Centrioles build centrosomes and cilia, but these microtubule-based structures are frequently abnormal in tumor cells and exhibit structural aberrations such as centriole elongation. Kong et al. demonstrate that centrioles don't have an elongation monitoring mechanism, which renders them prone to over-elongation, especially during prolonged mitosis.
Abstract
Centrioles are precisely built microtubule-based structures that assemble centrosomes and cilia. Aberrations in centriole structure are common in tumors, yet how these aberrations arise is unknown. Analysis of centriole structure is difficult because it requires demanding electron microscopy. Here we employ expansion microscopy to study the origins of centriole structural aberrations in large populations of human cells. We discover that centrioles do not have an elongation monitoring mechanism, which renders them prone to over-elongation, especially during prolonged mitosis induced by various factors, importantly including supernumerary centrioles. We identify that mitotic centriole over-elongation is dependent on mitotic Polo-like kinase 1, which we uncover as a novel regulator of centriole elongation in human cycling cells. While insufficient Plk1 levels lead to the formation of shorter centrioles lacking a full set of microtubule triplets, its overactivity results in over-elongated and structurally aberrant centrioles. Our data help explain the origin of structurally aberrant centrioles and why centriole numerical and structural defects coexist in tumors.
Introduction
Centrioles are built of microtubules (MTs) assembled in a ninefold symmetrical cylinder (Vorobjev and Chentsov, 1982). In mammals, the centrioles are ∼450 nm long. Their proximal end is the site of the accumulation of a proteinaceous material called pericentriolar material, and of centriole duplication. The distal end harbors subdistal and distal appendages, which anchor MTs and form cilia and flagella, respectively. New centrioles (procentrioles) are built perpendicularly and adjacent to the wall of existing (mother) centrioles in early S phase. After initiation, procentrioles elongate their MTs until they reach the length of a mother centriole. Unlike cytoplasmic MTs, which are dynamic, centriole MTs grow very slowly, are stable, and are passed on to new generations of cells. Centrosomal proteins such as centrosomal protein 4.1–associated protein (CPAP; Kohlmaier et al., 2009; Schmidt et al., 2009; Tang et al., 2009), Cep120 (Lin et al., 2013b; Mahjoub et al., 2010), centrobin, Cep135 (Gudi et al., 2015; Lin et al., 2013a), and SPICE (Comartin et al., 2013) are necessary for centriole elongation. However, how elongation of human centrioles is regulated throughout the cell cycle is still unclear.
Centriole structural aberrations, including over-elongation, have been well established in tumors (Chan, 2011). They can be observed already in pretumorigenic lesions and are present in almost all aggressive tumors (D’Assoro et al., 2002; Gönczy, 2015; Nigg and Raff, 2009). Structurally aberrant centrioles perturb tissue architecture and promote chromosome instability (Ganem et al., 2009) and invasive properties of cells (Godinho et al., 2014; Nigg et al., 2017). Over-elongated centrioles over-accumulate pericentriolar material, perturbing the symmetry of mitotic spindles. They can also contribute to centrosome amplification through fragmentation or the formation of multiple procentrioles along their elongated walls (Kohlmaier et al., 2009; Marteil et al., 2018). However, little is known about the origins of centriole structural defects in tumors, and it is not clear to which extent cells can tolerate variations in centriole length without harmful consequences.
Centrioles are of subdiffractional dimensions, so the analysis of centriolar structural characteristics requires high-resolution imaging techniques, which are laborious and unsuitable for the routine collection of large datasets to permit rigorous statistical analyses. Here, we take advantage of an adapted expansion microscopy method that enables analysis of centriole structural features in large populations of cells by conventional microscopy. We applied this method in combination with electron and super-resolution microscopy to explore the mechanisms that lead to the formation of structurally aberrant centrioles, as observed in tumor cells.
Results and discussion
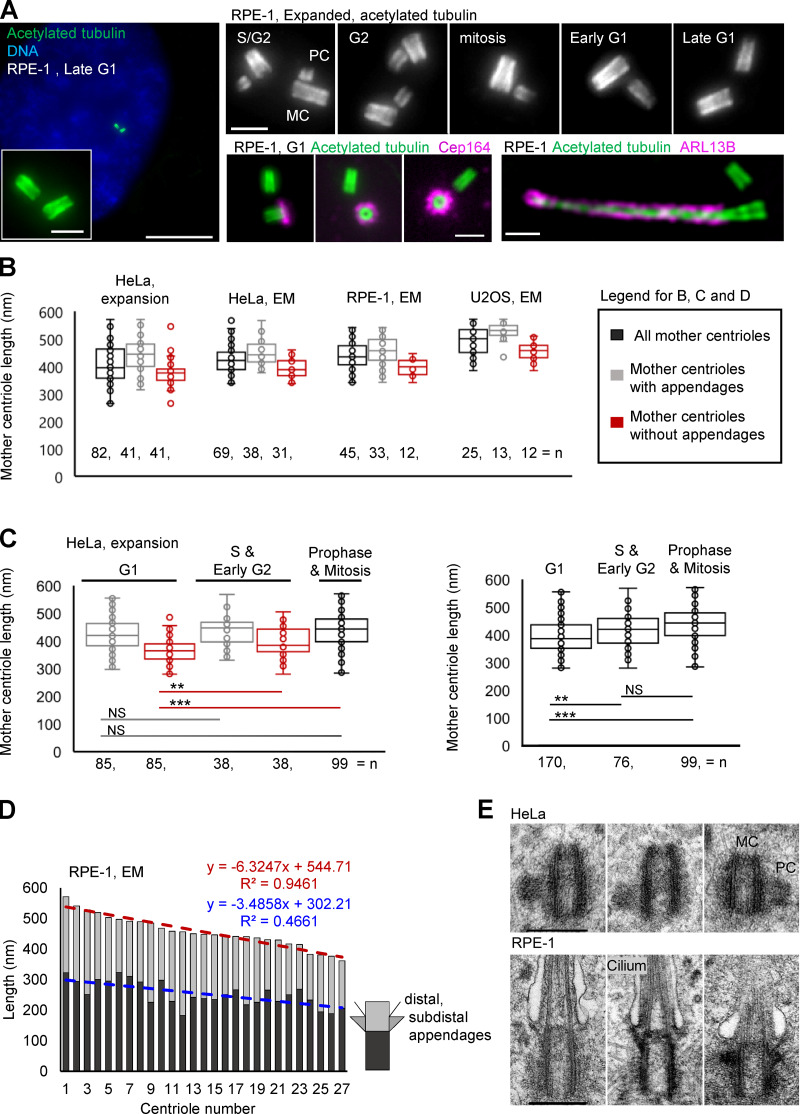
To determine centriole length within large populations of cultured cells, we used expansion microscopy, a recently pioneered imaging approach that allows the analysis of subresolutional structures by conventional optical microscopes (Chen et al., 2015; Gambarotto et al., 2019; Geertsema and Ewers, 2016; Wassie et al., 2019). We adapted the original expansion microscopy method “magnified analysis of the proteome” (Ku et al., 2016) to the analysis of centriole length, age, and ciliation (Fig. 1 A; Sahabandu et al., 2019). Our method is based on the fact that centriole MTs are ubiquitously acetylated and can be robustly labeled within fixed mammalian cells using a conventional antibody against acetylated tubulin. The increase in optical resolution achieved by fourfold isotropic expansion allowed us to study centriole elongation in a cell cycle–specific manner. In addition, after expansion, three generations of centrioles can be clearly distinguished. Older mother centrioles can be distinguished from younger centrioles by the presence of appendage proteins such as Cep164, and procentrioles can be distinguished from mother centrioles based on their orthogonal orientation to their mother centrioles (Fig. 1 A).
Figure 1.
Centriole length within a population of cycling cells is variable. (A) Examples of centrioles and one primary cilium after 4.2-fold expansion and imaging by conventional widefield microscope using 60× lens. Acetylated tubulin labels centriole MTs and ciliary axoneme. Cep164 is a component of centriole distal appendages present on older mother centriole. ARL13B is a ciliary membrane-associated protein. (B) Quantification of the length of mother centrioles in G1 and S phase. (C) The analysis of mother centriole length in specific phases of the cell cycle. Left: Older and younger mother centrioles plotted separately. Right: Older and younger mother centrioles plotted together. (D) Centrioles from electron micrographs were divided into proximal (dark gray) and distal portions (light gray) at the base of subdistal appendages. The length of these portions was measured and plotted for each centriole. Centrioles were plotted from the longest to the shortest. A linear regression trendline, a regression equation, and a correlation coefficient R2 are marked. (E) Examples of centrioles of variable length from HeLa and RPE-1 cells. MC, mother centriole; PC, procentriole. Scale bars, 20 (large) and 2 µm in A; 0.4 µm in E.
We first set out to determine the consistency of centriole length by measuring the length of mother centrioles in the commonly used human cell lines HeLa, RPE-1, and U2OS (Fig. 1 B). We found that mother centrioles showed significant variability in length in each cell type, even in untransformed RPE-1 cells (Fig. 1 B), with measured values ranging between a minimum extreme of ~300 and a maximum of ~500 nm. This variability was also apparent when older mother centrioles (with appendages) were analyzed separately (Fig. 1 B). Although younger mothers were shorter on average, they could exceed the length of older mother centrioles. A more detailed cell cycle–specific analysis revealed that the mean length of younger mother centrioles increases slightly between G1 phase and mitosis (Fig. 1 C), meaning that in HeLa cells, the elongation of younger mother centrioles continues during their second cell cycle. Our findings using expansion microscopy are similar to prior work analyzing centrioles isolated from human lymphoblastic cells (Chrétien et al., 1997).
