This Viewpoint proposes a simple way to highlight both experimental reproducibility and cell-to-cell variation, while avoiding pitfalls common in analysis of cell biology data.
Abstract
P values and error bars help readers infer whether a reported difference would likely recur, with the sample size n used for statistical tests representing biological replicates, independent measurements of the population from separate experiments. We provide examples and practical tutorials for creating figures that communicate both the cell-level variability and the experimental reproducibility.
Introduction
While far from perfect, the P value offers a pragmatic metric to infer whether an observed difference is reproducible and substantial relative to the noise in the measurements (Greenwald et al., 1996). The P value should be treated as a mere heuristic, interpreted as the degree of compatibility between the observed dataset and a given statistical model. A P value reports the probability that the observed data—or any more extreme values—would occur by chance (the “null hypothesis”). But a small P value does not actually tell us which assumption is incorrect, the null hypothesis or some other assumption of the statistical model (e.g., normal distribution, random sampling, equal variance, etc.). In the case of treating each cell as an n, the assumption that is violated is independent sampling, not necessarily the null hypothesis. The resulting P values are worse than useless: counting each cell as a separate n can easily result in false-positive rates of >50% (Aarts et al., 2015). For excellent practical guides to statistics for cell biologists, readers are referred to Lamb et al, (2008) and Pollard et al. (2019). In this paper, we specifically address simple ways to communicate reproducibility when performing statistical tests and plotting data.
Error bars and P values are often used to assure readers of a real and persistent difference between populations or treatments. P values are based on the difference between population means (or other summary metrics) as well as the number of measurements used to determine that difference. In general, increasing the number of measurements decreases the resulting P value. To convey experimental reproducibility, P values and standard error of the mean should be calculated using biological replicates—independent measurements of a population of interest, typically from independent samples or separate experiments (Hurlbert, 1984; Lazic, 2010; Vaux et al., 2012; Aarts et al., 2015; Naegle et al., 2015; Lazic et al., 2018). Limited time and resources often constrain cell biologists to repeat any particular experiment only a handful of times, so a typical sample size n is often in the single digits. However, if authors assign n as the number of cells observed during the experiment, n may be on the order of hundreds or thousands, resulting in small P values and error bars that do not convey the experimental reproducibility or the cell-level variability.
For example, if a researcher measures the length of 20 neurons in a zebrafish and 20 neurons in a fish exposed to a toxin, the correct n for each condition is 1, because the toxin exposure was only performed once. Without repeating the treatment multiple times with multiple fish, there is no way to know whether any observed difference was from the toxin or due to natural or otherwise uncontrolled differences between those two individual fish. The reader does not care that those two particular fish are different, but that treatments result in a consistent difference across multiple fish. The P value should be calculated to reflect the latter, not the former.
Well-designed studies embrace both cell-to-cell and sample-to-sample variation (Altman and Krzywinski, 2015). Repeatedly quantifying a biological parameter rarely converges on a single “true” value, due to the complexity of living cells or because many biological processes are intrinsically stochastic. Calculating standard error from thousands of cells conceals this expected variability. We have written this tutorial to help cell biologists plot data in a way that highlights both experimental robustness and cell-to-cell variability. Specifically, we propose the use of distribution–reproducibility “SuperPlots” that display the distribution of the entire dataset, and report statistics (such as means, error bars, and P values) that address the reproducibility of the findings.
What population is being sampled?
To clarify what your sample size n should be, ask yourself: What population are you trying to sample? The choice of n determines the population being evaluated or compared (Naegle et al., 2015; Lazic et al., 2018; Pollard et al., 2019). A typical cell biology experiment strives to draw general conclusions about an entire population of cells, so the sample selection should reflect the breadth of that population. For example, to test if a treatment changes the speed of crawling cells, you could split a flask of lymphocytes into two wells, treat one well with a drug of interest and one with a placebo, and then track individual cells in each of the two wells. If you use each cell as a sample (n = number of cells), the two populations you end up comparing are the cells in those two particular wells. Multiple observations within one well increase the precision for estimating the mean for that one sample, but do not reveal a truth about all cells in all wells. By repeating the experiment multiple times from new flasks, and using each experiment as a sample (n = number of independent experiments), you evaluate the effect of the treatment on any arbitrary flask of similar cells. (For more examples, see Table S1.)
