Desmosomes regulate intercellular adhesion, which controls tissue strength and plasticity. Bartle et al. show that desmosome adhesive strength is modulated through protein exchange in and out of desmosomes, not cadherin order. This exchange is tuned by desmoplakin phosphorylation, which acts as a switch between adhesive states.
Abstract
Desmosomes are cell–cell junctions that provide mechanical integrity to epithelial and cardiac tissues. Desmosomes have two distinct adhesive states, calcium-dependent and hyperadhesive, which balance tissue plasticity and strength. A highly ordered array of cadherins in the adhesive interface is hypothesized to drive hyperadhesion, but how desmosome structure confers adhesive state is still elusive. We employed fluorescence polarization microscopy to show that cadherin order is not required for hyperadhesion induced by pharmacologic and genetic approaches. FRAP experiments in cells treated with the PKCα inhibitor Gö6976 revealed that cadherins, plakoglobin, and desmoplakin have significantly reduced exchange in and out of hyperadhesive desmosomes. To test whether this was a result of enhanced keratin association, we used the desmoplakin mutant S2849G, which conferred reduced protein exchange. We propose that inside-out regulation of protein exchange modulates adhesive function, whereby proteins are “locked in” to hyperadhesive desmosomes while protein exchange confers plasticity on calcium-dependent desmosomes, thereby providing rapid control of adhesion.
Introduction
Many vital cellular processes including gene expression, cell division, and motility, are dependent on macromolecular complexes. Higher-level features of these complexes including protein architecture, order, organization, and dynamics, are all critical regulators of function. Importantly, complexes that appear static can adopt multiple conformational states (Vrabioiu and Mitchison, 2006), act as depots of regulatory proteins (Ray et al., 2007), and support exchange of protein components (Daigle et al., 2001; Griffis et al., 2003). Understanding this multifaceted regulation is key to deciphering the functions of macromolecular complexes in health and disease.
Cell junctions represent a class of plasma membrane–associated macromolecular complexes with roles in adhesion, force transmission, and electrical connections (Garcia et al., 2018; Goodenough and Paul, 2009; Parsons et al., 2010). To perform these myriad functions, cell junctions have complex architectures that are key in signal integration and dynamic regulation (Bertocchi et al., 2017; Kanchanawong et al., 2010; Kaufmann et al., 2012; Mehta et al., 2016; Nahidiazar et al., 2015; Stahley et al., 2016). Epithelial cells have two similar yet distinct adhesive junctions that span neighboring cells: desmosomes and adherens junctions. These junctions share the role of mediating cell–cell adhesion and are architecturally analogous, with adhesive cadherin cores linked to the cytoskeleton through a network of proteins. Despite these similarities, adherens junctions and desmosomes are molecularly and functionally distinct (Rübsam et al., 2018).
One key functional difference is the ability of desmosomes to adopt a calcium-independent, or hyperadhesive, state (Wallis et al., 2000). Whereas adherens junctions and calcium-dependent desmosomes disassemble and lose function upon chelation of extracellular Ca2+, hyperadhesive desmosomes maintain adhesion when Ca2+ has been removed (Garrod, 2010; Garrod et al., 2005; Wallis et al., 2000). These two functional states allow rapid and precise tuning of adhesion to balance tissue strength and plasticity in a multitude of processes. For example, during development and tissue remodeling, desmosomes are calcium-dependent and plastic, but ultimately become static and hyperadhesive in mature tissue (Kimura et al., 2012). In the epidermis, desmosomes have different adhesive strengths in basal versus suprabasal cells (Garrod and Kimura, 2008; Harmon and Green, 2013). During wound healing, desmosomes in suprabasal keratinocytes revert to a calcium-dependent state to promote cell migration and wound closure (Garrod et al., 2005; Owen et al., 2008). Conversion between these adhesive states is controlled by PKCα. Inhibition of PKCα induces hyperadhesion, likely owing to the loss of phosphorylation of desmoplakin (DP; Garrod et al., 2005; Wallis et al., 2000). Hyperadhesion can also be conferred by overexpression of the DP mutant S2849G, which cannot be phosphorylated at that site (Albrecht et al., 2015; Hobbs and Green, 2012). Conversely, hyperadhesive desmosomes can be converted to calcium-dependent by activation of PKCα (Wallis et al., 2000). In this way, regulation of PKCα allows for rapid and precise control of the desmosome adhesive state.
It is not known how desmosome architecture impacts the adhesive state. Because cadherins mediate adhesion by mechanically coupling neighboring cells, they are an obvious candidate for defining function. Classic and desmosomal cadherins are type I transmembrane proteins with five extracellular cadherin (EC) domains with interdomain Ca2+ binding sites. The cadherin tertiary structure is rigid when Ca2+ is bound and disorganized without Ca2+ (Harrison et al., 2016; Pokutta et al., 1994; Sotomayor and Schulten, 2008). Structurally, desmosomal cadherins have a more “bent” conformation (Harrison et al., 2016) and exhibit greater flexibility (Tariq et al., 2015) than classical cadherins, both features that have been proposed to play roles in accommodating hyperadhesion. In tissues, desmosomes have a characteristic dense midline bisecting the extracellular space, as characterized by EM. This dense midline corresponds with overlapping EC1 domains at the site of trans binding and is found solely in hyperadhesive desmosomes (Garrod et al., 2005; He et al., 2003; Shimizu et al., 2005). An ordered and periodic organization of cadherins in the extracellular space of hyperadhesive desmosomes has been proposed (Rayns et al., 1969; Al-Amoudi et al., 2005, 2007). These characteristics support the hypothesis that the arrangement of the cadherin EC domains into quasi-crystalline ordered arrays is a structural feature defining hyperadhesive desmosomes, which may drive function (Garrod, 2013).
Herein we determine the unique architectural features driving hyperadhesion by quantifying cadherin order, plaque organization, and protein dynamics in calcium-dependent and hyperadhesive states. We found that cadherin order and desmosome architecture did not correlate with adhesive state. Surprisingly, cadherins lost order upon Ca2+ chelation even in hyperadhesive desmosomes. Desmosomes with disordered cadherins maintained adhesion, eliminating cadherin order as the driving factor behind hyperadhesion. Using fluorescence recovery after photobleaching (FRAP), we identify protein exchange in and out of desmosomes as a key difference between calcium-dependent and hyperadhesive states. We show that protein exchange can be controlled via DP phosphorylation, providing a rapid switch in protein dynamics between static hyperadhesive and dynamic calcium-dependent desmosomes. These findings reveal a potential novel mechanism for conferring desmosome hyperadhesion and regulating cell adhesion.
Results
Cadherin order is not required for adhesion
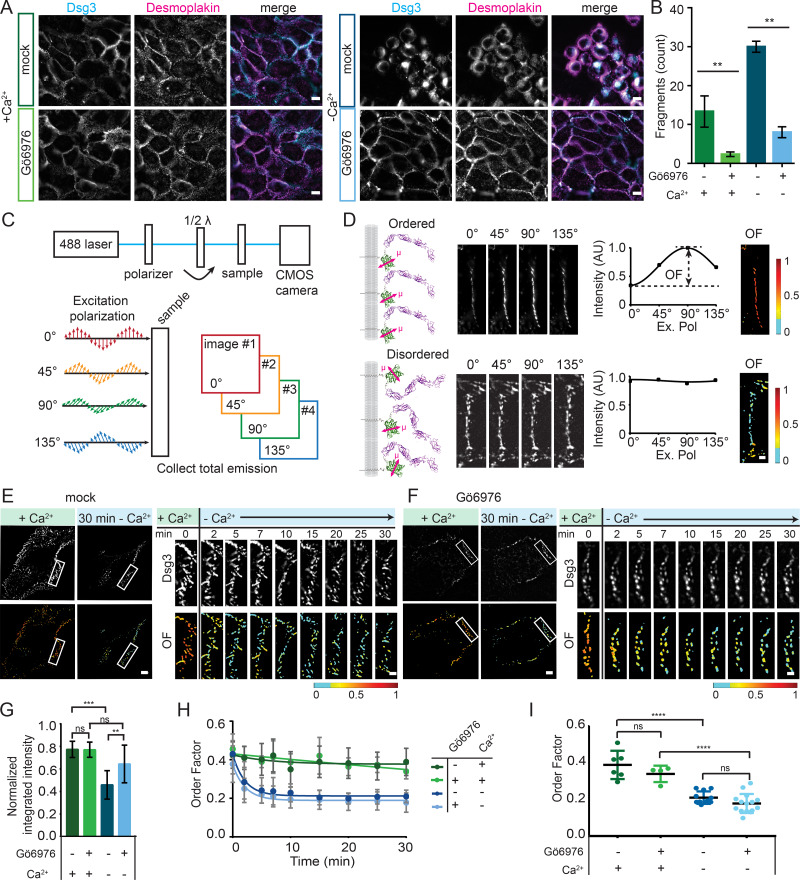
Desmosomes can be switched from calcium-dependent to hyperadhesive by inhibition of PKCα (Garrod et al., 2005; Kimura et al., 2007; Wallis et al., 2000). To study changes associated with hyperadhesion, we used the PKCα inhibitor Gö6976 (Hobbs and Green, 2012). Immortalized human keratinocyte (HaCaT) cells were treated with 50 nM Gö6976 or vehicle (DMSO; mock). To confirm hyperadhesion, cells were incubated in low-Ca2+ medium (Ca2+-free DMEM with 10% chelated FBS and 3 mM EGTA) for 90 min, following the protocol set forward by Garrod and colleagues (Garrod, 2013; Garrod et al., 2005). Hyperadhesive desmosomes persist following this treatment, whereas calcium-dependent desmosomes do not. To observe desmosomes and overall cellular morphology, cells were fixed and immunostained for desmoglein 3 (Dsg3), a desmosomal cadherin, and DP (Fig. 1 A). Dsg3 and DP colocalized in puncta at cell borders when cells were maintained in normal-Ca2+ medium (∼1.8 mM), indicating the presence of desmosomes at cell–cell junctions. After a 90-min incubation in low-Ca2+ medium, mock-treated cells had a rounded morphology, lack of cell–cell contacts, and loss of desmosomes. In contrast, Gö6976-treated cells retained their morphology, protein localization, and cell–cell contacts, suggesting they had acquired hyperadhesion. To test the adhesive function of these cells, intact epithelial sheets were lifted off the culture dish and subjected to mechanical stress to induce fragmentation in a dispase cell adhesion assay (Fig. 1 B). We observed that cells treated with Gö6976 resisted fragmentation compared with mock-treated cells in both high- and low-Ca2+ medium. Together, these data demonstrate that Gö6976 treatment leads to hyperadhesion in our experimental system.
Figure 1.
Hyperadhesion does not protect against loss of Dsg3 order in low-Ca2+ media. (A) Representative images of HaCaT cells treated with either vehicle (DMSO; mock) or Gö6976 and maintained in normal-Ca2+ medium (left) or switched into low-Ca2+ medium (right) for 90 min and labeled with antibodies for Dsg3 and DP. Scale bar = 20 µm. (B) Quantification of adhesive strength by dispase fragmentation assay in HaCaT cells with the same treatment conditions as in A (n = 3; representative of four independent experiments; mean ± SD). (C) Schematic of the fluorescence polarization microscope. Samples are sequentially illuminated with four different angles of polarized excitation, and the total emission is collected. (D) Ordered cadherins (top) are differentially excited by distinct excitation polarizations, resulting in modulated intensity across the four images. Disordered cadherins (bottom) are equally excited regardless of excitation polarization, resulting in equal intensity. Intensity was plotted as a function of excitation polarization, and the amplitude was used to calculate pixel-by-pixel order factor (OF). (E and F) HaCaT cells were transfected with Dsg3-ΔEA-GFP, mock (E) or Gö6976 (F) treated and switched from normal to low-Ca2+ medium. Dsg3 intensity and OF images of representative cells before and 30 min after the switch to low calcium with region of interest (ROI) indicated and intensity and Dsg3-ΔEA-GFP OR in the ROI over a 30-min time course after the switch to low-Ca2+ medium. Scale bar = 5 µm; ROI scale bar = 1 µm. (G–I) Population analysis of cells mock treated in normal-Ca2+ (n = 6) or low Ca2+ (n = 11) and Gö6976 treated in normal-Ca2+ (n = 4) or low Ca2+ (n = 15). (G) Normalized integrated intensity at t = 30 min (mean ± SD). (H) OF plotted as a function of time (mean ± SD). (I) OF at t = 30 min (mean ± SD) compared by one-way ANOVA with multiple comparisons. Data were acquired from three independent experiments (ns, not significant, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.001).
Our initial goal was to determine whether there were architectural features of desmosomes that promote hyperadhesion. The leading model is that the arrangement of cadherin extracellular domains in an ordered crystalline array is necessary for hyperadhesion (Garrod, 2013; Garrod et al., 2005). To test this, we used excitation-resolved fluorescence polarization microscopy to measure cadherin order (Bartle et al., 2017; DeMay et al., 2011; Kress et al., 2011). In this approach, the protein of interest is rigidly tagged with a fluorophore, such that the fluorophore orientation reflects the protein orientation. When illuminated with linearly polarized light, fluorophores with dipoles oriented parallel to the excitation polarization are excited, whereas those perpendicular are not (Fig. 1 C). If proteins within a complex are ordered, fluorescence intensity is modulated by changing the orientation of the excitation polarization (Fig. 1 D, top). If the proteins are not ordered, there is no modulation of intensity (Fig. 1 D, bottom). The order of many copies of the tagged protein within a diffraction limited (∼250-nm) spot can therefore be quantified from the fluorescence intensity recorded at four unique excitation polarizations.