Further, dividing centrioles into proximal and distal portions at the base of subdistal appendages showed that both portions can be of variable length (Fig. 1 D). Based on the low incidence of spindle and chromosome segregation defects in these cultures, the absence of centriole duplication errors, and the ability of centrioles of variable length to assemble cilia (Fig. 1 E), we concluded that the detected variability in length must be within physiological boundaries and inconsequential for centriole and centrosome functions. These results suggested that human cycling cells don’t have a stringent centriole elongation monitoring mechanism that ensures the formation of centrioles of a precise length. In the absence of such a mechanism, the formation of centrioles of inconsistent size should be expected and, to some degree, tolerated by cells, consistent with our observation. Without a mechanism to control centriole length, we speculated that centrioles, especially procentrioles, are prone to over-elongation past their physiologically tolerable range.
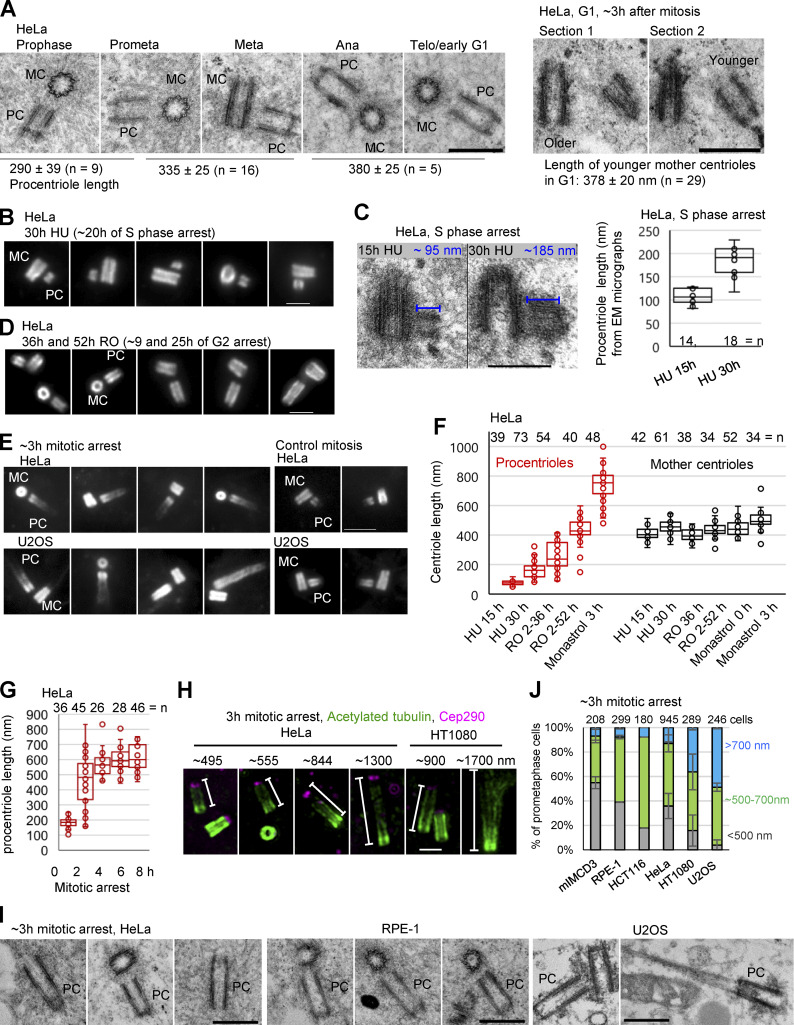
To test this hypothesis and to identify potential factors that lead to over-elongation, we analyzed the dynamics of centriole elongation under various perturbations of the cell cycle. First, we determined the dynamics of procentriole elongation in unperturbed cycling cells. The EM analysis of HeLa cell procentrioles revealed that from initiation in S phase until early mitosis, they elongate to 290 ± 39 nm. They further elongate to 335 ± 25 nm by prometaphase/metaphase, and to 380 ± 25 nm by telophase/early G1 (Fig. 2 A). These centrioles (hereafter referred to as younger mother centrioles) did not appear to elongate further within the first 3 h of G1 (Fig. 2 A, right panel). Thus, the dynamics of procentriole elongation vary during the cell cycle. During S and G2 phase, procentrioles elongate ∼290 nm in ∼10 h (0.5 nm/min), while between prophase and telophase, they elongate an additional ∼100 nm in only ∼50 min (2 nm/min). In parallel, younger mother centrioles only slightly elongate during their second cell cycle, while biochemically mature mother centrioles maintain their previously established length. Thus, under normal cell cycle conditions, the ability of centrioles to respond to elongation stimuli is inversely proportional to their maturation status.
Figure 2.
Procentrioles over-elongate during prolonged mitosis. (A) The analysis of procentriole length in cycling cells during mitosis and G1. Average PC length, SD, and centriole number are indicated below images. (B, D, and E) Examples of expanded centrioles arrested in S phase (by HU), G2 (by RO 3306, RO) and in mitosis (by monastrol). Centrioles were immunolabeled for acetylated tubulin. (C) EM analysis of procentriole length in cells arrested in S phase by HU. (F and G) Quantification of centriole length under various experimental conditions using expansion and acetylated tubulin labeling. (H) Examples of over-elongated procentrioles from mitotically arrested cells immunolabeled for acetylated tubulin and distal centriole protein Cep290. (I) Examples of over-elongated procentrioles analyzed by EM. (J) Quantification of centriole phenotypes in expanded mitotically arrested cells of various cell lines. Procentrioles were scored for the length of their acetylated tubulin signals. Cells were placed in the green or blue category by the presence of at least one procentriole of indicated length. Cells with both procentrioles shorter than ∼500 nm were placed in the gray category. Centriole counts are from 2, 2, 1, 5, 2, and 2 independent experiments, respectively. MC, mother centriole; PC, procentriole. Prometa, prometaphase; Meta, metaphase; Ana, anaphase; Telo, telophase. Scale bars, 0.5 µm in A and I; 2 µm in B, D, and H; 4 µm in E; 0.4 µm in C.
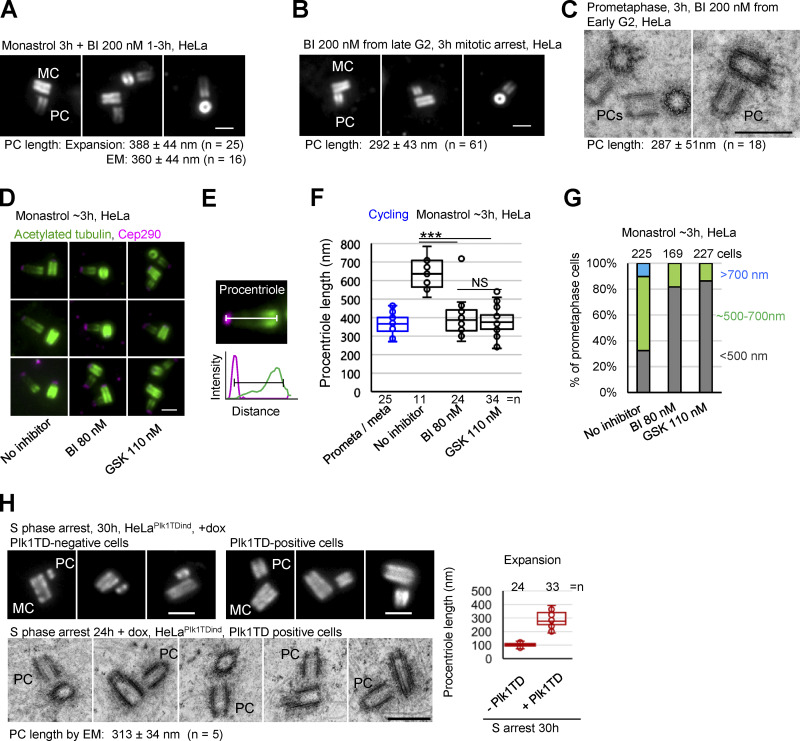
We next analyzed whether centrioles over-elongate during arrested cell cycle progression. To arrest cells in S phase, we collected mitotic cells by shake-off, replated them, and treated them with hydroxyurea 2 h later. Under these conditions, mother centrioles initiate duplication ∼9–10 h after shake-off and remain arrested in early S phase thereafter due to hydroxyurea. Cells were fixed and expanded 15 h and 30 h after shake-off (corresponding to ∼5 h and ∼20 h of arrest). Measurements of the length of procentriole acetylated tubulin signals indicated that during S phase arrest, procentrioles do not over-elongate (Fig. 2, B and F). This result was consistent with EM analysis, which showed ∼190-nm-long procentrioles at the 30 h time point (Fig. 2 C) and previous reports (Kuriyama and Borisy, 1981; Shukla et al., 2015). It is worth noting that using acetylated tubulin to determine the length of procentrioles may slightly underestimate their length, as procentriole MTs are acetylated gradually from proximal ends (Fig. S1; Sahabandu et al., 2019). We then tested whether centrioles over-elongate during G2 arrest. Cells were treated with Cdk1/cyclin B inhibitor RO 3306 from G1. Such cultures reached G2 ∼27 h after shake-off (Loncarek et al., 2010) and were incubated until 36 h or 52 h after shake-off (corresponding to ∼9 or 25 h of G2 arrest), when they were fixed and expanded. In cells arrested in late G2, procentrioles elongated to ∼400 nm (Fig. 2, D and F; Shukla et al., 2015), which exceeds the length of control procentrioles in G2.
