Abstract
RLR‐mediated type I IFN production plays a pivotal role in innate antiviral immune responses, where the signaling adaptor MAVS is a critical determinant. Here, we show that MAVS is a physiological substrate of SIRT5. Moreover, MAVS is succinylated upon viral challenge, and SIRT5 catalyzes desuccinylation of MAVS. Mass spectrometric analysis indicated that Lysine 7 of MAVS is succinylated. SIRT5‐catalyzed desuccinylation of MAVS at Lysine 7 diminishes the formation of MAVS aggregation after viral infection, resulting in the inhibition of MAVS activation and leading to the impairment of type I IFN production and antiviral gene expression. However, the enzyme‐deficient mutant of SIRT5 (SIRT5‐H158Y) loses its suppressive role on MAVS activation. Furthermore, we show that Sirt5‐deficient mice are resistant to viral infection. Our study reveals the critical role of SIRT5 in limiting RLR signaling through desuccinylating MAVS.
Keywords: desuccinylation, innate immunity, MAVS, SIRT5, viral infection
Subject Categories: Immunology; Post-translational Modifications, Proteolysis & Proteomics
Succinylation and desuccinylation of the key antiviral immunity component MAVS regulates its activity and downstream signaling, including type I interferon production and anti‐viral gene expression.
Introduction
Innate immunity, as the first line of defense against infection, senses pathogens, including RNA and DNA viruses, and subsequently triggers immune cells to secrete cytokines through germline‐encoded pattern recognition receptors (PRRs; Chen & Ichinohe, 2015; Tan et al, 2018). During RNA viral infection, cytosolic RNA species are recognized by retinoic acid‐inducible (RIG‐I)‐like receptors (RLRs), including RIG‐I and melanoma differentiation‐associated gene 5 (MDA5; Moore & Ting, 2008; Wu & Chen, 2014; Brubaker et al, 2015; Roers et al, 2016). The activation of RIG‐I and MDA5 leads to the recruitment of the signaling adaptor protein MAVS (also referred to as IPS‐1, VISA, or Cardif) to activate the downstream protein kinase TBK1, resulting in phosphorylation of the transcription factor interferon regulatory factor 3 (IRF3) to drive type I IFN production, a family of cytokines that are essential for host protection against viral infection (Kawai et al, 2005; Meylan et al, 2005; Seth et al, 2005; Xu et al, 2005; Xie et al, 2012; Tan et al, 2018). Increasing attention has been paid to post‐translational modifications (PTMs) of innate sensors and downstream signaling molecules, which influence their activity and function by inducing their covalent linkage to new functional groups (Liu et al, 2015; Liu et al, 2016a; Tan et al, 2018). Kdm6a promotes IL‐6 expression through demethylating H3K27me3 at promoter of IL‐6 (Li et al, 2017). HDAC9 deacetylates TBK1 for the activation of antiviral innate immunity (Li et al, 2016). HDAC6 modulates viral RNA sensing by deacetylating RIG‐I (Choi et al, 2016). Deamidation of cGAS by HSV UL37 protein enhances viral replication (Zhang et al, 2018). In addition, viral homologs of phosphoribosylformylglycinamidine synthase (PFAS) recruit cellular PFAS to deamidate and activate RIG‐I (He et al, 2015). As a critical adaptor protein in RLR signaling, the normal function of MAVS has been shown to be markedly influenced by its ubiquitination (You et al, 2009; Liu et al, 2017; He et al, 2019), phosphorylation (Liu et al, 2015), and O‐GlcNAcylation (Li et al, 2018a). Recently, more other modifications of MAVS that influence antiviral innate immunity by modulating MAVS function have been identified (Liu & Gao, 2018; Yang et al, 2019).
Accumulating evidence suggests that intermediates and derivatives of the Krebs cycle and glucose metabolism possess non‐metabolic signaling functions, such as regulation of cellular immunity, in addition to its traditional functions associated with bioenergetics or biosynthesis (Li et al, 2018a; Mills et al, 2018b; Williams & O'Neill, 2018; Ryan et al, 2019; Zhang et al, 2019). Krebs cycle‐derived metabolites have been shown to attribute both signaling functions and impact on multiple processes critical for the cellular immune response. These metabolites can either act as immune signaling molecules to inhibit specific enzymes or drive modification of proteins in the immune signaling pathway (Rubic et al, 2008). As the only metabolite directly linking the Krebs cycle and the mitochondrial respiratory chain, succinate acts as a signaling molecule to exert its inflammatory effects through several pathways (Rubic et al, 2008; Tannahill et al, 2013; Littlewood‐Evans et al, 2016; Lei et al, 2018; Peruzzotti‐Jametti et al, 2018). Recently, the critical role of succinate in autoimmune and autoinflammatory has been recognized (Tannahill et al, 2013, 2013; Littlewood‐Evans et al, 2016; Mills et al, 2016, 2018a; Li et al, 2018b). Dysregulation of succinate metabolism can cause accumulation of succinyl‐CoA, which may induce lysine succinylation (Peyssonnaux et al, 2007; Zhang et al, 2011; Xie et al, 2012; Park et al, 2013; Weinert et al, 2013). The enzyme responsible for succinylation has yet to be identified, and indeed, it is possible that this reaction occurs non‐enzymatically by direct reaction between succinyl‐CoA and the modified protein (Feng et al, 2017). It is evident that succinylation can modulate macrophage function. Succinylation of Lys311 of pyruvate kinase M2 (PKM2), a key glycolytic enzyme required for the shift to glycolysis in activated macrophages, has been shown to limit its activity by promoting its tetramer‐to‐dimer transition (Wang et al, 2017a). However, whether succinylation of innate immune sensors and downstream signaling molecules exists or succinylation influences innate immunity in response to viral infection is still largely unknown.
Sirtuin 5 (SIRT5) belongs to the sirtuin family of NAD+‐dependent deacetylases (Finkel et al, 2009). The deacetylase activity of SIRT5 is barely detected (Anderson et al, 2014). Of note, SIRT5 can promote acetylation of p65 through blocking the deacetylation of p65 catalyzed by SIRT2 in a deacetylase activity‐independent manner (Qin et al, 2017). In fact, SIRT5 has been shown to have potent desuccinylase activity (Finkel et al, 2009; Du et al, 2011; Park et al, 2013; Rardin et al, 2013; Tannahill et al, 2013; Wang et al, 2017a). Interestingly, LPS decreases SIRT5 expression in macrophages and increases protein succinylation (Tannahill et al, 2013). Consistent with the activation of PKM2 by SIRT5‐catalyzed desuccinylation, SIRT5‐deficient mice exhibit hypersuccinylation and increased IL‐1β production (Wang et al, 2017a). However, it has also been reported that Sirt5 deficiency does not compromise innate immune response to bacterial infections (Heinonen et al, 2018).
Due to lack of convincing data to support the enzymes responsible for succinylation existed (Weinert et al, 2013; Feng et al, 2017; Yang & Gibson, 2019), in order to address whether succinylation influences innate immunity in response to viral infection, we examined the impact of SIRT5, a well‐defined desuccinylase (Du et al, 2011; Park et al, 2013; Rardin et al, 2013; Anderson et al, 2014; Wagner & Hirschey, 2014; Wang et al, 2017a), on antiviral innate immunity. We found that SIRT5 negatively regulates the innate immunity in response to RNA viral infection. Further investigation shows that SIRT5 mediates desuccinylation of Lys7 of MAVS, leading to impairment of aggregation and activation of MAVS. These findings suggest a critical role of SIRT5 in limiting RLR signaling through desuccinylating MAVS.
Results
SIRT5 suppresses the MAVS‐mediated RLR signaling
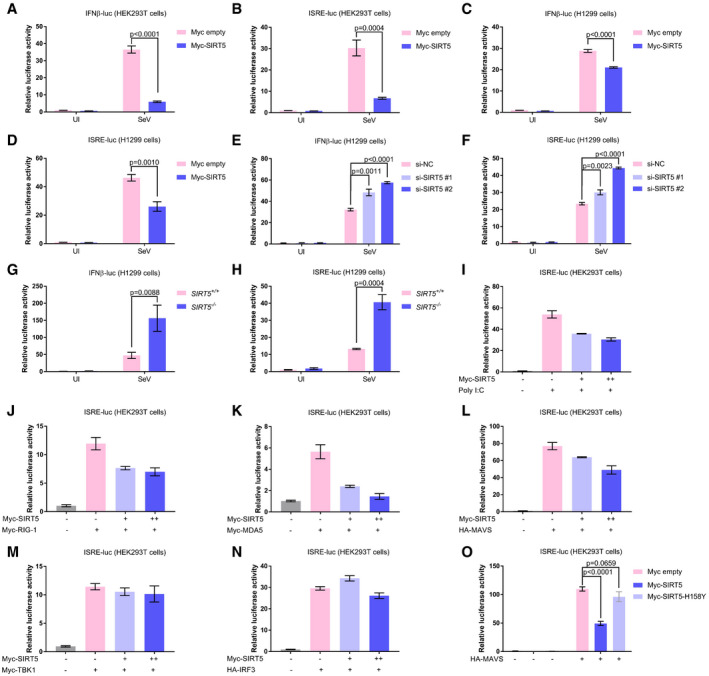
To investigate whether SIRT5 participates in regulating RLR signaling, we employed a promoter assay to examine the effect of SIRT5 on promoter activity. IFNβ promoter luciferase reporter and ISRE‐luciferase reporter (containing interferon stimulated response elements) are well‐defined reporters for monitoring RLR activation (Xu et al, 2005; Zhong et al, 2008; Liu et al, 2016b). Upon overexpression of SIRT5 in HEK293T cells or H1299 cells, Sendai virus (SeV)‐induced IFNβ promoter reporter (IFNβ‐luc.) activity and ISRE‐luciferase reporter activity were strongly inhibited (Fig 1A–D). Consistent with these observations, SeV‐induced IFNβ promoter reporter activity and ISRE‐luciferase reporter activity were enhanced upon SIRT5 knockdown by small interfering RNAs (si‐SIRT5#1 and si‐SIRT5#2; Wang et al, 2017a) in H1299 cells (Fig 1E and F). To further confirm the suppressive role of SIRT5 on RLR signaling, we knocked out SIRT5 in H1299 cells via CRISPR/Cas9. Consistently, SeV‐induced IFNβ promoter reporter activity and ISRE‐luciferase reporter activity were enhanced in SIRT5 −/− cells compared to the wild‐type H1299 cells (SIRT5 +/+; Fig 1G and H). In addition, poly(I:C) (RNA virus mimics)‐induced ISRE‐luciferase reporter activity was suppressed by the overexpression of SIRT5 in a dose‐dependent manner (Fig 1I). These findings indicate that SIRT5 inhibits RLR signaling.
Figure 1. SIRT5 suppresses the MAVS‐mediated type I IFN signaling.
-
A, BIFNβ promoter activity (A) and ISRE reporter activity (B) in Myc empty vector (200 ng) or Myc‐SIRT5 (200 ng)‐transfected HEK293T cells with or without SeV infection (SeV or UI) for 18–24 h.
-
C, DIFNβ promoter activity (C) and ISRE reporter activity (D) in Myc empty vector (200 ng) or Myc‐SIRT5 (200 ng)‐transfected H1299 cells with or without SeV infection (SeV or UI) for 18–24 h.
-
E, FIFNβ promoter activity (E) and ISRE reporter activity (F) in the indicated siRNA‐transfected H1299 cells (si‐NC, si‐SIRT5#1, and si‐SIRT5#2) with or without SeV infection (SeV or UI) for 18–24 h. NC, negative control.
-
G, HIFNβ promoter activity (G) and ISRE reporter activity (H) in SIRT5‐deficient H1299 cells (SIRT5−/−) or the wild‐type (WT) H1299 cells (SIRT5 +/+) with or without SeV infection (SeV or UI) for 18–24 h.
-
IISRE reporter activity in Myc‐SIRT5 (0, 100, or 200 ng)‐transfected HEK293T cells with or without poly(I:C) transfection for 18–24 h.
-
JISRE reporter activity in co‐transfection of Myc‐RIG‐I (200 ng) together with Myc‐SIRT5 (0, 100, or 200 ng)‐transfected HEK293T cells for 24 h.
-
KISRE reporter activity in co‐transfection of Myc‐MDA5 (200 ng) together with Myc‐SIRT5 (0, 100, or 200 ng) in HEK293T cells for 24 h.
-
LISRE reporter activity in co‐transfection of HA‐MAVS (200 ng) together with Myc‐SIRT5 (0, 100, or 200 ng) in HEK293T cells for 24 h.