If you are interested only in cell-to-cell variability within a particular sample, then n could be the number of cells observed. However, making inferences beyond that sample is difficult, because the natural variability of individual cells can be overshadowed by systematic differences between biological replicates. Whether caused by passage number, confluency, or location in the incubator, cells often vary from sample to sample and day to day. For example, an entire flask of cells can be described as “unhappy.” Accordingly, cells from experimental and control samples (e.g., tubes, flasks, wells, coverslips, rats, tissue samples, etc.) may differ from each other, regardless of the experimental treatment. When authors report the sample size as the number of cells, the statistical analysis cannot help the reader evaluate whether differences are due to the intended treatment or sample-to-sample variability. We are not prescribing any specific definition of n; researchers should consider what main source of variability they hope to overcome when designing experiments and statistical analyses (Altman and Krzywinski, 2015).
Statistics in cell biology typically assume independent tests of a hypothesis
Analysis becomes challenging when the experimental unit—the item that can be randomly assigned to a treatment—is different than the biological entity of interest. For example, we often care about how individual cells react to a treatment, but typically treat entire dishes of cells at a time. To test the hypothesis that two treatments or populations are different, the treatment must be applied or the populations sampled multiple times. Neighboring cells within one flask or well treated with a drug are not separate tests of the hypothesis, because the treatment was only applied once. But if individual cells are microinjected with a drug or otherwise randomly assigned to a different treatment, then each cell really can be a separate test of a hypothesis.
Finding truly independent groups and deciding what makes for a good biological replicate can be challenging (Vaux et al., 2012; Blainey et al., 2014; Naegle et al., 2015; Lazic et al., 2018). For example, is it acceptable to run multiple experiments from just one thawed aliquot of cells? Is it necessary to generate multiple knockout strains? Is it sufficient to test in one cell line? There’s no single right answer: each researcher must balance practicality with robust experimental design. At a minimum, researchers must perform an experiment multiple times if they want to know whether the results are robust.
Calculating P values from cell-level observations
Cell biologists often observe hundreds of cells per experiment and repeat an experiment multiple times. To leverage that work into robust statistics, one needs to take into account the hierarchy of the data. Combining the cell-level data from multiple independent experiments squanders useful information about run-to-run variability (Fig. 1). There is ample literature about the analysis of this type of hierarchical data (Galbraith et al., 2010), which takes into account both the variance within a sample and the clustering across multiple experimental runs (Aarts et al., 2015), or that propagate the error up the chain, such as a nested ANOVA (Krzywinski et al., 2014). Recently, statisticians have proposed a Bayesian approach to multilevel analysis (Lazic et al., 2020). For a detailed resource on hierarchical data analysis, see Gelman and Hill (2006).
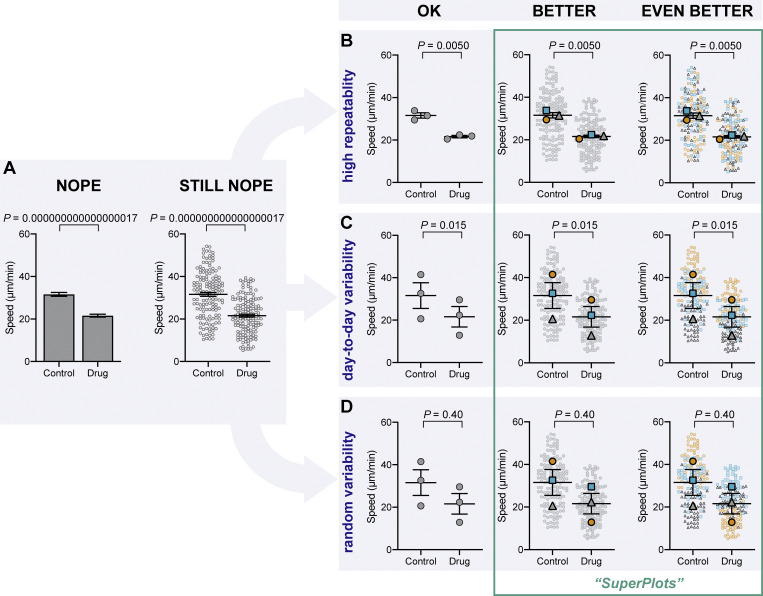
Figure 1.