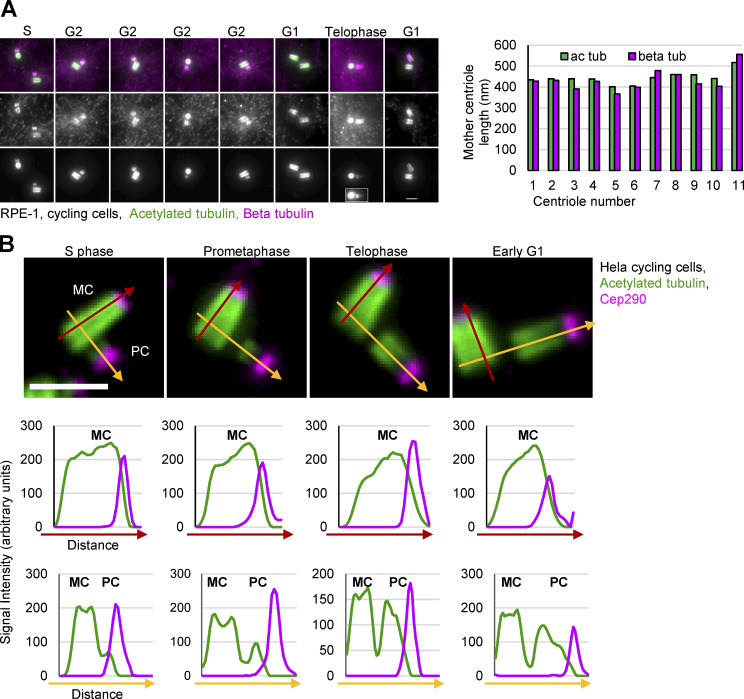
Figure S1.
Acetylation tubulin as a marker for centriole length. (A) RPE-1 cells were expanded ∼4.2× and labeled for acetylated tubulin (ac tub) and β tubulin. Left: Examples of centrioles in various cell cycle stages. Presented centrioles were selected for brighter and more even β tubulin signal, as β tubulin antibody, in our hands, does not evenly label all centrioles in the population. Right: The comparison of mother centriole length determined by measuring the length of acetylated and β tubulin signal. Acetylated tubulin is a reliable marker for the analysis of the length of mature centrioles. (B) The pattern of centriole acetylation during cell cycle progression. Cycling HeLa cells were expanded ∼4.2× and immunolabeled for acetylated tubulin and distal centriole protein Cep290. Four mother centrioles (MC) associated with procentrioles (PC) from different phases of the cell cycle are shown. The intensity plots of the fluorescent acetylated tubulin (green) and Cep290 signal (magenta) along red and yellow arrows are shown below. MC acetylated tubulin signal is present along microtubule walls and overlaps with Cep290 signal on centriole’s distal ends. Cep290 accumulates to procentriole distal ends from their early stages. Elongating procentrioles in mitosis and immediately after mitosis show a gradient of acetylated tubulin signal from proximal toward distal end. Due to such a gradient, the length of PCs, if determined by measuring only the length of acetylated tubulin signal, may be in some cells slightly underestimated. Scale bars, 2 µm.
To arrest cells in mitosis, logarithmically growing cultures were treated with the Eg5 inhibitor, monastrol. Mitotic cells were collected 1 h later and incubated for an additional 3 h, fixed, and analyzed by expansion. The analysis revealed drastically over-elongated procentrioles (Fig. 2, E and F). In HeLa cells, within ∼3 h of mitotic arrest, the average length of procentrioles exceeded the length of mother centrioles. Most centrioles elongated at a rate similar to control procentrioles in mitosis (∼2 nm/min; Fig. 2 A). However, some procentrioles gained ≥700 nm in length during the first 3 h of mitotic arrest, meaning that they elongated faster (∼3.9 nm/min) than control procentrioles. It was not unusual to find two procentrioles of different lengths in the same cell during arrest. This stochastic elongation pattern could signify a difference in the local concentration of elongation factors. Procentriole elongation plateaued after ∼6 h of mitotic arrest (Fig. 2 G), possibly due to the exhaustion of short-lived proteins required for elongation. The intensity of acetylated signal was lower toward the distal ends of elongated procentrioles, which were associated with distal centriole protein Cep290 (Fig. 2 H). EM analysis corroborated that elongated acetylated tubulin signals represent bona fide centrioles (Fig. 2 I). Over-elongation of procentrioles during mitotic arrest occurred in multiple cell lines (Fig. 2 J). However, the percentage of cells forming over-elongated and structurally abnormal procentrioles varied among cell lines (Fig. 2 J). It is not yet clear what causes this variability.
Occasionally, mother centrioles over-elongated during prolonged mitosis. However, the average length of mother centrioles increased only slightly during any arrest (Fig. 2 F), suggesting that the mechanisms that curb further elongation of mother centrioles remain active under these conditions.
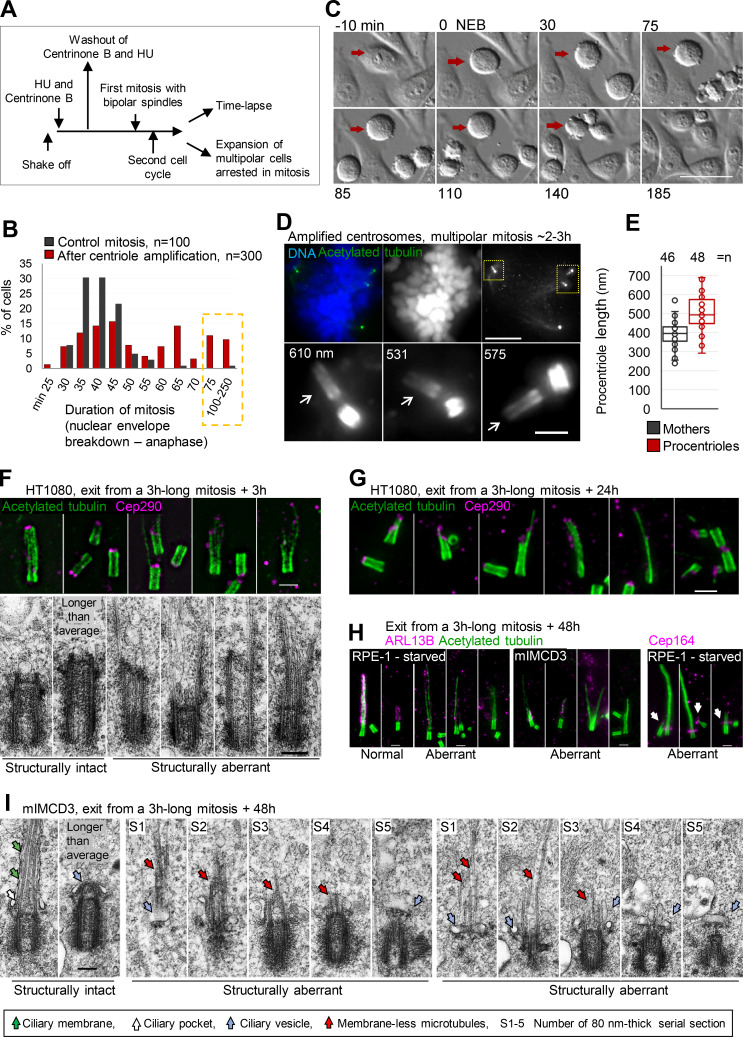
Over-elongation of procentrioles confirmed that cells lack a centriole elongation monitoring mechanism and that procentrioles are specifically prone to over-elongation, particularly during mitosis. The importance of this finding is that any event leading to mitotic delay might potentially promote centriole over-elongation. To test this idea, we induced the formation of multiple procentrioles by a transient treatment with centrinone B, an inhibitor of Plk4 kinase (Wong et al., 2015; Fig. 3 A). Such procentrioles formed multiple centrosomes in the subsequent cell cycle, which resulted in the formation of multipolar spindles, delaying mitotic exit due to unsatisfied mitotic checkpoint (Fig. 3, A–C). The analysis of multipolar mitotic cells delayed in mitosis for 2–3 h showed that they indeed harbored over-elongated procentrioles, the length of which exceeded the length of mother centrioles (Fig. 3, D and E).
Figure 3.