-
MISRE reporter activity in co‐transfection of Myc‐TBK1 (200 ng) together with Myc‐SIRT5 (0, 100, or 200 ng) in HEK293T cells for 24 h.
-
NISRE reporter activity in co‐transfection of HA‐IRF3 (200 ng) together with Myc‐SIRT5 (0, 100, or 200 ng) in HEK293T cells for 24 h.
-
OISRE reporter activity in co‐transfection of HA‐MAVS (200 ng) together with Myc‐SIRT5 (200 ng) or Myc‐SIRT5‐H158Y (200 ng), respectively, in HEK293T cells for 24 h.
Subsequently, we sought to determine which target in RLR signaling mediates SIRT5's suppressive function. The ISRE‐luciferase reporter assay revealed that overexpression of SIRT5 caused a reduction of luciferase activity, which was induced by RIG‐I, MDA5, or MAVS in a dose‐dependent manner (Fig 1J–L). However, overexpression of SIRT5 showed no effect on luciferase activity driven by TBK1, or IRF3, suggesting that SIRT5 functions at the MAVS level (Fig 1M and N). Of note, overexpression of enzyme‐deficient mutant of SIRT5 (SIRT5‐H158Y; Nakagawa et al, 2009; Zhang et al, 2017) had no effect on luciferase activity driven by MAVS, in contrast to overexpression of WT SIRT5 (Fig 1O). Expressions of the transfected plasmids, the efficacy of SIRT5‐knocked down by siRNA and SIRT5‐knocked out by CRISPR/Cas9 were confirmed by Western blot analysis (Appendix Fig S1A–O).
To further validate these results, we performed dose‐titration assays. Overexpression of SIRT5 suppressed dose‐dependent activation of IFNβ promoter activity by SeV infection in HEK293T cells (Appendix Fig S2A). Overexpression of SIRT5 also suppressed dose‐dependent activation of ISRE‐luciferase reporter activity by transfection of increasing amount of MDA5 in HEK293T cells (Appendix Fig S2B). However, overexpression of SIRT5 had no effect on dose‐dependent activation of ISRE reporter activity by transfection of increasing amount of IRF3 in HEK293T cells (Appendix Fig S2C). Protein expressions of transfected plasmids were confirmed by Western blot analysis (Appendix Fig S2D–F). Of note, overexpression of SIRT5 did not influence the ISRE‐luciferase reporter activity induced by co‐transfection of cGAS and STING, suggesting that SIRT5 might have no effect on the cytosolic DNA sensing pathway (Tan et al, 2018; Appendix Fig S2G). In addition, overexpression of SIRT2 did not affect ISRE‐luciferase reporter activity and IFNβ promoter activity induced by transfection of MAVS, indicating that SIRT5 might be the specific one in the sirtuin family to inhibit MAVS function (Appendix Fig S2H and I; Finkel et al, 2009). Protein expressions of transfected plasmids were confirmed by Western blot analysis (Appendix Fig S2J–L).
Taken together, these results suggest that SIRT5 regulates RLR signaling by influencing MAVS function.
SIRT5 interacts with MAVS
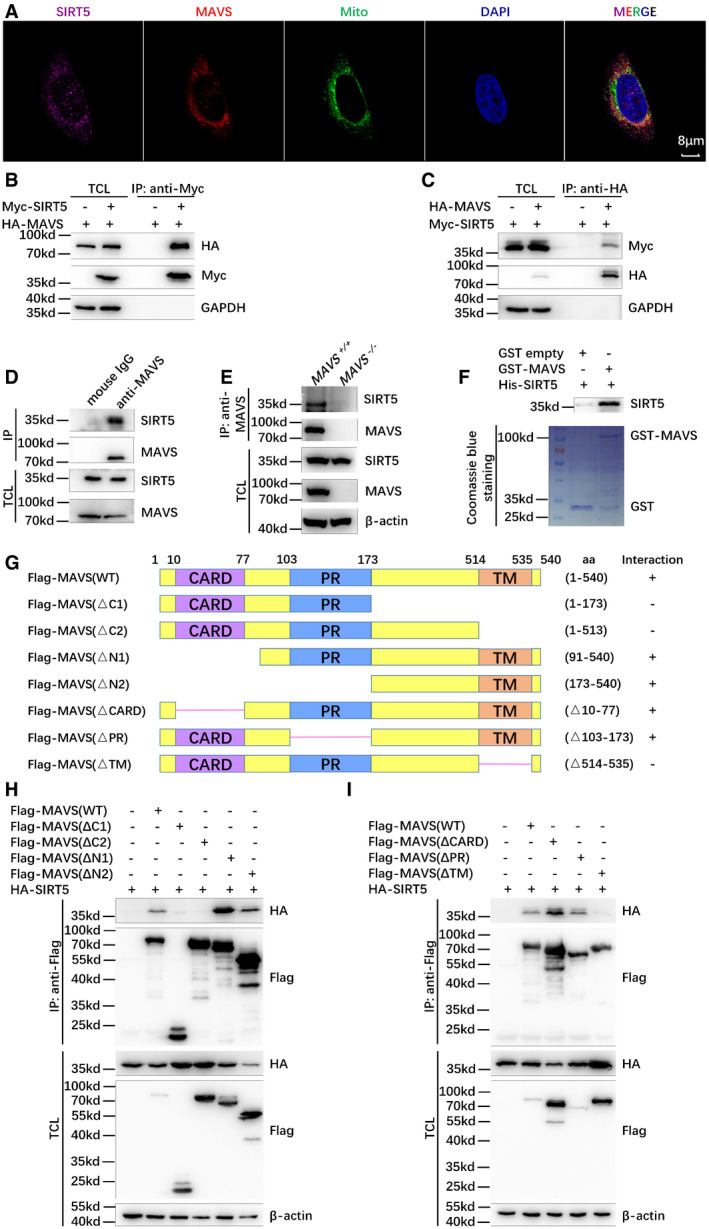
The observations that SIRT5 suppresses the activation of MAVS on RLR signaling prompted us to determine whether SIRT5 influences MAVS's function through protein–protein interaction. First, we examined their co‐localization by immunofluorescence staining using anti‐MAVS and anti‐SIRT5 antibodies. Fluorescence confocal microscopy showed that SIRT5 co‐localized with MAVS in the mitochondria in both H1299 cells and HeLa cells (Fig 2A, Appendix Figs S3A and B, and S4A–D; Anderson et al, 2014; Liu et al, 2017). Co‐immunoprecipitation assays showed that ectopically expressed SIRT5 pulled down ectopically expressed MAVS in HEK293T cells and vice versa (Fig 2B and C). In H1299 cells, endogenous MAVS was also co‐immunoprecipitated with endogenous SIRT5 (Fig 2D), while endogenous co‐immunoprecipitation between MAVS and SIRT5 was not detected in SIRT5‐deficient H1299 cells (SIRT5 −/−; Fig 2E). Escherichia coli‐expressed GST‐tagged MAVS interacted with E. coli‐expressed His‐tagged SIRT5 in vitro (Fig 2F). These data indicated that SIRT5 directly associated with MAVS. Further domain mapping of MAVS binding to SIRT5 indicated that the TM domain of MAVS was required for SIRT5 interaction (Fig 2G–I). However, SIRT5 did not influence protein stability of MAVS revealed by either overexpression of SIRT5 in HEK293T cells or knockout of SIRT5 in H1299 cells (Appendix Fig S5A and B).
Figure 2. SIRT5 interacts with MAVS.
-
AConfocal microscopy image of endogenous SIRT5 co‐localized with endogenous MAVS in H1299 cells detected by immunofluorescence staining using anti‐SIRT5 and anti‐MAVS antibodies. Mito, MitoTracker; Scale bar = 8 μm.
-
B, CCo‐immunoprecipitation of Myc‐SIRT5 with HA‐MAVS and vice versa. HEK293T cells were co‐transfected with indicated plasmids for 24 h. Anti‐Myc (B) or anti‐HA antibody‐conjugated agarose beads (C) were used for immunoprecipitation, and the interaction was detected by immunoblotting with the indicated antibodies.
-
DEndogenous interaction between MAVS and SIRT5. Anti‐MAVS antibody was used for immunoprecipitation, and normal mouse IgG was used as a control.
-
EEndogenous interaction between MAVS and SIRT5 in the wild‐type (WT) (MAVS +/+) or SIRT5‐deficient (MAVS −/−) H1299 cells. Anti‐MAVS antibody was used for immunoprecipitation, and the interaction was detected by immunoblotting with anti‐SIRT5 antibody.
-
FGST pull‐down assay for GST‐tagged MAVS and His‐tagged SIRT5. GST‐tagged MAVS and His‐tagged SIRT5 were expressed in Escherichia coli (BL21), respectively. The association of GST‐MAVS with His‐SIRT5 was detected by immunoblotting with anti‐SIRT5 antibody. GST and GST‐MAVS proteins were stained with Coomassie blue.
-
GSchematic of MAVS domains interacted with SIRT5. The interaction is indicated by plus (+) sign.
-
H, ICo‐immunoprecipitation analysis of HA‐SIRT5 with Flag‐MAVS‐truncated mutants. HEK293T cells were co‐transfected with the indicated plasmids. Anti‐Flag antibody‐conjugated agarose beads were used for immunoprecipitation, and the interaction was analyzed by immunoblotting with the indicated antibodies. Flag‐MAVS fragments (WT: full length; ΔC1, 1–173 aa; ΔC2, 1–513 aa; ΔN1, 91–540 aa; ΔN2, 173–540 aa; ΔCARD, Δ10–77 aa; ΔPR, Δ103–173 aa; ΔTM, Δ514–535 aa).
SIRT5 desuccinylates Lysine 7 of MAVS
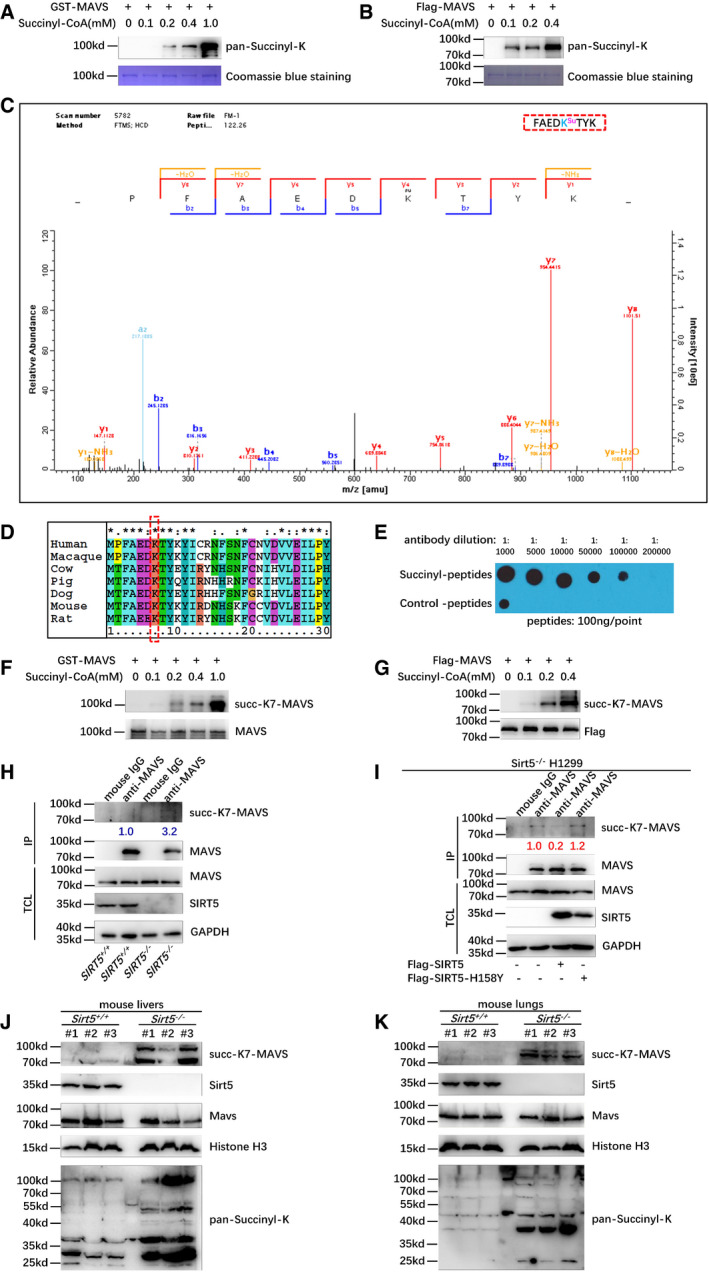
Given a well‐defined function of SIRT5 in desuccinylation, we sought to determine whether SIRT5 could influence MAVS succinylation (Du et al, 2011; Park et al, 2013; Rardin et al, 2013; Anderson et al, 2014; Wagner & Hirschey, 2014; Wang et al, 2017a). Initially, we examined whether succinylation of MAVS existed. Using in vitro succinylation assays, we found that MAVS expressed in either E. coli or HEK293T cells could be readily succinylated by adding succinyl‐CoA in a dose‐dependent manner (Fig 3A and B), suggesting that MAVS could be succinylated.
