Summary
Glycerol‐rich waste streams produced by the biodiesel, bioethanol and oleochemical industries can be treated and valorized by anaerobic microbial communities to produce methane. As current knowledge of the microorganisms involved in thermophilic glycerol conversion to methane is scarce, thermophilic glycerol‐degrading methanogenic communities were enriched. A co‐culture of Thermoanaerobacter and Methanothermobacter species was obtained, pointing to a non‐obligately syntrophic glycerol degradation. This hypothesis was further studied by incubating Thermoanaerobacter brockii subsp. finnii and T. wiegelii with glycerol (10 mM) in pure culture and with different hydrogenotrophic methanogens. The presence of the methanogen accelerated glycerol fermentation by the two Thermoanaerobacter strains up to 3.3 mM day−1, corresponding to 12 times higher volumetric glycerol depletion rates in the methanogenic co‐cultures than in the pure bacterial cultures. The catabolic pathways of glycerol conversion were identified by genome analysis of the two Thermoanaerobacter strains. NADH and reduced ferredoxin formed in the pathway are linked to proton reduction, which becomes thermodynamically favourable when the hydrogen partial pressure is kept low by the hydrogenotrophic methanogenic partner.
Glycerol is an important by‐product of the biodiesel and bioethanol industries, which results in a surplus of this compound. We investigated anaerobic glycerol fermentation coupled to methane production at high temperature (65°C), as a potential strategy for the valorization of this industrial by‐product. We discovered that glycerol fermentation by Thermoanaerobacter strains was much faster when performed in cooperative relationship with a hydrogenotrophic methanogenic partner. The methanogen facilitates glycerol conversion by consuming the hydrogen, thus assisting in the redox balance.
Introduction
Worldwide demand for biodiesel increased in the last decade, leading to a global biodiesel production of 36 × 109 l in 2016 (OECD/FAO, 2017). Glycerol is co‐produced in quantities that match approximately 10% of the total biodiesel production, leading to a surplus of this compound. Consequently, glycerol prices have decreased, changing glycerol from a commodity chemical to a surplus by‐product, and even a waste product (Viana et al., 2012; Clomburg and Gonzalez, 2013; Ciriminna et al., 2014). Glycerol is also generated in ethanol production by yeast (Navarrete et al., 2014) and is frequently present in different wastes/wastewater as e.g. from the oleochemical industry, where waste streams can contain up to 90% glycerol (Clomburg and Gonzalez, 2013).
Anaerobic microbial processes can provide a solution for these glycerol‐rich wastes producing a wide range of valuable compounds (Viana et al., 2012; Clomburg and Gonzalez, 2013). Since glycerol is a highly reduced compound, fermentative microorganisms must be able to dispose of the excess reducing equivalents, which is generally accomplished by the production of 1,3‐propanediol (1,3‐PDO), a product that is more reduced than glycerol (Clomburg and Gonzalez, 2013; Schindler et al., 2014). Microorganisms that lack the 1,3‐PDO formation pathway generally transfer the electrons to hydrogen or formate, as well as to pyruvate, generating organic compounds such as ethanol, butanol or succinate (Murarka et al., 2008; Scholten et al., 2009; Clomburg and Gonzalez, 2013). The problem of the release of excess electrons in glycerol fermentation has been studied with diverse mesophilic bacteria, including studies on electron transfer to electrodes (Emde et al., 1989; Emde and Schink, 1990) or medium sparging with inert gases for H2 removal (Dharmadi et al., 2006; Murarka et al., 2008).
The high energy content of glycerol makes it also an interesting substrate for biogas production, individually or in co‐digestion with different feedstocks – e.g. sewage sludge or the organic fraction of municipal solid wastes (Kolesárová et al., 2011; Yang et al., 2012). The generated biomethane may be stored or injected into the natural gas grid and used as biofuel (Beauchamp et al., 2014; Hengeveld et al., 2014).
The production of biodiesel and bioethanol typically generates waste streams at temperatures between 40 to 65°C. Therefore, thermophilic conditions are beneficial for valorization of glycerol. Moreover, anaerobic digestion is generally faster when performed by thermophilic than by mesophilic microorganisms (Ho et al., 2013). Some thermophilic bacteria were reported to grow with glycerol in pure culture, e.g. Thermoanaerobacter wiegelii (Cook et al., 1996), Moorella glycerini (Slobodkin et al., 1997) and Pseudothermotoga lettingae (Balk et al., 2002), but the thermophilic conversion of glycerol by mixed communities is only scarcely studied (Yang et al., 2008; Zhang et al., 2015).
This work aims to gain insight into the different microbial key players involved in glycerol degradation in mixed thermophilic anaerobic cultures. Thermophilic glycerol‐degrading cultures were enriched, and a co‐culture of Thermoanaerobacter brockii and a methanogenic partner was obtained, pointing to the possibility of facultatively syntrophic glycerol degradation. The influence of different methanogenic partners on glycerol degradation by two Thermoanaerobacter species was then investigated.
Results
Enrichment of glycerol‐degrading microbial cultures
A stable thermophilic (55°C) glycerol‐degrading enrichment (culture Gly(9)) was obtained through repeated transfers to fresh medium containing glycerol as sole substrate over a period of approximately one year (Fig. S1 and Table S1). This culture converted 6.5 ± 0.3 mM of glycerol mainly to methane (6.2 ± 0.1 mM) and acetate (6.7 ± 0.1 mM) during the first 6 days of incubation (Fig. 1). Propionate was also detected, but at concentrations lower than 1 mM (Fig. 1). No other fermentation products, such as lactate, ethanol, butanol, 1,3‐PDO, 1,2‐PDO or hydrogen, were detected.
Figure 1.
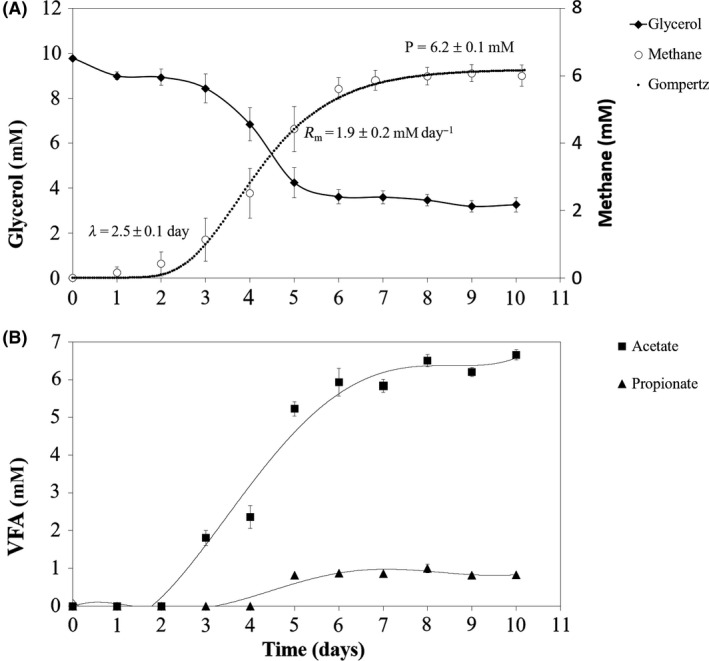
Glycerol consumption and product formation by enrichment culture Gly(9) at 55°C: glycerol concentration, experimental methane data and fitting with the modified Gompertz equation (Equation 1, R 2 = 0.984) (A); volatile fatty acids (B).