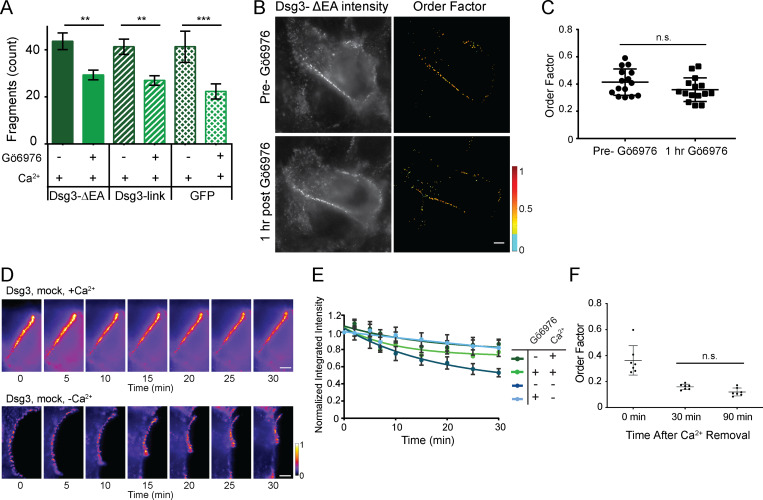
To measure the order of desmosomal cadherins, we replaced the Dsg3 extracellular anchor domain (EA) with GFP (Dsg3-ΔEA-GFP), as previously described (Bartle et al., 2017). This construct reports the order of the most membrane-proximal extracellular domain of Dsg3. To test whether expression of Dsg3-ΔEA-GFP disrupted desmosome function, cells were subjected to a dispase cell adhesion assay. Cells expressing Dsg3-ΔEA-GFP had increased adhesion following treatment with Gö6976 (Fig. S1 A). The degree of fragmentation was not significantly different from that in control cells expressing either Dsg3 with GFP attached to the C terminus with an unstructured linker or cytosolic GFP, indicating that Dsg3-ΔEA-GFP does not act as a dominant negative in desmosome adhesion.
Figure S1.
Fluorescence polarization microscopy controls. (A) Dispase fragmentation assay for cells expressing Dsg3-ΔEA-GFP, Dsg3-link-GFP, or GFP. Dsg3-ΔEA-GFP did not impact adhesive strength or the ability to acquire hyperadhesion (n = 3; mean ± SD). (B and C) HaCaT cells transfected with Dsg3-ΔEA-GFP were imaged by fluorescence polarization microscopy before and 1 h after treatment with Gö6976 while maintained in normal-Ca2+ medium. (B) Representative intensity and order factor images. Scale bar = 5 µm. (C) Order factor quantification (mean ± SD; n = 15 cells; n.s., not significant P > 0.05; Student’s t test). (D) HaCaT cells were transfected with Dsg3-ΔEA-GFP, mock treated, maintained in normal-Ca2+ medium (top) or switched to low-Ca2+ medium (bottom), and imaged for 30 min. Representative intensity images at each time point are shown. Scale bar = 5 µm. (E) Intensity over time was quantified by masking each image and calculating the integrated intensity. Integrated intensity was normalized and averaged across the population of cells (D and E are the same cells with analysis presented in Fig 1; normal-Ca2+ medium mock [n = 6] or Gö6976 [n = 6] treated; low-Ca2+ medium mock [n = 11] or Gö6976 [n = 15] treated; mean ± SEM). (F) Order factor from Gö6976-treated cells fixed at 0, 30, or 90 min after the switch to low-Ca2+ medium (mean ± SD). Order factor was not significantly different at 30 or 90 min in low Ca2+ (n = 7 cells each condition; n.s., not significant P > 0.05; ANOVA).
Dsg3-ΔEA-GFP was transfected into HaCaT cells and imaged with fluorescence polarization microscopy. Cells were imaged in normal-Ca2+ medium before and after Gö6976 treatment, which was found to have no effect on Dsg3-ΔEA-GFP order factor (Fig. S1, B and C). In mock-treated cells with calcium-dependent desmosomes, Dsg3-ΔEA-GFP was ordered in the presence of Ca2+, and order was rapidly lost when cells were switched into low-Ca2+ medium (Fig. 1 E; Bartle et al., 2017). The switch to low-Ca2+ medium also led to desmosome disassembly, as shown by a reduction of fluorescence intensity, rearrangement of the cell junctions, and changes in cell morphology (Fig. 1 E).
We next wanted to know whether cadherins remain ordered in hyperadhesive desmosomes without Ca2+. Surprisingly, when Gö6976-treated cells were switched to low-Ca2+ medium, order factor rapidly decreased (Fig. 1 F). Desmosomes in Gö6976-treated cells had a consistent morphology before and after the switch to low Ca2+, and we were able to track individual desmosomes throughout the time course. This was in contrast to the changing morphology and loss of desmosomes observed in the mock-treated cells. To examine whether proteins were lost from desmosomes over the experiment, we quantified the integrated fluorescence intensity of desmosomal puncta. The normalized fluorescence intensity at t = 30 min did not change significantly for Gö6976-treated cells between high and low Ca2+, whereas in mock-treated cells, the intensity was significantly reduced in low Ca2+ (Figs. 1 G and S1, D and E). This indicates that the amount of Dsg3-ΔEA-GFP in desmosomes was decreased in mock- but not Gö6976-treated cells 30 min after the switch to low-Ca2+ medium. The retention of protein and maintenance of morphology for Gö6976-treated cells in low-Ca2+ medium suggests that these represent hyperadhesive desmosomes.
The dynamics of the decreasing order factors were similar between mock- and Gö6976-treated cells in low-Ca2+ medium (Fig. 1 H). In contrast, order factor remained stable for cells maintained in normal-Ca2+ in both treatments. The mean order factor 30 min after the change to low-Ca2+ medium was not significantly different between mock- and Gö6976-treated cells (Fig. 1 I). To confirm that this reduced order factor was persistent, Gö6976-treated cells were fixed either 30 or 90 min after a switch to low-Ca2+ medium. When imaged with fluorescence polarization, the Dsg3-ΔEA-GFP order factor was not significantly different at 30 or 90 min in low-Ca2+ medium (Fig. S1 F). These experiments reveal that cadherin order, specifically the order of the most membrane-proximal extracellular domain of Dsg3, is not determined by adhesive state, but by Ca2+. Intriguingly our data suggest that cadherins in hyperadhesive desmosomes can be simultaneously disordered and adhesive. Therefore, a quasi-crystalline arrangement of cadherins cannot be the driving mechanism of hyperadhesion.
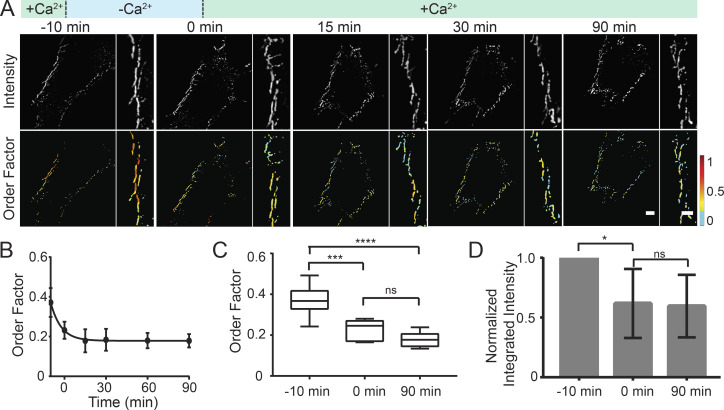
Given the unexpected result that cadherin order was dependent on Ca2+, not adhesive state, we wanted to further investigate this relationship. Because of the importance of Ca2+ in cadherin tertiary structure, we hypothesized that order is indicative of Ca2+ binding, and that if Ca2+ is reintroduced, order will be restored. To test this, we used fluorescence polarization microscopy to measure the dynamics of Dsg3-ΔEA-GFP order in Ca2+-dependent desmosomes after a “low-Ca2+ pulse.” We chose to use calcium-dependent desmosomes for this assay because the cadherins are disordered after 10 min in low Ca2+, but desmosomes are still present. This allowed us to ask whether reintroducing Ca2+ will restore cadherin order or halt desmosome disassembly. For the low-Ca2+ pulse, cells in normal-Ca2+ medium were switched into low-Ca2+ medium for 10 min and then switched back to normal-Ca2+ medium. Fluorescence polarization imaging revealed that Dsg3-ΔEA-GFP order was lost following the switch to low-Ca2+ medium as anticipated, yet switching the cells back to normal-Ca2+ medium did not rescue order (Fig. 2 A). We tracked individual junctions and found that the majority were present for the duration of the time course and did not represent newly formed junctions. The decreased order factor persisted for 90 min after the low-Ca2+ pulse (Fig. 2 B). Loss of order was further quantified by comparing the order factor from the cells in normal-Ca2+ medium before the low-Ca2+ pulse (0.37 ± 0.07; mean ± SD), after the 10 min low-Ca2+ pulse (0.23 ± 0.04), and 90 min after the pulse in normal-Ca2+ medium (0.18 ± 0.03; Fig. 2 C). We found that there was an initial loss of Dsg3-ΔEA-GFP fluorescence intensity from puncta after the switch to low-Ca2+ medium. However, no further loss of intensity occurred over the 90-min experiment, indicating that the amount of Dsg3-ΔEA-GFP in the puncta was consistent once the cells were returned to normal-Ca2+ medium (Fig. 2 D). This revealed that loss of cadherin order is not reversable by reintroducing Ca2+. Once the membrane-proximal domain of Dsg3 was disordered following removal of Ca2+, it did not return to its original ordered arrangement. These results show that desmosomes can be manipulated to have disordered cadherins in both calcium-dependent and hyperadhesive states.
Figure 2.
Dsg3 order is not restored following a low-Ca2+ pulse. (A) HaCaT cells were transfected with Dsg3-ΔEA-GFP and were not treated (calcium-dependent desmosomes). Cells were subjected to a low-Ca2+ pulse including incubation in normal-Ca2+ medium, a switch to low-Ca2+ medium at t = −10 min, and a return to normal-Ca2+ medium at t = 0 min. Cells were imaged with fluorescence polarization microscopy and Dsg3-ΔEA-GFP intensity, and order factor images are shown over the time course. Scale bar = 5 µm; region of interest scale bar = 1 µm. (B) Order factor (mean ± SD) as a function of time (n = 6 cells from three independent experiments). (C) Order factor at −10, 0, and 90 min, where whiskers indicate the range of the data, and the line and box represent the median ± quartile. (D) Quantification of normalized integrated intensity at −10, 0, and 90 min (ns, not significant, P > 0.05; *, P ≤ 0.05; ***, P ≤ 0.001; ****, P ≤ 0.0001; ANOVA).
Desmosome architecture is maintained in Gö6976-induced hyperadhesion
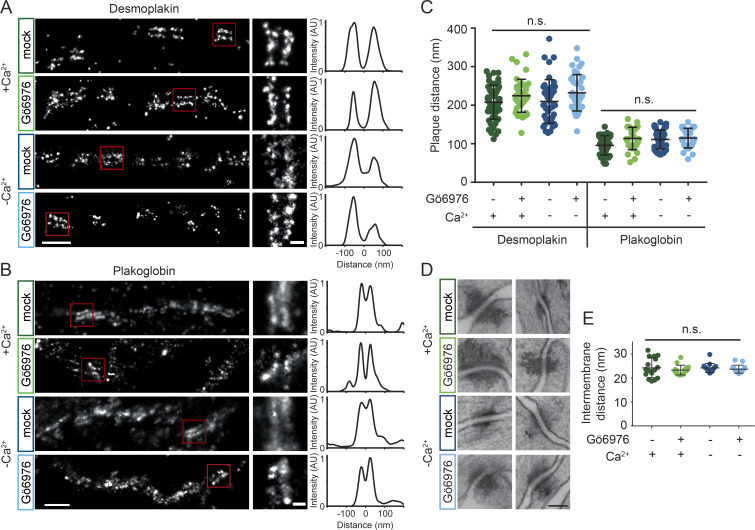
Given that cadherin order did not correlate with adhesive state, we hypothesized that changes in the nanoscale architecture of the desmosomal plaque may provide additional insight toward the mechanism of hyperadhesion. We used superresolution direct stochastic optical reconstruction microscopy (dSTORM; Heilemann et al., 2008) to quantify desmosome protein architecture in different adhesive states (Stahley et al., 2016). We conducted dSTORM on HaCaT cells with calcium-dependent (mock-treated) or hyperadhesive (Gö6976-treated) desmosomes. To test whether extracellular Ca2+ impacted plaque architecture, cells were either maintained in normal-Ca2+ medium or switched to low-Ca2+ medium. Mock-treated samples were switched to low-Ca2+ medium for only 10 min, which preserved junctions for imaging, whereas Gö6976 treated cells were incubated in low-Ca2+ medium for 90 min. Cells were fixed and labeled with antibodies for either the C-terminal keratin binding domain of DP or plakoglobin (PG), an armadillo protein that binds to the cadherin tails, allowing us to assess architecture throughout the different regions of the plaque. After dSTORM imaging, protein localization and architecture were quantified by measuring plaque-to-plaque distance (Fig. 3, A and B). The quantification of plaque-to-plaque distance for both DP C-terminus and PG showed no significant difference across treatment conditions (Fig. 3 C). These results show that the localization of PG in the membrane proximal outer dense plaque and the localization of the C-terminus of DP in the inner dense plaque are not altered within the resolution of dSTORM microscopy.