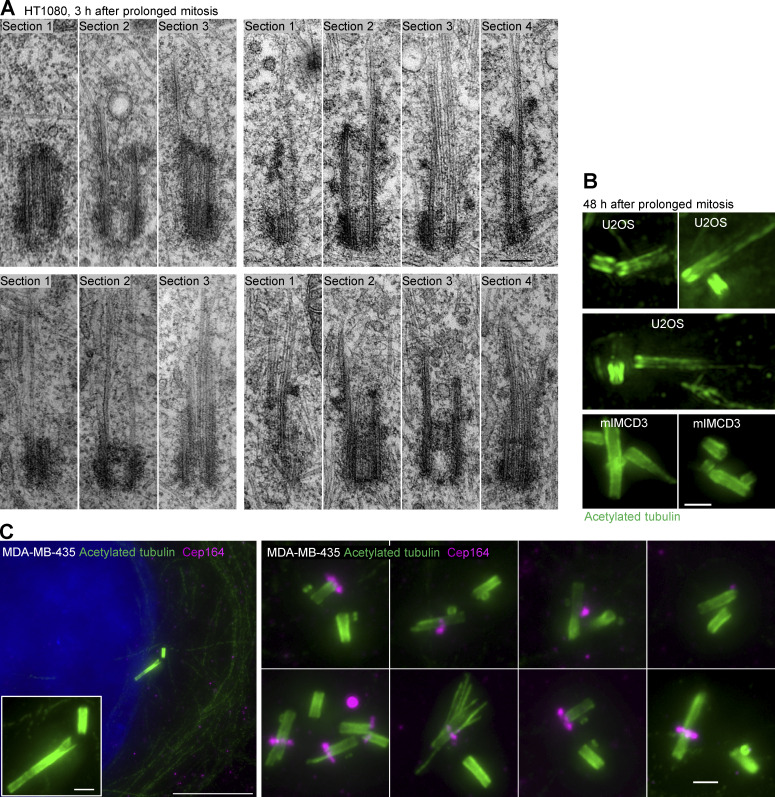
Prolonged mitosis leads to the formation of over-elongated and structurally and functionally aberrant centrioles. (A) A scheme delineating experimental design used to generate cells with amplified centrosomes leading to prolonged mitosis. (B) Quantification of duration of mitosis in control HeLa cells and HeLa cells containing amplified centrosomes. Cells were recorded by time-lapse in 5-min time intervals. (C) Example of a cell arrested in prometaphase for 140 min (red arrow). (D and E) Cells arrested in mitosis for ∼2–3 h were collected and analyzed by expansion. (D) Cell arrested in mitosis with multipolar spindle and over-elongated procentrioles (arrows). Numbers indicate procentriole length. (E) Quantification of centriole length in mitotically arrested cells. (F and G) Examples of aberrant centrioles analyzed by expansion and EM 3 h and 24 h after release from prolonged mitosis. (H) Representative examples of expanded structurally intact and aberrant centrioles/cilia 48 h after the release form a 3-h-long mitosis. ARL13B staining was used to distinguish normal looking from aberrant cilia. Arrows point to aberrantly localized Cep164. (I) Electron micrographs of structurally intact and aberrant centrioles in ciliation, 48 h after the release from prolonged mitosis. Scale bars, 20 µm in C; 20 µm for mitotic cell; 2 µm for enlarged centrioles in D, and for F, G, and H; 0.2 µm for EM in F and I.
Previous studies have demonstrated that over-elongated centrioles can lead to centrosome amplification (Kohlmaier et al., 2009; Marteil et al., 2018). Here, we establish for the first time that the inverse is possible and that centriole numerical errors can directly instigate centriole structural aberrations. Centrosome numerical errors can originate from aborted mitosis, from cell fusion, or from centriole amplification. Our data imply that all of these events can promote the formation of centriolar structural aberrations. This can explain the origin of aberrantly structured centrioles in tumors and why centriolar structural defects are frequently found in tumor cells along with numerical defects. To the best of our knowledge, this is the first report showing how centriole over-elongation can occur in cells without chemical or genetic manipulation of centrosomal proteins. In addition, our data show that any perturbation leading to prolonged mitotic arrest might result in over-elongated centrioles.
To understand the fate of over-elongated procentrioles, monastrol was washed out after 3 h of mitotic arrest to allow mitotic exit. Cells were fixed 3 h and 24 h later and analyzed by expansion and EM. Immunolabeling of acetylated tubulin and Cep290 showed that over-elongated centrioles remained in the population after mitosis (Fig. 3, F and G; and Fig. S2). They appeared stable and could duplicate and form mitotic spindles, indicating that their proximal ends were functional. However, their distal ends were characterized by an uneven acetylated tubulin signal decorated with disorderly positioned Cep290 (Fig. 3, F and G). EM analysis (Fig. 3 F and Fig. S2 A) revealed centrioles with aberrant distal ends and elongated MT singlets, doublets, or triplets. Abnormal centriolar phenotypes found in populations of cells exposed to mitotic arrest resembled centrioles detected in melanoma-derived MDA-MB-435 cells, 40% of which have been shown to harbor over-elongated and structurally abnormal centrioles (Fig. S2 C; Marteil et al., 2018).
Figure S2.
Examples of structurally aberrant centrioles. (A) Electron micrographs of aberrant centrioles from HT1080 cells, fixed ∼3 h after they exit from 3-h-long mitosis. Consecutive 80-nm-thick serial sections (S1–4) are shown. One section of each centriole is presented in Fig. 3 F. (B) Examples of expanded aberrant centrioles fixed 48 h after the exit from ∼3-h-long mitosis. (C). Examples of over-elongated and structurally aberrant centrioles from cycling MDA-MB-435 cell line. Cells were expanded ∼4.2×, and centrioles were labeled for acetylated tubulin and distal appendage protein Cep164. Left: A cell showing over-elongated centriole. Right: More examples of aberrant centrioles from the same cell population. Overly long and aberrant centrioles often lack or have an irregular or diminished Cep164 signal. Scale bars, 0.2 µm in A; 20 µm for low magnification in C; 2 µm for B and for the insert and the right panel in C.
To understand whether the detected defects on the distal ends of centrioles affect ciliation, we used mIMCD3 cells, which form cilia in the presence of serum, and RPE-1 cells, which form cilia upon serum starvation. Cells were fixed 48 h after exit from prolonged mitosis, which allowed the over-elongated procentrioles to mature and form cilia. In control cells, ∼80% of mature centrioles formed ciliary axonemes of various lengths, which were associated with ciliary component ARL13B (Fig. 3 H). The analysis of cultures 48 h after prolonged mitosis showed that structurally intact-looking centrioles formed axonemes associated with ARL13B (Fig. 3 H). Centrioles with over-elongated but uneven acetylated signal on their distal ends lacked, or had diminished, ARL13B signal (Fig. 3 H). In addition, aberrant centrioles showed diminished localization of the distal appendage protein Cep164 (Fig. 3 H). Out of the 25 mIMCD3 cells analyzed by EM, 2 showed aberrantly structured centrioles (consistent with the low frequency of over-elongated centrioles in this cell line observed by expansion; Fig. 2 J). Both aberrant centrioles were associated with ciliary vesicles, but their extended MTs were intracellular and devoid of ciliary membrane, suggesting that ciliation was initiated but that ciliary membrane growth was impaired (Fig. 3 I), consistent with the lack of ciliary membrane–associated protein ARL13B around extended centriole MTs detected with expansion (Fig. 3 H). A similar defect in ciliary membrane growth was reported in the fibroblasts derived from patients carrying a Cep290 mutation (Shimada et al., 2017). In 23 cells containing intact-looking centrioles (found at various stages of ciliation within the same cell population) and in 6 control ciliated mIMCD3 centrioles, axoneme MTs were never found outside ciliary vesicles or lacking ciliary membranes. These data suggest that over-elongated centrioles formed during prolonged mitosis have an impaired ability to form cilia.
The notion that procentrioles do not continue elongation if early S phase is prolonged, but over-elongate if cells are arrested in G2 or in mitosis, pointed to the presence of a cell cycle–specific regulator that promotes centriole elongation in later phases of the cell cycle. We turned our attention toward Polo-like kinase (Plk) 1, which has been shown to associate with the centriole elongation factor CPAP (Firat-Karalar et al., 2014; Novak et al., 2016) and whose activity peaks in G2 and mitosis (Martin and Strebhardt, 2006), coincident with the elongation of procentriole distal ends. To examine the potential role of Plk1 in procentriole over-elongation, we collected cells freshly arrested in mitosis by monastrol, treated them with the Plk1 inhibitor BI2536, incubated them for an additional 3 h, and expanded them to analyze centriole length. Indeed, Plk1 inhibition during mitotic arrest prevented procentriole over-elongation (Fig. 4 A). Inhibition of Plk1 alone leads to prometaphase arrest. In such prometaphase-arrested cells, procentrioles did not over-elongate either (Fig. 4, B and C). At higher concentrations, BI2536 also inhibits Plk2 (Steegmaier et al., 2007), which has been previously implicated in centriole initiation and elongation (Chang et al., 2010). So, we additionally employed a more specific Plk1 inhibitor, GSK461364 (GSK; Gilmartin et al., 2009), and repeated the experiment with the lowest concentration of BI2536 that we found to prevent mitotic exit of HeLa cells (80 nM). Both treatments blocked procentriole over-elongation during mitosis, as determined by the distance between the proximal end of acetylated tubulin and Cep290 signals (Fig. 4, D–G).
Figure 4.