The importance of displaying reproducibility. Drastically different experimental outcomes can result in the same plots and statistics unless experiment-to-experiment variability is considered. (A) Problematic plots treat n as the number of cells, resulting in tiny error bars and P values. These plots also conceal any systematic run-to-run error, mixing it with cell-to-cell variability. (B–D) To illustrate this, we simulated three different scenarios that all have identical underlying cell-level values but are clustered differently by experiment: B shows highly repeatable, unclustered data, C shows day-to-day variability, but a consistent trend in each experiment, and D is dominated by one random run. Note that the plots in A that treat each cell as its own n fail to distinguish the three scenarios, claiming a significant difference after drug treatment, even when the experiments are not actually repeatable. To correct that, “SuperPlots” superimpose summary statistics from biological replicates consisting of independent experiments on top of data from all cells, and P values were calculated using an n of three, not 300. In this case, the cell-level values were separately pooled for each biological replicate and the mean calculated for each pool; those three means were then used to calculate the average (horizontal bar), standard error of the mean (error bars), and P value. While the dot plots in the “OK” column ensure that the P values are calculated correctly, they still fail to convey the experiment-to-experiment differences. In the SuperPlots, each biological replicate is color-coded: the averages from one experimental run are yellow dots, another independent experiment is represented by gray triangles, and a third experiment is shown as blue squares. This helps convey whether the trend is observed within each experimental run, as well as for the dataset as a whole. The beeswarm SuperPlots in the rightmost column represent each cell with a dot that is color-coded according to the biological replicate it came from. The P values represent an unpaired two-tailed t test (A) and a paired two-tailed t test (B–D). For tutorials on making SuperPlots in Prism, R, Python, and Excel, see the supporting information.
A simple approach—which permits conventional t test or ANOVA calculations—is to pool the cell-level data from each experiment separately and compare the subsequent sample-level means (Altman and Bland, 1997; Galbraith et al., 2010; Lazic, 2010). For example, if you have three biological replicates of control and treated samples, and you measure the cell diameter of 200 cells in each sample, first calculate the mean of those 200 measurements for each sample, then run a t test on those sample means (three control, three treated). When using this simplified method, it is best to keep the number of observations per sample similar, because each sample gets the same weighting in the analysis.
While pooling dependent observations together avoids false positives (Galbraith et al., 2010; Aarts et al., 2015), this simple approach might fail to detect small but real differences between groups, where more advanced techniques may prove to be more powerful. However, increasing the number of biological replicates usually has a larger influence on the statistical power than measuring many more cells in each sample (Blainey et al., 2014; Aarts et al., 2015). While n of 3 is often considered a pragmatic minimum in cell biology (Naegle et al., 2015), distinguishing more subtle observed differences will require planning for more biological replicates and/or harnessing the power of more robust statistical analyses.
Communicating variability with SuperPlots
After analyzing hundreds of cells across multiple rounds of experimentation, it would be useful to incorporate both the cell-level variability and experimental repeatability into a single diagram. In Fig. 1 A, the plots have small error bars and P values, which should raise red flags given how difficult it would be to replicate a cell biology experiment with identical results and/or to repeat it hundreds of times, which such miniscule P values imply. Bar graphs are problematic because they obscure the distribution of cell-level data as well as the sample-to-sample repeatability (Weissgerber et al., 2015). While beeswarm, box-and-whisker, and violin plots are great at conveying information about the range and distribution of the underlying data, plotting the entire dataset does not make it appropriate to treat repeated measurements on the same sample as independent experiments.