Figure 3.
Desmosome architecture is maintained in Gö6976-induced hyperadhesion. (A and B) HaCaT cells were mock or Gö6976 treated and incubated in normal or low-Ca2+ medium. Cells were stained with antibodies for DP (A) or PG (B) and imaged by dSTORM. Representative cell borders are shown for each condition with single desmosome regions of interest (ROIs) indicated by the red squares. Scale bar = 500 nm; ROI scale bar = 100 nm. Linescans of fluorescence intensity perpendicular to the desmosome axis were used to quantify plaque-to-plaque distance; examples are shown for the ROIs. (C) Quantification of plaque-to-plaque distance shows no significant difference across treatment groups for DP or PG (DP left to right, n = 58, 40, 44, 41; PG left to right n = 41, 30, 48, 31, each from three independent experiments). (D) Electron micrographs of desmosomes under the same treatment conditions. Scale bar = 100 nm. (E) Quantification of intermembrane distance shows no difference across treatment groups (n = 15 desmosomes per group). Bars represent mean ± SD (n.s., not significant, P > 0.05; one-way ANOVA followed by Tukey’s multiple comparison test).
To further investigate desmosome ultrastructure, transmission EM was conducted on cells following the same treatment paradigms (Fig. 3 D). There were no obvious alterations of the plaques by treatment with Gö6976 or incubation in low Ca2+. Quantification of the width of the intermembrane space showed no significant difference between groups (Fig. 3 E). We conclude that plaque architecture and desmosome ultrastructure are maintained under the conditions tested and thus cannot explain the mechanism of hyperadhesion.
Desmosomal cadherins are stabilized by hyperadhesion
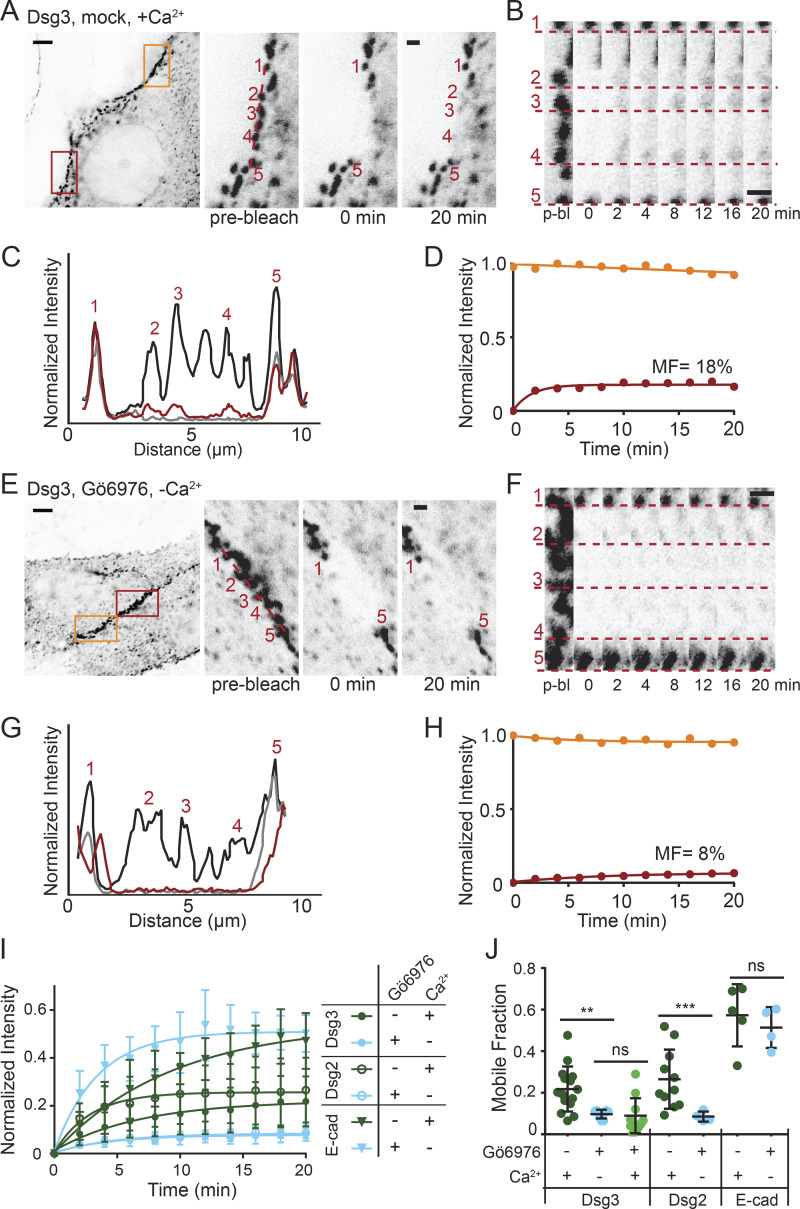
To further investigate architectural changes that could drive the switch between adhesive states, we next explored cadherin exchange in and out of desmosomes using FRAP. First, we measured cadherin exchange in cells with calcium-dependent desmosomes transfected with Dsg3-ΔEA-GFP in normal-Ca2+ medium. A region along the cell border containing multiple desmosomes was photobleached, and fluorescence recovery was measured over 20 min (Fig. 4 A). We found that fluorescence recovery localized to desmosomes, as highlighted by tracking individual puncta within the bleach region over the time course (Fig. 4, B and C). The mobile fraction, calculated from the fluorescence recovery curve, represents the amount of Dsg3-ΔEA-GFP exchange in and out of desmosomes (Fig. 4 D). Next, FRAP was conducted on cells treated with Gö6976 and incubated in low-Ca2+ medium for 90 min to select for hyperadhesive desmosomes (Fig. 4 E). There was minimal recovery of Dsg3-ΔEA-GFP fluorescence localized to desmosomal puncta within the bleach region (Fig. 4, F–H). To test whether this reduced Dsg3 exchange in hyperadhesion was conserved for different cadherin isotypes, we performed FRAP on cells transfected with Dsg2-GFP under the same treatment conditions (Fig. S2). The mean normalized intensity was fitted to a one-phase association, revealing reduced exchange of Dsg2 and Dsg3 in hyperadhesive desmosomes (Fig. 4 I).
Figure 4.
Desmosomal cadherin exchange is reduced in hyperadhesive desmosomes. (A–D) FRAP experiments were conducted on HaCaT cells transfected with Dsg3-ΔEA-GFP and mock treated in normal-Ca2+ medium. (A) Representative cell (inverted intensity) with bleach (red) and reference (orange) regions of interest (ROIs). Bleach ROI shown with individual desmosomal puncta indicated. Scale bar = 5 µm; ROI scale bar = 1 µm. (B) Bleach ROI over time with dashed lines underscoring individual puncta, highlighting recovery. Scale bar = 1 µm. (C) Linescans through the bleach region indicate relative intensities before bleaching (black), immediately after bleaching (gray), and at 20 min after bleaching (red). (D) FRAP recovery curves of the bleach (red) and reference (orange) ROIs. The bleach ROI intensity was fitted to a one-phase association to determine mobile fraction (MF). (E–H) FRAP experiments were conducted on HaCaT cells transfected with Dsg3-ΔEA-GFP, Gö6976 treated, and switched to low-Ca2+ medium for 90 min. (E) Representative cell (inverted intensity) with bleach (red) and reference (orange) ROIs. Bleach ROI shown with individual desmosomal puncta indicated. Scale bar = 5 µm; ROI scale bar = 1 µm. (F) Bleach region over time with dashed lines underscoring individual puncta. Scale bar = 1 µm. (G) Linescans through the bleach region indicate relative intensities before bleaching (black), immediately after bleaching (gray), and at 20 min after bleaching (red). (H) FRAP recovery curves of the bleach (red) and reference (orange) ROIs. The bleach ROI intensity was fitted to a one-phase exponential association to determine the MF. (I) FRAP recovery curves (mean ± SD) for cells transfected with Dsg3-ΔEA-GFP, Dsg2-GFP, or E-Cad-GFP and mock treated in normal Ca2+ (n = 15, 11, 5) or Gö6976 treated in low Ca2+ (n = 11, 10, 4). (J) Mean mobile fraction ± SD for treatments in I and Dsg3-ΔEA-GFP Gö6976 treated in normal Ca2+ (n = 11). All data were acquired from three independent experiments (ns, not significant, P > 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ANOVA).
Figure S2.
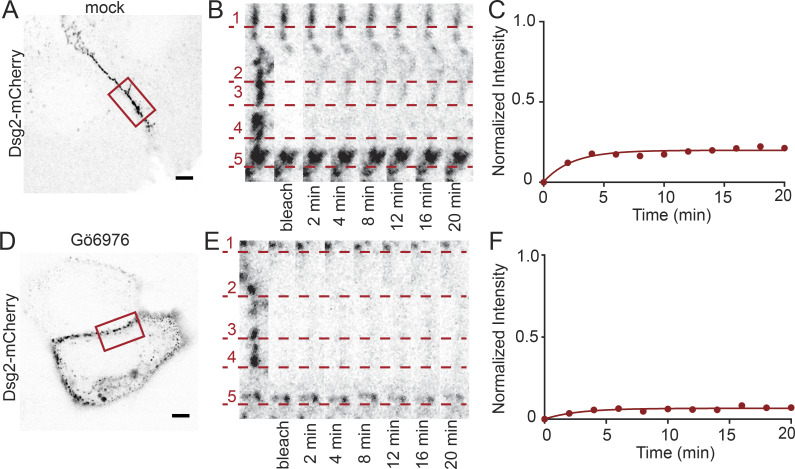
Representative images of cells expressing Dsg2-mCherry showing that exchange is reduced in hyperadhesion. Representative cells from data presented in Fig 4. (A) HaCaT cell transfected with Dsg2-mCherry, mock treated, and maintained in normal Ca2+ with bleach region of interest (ROI) indicated. Scale bar = 5 µm. (B) Zoom-in of cell border in bleach ROI over time. Recovery of individual puncta is underscored by the dashed lines. (C) Fluorescence intensity plotted as a function of time. Mobile fraction = 20%. (D) Representative HaCaT cell transfected with Dsg2-mCherry, Gö6976 treated, and maintained in low-Ca2+ medium for 90 min. Scale bar = 5 µm. (E) Zoom-in of cell border in bleach ROI over time with individual puncta underscored by the dashed lines. (F) Fluorescence intensity plotted as a function of time. Mobile fraction = 7%.
To quantify this difference the recovery curve from each cell was fitted, and the resulting mobile fractions were averaged to determine the mean mobile fraction. The mean mobile fraction of Dsg3-ΔEA-GFP in cells with calcium-dependent desmosomes (22 ± 11%) was significantly higher than in hyperadhesion (10 ± 2%). Similarly the mean mobile fraction of Dsg2-mCherry in cells with calcium-dependent desmosomes (27 ± 14%) was significantly higher than in cells with hyperadhesive desmosomes (9 ± 3%; Fig. 4 J). There was no significant difference in the mobile fraction between cadherin isotypes within the same treatment group.
To confirm that removal of Ca2+ from the medium was not responsible for the reduced protein exchange, we performed FRAP of Dsg3-ΔEA-GFP in cells treated with Gö6976 and maintained in normal-Ca2+ medium. The mean mobile fraction (9 ± 8%) was not significantly different from hyperadhesive desmosomes in low-Ca2+ medium (Fig. 4 J). This significant decrease in desmosomal cadherin mobility in hyperadhesive cells represents a reduced exchange of proteins in and out of desmosomes.
To test whether the reduced mobile fraction of desmogleins in response to Gö6976 treatment is specific to desmosomal cadherins, we measured the mobility of the adherens junction protein E-cadherin (E-cad). FRAP experiments were conducted on cells transfected with E-cad-GFP. The mobile fraction did not depend on treatment (57 ± 15% mock and 51 ± 10% Gö6976) and was significantly higher than that of the desmosomal cadherins (Fig. 4, I and J).