Plk1 is required for procentriole over-elongation in mitosis. (A) Expanded centrioles from cells arrested in mitosis for 3 h and treated with Plk1 inhibitor BI. (B) Expanded centrioles from cells arrested in mitosis by BI. (C) Electron micrographs of centrioles from cells arrested in mitosis by BI. (D) Expanded centrioles from cells arrested in mitosis and treated with Plk1 inhibitors BI and GSK. (E) Approach used to quantify procentriole length using acetylate tubulin and Cep290 signals. (F) Quantification of procentriole length using method in E. ***, P ≤ 0.001. (G) Quantification of procentriole phenotypes in expanded mitotically arrested cells. Procentrioles were scored for the length of their acetylated tubulin signals. Cells were placed in the green or blue category by the presence of at least one procentriole of indicated length. Cells with both procentrioles shorter than ∼500 nm were placed in the gray category. (H) Examples of centrioles from S phase arrested cells with/without active Plk1 (Plk1TD) analyzed by correlative conventional/expansion microscopy and EM. Plot: quantification of the length of procentriole acetylated tubulin signal from expanded samples. MC, mother centriole; PC, procentriole; average procentriole (PC) length, SD, and centriole number are indicated below images; scale bars, 0.5 µm for EM; 2 µm for expansion.
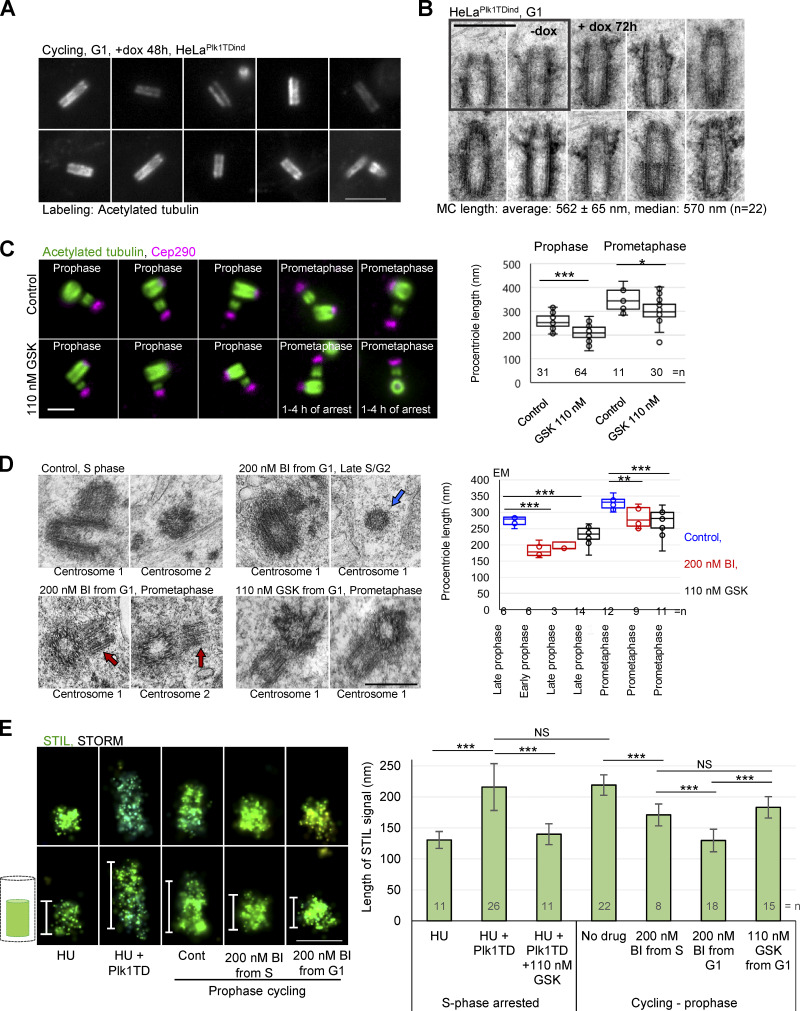
To further assess whether Plk1 can stimulate procentriole elongation in interphase, we expressed active Plk1 (carrying the activation 210TD mutation, Plk1TD) in S phase–arrested HeLa cells, which are characterized by lower activity of endogenous Plk1 than mitotic cells. Plk1TD expression indeed resulted in procentriole elongation beyond their S phase length (Fig. 4 H). Consistently, overexpression of Plk1TD in cycling cells increased the average length of mother centrioles (Fig. 5 A). EM analysis revealed that such elongated centrioles had longer than average proximal or distal ends and incorrectly positioned appendages (Fig. 5 B).
Figure 5.
Plk1 regulates centriole length in human cells. (A) Examples of expanded G1 centrioles from cycling cells expressing active Plk1 (Plk1TD). dox, doxycycline. (B) Electron micrographs of centrioles from cycling cells expressing Plk1TD. Centrioles are longer than average with aberrantly positioned appendages. (C) Expanded centrioles from control cycling cells and from cells treated with Plk1 inhibitor GSK from G1. Plot: Quantification of procentriole length using the method described in Fig. 4 E. (D) Electron micrographs of centrioles. Red arrows point to missing and unevenly elongated procentriole MTs. Blue arrow: Procentriole in cross-section lacking a full set of MTs. For more examples of shorter and narrower procentrioles, see Fig. S3. (E) The analysis of cartwheel length by 3D STORM using STIL signal. Cells were immunolabeled for cartwheel protein STIL and imaged by 3D STORM. Cont, control. The length of STIL signals was measured from 3D STORM recordings. Left: Examples of STIL signals of various length. Histogram shows the average length of 3D STORM STIL signals ± SD. Scale bars, 3 µm in A; 0.5 µm in B; 2 µm in C; 0.4 µm in D; 0.2 µm in E. NS, nonsignificant; *, P ≤ 0.05; **, P ≤ 0.001; ***, P ≤ 0.001.
To understand whether endogenous Plk1 is essential for the formation of properly structured centrioles in interphase, we inhibited Plk1 from G1 or early S phase, allowed cells to progress to prophase/mitosis (∼21 h after shake-off), fixed them, and analyzed procentrioles by expansion and EM. The analysis of prophase and prometaphase procentrioles by expansion showed that Plk1 inhibition decreases the average length of procentrioles (Fig. 5 C). EM analysis confirmed the result obtained by expansion and revealed procentrioles of shorter-than-average length with variability in the length of their MTs (Fig. 5 D and Fig. S3). Late S/G2 procentrioles of Plk1-inhibited cells were frequently narrower than the late S/G2 procentrioles of the control samples, consistent with the absence of a full set of MT triplets detected in three procentrioles favorably oriented during sectioning (Fig. 5 D and Fig. S3). These data indicate that, in human cycling cells, Plk1 activity is not necessary for procentriole initiation, but is important for procentriole MT assembly and elongation, especially during mitosis. Mature centrioles remained intact in the absence of Plk1 activity, meaning that Plk1 activity is not required for the maintenance of their MTs, at least within the boundaries of our experiments. Since even 200 nM of BI2356 could not completely abrogate procentriole elongation during interphase, it is plausible that, in human cells, multiple regulators drive procentriole assembly. In agreement, Plk4 and 2 have both been implicated in centriole elongation (Aydogan et al., 2018; Chang et al., 2010).
Figure S3.
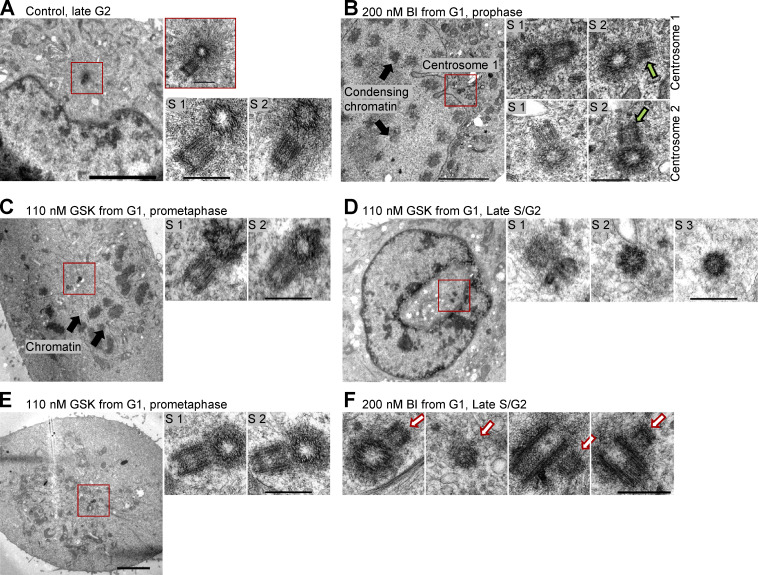
Electron micrographs of centrioles from Plk1-inhibited cells. Synchronous population of HeLa cells was treated with Plk1 inhibitors BI and GSK in G1. Cells were allowed to progress through the cell cycle and were fixed at the peak of mitotic entry (21 h after shake-off). (A, B, D, and E) Low magnification images are illustrating phases of the cell cycle. In addition, two or three 80-nm-thick serial sections (S1–3) through mother centrioles associated with a procentriole are shown. (A) Control late G2 cell with duplicated mother centriole surrounded with expanding pericentriolar material and nucleated microtubules (insert). (B) Examples of shorter-than average procentrioles in Plk1-inhibited prophase cell. Green arrows point to the regions with uneven length of procentriole microtubules. (C) Example of shorter-than-average procentrioles in Plk1-inhibited prometaphase cell. (D) Example of a cross-sectioned short procentriole lacking the third set of microtubules. (E) Example of shorter-than-average procentrioles in prometaphase cell. (F) Examples of procentrioles (white arrows) from late S and G2 cells. One section is shown for each centriole. Procentrioles appear narrower and shorter than the typical control procentrioles in late S and G2. Procentriole in cross-section lacks the full set of triplet microtubules. Some serial sections in B and C are shown in Fig. 5 D of the main text. Scale bars, 4 µm for low magnification, 0.4 µm nm for serial sections and insert in A.