Therefore, we suggest authors incorporate information about distribution and reproducibility by creating “SuperPlots,” which superimpose summary statistics from repeated experiments on a graph of the entire cell-level dataset (Fig. 1, right columns). SuperPlots convey more information than a conventional bar graph or beeswarm plot, and they make it clear that statistical analyses (e.g., error bars and P values) are calculated across separate experiments, not individual cells—even when each cell is represented on the plot. For example, the mean from each experiment could be listed in the caption or plotted as a larger dot on top of the many smaller dots that denote individual measurements.
When possible, it is best to link samples by run, for instance, by color-coding the dots by experiment or a line linking paired measurements together (Fig. S1 D). These linkages convey the repeatability of the work: readers learn more if they know that one experiment exhibited high readings across the board than if they have to guess the trend in each sample. Linking data can also eliminate the need to normalize data in order to directly compare different experimental runs. Often, multiple experiments might all exhibit the same trend, but different absolute values (Fig. 1 C). By encoding the biological replicate into the data, such trends can be revealed without normalizing to a control group: P values can then be calculated using statistical tests that take into account linkages among samples (e.g., a paired or ratio t test). In fact, not taking into account linkages can make the t test too conservative, yielding false negatives (Galbraith et al., 2010).
Figure S1.
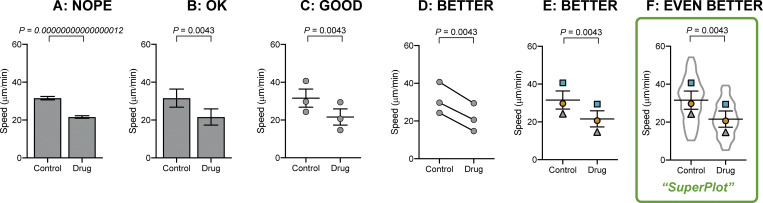
Other plotting examples. Bar plots can be enhanced even without using beeswarm plots. (A) Bar plots that calculate P and standard error of the mean using the number of cells as n are unhelpful. (B) A bar graph can be corrected by using biological replicates to calculate P value and standard error of the mean. (C) Dot plots reveal more than a simple bar graph. (D and E) Linking each pair by the replicate conveys important information about the trend in each experiment. (F) A SuperPlot not only shows information about each replicate and the trends, but also superimposes the distribution of the cell-level data, here using a violin plot.
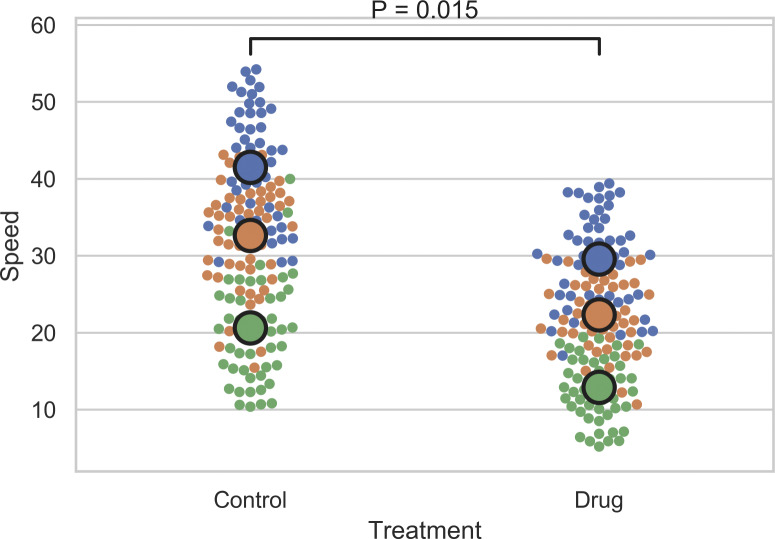
An impressive amount of information can be depicted by color-coded beeswarm SuperPlots (see Fig. 1, rightmost plots), where each cell-level datapoint divulges which experiment it came from (Galbraith et al., 2010; Weissgerber et al., 2017). This helps convey to the reader whether each experimental round gave similar results or if one run biases the conclusion (Fig. 1 D). The summary statistics and P values in beeswarm SuperPlots are overlaid on the color-coded scatter. (See Fig. S2, Fig. S3, Fig. S4, and Fig. S5 for tutorials on how to make beeswarm SuperPlots in Prism, Python, R, and Excel using Data S1.)