Plaque proteins are stabilized in hyperadhesion
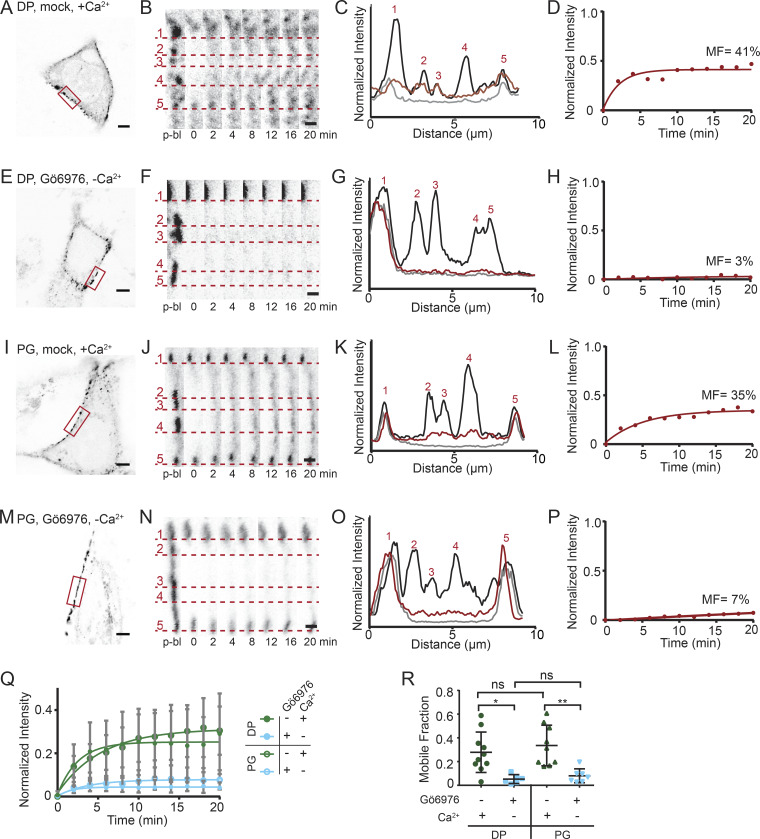
We next asked whether the mobility of desmosomal plaque proteins was also reduced in hyperadhesive desmosomes. DP is an intermediate filament binding protein that anchors the desmosomal plaque to the cytoskeleton. First, FRAP experiments were conducted on HaCaT cells transfected with DP tagged with mCherry (DP-mCherry). In mock-treated cells maintained in normal-Ca2+ medium, there was significant recovery of DP-mCherry fluorescence within desmosomal puncta (Fig. 5, A–D). In contrast, the recovery of DP-mCherry fluorescence was negligible in cells treated with Gö6976 and incubated in low-Ca2+ medium for 90 min (Fig. 5, E–H). PG binds both DP and cadherins (Kowalczyk et al., 1994, 1997), suggesting that its mobility would also be reduced in hyperadhesion. To test this, we conducted FRAP experiments on cells transfected with PG-mEmerald. In mock-treated cells with calcium-dependent desmosomes, there was recovery of PG-mEmerald fluorescence to desmosomal puncta, indicating protein exchange (Fig. 5, I–L). In contrast, in cells treated with Gö6976 and incubated in low-Ca2+ medium for 90 min, there was minimal recovery of PG-mEmerald fluorescence, demonstrating reduced protein exchange (Fig. 5, M–P).
Figure 5.
Plaque protein exchange is reduced in hyperadhesive desmosomes. (A–D) FRAP experiments were conducted on HaCaT cells transfected with DP-mCherry and mock treated in normal-Ca2+ medium. (A) Representative cell (inverted intensity) with bleach region of interest (ROI; red). (B) Individual puncta over the time course underscored by dashed lines. (C) Linescans through the bleach region indicate relative intensities before bleaching (black), immediately after bleaching (gray), and at 20 min after bleaching (red). (D) Fluorescence intensity over time and mobile fraction of the bleach ROI (MF). (E–H) Representative cell from FRAP experiments conducted on HaCaT cells transfected with DP-mCherry, treated with Gö6976, and switched into low-Ca2+ medium for 90 min. (I–L) Representative cell from FRAP experiments conducted on HaCaT cells transfected with PG-mEmerald and mock treated in normal-Ca2+ medium. (M–P) Representative cell from FRAP experiments conducted on HaCaT cells transfected with PG-mEmerald, treated with Gö6976, and switched into low-Ca2+ medium for 90 min. (A, E, I, M) Scale bar = 5 µm. (B, F, J, N) Scale bar = 1 µm. (Q and R) FRAP recovery and fit to one-phase association curves (mean ± SD; Q) and mobile fraction (mean ± SD; R) of PG-mEmerald in mock (n = 9) and Gö6976 (n = 8) treated cells and DP-mCherry in mock (n = 14) and Gö6976 (n = 7) treated cells. All data from three independent experiments. (ns, not significant, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ANOVA).
These experiments revealed that the mobility of both DP and PG was reduced in hyperadhesive desmosomes (Fig. 5 Q). The mobile fractions of DP and PG in cells with hyperadhesive desmosomes (DP, 5 ± 4%; PG, 8 ± 6%) was significantly lower than in calcium-dependent desmosomes (DP, 28 ± 17%; PG, 34 ± 17%). There was no significant difference in mobile fraction between PG and DP within each adhesive state (Fig. 5 R). These results demonstrate that there is a loss of plaque protein mobility in hyperadhesive desmosomes.
Trans binding contributes to cadherin order and reduced exchange in hyperadhesion
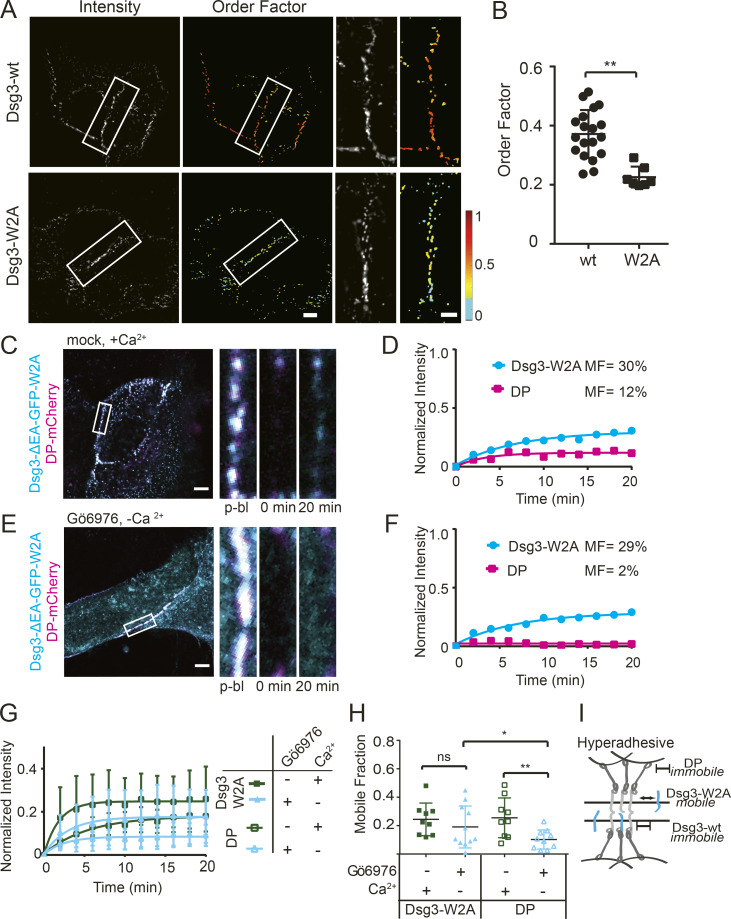
Because cadherin trans binding mediates desmosome adhesion, we next tested whether trans interactions influence cadherin order or exchange. Cadherin trans binding occurs through a strand-swap mechanism involving insertion of a tryptophan residue on the EC1 domain (W2) into a hydrophobic binding pocket of a cadherin on the opposing cell (Haüssinger et al., 2004; Overduin et al., 1995; Shapiro et al., 1995). Site-directed mutagenesis was used to mutate this key tryptophan residue to alanine (W2A) in Dsg3-ΔEA-GFP (Dsg3-W2A), a mutation that has been demonstrated to abrogate trans binding (Harrison et al., 2016; Lowndes et al., 2014). Dsg3-W2A was transfected into HaCaT cells and observed to localize to puncta at the plasma membrane, indicating incorporation into desmosomes. The order factor of Dsg3-W2A (0.22 ± 0.04) was significantly lower than wild-type Dsg3 (0.37 ± 0.08) in calcium-dependent desmosomes maintained in normal-Ca2+ medium (Fig. 6, A and B). How Dsg3-W2A is arranged in the intermembrane space is not known, but the decreased order factor could be attributed to a range of Dsg3-W2A orientations, perhaps restricted by the ordering of wild-type cadherins or a mixed population of disordered and ordered Dsg3-W2A. We conclude that Dsg3-W2A is recruited to desmosomes but does not adopt the same ordered conformation as wild type, indicating that trans binding contributes to establishing cadherin order.
Figure 6.
Dsg3 trans binding is critical in cadherin order and dynamics. (A) HaCaT cells were transfected with Dsg3-ΔEA-GFP (Dsg3-wt) or Dsg3-ΔEA-GFP-W2A (Dsg3-W2A) and imaged with fluorescence polarization microscopy. (B) Mean order factor (Dsg3-W2A n = 7; Dsg3-wt n = 19; mean ± SD; **, P ≤ 0.01; Student’s t test). (C and D) FRAP experiments were conducted on HaCaT cells cotransfected with Dsg3-W2A and DP-mCherry and mock treated in normal-Ca2+ medium. (C) Representative cell and time points of region of interest (ROIs). (D) Fluorescence recovery over time and mobile fraction of the ROI. (E and F) FRAP experiments were conducted on HaCaT cells cotransfected Dsg3-W2A and DP-mCherry, treated with Gö6976, and switched to low-Ca2+ medium. (E) Representative cell and time points of ROI. (F) Fluorescence recovery over time and mobile fraction of the ROI. (G and H) FRAP recovery curves (mean ± SD; G) and mobile fractions (mean ± SD; H) for cells cotransfected with both Dsg3-W2A and DP-mCherry with mock (n = 9) or Gö6976 (n = 11) treatment (ns, not significant, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ANOVA). (I) Schematic of how trans binding changes cadherin mobility in hyperadhesive desmosomes. Scale bars = 5 µm; ROI scale bars =1 µm.
To investigate whether cadherin mobility depends on trans binding, we conducted FRAP experiments on cells cotransfected with Dsg3-W2A and DP-mCherry. In cells with calcium-dependent desmosomes, Dsg3-W2A and DP-mCherry fluorescence recovered and was localized to desmosomal puncta (Fig. 6, C and D). In Gö6976-treated cells incubated in low-Ca2+ medium for 90 min, there was no recovery of DP-mCherry, as seen in previous experiments. However, Dsg3-W2A fluorescence recovered as much as in the calcium-dependent desmosomes (Fig. 6, E and F). Quantification showed that hyperadhesion did not significantly change the Dsg3-W2A mobile fraction (calcium-dependent, 24 ± 11%; hyperadhesive, 19 ± 15%), while in the same cells, the mean mobile fraction of DP-mCherry was significantly reduced in hyperadhesive desmosomes (10 ± 7%) compared with calcium-dependent desmosomes (26 ± 14%; Fig. 6, G and H). These data show that expression of Dsg3-W2A does not have a dominant negative effect on DP exchange. These results indicate that trans binding is an essential component of reduced cadherin mobility in hyperadhesion (Fig. 6 I).
DP phosphorylation is a molecular switch controlling protein exchange
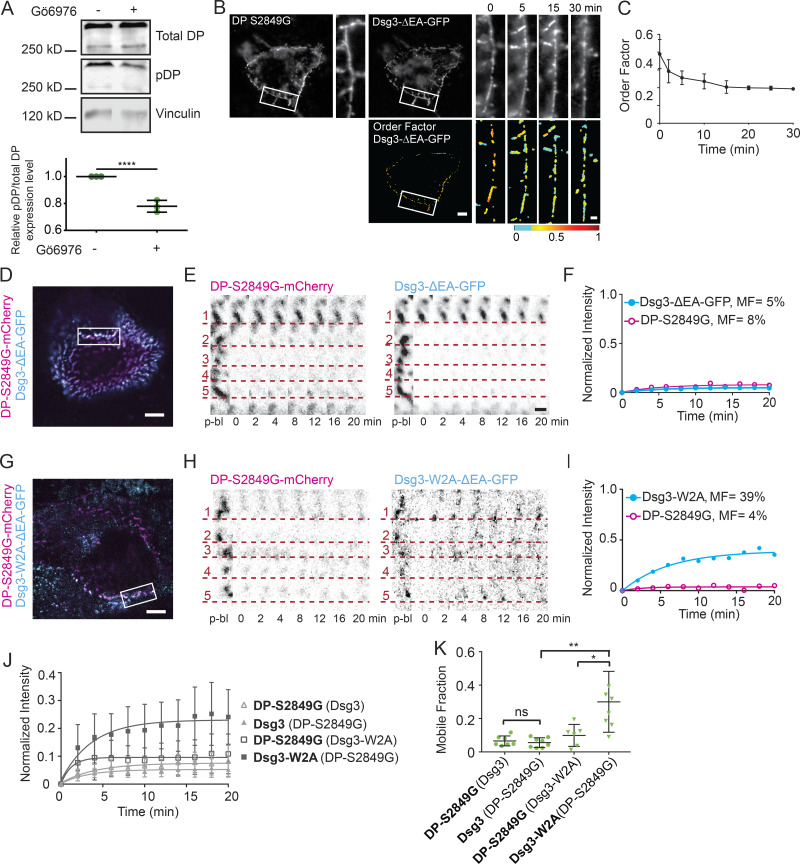
Gö6976 treatment is hypothesized to block DP phosphorylation at S2849, a PKCα consensus site. We confirmed this by immunoblot, which revealed that the percentage of total DP phosphorylated at S2849 decreased by 22.4% in HaCaT cells after treatment with Gö6976 (Fig. 7 A). Lack of phosphorylation at this serine has been shown to increase the affinity of DP for keratin (Godsel et al., 2005). Blocking this phosphorylation with a point mutant (S2849G) has been shown to increase the affinity of DP for keratin and induce hyperadhesion (Albrecht et al., 2015; Hobbs and Green, 2012; Meng et al., 1997). We set out to use the DP mutant S2849G to test whether the order and mobility results obtained when Gö6976 was used to induce hyperadhesion could be recapitulated by blocking phosphorylation of DP at S2849.
