed downstream of 2,4‐DNT reduction, we could not precisely quantitate the flux through the oxidative pathway with our data, but we do see a ca. 25% reduction in the accumulation of the reductive products 2A4NT and 4A2NT. Fitting our kinetic data of 2,4‐DNT transformation (Fig. 4 and Fig. S5A) to a simple mass‐action model of a branched metabolic pathway suggests that up to 40% of 2,4‐DNT may be fully mineralized via the oxidative pathway (see Experimental Procedures). Further engineering efforts may be required to increase the fraction of 2,4‐DNT that is completely catabolized and/or incorporated into biomass, e.g. by improving the affinity of enzymes in the oxidative pathway for 2,4‐DNT (Monti et al., 2005; Ju and Parales, 2010) or by coupling the growth of engineered strains to fully oxidative catabolism of the aromatic substrate (Durante‐Rodríguez et al., 2018; Fernández‐Cabezón et al., 2019).
In tandem with the transformation of 2,4‐DNT, co‐cultures were programmed to synthesize a valuable by‐product in the form of PHA. Co‐cultures of engineered P. putida and S. elongatus accumulated the polymer with specific volumetric productivities of ca. 5 mg PHA l−1 day−1 using gaseous CO2 as the primary carbon source – which is lower than the productivity reported in recent publications describing bacterial co‐cultures or mono‐cultures of cyanobacteria directly engineered to express PHA‐producing pathways (Löwe et al., 2017a,b). Nevertheless, we have demonstrated that polymer accumulation can occur in P. putida the presence of 2,4‐DNT and that PHA synthesis could be coupled to the removal of the nitroaromatic. Therefore, PHA produced from these co‐cultures represents a value‐added by‐product – and a first‐case example of polymer accumulation coupled to bioremediation of an environmental pollutant. The economic prospects of the process can be improved by optimizing PHA yields (e.g. by modifying the regulation of the PHA synthesis to render it insensitive to nitrogen availability) or by incorporating pathways leading to higher‐value bioproducts in the system. Further modifications could include the addition of other microbial partners in the consortium, thereby multiplying the range of products that can be obtained in these co‐culture setups. One way or the other, the rational pairing of cyanobacteria and P. putida, together with the plethora of synthetic biology approaches created to engineer novel properties in whole‐cell biocatalysts, allows for the design of systems with the potential to transform current production platforms.
Experimental procedures
Bacterial strains and culture conditions
Planktonic S. elongatus strains were grown in a Multitron Pro (Infors HT, Annapolis Junction, MD, USA) photobiological shaker at 32°C, 2% (v/v) CO2, and a constant illumination of ~ 70 μmol m−2 s−1 (15 W Grow‐Lux; OSRAM Sylvania Inc., Wilmington, MA, USA) as previously described (Weiss et al., 2017). Selection for the genomically integrated cscB module was achieved by addition of 12.5 μg ml−1 chloramphenicol to the culture medium. Engineered P. putida strains were constructed essentially as reported previously (Martínez‐García et al., 2014; Löwe et al., 2017a,b, 2019; Akkaya et al., 2018). During routine cultivation, P. putida strains carrying an integrated dnt gene cluster were maintained in the presence of 25 μg ml−1 gentamicin, while strains transformed with plasmid pSEVA221·cscRABY were maintained with 50 μg ml−1 kanamycin. Lysogeny broth (LB) medium cultures were at 32°C in a Multitron (Infors HT) incubator with rotary agitation at 150 rpm. For co‐culture experiments, P. putida cultures grown in LB medium were re‐suspended in M3 minimal medium (Table S1) with either 20 g l−1 or 2 g l−1 sucrose, as indicated in the main text. These cultures were then used as inoculum for subsequent experiments after pelleting cells, rinsing the pellet, and re‐suspending cells in fresh media to prevent the carryover of carbohydrates and other soluble metabolites. Antibiotic selection was omitted for all co‐culture experiments.