Figure S2.
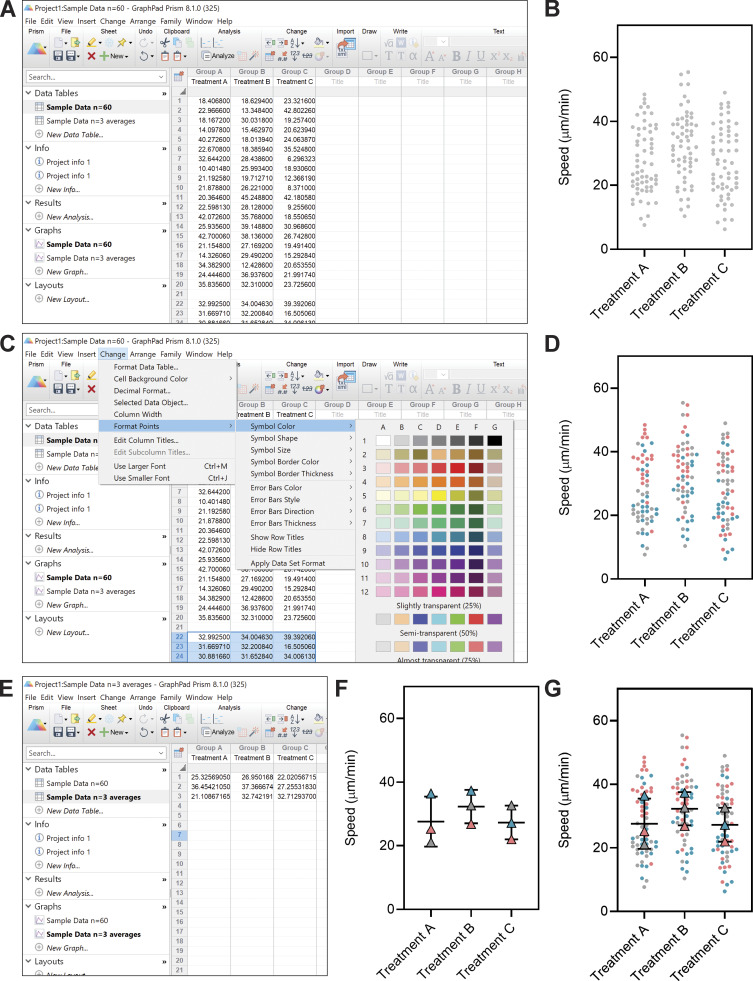
Tutorial for making SuperPlots in Prism. We describe how to make SuperPlots in GraphPad Prism 8 (version 8.1.0) graphing software. If using other graphing software, one may create a separate, different colored plot for each replicate, then overlay those plots in software like Adobe Illustrator. (A) When adding data to the table, leave a blank row between replicates. (B) Create a new graph of this existing data; under type of graph select “Column” and “Individual values,” and select “No line or error bar.” (C) After formatting the universal features of plot from B (e.g., symbol size, font, axes), go back to the data table and highlight the data values that correspond to one of the replicates. Under the “Change” menu, select “Format Points” and change the color, shape, etc. of the subset of points that correspond to that replicate. (D) Repeat for the other replicates to produce a graph with each trial color coded. (E and F) To display summary statistics, take the average of the technical replicates in each biological replicate (so you will have one value for each condition from each biological replicate), and enter those averages into another data table and graph. Use this data sheet that contains only the averages to run statistical tests. (G) To make a plot that combines the full dataset with the correct summary statistics, format this graph and overlay it with the above scatter SuperPlots (in Prism, this can be done on a “Layout”). This process could be tweaked to display other overlaid, color-coded plots (e.g., violin).
Figure S3.