Culture Gly(9) was mainly composed by microorganisms of the genera Methanothermobacter, Thermoanaerobacter, Pseudothermotoga and Acetomicrobium, as shown in Table 1. Taxonomic identification was not possible for approximately 25% of the retrieved sequences.
Table 1.
Microbial composition of the glycerol‐degrading enrichments Gly(9) and Col‐Gly.
| Taxonomic identificationa | Relative abundance (%)b | Closest relativesc | Identity of 16S | |||
|---|---|---|---|---|---|---|
| Gly(9) | Col‐Gly | rRNA genes (%)c | ||||
| Methanothermobacter | 36.7 | 31.7 | 30.1 | 23.7 | Methanothermobacter wolfeii strain SIV6 16S ribosomal RNA gene, partial sequence | 100 |
| Thermoanaerobacter | 16.9 | 20.1 | 69.8 | 76.2 | Thermoanaerobacter brockii subsp. finnii strain Ako‐1 16S ribosomal RNA gene, complete sequence | 100 |
| Thermotoga | 12.0 | 12.6 | 0.0 | 0.0 | Pseudothermotoga profunda AZM34c06 DNA, complete genome | 98 |
| Acetomicrobium | 9.6 | 10.6 | 0.0 | 0.0 | Acetomicrobium mobile strain NGA 16S ribosomal RNA gene, partial sequence | 99 |
| Other taxad | 24.9 | 25.1 | 0.0 | 0.0 | – | – |
Taxonomic identification at the genus level based on 16S rRNA genes sequences of approximately 291 bp length by Illumina MiSeq.
Results of duplicate samples.
Results of sequence alignment by using BLAST towards the NCBI nucleotide database.
Taxa with relative abundance < 1% and taxa with classification above the order level were included in Other taxa.
This culture was further incubated in agar‐shake cultures at 70 and 40ºC, considering that several members of the Thermoanaerobacter genus and all the known Acetomicrobium species can grow at this last temperature. A methanogenic glycerol‐degrading culture designated Col‐Gly was obtained at 40ºC, which presented very low diversity when examined by phase contrast microscopy (Fig. S2). Microbial community analysis showed the presence of only two microorganisms belonging to Methanothermobacter and Thermoanaerobacter genera, with relative abundances of 24–30% and 70–76% respectively (Table 1).
When culture Col‐Gly was incubated at 65°C (the optimal growth temperature for both identified microorganisms), 10 mM glycerol was completely degraded within 6 days of incubation (data not shown), associated with the formation of methane (8.0 ± 0.2 mM), acetate (8.7 ± 1.8 mM) and lactate (2.8 ± 0.3 mM) (Fig. 2). Hydrogen was detected at residual concentrations (< 0.01 mM) during the experiment (data not shown). Similar glycerol consumption (i.e. glycerol was not detectable after 7 days of incubation) and products profile (Fig. S3) were obtained in the incubations at 55°C (the original incubation temperature of the enrichment Gly(9)), and therefore further experiments were performed at 65°C.
Figure 2.
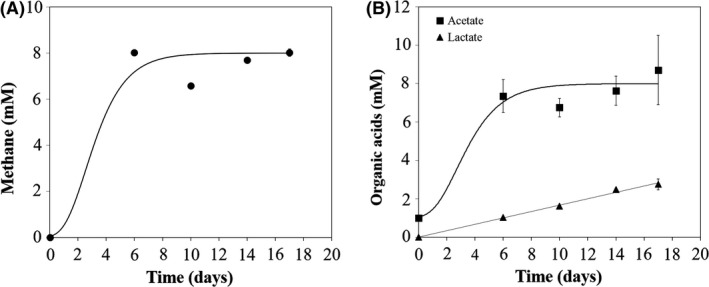
Methane (A) and organic acids (B) production by culture Col‐Gly at 65°C.
When the enriched co‐culture Col‐Gly was incubated with BrES, a selective inhibitor of methane‐producing archaea (DiMarco et al., 1990), no methane was detected in the headspace of the bottles and only vestigial glycerol consumption was observed during 7 days of incubation (Fig. 3). Hydrogen accumulated at very low amounts (< 0.1 mM), while ethanol and lactate were not detected (Fig. 3). No noteworthy effect of BrES could be detected in glycerol consumption and products formation by the T. brockii subsp. finnii type strain (Fig. S4). These results raised the hypothesis that the presence of the methanogen could influence the observed glycerol conversion rates in the enriched co‐culture Col‐Gly.
Figure 3.
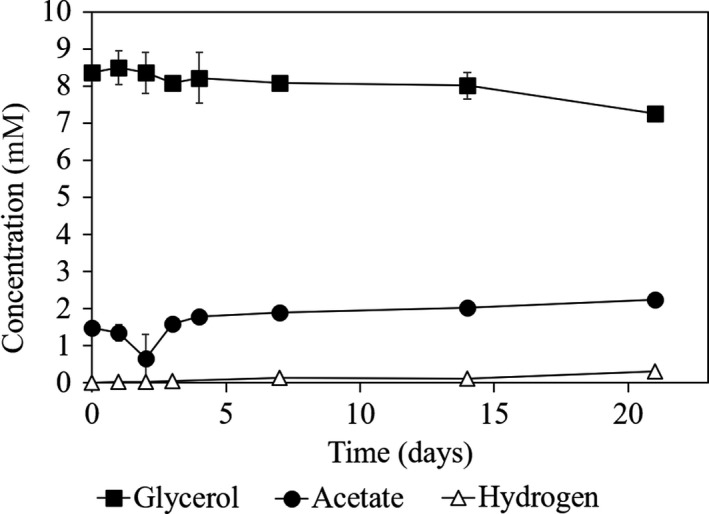
Glycerol, acetate and methane concentrations measured during the incubation of Col‐Gly with BrES.
Glycerol degradation by Thermoanaerobacter species, in pure culture or in co‐culture with methanogens
Within the Thermoanaerobacter genus, only T. wiegelii, T. siderophilus, T. brockii subsp. finnii and T. subterraneus were reported to grow with glycerol (Cook et al., 1996; Slobodkin et al., 1999; Fardeau et al., 2000; Alves et al., 2016), although growth of T. brockii subsp. finnii was described as poor (Alves et al., 2016). Therefore, to further assess the possible positive effect of methanogens on glycerol fermentation, T. brockii subsp. finnii DSM 3389T and T. wiegelii DSM 10319T were grown individually in pure culture or with a methanogenic partner. Methanothermobacter sp. strain GH (the culture obtained after incubation of the enrichment Col‐Gly with H2/CO2 for 15 transfers) and M. marburgensis were the selected methanogens. The results obtained are shown in Figs 4 and 5.
Figure 4.