Figure 7.
DP S2849G conferred reduced protein exchange. (A) Representative immunoblot and densitometry analysis of total DP and phospho-DP in HaCaT cells mock or Gö6976 treated. For densitometry, the phospho-DP/total DP ratio was normalized to mock treated (****, P < 0.0001 by unpaired t test; data are mean ± SD of three independent experiments). (B and C) HaCaT cells were cotransfected with Dsg3-ΔEA-GFP and DP-S2849G-mCherry, switched from normal to low-Ca2+ medium, and imaged with fluorescence polarization microscopy. (B) Representative cell fluorescence and order factor images over time (scale bar = 5 µm; region of interest [ROI] scale bar = 1 µm). (C) Mean Dsg3-ΔEA-GFP order factor over time (mean ± SD; n = 8 cells). (D) Representative cell expressing DP-S2849G-mCherry (magenta) and Dsg3-ΔEA-GFP (cyan) with bleach ROI indicated. Scale bar = 5 µm. (E) ROI (inverted intensity) with dotted lines underscoring individual puncta of DP-S2849G-mCherry and Dsg3-ΔEA-GFP over the time course (scale bar = 1 µm). (F) Bleach ROI of DP-S2849G-mCherry and Dsg3-ΔEA-GFP intensity plotted as a function of time and fitted to a one-phase association. (G) Representative cell expressing DP-S2849G-mCherry (magenta) and Dsg3-W2A-ΔEA-GFP (cyan) with bleach ROI indicated. Scale bar = 5 µm. (H) ROI (inverted intensity) with dotted lines underscoring individual puncta of DP-S2849G-mCherry and Dsg3-W2A-ΔEA-GFP over the time course (scale bar = 1 µm). (I) Bleach ROI of DP-S2849G-mCherry and Dsg3-W2A-ΔEA-GFP intensity plotted as a function of time and fitted to a one-phase association. (J and K) Normalized intensity over time fitted to a one-phase association (mean ± SD; J) and mobile fraction (mean ± SD; K) for cotransfections with quantified protein in bold and coexpressed protein in parentheses. DP-S2849G-mCherry (Dsg3-ΔEA-GFP; n = 8) and Dsg3-ΔEA-GFP (DP-S2849G-mCherry; n = 8) and DP-S2849G-mCherry (Dsg3-W2A-ΔEA-GFP; n = 6) and Dsg3-W2A-ΔEA-GFP (DP-S2849G-mCherry; n = 8). All data from three independent experiments (ns, not significant, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ANOVA).
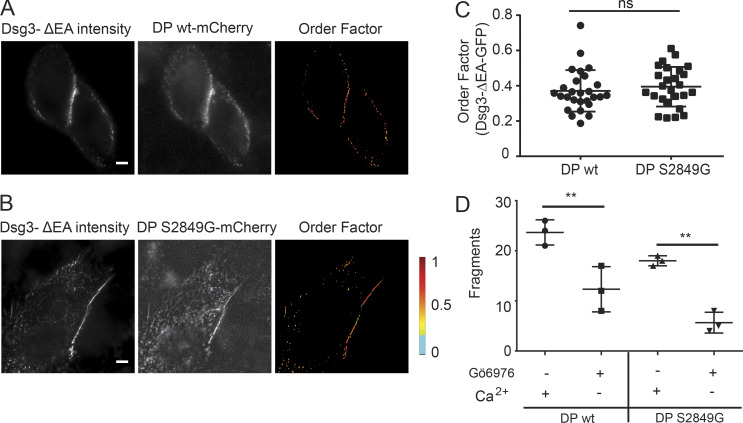
We first wanted to see whether mutant DP-S2849G-mCherry impacted the order of the Dsg3-ΔEA-GFP extracellular domain. Cotransfection with DP-mCherry caused no significant difference in Dsg3-ΔEA-GFP order factor while maintained in normal-Ca2+ medium (wild-type DP, 0.37 ± 0.12; mutant DP, 0.39 ± 0.11; Fig. S3, A–C). Cells transfected with DP-S2849G-mCherry had increased adhesion compared with wild-type DP-mCherry (Fig. S3 D). After a switch into low-Ca2+ medium, cell morphology was unchanged, and puncta were maintained at cell borders over the 30-min imaging window, indicating the presence of hyperadhesive desmosomes. Dsg3-ΔEA-GFP order decreased rapidly following the switch to low-Ca2+ medium (Fig. 7, B and C). The final order factor after 30 min in low-Ca2+ medium (0.19 ± 0.07) was not significantly different from that measured in cells treated with Gö6976 (0.18 ± 0.05; see Fig. 1; Table S1).
Figure S3.
Dsg3 order factor is not impacted by DP-S8249G-mCherry transfection. (A and B) Representative images of HaCaT cells cotransfected with (A) Dsg3-ΔEA-GFP and DP-wt-mCherry or (B) Dsg3-ΔEA-GFP and DP-S2849G-mCherry and maintained in normal-Ca2+ medium. Scale bar = 5 µm. (C) Order factor of Dsg3-ΔEA-GFP in cells expressing DP-wt-mCherry (n = 27) or DP S2849G-mCherry (n = 26; ns, not significant, P > 0.05; Student’s t test). (D) Quantification of adhesive strength by dispase fragmentation assay in HaCaT cells expressing either wt or mutant DP with Gö6976 and Ca2+ conditions indicated (n = 3; mean ± SD; **, P ≤ 0.01; Student’s t test).
Next, we tested whether expression of DP-S2849G was sufficient to block protein exchange. FRAP experiments conducted on cells cotransfected with Dsg3-ΔEA-GFP and DP-S2849G-mCherry and maintained in normal-Ca2+ medium showed minimal exchange of either protein (Fig. 7, D–F). The mobility of DP-S2849G was significantly lower than that established earlier for wild-type DP ( Student’s t test; Table S2). We next wanted to investigate whether cadherin trans binding was required for this reduced exchange. The trans-binding mutant Dsg3-W2A retained mobility when cotransfected with DP-S2849G-mCherry (Fig. 7, G–I). The mean mobile fractions of DP-S2849G-mCherry (7 ± 3%) and Dsg3-ΔEA-GFP (6 ± 3%) were not significantly different from each other in the same cells (Fig. 7, J and K). In contrast, the mean mobile fraction of Dsg3-W2A (30 ± 18%) was significantly higher than that of the DP-S2849G-mCherry (10 ± 7%) it was cotransfected with. The mobility of DP-S2849G-mCherry was similar to that of wild-type DP in Gö6976-treated cells with hyperadhesive desmosomes (Table S2).
These findings demonstrate that blocking DP phosphorylation at S2849 is sufficient to prevent protein exchange in desmosomes. However, this reduced exchange requires cadherin trans binding. Together these results show that DP phosphorylation at S2849 acts as a key molecular switch in regulating desmosome adhesive state by controlling protein dynamics.
Discussion
Modulating the adhesive state allows desmosomes to adjust their strength to facilitate modulation of tissue integrity and plasticity in processes such as embryonic development and wound healing. A key hypothesis is that the arrangement of cadherins into an ordered array at the adhesive interface is a defining property of hyperadhesion (Al-Amoudi et al., 2007; Garrod, 2013; Rayns et al., 1969). Our data show that order of the most membrane-proximal extracellular domain of Dsg3 is lost upon removal of Ca2+ in hyperadhesive desmosomes. Order was also shown to be independent of Ca2+ binding, as cadherin order was not restored by reintroduction of Ca2+ to calcium-dependent desmosomes following a low-Ca2+ pulse. Our data uncouple cadherin order and desmosome adhesion, suggesting that cadherin order is not a defining structural feature of hyperadhesion. It is interesting to consider how hyperadhesive desmosomes maintain function in the absence of Ca2+. It is possible that interdomain flexibility could allow disordered cadherins to engage in trans binding in a hyperadhesive state (Tariq et al., 2015). An important consideration is that this approach measures the order of the tagged protein domain, in this case the most membrane-proximal extracellular domain of Dsg3. This leaves open the possibility that cadherins could retain order in other domains, closer to the adhesive interface. However, if a crystalline ectodomain were driving hyperadhesion, we would expect no loss of order from any cadherin domain when Ca2+ is removed from hyperadhesive desmosomes. Given this uncoupling of order and adhesion, why and how cadherin order is conferred to desmosomes is an intriguing question for future investigation.
Fluorescence polarization microscopy is a powerful approach with the unique ability to probe the order of proteins within subcellular structures (DeMay et al., 2011; Rocheleau et al., 2003; Swaminathan et al., 2017; Valades Cruz et al., 2016). This unique insight has allowed us to investigate critical questions about cadherin organization in this work. An important limitation of our approach is that we are unable to determine the type of order or identify specific structural changes. In addition, dynamics, disorder, and some unfavorable GFP orientations can all contribute to lowering the order factor (Bartle et al., 2017). Ultimately, order factor offers a readout for structural organization within the desmosome, but it does not currently indicate the ultrastructure conveying order.
In light of our discovery that cadherin order is lost with the removal of Ca2+, it is interesting to consider the role of Ca2+ in hyperadhesion. Hyperadhesion has been defined by resistance to incubation in low-Ca2+ medium for 90 min (Garrod and Kimura, 2008; Kimura et al., 2007). However, we present here the destructive effect removal of Ca2+ has on cadherin order, regardless of adhesive state. After treatment with Gö6976, Dsg3 mobility was significantly reduced in cells that were subjected to the 90-min incubation in low-Ca2+ medium or maintained in normal-Ca2+ medium. Interestingly, when Ca2+ was kept in the medium, we saw a larger distribution of mobile fractions, possibly because there was a mix of calcium-dependent and hyperadhesive desmosomes resulting from an incomplete effect of Gö6976 on conferring that state. The dynamic, single-cell sensitivity of FRAP experiments could prove helpful in future work investigating hyperadhesion under native Ca2+ conditions.
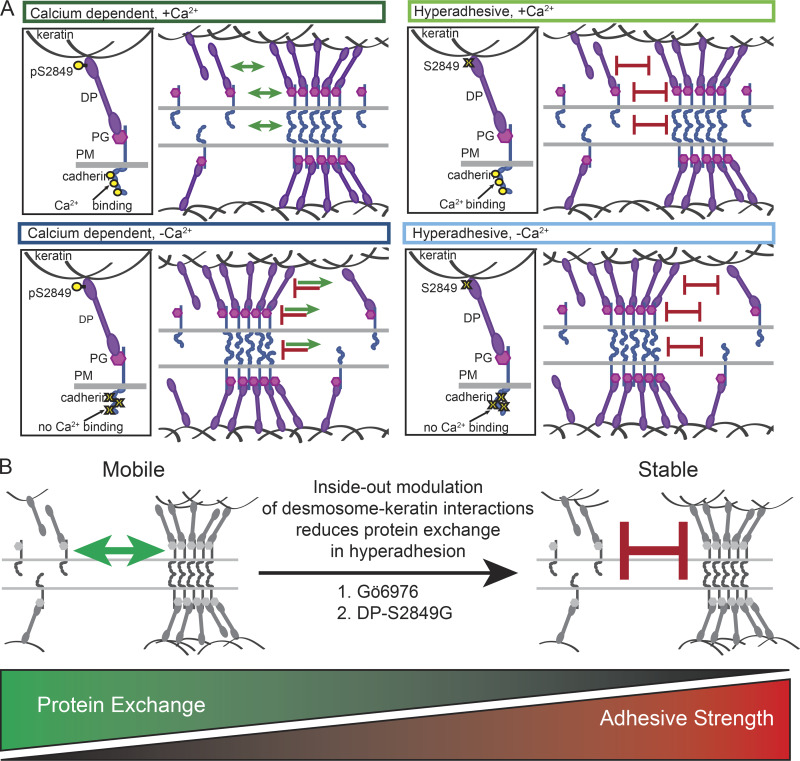
In this work, we show that a defining physical characteristic correlating with adhesive state is protein exchange, not cadherin order (Fig. 8 A). Cadherins and plaque proteins in desmosomes engaged in calcium-dependent adhesion exhibited significant mobility. This indicates that there is exchange between two pools of proteins: desmosomal and non-desmosomal. We propose that sequestration, degradation, or endocytosis of cadherins not associated with desmosomes could “tip the balance” of the ratio between these pools, with the potential to promote disassembly and impact desmosome retention in cells. Assuming that cadherin mobility out of the complex remains unchanged, a reduction in the ability to recruit or retain new cadherins would ultimately result in desmosome disassembly. This state is represented by our low-Ca2+ treatment, in which the ability of cadherins to engage in trans binding is reduced by removal of Ca2+. In this case, the diffusion of cadherins out of desmosomes is unchanged, but the ability to incorporate new protein is reduced. Over time, cadherins are extracted from desmosomes and not replaced, leading to dissolution of the structures. A similar state could be achieved by depleting the free pool of proteins, reducing the availability of proteins to incorporate into desmosomes, while not altering the rate of loss. In this way, protein exchange provides a mechanism for desmosomes to rapidly adapt to changing conditions by either maintaining or disassembling cell adhesion structures.