Consistent with the positive role of Plk1 in procentriole elongation, stochastic optical reconstruction microscopy (STORM) analysis of the cartwheel protein STIL revealed elongated cartwheels in S phase–arrested cells expressing Plk1TD that was abrogated by the addition of Plk1 inhibitor (Fig. 5 E). Conversely, inhibition of Plk1 from G1 or S phase resulted in significantly shorter cartwheels than in control prophase cells (Fig. 5 E).
Currently, we do not understand how Plk1 regulates procentriole elongation. In addition to the previously proposed Plk2 (Chang et al., 2010), our data suggest that Plk1 is yet another member of the polo family involved in procentriole assembly. Whether, and to what extent, Plk1’s functions in procentriole assembly overlap with those of other polo kinases is currently not clear, but Plk1 overexpression and inhibition can clearly influence procentriole elongation. An exciting possibility is that Plk2, most active in G1 and early S phase (Ma et al., 2003), stimulates procentriole elongation at the beginning of the cell cycle, while Plk1, the activity of which increases from S phase until mitosis, stimulates procentriole elongation at later stages of the cell cycle. Indeed, our data so far indicate that treatment from G1 with BI3536, which presumably at least partially inhibits Plk2 activity in addition to Plk1 activity, results in more penetrant procentriole and cartwheel elongation defects compared with those defects observed using the more specific Plk1 inhibitor GSK. It is noteworthy that both Plk2 and Plk1 interact with centriole elongation factor CPAP (Chang et al., 2010; Novak et al., 2016).
During their maturation, centrioles acquire biochemical modifications that somehow prevent their further elongation. Plk1 has previously been shown to promote the biochemical maturation of centrioles (Kong et al., 2014; Loncarek et al., 2010; Tsou et al., 2009; Wang et al., 2011). We propose that Plk1 plays a dual role in the centriole cycle. It stimulates elongation of procentrioles, while at the same time it promotes their gradual maturation, which ultimately reduces their ability to elongate. Thus, a dual role of Plk1 in procentriole elongation and maturation could represent an efficient way to mechanistically couple these two opposing processes and, in turn, assert some consistency in centriole size during unperturbed cell cycles.
In conclusion, our data can explain the origin of centriole variability within cell populations and why overactive Plk1 is frequently associated with centriolar structural abnormalities, and demonstrates why numerical and structural centriole abnormalities often coincide in tumors.
Materials and methods
Cell culture
HeLa (Piel et al., 2000), U2OS, and RPE-1 (Uetake et al., 2007) cells constitutively express centrin1-GFP (C1-GFP). HeLa-C1-GFP cells with doxycycline-inducible active Plk1 (Plk1TD, carrying activating T210D mutation, HeLaPlk1TDind; Shukla et al., 2015) was used to introduce active Plk1 to S phase cells. MDA-MB-435 human cells are from the Division of Cancer Treatment and Diagnosis/Developmental Therapeutics Program Tumor Repository. Mouse inner-medullary collecting duct cell line mIMCD3, human colon cancer cell line HCT116, and human fibrosarcoma cell line HT1080 are from the American Type Culture Collection. mIMCD3 cells were grown in F12/DMEM medium (Thermo Fisher Scientific; 10565018), and other cells were grown in DMEM medium (Thermo Fisher Scientific; 10569010) supplemented with 10% FBS (Omega Scientific; FB-12) at 37°C in a humified environment with 5% CO2. For live-cell imaging, coverslips with cells were mounted in Attofluor Cell chambers (Thermo Fisher Scientific; A7816) in complete CO2 independent medium (Thermo Fisher Scientific; 18045088). To induce the formation of cilia in RPE-1, cells were grown in DMEM containing 1% penicillin/streptomycin (Thermo Fisher Scientific; 15140122) and no serum for 24 h.
Cell synchronization and drug treatments
Cells were synchronized by mitotic shake-off. The medium was aspirated from tissue flasks containing logarithmically growing cells. Cells were gently detached by tapping on the flask, collected, and replated on round 25-mm #1.5 high tolerance cover glasses (Warner Instruments; 64–0735). After shake-off, cells enter G1 phase within ∼1 h, S phase within ∼9–10 h, G2 phase within ∼18–19 h, and mitosis within ∼20–22 h. Alternatively, mitotic cells were spun down by centrifugation at 1,000 g for 3 min, the pellet was resuspended in 300 µl of DMEM, and cells were pipetted to the coverslips coated with 1 mg/ml of poly-L-lysine (Sigma-Aldrich; P5899) and fixed.
To arrest cells in S phase, 2 mM hydroxyurea (HU; Sigma-Aldrich; H8627) was added 2 h after the shake-off. To induce G2 arrest, 10 µM of RO 3306 (Tocris; 4181) was added to G1 cells. To inhibit Plk4, cells were treated with 0.5 µM centrinone B (Tocris; 5690). To arrest cells in prometaphase, cells were treated for 1 h with 100 µM monastrol (Tocris; 1305). 80 and 200 nM of Plk1 inhibitor BI2536 (BI; Selleckchem; S1109) and 110 nM of GSK (Selleckchem; S2193) were used to inhibit Plk1 activity.
Identification of cell cycle stages
During microscopy, cell cycle phases were identified as follows: G1 was recognized by the presence of two single C1-GFP signals and/or the presence of the midbody between two sister cells. Early G1 was recognized by the proximity of the sister cells and their round shape. In S phase, cells contained duplicated centrioles (four C1-GFP signals) and closely positioned centrosomes. G2 was identified by two separated and duplicated centrioles with developed procentrioles. Prophase was identified by DNA condensation. Stages of mitosis were recognized by characteristic DNA morphology.
Immunostaining of nonexpanded samples
Cells were fixed for 4 min in 1.5% formaldehyde at RT and post-fixed in methanol for 4 min at −20°C, washed in 1× PBS for 30 min, and incubated with immunofluorescence (IF) buffer (1% BSA [Sigma-Aldrich; A9647] and 0.05% Tween-20 [Sigma-Aldrich; P9416] in 1× PBS) for 15 min. Cells were labeled with primary rabbit anti-STIL antibody (a generous gift from the A. Holland laboratory [Johns Hopkins University School of Medicine, Baltimore, MD]; Moyer et al., 2015), diluted in IF buffer 1:500, and incubated for 1 h at 37°C. DNA was labeled with Hoechst 33342 (Thermo Fisher Scientific; H3570). Secondary CF647 anti-rabbit (Biotium; 20045) antibodies were used at a 1:800 dilution to label primary antibodies.
Expansion protocol for approximately fourfold expansion of centrioles and cilia
Cells growing on glass coverslips were fixed (or post-fixed after immunostaining) with 4% formaldehyde (Electron Microscopy Sciences; 15686) in 1× PBS at RT for 1 h. After fixation, the coverslips were incubated at 40°C for 16 h in a solution containing 30% acrylamide (Sigma-Aldrich; A4058) and 4% formaldehyde in 1× PBS. Cells were washed three times in 1× PBS (10 min each wash at RT). Coverslips were placed on a parafilm-covered petri dish floating in an ice water bath. Precooled gelling mixture (20% acrylamide, 7% sodium acrylate [Sigma-Aldrich; 408220], 0.04% bis-acrylamide [Sigma-Aldrich; A9926], 0.5% ammonium persulfate [Sigma-Aldrich; 248614], and 0.5% Tetramethylethylenediamine [Sigma-Aldrich; 411019]) was pipetted to the coverslips and incubated on ice for 20 min and additionally 1–2 h at RT. Following gel polymerization, a 4-mm biopsy puncher (Integra Miltex; 33–34-P/25) was used to excise several punches from each gelled sample. Dry punches were placed in a 50-ml tube and dry-preheated at >90°C for 10 min. SDS solution (200 mM SDS, 200 mM NaCl, 50 mM Tris, pH 9.0), preheated to >90°C, was added to the punches. Punches were boiled for 1 h at >90°C, with swirling every 10 min. After boiling, SDS solution containing punches was cooled to RT, and SDS was removed by exchanging 1× PBS every 20 min for the first 2–3 h, followed by an overnight wash in 1× PBS at 4°C.