Tutorial for making SuperPlots in Excel. (A) To make a SuperPlot using Excel (Microsoft Office 365 ProPlus for Windows; version 1912; Build 12325.20172), enter the values for the first replicate for the first condition into column B (highlighted in yellow), the second condition into column D (highlighted in yellow), and continue to skip columns between datasets for the remaining conditions and replicates (in this example, replicate 2 is highlighted in green and replicate 3 is in orange). For example, “Treatment A” could be control cells and “Treatment B” could be drug-treated cells. Label the empty columns as “x” and, starting with column A, enter random values to generate the scatter effect by using the formula “=RANDBETWEEN(25, 100)”. To create a gap between the datasets A and B, use larger X values for treatment B by entering the formula “=RANDBETWEEN(225, 300)”. (B) Highlight all the data and headings. In the insert menu, expand the charts menu to open the “Insert Chart” dialog box. Select “All Charts,” and choose “X Y Scatter.” Select the option that has Y values corresponding to your datasets. (In Excel for Mac, there is not a separate dialog box. Instead, make a scatter plot, right click on the plot and select “Select Data,” remove the “x” columns from the list, then manually select the corresponding “X values =” for each dataset.) (C) Change the general properties of the graph to your liking. In this example, we removed the chart title and the gridlines, added a black outline to the chart area, resized the graph, adjusted the x axis range to 0–325, removed the x axis labels, added a y axis title and tick marks, changed the font to Arial, and changed the font color to black. This style can be saved as a template for future use by right clicking. We recommend keeping the figure legend until the next step. (D) Next, double click the graph to open the “Format Plot Area” panel. Under “Chart Options,” select your first dataset, “Series Treatment A (replicate 1).” (On a Mac, click on a datapoint from one of the replicates, right click and select “Format Data Series.”) Select “Marker” and change the color and style of the data points. Repeat with the remaining datasets so that the colors, shapes, etc. correspond to the biological replicate the data points came from. Delete the chart legend and add axis labels with the text tool if desired. (E) Calculate the average for each replicate for each condition, and pair this value with the X coordinate of 62.5 for the first treatment, and 262.5 for the second treatment to center the values in the scatterplot. Then, click the graph, and under the “Chart Design” menu, click “Select Data.” Under “Legend Entries (Series),” select “Add” and under series name, select the three trial names, then select all three X and Y values for first treatment condition for “Series X Values” and “Series Y Values,” respectively. Repeat for the second treatment condition, and hit “OK.” (F) On the chart, select the data point corresponding to the first average and double click to isolate the data point. Format the size, color, etc. and repeat for remaining data points. (G) Optional: To add an average and error bars, either generate a second graph and overlay the data, or calculate the average and standard deviation using Excel and add the data series to the graph as was done in E and F, using the “-” symbol for the data point.
Figure S4.
Tutorial for making SuperPlots in R. Here is some simple code to help make SuperPlots in R using the ggplot2, ggpubr, dplyr, and ggbeeswarm packages. Dataset that can be renamed “combined” is included in the supporting information. Lines separated by semicolons: ReplicateAverages <- combined %>% group_by(Treatment, Replicate) %>% summarise_each(list(mean)); ggplot(combined, aes(x=Treatment,y=Speed,color=factor(Replicate))) + geom_beeswarm(cex=3) + scale_colour_brewer(palette = "Set1") + geom_beeswarm(data=ReplicateAverages, size=8) + stat_compare_means(data=ReplicateAverages, comparisons = list(c("Control", "Drug")), method="t.test", paired=TRUE) + theme(legend.position="none").
Figure S5.
Tutorial for making SuperPlots in Python. Here is some simple code to help make SuperPlots in Python using the Matplotlib, Pandas, Numpy, Scipy, and Seaborn packages. Dataset that can be renamed “combined.csv” is included in the supporting information. Lines separated by semicolons: combined = pd.read_csv("combined.csv"); sns.set(style="whitegrid"); ReplicateAverages = combined.groupby(['Treatment','Replicate'], as_index=False).agg({'Speed': "mean"}); ReplicateAvePivot = ReplicateAverages.pivot_table(columns='Treatment', values='Speed', index="Replicate"); statistic, pvalue = scipy.stats.ttest_rel(ReplicateAvePivot['Control'], ReplicateAvePivot['Drug']); P_value = str(float(round(pvalue, 3))); sns.swarmplot(x="Treatment", y="Speed", hue="Replicate", data=combined); ax = sns.swarmplot(x="Treatment", y="Speed", hue="Replicate", size=15, edgecolor="k", linewidth=2, data=ReplicateAverages); ax.legend_.remove(); x1, x2 = 0, 1; y, h, col = combined['Speed'].max() + 2, 2, 'k'; plt.plot([x1, x1, x2, x2], [y, y+h, y+h, y], lw=1.5, c=col); plt.text((x1+x2)*.5, y+h*2, "P = "+P_value, ha='center', va='bottom', color=col).