Figure 8.
Protein exchange controls adhesive state. Models illustrating how reduced protein exchange confers hyperadhesion. (A) Model illustrating dynamics of DP phosphorylation, protein exchange, and cadherin order in calcium-dependent adhesion (left) and hyperadhesion (right) with (top) and without (bottom) Ca2+. In calcium-dependent desmosomes with Ca2+, cadherins are ordered, and desmosomal proteins can diffuse in and out of the complex. In the absence of Ca2+, cadherin order is lost, proteins diffuse out but do not reenter, and desmosomes disassemble. In hyperadhesive desmosomes with Ca2+, cadherins are ordered, but desmosomal proteins are unable to diffuse in or out of the complex. In the absence of Ca2+, cadherin order is lost, but adhesion is maintained because proteins are unable to diffuse out of the complex. (B) We suggest that protein mobility is blocked by Gö6976 or DPS2849G though modulation of DP–keratin interactions via an inside-out mechanism. PM, plasma membrane.
In contrast, we found that there was limited exchange of proteins in hyperadhesive desmosomes. Because there is limited exchange, there is less interaction between the two pools of desmosomal proteins. Therefore, in hyperadhesion, alterations to the non-desmosomal pool of proteins do not have the opportunity to impact the stability of cell adhesion. When treated with low-Ca2+ medium, cadherins became disordered, but desmosomes remained, and adhesion was not lost. We postulate that this is because the proteins are “locked” within the complex by the combination of trans interactions and plaque associations. This is supported by our data showing that when trans binding is abrogated, cadherins are no longer locked in to hyperadhesive desmosomes. This suggests that in the absence of Ca2+, the balance between the probability of cadherins engaging in trans binding and diffusing out of the complex favors retention within desmosomes and maintenance of functional integrity.
Evidence suggests that PKCα endogenously regulates desmosome adhesive state (Thomason et al., 2012), facilitated by its localization to keratin and trafficking with DP (Bass-Zubek et al., 2008; Kröger et al., 2013). It has been previously established that the phospho-null mutant DP S2849G has an increased affinity for keratin, resulting in delayed DP trafficking and increased keratin integration at desmosomes (Godsel et al., 2005). This behavior, and the evidence presented here and by others showing that PKCα is responsible for phosphorylating DP at S2849 (Wallis et al., 2000), suggests that both PKCα inhibition and DP S2849G act through similar means. We posit that increased integration of DP with keratin due to reduced DP phosphorylation is responsible for the loss of protein mobility, but do not eliminate the possibility of other downstream posttranslational modifications of DP or other desmosomal proteins acting in the same pathway. We also leave open a role for cellular mechanics, which could be similarly altered by the change in DP and keratin association. Increasing the affinity of DP for keratin resulted in reduced protein exchange transmitted throughout the complex. This inside-out pathway results in a desmosome structure in which proteins are essentially locked in. In this way, DP phosphorylation can act as a fast and powerful molecular switch for regulating desmosome adhesion by controlling protein exchange (Fig. 8 B).
Our findings shed new light on the cadherin diffusion rates found in the literature. The mobility of Dsg2 and Dsg3 reported from cell culture models is generally consistent with the values we report here in calcium-dependent desmosomes (Gloushankova et al., 2003; Lowndes et al., 2014; Vielmuth et al., 2018; Windoffer et al., 2002), while the lower mobile fraction reported in mouse epidermis is consistent with what we measured in hyperadhesion (Foote et al., 2013). The reduced mobility of desmosomal cadherins is in direct contrast to adherens junction proteins, which have been found to be mobile in mature epidermis (Cao and Schnittler, 2019; Foote et al., 2013). These data agree with our finding that the mobile fraction of E-cad was higher than that of the desmosomal cadherins and not altered by hyperadhesion. Interestingly, the mobile fraction of E-cad in vivo has been shown to correlate with junction integrity and cell invasiveness (Canel et al., 2010; Erami et al., 2016). Vascular endothelial cadherin has also been shown to have high mobility, which could be restricted by protein–protein interactions (Nanes et al., 2012). Together, these data point to a global mechanism in which protein mobility in and out of cell junctions plays a central role in defining stability versus plasticity.
This work provides a novel framework for understanding the regulation of cell–cell adhesion and raises new and exciting questions. Our results demonstrate that hyperadhesion is established by a loss of protein dynamics and plasticity. This mechanism allows cells to rapidly and precisely control cell cohesion and tissue integrity by modulating protein exchange and desmosome stability through posttranslational modifications. Epithelial tissues can thus rapidly respond to both intrinsic and extrinsic cues to control processes such as cell migration and proliferation that require tuning of adhesive function. This regulation is critical for maintaining tissue homeostasis, with aberrant adhesive function contributing to cancer invasiveness, skin fragility, and cardiac arrhythmias. Assessing adhesive function by measuring cell junction protein dynamics is a potential avenue for investigation of adhesion in situ, which would provide a platform for integrating adhesion with the broader context of signaling events, morphological changes, and other regulatory mechanisms. Future work focused on measuring and modulating junctional protein mobility could shed light on the function and dysfunction of junctions, possibly resulting in new therapeutic avenues.
Materials and methods
Cell culture
The human immortalized keratinocyte cell line HaCaT (Addex Bio) was cultured in optimized DMEM (Addex Bio) supplemented with 10% FBS (Corning) and 100 U/ml penicillin/streptomycin (Corning) and maintained at 37°C and 5% CO2. For imaging, cells were seeded into either eight-well #1.5 coverslip bottom dishes (Ibidi) or 25-mm #1.5 coverslips (Electron Microscopy Sciences).
Cloning and constructs
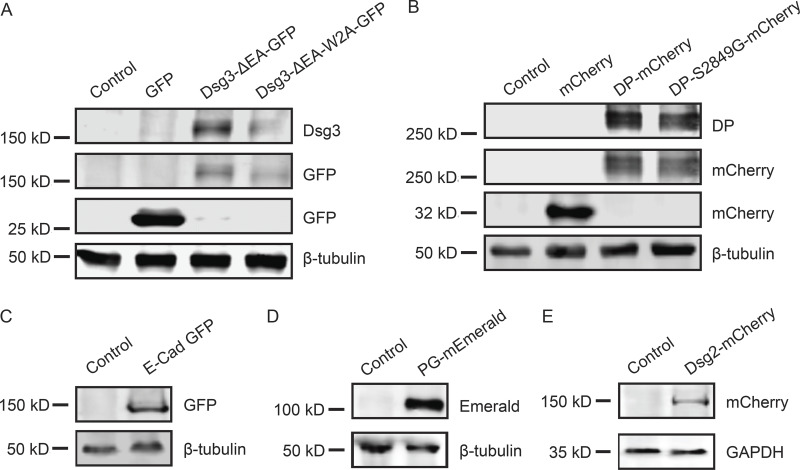
Dsg3-ΔEA-GFP, Dsg3-link-GFP, and DP-mCherry were previously published (Bartle et al., 2017). PG-mEmerald was a gift from Michael Davidson (Florida State University, Tallahassee, FL; Addgene plasmid 54133), and E-cad-GFP was a gift from Jennifer Stow (University of Queensland, Brisbane, Australia; Addgene plasmid 28009; Miranda et al., 2001). Site-directed mutagenesis was conducted on the Dsg3-ΔEA-GFP backbone using QuikChange II (Agilent) to generate Dsg3-ΔEA-W2A using sense primer 5′-GGTACAAACGTGAAGCGGTGAAATTTGC-3′ and antisense primer 5′-GCAAATTTCACCGCTTCACGTTTGTACC-3′. Site-directed mutagenesis was conducted on the DP-mCherry backbone using QuikChange II to generate DP-S2849G-mCherry mutants using sense primer 5′-GGCGTCAAAGCCTCCTCTCCGGGACCC-3′ and antisense primer 5′-GGGTCCCGGAGAGGAGGCTTTGACGCC-3′. All constructs were verified by sequencing and demonstrated to be expressed at the expected molecular weight (Fig. S4).
Figure S4.
Tagged proteins are expressed at the expected molecular weights. Cells were transiently transfected with plasmids as indicated. 48 h after transfection, cells were harvested for SDS-PAGE and Western blot analysis. Each blot includes an untransfected control. (A) Cos7 cells transfected with GFP, Dsg3-ΔEA-GFP, or Dsg3-ΔEA-W2A-GFP blot were probed for Dsg3 or GFP. Tubulin was used as a loading control. (B) Cos7 cells transfected with mCherry, DP-mCherry, or DP-S2849-mCherry. Blot was probed for DP or mCherry with tubulin as a loading control. (C) A431 cells transfected with E-cad-GFP. Blot was probed with GFP and tubulin. (D) A431 cells transfected with PG-mEmerald. Blot was probed with GFP, and tubulin was used as a loading control. (E) A431 cells transfected with Dsg2-mCherry. Blot was probed with mCherry, and GAPDH was used as a loading control.
Transfection
HaCaT cells were transfected with Lipofectamine 3000 (Thermo Fisher Scientific) at 50% confluence following the manufacturer’s instructions. DNA concentration was optimized to 110 ng per 1 cm2. Cells were imaged in FluoroBrite DMEM (Thermo Fisher Scientific) supplemented with 10% FBS and 100 U/ml penicillin/streptomycin 40–48 h after transfection.
Inhibitor treatment
Gö6976 inhibitor (Abcam or Biovision) was diluted in DMSO to 50 µM and used at 1 µl/ml in medium to achieve a 50-nM final concentration. Mock treatment used 1 µl/ml of DMSO in medium. Cells were incubated with the inhibitor or DMSO for 1 h at 37°C and 5% CO2.
Calcium switch
Low-Ca2+ medium was generated by incubating FluoroBrite DMEM (Thermo Fisher Scientific) with Chelex resin (Bio-Rad) for 1 h, rotating at 4°C, followed by sterile filtration and supplementation with 3 mM EGTA (Millipore). For live-cell fluorescence polarization experiments, cells were switched from normal-Ca2+ FluoroBrite DMEM (∼1.8 mM) to low-Ca2+ medium while on the microscope stage. For all other experiments, the cells were incubated with low-Ca2+ medium at 37°C and 5% CO2 for 90 min while the treatment (mock or Gö6976) was maintained if applicable.
Dispase fragmentation assay
Cells were seeded, transfected (if applicable), and cultured to confluence in 12-well tissue culture plates. After drug and Ca2+ treatment (as described above), cells were incubated with 1 U/ml dispase (Sigma-Aldrich) for 30–45 min. The released monolayer was gently rinsed with PBS and subjected to mechanical stress by pipetting. Fragments were fixed in PFA and counted using a dissecting microscope.
EM
HaCaT cells were plated on 10-mm Transwell inserts with 0.4-µm pores (Corning). Once grown to 80% confluence, cells were either treated with 50 nM Gö6976 or mock treated for 1 h at 37°C. Cells were switched into either normal-Ca2+ medium (for 90 min) or low-Ca2+ medium (for 10 min [mock] or 90 min [Gö6976]) at 37°C. After treatment, cells were fixed with EM-grade fixative (2.0% glutaraldehyde in 0.2 M NaCacodylate, pH 7.2) postfixed in 0.2 M Na Cacodylate buffer containing 1% OsO4, washed in 0.1 M NaCacodylate buffer (3×), treated with 1% low molecular weight tannic acid in 0.05 M NaCacodylate for 30 min at RT, washed in 0.05 M NaCacodylate containing 1% NaSO4 for 5 min at RT, and dehydrated using a graded EtOH series. After dehydration, samples were embedded (Embed 812, Electron Microscopy Sciences), thin sectioned at 70–100 nm, and placed on formvar-coated copper mesh grids for transmission EM imaging. After drying, the sections were poststained with a 1:1 mixture of 1% uranyl acetate/EtOH for 5 min in the dark, followed by 5 min in Reynold’s lead citrate solution.