Immunolabeling of expanded samples
Punches were blocked in IF buffer (1% BSA; [Sigma-Aldrich; A9647] and 0.05% Tween-20 [Sigma-Aldrich; P9416] in 1× PBS) for 1–2 h at RT, and incubated with primary antibody diluted in IF buffer for 24 h at 4°C. Punches were washed in 1× PBS for 1–2 h and incubated with secondary antibody and DAPI (1:10,000 final dilution; Thermo Fisher Scientific; D1306) in IF buffer for 24 h at 4°C. The following primary antibodies were used for immunostaining: mouse anti-acetylated tubulin (Sigma-Aldrich; T7451) at 1:4,000, rabbit anti-β tubulin (Abcam; ab15568) at 1:250, rabbit anti-ARL13B (Abcam; ab83879) at 1:500, rabbit anti-Cep290 (Abcam; ab84870) at 1:600, and rabbit anti-Cep164 (Proteintech; 22227–1-AP) at 1:500. Secondary antibodies Alexa Fluor 488 anti-mouse (Thermo Fisher Scientific; A11029) and Alexa Fluor 555 anti-rabbit (Thermo Fisher Scientific; A21429) were used at a 1:800 dilution to label primary antibodies for 24 h. After immunostaining, the samples were expanded in deionized H2O for 2 h at RT with deionized H2O exchanged every 10 min and additionally overnight at 4°C. Prior to imaging, expanded punches were mounted in Rose chambers, Attofluor cell chambers, or glass-bottom Microwell dishes (MatTek; P35G-1.5-14-C).
Widefield microscopy
Widefield imaging of expanded samples was performed with an inverted Eclipse Ti microscope (Nikon), equipped with OrcaFlash4 camera (Hamamatsu), Intensilight C-HGFIE illuminator, 60× NA 1.45, Plan Apo objective, using 1.5× magnifying tube lens. 200- or 300-nm-thick Z-sections spanning the entire centriole or central centriole region were acquired as needed. For live cell imaging, cells growing on the coverslip were mounted in Attofluor cell chambers and imaged in differential interference contrast mode using a Nikon Eclipse Ti inverted microscope, using 20× NA 0.75 objective and DS-Qi1Mc camera (Nikon). For time-lapse recordings, one 2D image was recorded every 5 min.
Structured illumination microscopy (SIM)
SIM was performed using N-SIM (Nikon), equipped with 405-, 488-, 561-, and 640-nm excitation lasers, Apo TIRF 100× NA 1.49 Plan Apo oil objective, and a back-illuminated 16 µm pixel electron multiplying CCD camera (Andor; DU897). 100-nm-thick Z-sections were acquired in 3D SIM mode and reconstructed to generate a final image using Nikon NIS-Elements software.
STORM of cartwheel protein
Before STORM imaging, coverslips with immunolabeled cells were layered with 100-nm tetra-spectral fluorescent microspheres (Thermo Fisher Scientific; T7279), which served as fiducial markers. Coverslips were mounted in Attofluor Cell chambers (Thermo Fisher Scientific; A7816), filled with STORM buffer (25 mM β-mercaptoethylamine [Sigma-Aldrich; 30070], 0.5 mg/ml glucose oxidase [Sigma-Aldrich; G2133], 67 µg/ml catalase [Sigma-Aldrich; C40], and 10% dextrose [Sigma-Aldrich; D9434], in 100 mM Tris, pH 8.0) and covered with an empty coverslip. 3D STORM imaging was performed on Nikon N-STORM4.0 system using an Eclipse Ti2 inverted microscope, Apo TIRF 100X SA NA 1.49 Plan Apo oil objective, 405-, 561-, and 647-nm excitation laser launch, and a back-illuminated EMCCD camera (Andor; DU897). The 647-nm laser line (∼150 mW out of the fiber and ∼90 mW before the objective lens) was used to promote fluorophore blinking. The 405-nm laser was used to reactivate fluorophores. The 561-nm laser was used to record the signals of fiducial markers. 20,000 to 30,000 time points were acquired every 20 ms at a 50-Hz frame rate. NIS Elements (Nikon) was used to analyze the data.
Prior to STORM imaging, centrioles labeled with C1-GFP and CF647-labeled STIL were recorded in widefield mode. A rainbow Z-color coding scheme, which typically spanned 650 nm of a working Z-imaging range, was used for signal presentation. The signals closer to the coverslip were presented in red and those further from the coverslip in blue. The original Z-color coding scheme is preserved on image panels. 3D STORM data are presented as a projection of the entire 3D volume containing STIL signal. Each recording was analyzed in a 3D volume viewer to confirm the orientation of a procentriole. Only procentrioles oriented horizontally or near horizontally were used for cartwheel length measurements.
EM
For EM analysis, cells grown on coverslips were fixed in 2.5% glutaraldehyde (Sigma-Aldrich; G5882) and 0.25% formaldehyde in 1× PBS (pH 7.4) for 1 h at RT, washed in 1× PBS for 30 min (10 min each wash), prestained with 1% osmium tetroxide (Electron Microscopy Sciences; 19100) and 1% uranyl acetate (Electron Microscopy Sciences; 22400), dehydrated in graded ethanol series, and then embedded in EMbed-812 resin (Electron Microscopy Sciences; 13940). 80-nm serial sections were sectioned, placed on the formvar-coated copper grids (SPI Supplies; 2330P-XA), and further contrasted with uranyl acetate and lead citrate. Imaging was performed using a FEI Spirit or H-7650 Hitachi transmission electron microscope, operating at 80 kV.
Image analysis, processing, and data presentation
Image analysis and assembly were performed in Photoshop (Adobe) and Fiji (National Institutes of Health). Fiji and NIS-Elements were used to measure the intensities and the length of fluorescent signals. AutoQuant X3 software (MediaCybernetics) was used for deconvolution of widefield images. For image presentation purposes, the levels of fluorescent signals were sometimes differentially adjusted between different image panels to improve the visibility of the dimmer signals. In some image panels, maximum intensity projections of all acquired Z-slices spanning entire centrioles were presented. In some instances, to better illustrate centriole width and length or a specific point discussed in text, only a central Z-section throughout a centriole is presented.
Measurement of centriole length and width
The length of expanded mother centrioles was determined by measuring the length of acetylated tubulin signal along centriole longitudinal axis at half-width-half maxima on both centriole ends. Procentriole length in S and G2 arrested cells was measured using the same criteria. The length of procentrioles in mitosis was determined by measuring the length of acetylated tubulin signal along procentriole longitudinal axes from half-width at half maximum on procentriole proximal end until the end of the acetylated tubulin signal on procentriole distal ends. Alternatively, procentriole length was measured as the distance of the proximal end of acetylated tubulin signal, until the maximum of the Cep290 signal on the distal procentriole end (Fig. 4 E). To measure centriole length from electron micrographs, the length of centriole MTs was measured from longitudinally or nearly longitudinally sectioned centrioles.
Statistical analysis
Statistical difference between two sets of data was determined in Excel using an unpaired, two-tailed Student’s t test. NS, nonsignificant; *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001. Box-and-whisker plots show the minimum, median line, upper and lower quartile, maximum, and inner and outer points. Histograms show average values ± SD. Sample sizes (the number of counted or measured centrioles and experiment numbers) are indicated above the plots or in the figure legends.
Online supplemental material
Fig. S1 shows a comparison of β tubulin and acetylated tubulin immunolabeling of expanded samples and a distribution of acetylated tubulin on centrosomes in different phases of the cell cycle. Fig. S2 shows analysis of aberrant procentrioles formed during prolonged mitosis in HT1080 and spontaneously in the MDA-MB-435 cell line. Fig. S3 shows EM analysis of procentrioles in cycling cells treated with Plk1 inhibitors from the G1 phase of the cell cycle.
Acknowledgments
We thank Dr. Meredith Metzger for critical reading of the manuscript.
This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute to J. Loncarek.
The authors declare no competing financial interests.
Author contributions: D. Kong and N. Sahabandu conducted most experiments and analyzed the data. D. Luvsanjav prepared some EM samples. K. Lukasik and A. Vasquez-Limeta analyzed the effect of Plk1 on centriole elongation. C. Sullenberger conducted STORM analysis. J. Loncarek supervised the project, conducted some EM/light microscopy, and analyzed the data. All authors discussed the data and prepared the manuscript.
