Figure 4. Two types of NADs in F121-9 mESC.
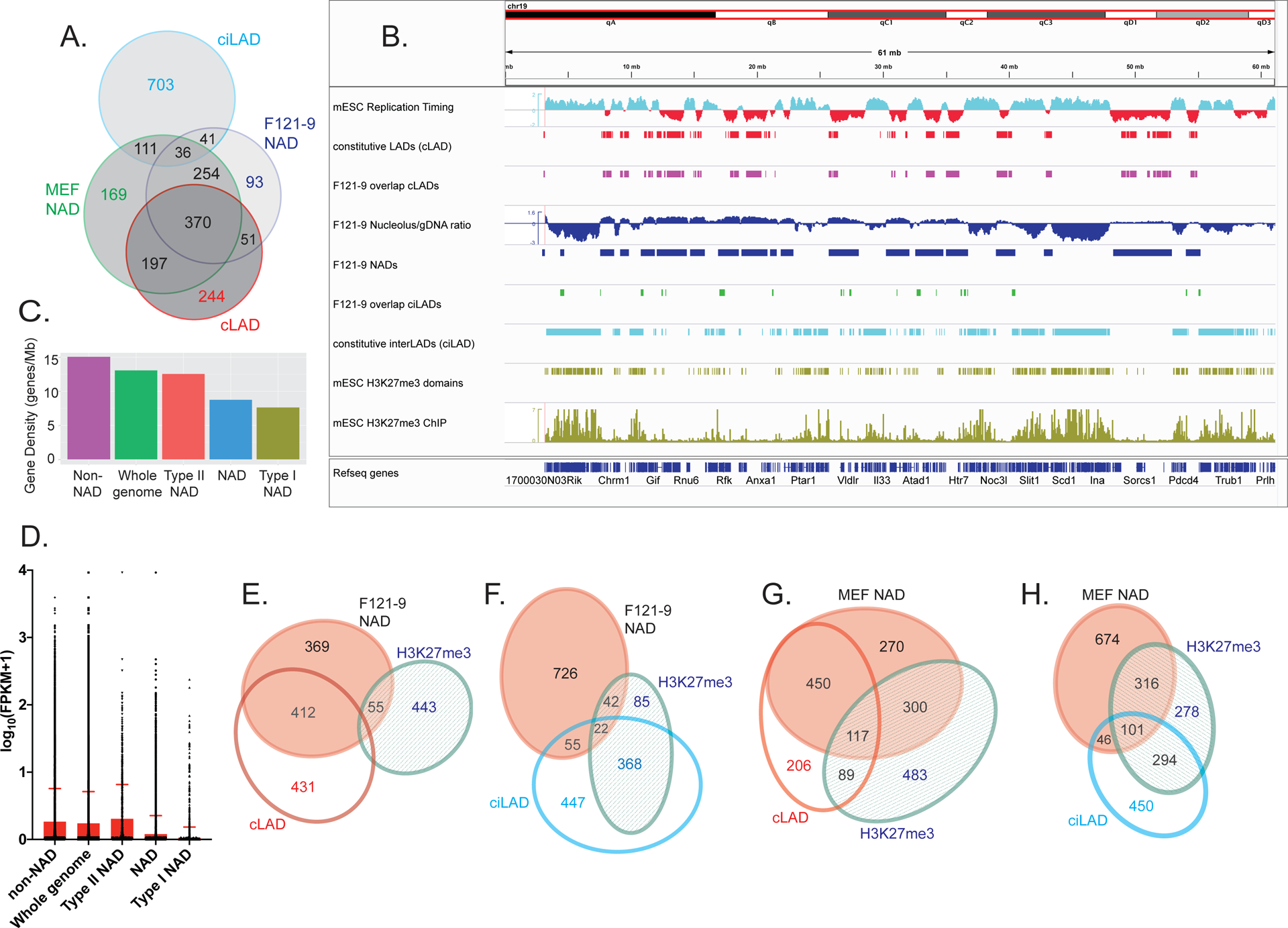
A. Venn diagram illustrating the overlaps among F121-9 NADs, MEF NADs (Vertii et al. 2019), cLAD, and ciLAD regions (Peric-Hupkes et al. 2010). Numbers show the size of the indicated regions in Mb.
B. Chromosomal view of F121-9 NADs overlapping cLADs and ciLADs. The entire chromosome 19 is shown. Euchromatic features (early replication timing, ciLAD) are displayed in cyan, and heterochromatic features (late replication timing, cLAD) are shown in red. From the top, displayed tracks are mESC replication timing (Hiratani et al. 2010), cLAD (Peric-Hupkes et al. 2010), NAD overlap with cLAD (i.e. Type I NADs, magenta), nucleolar/genomic ratio and NAD peaks (blue), NAD overlap with ciLAD (i.e. Type II NADs, green), ciLAD (Peric-Hupkes et al. 2010), H3K27me3 domains, and mESC H3K27me3 ChIP-seq data (Cruz-Molina et al. 2017) used for H3K27me3 domain identification (olive green).
C. Gene densities (genes/Mb) of the indicated regions, ranked left to right. “NAD” indicates all F121-9 NADs.
D. A box plot of gene expression levels from F121-9 RNA-seq data, expressed as log10(FPKM+1) for the same indicated genomic regions as in panel C. The top of the red box indicates the mean value for each population, and the standard deviation is marked by the red error bar.
E. Venn diagram illustrating the overlaps among F121-9 NADs, cLADs (Peric-Hupkes et al. 2010) and mESC H3K27me3 domains (Cruz-Molina et al. 2017). Numbers indicate the size of regions in Mb. The overlaps among all three sets (9 Mb) and between the cLAD and H3K27me3 sets (10 Mb) are left off the diagram because of their small sizes. Diagram was generated using eulerAPE 3.0 .
F. As in panel E, except here the overlap analysis includes ciLADs (Peric-Hupkes et al. 2010) instead of cLADs.
G. As in panel E, except here Venn diagram illustrates the overlaps among crosslinked MEF NADs (Vertii et al. 2019), cLADs (Peric-Hupkes et al. 2010) and MEF H3K27me3 domains (Delbarre et al. 2017).
H. As in panel G, except here the overlap analysis includes ciLADs (Peric-Hupkes et al. 2010) instead of cLADs.