Images were acquired with a Tecnai Spirit T12 120-kV transmission electron microscope (FEI) operating at 80 kV at 42,000× nominal magnification on a BioSprint 29 charge-coupled device camera (AMT). Desmosomes in which both leaflets of the plasma membrane were clearly visible were used for quantification of intermembrane distance. Intermembrane distance values were determined by taking the average of five measurements made at different points along the length of each desmosome in ImageJ (National Institutes of Health).
Fluorescence polarization microscopy
Fluorescence polarization microscopy was conducted as described previously (Bartle et al., 2017). Experiments were conducted using a Nikon Ti-2 microscope, equipped with a motorized stage, stage-top incubator to maintain 37°C and 5% CO2 (Tokai Hit, INUBG2SF-TIZB), and 60× 1.49-NA objective. A cleanup polarizer, half-wave plate, and lens were used to polarize and focus a 488-nm laser (Coherent) at the microscope back focal plane (ThorLabs). A motorized mount (PRM1Z8; ThorLabs) controlled half-wave plate rotation and the orientation of the excitation polarization. Images were captured with an ORCA-Flash 4.0 v3 complementary metal–oxide–semiconductor camera (Hamamatsu). The system was controlled with Nikon Elements software. A series of four images were collected at 0°, 45°, 90°, and 135° excitation polarization for each order factor image.
Fluorescence polarization microscopy analysis
Fluorescence polarization images were analyzed using custom-built Polarized Order Detection Software (PODS) v1.0 for Matlab (MathWorks), as previously published (Bartle et al., 2017). Briefly, data images were corrected with the mean of three flatfield images acquired by imaging a green fluorescent slide (Chroma) to control for uneven laser illumination and polarization-dependent intensity changes. A binary mask was then generated on the average intensity image to identify desmosomes. Pixel-by-pixel order factor was calculated as previously described and displayed within the mask.
FRAP microscopy
FRAP experiments were conducted on a Nikon Ti-2 A1R confocal microscope, equipped with a motorized stage, stage-top incubator to maintain 37°C and 5% CO2 (Tokai Hit), and 60× 1.49-NA objective. Images were acquired in Nikon Elements at Nyquist resolution with 488- and 568-nm excitation lasers, gallium arsenide phosphide detectors, and 525/50 and 595/30 emission filters. Five prebleach images were acquired, and postbleach images were acquired every 2 min for 20 min.
FRAP analysis
Using Fiji, intensity was measured for the bleach, reference, photobleaching correction, and background regions for each cell at each time point. The bleach and reference intensities were corrected for background and photobleaching. GraphPad Prism (GraphPad Software) was used to fit the intensities to an exponential and calculate the mobile fraction and t1/2 for each bleach region. For population analysis, mean normalized intensity and SD were calculated at each time point after bleaching for n > 10 cells from three technical replicates. GraphPad Prism was used to fit the mean normalized intensity to an exponential and calculate the mobile fraction for the population. In the case of minimal recovery, where an exponential curve had an ambiguous fit, the recovery was fitted to a linear regression, and the mobile fraction (y) was calculated at x = 20 min.
Immunofluorescence staining
Fixation and labeling protocols were done as described previously (Stahley et al., 2016). Briefly, cells were preextracted for 60 s with 0.2% Triton X-100 and 300 mM sucrose in PBS with calcium and magnesium at 37°C. Cells were then fixed in 4% PFA prepared fresh from 16% EM-grade material (Electron Microscopy Sciences) for 12 min, followed by 30-min blocking and permeabilization with 5% normal horse serum, 5% normal goat serum, 1% BSA, and 0.25% Triton X-100, with multiple washes between steps. Cells were incubated in primary antibody for 3 h and secondary antibody for 1 h.
Cell lysate preparation, immunoblots, and antibodies
Cells were homogenized in radioimmunoprecipitation assay buffer containing SDS, NP-40 (EMD Millipore), and protease/phosphatase inhibitor (Complete, Roche). All lysates were clarified by centrifugation. Protein concentrations were measured by the bicinchoninic acid method (Pierce, Thermo Fisher Scientific). A quantity of 65 µg per sample was loaded in 8% SDS-polyacrylamide gels under denaturing conditions. The size-fractionated proteins were electroblotted to polyvinylidene difluoride (Bio-Rad) membranes. The membranes were treated with Intercept Blocking Buffer (LI-COR) for 1 h and incubated overnight with appropriate primary antibodies sequentially in Intercept Antibody Diluent (LI-COR). Membranes were washed and incubated with secondary antibodies (goat anti-rabbit RDye 800 CW and goat anti-mouse 680 LT; LI-COR) for 1 h and washed with TBS with Tween. Images were captured using the Odyssey CLx Imaging system (LI-COR). Primary antibodies were Dsg3 (clone 5G11, Thermo Fisher Scientific), DP I+II (ab127941, Abcam), DP 1&2 (CBL173, Millipore), DP pS2849 (600-401-J65, Rockland), Vinculin (V9131, Millipore Sigma), β-tubulin (2146, Cell Signaling Technology), GFP (ab290, Abcam), and mCherry (ab167453, Abcam).
Antibodies
Primary antibodies for microscopy were anti-DP (DPI/II, Bethyl Labs), anti-γ-catenin (H-80, Santa Cruz Biotechnology), and anti-Dsg3 (G194, Progen Biotechnik). Secondary antibodies were Alexa Fluor 488– and 647–conjugated goat anti-mouse or goat anti-rabbit (IgG H&L) from Invitrogen.
dSTORM
dSTORM experiments were conducted on a Nikon Ti-2 N-STORM microscope equipped with a 100× 1.49-NA oil-immersion objective, 488- and 647-nm lasers, and an iXon ultra electron-multiplying charge-coupled device camera (Andor). 60,000-100,000 frames were collected with subcritical inclined excitation and reconstructed in Nikon Elements. Photoswitching imaging buffer included glucose oxidase (Sigma-Aldrich), glucose (Sigma-Aldrich), catalase (Roche), and β-mercaptoethanol (Sigma-Aldrich; Rust et al., 2006).
dSTORM analysis
Reconstructed dSTORM images were rendered in Nikon Elements, exported at 4 nm/pixel, and saved as 16-bit TIFF images for analysis in Matlab as previously described (Stahley et al., 2016). Briefly, linescans were generated from intensity averaged along the length of the desmosome and then plotted across the desmosome axis. Linescans were normalized and smoothed, and the peak finder function was used to measure the peak-to-peak distance. Custom Matlab scripts for desmosome dSTORM processing are available upon request.
Statistics
All statistical analysis was performed with GraphPad Prism. P values were determined by Student’s t test when measuring significance between two conditions or by one-way ANOVA with multiple comparisons when measuring significance between more than two conditions. Curve fitting for FRAP analysis was done using one-phase association-fitting algorithms, or linear regression in the absence of recovery, where the plateau determined the mobile fraction.
Online supplemental material
Fig. S1 contains controls for fluorescence polarization microscopy. Fig. S2 has representative images of Dsg2-mCherry–expressing cells showing that exchange is reduced in hyperadhesion. Fig. S3 shows that Dsg3 order factor is not impacted by cotransfection with DP-S2849G-mCherry. Fig. S4 contains blots showing expression of tagged proteins. Table S1 is a summary of all Dsg3-ΔEA-GFP order factors, and Table S2 is a summary of all mobile fractions.
Supplementary Material
is a summary of Dsg3-ΔEA-GFP order factors.
is a summary of mobile fractions.
Acknowledgments
This work was supported by funding to A.L. Mattheyses from the National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01AR072697) and the National Science Foundation Career award (1832100).
The authors declare no competing financial interests.
Author contributions: E.I Bartle carried out the majority of experiments and data analysis. T.C. Rao carried out the biochemical experiments and analysis, generated reagents, and provided cell culture support. R.R. Beggs and W.F. Dean carried out additional key experiments and analysis. T.M. Urner collected data, developed dSTORM and fluorescence polarization analysis methods, and provided critical discussions of the results. A.P. Kowalczyk provided critical discussions of the results and experimental design. E.I. Bartle and A.L. Mattheyses conceived and designed the study, interpreted results, and wrote the manuscript. All authors provided intellectual input, revised, edited, and approved the manuscript.
References
- Al-Amoudi A., Díez D.C., Betts M.J., and Frangakis A.S.. 2007. The molecular architecture of cadherins in native epidermal desmosomes. Nature. 450:832–837. 10.1038/nature05994 [DOI] [PubMed] [Google Scholar]
- Al-Amoudi A., Dubochet J., and Norlén L.. 2005. Nanostructure of the epidermal extracellular space as observed by cryo-electron microscopy of vitreous sections of human skin. J. Invest. Dermatol. 124:764–777. 10.1111/j.0022-202X.2005.23630.x [DOI] [PubMed] [Google Scholar]
- Albrecht L.V., Zhang L., Shabanowitz J., Purevjav E., Towbin J.A., Hunt D.F., and Green K.J.. 2015. GSK3- and PRMT-1-dependent modifications of desmoplakin control desmoplakin-cytoskeleton dynamics. J. Cell Biol. 208:597–612. 10.1083/jcb.201406020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartle E.I., Urner T.M., Raju S.S., and Mattheyses A.L.. 2017. Desmoglein 3 Order and Dynamics in Desmosomes Determined by Fluorescence Polarization Microscopy. Biophys. J. 113:2519–2529. 10.1016/j.bpj.2017.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass-Zubek A.E., Hobbs R.P., Amargo E.V., Garcia N.J., Hsieh S.N., Chen X., Wahl J.K. III, Denning M.F., and Green K.J.. 2008. Plakophilin 2: a critical scaffold for PKC alpha that regulates intercellular junction assembly. J. Cell Biol. 181:605–613. 10.1083/jcb.200712133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertocchi C., Wang Y., Ravasio A., Hara Y., Wu Y., Sailov T., Baird M.A., Davidson M.W., Zaidel-Bar R., Toyama Y., et al. 2017. Nanoscale architecture of cadherin-based cell adhesions. Nat. Cell Biol. 19:28–37. 10.1038/ncb3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canel M., Serrels A., Miller D., Timpson P., Serrels B., Frame M.C., and Brunton V.G.. 2010. Quantitative in vivo imaging of the effects of inhibiting integrin signaling via Src and FAK on cancer cell movement: effects on E-cadherin dynamics. Cancer Res. 70:9413–9422. 10.1158/0008-5472.CAN-10-1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., and Schnittler H.. 2019. Putting VE-cadherin into JAIL for junction remodeling. J. Cell Sci. 132 jcs222893 10.1242/jcs.222893 [DOI] [PubMed] [Google Scholar]
- Daigle N., Beaudouin J., Hartnell L., Imreh G., Hallberg E., Lippincott-Schwartz J., and Ellenberg J.. 2001. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 154:71–84. 10.1083/jcb.200101089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMay B.S., Noda N., Gladfelter A.S., and Oldenbourg R.. 2011. Rapid and quantitative imaging of excitation polarized fluorescence reveals ordered septin dynamics in live yeast. Biophys. J. 101:985–994. 10.1016/j.bpj.2011.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erami Z., Herrmann D., Warren S.C., Nobis M., McGhee E.J., Lucas M.C., Leung W., Reischmann N., Mrowinska A., Schwarz J.P., et al. 2016. Intravital FRAP Imaging using an E-cadherin-GFP Mouse Reveals Disease- and Drug-Dependent Dynamic Regulation of Cell-Cell Junctions in Live Tissue. Cell Rep. 14:152–167. 10.1016/j.celrep.2015.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote H.P., Sumigray K.D., and Lechler T.. 2013. FRAP analysis reveals stabilization of adhesion structures in the epidermis compared to cultured keratinocytes. PLoS One. 8 e71491 10.1371/journal.pone.0071491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M.A., Nelson W.J., and Chavez N.. 2018. Cell-Cell Junctions Organize Structural and Signaling Networks. Cold Spring Harb. Perspect. Biol. 10 a029181 10.1101/cshperspect.a029181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrod D. 2010. Desmosomes in vivo. Dermatol. Res. Pract. 2010 212439 10.1155/2010/212439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrod D., and Kimura T.E.. 2008. Hyper-adhesion: a new concept in cell-cell adhesion. Biochem. Soc. Trans. 36:195–201. 10.1042/BST0360195 [DOI] [PubMed] [Google Scholar]