References
- Aydogan M.G., Wainman A., Saurya S., Steinacker T.L., Caballe A., Novak Z.A., Baumbach J., Muschalik N., and Raff J.W.. 2018. A homeostatic clock sets daughter centriole size in flies. J. Cell Biol. 217:1233–1248. 10.1083/jcb.201801014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.Y. 2011. A clinical overview of centrosome amplification in human cancers. Int. J. Biol. Sci. 7:1122–1144. 10.7150/ijbs.7.1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J., Cizmecioglu O., Hoffmann I., and Rhee K.. 2010. PLK2 phosphorylation is critical for CPAP function in procentriole formation during the centrosome cycle. EMBO J. 29:2395–2406. 10.1038/emboj.2010.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Tillberg P.W., and Boyden E.S.. 2015. Optical imaging. Expansion microscopy. Science. 347:543–548. 10.1126/science.1260088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrétien D., Buendia B., Fuller S.D., and Karsenti E.. 1997. Reconstruction of the centrosome cycle from cryoelectron micrographs. J. Struct. Biol. 120:117–133. 10.1006/jsbi.1997.3928 [DOI] [PubMed] [Google Scholar]
- Comartin D., Gupta G.D., Fussner E., Coyaud É., Hasegan M., Archinti M., Cheung S.W., Pinchev D., Lawo S., Raught B., et al. 2013. CEP120 and SPICE1 cooperate with CPAP in centriole elongation. Curr. Biol. 23:1360–1366. 10.1016/j.cub.2013.06.002 [DOI] [PubMed] [Google Scholar]
- D’Assoro A.B., Lingle W.L., and Salisbury J.L.. 2002. Centrosome amplification and the development of cancer. Oncogene. 21:6146–6153. 10.1038/sj.onc.1205772 [DOI] [PubMed] [Google Scholar]
- Firat-Karalar E.N., Rauniyar N., Yates J.R. III, and Stearns T.. 2014. Proximity interactions among centrosome components identify regulators of centriole duplication. Curr. Biol. 24:664–670. 10.1016/j.cub.2014.01.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambarotto D., Zwettler F.U., Le Guennec M., Schmidt-Cernohorska M., Fortun D., Borgers S., Heine J., Schloetel J.G., Reuss M., Unser M., et al. 2019. Imaging cellular ultrastructures using expansion microscopy (U-ExM). Nat. Methods. 16:71–74. 10.1038/s41592-018-0238-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N.J., Godinho S.A., and Pellman D.. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature. 460:278–282. 10.1038/nature08136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geertsema H., and Ewers H.. 2016. Expansion microscopy passes its first test. Nat. Methods. 13:481–482. 10.1038/nmeth.3872 [DOI] [PubMed] [Google Scholar]
- Gilmartin A.G., Bleam M.R., Richter M.C., Erskine S.G., Kruger R.G., Madden L., Hassler D.F., Smith G.K., Gontarek R.R., Courtney M.P., et al. 2009. Distinct concentration-dependent effects of the polo-like kinase 1-specific inhibitor GSK461364A, including differential effect on apoptosis. Cancer Res. 69:6969–6977. 10.1158/0008-5472.CAN-09-0945 [DOI] [PubMed] [Google Scholar]
- Godinho S.A., Picone R., Burute M., Dagher R., Su Y., Leung C.T., Polyak K., Brugge J.S., Théry M., and Pellman D.. 2014. Oncogene-like induction of cellular invasion from centrosome amplification. Nature. 510:167–171. 10.1038/nature13277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P. 2015. Centrosomes and cancer: revisiting a long-standing relationship. Nat. Rev. Cancer. 15:639–652. 10.1038/nrc3995 [DOI] [PubMed] [Google Scholar]
- Gudi R., Haycraft C.J., Bell P.D., Li Z., and Vasu C.. 2015. Centrobin-mediated regulation of the centrosomal protein 4.1-associated protein (CPAP) level limits centriole length during elongation stage. J. Biol. Chem. 290:6890–6902. 10.1074/jbc.M114.603423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmaier G., Loncarek J., Meng X., McEwen B.F., Mogensen M.M., Spektor A., Dynlacht B.D., Khodjakov A., and Gönczy P.. 2009. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr. Biol. 19:1012–1018. 10.1016/j.cub.2009.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D., Farmer V., Shukla A., James J., Gruskin R., Kiriyama S., and Loncarek J.. 2014. Centriole maturation requires regulated Plk1 activity during two consecutive cell cycles. J. Cell Biol. 206:855–865. 10.1083/jcb.201407087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku T., Swaney J., Park J.Y., Albanese A., Murray E., Cho J.H., Park Y.G., Mangena V., Chen J., and Chung K.. 2016. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat. Biotechnol. 34:973–981. 10.1038/nbt.3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama R., and Borisy G.G.. 1981. Centriole cycle in Chinese hamster ovary cells as determined by whole-mount electron microscopy. J. Cell Biol. 91:814–821. 10.1083/jcb.91.3.814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.C., Chang C.W., Hsu W.B., Tang C.J., Lin Y.N., Chou E.J., Wu C.T., and Tang T.K.. 2013a Human microcephaly protein CEP135 binds to hSAS-6 and CPAP, and is required for centriole assembly. EMBO J. 32:1141–1154. 10.1038/emboj.2013.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.N., Wu C.T., Lin Y.C., Hsu W.B., Tang C.J., Chang C.W., and Tang T.K.. 2013b CEP120 interacts with CPAP and positively regulates centriole elongation. J. Cell Biol. 202:211–219. 10.1083/jcb.201212060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J., Hergert P., and Khodjakov A.. 2010. Centriole reduplication during prolonged interphase requires procentriole maturation governed by Plk1. Curr. Biol. 20:1277–1282. 10.1016/j.cub.2010.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Charron J., and Erikson R.L.. 2003. Role of Plk2 (Snk) in mouse development and cell proliferation. Mol. Cell. Biol. 23:6936–6943. 10.1128/MCB.23.19.6936-6943.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub M.R., Xie Z., and Stearns T.. 2010. Cep120 is asymmetrically localized to the daughter centriole and is essential for centriole assembly. J. Cell Biol. 191:331–346. 10.1083/jcb.201003009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteil G., Guerrero A., Vieira A.F., de Almeida B.P., Machado P., Mendonça S., Mesquita M., Villarreal B., Fonseca I., Francia M.E., et al. 2018. Over-elongation of centrioles in cancer promotes centriole amplification and chromosome missegregation. Nat. Commun. 9:1258 10.1038/s41467-018-03641-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B.T., and Strebhardt K.. 2006. Polo-like kinase 1: target and regulator of transcriptional control. Cell Cycle. 5:2881–2885. 10.4161/cc.5.24.3538 [DOI] [PubMed] [Google Scholar]
- Moyer T.C., Clutario K.M., Lambrus B.G., Daggubati V., and Holland A.J.. 2015. Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. J. Cell Biol. 209:863–878. 10.1083/jcb.201502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A., and Raff J.W.. 2009. Centrioles, centrosomes, and cilia in health and disease. Cell. 139:663–678. 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- Nigg E.A., Schnerch D., and Ganier O.. 2017. Impact of Centrosome Aberrations on Chromosome Segregation and Tissue Architecture in Cancer. Cold Spring Harb. Symp. Quant. Biol. 82:137–144. 10.1101/sqb.2017.82.034421 [DOI] [PubMed] [Google Scholar]
- Novak Z.A., Wainman A., Gartenmann L., and Raff J.W.. 2016. Cdk1 Phosphorylates Drosophila Sas-4 to Recruit Polo to Daughter Centrioles and Convert Them to Centrosomes. Dev. Cell. 37:545–557. 10.1016/j.devcel.2016.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M., Meyer P., Khodjakov A., Rieder C.L., and Bornens M.. 2000. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 149:317–330. 10.1083/jcb.149.2.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahabandu N., Kong D., Magidson V., Nanjundappa R., Sullenberger C., Mahjoub M.R., and Loncarek J.. 2019. Expansion microscopy for the analysis of centrioles and cilia. J. Microsc. 276:145–159. 10.1111/jmi.12841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T.I., Kleylein-Sohn J., Westendorf J., Le Clech M., Lavoie S.B., Stierhof Y.D., and Nigg E.A.. 2009. Control of centriole length by CPAP and CP110. Curr. Biol. 19:1005–1011. 10.1016/j.cub.2009.05.016 [DOI] [PubMed] [Google Scholar]
- Shimada H., Lu Q., Insinna-Kettenhofen C., Nagashima K., English M.A., Semler E.M., Mahgerefteh J., Cideciyan A.V., Li T., Brooks B.P., et al. 2017. In Vitro Modeling Using Ciliopathy-Patient-Derived Cells Reveals Distinct Cilia Dysfunctions Caused by CEP290 Mutations. Cell Rep. 20:384–396. 10.1016/j.celrep.2017.06.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A., Kong D., Sharma M., Magidson V., and Loncarek J.. 2015. Plk1 relieves centriole block to reduplication by promoting daughter centriole maturation. Nat. Commun. 6:8077 10.1038/ncomms9077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegmaier M., Hoffmann M., Baum A., Lénárt P., Petronczki M., Krssák M., Gürtler U., Garin-Chesa P., Lieb S., Quant J., et al. 2007. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr. Biol. 17:316–322. 10.1016/j.cub.2006.12.037 [DOI] [PubMed] [Google Scholar]
- Tang C.J., Fu R.H., Wu K.S., Hsu W.B., and Tang T.K.. 2009. CPAP is a cell-cycle regulated protein that controls centriole length. Nat. Cell Biol. 11:825–831. 10.1038/ncb1889 [DOI] [PubMed] [Google Scholar]
- Tsou M.F., Wang W.J., George K.A., Uryu K., Stearns T., and Jallepalli P.V.. 2009. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev. Cell. 17:344–354. 10.1016/j.devcel.2009.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y., Loncarek J., Nordberg J.J., English C.N., La Terra S., Khodjakov A., and Sluder G.. 2007. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J. Cell Biol. 176:173–182. 10.1083/jcb.200607073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev I.A., and Chentsov Y.u.S.. 1982. Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 93:938–949. 10.1083/jcb.93.3.938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.J., Soni R.K., Uryu K., and Tsou M.F.. 2011. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J. Cell Biol. 193:727–739. 10.1083/jcb.201101109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassie A.T., Zhao Y., and Boyden E.S.. 2019. Expansion microscopy: principles and uses in biological research. Nat. Methods. 16:33–41. 10.1038/s41592-018-0219-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Y.L., Anzola J.V., Davis R.L., Yoon M., Motamedi A., Kroll A., Seo C.P., Hsia J.E., Kim S.K., Mitchell J.W., et al. 2015. Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science. 348:1155–1160. 10.1126/science.aaa5111 [DOI] [PMC free article] [PubMed] [Google Scholar]