Encapsulation of S . elongatus CscB cells in alginate beads
Alginate encapsulation was performed as previously described (Weiss et al., 2017) with minor adjustments. Briefly, planktonic S. elongatus CscB cells grown in minimal BG‐11 medium containing 1 g l−1 HEPES (pH = 8.3; Sigma‐Aldrich Co., St. Louis, MO, USA) were harvested at an optical density at 750 nm (OD750) of 2.0 via centrifugation at 3500 g for 30 min and concentrated in 3 ml of sulfur‐free BG‐11 minimal medium. Concentrated cells were mixed into a sterile and degassed volume of 3% (w/v) sodium alginate at a final OD750 = 5.0, thereby resulting in a 2.75% (w/v) sodium alginate‐S. elongatus CscB suspension. In a sterile biosafety cabinet, a vertically oriented syringe pump (KD Scientific, Holliston, MA, USA) was used to dispense the solution dropwise through a 30 G needle into a ≥ 20‐fold larger volume of 20 mM BaCl2. The drops travelled ~ 35 cm from needle to the slowly stirred BaCl2 solution and were allowed to equilibrate in the solution for at least 20 min. Solidified beads were rinsed once with BG‐11 medium and incubated overnight in M3 minimal medium (without NaCl); the mean diameter of cured alginate beads used in these experiments was ~ 1.8 mm. To acclimate cells within the alginate hydrogel and precipitate residual Ba2+, beads were transferred through a series of media washes in the following days. The first day post‐encapsulation, beads were rinsed and re‐suspended in fresh M3 minimal medium (without NaCl), transferred to 250‐ml baffled Erlenmeyer flasks, and placed into a Multitron Pro incubator (Infors HT) with rotary shaking (125 rpm) for 24 h. Subsequently, the M3 minimal medium was exchanged, and the alginate bead suspension were portioned in ~ 10‐ml aliquots into 125‐ml baffled Erlenmeyer flasks. To acclimate cells to the final working salt concentrations, the spent medium was replaced with M3 minimal medium with only 25 mM NaCl for 24 h, then M3 minimal medium with 50 mM NaCl for 24 h, and finally to complete M3 minimal medium (containing 100 mM NaCl, Table S1). The culture medium was subsequently refreshed daily for 3–5 days prior to initiation of experiments. In the cases where the beads were utilized in co‐culture, cscB expression was induced with the addition of IPTG at 1 mM 24 h prior to the addition of P. putida cells. The culture medium was exchanged a final time coincident with addition of P. putida cells.
Analytical methods
Culture optical densities were measured with a Genesys 20 (Thermo Fisher Scientific, Waltham, MA, USA) spectrophotometer. The density of planktonic S. elongatus CscB was measured at OD750, and both co‐culture experiments and P. putida mono‐cultures were measured at OD600. This spectrophotometer was also used to measure Chl(a) concentrations of planktonic S. elongatus cultures (Zavřel et al., 2015). For Chl(a) measurements in alginate‐encapsulated S. elongatus, 1 ml of pre‐chilled methanol was added to four alginate beads in three technical replicates. Beads were vortexed and incubated at 4°C for 30 min in the dark to extract Chl(a). The absorbance of the supernatant was measured in cuvettes at 720 nm and 665 nm to calculate the final Chl(a) concentration as previously described (Zavřel et al., 2015).
Sucrose concentration of beaded cultures was measured as indicated previously (Weiss et al., 2017). Briefly, 1 ml of culture supernatant was withdrawn at the indicated time points, cells were pelleted at 17 000 g for 10 min, and sucrose was quantified via a sucrose/d‐glucose assay kit (Megazyme, Bray, Ireland) with three technical replicates for each sample. Culture supernatant spectra were scanned using a DU800 Spectrophotometer (Beckman Coulter Inc., Brea, CA, USA) to assess the presence of coloured metabolites within the 2,4‐DNT degradation pathway. Cell‐free culture supernatant was obtained by centrifugation for 10 min at 17 000 g. The supernatant was then transferred to a cuvette for spectral analysis.
Direct 2,4‐DNT measurements were made using an Agilent 5975 GC/single quadrupole MS (Agilent Technologies Inc., Santa Clara, CA, USA). Culture samples were centrifuged at 17 000 g for 10 min and 10 μl of cell‐free supernatant were removed to a new tube. A 50‐μl aliquot of ethyl acetate was added to the supernatant and incubated at room temperature for 30 min. The ethyl acetate phase was then transferred to a GC vial and injected (the injection volume was 1 μl). Samples were separated with a 5% (w/w) phenyl‐methyl capillary column (Agilent Technologies Inc.) and measured by mass selective detector (MSD). A temperature of 275°C was set for the split/splitless injector (ratio of 10:1). Helium gas was used as the carrier gas at a flow rate of 1 ml min−1. The release of inorganic nitrite (NO2 –) from nitroaromatic compounds was assessed in culture supernatants by a colorimetric method essentially as described by Zhang et al . (2001).
