Microtubule-severing enzymes are evolutionarily conserved proteins that remodel microtubules and regulate microtubule dynamics in diverse cellular and developmental processes. Using a combination of biochemical, genetic, and live-imaging approaches, Joly et al. show how phosphorylation of the AAA+ microtubule-severing enzyme Katanin regulates its activity and stability during C. elegans development.
Abstract
The evolutionarily conserved microtubule (MT)-severing AAA-ATPase enzyme Katanin is emerging as a critical regulator of MT dynamics. In Caenorhabditis elegans, Katanin MT-severing activity is essential for meiotic spindle assembly but is toxic for the mitotic spindle. Here we analyzed Katanin dynamics in C. elegans and deciphered the role of Katanin phosphorylation in the regulation of its activity and stability. Katanin is abundant in oocytes, and its levels drop after meiosis, but unexpectedly, a significant fraction is present throughout embryogenesis, where it is dynamically recruited to the centrosomes and chromosomes during mitosis. We show that the minibrain kinase MBK-2, which is activated during meiosis, phosphorylates Katanin at multiple serines. We demonstrate unequivocally that Katanin phosphorylation at a single residue is necessary and sufficient to target Katanin for proteasomal degradation after meiosis, whereas phosphorylation at the other sites only inhibits Katanin ATPase activity stimulated by MTs. Our findings suggest that cycles of phosphorylation and dephosphorylation fine-tune Katanin level and activity to deliver the appropriate MT-severing activity during development.
Introduction
The active remodeling of the microtubule (MT) cytoskeleton is instrumental for multiple dynamic cellular processes. MT-Severing Enzymes (MSEs), which include Katanin, Fidgetin, and Spastin, are emerging as an important class of evolutionarily conserved MT remodelers, with critical functions in the regulation of MT dynamics in diverse cellular and biological contexts (McNally and Roll-Mecak, 2018; Sharp and Ross, 2012). Instead of regulating MT dynamics by interacting with the plus or minus ends of MTs, these enzymes interact with the MT lattice and might facilitate the extraction of tubulin dimers, eventually leading to MT severing (Vemu et al., 2018; Zehr et al., 2020). Depending on the amount of GTP-tubulin available, these severing events, leading to the formation of new MT extremities, can be used as seeds for the nucleation of new MTs or can induce the rapid depolymerization of MTs (Kuo et al., 2019; Roll-Mecak and Vale, 2006; Vemu et al., 2018). Despite considerable progress in deciphering the function of these enzymes, how these molecular machines are regulated in space and time to deliver the adequate level of MT-severing activity remains poorly understood.
The nematode Caenorhabditis elegans provides a dynamic developmental context for the study of Katanin function and regulation during meiotic and mitotic cell divisions (Bowerman and Kurz, 2006; DeRenzo and Seydoux, 2004; Pintard and Bowerman, 2019). In C. elegans, the mei-1 and mei-2 genes (meiosis defective 1 and 2), which encode the catalytic (p60, -ATPases Associated with diverse cellular Activities [AAA ATPase]) and regulatory (p80-like) subunits, respectively, of Katanin, are essential for meiotic spindle assembly (Mains et al., 1990; McNally and Vale, 1993; Roll-Mecak and McNally, 2010; Sharp and Ross, 2012; Srayko et al., 2000). Katanin MT-severing activity is required to produce seeds for the MT nucleation essential for meiotic spindle formation (Joly et al., 2016; Srayko et al., 2006). Katanin also keeps the meiotic spindles short, triggers the anaphase shortening of the spindle, and severs MTs between the polar bodies and the female pronucleus during meiosis II (Gomes et al., 2013; McNally et al., 2006). While essential for meiosis, Katanin must be rapidly inactivated before the first mitotic division (Clark-Maguire and Mains, 1994). Failure to down-regulate Katanin in mitosis results in embryonic lethality, presumably as a consequence of inappropriate MT severing during the early mitotic divisions (Clark-Maguire and Mains, 1994; Kurz et al., 2002; Müller-Reichert et al., 2010; Pintard et al., 2003a,b). Over the past two decades, significant progress has been made in identifying the pathways responsible for Katanin down-regulation after meiosis. First, an E3 ubiquitin ligase nucleated by Cullin 3 (CUL-3) and using the substrate adaptor protein Maternal Effect Lethal 26 (MEL-26; Dow and Mains, 1998) triggers MEIosis defective 1 (MEI-1) ubiquitylation and its subsequent degradation by the 26S proteasome (Furukawa et al., 2003; Pintard et al., 2003b; Xu et al., 2003. Second, MiniBrain Kinase 2 (MBK-2), a member of the Dual-specificity and tyrosine (Y)-Regulated Kinase (DYRK) family of protein kinases, which is specifically activated during female meiosis to orchestrate the oocyte-to-embryo transition (Cheng et al., 2009), contributes to MEI-1 degradation after meiosis (Pellettieri et al., 2003; Quintin et al., 2003; Stitzel et al., 2006, 2007). However, whether MBK-2 is directly required for MEI-1 degradation by the CRL3MEL-26 E3 ligase or acts in a parallel degradation pathway with an unknown ubiquitin ligase is still unclear (Beard et al., 2016; Lu and Mains, 2007). Reducing mbk-2 function enhances the incomplete lethality of mel-26(null) at 15°C, arguing that MEL-26 acts in parallel to MBK-2 (Lu and Mains, 2007). Alternatively, MBK-2 might regulate both Katanin activity and stability, which could also explain this genetic interaction. Additional layers of Katanin regulation involve a Protein Phosphatase 4 (PP4) complex, using Protein Phosphatase Four Regulatory subunit 1 (PPFR-1) as a regulatory subunit (henceforth PP4PPFR-1), which genetically acts as an activator of Katanin during meiosis (Gomes et al., 2013; Han et al., 2009). Consistent with kinases and phosphatases regulating Katanin activity and/or stability, MEI-1 exhibits a complex phosphorylation pattern by 2D gel analysis, and this pattern is altered upon inactivation of the PP4PPFR-1 phosphatase complex (Gomes et al., 2013). Finally, postmeiotic Katanin down-regulation has been shown to involve inhibition of mei-1 mRNA translation (Li et al., 2009).