Figure 3. MAVS is succinylated at Lysine 7, and SIRT5 mediates its desuccinylation.
-
A, BMAVS could be succinylated in vitro. GST‐MAVS (A) extracted from Escherichia coli or Flag‐tagged MAVS (B) protein purified from HEK293T cells was incubated with the indicated concentrations of succinyl‐CoA. Protein succinylation was detected with anti‐pan‐succinyl‐lysine antibody; GST‐MAVS or Flag‐MAVS was stained by Coomassie blue.
-
CThe succinylated residue in MAVS was identified by mass spectrometry analysis.
-
DSequence alignment of partial MAVS (1–30 amino acids) from human, macaque, cow, pig, dog, mouse, and rat. The red box indicates a conserved lysine (K7).
-
EDot blot assay for the specificity of anti‐succ‐K7‐MAVS antibody. Equal amounts of succinyl‐peptides or the control peptides were immunoblotted with the indicated dilutions of anti‐succ‐K7‐MAVS antibody.
-
F, GGST‐MAVS (F) extracted from E. coli or Flag‐MAVS (G) purified from HEK293T cells was incubated with the indicated concentrations of succinyl‐CoA. The succinylation of MAVS at Lysine 7 was detected by anti‐succ‐K7‐MAVS antibody.
-
HDisruption of SIRT5 in H1299 cells enhanced succinylation of MAVS at Lys 7 compared to that in the SIRT5‐intact H1299 cells (SIRT5 +/+) (3.2 vs. 1.0). The cell lysates from SIRT5‐deficient H1299 cells or the SIRT5‐intact H1299 cells were immunoprecipitated with anti‐MAVS antibody or mouse IgG control, followed by immunoblotting with anti‐succ‐K7‐MAVS antibody.
-
IReconstitution of WT SIRT5 in SIRT5‐deficient H1299 cells (SIRT5 −/−) caused a significant reduction in succinylation of MAVS at Lys 7 (1.0 vs. 0.2); but overexpression of SIRT5‐H158Y in SIRT5 −/− H1299 cells has no effect on the reduction in succinylation of MAVS at Lys 7 (1.0 vs. 1.2). The SIRT5 −/− H1299 cells were transfected with Flag‐SIRT5 or Flag‐SIRT5‐H158Y, followed by immunoprecipitating with anti‐MAVS antibody or mouse IgG control, and immunoblotting with anti‐succ‐K7‐MAVS antibody.
-
J, KKnockout of Sirt5 increased MAVS succinylation in mouse livers (J) and lungs (K). Proteins extracted from livers (J) and lungs (K) of Sirt5 KO and the wild‐type littermates (n = 3 per group) were detected by the indicated antibodies. MAVS succinylation was determined by anti‐succ‐K7‐MAVS antibody, and anti‐pan‐succinyl‐lysine antibody was used as positive controls.
Subsequently, we examined the succinylation site (s) on MAVS through mass spectrometry (MS) analysis. One succinylation site (Lysine 7) in MAVS was identified (Fig 3C). Lysine 7 of MAVS is evolutionary conserved across species (Fig 3D). To further confirm this succinylated site in MAVS, we developed a specific antibody against Lysine 7 of MAVS (anti‐succ‐K7‐MAVS antibody). The specificity of this antibody was validated by dot blot assay (Fig 3E). The addition of succinyl‐CoA caused succinylation of MAVS at Lysine 7 in a dose‐dependent manner revealed by the anti‐succ‐K7‐MAVS antibody (Fig 3F and G). In SIRT5‐deficient H1299 cells (SIRT5 −/−), succinylation of MAVS at Lysine 7 was significantly higher than that of the SIRT5‐intact H1299 cells (SIRT5 +/+; 3.2 vs. 1.0; Fig 3H). Reconstitution of SIRT5 in SIRT5‐deficient H1299 cells (SIRT5 −/−) caused a significant reduction in succinylation of MAVS at Lysine 7 (1.0 vs. 0.2; Fig 3I). However, overexpression of SIRT5‐H158Y had no effect on the reduction in succinylation of MAVS at Lysine 7 (1.0 vs. 1.2; Fig 3I). These findings indicated that SIRT5 desuccinylated Lysine 7 of MAVS. Immunoprecipitation assays showed that MAVS‐K7R still interacted with SIRT5 and SIRT5‐H158Y also interacted with MAVS as well (Appendix Fig S5C and D).
To further determine the effect of Sirt5 on desuccinylation of MAVS in vivo, we obtained Sirt5‐knocked‐out mice (KO) from the Jackson Laboratory (https://www.jax.org/strain/012757). In liver and lung, succinylation of MAVS at Lysine 7 was significantly higher in Sirt5 −/− mice compared to WT (Sir5 +/+) mice detected by anti‐succ‐K7‐MAVS antibody (Fig 3J and K). In addition, in the whole tissue lysate, succinylation levels were higher in Sirt5 −/− mice compared to WT mice detected by a pan‐succinyl‐K antibody (used as a positive control; Fig 3J and K). These findings suggest that Sirt5 desuccinylates MAVS at Lysine 7 in vivo.
SIRT5 negatively regulates cellular innate antiviral immune response
To further investigate the effect of SIRT5 in innate antiviral immunity, we initially examined whether viral infection had impacts on SIRT5 expression, localization, and binding ability to MAVS. VSV infection did not affect SIRT5 protein stability and localization (Fig EV1A and B), but did attenuate SIRT5 binding to MAVS (Fig EV1C). Interestingly, VSV infection diminished cellular succinate level (Fig EV1D). Furthermore, SIRT5 indeed impaired K63‐linked polyubiquitination of MAVS (Fig EV1E–G), implying that SIRT5 may have a suppressive role on MAVS activation (Liu et al, 2017).
Figure EV1. VSV infection does not influence protein level, translocation of SIRT5, but attenuates SIRT5 binding to MAVS; VSV infection diminishes K63‐linked polyubiquitination of MAVS .
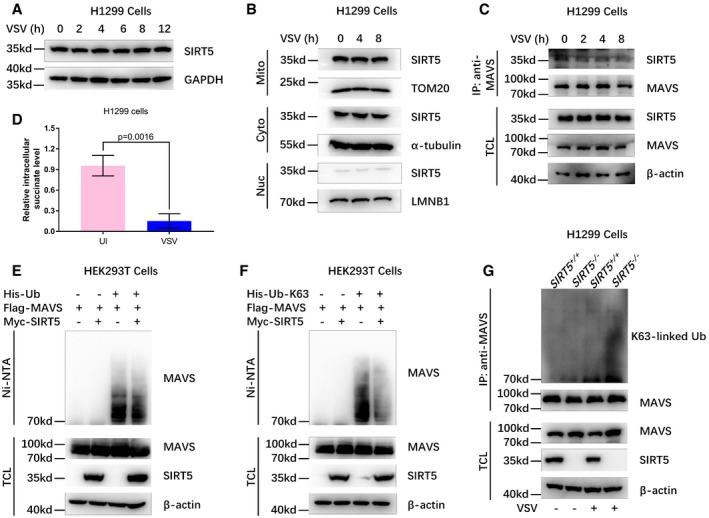
- Western blot analysis of SIRT5 in H1299 cells infected with VSV at different time points (0, 2, 4, 6, 8, 12 h).
- Western blot analysis of SIRT5 in mitochondria, cytoplasm, and nuclei of H1299 cell infected with VSV at different time points (0, 4, 8 h).
- Western blot analysis for SIRT5 binding to MAVS in H1299 cells infected with VSV at different time points (0, 2, 4, 8 h).
- VSV infection caused a reduction of intracellular succinate level.
- Overexpression of SIRT5 attenuated polyubiquitination of MAVS in HEK293T cells.
- Overexpression of SIRT5 attenuated K63‐linked polyubiquitination of MAVS in HEK293T cells.
- Knockout of SIRT5 in H1299 cells enhanced K63‐linked polyubiquitination of endogenous MAVS in response to VSV infection.
Source data are available online for this figure.
Subsequently, we examined expression of antiviral response genes in RLR signaling. Consistent with the promoter assays, overexpression of SIRT5 in HEK293T cells suppressed expression of IFNβ, CXCL10, and IFIT1 induced by SeV challenge (Figs 4A–C and EV2A). On the contrary, knockout of SIRT5 in H1299 cells (SIRT5 −/−) enhanced expression of IFNβ, CXCL10, and IFIT1 induced by either SeV or VSV challenge (Figs 4D–I and EV2B–G). Moreover, overexpression of SIRT5 suppressed IFNβ expression induced by poly(I:C) treatment in HEK293T cells (Fig 4J). However, overexpression of SIRT5‐H158Y, an enzyme‐deficient mutant of SIRT5 (Rardin et al, 2013), showed no obvious effect on suppression of IFNβ expression induced by poly(I:C) treatment (Fig 4J), indicating that the enzymatic activity of SIRT5 was required for the suppressive function of SIRT5 on RLR signaling. Similar results were obtained in SIRT5‐deficient H1299 cells in response to SeV and VSV infection (Figs 4K, and EV2H and I). Furthermore, by SDS–PAGE assays, overexpression of WT SIRT5, but not of SIRT5‐H158Y, attenuated aggregation of MAVS in response to SeV infection, suggesting that SIRT5 might impair the formation of MAVS aggregation in response to viral infection (Fig 4L).
Figure 4. SIRT5 negatively regulates cellular antiviral response.
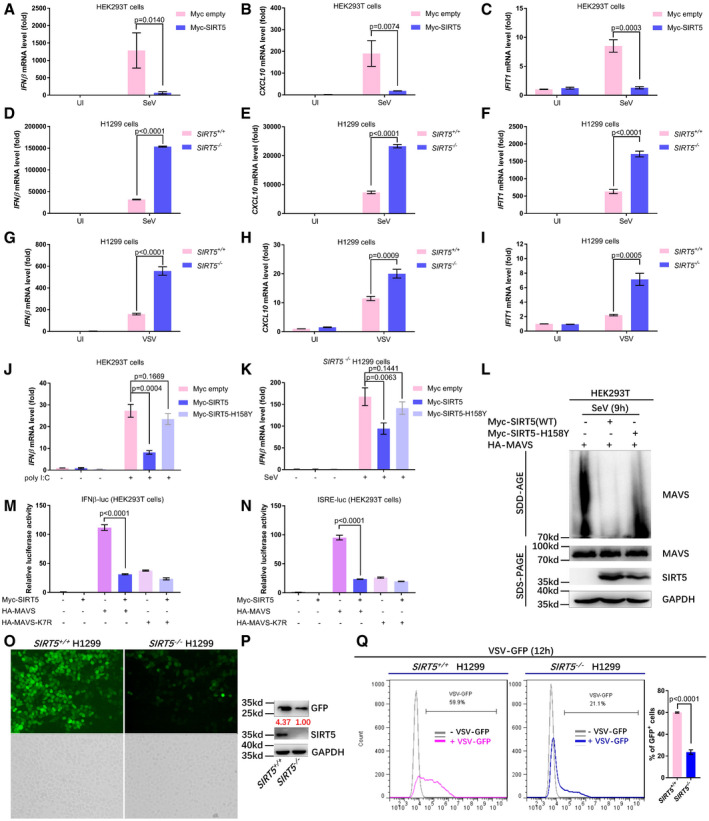
-
A–CQuantitative real‐time PCR analysis (qPCR) of IFNβ (A), CXCL10 (B), and IFIT1 (C) mRNA in HEK293T cells transfected with the control plasmid (Myc empty) or the plasmid expressing Myc‐SIRT5 (Myc‐SIRT5) for 24 h, followed by infection with or without SeV (SeV or UI) for 8 h.
-
D–FqPCR analysis of IFNβ (D), CXCL10 (E), and IFIT1 (F) mRNA in SIRT5‐deficient or WT H1299 cells (SIRT5 −/− or SIRT5 +/+) infected with or without SeV (SeV or UI) for 8 h.
-
G–IqPCR analysis of IFNβ (G), CXCL10 (H), and IFIT1 (I) mRNA in SIRT5‐deficient or WT H1299 cells (SIRT5 −/− or SIRT5 +/+) infected with or without VSV (VSV or UI) for 8 h.