LC‐MS measurements of metabolites within the 2,4‐DNT degradation pathway were made using a Waters Xevo G2‐XS UPLC‐MS/MS (Waters Corp., Milford, MA, USA). Culture samples were harvested and pelleted via centrifugation (17 000 g for 10 min), and a 100‐μl aliquot of the supernatant was transferred to a clean tube, lyophilized, and re‐suspended in 1 ml of deionized water for measurements. HPLC measurements of crotonic acid (as a proxy of PHA accumulation) were made with a Waters 2695 HPLC. Samples were processed and analysed as previously described (Weiss et al., 2017). Briefly, cells were centrifuged at 17 000 g for 10 min, the supernatant was decanted, and the pellet was lyophilized. The cell biomass was suspended in 1 ml of concentrated H2SO4 and heated to 90°C for 1 h to promote dehydration of 3‐hydroxybutyrate into crotonic acid. After cooling the reaction mixture to room temperature, particulate material was removed by filtration and the mixture was diluted 100‐fold with deionized water. A 20‐μl aliquot of the resulting solution was injected onto an Aminex 300‐mm HPX‐87H column (Bio‐Rad Laboratories, Hercules, CA, USA), and 0.028 N H2SO4 was used as the mobile phase at a flow of 1 ml min−1. The column temperature was maintained at 60°C and the UV absorption of crotonic acid was monitored at 210 nm. Two standards of commercial PHA (Sigma‐Aldrich Co.) were similarly treated and used for quantification purposes.
A simple mass‐action model was used to explore the parameter space that could sufficiently fit our kinetic data sets of 2,4‐DNT transformation (Fig. 4 and Fig. S5; Voit, 2000, 2017). Briefly, metabolite flux through both reductive and oxidative branches was modelled for the initial steps of 2,4‐DNT transformation as a first‐order kinetic dependent upon the concentrate of each substrate (d[P]/dt = k1[S] – k2[P]); where P = product, S = substrate, and k1 and k2 are coefficients related to each branch point within the metabolic network. Reactions were modelled to be irreversible based on the unidirectionality of the early steps of 2,4‐DNT transformation. Models were capable of producing simulated metabolite profiles that fit our kinetic profiles (within an allowed 25% error of our experimentally measured values), including the loss of 2,4‐DNT from solution, and gain of the colorimetric products 4M5NC and 2H5MQ across a range of parameter space without making further assumptions of more complex pathway regulation (e.g. feedback or feed‐forward regulatory steps), including oxidative branch flux between 15 and 40% of the competing reductive pathway.
Conflict of interest
None declared.
Author contributions
D.T.F., P.I.N. and D.C.D. designed and directed the project and experiments. P.C. designed and engineered the P. putida strains utilized in this work. D.T.F. and P.S. performed the experiments and recorded the results. D.T.F., P.S., P.C., P.I.N. and D.C.D. analysed and interpreted the data. D.T.F. drafted the manuscript and figures. D.T.F., P.S., P.C., P.I.N. and D.C.D. provided commentary and edits to the manuscript and figures and prepared the final version of the article.
Supporting information
Table S1. Chemical composition of BG‐11, M3, and M3‐N minimal media used in this study.
Fig. S1. Influence of 2,4‐DNT on growth of S. elongatus PCC 7942.
Fig. S2. Effect of 2,4‐DNT on alginate‐encapsulated S. elongatus CscB.
Fig. S3. Chlorophyll a content per cell comparison in planktonic and alginate‐encapsulated S. elongatus CscB.
Fig. S4. Spectral analysis of supernatants of P. putida EM·S cultures.
Fig. S5. Kinetics of 2,4‐DNT transformation by engineered P. putida through the oxidative and reductive pathway.
Acknowledgements
This work was supported by the National Science Foundation (Award Number # 1437657) to D.C.D. Additional institutional and equipment support was provided by the Office of Basic Energy Sciences, Office of Science, US Department of Energy (DE–FG02–91ER20021) to D.C.D. This study was also supported by The Novo Nordisk Foundation (Grant NNF10CC1016517) and the Danish Council for Independent Research (SWEET, DFF‐Research Project 8021‐00039B) to P.I.N. The authors would like to thank Dan Jones, Anthony Schilmiller, and Scott Smith from the MSU Mass Spectrometry and Metabolomics Core for their assistance in data acquisition and analysis during this project, as well as Cheryl Kerfeld for the use of the HPLC. The authors are also indebted to Hannes Löwe and Katharina Pflüger‐Grau (Technische Universität München, Germany) for generously sharing research materials.
Microbial Biotechnology (2020) 13(4), 997–1011
Funding Information
This work was supported by the National Science Foundation (Award Number # 1437657) to D.C.D. Additional institutional and equipment support was provided by the Office of Basic Energy Sciences, Office of Science, US Department of Energy (DE–FG02–91ER20021) to D.C.D. This study was also supported by The Novo Nordisk Foundation (Grant NNF10CC1016517) and the Danish Council for Independent Research (SWEET, DFF‐Research Project 8021‐00039B) to P.I.N.
Contributor Information
Pablo Iván Nikel, Email: pabnik@biosustain.dtu.dk.
Daniel C. Ducat, Email: ducatdan@msu.edu.