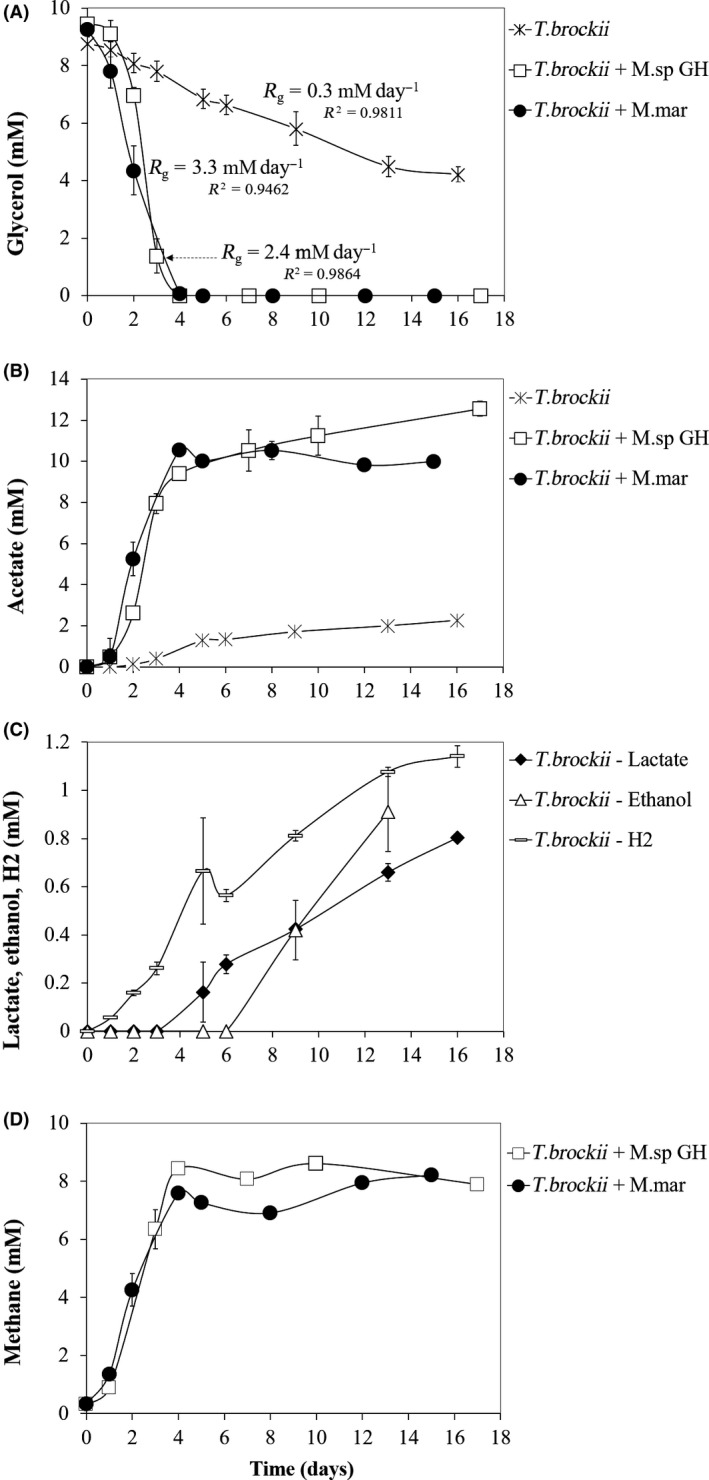
Glycerol consumption (A) and production of acetate (B) by Thermoanaerobacter brockii subsp. finnii (DSM 3389T) when incubated in pure culture or in co‐culture with methanogens. Lactate, ethanol and H2 production by T. brockii subsp. finnii in pure culture (C) and methane production in co‐culture with methanogens (D). M. sp. GH, culture obtained after 15 transfers of the enriched culture Col‐Gly with H2/CO2; M. mar, Methanothermobacter marburgensis DSM 2133T. R g, Volumetric glycerol depletion rate.
Figure 5.
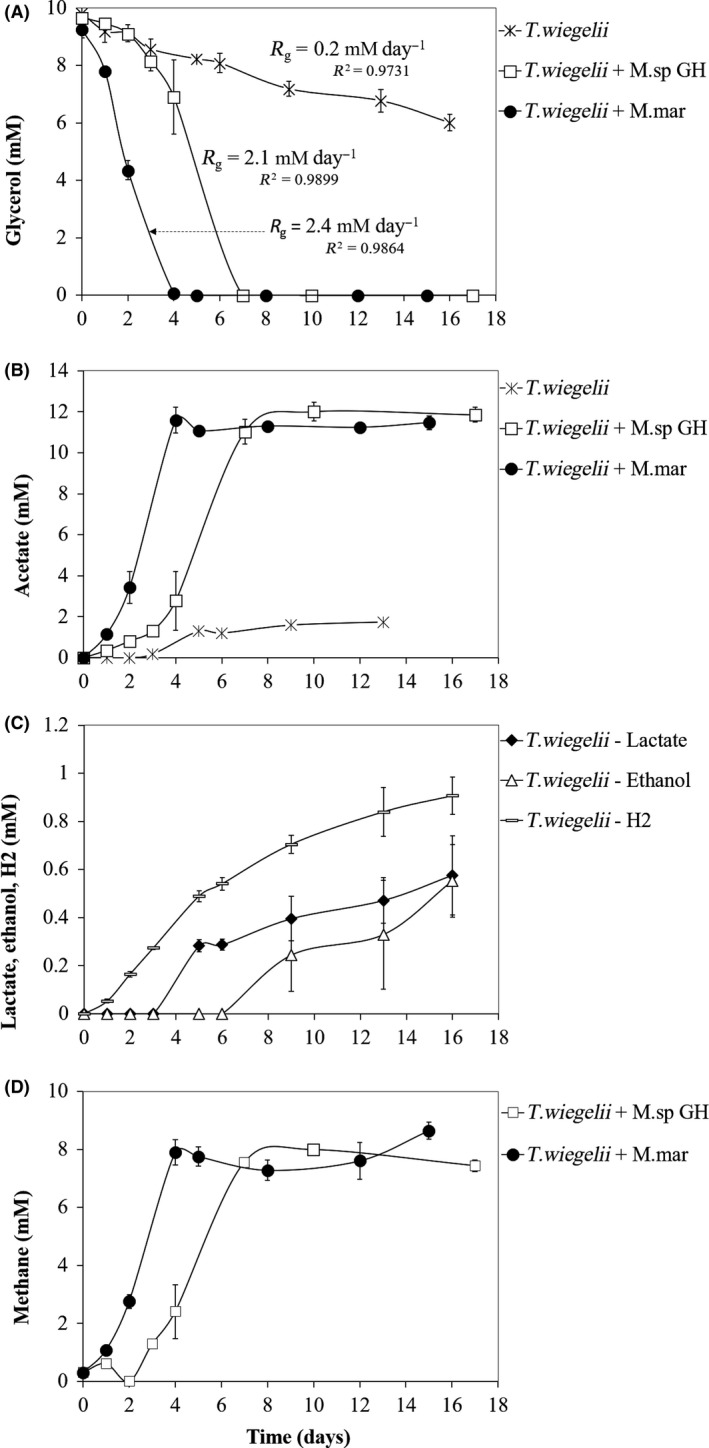
Glycerol consumption (A) and production of acetate (B) by Thermoanaerobacter wiegelii (DSM 10319T) when incubated in pure culture or in co‐culture with methanogens. Lactate, ethanol and H2 production by T. wiegelii in pure culture (C) and methane production in co‐culture with methanogens (D). M. sp. GH, culture obtained after 15 transfers of the enriched culture Col‐Gly with H2/CO2; M. mar, Methanothermobacter marburgensis DSM 2133T. R g, Volumetric glycerol depletion rate.