-
JqPCR analysis of IFNβ mRNA in HEK293T cells transfected with the control plasmid (Myc empty), or the plasmid expressing Myc‐SIRT5 (Myc‐SIRT5), or its enzyme‐deficient mutant H158Y (Myc‐SIRT5‐H158Y) for 24 h, followed by stimulation with or without poly(I:C) for 8 h.
-
KqPCR analysis of IFNβ mRNA in SIRT5‐deficient H1299 cells (SIRT5 −/−) transfected with the control plasmid (Myc empty) or the plasmid expressing Myc‐SIRT5 (Myc‐SIRT5) or its enzyme‐deficient mutant H158Y (Myc‐SIRT5‐H158Y) for 24 h, followed by infection with (+) or without (−) SeV for 8 h.
-
LSDD‐AGE analysis of MAVS aggregates in HEK293T cells transfected HA‐MAVS together with WT Myc‐SIRT5 or its enzyme‐deficient mutant (Myc‐SIRT5‐H158Y) after infected with SeV for 12 h. SDS–PAGE immunoblotting was used as a loading control.
-
M, NIFNβ promoter activity (M) and ISRE reporter activity (N) in HEK293T cells co‐transfected Myc‐SIRT5 together with HA‐MAVS or HA‐MAVS‐K7R.
-
O–QSIRT5‐deficient or WT H1299 cells (SIRT5 −/− or SIRT5 +/+) were infected with VSV‐GFP viruses (MOI = 0.1) for 12 h, and viral infectivity was detected by fluorescence microscopy (O), Western blot analysis (P), or flow cytometry analysis (n = 3) (Q).
Figure EV2. SIRT5 negatively regulates cellular antiviral response.
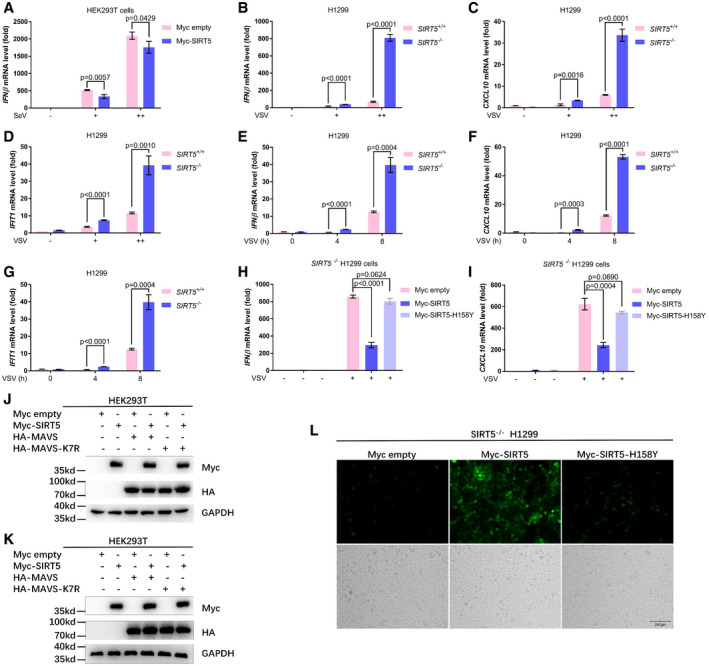
-
AqRT–PCR analysis of IFNβ mRNA in HEK293T cell transfected with Myc empty vector or Myc‐SIRT5 for 24 h, followed by infected with SeV in an increasing titration (+, ++) for 8 h. −, without infection.
-
B–DqRT–PCR analysis of IFNβ (B), CXCL10 (C), and IFIT1 (D) mRNA in SIRT5‐deficient (SIRT5 −/−) or WT (SIRT5 +/+) H1299 cells infected with SeV in an increasing titration (+, ++) for 8 h. −, without infection.
-
E–GqPCR analysis of IFNβ (E), CXCL10 (F), and IFIT1 (G) mRNA in SIRT5‐deficient (SIRT5 −/−) or WT (SIRT5 +/+) H1299 cells infected with SeV for different time points (0, 4, 8 h).
-
H, IqPCR analysis of IFNβ (H) and CXCL10 (I) mRNA in SIRT5‐deficient H1299 cells (SIRT5 −/−) transfected with the control plasmid (Myc empty), the plasmid expressing Myc‐SIRT5 (Myc‐SIRT5), or the plasmid expressing the enzyme‐deficient mutant H158Y (Myc‐SIRT5‐H158Y) for 24 h, followed by infection with (+) or without (−) VSV for 8 h.
-
JWestern blot analysis for Fig 4M.
-
KWestern blot analysis for Fig 4N.
-
LFluorescence microscopy images of VSV‐GFP virus replication in SIRT5‐deficient H1299 cells (SIRT5 −/−) transfected with the control plasmid (Myc empty), the plasmid expressing Myc‐SIRT5 (Myc‐SIRT5), or the plasmid expressing the enzyme‐deficient mutant H158Y (Myc‐SIRT5‐H158Y) for 24 h, followed by infection with VSV‐GFP (MOI = 0.1) for 12 h.
Next, we compared the activity on RLR signaling between the WT MAVS and the succinylated site‐mutated MAVS (MAVS‐K7R). The induction on IFN signaling by MAVS‐K7R significantly decreased compared to WT MAVS in promoter assays, indicating the importance of Lysine 7 in MAVS for MAVS activation (Fig 4M and N). Overexpression of SIRT5 dramatically suppressed MAVS‐induced activity of IFNβ promoter reporter and ISRE‐luciferase reporter (Fig 4M and N). But, the inhibitory effect of SIRT5 on MAVS‐K7R‐induced activity of IFNβ promoter reporter and ISRE‐luciferase reporter was not as dramatic as that on WT MAVS‐induced activity of IFNβ promoter reporter and ISRE‐luciferase reporter, even though it still existed (Fig 4M and N), suggesting that succinylation of Lysine 7 of MAVS was critical for MAVS activation. Protein expressions of transfected plasmids were confirmed by Western blot analysis (Fig EV2J and K).
To determine whether SIRT5 could influence cellular antiviral response, we examined virus replication in the SIRT5‐deficient (SIRT5 −/−) or the SIRT5‐intact H1299 cells (SIRT5 +/+) via infection of VSV‐GFP viruses. Fluorescence microscopy showed that VSV‐GFP virus replication was substantially inhibited in SIRT5‐deficient H1299 cells (Fig 4O), which was further confirmed by Western blot analysis using anti‐GFP antibody (4.37 vs. 1.00; Fig 4P) and flow cytometry analysis (59.9% vs. 21.1%). In the SIRT5‐deficient (SIRT5 −/−) H1299 cells, reconstitution of WT SIRT5 promoted VSV replication, but overexpression of SIRT5‐H158Y did not do so (Fig EV2L). Similar results were obtained in HCT116 cells (Fig EV3A–E).
Figure EV3. SIRT5 deficiency enhances cellular antiviral response in HCT116 cells.
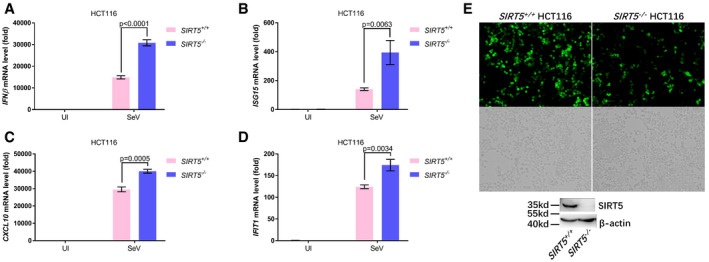
-
A–DqPCR analysis of IFNβ (A), ISG15 (B), CXCL10 (C), and IFIT1 (D) mRNA in SIRT5‐deficient (SIRT5 −/−) or WT (SIRT5 +/+) HCT116 cells infected with or without SeV (Sev or UI) for 8 h. UI, uninfected; the graphs represent the fold induction relative to the untreated control cells. All data are presented as the mean values based on three independent experiments, and error bars indicate s.e.m.
-
EFluorescence microscopy images of VSV‐GFP virus replication in SIRT5‐deficient (SIRT5 −/−) or WT (SIRT5 +/+) HCT116 cells after infected with VSV‐GFP viruses (MOI = 0.1) for 12 h (fluorescence, upper; bright‐field, bottom). The expression of SIRT5 in SIRT5‐deficient (SIRT5 −/−) or WT (SIRT5 +/+) HCT116 cells was examined by Western blot analysis.
Taken together, these results suggest that SIRT5 negatively regulates cellular antiviral immune response.
Disruption of Sirt5 potentiates cellular antiviral immune response in MEF cells
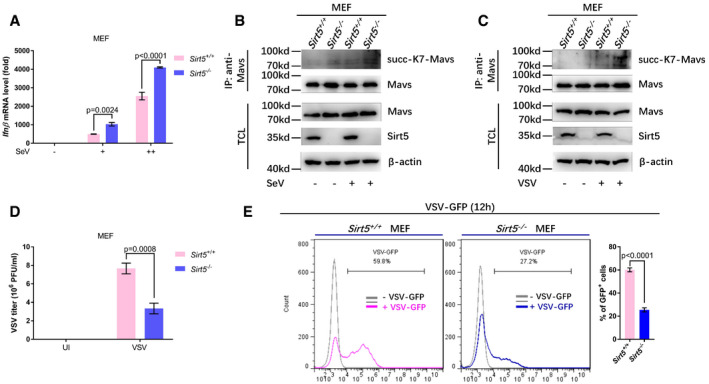
To investigate the effects of Sirt5 on innate immunity in vivo, we made use of Sirt5‐null mice. Initially, we established mouse embryonic fibroblast (MEF) cell lines from WT (Sirt5 +/+) and Sirt5 −/− mice and stimulated them with SeV, VSV, or poly(I:C). Compared to WT (Sirt5 +/+) MEFs, expressions of Ifnβ, Ifna1, ifna4, Ifit1, Cxcl10, and Cxcl11 were significantly up‐regulated in Sirt5 −/− MEF cells upon SeV challenge (Figs 5A–F and EV4A). In agreement with this notion, dimerization and phosphorylation of Irf3 were enhanced in Sirt5 −/− MEF cells compared to Sirt5 +/+ MEF cells upon SeV infection (Fig 5G). Moreover, Sirt5 deficiency enhanced the interaction between MAVS and Rig‐I (Fig 5H) and promoted the formation of MAVS aggregation in response to SeV infection (Fig 5I), which were consistent with the suppressive role of Sirt5 on RLR signaling. As expected, the succinylation levels of MAVS were enhanced in Sirt5 −/− MEF cells compared to Sirt5 +/+ MEF cells upon SeV or VSV infection (Fig EV4B and C). In addition, compared to WT (Sirt5 +/+) MEF cells, expressions of Ifnβ, Ifnα1, Ifnα4, Ifit1, Cxcl10, and Cxcl11 were significantly up‐regulated in Sirt5 −/− MEF cells upon VSV infection (Fig 5J–O).
Figure 5. Sirt5 deficiency potentiates the host innate antiviral immune response.
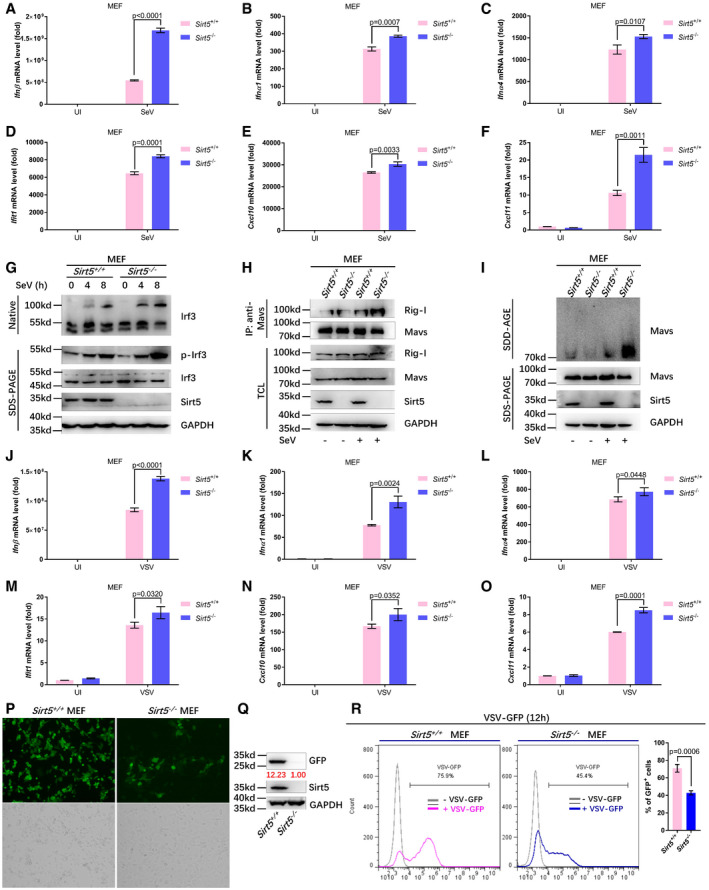
-
A–FqPCR analysis of Ifnβ (A), Ifnα1 (B), Ifnα4 (C), Ifit1 (D), Cxcl10 (E), and Cxcl11 (F) mRNA in WT or Sirt5‐deficient MEF cells (Sirt5 +/+ or Sirt5 −/−) infected with or without SeV (SeV or UI) for 8 h.