References
- Aburto‐Medina, A. , Taha, M. , Shahsavari, E. , and Ball, A.S. (2017) Degradation of the dinitrotoluene isomers 2,4‐ and 2,6‐DNT: Appraising the role of microorganisms In Enhancing Cleanup of Environmental Pollutants. Anjum N.A., Gill S.S., and Tuteja N. (eds). Berlin, Germany: Springer, pp. 5–20. [Google Scholar]
- Achtnich, C. , Sieglen, U. , Knackmuss, H.J. , and Lenke, H. (1999) Irreversible binding of biologically reduced 2,4,6‐trinitrotoluene to soil. Environ Toxicol Chem 18: 2416–2423. [Google Scholar]
- Akkaya, Ö. , Pérez‐Pantoja, D.R. , Calles, B. , Nikel, P.I. , and de Lorenzo, V. (2018) The metabolic redox regime of Pseudomonas putida tunes its evolvability toward novel xenobiotic substrates. mBio 9: e01512‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkaya, Ö. , Nikel, P.I. , Pérez‐Pantoja, D. , and de Lorenzo, V. (2019) Evolving metabolism of 2,4‐dinitrotoluene triggers SOS‐independent diversification of host cells. Environ Microbiol 21: 314–326. [DOI] [PubMed] [Google Scholar]
- Bailliez, C. , Largeau, C. , and Casadevall, E. (1985) Growth and hydrocarbon production of Botryococcus braunii immobilized in calcium alginate gel. Appl Microbiol Biotechnol 23: 99–105. [Google Scholar]
- Barry, A. , Wolfe, A. , English, C. , Ruddick, C. , and Lambert, D. (2016) DOE (U.S. Department of Energy). National Algal Biofuels Technology Review. U.S. Department of Energy, Office of Energy Efficiency and Renewable Energy, Bioenergy Technologies Office. [Google Scholar]
- Blasche, S. , Kim, Y. , Oliveira, A.P. , and Patil, K.R. (2017) Model microbial communities for ecosystems biology. Curr Opin Syst Biol 6: 51–57. [Google Scholar]
- Calero, P. , and Nikel, P.I. (2019) Chasing bacterial chassis for metabolic engineering: A perspective review from classical to non‐traditional microorganisms. Microb Biotechnol 12: 98–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrieri, D. , Paddock, T. , Maness, P.C. , Seibert, M. , and Yu, J. (2012) Photo‐catalytic conversion of carbon dioxide to organic acids by a recombinant cyanobacterium incapable of glycogen storage. Energy Environ Sci 5: 9457–9461. [Google Scholar]
- de las Heras, A. , Chavarría, M. , and de Lorenzo, V. (2011) Association of dnt genes of Burkholderia sp. DNT with the substrate‐blind regulator DntR draws the evolutionary itinerary of 2,4‐dinitrotoluene biodegradation. Mol Microbiol 82: 287–299. [DOI] [PubMed] [Google Scholar]
- Dolinšek, J. , Goldschmidt, F. , and Johnson, D.R. (2016) Synthetic microbial ecology and the dynamic interplay between microbial genotypes. FEMS Microbiol Rev 40: 961–979. [DOI] [PubMed] [Google Scholar]
- Ducat, D.C. , Avelar‐Rivas, J.A. , Way, J.C. , and Silver, P.A. (2012) Rerouting carbon flux to enhance photosynthetic productivity. Appl Environ Microbiol 78: 2660–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durante‐Rodríguez, G. , de Lorenzo, V. , and Nikel, P.I. (2018) A post‐translational metabolic switch enables complete decoupling of bacterial growth from biopolymer production in engineered Escherichia coli . ACS Synth Biol 7: 2686–2697. [DOI] [PubMed] [Google Scholar]
- Fernández‐Cabezón, L. , Cros, A. , and Nikel, P.I. (2019) Evolutionary approaches for engineering industrially‐relevant phenotypes in bacterial cell factories. Biotechnol J 14: 1800439. [DOI] [PubMed] [Google Scholar]
- French, C.E. , Rosser, S.J. , and Bruce, N.C. (2001) Biotransformations of explosives. Biotechnol Genet Eng Rev 18: 171–217. [DOI] [PubMed] [Google Scholar]
- Gilleta, F. , Roisin, C. , Fliniaux, M. , Jacquin‐Dubreuil, A. , Barbotin, J. , and Nava‐Saucedo, J. (2000) Immobilization of Nicotiana tabacum plant cell suspensions within calcium alginate gel beads for the production of enhanced amounts of scopolin. Enzyme Microb Technol 26: 229–234. [DOI] [PubMed] [Google Scholar]