Whatever way authors choose to display their data, it is critical to list the number of independent experiments in the figure or caption, as well as how the means and statistical tests were calculated.
Error bars that communicate reproducibility
The choice of error bars on a SuperPlot depends on what you hope to communicate: descriptive error bars characterize the distribution of measurements (e.g., standard deviation), while inferential error bars evaluate how likely it is that the same result would occur if the experiment were to be repeated (e.g., standard error of the mean or confidence intervals; Cumming et al., 2007). To convey how repeatable an experiment is, it is appropriate to choose inferential error bars calculated using the number of independent experiments as the sample size. However, calculating standard error of the mean by inputting data from all cells individually fails in two ways: first, the natural variability we expect from biology would be better summarized with a descriptive measure, like standard deviation; and second, the inflated n produces error bars that are artificially small (due to √n in the denominator) and do not communicate the repeatability of the experiment.
The problems with calculating error bars using cell count as the sample size are illustrated by comparing the error bars in Fig. 1 A to those in Fig. 1, B–D: when each cell measurement is treated as an independent sample, the standard error of the mean is always tiny, whether or not there is variability among experimental replicates. In contrast, the error bars calculated using biological replicates grow when the results vary day to day. In cases where displaying every data point is not practical, authors should consider some way of representing the cell-to-cell variability as well as the run-to-run repeatability. This could be error bars that represent the standard deviation of the entire dataset, but with P values calculated from biological replicates.
Conclusions
When calculating your P value, take a moment to consider these questions: What variability does your P value represent? How many independent experiments have you performed, and does this match with your n? (See Table S1 for practical examples of this analysis.) We encourage authors and editors to focus less on reporting satisfying yet superficial statistical tests such as P values, and more on presenting the data in a manner that conveys both the variability and the reproducibility of the work.
Online supplemental material
Fig. S1 shows other plotting examples. Fig. S2 is a tutorial for making SuperPlots in Prism. Fig. S3 is a tutorial for making SuperPlots in Excel. Fig. S4 is a tutorial for making SuperPlots in R. Fig. S5 is a tutorial for making SuperPlots in Python. Table S1 shows how the choice of n influences conclusions. Data S1 is the raw data used to generate Figs. S4 and S5.
Supplementary Material
shows how the choice of n influences conclusions.
is the raw data used to generate Figs. S4 and S5.
Acknowledgments
We acknowledge that our past selves are not innocent of the mistakes described in this manuscript. We are grateful to several colleagues who provided feedback on our preprint, including Kenneth Campellone, Adam Zweifach, William Velle, Geoff O’Donoghue, and Nico Stuurman. We also thank Natalie Petek for providing some hints on using Prism and Jiongyi Tan for reviewing the Python code.
This work was supported by grants to L.K. Fritz-Laylin from the National Institutes of Health (from the National Institute of Allergy and Infectious Diseases grant 1R21AI139363), from the National Science Foundation (grant IOS-1827257), and from the Pew Scholars Program in the Biomedical Sciences; and by grants to R.D. Mullins from the National Institutes of Health (R35-GM118119) and Howard Hughes Medical Institute.
The authors declare no competing financial interests.