-
GWT or Sirt5‐deficient MEF cells (Sirt5 +/+ or Sirt5 −/−) were infected with or without SeV for the indicated times, and the cell lysates were analyzed by immunoblotting for monomeric (Monomer) and dimeric (Dimer) Irf3 (top; native PAGE), phosphorylated Irf3 (p‐Irf3), total Irf3, Sirt5, and GAPDH (bottom).
-
HWT or Sirt5‐deficient MEF cells (Sirt5 +/+ or Sirt5 −/−) were infected with or without SeV for 8 h, and the interaction between endogenous MAVS and Rig‐I was revealed by immunoprecipitation using anti‐MAVS antibody, followed by immunoblotting with anti‐Rig‐I antibody.
-
ISDD‐AGE analysis of MAVS aggregates in WT or Sirt5‐deficient MEF cells (Sirt5 +/+ or Sirt5 −/−) infected with SeV for 12 h. SDS–PAGE immunoblotting was used as a loading control.
-
J–OqPCR analysis of Ifnβ (J), Ifnα1 (K), Ifnα4 (L), Ifit1 (M), Cxcl10 (N), and Cxcl11 (O) mRNA in WT or Sirt5‐deficient MEF cells (Sirt5 +/+ or Sirt5 −/−) infected with or without VSV (VSV or UI) for 8 h.
-
P–RWT or Sirt5‐deficient MEF cells (Sirt5 +/+ or Sirt5 −/−) were infected with or without VSV‐GFP viruses for 12 h, and viral infectivity was detected by fluorescence microscopy (P), Western blot analysis (Q), and flow cytometry analysis (n = 3) (R).
Figure EV4. Sirt5 deficiency potentiates the host innate antiviral immune response.
- qPCR analysis of Ifnβ mRNA in WT (Sirt5 +/+) or Sirt5‐deficient (Sirt5 −/−) MEF cells infected with an increasing titration of SeV (+, ++) for 8 h. −, without infection. The graphs represent fold induction relative to the uninfected cells.
- WT (Sirt5 +/+) or Sirt5‐deficient (Sirt5 −/−) MEF cells were infected with (+) or without (−) SeV for 8 h, and the cell lysates were immunoprecipitated with anti‐MAVS antibody, followed by immunoblotting with anti‐succ‐K7‐MAVS antibody.
- WT (Sirt5 +/+) or Sirt5‐deficient (Sirt5 −/−) MEF cells were infected with (+) or without (−) VSV for 8 h, and the cell lysates were immunoprecipitated with anti‐MAVS antibody, followed by immunoblotting with anti‐succ‐K7‐MAVS antibody.
- VSV titer was determined in WT (Sirt5 +/+) or Sirt5‐deficient (Sirt5 −/−) MEF cells after infected with or without VSV for 12 h. UI, uninfected.
- WT (Sirt5 +/+) or Sirt5‐deficient (Sirt5 −/−) MEF cells were infected with (+) or without (−) VSV‐GFP viruses (relative lower titration) for 12 h, and viral infectivity was detected by flow cytometry analysis (n = 3).
To further determine whether loss of Sirt5 could indeed influence cellular antiviral response, we examined virus replication in Sirt5 +/+ or Sirt5 −/− MEFs via infection with VSV‐GFP viruses. In fact, the viral titer of VSV viruses in Sirt5 −/− MEF cells was lower than that in Sirt5 +/+ MEF cells after VSV infection (Fig EV4D). Fluorescence microscopy showed that VSV‐GFP virus replication was substantially inhibited in Sirt5 −/− MEFs (Fig 5P), which was further confirmed by Western blot analysis using anti‐GFP antibody (12.23 vs. 1.00; Fig 5Q) and flow cytometry analysis (79.9% vs. 45.4%; 59.8% vs. 27.2%; Figs 5R and EV4E). In response to poly(I:C) treatment, expressions of Ifnβ, Ifnα1, Ifnα4, Ifit1, Cxcl10, and Cxcl11 were significantly up‐regulated in Sirt5 −/− MEF cells compared to WT (Sirt5 +/+) MEF cells (Appendix Fig S6A–F). However, upon HSV‐1 infection, Ifnβ was down‐regulated and Ifit1 was not changed in Sirt5 −/− MEF cells compared to WT (Sirt5 +/+) MEF cells (Appendix Fig S6G and H), further suggesting that Sirt5 has no suppressive role on the cytosolic DNA sensing pathway.
Collectively, these data suggest that loss of Sirt5 potentiates cellular antiviral immune response in MEF cells.
Disruption of Sirt5 potentiates cellular antiviral immune response in BMDCs and BMDM cells
To further verify the above observations, we isolated both bone marrow‐derived dendritic cells (BMDCs) and bone marrow‐derived macrophages (BMDMs) and performed a similar set of experiments. SeV‐ and VSV‐induced expressions of Ifnβ, Ifnα1, Ifnα4, Ifit1, Rig‐I, Cxcl10, and Cxcl11 were significantly up‐regulated in Sirt5 −/− BMDCs compared to the induction in WT counterparts (Fig 6A–N). In addition, we compared the virus replication in Sirt5‐deficient (Sirt5 −/−) BMDCs or WT counterparts (Sirt5 +/+) by infected with VSV‐GFP viruses. Fluorescence microscopy showed that VSV‐GFP virus replication was substantially inhibited in Sirt5‐deficient BMDCs (Fig 6O). Similarly, SeV‐ and VSV‐induced expressions of Ifnβ, Ifnα1, Ifnα4, Cxcl10, and Ccl5 or Cxcl11 were significantly up‐regulated in Sirt5 −/− BMDMs compared to the induction in the WT counterparts (Appendix Fig S7). These observations suggest that Sirt5 suppresses innate antiviral response in bone marrow‐derived dendritic cells and macrophages.
Figure 6. Sirt5 deficiency potentiates antiviral immune response in bone marrow‐derived dendritic (BMDC) cells.
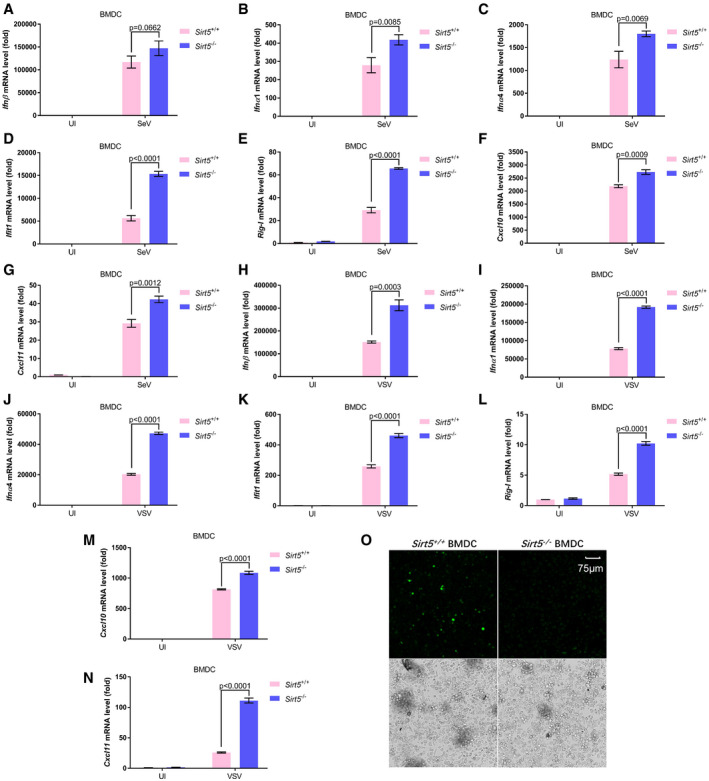
-
A–GqPCR analysis of Ifnβ (A), Ifnα1 (B), Ifnα4 (C), Ifit1 (D), Rig‐I (E), Cxcl10 (F), and Cxcl11 (G) mRNA in WT or Sirt5‐deficient BMDCs (Sirt5 +/+ or Sirt5 −/−) infected with or without SeV (Sev or UI) for 8 h.
-
H–NqPCR analysis of Ifnβ (H), Ifnα1 (I), Ifnα4 (J), Ifit1 (K), Rig‐I (L), Cxcl10 (M), and Cxcl11 (N) mRNA in WT or Sirt5‐deficient BMDCs (Sirt5 +/+ or Sirt5 −/−) infected with or without VSV viruses (VSV or UI) for 8 h.
-
OConfocal fluorescence microscopy image of replication in WT or Sirt5‐deficient BMDCs (Sirt5 +/+ or Sirt5 −/−) after infected with VSV‐GFP viruses for 12 h (MOI = 1) (fluorescence, upper; bright‐field, bottom). Scale bar = 75 μm.
Disruption of Sirt5 potentiates host innate antiviral immune responses
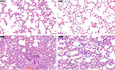
To further determine the physiological function of Sirt5 in antiviral immunity, we challenged WT littermates (Sirt5 +/+) and Sirt5‐deficient (Sirt5 −/−) mice with VSV. After intraperitoneal injection (i.p.) with VSV, succinylation of MAVS in livers and lungs of WT mice was increased revealed by anti‐succ‐K7‐MAVS antibody (Fig 7A and B). Moreover, after i.p. injection with VSV, succinylation of MAVS in livers and lungs was significantly higher in Sirt5 −/− mice than in Sirt5 +/+ mice (Fig 7C and D). Sirt5 −/− mice were also more resistant to infection with VSV than Sirt5 +/+ mice (Fig 7E). Consistently, the Ifnβ level in serum was higher in Sirt5 −/− mice than in Sirt5 +/+ mice (Fig 7F). In addition, after infection with VSV, H & E staining showed less infiltration of immune cells and less injury in the lungs of Sirt5 −/− mice, compared to the lungs of Sirt5 +/+ mice.
Figure 7. Pivotal role of Sirt5 in MAVS desuccinylation and antiviral immune response in vivo .
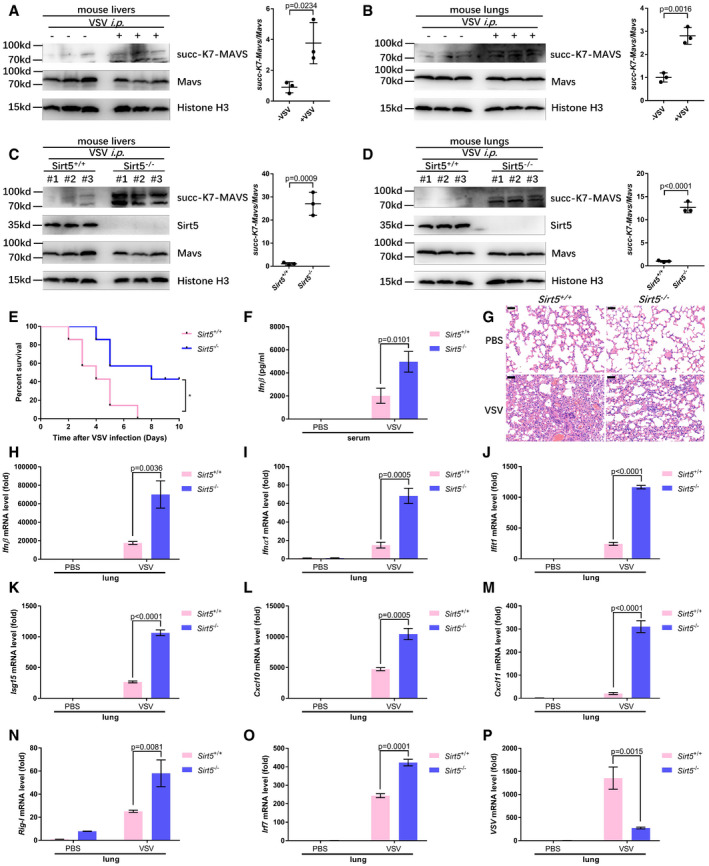
-
A, BVSV infection increased MAVS succinylation in mouse livers (A) and lungs (B). Proteins in the livers (A) and lungs (B) of WT littermates injected intraperitoneally with (+) VSV (1 × 107 plaque‐forming units (PFU) per mouse) or PBS control (−) (n = 3 per group) were detected with the indicated antibodies. MAVS succinylation was determined by anti‐succ‐K7‐MAVS antibody. Relative succinylation level was quantified (right panel).