After approximately 16 days of incubation, T. brockii subsp. finnii and T. wiegelii consumed 52 ± 4% and 39 ± 5% of the glycerol added, respectively, with corresponding volumetric substrate depletion rates (R g) of 0.3 and 0.2 mM day−1 (Figs 4A and 5A). Acetate was the main product (around 2 mM in both cases, Figs 4B and 5B) and hydrogen, lactate and ethanol were obtained in small amounts (0.5–1.2 mM, Figs 4C and 5C). Product yields were calculated relatively to the amount of glycerol consumed, and were similar for both species, i.e. around 0.5 mmol mmol−1 for acetate, 0.25 mmol mmol−1 for hydrogen and between 0.15 and 0.20 mmol mmol−1 for lactate and ethanol (Table 2). Considering the stoichiometry of the possible reactions involved (Table 3), the products measured accounted for 87% and 75% of the glycerol consumed by T. brockii and T wiegelii, respectively. When the Thermoanaerobacter type strains were incubated in co‐culture with the methanogens, glycerol consumption rate was substantially accelerated, i.e. all the constructed co‐cultures (Thermoanaerobacter strain + methanogen) completely degraded the added glycerol in 4–7 days with volumetric substrate depletion rates (R g) 8–12 times higher than the bacterial pure cultures (i.e. between 2.1 and 3.3 mM day−1, Figs 4A and 5A). Acetate and methane were the main products obtained (Figs 4B, D and 5B, D), with respective yield of 1.0 and 0.63–0.82 mmol mmol−1 relatively to the amount of glycerol consumed, which are close to the theoretically expected values (Tables 2 and 3).
Table 2.
Product yields of glycerol fermentation, calculated relatively to the amount of glycerol consumed (mmol mmol−1), by T. brockii subsp. finnii (DSM 3389T) and T. wiegelii (DSM 10319T), when incubated in pure culture or in co‐culture with methanogens.
| Culture | Acetate | Lactate | Ethanol | H2 | Methane |
|---|---|---|---|---|---|
| T. brockii | 0.50 ± 0.05 | 0.18 ± 0.01 | 0.20 ± 0.04 | 0.25 ± 0.02 | n.a. |
| T. brockii + M. sp. GH | 1.00 ± 0.04 | n.d. | n.d. | n.d. | 0.63 ± 0.02 |
| T. brockii + M. mar | 1.00 ± 0.00 | n.d. | n.d. | n.d. | 0.82 ± 0.01 |
| T. wiegelli | 0.46 ± 0.06 | 0.15 ± 0.05 | 0.15 ± 0.04 | 0.24 ± 0.04 | n.a. |
| T. wiegelli + M. sp. GH | 1.00 ± 0.04 | n.d. | n.d. | n.d. | 0.63 ± 0.03 |
| T. wiegelli + M. mar | 1.00 ± 0.04 | n.d. | n.d. | n.d. | 0.75 ± 0.03 |
M. sp. GH, culture obtained after 15 transfers of the enriched culture Col‐Gly with H2/CO2; M. mar, Methanothermobacter marburgensis DSM 2133T; n.a., not applicable. n.d., not determined.
Table 3.
Possible reactions involved in glycerol degradation by the enrichment cultures Gly(9) and Col‐Gly and their corresponding Gibbs free energy changes at 25°C.
| Reaction | Reactant | Main products | Equation |
ΔG 0’ (kJ reaction−1)a |
|---|---|---|---|---|
| (1) | Glycerol | Acetate | C3H8O3 + 2 H2O → C2H3O2 ‐ + HCO3 ‐ + 3 H2 + 2 H+ | −73.1 |
| (2) | H2 + CO2 | Methane | 4 H2 + + H+ → CH4 + 3 H2O | −135.6 |
| (3) = (1 + 2) | Glycerol | Acetate + Methane | C3H8O3 → + 0.75 CH4 + 0.25 + 0.25 H2O + 1.25 H+ | −174.7 |
| (4) | Glycerol | Lactate | C3H8O3 → + H2 + H+ | −69.1 |
| (5) | Glycerol | Ethanol | C3H8O3 + H2O → C2H4OH + + H2 + H+ | −82.7 |
Discussion
Glycerol (1,2,3‐propanetriol) can sustain growth of a diverse microbial community, as shown by the composition and activity of culture Gly(9) enriched at 55°C (Table 1, Fig. 1). The main bacterial genera identified were probably involved in glycerol conversion, since some of the characterized strains within the genera Pseudothermotoga, Acetomicrobium and Thermoanaerobacter have been reported as glycerol degraders (Rees et al., 1997; Menes and Muxí, 2002; Maru et al., 2013; Alves et al., 2016). Acetate and methane were the main products of glycerol conversion, indicating that methane was produced from formate or H2/CO2 (Fig. 1). This was reinforced by the identification of Methanothermobacter sp., a hydrogenotrophic methanogen, as the only archaeon in this community (Table 1).
When applying a lower temperature (40ºC) as selective factor, a co‐culture composed by Thermoanaerobacter and Methanothermobacter was enriched (culture Col‐Gly). This co‐culture was capable of fast glycerol degradation (< 6 days) coupled to good growth evaluated by visual inspection. Also in the work of Zhang et al. (2015), Thermoanaerobacter spp. and hydrogenotrophic methanogens (mainly Methanothermobacter thermautotrophicus) were the dominant microorganisms in the community developed in a continuous bioreactor operated with glycerol at 70°C. However, as previously mentioned, Alves et al. (2016) reported that glycerol was only poorly utilized by T. brockii subsp. finnii. When we incubated pure cultures of T. brockii subsp. finnii or T. wiegelii with glycerol at 10 mM, glycerol was hardly fermented, i.e. more than 16 days were required to convert 40–50% of the added glycerol (Figs 4 and 5). In spite of that, these two strains easily ferment glucose in pure culture, e.g. Alves et al. (2016) reported degradation of 20 mM of glucose by T. brockii subsp. finnii and T. wiegelli in 7 and 3 days respectively. This difference is probably related to the more reduced nature of glycerol, which leads to the generation of twice the number of reducing equivalents per pyruvate molecule formed, compared with glucose (Clomburg and Gonzalez, 2013).
The two Thermoanaerobacter strains studied in this work did not produce 1,3‐PDO, and the analysis of their genomes confirmed that these bacteria lack the genes encoding for the enzymes involved in the pathway of 1,3‐PDO formation (enzymes 1 and 2, Fig. 6). Therefore, these bacteria are not able to easily dispose of the excess of reducing equivalents generated from glycerol; as lactate and ethanol (+CO2) are more oxidized than glycerol, formation of these compounds cannot balance the surplus of electrons formed in the conversion of glycerol to acetate, and hydrogen production is constrained by thermodynamics (Figs 4, 5, 6, Table 3). The two Thermoanaerobacter strains oxidize and decarboxylate pyruvate to acetyl‐coenzyme A with ferredoxin as redox mediator, as indicated by the presence in their genomes of the genes coding for pyruvate:ferredoxin oxidoreductase (enzyme 11, Fig. 6), and by the absence of pyruvate formate lyase and formate hydrogen lyase encoding genes (Fig. 6 – enzymes 12 and 13 respectively). Subsequently, a trimeric or a monomeric hydrogenase (enzyme 16) oxidizes ferredoxin and, using protons as final electron acceptors, leads to hydrogen production. In the reoxidation of NADH+ + H+, electrons are transferred by a NADH:ferredoxin oxidoreductase (enzyme 17) to a hydrogenase (trimeric or monomeric, enzyme 16) and thus hydrogen can also be produced (Vardar‐Schara et al., 2006; Calusinska et al., 2010).