References
- Aarts E., Dolan C.V., Verhage M., and van der Sluis S.. 2015. Multilevel analysis quantifies variation in the experimental effect while optimizing power and preventing false positives. BMC Neurosci. 16:94 10.1186/s12868-015-0228-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman D.G., and Bland J.M.. 1997. Statistics notes. Units of analysis. BMJ. 314:1874 10.1136/bmj.314.7098.1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman N., and Krzywinski M.. 2015. Points of significance: Sources of variation. Nat. Methods. 12:5–6. 10.1038/nmeth.3224 [DOI] [PubMed] [Google Scholar]
- Blainey P., Krzywinski M., and Altman N.. 2014. Points of significance: replication. Nat. Methods. 11:879–880. 10.1038/nmeth.3091 [DOI] [PubMed] [Google Scholar]
- Cumming G., Fidler F., and Vaux D.L.. 2007. Error bars in experimental biology. J. Cell Biol. 177:7–11. 10.1083/jcb.200611141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith S., Daniel J.A., and Vissel B.. 2010. A study of clustered data and approaches to its analysis. J. Neurosci. 30:10601–10608. 10.1523/JNEUROSCI.0362-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A., and Hill J.. 2006. Data Analysis Using Regression and Multilevel/Hierarchical Models. First edition Cambridge University Press, Cambridge: 10.1017/CBO9780511790942 [DOI] [Google Scholar]
- Greenwald A.G., Gonzalez R., Harris R.J., and Guthrie D.. 1996. Effect sizes and p values: what should be reported and what should be replicated? Psychophysiology. 33:175–183. 10.1111/j.1469-8986.1996.tb02121.x [DOI] [PubMed] [Google Scholar]
- Hurlbert S.H. 1984. Pseudoreplication and the Design of Ecological Field Experiments. Ecol. Monogr. 54:187–211. 10.2307/1942661 [DOI] [Google Scholar]
- Krzywinski M., Altman N., and Blainey P.. 2014. Points of significance: nested designs. For studies with hierarchical noise sources, use a nested analysis of variance approach. Nat. Methods. 11:977–978. 10.1038/nmeth.3137 [DOI] [PubMed] [Google Scholar]
- Lamb T.J., Graham A.L., and Petrie A.. 2008. T testing the immune system. Immunity. 28:288–292. 10.1016/j.immuni.2008.02.003 [DOI] [PubMed] [Google Scholar]
- Lazic S.E. 2010. The problem of pseudoreplication in neuroscientific studies: is it affecting your analysis? BMC Neurosci. 11:5 10.1186/1471-2202-11-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazic S.E., Clarke-Williams C.J., and Munafò M.R.. 2018. What exactly is ‘N’ in cell culture and animal experiments? PLoS Biol. 16:e2005282 10.1371/journal.pbio.2005282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazic S.E., Mellor J.R., Ashby M.C., and Munafo M.R.. 2020. A Bayesian predictive approach for dealing with pseudoreplication. Sci. Rep. 10:2366 10.1038/s41598-020-59384-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naegle K., Gough N.R., and Yaffe M.B.. 2015. Criteria for biological reproducibility: what does “n” mean? Sci. Signal. 8:fs7 10.1126/scisignal.aab1125 [DOI] [PubMed] [Google Scholar]
- Pollard D.A., Pollard T.D., and Pollard K.S.. 2019. Empowering statistical methods for cellular and molecular biologists. Mol. Biol. Cell. 30:1359–1368. 10.1091/mbc.E15-02-0076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux D.L., Fidler F., and Cumming G.. 2012. Replicates and repeats--what is the difference and is it significant? A brief discussion of statistics and experimental design. EMBO Rep. 13:291–296. 10.1038/embor.2012.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissgerber T.L., Milic N.M., Winham S.J., and Garovic V.D.. 2015. Beyond bar and line graphs: time for a new data presentation paradigm. PLoS Biol. 13:e1002128 10.1371/journal.pbio.1002128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissgerber T.L., Savic M., Winham S.J., Stanisavljevic D., Garovic V.D., and Milic N.M.. 2017. Data visualization, bar naked: A free tool for creating interactive graphics. J. Biol. Chem. 292:20592–20598. 10.1074/jbc.RA117.000147 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows how the choice of n influences conclusions.
is the raw data used to generate Figs. S4 and S5.