-
C, DKnockout of Sirt5 increased MAVS succinylation in mouse livers (C) and lungs (D) upon VSV infection. Proteins in the livers (C) and lungs (D) of Sirt5‐deficient or WT littermates (Sirt5 −/− or Sirt5 +/+) injected intraperitoneally with VSV (1 × 107 plaque‐forming units (PFU) per mouse) (n = 3 per group) for 24 h were detected with the indicated antibodies. MAVS succinylation was determined by anti‐succ‐K7‐MAVS antibody. Relative succinylation level was quantified (right panel).
-
ESurvival (Kaplan–Meier curve) of WT mice (n = 7) and Sirt5‐deficient mice (n = 7) injected intraperitoneally with a high dose of VSV (5 × 107 PFU per mouse) and monitored for 10 days.
-
FELISA assay of Ifnβ in serum from the WT mice (n = 4) and Sirt5‐deficient mice (n = 4) injected intraperitoneally with VSV (1 × 107 plaque‐forming units (PFU) per mouse) or PBS control for 24 h.
-
GHematoxylin‐and‐eosin‐stained images of lung sections from mice in F. Scale bar = 50 μm.
-
H–OqPCR analysis of Ifnβ (H), Ifnα1 (I), Ifit1 (J), Isg15 (K), Cxcl10 (L), Cxcl11 (M), Rig‐I (N), and Irf7 (O) mRNA in the lungs of WT or Sirt5‐deficient mice (Sirt5 +/+ or Sirt5 −/−) injected intraperitoneally with VSV (1 × 107 plaque‐forming units (PFU) per mouse) or PBS control for 24 h.
-
PqPCR analysis of VSV mRNA in the lungs of WT or Sirt5‐deficient mice (Sirt5 +/+ or Sirt5 −/−) injected intraperitoneally with VSV (1 × 107 plaque‐forming units (PFU) per mouse) or PBS control for 24 h.
Consistent with the increased production of Ifnβ, Ifnβ mRNA, Ifnα1 mRNA, Ifit1 mRNA, Isg15 mRNA, Cxcl10 mRNA, Cxcl11 mRNA, Rig‐I mRNA, and Irf7 mRNA were higher in lungs and spleens of Sirt5 −/− mice (Figs 7H–O and EV5A–E). As expected, VSV‐specific mRNA was lower in lungs and spleens of Sirt5 −/− mice (Figs 7P and EV5F).
Figure EV5. Sirt5 deficiency potentiates antiviral immune response in spleen.
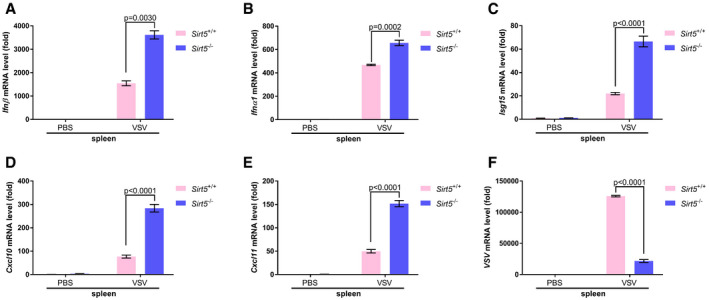
-
A–EqPCR analysis of Ifnβ (A), Ifnα1 (B), Isg15 (C), Cxcl10 (D), and Cxcl11 (E) mRNA in the spleens of WT (Sirt5 +/+) or Sirt5‐deficient (Sirt5 −/−) mice injected intraperitoneally with VSV (1 × 107 plaque‐forming units (PFU) per mouse) or PBS control for 24 h.
-
FqPCR of VSV mRNA in the spleens of WT (Sirt5 +/+) or Sirt5‐deficient (Sirt5 −/−) mice injected intraperitoneally with VSV (1 × 107 plaque‐forming units (PFU) per mouse) or PBS control for 24 h.
These in vivo data indicate that Sirt5 is an important negative regulator of antiviral immune response against RNA viruses.
Discussion
MAVS is the central molecule that links viral recognition to downstream antiviral kinase cascades (Kawai et al, 2005; Meylan et al, 2005; Seth et al, 2005; Xu et al, 2005; Moore & Ting, 2008; Hou et al, 2011; Tan et al, 2018). Regulation of MAVS activation through PTM has been extensively investigated (Liu et al, 2015, 2016a; Li et al, 2018a; Liu & Gao, 2018; He et al, 2019). In this study, we found that MAVS could be succinylated upon viral challenge, uncovering a novel PTM of MAVS. In addition, we found that SIRT5 desuccinylates MAVS at Lysine 7, leading to the impairment of type I IFN production and antiviral gene expression. This finding revealed a novel function of SIRT5 in antiviral innate immunity.
Protein succinylation caused by succinyl‐CoA is a recently identified PTM of proteins, which links metabolism to protein function (Weinert et al, 2013; Yang & Gibson, 2019). Here, we identified that MAVS is succinylated in response to viral infection and succinylation of MAVS appears to facilitate its activity in antiviral innate immunity. It is evident that even though succinyl‐CoA can be generated in cells through different metabolic pathways (Weinert et al, 2013; Yang & Gibson, 2019), succinyl‐CoA formed in the Krebs cycle is considered the main succinyl donor to lysine in protein succinylation (Weinert et al, 2013). Therefore, our findings suggest a critical role for succinyl‐CoA mainly derived from the mitochondrial Krebs cycle in antiviral innate immunity, connecting Krebs cycle to RLR signaling pathway.
Substantial evidence that has accumulated in recent years has highlighted that intermediates and derivatives of the mitochondrial Krebs cycle possess immune signaling functions (Tannahill et al, 2013; Mills et al, 2016; Lei et al, 2018; Murphy & O'Neill, 2018; Williams & O'Neill, 2018; Peruzzotti‐Jametti et al, 2018; Ryan et al, 2019). SDH inhibition and succinate accumulation resulting from dysregulation of succinate metabolism can cause accumulation of succinyl‐CoA, which causes lysine succinylation (Weinert et al, 2013; Yang & Gibson, 2019). Here, our findings that succinylation of MAVS modulates antiviral innate immunity extend the importance of intermediates and derivatives of the mitochondria Krebs cycle in immune signaling. However, the de novo process and the underlying molecular mechanisms of succinylation in response to viral infection are still unclear. In addition, the contribution of succinylation to antiviral innate immunity remains poorly understood. Resolving these issues may improve our understanding of the role of protein succinylation in innate immunity.
MAVS aggregation is considered to be a hallmark of its activation and to be essential for its antiviral function (Hou et al, 2011; Cai et al, 2014). The N‐terminal CARD domain of MAVS is necessary and sufficient for forming active MAVS aggregates (Hou et al, 2011; Xu et al, 2014). Therefore, to date, some factors reported affect MAVS activation usually through modifying its CARD domain directly or indirectly (Liu et al, 2015, 2017; Dai et al, 2018). In this study, we identified that Lysine 7 of MAVS is succinylated, which is proximal, but does not exactly localize to the CARD domain. However, succinylation or desuccinylation of Lysine 7 in MAVS still influences the formation of aggregates of MAVS after viral infection, suggesting that the modification of other sites not located in CARD domain also has impacts on MAVS aggregation. Notably, compared to other modifications, succinylation causes a relatively large increase in mass (100.02 Daltons) in addition to a protein charge flip from positive to negative (Zhang et al, 2011; Alleyn et al, 2018; Yang & Gibson, 2019). This may cause it to have a large impact on the structure as well as function of the proteins that it modifies, particularly on dramatic conformational alternations of its targeted substrates (Alleyn et al, 2018). These notions may provide a possible explanation for the effect of succinylation or desuccinylation of Lysine 7 on MAVS aggregation even though Lysine 7 of MAVS is far from the MAVS‐MAVS interaction interface (Hou et al, 2011; Xu et al, 2014; Yang & Gibson, 2019). Consciously, the mechanistic insights beyond this phenomenon are still mystical. To further demonstrate how succinylation or desuccinylation of Lysine 7 affects MAVS aggregation will get insights to the mechanism of SIRT5 in regulating antiviral innate immunity.
Notably, ubiquitination of Lysine 7 in MAVS catalyzed by TRIM25 and MARCH5 has been identified, which promotes proteasome‐mediated degradation of MAVS (Castanier et al, 2012; Yoo et al, 2015). In this study, we found that desuccinylation of Lysine 7 in MAVS by SIRT5 apparently had no impact on MAVS protein stability. Thus, succinylation of MAVS Lysine 7 might not influence MAVS ubiquitination at Lysine 7. However, we observed that SIRT5 attenuated K63‐linked polyubiquitination of MAVS, which is consistent with the suppressive role of SIRT5 on MAVS activation (Liu et al, 2017).
Whether the enzyme responsible for succinylation existed is still debatable, but SIRT5 has been identified as an important desuccinylase, which plays pivotal functions in diverse pathways (Park et al, 2013; Rardin et al, 2013; Anderson et al, 2014; Wang et al, 2017a; Heinonen et al, 2018). However, to date, the connection between protein desuccinylation catalyzed by SIRT5 and RLR‐mediated innate immune activation has remained elusive. Our study identified that SIRT5 catalyzes desuccinylation of MAVS at Lysine 7, thereby attenuating RLR signaling and downstream type I IFN production. This finding suggests a critical role of MAVS desuccinylation in antiviral innate immunity.
Since the desuccinylase activity of SIRT5 has been reported, increasing evidence supports its pivotal role in desuccinylation (Du et al, 2011; Park et al, 2013; Wang et al, 2017a; Yang & Gibson, 2019). However, in addition to desuccinylase activity, demalonylase, and deglutarylase activity have been revealed for SIRT5 (Du et al, 2011; Tan et al, 2014; Nishida et al, 2015). To further investigate whether these activities owned by SIRT5 can affect MAVS activation and antiviral response will be interesting.
SIRT5 mainly localizes in mitochondrial (Finkel et al, 2009). In fact, MAVS localizes in mitochondria to function (Seth et al, 2005; Liu et al, 2017; Dai et al, 2018). Here, we identified that SIRT5 co‐localizes with MAVS in mitochondria and catalyzes MAVS desuccinylation, re‐enforcing the impact of SIRT5 on MAVS function.
In this study, we noticed that SIRT5 still suppresses MAVS‐K7R mutant activity even though the effect is not as dramatic as that on WT MAVS activity. Thus, SIRT5 might modify other lysine (s) in addition to Lysine 7 of MAVS. To further determine whether other lysines of MAVS are desuccinylated by SIRT5 and subsequently affect MAVS function will gain a full picture of the role of SIRT5 in RLR signaling.
Materials and Methods
Cell culture
HEK293T, H1299, and HCT116 cells were cultured in Dulbecco's modified Eagle medium (DMEM) (HyClone) with 10% fetal bovine serum (FBS). WT Sirt5 and Sirt5‐deficient mouse embryo fibroblasts (MEFs) were maintained in DMEM supplemented with sodium pyruvate (110 mg/l), 10% FBS, 1× nonessential amino acids (Sigma) and 1% penicillin–streptomycin. The cells were grown at 37°C in a humidified incubator containing 5% CO2.
Mice
Sirt5 knockout (KO) mice (B6; 129 background) were purchased from the Jackson Laboratory (https://www.jax.org/strain/012757). All animal procedures were approved by Institutional Animal Care and Use Committee (IACUC) at Institute of Hydrobiology, Chinese Academy of Sciences. The Sirt5‐deficient mice were backcrossed seven generations onto a C57BL/6J background before conducting this study. Mice were housed (12‐h light/dark cycle, 22°C) and given unrestricted access to standard diet and tap water under specific pathogen‐free conditions in Animal Research Center of Wuhan University. Littermates of the same sex were randomly assigned to experimental groups. All the analyses were performed blindly.
Viruses
Sendai viruses (SeV) (provided by Dr. Bo Zhong; Wuhan University, China), VSV‐GFP viruses (VSV) (provided by Dr. Mingzhou Chen; Wuhan University, China), and HSV‐1 (provided by Dr. Chunfu Zheng; Fujian Medical University, Fuzhou, China) were propagated and tittered by plague assay on Vero cells.
Reagents and resources
The reagents and resources are listed in Appendix Table S1.