- Goers, L. , Freemont, P. , and Polizzi, K.M. (2014) Co‐culture systems and technologies: Taking synthetic biology to the next level. J R Soc Interface 11: 20140065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldford, J.E. , Lu, N. , Bajić, D. , Estrela, S. , Tikhonov, M. , Sánchez‐Gorostiaga, A. , et al (2018) Emergent simplicity in microbial community assembly. Science 361: 469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griest, W.H. , Tyndall, R.L. , Stewart, A.J. , Caton, J.E. , Vass, A.A. , Ho, C.H. , and Caldwell, W.M. (1995) Chemical characterization and toxicological testing of windrow composts from explosives‐contaminated sediments. Environ Toxicol Chem 14: 51–59. [Google Scholar]
- Griest, W.H. , Stewart, A.J. , Vass, A.A. , and Ho, C.H. (1998) Chemical and Toxicological Characterization of Slurry Reactor Biotreatment of Explosives‐contaminated Soils. Oak Ridge TN: National Lab. [Google Scholar]
- Haigler, B.E. , Nishino, S.F. , and Spain, J.C. (1994) Biodegradation of 4‐methyl‐5‐nitrocatechol by Pseudomonas sp. strain DNT. J Bacteriol 176: 3433–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays, S.G. , and Ducat, D.C. (2015) Engineering cyanobacteria as photosynthetic feedstock factories. Photosynth Res 123: 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays, S.G. , Yan, L.L.W. , Silver, P.A. , and Ducat, D.C. (2017) Synthetic photosynthetic consortia define interactions leading to robustness and photoproduction. J Biol Eng 11: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, N. , and Rehm, B.H. (2004) Regulation of polyhydroxyalkanoate biosynthesis in Pseudomonas putida and Pseudomonas aeruginosa . FEMS Microbiol Lett 237: 1–7. [DOI] [PubMed] [Google Scholar]
- Hooker, B.S. , and Skeen, R.S. (1999) Transgenic phytoremediation blasts onto the scene. Nat Biotechnol 17: 428. [DOI] [PubMed] [Google Scholar]
- Ives, A.R. , and Carpenter, S.R. (2007) Stability and diversity of ecosystems. Science 317: 58–62. [DOI] [PubMed] [Google Scholar]
- Jessup, C.M. , Kassen, R. , Forde, S.E. , Kerr, B. , Buckling, A. , Rainey, P.B. , and Bohannan, B.J.M. (2004) Big questions, small worlds: Microbial model systems in ecology. Trends Ecol Evol 19: 189–197. [DOI] [PubMed] [Google Scholar]
- Jessup, C.M. , Forde, S.E. , and Bohannan, B.J. M. (2005) Microbial experimental systems in ecology. Adv Ecol Res 37: 273–307. [Google Scholar]
- Johnson, G.R. , Jain, R.K. , and Spain, J.C. (2000) Properties of the trihydroxytoluene oxygenase from Burkholderia cepacia R34: An extradiol dioxygenase from the 2,4‐dinitrotoluene pathway. Arch Microbiol 173: 86–90. [DOI] [PubMed] [Google Scholar]
- Johnson, G.R. , Jain, R.K. , and Spain, J.C. (2002) Origins of the 2,4‐dinitrotoluene pathway. J Bacteriol 184: 4219–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju, K.S. , and Parales, R.E. (2010) Nitroaromatic compounds, from synthesis to biodegradation. Microbiol Mol Biol Rev 74: 250–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korosh, T.C. , Dutcher, A. , Pfleger, B.F. , and McMahon, K.D. (2018) Inhibition of cyanobacterial growth on a municipal wastewater sidestream is impacted by temperature. mSphere 3: e00538‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperman, R.G. , Simini, M. , Siciliano, S. , and Gong, P. (2009) Effects of energetic materials on soil organisms In Ecotoxicology of Explosives. Boca Raton, FL: CRC Press, pp. 35–76. [Google Scholar]
- Latifi, A. , Ruiz, M. , and Zhang, C.C. (2009) Oxidative stress in cyanobacteria. FEMS Microbiol Rev 33: 258–278. [DOI] [PubMed] [Google Scholar]
- Leino, H. , Kosourov, S.N. , Saari, L. , Sivonen, K. , Tsygankov, A.A. , Aro, E.M. , and Allahverdiyeva, Y. (2012) Extended H2 photoproduction by N2‐fixing cyanobacteria immobilized in thin alginate films. Intern J Hydrogen Energy 37: 151–161. [Google Scholar]
- Li, T. , Li, C.T. , Butler, K. , Hays, S.G. , Guarnieri, M.T. , Oyler, G.A. , and Betenbaugh, M.J. (2017) Mimicking lichens: Incorporation of yeast strains together with sucrose‐secreting cyanobacteria improves survival, growth, ROS removal, and lipid production in a stable mutualistic co‐culture production platform. Biotechnol Biofuels 10: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little, A.E.F. , Robinson, C.J. , Peterson, S.B. , Raffa, K.F. , and Handelsman, J. (2008) Rules of engagement: Interspecies interactions that regulate microbial communities. Annu Rev Microbiol 62: 375–401. [DOI] [PubMed] [Google Scholar]