Figure 6.
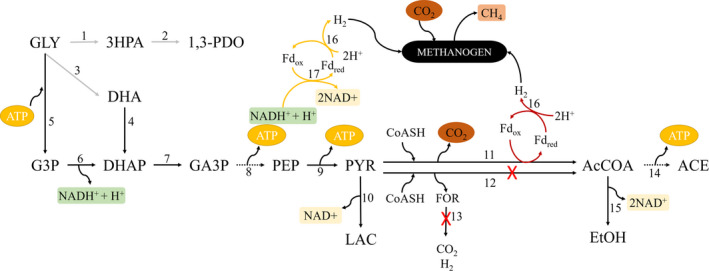
Main metabolic pathway for glycerol conversion by Thermoanaerobacter brockii subsp. finnii (DSM 3389T) and Thermoanaerobacter wiegelii (DSM 10319T). Grey lines: absent in Thermoanaerobacter. Broken lines mean multiple steps. Abbreviations: GLY, glycerol; 3HPA, 3‐hydroxypropionaldehyde; 1,3‐PDO, 1,3‐propanediol; DHA, dihydroxyacetone; DHAP, dihydroxyacetone phosphate; G3P, glycerol‐3‐phosphate; GA3P, glyceraldehyde‐3‐phosphate; PEP, phosphoenolpyruvate; PYR, pyruvate; LAC, lactate; AcCOA, acetyl‐coenzyme A; FOR, formate; ACE, acetate; EtOH, ethanol; Fdox, oxidized ferredoxin; Fdred, reduced ferredoxin. 1, glycerol dehydratase; 2, 1,3‐PDO dehydrogenase; 3, glycerol dehydrogenase; 4, dihydroxyacetone kinase; 5, glycerol kinase; 6, glycerol‐3‐phosphate dehydrogenase; 7, triosephosphate isomerase; 8, glyceraldehyde‐3‐phosphate dehydrogenase, phosphoglycerate kinase, phosphoglycerate mutase and enolase; 9, pyruvate kinase; 10, L‐lactate dehydrogenase; 11, pyruvate:ferredoxin oxidoreductase; 12, pyruvate formate lyase; 13, formate hydrogen lyase; 14, phosphate acetyltransferase and acetate kinase; 15, acetaldehyde dehydrogenase; 16, hydrogenase; and 17, ferredoxin‐NADP(+) reductase. Additional information of EC number and genome location of the enzymes of this metabolic pathway can be found at Table S2.
The oxidation of reduced ferredoxin and especially NADH coupled to proton reduction only becomes thermodynamically feasible at low hydrogen partial pressure (Sousa et al., 2009; Stams and Plugge, 2009). Therefore, Thermoanaerobacter strains surpass the metabolic dilemma of redox balancing and energy acquisition when in the presence of a methanogen, which consumes the hydrogen produced during glycerol fermentation and functions as biological electron acceptor (Fig. 6). In fact, glycerol conversion to acetate becomes more exergonic and thermodynamically more favourable if the hydrogen produced is used by hydrogenotrophic methanogens to produce methane, as shown by the Gibbs free energy changes of ΔG 0’ = −73.1 and −174.7 kJ reaction−1 respectively (reactions 1 and 3, Table 3). In these co‐cultures, ethanol production could not be detected, and only small amounts of lactate were produced by the enriched culture Col‐Gly (Figs 4 and 5).
The positive effect of the methanogen was also experimentally confirmed when the methanogenic activity in culture Col‐Gly was inhibited by BrES, which caused glycerol degradation to proceed at a much lower rate (Fig. 3). The relationship between the Thermoanaerobacter strains and the methanogen points to a facultative syntrophy, since glycerol fermentation can be performed by these bacteria in pure culture but their growth and metabolic products are directly influenced by the hydrogen scavenger (Stams and Plugge, 2009). This syntrophic relationship is energetically advantageous for the Thermoanaerobacter bacteria, compared with glycerol fermentation in pure culture, since it allows higher ATP gain, i.e. 2 ATP are formed from glycerol to acetate instead of 1 from glycerol to lactate and/or ethanol (Fig. 6). Since syntrophic glycerol fermentation by the two Thermoanaerobacter strains does not involve a pyruvate formate lyase, the ability of the methanogenic partner to consume formate is not needed for this interspecies relationship. Only a slight delay of approximately 2 days was observed in the incubations of T. wiegellii and the Methanothermobacter. sp. strain GH (Fig. 5).
The importance of an external electron acceptor for improving glycerol conversion has been previously reported, for example in cultures of Actinobacillus succinogenes grown in the presence of dimethylsulfoxide (DMSO) as external electron acceptor (Carvalho et al., 2014; Schindler et al., 2014). Moreover, glycerol fermentation by Escherichia coli was shown to be impaired by hydrogen accumulation (Dharmadi et al., 2006; Gonzalez et al., 2008), which could be overcome by co‐cultivation with the methanogen Methanobacterium formicicum (Richter and Gescher, 2014; Kim et al., 2017). Likewise, glycerol fermentation by E. coli and Propionibacterium freudenreichii can be supported through electron transfer to electrodes mediated by potassium ferricyanide (Emde et al., 1989; Emde and Schink, 1990). For Thermoanaerobacter brockii subsp. brockii, the addition of thiosulphate or Methanobacterium sp. as electron acceptors improved the oxidative deamination of aminoacids (Fardeau et al., 1997). The consumption of glucose and pyruvate by Thermoanaerobium brockii was enhanced as well by using acetone as electron acceptor (Ben‐Bassat et al., 1981). Also, Vipotnik et al. (2016) showed inhibition of glucose consumption by Thermoanaerobacter strain AK68 when exposed to high hydrogen partial pressure and that the addition of thiosulphate or co‐cultivation with Methanothermobacter strain M39 (as electron scavenger) increased the utilization of glucose and acetate production (Vipotnik et al., 2016). In summary, we show that the presence of a hydrogenotrophic methanogenic partner, acting as biological electron acceptor, enhances glycerol conversion by Thermoanaerobacter species, since it facilitates the redox balance and contributes to a higher energy gain by these bacteria. Therefore, syntrophic glycerol fermentation promotes faster anaerobic treatment of glycerol‐rich waste streams coupled to methane production.
Experimental procedures
Biomass source
Thermophilic anaerobic sludge was collected from a lab‐scale up‐flow anaerobic column reactor operated at 55°C, fed with a mixture of skim milk and sodium oleate (50:50% of the chemical oxygen demand, COD) at a COD concentration of 10 g l−1 and hydraulic retention time of 1 day. Additional details of the reactor operation are provided in Supporting Information. Degradation of the substrate accumulated during the reactor operation was promoted by incubation in batch at 55°C for 18 days, before starting the enrichments.
Medium composition and cultivation
All the experiments were performed using a bicarbonate‐buffered mineral salt medium (basal medium, BM) prepared as described by Stams et al. (1993). BM was dispensed in serum bottles which were sealed with butyl rubber septa and aluminum crimp caps. The headspace of the bottles was flushed with a gas mixture of N2 and CO2 (80:20% v/v), at a final pressure of 1.7 × 105 Pa. Before incubation, the medium was reduced with 0.8 mM sodium sulfide and supplemented with salts and vitamins. All inoculations and transfers were done aseptically using sterile syringes and needles.