Luciferase reporter assays
Cells were grown in 24‐well plates and transfected with various amounts of plasmids by VigoFect (Vigorous Biotech, Beijing, China), as well as with pCMV‐Renilla used as an internal control. After the cells were transfected for 18–24 h, the luciferase activity was determined by the dual‐luciferase reporter assay system (Promega). Data were normalized to Renilla luciferase. Data are reported as mean ± s.e.m., which are representative of at least three independent experiments, each performed in triplicate.
Quantitative real‐time PCR (qPCR)
Total RNAs were extracted using RNAiso Plus (TaKaRa Bio., Beijing, China) following the protocol provided by the manufacturer. cDNAs were synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA). MonAmp™ SYBR® Green qPCR Mix (high Rox) (Monad Bio., Shanghai, China) was used for quantitative RT–PCR assays. The primers for quantitative real‐time PCR assays are listed in Appendix Table S2.
SiRNA‐mediated knockdown cell lines
The siRNA sequences targeting SIRT5 were described previously (Wang et al, 2017a). The siRNA oligo was obtained from GenePharma Company and used at a final concentration of 100 μM.
CRISPR‐Cas9 knockout cell lines
To generate H1299 or HCT116 SIRT5‐knocked‐out cell lines, sgRNA sequence were ligated into LentiCRISPRv2 plasmid and then co‐transfected with viral packaging plasmids (psPAX2 and pMD2G) into HEK293T cells. Six hours after transfection, medium was changed, and viral supernatant was filtered through 0.45‐μm strainer 42 h later. Targeted cells were infected by viral supernatant and selected by 1 μg/ml puromycin for 2 weeks. The sgRNA sequences targeting SIRT5 and MAVS were described as previously (Fang et al, 2017; Yang et al, 2018).
Immunofluorescence confocal microscopy
Cells grown on glass coverslips were fixed with 4% paraformaldehyde in PBS for 20 min, permeabilized with 0.1% Triton X‐100, and blocked with 1% bovine serum albumin. Then, the cells were stained with the indicated primary antibodies followed by incubation with fluorescent‐dye‐conjugated secondary antibodies. Nuclei were counterstained with DAPI (Sigma‐Aldrich). For mitochondrial staining, living cells were incubated with 300 nM MitoTracker (Invitrogen) for 30 min at 37°C. Imaging of the cells was carried out using a Leica laser‐scanning confocal microscope.
Immunoprecipitation and Western blot
Co‐immunoprecipitation and Western blot analysis were performed as described previously (Wang et al, 2017b). The Fuji Film LAS4000 mini‐luminescent image analyzer was used to photograph the blots. Multi Gauge V3.0 or ImageJ software (National Institutes of Health) was used for quantifying the protein levels based on the band density obtained in Western blot analysis.
In vitro succinylation assay
Reactions contain succinylation buffer (20 mmol/l pH 8.0 HEPES, 1 mmol/l dithiothreitol, 1 mmol/l phenylmethylsulfonyl fluoride, and 0.1 mg/ml BSA), purified MAVS proteins, and different concentrations of succinyl‐CoA (S1129, Sigma‐Aldrich). Reaction mixture was incubated at 30°C for 15 min. The reaction was stopped by adding loading buffer and subjected to SDS–PAGE. Proteins were analyzed by Western blot analysis and Coomassie blue staining.
Identification of MAVS succinylation site(s) by mass spectrometry
HEK293T cells were transfected with Flag‐MAVS plasmid. Cell lysate was immunoprecipitated with anti‐Flag Ab‐conjugated agarose beads overnight. Immunoprecipitated MAVS proteins were subjected to 8% SDS–PAGE gel, and MAVS bands were excised from the gel and analyzed by mass spectrometry in Protein Gene Biotech, Wuhan, Hubei, China.
Generation of anti‐succ‐K7‐MAVS antibody
MAVS K7 site‐specific succinylation antibody (anti‐succ‐K7‐MAVS) was generated by using a human MAVS succinylated peptide (MPFAED(succ‐K)TYKYIC) as an antigen. After purifying the antibodies with excess unmodified peptide (MPFAEDKTYKYIC), antibodies recognizing site‐specific succinylation were enriched by biotin labeled MAVS succinylated peptides. The specificity of anti‐succ‐K7‐MAVS antibody was verified by dot blot assay.
MEF cells, BMDM cells, and BMDCs
Primary mouse embryonic fibroblasts (MEFs) were prepared from embryos at embryonic days 12.5–14.5 and were cultured in DMEM containing 20% FBS, 1% streptomycin–penicillin, and 10 μM β‐mercaptoethanol. Bone marrow cells were isolated from mouse femur. The cells were cultured in DMEM containing 10% FBS, 1% streptomycin–penicillin, and 10 μM β‐mercaptoethanol, with M‐CSF (10 ng/ml, Peprotech) for BMDM differentiation or GM‐CSF (20 ng/ml, Peprotech) for BMDC differentiation, respectively. On day 6, the cells were re‐seeded and used for subsequent experiments.
Native PAGE
Cells were harvested and lysed with lysis buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP‐40, and protease inhibitor), and 20 μg total protein was applied for native PAGE after centrifugation at 11,950 g. Native PAGE was performed with an 8% gel without SDS. The gel was pre‐run with the running buffer (25 mM Tris–HCl, pH 8.4, 192 mM glycine, with or without 0.2% deoxycholate in the cathode and anode buffer, respectively) at 75 V for 30 min. The samples were electrophoresed at 75 V for 3 h in a cold temperature and followed by transferring onto membrane for immunoblot analysis.
Semi‐denaturing detergent agarose gel electrophoresis (SDD‐AGE)
Crude mitochondria and cytosolic extracts were separated from HEK293T or MEF cells using differential centrifugation. After infected with SeV for 9 h, the cells were harvested and resuspended in buffer A (10 mM Tris–HCl pH 7.5, 1.5 mM MgCl2, 10 mM KCl, 0.25 M d‐mannitol) and then lysed by grinding; cell debris and the supernatant were separated by centrifugation at 700 g for 10 min. The supernatants were then removed to a new EP tube and centrifuged at 10,000 g for 10 min at 4°C to separate crude mitochondria and cytosolic extracts. The crude mitochondria were resuspended in 1× sample buffer (0.5 × TBE, 10% glycerol, 2% SDS, and 0.0025% bromophenol blue) and loaded onto a 1.5% agarose gel. Then, the samples were electrophoresed at a constant voltage of 100 V at 4°C until the bromophenol blue dye reached the bottom of the agarose gel. The proteins were transferred to PVDF membranes (Millipore) for immunoblotting.
Flow cytometry assay
H1299 and MEF cells were harvested and washed with PBS. Total cells were analyzed using Beckman CytoFLEX S. The data were analyzed and generated with FlowJo software.
Viral infection in vivo
For in vivo viral infection studies, age‐ and sex‐matched Sirt5 +/+ and Sirt5 −/− mice (6–8 weeks old) were infected with VSV (1 × 107 pfu/mouse) by intraperitoneal injection. IFNβ concentration in supernatants of serum from Sirt5 +/+ and Sirt5 −/− mice was determined by using the mouse IFNβ ELASA kit (BioLegend, 439408) according to the manufacturer's protocol. The VSV mRNA levels in the lung and spleen were determined by quantitative real‐time PCR assays (qPCR). For the survival experiments, mice were monitored for survival after VSV (5 × 107 pfu/mouse) infection by intraperitoneal injection.
Lung histology
Age‐ and sex‐matched Sirt5 +/+ and Sirt5 −/− mice were infected with VSV (1 × 107 pfu/mouse) by intraperitoneal injection. Twenty‐four h post‐infection, lungs from control or virus‐infected mice were dissected, fixed in 10% phosphate‐buffered formalin, embedded into paraffin, sectioned, stained with hematoxylin and eosin (H & E) solution, examined, and photographed under a microscope.
Cellular succinate measurement
Intracellular succinate levels were measured using a commercial assay kit (ab204718, Abcam) following the protocol provided by the manufacturer. Briefly, 2 × 106 cells/each group were collected and cell pellets were homogenized with assay buffer; then, the debris was removed by centrifugation. The supernatant was further deproteinized using Amicon Ultra‐10k centrifugal filter (Millipore) and subjected to succinate measurement.
Ubiquitination assay
Ubiquitination assays were followed the protocol described previously with some modifications (Liu et al, 2017). Briefly, HEK293T cells were transfected with the plasmids expressing Flag‐MAVS, His‐ubiquitin(WT), His‐ubiquitin(K63), and Myc‐SIRT5 for 24 h and then lysed by denatured buffer (6 M guanidine‐HCl, 0.1 M Na2HPO4/NaH2PO4, 10 mM imidazole), followed by nickel bead purification and immunoblotting with the indicated antibodies.
For ubiquitination assay in MEF cells, MEF cells were infected with or without VSV for 8 h, then collected, and lysed with the lysis buffer (100 μl). The supernatants were denatured at 95°C for 5 min in the presence of 1% SDS. The denatured lysates were diluted with lysis buffer to reduce the concentration of SDS (less than 0.1%). Immunoprecipitation (denature‐IP) was conducted using anti‐MAVS antibody and then subjected to immunoblotting with anti‐K63‐linked ubiquitin antibody.
Viral titer determination
Viral titer determination was followed the protocol described previously with some modifications (Liu et al, 2017). Briefly, WT or Sirt5‐deficient MEF cells (Sirt5 +/+ or Sirt5 −/−) cells were plated 24 h before infection. The cells were infected with VSV (MOI = 0.1). One hour later, the cells were washed with PBS and then added with fresh medium. After the cells were incubated for 12 h, the supernatants were collected and the plaque assay was conducted for determining VSV titers.
Statistical analysis
GraphPad Prism 6 software (GraphPad Software, San Diego, CA) was used for all statistical analyses. Differences between experimental and control groups were determined by unpaired two‐tailed Student's t‐test (where two groups of data were compared) or two‐way ANOVA analysis (where more than two groups of data were compared). P values less than 0.05 were considered statistically significant. For animal survival analysis, the Kaplan–Meier method was adopted to generate graphs, and the survival curves were analyzed by log‐rank analysis.
Author contributions
XL, CZ, and WX designed the experiments and analyzed the data. XL, CZ, HZ, JT, FR, XCh, SF, CX, JD, JZ, JW, GO, GY, XCa, and ZC performed the experiments. XL and WX wrote the manuscript. All authors discussed the results and commented on the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 7
Acknowledgements
We are grateful to Drs. Hongbing Shu, Bo Zhong, Mingzhou Chen, and Chunfu Zheng for reagents and also grateful to Yan Wang for helping in flow cytometry assay and Fang Zhou for helping in fluorescent microscope. This work was supported by National Key R & D Program of China 2018YFD0900602; National Natural Science Foundation of China Grants 31830101, 31721005, 31671315, and 31772872; the Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDA24010308; and Youth Innovation Promotion Association (CAS).