- López, N.I. , Pettinari, M.J. , Nikel, P.I. , and Méndez, B.S. (2015) Polyhydroxyalkanoates: Much more than biodegradable plastics. Adv Appl Microbiol 93: 93–106. [DOI] [PubMed] [Google Scholar]
- Löwe, H. , Hobmeier, K. , Moos, M. , Kremling, A. , and Pflüger‐Grau, K. (2017a) Photoautotrophic production of polyhydroxyalkanoates in a synthetic mixed culture of Synechococcus elongatus cscB and Pseudomonas putida cscAB . Biotechnol Biofuels 10: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwe, H. , Schmauder, L. , Hobmeier, K. , Kremling, A. , and Pflüger‐Grau, K. (2017b) Metabolic engineering to expand the substrate spectrum of Pseudomonas putida toward sucrose. MicrobiologyOpen 6: e00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwe, H. , Sinner, P. , Kremling, A. , and Pflüger‐Grau, K. (2019) Engineering sucrose metabolism in Pseudomonas putida highlights the importance of porins. Microb Biotechnol 13.: 97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐García, E. , Aparicio, T. , de Lorenzo, V. , and Nikel, P.I. (2014) New transposon tools tailored for metabolic engineering of Gram‐negative microbial cell factories. Front Bioeng Biotechnol 2: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐García, E. , Jatsenko, T. , Kivisaar, M. , and de Lorenzo, V. (2015) Freeing Pseudomonas putida KT2440 of its proviral load strengthens endurance to environmental stresses. Environ Microbiol 17: 76–90. [DOI] [PubMed] [Google Scholar]
- McCann, K.S. (2000) The diversity‐stability debate. Nature 405: 228–233. [DOI] [PubMed] [Google Scholar]
- McCarty, N.S. , and Ledesma‐Amaro, R. (2019) Synthetic biology tools to engineer microbial communities for biotechnology. Trends Biotechnol 37: 181–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick, N.G. , Cornell, J.H. , and Kaplan, A.M. (1978) Identification of biotransformation products from 2,4‐dinitrotoluene. Appl Environ Microbiol 35: 945–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momeni, B. , Chen, C.C. , Hillesland, K.L. , Waite, A. , and Shou, W. (2011) Using artificial systems to explore the ecology and evolution of symbioses. Cell Mol Life Sci 68: 1353–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti, M.R. , Smania, A.M. , Fabro, G. , Álvarez, M.E. , and Argaraña, C.E. (2005) Engineering Pseudomonas fluorescens for biodegradation of 2,4‐dinitrotoluene. Appl Environ Microbiol 71: 8864–8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikel, P.I. , and de Lorenzo, V. (2018) Pseudomonas putida as a functional chassis for industrial biocatalysis: From native biochemistry to trans‐metabolism. Metab Eng 50: 142–155. [DOI] [PubMed] [Google Scholar]
- Nikel, P.I. , Silva‐Rocha, R. , Benedetti, I. , and de Lorenzo, V. (2014) The private life of environmental bacteria: Pollutant biodegradation at the single cell level. Environ Microbiol 16: 628–642. [DOI] [PubMed] [Google Scholar]
- Nikel, P.I. , Chavarría, M. , Danchin, A. , and de Lorenzo, V. (2016) From dirt to industrial applications: Pseudomonas putida as a synthetic biology chassis for hosting harsh biochemical reactions. Curr Opin Chem Biol 34: 20–29. [DOI] [PubMed] [Google Scholar]
- Nishino, S.F. , Paoli, G.C. , and Spain, J.C. (2000) Aerobic degradation of dinitrotoluenes and pathway for bacterial degradation of 2,6‐dinitrotoluene. Appl Environ Microbiol 66: 2139–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortíz‐Marquez, J. C. F. , do Nascimento, M. , Zehr, J.P. , and Curatti, L. (2013) Genetic engineering of multispecies microbial cell factories as an alternative for bioenergy production. Trends Biotechnol 31: 521–529. [DOI] [PubMed] [Google Scholar]
- Padda, R.S. , Wang, C. , Hughes, J.B. , Kutty, R. , and Bennett, G.N. (2003) Mutagenicity of nitroaromatic degradation compounds. Environ Toxicol Chem 22: 2293–2297. [DOI] [PubMed] [Google Scholar]
- Parales, J.V. , Kumar, A. , Parales, R.E. , and Gibson, D.T. (1996) Cloning and sequencing of the genes encoding 2‐nitrotoluene dioxygenase from Pseudomonas sp. JS42. Gene 181: 57–61. [DOI] [PubMed] [Google Scholar]