Enrichment of glycerol‐degrading microbial cultures
Enrichments (coded Gly(x), where × represents the number of transfers) were started by inoculating 120 ml serum bottles, containing 50 ml BM, with 10% (v/v) of the sludge. Glycerol was added from a sterile stock solution to a final concentration of 10 mM, based on the works of Fardeau et al. (2000) and Alves et al. (2016). Successive transfers of the cultures to new medium (10% v/v) and serial dilutions were made after confirming microbial growth and activity, based on microscopic observations and methane measurements (more than 30% of the theoretical value expected). All cultures were incubated at 55°C, statically and in the dark. Schematic representation of the experimental procedure applied is shown in Fig. S1.
Physiological characterization was performed after nine successive transfers (enrichment Gly(9)), in triplicate 500 ml bottles containing 250 ml BM (Fig. S1) and glycerol (10 mM). Methane content in the headspace, volatile fatty acids (VFA), lactate, glycerol, ethanol, butanol, 1,3‐PDO and 1,2‐PDO were measured daily. The final hydrogen content of the headspace was also measured. In addition, duplicate samples were collected at the end of the incubation period for DNA extraction and 16S rRNA genes sequencing by Illumina MiSeq. The experimental methane production data was fitted by the modified Gompertz equation (equation 1) for estimation of the methane production kinetics (Zwietering et al., 1990).
| (1) |
where M (t) = cumulative methane production (mM), P = maximum methane production (mM), Rm = methane production rate (mM day−1), e = 2.7182818 and = lag phase (days). The standard error for each variable and the coefficient of determination (R 2) were calculated.
Further microbial selection was then performed by serially diluting the enrichment Gly(9) in agar‐shake cultures, containing 50 ml BM solidified with 1.5% (w/v) agar. Incubations were made at 40 and 70°C, in the dark and without agitation. Colonies were picked and transferred to the same medium without agar. Growth (verified by visual inspection of the bottles and by microscopic observations) was only observed in the cultures incubated at 40°C, and thus the enrichments at 70°C were not continued. At 40°C, after four successive transfers in liquid medium, microbial community composition was analysed by sequencing of 16S rRNA genes (Illumina MiSeq). This culture was coded Col‐Gly. Its ability to consume glycerol (10 mM) and the products formed (methane, hydrogen, VFA, lactate, alcohols) were monitored in triplicate assays, incubated at two different temperatures (55 and 65°C). Incubation with glycerol in the presence of 20 mM of 2‐bromoethanesulfonate (BrES) was also performed in triplicate, and glycerol, acetate, methane and hydrogen concentrations were measured with time.
Glycerol degradation by Thermoanaerobacter species, in pure culture or in co‐culture with methanogens
Thermoanaerobacter brockii subsp. finnii (DSM 3389T), Thermoanaerobacter wiegelii (DSM 10319T) and Methanothermobacter marburgensis (DSM 2133T) were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ, Braunschweig, Germany). The two Thermoanaerobacter strains were grown with 20 mM glucose, and M. marburgensis was cultured using a gas phase of H2/CO2 (80:20% v/v, 1.7 × 105 Pa). Enrichment Col‐Gly was also incubated with H2/CO2 (80:20% v/v, 1.7 × 105 Pa) for 15 transfers, with the aim of retrieving the methanogen from this culture. The culture obtained was named Methanothermobacter sp. strain GH.
An assay was then set up to study glycerol degradation by the two Thermoanaerobacter species, in pure culture or in co‐culture with methanogens. For this, the two cultures of methanogens (Methanothermobacter sp. strain GH and M. marburgensis) were pre‐grown until complete hydrogen consumption, after which the headspace of the bottles was flushed and pressurized with N2/CO2 (80:20% v/v, 1.7 × 105 Pa) under sterile conditions. The two Thermoanaerobacter type strains were also pre‐grown with glycerol and were then transferred (10% v/v) to bottles containing fresh medium or mixed with the pre‐grown cultures of methanogens. Glycerol was added at 10 mM. The concentrations of glycerol, other alcohols, VFA, lactate, methane and hydrogen were measured over time. All tests were performed in triplicate. Incubations were made at 65°C, statically and in the dark.
Analytical methods
Phase contrast microscopy was performed using an Olympus CX41 RF microscope, and micrographs were obtained with an Olympus Altra 20 image acquisition system. The software used with this setup was the AnalySIS getIT (Olympus soft imaging solutions GmbH). Methane and hydrogen were quantified by gas chromatography. For methane quantification, a GC‐2014 Shimadzu gas chromatograph was used with a Porapak Q column and a flame ionization detector. N2 was used as carrier gas. Injection port, column and detector temperatures were 110, 35 and 220°C respectively. Hydrogen was analysed using a Molsieve column (MS‐13x 80/100 mesh) and a thermal conductivity detector Bruker Scion 456 Chromatograph (Bruker, Billerica, MA, USA) with argon (60 ml min−1) as the carrier gas. The injector, column and detector temperatures were 100, 35 and 130°C respectively. Volatile fatty acids (VFA), lactate, glycerol and other alcohols were analysed by high‐performance liquid chromatography (HPLC; Jasco, Tokyo, Japan). For organic acids quantification, an Agilent Hi‐Plex H (300 × 7.7 mm) column was used, with a mobile phase of 2.5 mM H2SO4 at a flow rate of 0.6 ml min−1. The column temperature was set at 60°C and spectrophotometric ultraviolet (UV) detection was performed at 210 nm. Glycerol and other alcohols were analysed using a Varian Aminex 87H (300 × 7.8 mm) with a mobile phase of 5 mM H2SO4 at a flow rate of 0.7 ml min−1, with the column temperature set at 60°C and refractive index (RI) detection.
Microbial composition of the glycerol‐degrading enrichment cultures
Aliquots of well‐homogenized sludge were collected from cultures Gly(9) and Col‐Gly, and immediately frozen at −20°C. Total genomic DNA was extracted using the FastDNA SPIN Kit for Soil (MP Biomedicals, Solon, OH) and purified by ethanol precipitation. DNA amplification, Illumina library preparation, amplicon sequencing (Illumina MiSeq, Inc. San Diego, CA, USA) and bioinformatics analysis of the data were performed at Research and Testing Laboratory (Lubbock, TX, USA). Samples were amplified for sequencing using the universal primer pair 515f and 806r (Caporaso et al., 2011), targeting the prokaryotic 16S rRNA gene. Details on the sequencing and bioinformatics data analysis can be found elsewhere (Paulo et al., 2017). All sequencing reads were submitted to the European Nucleotide Archive (ENA) under the study accession number PRJEB30535 (http://www.ebi.ac.uk/ena/data/view/PRJEB30535). A comparison of 16S rRNA gene sequences of OTU to the NCBI database was also performed using the BLASTN alignment tool (http://ncbi.nlm.nih.gov/blast).
Genome analysis of Thermoanaerobacter strains
Analysis of Thermoanaerobacter brockii subsp. finnii (DSM 3389T) and Thermoanaerobacter wiegelii (DSM 10319T) genomes, was performed using the Integrated Microbial Genomes (IMG) (https://img.jgi.doe.gov/) and The National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/) genomic platforms.
Conflict of interest
None declared.
Supporting information
Figure S1 . Experimental procedure applied for the enrichment of thermophilic glycerol‐degrading microbial cultures at 55ºC.