- Garrod D.R. 2013. The assay that defines desmosome hyper-adhesion. J. Invest. Dermatol. 133:576–577. 10.1038/jid.2012.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrod D.R., Berika M.Y., Bardsley W.F., Holmes D., and Tabernero L.. 2005. Hyper-adhesion in desmosomes: its regulation in wound healing and possible relationship to cadherin crystal structure. J. Cell Sci. 118:5743–5754. 10.1242/jcs.02700 [DOI] [PubMed] [Google Scholar]
- Gloushankova N.A., Wakatsuki T., Troyanovsky R.B., Elson E., and Troyanovsky S.M.. 2003. Continual assembly of desmosomes within stable intercellular contacts of epithelial A-431 cells. Cell Tissue Res. 314:399–410. 10.1007/s00441-003-0812-3 [DOI] [PubMed] [Google Scholar]
- Godsel L.M., Hsieh S.N., Amargo E.V., Bass A.E., Pascoe-McGillicuddy L.T., Huen A.C., Thorne M.E., Gaudry C.A., Park J.K., Myung K., et al. 2005. Desmoplakin assembly dynamics in four dimensions: multiple phases differentially regulated by intermediate filaments and actin. J. Cell Biol. 171:1045–1059. 10.1083/jcb.200510038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough D.A., and Paul D.L.. 2009. Gap junctions. Cold Spring Harb. Perspect. Biol. 1 a002576 10.1101/cshperspect.a002576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis E.R., Xu S., and Powers M.A.. 2003. Nup98 localizes to both nuclear and cytoplasmic sides of the nuclear pore and binds to two distinct nucleoporin subcomplexes. Mol. Biol. Cell. 14:600–610. 10.1091/mbc.e02-09-0582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon R.M., and Green K.J.. 2013. Structural and functional diversity of desmosomes. Cell Commun. Adhes. 20:171–187. 10.3109/15419061.2013.855204 [DOI] [PubMed] [Google Scholar]
- Harrison O.J., Brasch J., Lasso G., Katsamba P.S., Ahlsen G., Honig B., and Shapiro L.. 2016. Structural basis of adhesive binding by desmocollins and desmogleins. Proc. Natl. Acad. Sci. USA. 113:7160–7165. 10.1073/pnas.1606272113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häussinger D., Ahrens T., Aberle T., Engel J., Stetefeld J., and Grzesiek S.. 2004. Proteolytic E-cadherin activation followed by solution NMR and X-ray crystallography. EMBO J. 23:1699–1708. 10.1038/sj.emboj.7600192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Cowin P., and Stokes D.L.. 2003. Untangling desmosomal knots with electron tomography. Science. 302:109–113. 10.1126/science.1086957 [DOI] [PubMed] [Google Scholar]
- Heilemann M., van de Linde S., Schüttpelz M., Kasper R., Seefeldt B., Mukherjee A., Tinnefeld P., and Sauer M.. 2008. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. Engl. 47:6172–6176. 10.1002/anie.200802376 [DOI] [PubMed] [Google Scholar]
- Hobbs R.P., and Green K.J.. 2012. Desmoplakin regulates desmosome hyperadhesion. J. Invest. Dermatol. 132:482–485. 10.1038/jid.2011.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchanawong P., Shtengel G., Pasapera A.M., Ramko E.B., Davidson M.W., Hess H.F., and Waterman C.M.. 2010. Nanoscale architecture of integrin-based cell adhesions. Nature. 468:580–584. 10.1038/nature09621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann R., Piontek J., Grüll F., Kirchgessner M., Rossa J., Wolburg H., Blasig I.E., and Cremer C.. 2012. Visualization and quantitative analysis of reconstituted tight junctions using localization microscopy. PLoS One. 7 e31128 10.1371/journal.pone.0031128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T.E., Merritt A.J., and Garrod D.R.. 2007. Calcium-independent desmosomes of keratinocytes are hyper-adhesive. J. Invest. Dermatol. 127:775–781. 10.1038/sj.jid.5700643 [DOI] [PubMed] [Google Scholar]
- Kimura T.E., Merritt A.J., Lock F.R., Eckert J.J., Fleming T.P., and Garrod D.R.. 2012. Desmosomal adhesiveness is developmentally regulated in the mouse embryo and modulated during trophectoderm migration. Dev. Biol. 369:286–297. 10.1016/j.ydbio.2012.06.025 [DOI] [PubMed] [Google Scholar]
- Kowalczyk A.P., Bornslaeger E.A., Borgwardt J.E., Palka H.L., Dhaliwal A.S., Corcoran C.M., Denning M.F., and Green K.J.. 1997. The amino-terminal domain of desmoplakin binds to plakoglobin and clusters desmosomal cadherin-plakoglobin complexes. J. Cell Biol. 139:773–784. 10.1083/jcb.139.3.773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk A.P., Palka H.L., Luu H.H., Nilles L.A., Anderson J.E., Wheelock M.J., and Green K.J.. 1994. Posttranslational regulation of plakoglobin expression. Influence of the desmosomal cadherins on plakoglobin metabolic stability. J. Biol. Chem. 269:31214–31223. [PubMed] [Google Scholar]
- Kress A., Ferrand P., Rigneault H., Trombik T., He H.T., Marguet D., and Brasselet S.. 2011. Probing orientational behavior of MHC class I protein and lipid probes in cell membranes by fluorescence polarization-resolved imaging. Biophys. J. 101:468–476. 10.1016/j.bpj.2011.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger C., Loschke F., Schwarz N., Windoffer R., Leube R.E., and Magin T.M.. 2013. Keratins control intercellular adhesion involving PKC-α-mediated desmoplakin phosphorylation. J. Cell Biol. 201:681–692. 10.1083/jcb.201208162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowndes M., Rakshit S., Shafraz O., Borghi N., Harmon R.M., Green K.J., Sivasankar S., and Nelson W.J.. 2014. Different roles of cadherins in the assembly and structural integrity of the desmosome complex. J. Cell Sci. 127:2339–2350. 10.1242/jcs.146316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S.B., McQuilken M., La Riviere P.J., Occhipinti P., Verma A., Oldenbourg R., Gladfelter A.S., and Tani T.. 2016. Dissection of molecular assembly dynamics by tracking orientation and position of single molecules in live cells. Proc. Natl. Acad. Sci. USA. 113:E6352–E6361. 10.1073/pnas.1607674113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J.J., Bornslaeger E.A., Green K.J., Steinert P.M., and Ip W.. 1997. Two-hybrid analysis reveals fundamental differences in direct interactions between desmoplakin and cell type-specific intermediate filaments. J. Biol. Chem. 272:21495–21503. 10.1074/jbc.272.34.21495 [DOI] [PubMed] [Google Scholar]
- Miranda K.C., Khromykh T., Christy P., Le T.L., Gottardi C.J., Yap A.S., Stow J.L., and Teasdale R.D.. 2001. A dileucine motif targets E-cadherin to the basolateral cell surface in Madin-Darby canine kidney and LLC-PK1 epithelial cells. J. Biol. Chem. 276:22565–22572. 10.1074/jbc.M101907200 [DOI] [PubMed] [Google Scholar]
- Nahidiazar L., Kreft M., van den Broek B., Secades P., Manders E.M.M., Sonnenberg A., and Jalink K.. 2015. The molecular architecture of hemidesmosomes, as revealed with super-resolution microscopy. J. Cell Sci. 128:3714–3719. 10.1242/jcs.171892 [DOI] [PubMed] [Google Scholar]
- Nanes B.A., Chiasson-MacKenzie C., Lowery A.M., Ishiyama N., Faundez V., Ikura M., Vincent P.A., and Kowalczyk A.P.. 2012. p120-catenin binding masks an endocytic signal conserved in classical cadherins. J. Cell Biol. 199:365–380. 10.1083/jcb.201205029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overduin M., Harvey T.S., Bagby S., Tong K.I., Yau P., Takeichi M., and Ikura M.. 1995. Solution structure of the epithelial cadherin domain responsible for selective cell adhesion. Science. 267:386–389. 10.1126/science.7824937 [DOI] [PubMed] [Google Scholar]
- Owen G.R., Acehan D., Derr K.D., Rice W.J., and Stokes D.L.. 2008. Cryoelectron tomography of isolated desmosomes. Biochem. Soc. Trans. 36:173–179. 10.1042/BST0360173 [DOI] [PubMed] [Google Scholar]
- Parsons J.T., Horwitz A.R., and Schwartz M.A.. 2010. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11:633–643. 10.1038/nrm2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokutta S., Herrenknecht K., Kemler R., and Engel J.. 1994. Conformational changes of the recombinant extracellular domain of E-cadherin upon calcium binding. Eur. J. Biochem. 223:1019–1026. 10.1111/j.1432-1033.1994.tb19080.x [DOI] [PubMed] [Google Scholar]
- Ray P.S., Arif A., and Fox P.L.. 2007. Macromolecular complexes as depots for releasable regulatory proteins. Trends Biochem. Sci. 32:158–164. 10.1016/j.tibs.2007.02.003 [DOI] [PubMed] [Google Scholar]
- Rayns D.G., Simpson F.O., and Ledingham J.M.. 1969. Ultrastructure of desmosomes in mammalian intercalated disc; appearances after lanthanum treatment. J. Cell Biol. 42:322–326. 10.1083/jcb.42.1.322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocheleau J.V., Edidin M., and Piston D.W.. 2003. Intrasequence GFP in class I MHC molecules, a rigid probe for fluorescence anisotropy measurements of the membrane environment. Biophys. J. 84:4078–4086. 10.1016/S0006-3495(03)75133-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rübsam M., Broussard J.A., Wickström S.A., Nekrasova O., Green K.J., and Niessen C.M.. 2018. Adherens Junctions and Desmosomes Coordinate Mechanics and Signaling to Orchestrate Tissue Morphogenesis and Function: An Evolutionary Perspective. Cold Spring Harb. Perspect. Biol. 10 a029207 10.1101/cshperspect.a029207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust M.J., Bates M., and Zhuang X.. 2006. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods. 3:793–795. 10.1038/nmeth929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L., Fannon A.M., Kwong P.D., Thompson A., Lehmann M.S., Grübel G., Legrand J.F., Als-Nielsen J., Colman D.R., and Hendrickson W.A.. 1995. Structural basis of cell-cell adhesion by cadherins. Nature. 374:327–337. 10.1038/374327a0 [DOI] [PubMed] [Google Scholar]
- Shimizu A., Ishiko A., Ota T., Saito H., Oka H., Tsunoda K., Amagai M., and Nishikawa T.. 2005. In vivo ultrastructural localization of the desmoglein 3 adhesive interface to the desmosome mid-line. J. Invest. Dermatol. 124:984–989. 10.1111/j.0022-202X.2005.23706.x [DOI] [PubMed] [Google Scholar]
- Sotomayor M., and Schulten K.. 2008. The allosteric role of the Ca2+ switch in adhesion and elasticity of C-cadherin. Biophys. J. 94:4621–4633. 10.1529/biophysj.107.125591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahley S.N., Bartle E.I., Atkinson C.E., Kowalczyk A.P., and Mattheyses A.L.. 2016. Molecular organization of the desmosome as revealed by direct stochastic optical reconstruction microscopy. J. Cell Sci. 129:2897–2904. 10.1242/jcs.185785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan V., Kalappurakkal J.M., Mehta S.B., Nordenfelt P., Moore T.I., Koga N., Baker D.A., Oldenbourg R., Tani T., Mayor S., et al. 2017. Actin retrograde flow actively aligns and orients ligand-engaged integrins in focal adhesions. Proc. Natl. Acad. Sci. USA. 114:10648–10653. 10.1073/pnas.1701136114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq H., Bella J., Jowitt T.A., Holmes D.F., Rouhi M., Nie Z., Baldock C., Garrod D., and Tabernero L.. 2015. Cadherin flexibility provides a key difference between desmosomes and adherens junctions. Proc. Natl. Acad. Sci. USA. 112:5395–5400. 10.1073/pnas.1420508112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason H.A., Cooper N.H., Ansell D.M., Chiu M., Merrit A.J., Hardman M.J., and Garrod D.R.. 2012. Direct evidence that PKCα positively regulates wound re-epithelialization: correlation with changes in desmosomal adhesiveness. J. Pathol. 227:346–356. 10.1002/path.4016 [DOI] [PubMed] [Google Scholar]
- Valades Cruz C.A., Shaban H.A., Kress A., Bertaux N., Monneret S., Mavrakis M., Savatier J., and Brasselet S.. 2016. Quantitative nanoscale imaging of orientational order in biological filaments by polarized superresolution microscopy. Proc. Natl. Acad. Sci. USA. 113:E820–E828. 10.1073/pnas.1516811113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielmuth F., Wanuske M.T., Radeva M.Y., Hiermaier M., Kugelmann D., Walter E., Buechau F., Magin T.M., Waschke J., and Spindler V.. 2018. Keratins Regulate the Adhesive Properties of Desmosomal Cadherins through Signaling. J. Invest. Dermatol. 138:121–131. 10.1016/j.jid.2017.08.033 [DOI] [PubMed] [Google Scholar]
- Vrabioiu A.M., and Mitchison T.J.. 2006. Structural insights into yeast septin organization from polarized fluorescence microscopy. Nature. 443:466–469. 10.1038/nature05109 [DOI] [PubMed] [Google Scholar]
- Wallis S., Lloyd S., Wise I., Ireland G., Fleming T.P., and Garrod D.. 2000. The alpha isoform of protein kinase C is involved in signaling the response of desmosomes to wounding in cultured epithelial cells. Mol. Biol. Cell. 11:1077–1092. 10.1091/mbc.11.3.1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windoffer R., Borchert-Stuhlträger M., and Leube R.E.. 2002. Desmosomes: interconnected calcium-dependent structures of remarkable stability with significant integral membrane protein turnover. J. Cell Sci. 115:1717–1732. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
is a summary of Dsg3-ΔEA-GFP order factors.
is a summary of mobile fractions.