- Park, W. , Pena‐Llopis, S. , Lee, Y. , and Demple, B. (2006) Regulation of superoxide stress in Pseudomonas putida KT2440 is different from the SoxR paradigm in Escherichia coli . Biochem Biophy Res Commun 341: 51–56. [DOI] [PubMed] [Google Scholar]
- Pérez‐Pantoja, D. , Nikel, P.I. , Chavarría, M. , and de Lorenzo, V. (2013) Endogenous stress caused by faulty oxidation reactions fosters evolution of 2,4‐dinitrotoluene‐degrading bacteria. PLoS Genet 9: e1003764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poblete‐Castro, I. , Borrero de Acuña, J.M. , Nikel, P.I. , Kohlstedt, M. , and Wittmann, C. (2017) Host organism: Pseudomonas putida In Industrial Biotechnology: Microorganisms. Wittmann C., and Liao J. C. (eds). Weinheim: Wiley‐VCH Verlag GmbH & Co. KGaA. [Google Scholar]
- Prieto, M.A. , Escapa, I. F. , Martínez, V. , Dinjaski, N. , Herencias, C. , de la Peña, F. , et al (2016) A holistic view of polyhydroxyalkanoate metabolism in Pseudomonas putida . Environ Microbiol 18: 341–357. [DOI] [PubMed] [Google Scholar]
- Rocheleau, S. , Kuperman, R.G. , Simini, M. , Hawari, J. , Checkai, R.T. , Thiboutot, S. , et al (2010) Toxicity of 2,4‐dinitrotoluene to terrestrial plants in natural soils. Sci Total Environ 408: 3193–3199. [DOI] [PubMed] [Google Scholar]
- Roell, G.W. , Zha, J. , Carr, R.R. , Koffas, M.A. , Fong, S.S. , and Tang, Y.J. (2019) Engineering microbial consortia by division of labor. Microb Cell Fact 18: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz‐Güereca, D.A. , and Sánchez‐Saavedra, M.D.P. (2016) Growth and phosphorus removal by Synechococcus elongatus co‐immobilized in alginate beads with Azospirillum brasilense . J Appl Phycol 28: 1501–1507. [Google Scholar]
- Santos‐Merino, M. , Singh, A.K. , and Ducat, D.C. (2019) New applications of synthetic biology tools for cyanobacterial metabolic engineering. Front Bioeng Biotechnol 7: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, J. , Schreiber, U. , Schmid, R. , Völker, U. , and Forchhammer, K. (2001) Nitrogen starvation‐induced chlorosis in Synechococcus PCC7942. Low‐level photosynthesis as a mechanism of long‐term survival. Plant Physiol 126: 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena, S. (2015) Diversity of industrially relevant microbes In Applied Microbiology. New Delhi, India: Springer, pp. 1–11. [Google Scholar]
- Shemer, B. , Yagur‐Kroll, S. , Hazan, C. , and Belkin, S. (2018) Aerobic transformation of 2,4‐dinitrotoluene by Escherichia coli and its implications for the detection of trace explosives. Appl Environ Microbiol 84: e01729‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva‐Rocha, R. , Martínez‐García, E. , Calles, B. , Chavarría, M. , Arce‐Rodríguez, A. , de Las Heras, A. , et al (2013) The Standard European Vector Architecture (SEVA): A coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res 41: D666–D675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M.J. , and Francis, M.B. (2016) A designed A. vinelandii–S. elongatus coculture for chemical photoproduction from air, water, phosphate, and trace metals. ACS Synth Biol 5: 955–961. [DOI] [PubMed] [Google Scholar]
- Smith, M.J. , and Francis, M.B. (2017) Improving metabolite production in microbial co‐cultures using a spatially constrained hydrogel. Biotechnol Bioeng 114: 1195–1200. [DOI] [PubMed] [Google Scholar]
- Song, K. , Tan, X. , Liang, Y. , and Lu, X. (2016) The potential of Synechococcus elongatus UTEX 2973 for sugar feedstock production. Appl Microbiol Biotechnol 100: 7865–7875. [DOI] [PubMed] [Google Scholar]
- Spain, J.C. (1995) Biodegradation of nitroaromatic compounds. Annu Rev Microbiol 49: 523–555. [DOI] [PubMed] [Google Scholar]
- Spanggord, R.J. , Spain, J.C. , Nishino, S.F. , and Mortelmans, K.E. (1991) Biodegradation of 2,4‐dinitrotoluene by a Pseudomonas sp. Appl Environ Microbiol 57: 3200–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen, W.C. , and Spain, J.C. (1993) Cloning and characterization of Pseudomonas sp. strain DNT genes for 2,4‐dinitrotoluene degradation. J Bacteriol 175: 1831–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen, W.C. , Haigler, B.E. , and Spain, J.C. (1996) 2,4‐Dinitrotoluene dioxygenase from Burkholderia sp. strain DNT: Similarity to naphthalene dioxygenase. J Bacteriol 178: 4926–4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons, Z.C. , and Bruce, N.C. (2006) Bacterial pathways for degradation of nitroaromatics. Nat Prod Rep 23: 845–850. [DOI] [PubMed] [Google Scholar]