Figure S2 . Phase contrast micrograph of culture Col‐Gly.
Figure S3 . Methane (A) and organic acids (B) production by enriched culture Col‐Gly at 55°C.
Figure S4 . Glycerol consumption (A) and production of acetate (B) and H2 (C) by T. brockii subsp. finnii type strain, when incubated with BrES (w/BrES) and without BrES (w/o BrES).
Table S1 . Methane production from glycerol by the different generations (coded Gly(x), where × represents the number of transfers), during the enrichment process.
Table S2 . Additional information about the enzymes involved in the metabolic pathway for glycerol conversion of Thermoanaerobacter brockii subsp. finnii (DSM 3389T) and Thermoanaerobacter wiegelii (DSM 10319T). The data were retrieved from NCBI genomic platform.
Acknowledgements
The authors thank Ruben Gonçalves for preparing the thermophilic biomass and Andreia Salvador for the support with the microbial communities’ analysis. This study was supported by the Portuguese Foundation for Science and Technology (FCT) under the scope of the strategic funding of UID/BIO/04469/2019 unit, Project SAICTPAC/0040/2015 (POCI‐01‐0145‐FEDER‐016403) and BioTecNorte operation (NORTE‐01‐0145‐FEDER‐000004) funded by the European Regional Development Fund under the scope of Norte2020 – Programa Operacional Regional do Norte. The authors also acknowledge the financial support of FCT and European Social Fund through the grants attributed to C.P. Magalhães (SFRH/BD/132845/2017) and A.L. Arantes (PD/BD/128030/2016).
Microbial Biotechnology (2020) 13(4), 962–973
Funding Information
This study was supported by the Portuguese Foundation for Science and Technology (FCT) under the scope of the strategic funding of UID/BIO/04469/2019 unit, Project SAICTPAC/0040/2015 (POCI‐01‐0145‐FEDER‐016403) and BioTecNorte operation (NORTE‐01‐0145‐FEDER‐000004) funded by the European Regional Development Fund under the scope of Norte2020 – Programa Operacional Regional do Norte. The authors also acknowledge the financial support of FCT and European Social Fund through the grants attributed to C.P. Magalhães (SFRH/BD/132845/2017) and A.L. Arantes (PD/BD/128030/2016).
References
- Alves, J.I. , Alves, M.M. , Plugge, C.M. , Stams, A.J.M. , and Sousa, D.Z. (2016) Comparative analysis of carbon monoxide tolerance among Thermoanaerobacter species. Front Microbiol 7: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk, M.M. , Weijma, J. , and Stams, A.J.M. (2002) Thermotoga lettingae sp. nov., a novel thermophilic, methanol‐degrading bacterium isolated from a thermophilic anaerobic reactor. Int J Syst Evol Microbiol 52: 1361–1368. [DOI] [PubMed] [Google Scholar]
- Beauchamp, J. , Gross, P.G. , and Vieille, C. (2014) Characterization of Thermotoga maritima glycerol dehydrogenase for the enzymatic production of dihydroxyacetone. Appl Microbiol Biotechnol 98: 7039–7050. [DOI] [PubMed] [Google Scholar]
- Ben‐Bassat, A. , Lamed, R. , and Zeikus, J.G. (1981) Ethanol production by thermophilic bacteria: metabolic control of end product formation in Thermoanaerobium brockii . J Bacteriol 146: 192–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calusinska, M. , Happe, T. , Joris, B. , and Wilmotte, A. (2010) The surprising diversity of clostridial hydrogenases: a comparative genomic perspective. Microbiology 156: 1575–1588. [DOI] [PubMed] [Google Scholar]
- Caporaso, J.G. , Lauber, C.L. , Walters, W.A. , Berg‐Lyons, D. , Lozupone, C.A. , Turnbaugh, P.J. , et al (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108: 4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, M. , Matos, M. , Roca, C. , and Reis, M.A.M. (2014) Succinic acid production from glycerol by Actinobacillus succinogenes using dimethylsulfoxide as electron acceptor. N Biotechnol 31: 133–139. [DOI] [PubMed] [Google Scholar]
- Ciriminna, R. , Pina, C.D. , Rossi, M. , and Pagliaro, M. (2014) Understanding the glycerol market. Eur J Lipid Sci Technol 116: 1432–1439. [Google Scholar]
- Clomburg, J.M. , and Gonzalez, R. (2013) Anaerobic fermentation of glycerol: a platform for renewable fuels and chemicals. Trends Biotechnol 31: 20–28. [DOI] [PubMed] [Google Scholar]
- Cook, G.M. , Rainey, F.A. , Patel, B.K.C. , and Morgan, H.W. (1996) Characterization of a new obligately anaerobic thermophile, Thermoanaerobacter wiegelii sp. nov. Int J Syst Bacteriol 46: 123–127. [DOI] [PubMed] [Google Scholar]
- Dharmadi, Y. , Murarka, A. , and Gonzalez, R. (2006) Anaerobic fermentation of glycerol by Escherichia coli: a new platform for metabolic engineering. Biotechnol Bioeng 9: 821–829. [DOI] [PubMed] [Google Scholar]
- DiMarco, A.A. , Bobik, T.A. , and Wolf, R.S. (1990) Unusual coenzymes of methanogenesis. Annu Rev Biochem 59: 355–394. [DOI] [PubMed] [Google Scholar]
- Emde, R. , and Schink, B. (1990) Oxidation of glycerol, lactate, and propionate by Propionibacterium freudenreichii in a poised‐potential amperometric culture system. Arch Microbiol 02: 506–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emde, R. , Swain, A. , and Schink, B. (1989) Anaerobic oxidation of glycerol by Escherichia coli in an amperometric poised‐potential culture system. Appl Microbiol Biotechnol 32: 170–175. [Google Scholar]
- Fardeau, M.L. , Patel, B.K. , Magot, M. , and Ollivier, B. (1997) Utilization of serine, leucine, isoleucine, and valine by Thermoanaerobacter brockii in the presence of thiosulfate or Methanobacterium sp. as electron acceptors. Anaerobe 3: 405–10. [DOI] [PubMed] [Google Scholar]
- Fardeau, M.L. , Magot, M. , Patel, B.K.C. , Thomas, P. , Garcia, J.L. , and Ollivier, B. (2000) Thermoanaerobacter subterraneus sp. nov., a novel thermophile isolated from oilfield water. Int J Syst Evol Microbiol 50: 2141–2149. [DOI] [PubMed] [Google Scholar]
- Gonzalez, R. , Murarka, A. , Dharmadi, Y. , and Yazdani, S.S. (2008) A new model for the anaerobic fermentation of glycerol in enteric bacteria: trunk and auxiliary pathways in Escherichia coli . Metab Eng 10: 234–245. [DOI] [PubMed] [Google Scholar]
- Hengeveld, E.J. , van Gemert, W.J.T. , Bekkering, J. , and Broekhuis, A.A. (2014) When does decentralized production of biogas and centralized upgrading and injection into the natural gas grid make sense? Biomass Bioenerg 67: 363–371. [Google Scholar]