The EMBO Journal (2020) 39: e103285
References
- Alleyn M, Breitzig M, Lockey R, Kolliputi N (2018) The dawn of succinylation: a posttranslational modification. Am J Physiol Cell Physiol 314: C228–C232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KA, Green MF, Huynh FK, Wagner GR, Hirschey MD (2014) SnapShot: mammalian sirtuins. Cell 159: 956 e951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brubaker SW, Bonham KS, Zanoni I, Kagan JC (2015) Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol 33: 257–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Chen J, Xu H, Liu S, Jiang QX, Halfmann R, Chen ZJ (2014) Prion‐like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell 156: 1207–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanier C, Zemirli N, Portier A, Garcin D, Bidere N, Vazquez A, Arnoult D (2012) MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG‐I‐like receptors. BMC Biol 10: 44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IY, Ichinohe T (2015) Response of host inflammasomes to viral infection. Trends Microbiol 23: 55–63 [DOI] [PubMed] [Google Scholar]
- Choi SJ, Lee HC, Kim JH, Park SY, Kim TH, Lee WK, Jang DJ, Yoon JE, Choi YI, Kim S et al (2016) HDAC6 regulates cellular viral RNA sensing by deacetylation of RIG‐I. EMBO J 35: 429–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai T, Wu L, Wang S, Wang J, Xie F, Zhang Z, Fang X, Li J, Fang P, Li F et al (2018) FAF1 regulates antiviral immunity by inhibiting MAVS but is antagonized by phosphorylation upon viral infection. Cell Host Microbe 24: 776–790 e775 [DOI] [PubMed] [Google Scholar]
- Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH et al (2011) Sirt5 is a NAD‐dependent protein lysine demalonylase and desuccinylase. Science 334: 806–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R, Jiang QF, Zhou X, Wang CG, Guan YK, Tao JL, Xi JZ, Feng JM, Jiang ZF (2017) MAVS activates TBK1 and IKK epsilon through TRAFs in NEMO dependent and independent manner. PLoS Pathog 13: e1006720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Jiao K, Guo H, Jiang M, Hao J, Wang H, Shen C (2017) Succinyl‐proteome profiling of Dendrobium officinale, an important traditional Chinese orchid herb, revealed involvement of succinylation in the glycolysis pathway. BMC Genom 18: 598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Deng CX, Mostoslavsky R (2009) Recent progress in the biology and physiology of sirtuins. Nature 460: 587–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Zhao J, Song S, He X, Minassian A, Zhou Y, Zhang J, Brulois K, Wang Y, Cabo J et al (2015) Viral pseudo‐enzymes activate RIG‐I via deamidation to evade cytokine production. Mol Cell 58: 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhu Y, Zhang Y, Geng Y, Gong J, Geng J, Zhang P, Zhang X, Liu N, Peng Y et al (2019) RNF34 functions in immunity and selective mitophagy by targeting MAVS for autophagic degradation. EMBO J 38: e100978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinonen T, Ciarlo E, Theroude C, Pelekanou A, Herderschee J, Le Roy D, Roger T (2018) Sirtuin 5 deficiency does not compromise innate immune responses to bacterial infections. Front Immunol 9: 2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ (2011) MAVS forms functional prion‐like aggregates to activate and propagate antiviral innate immune response. Cell 146: 448–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S (2005) IPS‐1, an adaptor triggering RIG‐I‐ and Mda5‐mediated type I interferon induction. Nat Immunol 6: 981–988 [DOI] [PubMed] [Google Scholar]
- Lei W, Ren W, Ohmoto M, Urban JF Jr, Matsumoto I, Margolskee RF, Jiang P (2018) Activation of intestinal tuft cell‐expressed Sucnr1 triggers type 2 immunity in the mouse small intestine. Proc Natl Acad Sci USA 115: 5552–5557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang Q, Ding Y, Liu Y, Zhao D, Zhao K, Shen Q, Liu X, Zhu X, Li N et al (2016) Methyltransferase Dnmt3a upregulates HDAC9 to deacetylate the kinase TBK1 for activation of antiviral innate immunity. Nat Immunol 17: 806–815 [DOI] [PubMed] [Google Scholar]
- Li X, Zhang Q, Shi QZ, Liu Y, Zhao K, Shen QC, Shi Y, Liu XG, Wang CM, Li N et al (2017) Demethylase Kdm6a epigenetically promotes IL‐6 and IFN‐beta production in macrophages. J Autoimmun 80: 85–94 [DOI] [PubMed] [Google Scholar]
- Li T, Li X, Attri KS, Liu C, Li L, Herring LE, Asara JM, Lei YL, Singh PK, Gao C et al (2018a) O‐GlcNAc transferase links glucose metabolism to MAVS‐mediated antiviral innate immunity. Cell Host Microbe 24: 791–803 e796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu Y, Wang C, Xia WR, Zheng JY, Yang J, Liu B, Liu JQ, Liu LF (2018b) Succinate induces synovial angiogenesis in rheumatoid arthritis through metabolic remodeling and HIF‐1 alpha/VEGF axis. Free Radical Bio Med 126: 1–14 [DOI] [PubMed] [Google Scholar]
- Littlewood‐Evans A, Sarret S, Apfel V, Loesle P, Dawson J, Zhang J, Muller A, Tigani B, Kneuer R, Patel S et al (2016) GPR91 senses extracellular succinate released from inflammatory macrophages and exacerbates rheumatoid arthritis. J Exp Med 213: 1655–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV et al (2015) Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347: aaa2630 [DOI] [PubMed] [Google Scholar]
- Liu J, Qian C, Cao XT (2016a) Post‐translational modification control of innate immunity. Immunity 45: 15–30 [DOI] [PubMed] [Google Scholar]
- Liu X, Cai XL, Zhang DW, Xu CX, Xiao WH (2016b) Zebrafish foxo3b negatively regulates antiviral response through suppressing the transactivity of irf3 and irf7. Journal of Immunology 197: 4736–4749 [DOI] [PubMed] [Google Scholar]
- Liu BY, Zhang M, Chu HL, Zhang HH, Wu HF, Song GH, Wang P, Zhao K, Hou JX, Wang X et al (2017) The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63‐linked polyubiquitination. Nat Immunol 18: 214–224 [DOI] [PubMed] [Google Scholar]
- Liu B, Gao C (2018) Regulation of MAVS activation through post‐translational modifications. Curr Opin Immunol 50: 75–81 [DOI] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J (2005) Cardif is an adaptor protein in the RIG‐I antiviral pathway and is targeted by hepatitis C virus. Nature 437: 1167–1172 [DOI] [PubMed] [Google Scholar]
- Mills EL, Kelly B, Logan A, Costa ASH, Varma M, Bryant CE, Tourlomousis P, Dabritz JHM, Gottlieb E, Latorre I et al (2016) Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell 167: 457–470 e413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EL, Pierce KA, Jedrychowski MP, Garrity R, Winther S, Vidoni S, Yoneshiro T, Spinelli JB, Lu GZ, Kazak L et al (2018a) Accumulation of succinate controls activation of adipose tissue thermogenesis. Nature 560: 102–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EL, Ryan DG, Prag HA, Dikovskaya D, Menon D, Zaslona Z, Jedrychowski MP, Costa ASH, Higgins M, Hams E et al (2018b) Itaconate is an anti‐inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556: 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CB, Ting JP (2008) Regulation of mitochondrial antiviral signaling pathways. Immunity 28: 735–739 [DOI] [PubMed] [Google Scholar]
- Murphy MP, O'Neill LAJ (2018) Krebs cycle reimagined: the emerging roles of succinate and itaconate as signal transducers. Cell 174: 780–784 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L (2009) SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell 137: 560–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y, Rardin MJ, Carrico C, He W, Sahu AK, Gut P, Najjar R, Fitch M, Hellerstein M, Gibson BW et al (2015) SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol Cell 59: 321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, Xie Z, Zhang Y, Zwaans BM, Skinner ME et al (2013) SIRT5‐mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell 50: 919–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peruzzotti‐Jametti L, Bernstock JD, Vicario N, Costa ASH, Kwok CK, Leonardi T, Booty LM, Bicci I, Balzarotti B, Volpe G et al (2018) Macrophage‐derived extracellular succinate licenses neural stem cells to suppress chronic neuroinflammation. Cell Stem Cell 22: 355–368 e313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyssonnaux C, Cejudo‐Martin P, Doedens A, Zinkernagel AS, Johnson RS, Nizet V (2007) Cutting edge: essential role of hypoxia inducible factor‐1alpha in development of lipopolysaccharide‐induced sepsis. J Immunol 178: 7516–7519 [DOI] [PubMed] [Google Scholar]
- Qin K, Han C, Zhang H, Li T, Li N, Cao X (2017) NAD(+) dependent deacetylase Sirtuin 5 rescues the innate inflammatory response of endotoxin tolerant macrophages by promoting acetylation of p65. J Autoimmun 81: 120–129 [DOI] [PubMed] [Google Scholar]
- Rardin MJ, He W, Nishida Y, Newman JC, Carrico C, Danielson SR, Guo A, Gut P, Sahu AK, Li B et al (2013) SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab 18: 920–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roers A, Hiller B, Hornung V (2016) Recognition of endogenous nucleic acids by the innate immune system. Immunity 44: 739–754 [DOI] [PubMed] [Google Scholar]
- Rubic T, Lametschwandtner G, Jost S, Hinteregger S, Kund J, Carballido‐Perrig N, Schwarzler C, Junt T, Voshol H, Meingassner JG et al (2008) Triggering the succinate receptor GPR91 on dendritic cells enhances immunity. Nat Immunol 9: 1261–1269 [DOI] [PubMed] [Google Scholar]
- Ryan DG, Murphy MP, Frezza C, Prag HA, Chouchani ET, O'Neill LA, Mills EL (2019) Coupling Krebs cycle metabolites to signalling in immunity and cancer. Nat Metab 1: 16–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ (2005) Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF‐kappaB and IRF 3. Cell 122: 669–682 [DOI] [PubMed] [Google Scholar]
- Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, Park J, Chen Y, Huang H, Zhang Y et al (2014) Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab 19: 605–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Sun L, Chen J, Chen ZJ (2018) Detection of microbial infections through innate immune sensing of nucleic acids. Annu Rev Microbiol 72: 447–478 [DOI] [PubMed] [Google Scholar]
- Tannahill GM, Curtis AM, Adamik J, Palsson‐McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH et al (2013) Succinate is an inflammatory signal that induces IL‐1beta through HIF‐1alpha. Nature 496: 238–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GR, Hirschey MD (2014) Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol Cell 54: 5–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Wang K, Xu W, Zhao S, Ye D, Wang Y, Xu Y, Zhou L, Chu Y, Zhang C et al (2017a) SIRT5 desuccinylates and activates pyruvate kinase M2 to block macrophage il‐1beta production and to prevent DSS‐induced colitis in mice. Cell Rep 19: 2331–2344 [DOI] [PubMed] [Google Scholar]
- Wang J, Zhang D, Du J, Zhou C, Li Z, Liu X, Ouyang G, Xiao W (2017b) Tet1 facilitates hypoxia tolerance by stabilizing the HIF‐alpha proteins independent of its methylcytosine dioxygenase activity. Nucleic Acids Res 45: 12700–12714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert BT, Scholz C, Wagner SA, Iesmantavicius V, Su D, Daniel JA, Choudhary C (2013) Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep 4: 842–851 [DOI] [PubMed] [Google Scholar]
- Williams NC, O'Neill LAJ (2018) A role for the Krebs cycle intermediate citrate in metabolic reprogramming in innate immunity and inflammation. Front Immunol 9: 141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Chen ZJ (2014) Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol 32: 461–488 [DOI] [PubMed] [Google Scholar]
- Xie Z, Dai J, Dai L, Tan M, Cheng Z, Wu Y, Boeke JD, Zhao Y (2012) Lysine succinylation and lysine malonylation in histones. Mol Cell Proteomics 11: 100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB (2005) VISA is an adapter protein required for virus‐triggered IFN‐beta signaling. Mol Cell 19: 727–740 [DOI] [PubMed] [Google Scholar]
- Xu H, He X, Zheng H, Huang LJ, Hou F, Yu Z, de la Cruz MJ, Borkowski B, Zhang X, Chen ZJ et al (2014) Structural basis for the prion‐like MAVS filaments in antiviral innate immunity. Elife 3: e01489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wang Z, Li X, Liu B, Liu M, Liu L, Chen S, Ren M, Wang Y, Yu M et al (2018) SHMT2 desuccinylation by SIRT5 drives cancer cell proliferation. Can Res 78: 372–386 [DOI] [PubMed] [Google Scholar]
- Yang Y, Gibson GE (2019) Succinylation links metabolism to protein functions. Neurochem Res 44: 2346–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Harding AT, Sweeney C, Miao D, Swan G, Zhou C, Jiang Z, Fitzgerald KA, Hammer G, Bergo MO et al (2019) Control of antiviral innate immune response by protein geranylgeranylation. Science advances 5: eaav7999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo YS, Park YY, Kim JH, Cho H, Kim SH, Lee HS, Kim TH, Sun Kim Y, Lee Y, Kim CJ et al (2015) The mitochondrial ubiquitin ligase MARCH5 resolves MAVS aggregates during antiviral signalling. Nat Commun 6: 7910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You F, Sun H, Zhou X, Sun W, Liang S, Zhai Z, Jiang Z (2009) PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat Immunol 10: 1300–1308 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Tan M, Xie Z, Dai L, Chen Y, Zhao Y (2011) Identification of lysine succinylation as a new post‐translational modification. Nat Chem Biol 7: 58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Bharathi SS, Rardin MJ, Lu J, Maringer KV, Sims‐Lucas S, Prochownik EV, Gibson BW, Goetzman ES (2017) Lysine desuccinylase SIRT5 binds to cardiolipin and regulates the electron transport chain. J Biol Chem 292: 10239–10249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhao J, Xu S, Li J, He S, Zeng Y, Xie L, Xie N, Liu T, Lee K et al (2018) Species‐specific deamidation of cGAS by herpes simplex virus UL37 protein facilitates viral replication. Cell Host Microbe 24: 234–248 e235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wang G, Xu ZG, Tu H, Hu F, Dai J, Chang Y, Chen Y, Lu Y, Zeng H et al (2019) Lactate is a natural suppressor of rlr signaling by targeting MAVS. Cell 178: 176–189 e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao FC, Lei CQ, He X, Zhang L, Tien P et al (2008) The adaptor protein MITA links virus‐sensing receptors to IRF3 transcription factor activation. Immunity 29: 538–550 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 7