- Tan, J. , Zuniga, C. , and Zengler, K. (2015) Unraveling interactions in microbial communities‐from co‐cultures to microbiomes. J Microbiol 53: 295–305. [DOI] [PubMed] [Google Scholar]
- Tecon, R. , and Or, D. (2017) Cooperation in carbon source degradation shapes spatial self‐organization of microbial consortia on hydrated surfaces. Sci Rep 7: 43726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therien, J.B. , Zadvornyy, O.A. , Posewitz, M.C. , Bryant, D.A. , and Peters, J.W. (2014) Growth of Chlamydomonas reinhardtii in acetate‐free medium when co‐cultured with alginate‐encapsulated, acetate‐producing strains of Synechococcus sp. PCC7002. Biotechnol Biofuels 7: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi, R. , Dai, Z. , and You, L. (2019) Emerging strategies for engineering microbial communities. Biotechnol Adv 37: 107372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay, D. , Akhtar, M.K. , and Hess, W.R. (2019) Genetic and metabolic advances in the engineering of cyanobacteria. Curr Opin Biotechnol 59: 150–156. [DOI] [PubMed] [Google Scholar]
- Voit, E.O. (2000) Computational Analysis of Biochemical Systems: A Practical Guide for Biochemists and Molecular Biologists. Cambridge: Cambridge University Press. [Google Scholar]
- Voit, E.O. (2017) The best models of metabolism. Wiley Interdiscip Rev Syst Biol Med 9: e1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volke, D.C. , and Nikel, P.I. (2018) Getting bacteria in shape: Synthetic morphology approaches for the design of efficient microbial cell factories. Adv Biosyst 2: 1800111. [Google Scholar]
- Weiss, T.L. , Young, E.J. , and Ducat, D.C. (2017) A synthetic, light‐driven consortium of cyanobacteria and heterotrophic bacteria enables stable polyhydroxybutyrate production. Metab Eng 44: 236–245. [DOI] [PubMed] [Google Scholar]
- Wilkinson, T.G. , Topiwala, H.H. , and Hamer, G. (1974) Interactions in a mixed bacterial population growing on methane in continuous culture. Biotechnol Bioeng 16: 41–59. [DOI] [PubMed] [Google Scholar]
- Wirth, N.T. , Kozaeva, E. , and Nikel, P.I. (2020) Accelerated genome engineering of Pseudomonas putida by I‐SceI‐mediated recombination and CRISPR‐Cas9 counterselection. Microb Biotechnol 13: 233–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y. , Guerra, T.L. , Li, Z. , Ludwig, M. , Dismukes, C.G. , and Bryant, D.A. (2013) Altered carbohydrate metabolism in glycogen synthase mutants of Synechococcus sp. strain PCC7002: Cell factories for soluble sugars. Metab Eng 16: 56–67. [DOI] [PubMed] [Google Scholar]
- Yinon, J. (1990) Toxicity and Metabolism of Explosives. Boston, MA: CRC Press. [Google Scholar]
- Yoon, J.M. , Oliver, D.J. , and Shanks, J.V. (2006) Phytotransformation of 2,4‐dinitrotoluene in Arabidopsis thaliana: Toxicity, fate, and gene expression studies in vitro . Biotechnol Prog 22: 1524–1531. [DOI] [PubMed] [Google Scholar]
- Zavřel, T. , Sinetova, M.A. , and Červený, J. (2015) Measurement of chlorophyll a and carotenoids concentration in cyanobacteria. BioProtocol 5: e1467. [Google Scholar]
- Zhang, C. , Daprato, R.C. , Nishino, S.F. , Spain, J.C. , and Hughes, J.B. (2001) Remediation of dinitrotoluene contaminated soils from former ammunition plants: Soil washing efficiency and effective process monitoring in bioslurry reactors. J Hazard Mat B87: 139–154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Chemical composition of BG‐11, M3, and M3‐N minimal media used in this study.
Fig. S1. Influence of 2,4‐DNT on growth of S. elongatus PCC 7942.
Fig. S2. Effect of 2,4‐DNT on alginate‐encapsulated S. elongatus CscB.
Fig. S3. Chlorophyll a content per cell comparison in planktonic and alginate‐encapsulated S. elongatus CscB.
Fig. S4. Spectral analysis of supernatants of P. putida EM·S cultures.
Fig. S5. Kinetics of 2,4‐DNT transformation by engineered P. putida through the oxidative and reductive pathway.