- Ho, D.P. , Jensen, P.D. , and Batstone, D.J. (2013) Methanosarcinaceae and acetate‐oxidizing pathways dominate in high‐rate thermophilic anaerobic digestion of waste‐activated sludge. Appl Environ Microbiol 79: 6491–6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, N.Y. , Kim, S.N. , and Kim, O.B. (2017) Long‐term adaptation of Escherichia coli to methanogenic co‐culture enhanced succinate production from crude glycerol. J Ind Microbiol Biotechnol 45: 71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesárová, N. , Hutan, M. , Bodík, I. , and Špalková, V. (2011) Utilization of biodiesel by‐products for biogas production. J Biomed Biotechnol 2011: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maru, B.T. , Bielen, A.A.M. , Constantí, M. , Medina, F. , and Kengen, S.W.M. (2013) Glycerol fermentation to hydrogen by Thermotoga maritima: proposed pathway and bioenergetic considerations. Int J Hydrogen Energy 38: 5563–5572. [Google Scholar]
- Menes, R.J. , and Muxí, L. (2002) Anaerobaculum mobile sp. nov., a novel anaerobic, moderately thermophilic, peptide‐fermenting bacterium that uses crotonate as an electron acceptor, and emended description of the genus Anaerobaculum . Int J Syst Evol Microbiol 52: 157–164. [DOI] [PubMed] [Google Scholar]
- Murarka, A. , Dharmadi, Y. , Yazdani, S.S. , and Gonzalez, R. (2008) Fermentative utilization of glycerol by Escherichia coli and its implications for the production of fuels and chemicals. Appl Environ Microbiol 74: 1124–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarrete, C. , Nielsen, J. , and Siewers, V. (2014) Enhanced ethanol production and reduced glycerol formation in fps1∆ mutants of Saccharomyces cerevisiae engineered for improved redox balancing. AMB Express 4: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OECD, FAO (2017) “OECD-FAO Agricultural Outlook (Edition 2017)”, OECD Agriculture Statistics (database). 10.1787/d9e81f72-en (accessed on 23 January 2018). [DOI]
- Paulo, A.M.S. , Salvador, A.F. , Alves, J.I. , Castro, R. , Langenhoff, A.A.M. , Stams, A.J.M. , and Cavaleiro, A.J. (2017) Enhancement of methane production from 1‐hexadecene by additional electron donors. Microb Biotechnol 11: 657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees, G.N. , Patel, B.K. , Grassia, G.S. , and Sheehy, A.J. (1997) Anaerobaculum thermoterrenum gen. nov., sp. nov., a novel, thermophilic bacterium which ferments citrate. J Syst Bacteriol 47: 150–154. [DOI] [PubMed] [Google Scholar]
- Richter, K. , and Gescher, J. (2014) Accelerated glycerol fermentation in Escherichia coli using methanogenic formate consumption. Bioresour Technol 162: 389–391. [DOI] [PubMed] [Google Scholar]
- Schindler, B.D. , Joshi, R.V. , and Vieille, C. (2014) Respiratory glycerol metabolism of Actinobacillus succinogenes 130Z for succinate production. J Ind Microbiol Biotechnol 41: 1339–1352. [DOI] [PubMed] [Google Scholar]
- Scholten, E. , Renz, T. , and Thomas, J. (2009) Continuous cultivation approach for fermentative succinic acid production from crude glycerol by Basfia succiniciproducens DD1. Biotechnol Lett 31: 1947–1951. [DOI] [PubMed] [Google Scholar]
- Slobodkin, A. , Reysenbach, A.L. , Mayer, F. , and Wiegel, J. (1997) Isolation and characterization of the homoacetogenic thermophilic bacterium Moorella glycerini sp. nov. Int J Syst Bacteriol 47: 969–974. [DOI] [PubMed] [Google Scholar]
- Slobodkin, A.I. , Tourova, T.P. , Kuznetsov, B.B. , Kostrikina, N.A. , and Chernyh, N.A. (1999) Thermoanaerobacter siderophilus sp. nov., a novel dissimilatory Fe(III)‐reducing, anaerobic thermophilic bacterium. Int J Syst Bacteriol 49: 1471–1478. [DOI] [PubMed] [Google Scholar]
- Sousa, D.Z. , Smidt, H. , Alves, M.M. , and Stams, A.J.M. (2009) Ecophysiology of syntrophic communities that degrade saturated and unsaturated long‐chain fatty acids. FEMS Microbiol Ecol 68: 257–272. [DOI] [PubMed] [Google Scholar]
- Stams, A.J.M. , and Plugge, C.M. (2009) Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat Rev Microbiol 7: 568–577. [DOI] [PubMed] [Google Scholar]
- Stams, A.J.M. , Van Dijk, J.B. , Dijkema, C. , and Plugge, C.M. (1993) Growth of syntrophic propionate‐oxidizing bacteria with fumarate in the absence of methanogenic bacteria. Appl Environ Microbiol 59: 1114–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer, R.K. , Jungermann, K. , and Decker, K. (1977) Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev 41: 100–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardar‐Schara, G. , Maeda, T. , and Wood, T.K. (2006) Metabolically engineered bacteria for producing hydrogen via fermentation. Microb Biotechnol 1: 107–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana, M.B. , Freitas, A.V. , Leitão, R.C. , Pinto, G.A.S. , and Santaella, S.T. (2012) Anaerobic digestion of crude glycerol: a review. Environ Technol Rev 1: 81–92. [Google Scholar]
- Vipotnik, Z. , Jessen, J.E. , Scully, S.M. , and Orlygsson, J. (2016) Effect of culture conditions on hydrogen production by Thermoanaerobacter strain AK68. Int J Hydrogen Energy 41: 181–189. [Google Scholar]
- Yang, Y. , Tsukahara, K. , and Sawayama, S. (2008) Biodegradation and methane production from glycerol‐containing synthetic wastes with fixed‐bed bioreactor under mesophilic and thermophilic anaerobic conditions. Process Biochem 43: 362–367. [Google Scholar]
- Yang, F. , Hanna, M.A. , and Sun, R. (2012) Value‐added uses for crude glycerol‐a byproduct of biodiesel production. Biotechnol Biofuels 5: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F. , Zhang, Y. , Chen, Y. , Dai, K. , van Loosdrecht, M.C.M. , and Zeng, R.J. (2015) Simultaneous production of acetate and methane from glycerol by selective enrichment of hydrogenotrophic methanogens in extreme‐thermophilic (70°C) mixed culture fermentation. Appl Energy 148: 326–333. [Google Scholar]
- Zwietering, M.H. , Jongenburger, I. , Rombouts, F.M. , and Van’t Riet, K., (1990) Modeling of the bacterial growth curve. Appl Environ Microbiol 56: 1875–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 . Experimental procedure applied for the enrichment of thermophilic glycerol‐degrading microbial cultures at 55ºC.
Figure S2 . Phase contrast micrograph of culture Col‐Gly.
Figure S3 . Methane (A) and organic acids (B) production by enriched culture Col‐Gly at 55°C.
Figure S4 . Glycerol consumption (A) and production of acetate (B) and H2 (C) by T. brockii subsp. finnii type strain, when incubated with BrES (w/BrES) and without BrES (w/o BrES).
Table S1 . Methane production from glycerol by the different generations (coded Gly(x), where × represents the number of transfers), during the enrichment process.
Table S2 . Additional information about the enzymes involved in the metabolic pathway for glycerol conversion of Thermoanaerobacter brockii subsp. finnii (DSM 3389T) and Thermoanaerobacter wiegelii (DSM 10319T). The data were retrieved from NCBI genomic platform.