In summary, the current view is that Katanin is eliminated after meiosis (Bowerman and Kurz, 2006; DeRenzo and Seydoux, 2004; Verlhac et al., 2010) and therefore does not participate in mitosis. Here we revise this model and show, using an endogenously tagged GFP line generated by CRISPR/Cas9, that Katanin is readily expressed during embryogenesis, where it is actively recruited to the centrosomes and chromosomes during mitosis. To understand how Katanin is tolerated during mitosis, we investigated the precise role of MEI-1 phosphorylation in the regulation of Katanin activity and stability. By combining biochemical approaches with structure–function analysis in vivo, we show that MBK-2 phosphorylates MEI-1 and MEI-2 at multiple residues to inhibit MT-stimulated ATPase Katanin activity. In addition, we demonstrate unequivocally that phosphorylation of MEI-1 by MBK-2 at a single serine (S92) is both necessary and sufficient to target MEI-1 for degradation after meiosis but probably also during mitosis. Overall, our findings suggest that Katanin stability and activity is finely tuned by cycles of phosphorylation and dephosphorylation to deliver the adequate level of MT-severing activity at the right time and place during C. elegans development.
Results
Spatiotemporal analysis of Katanin dynamics during C. elegans development
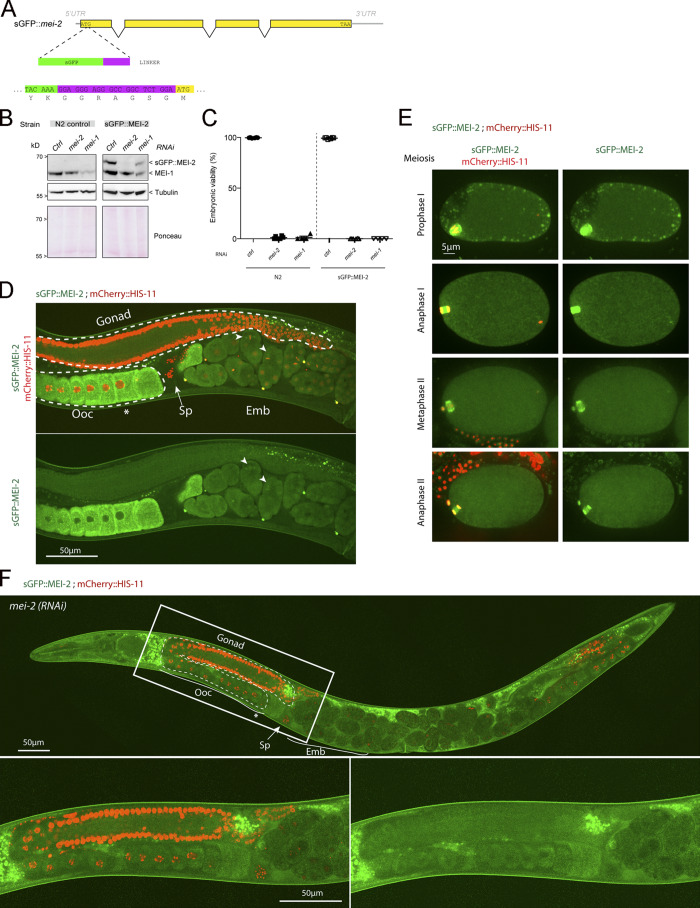
The analysis of Katanin dynamics in C. elegans has so far been limited to the analysis of a strain generated by microparticle bombardment expressing a nonfunctional GFP::MEI-1 version driven by the germline-specific promoter pie-1 (McNally et al., 2006; Pellettieri et al., 2003; Pintard et al., 2003a; Stitzel et al., 2006). To visualize the spatiotemporal expression of a functional C. elegans Katanin by live-imaging approaches, we generated a superfolder GFP (sGFP) knock-in mei-2 allele (sGFP::mei-2) using the CRISPR/Cas9 system (Materials and methods; Fig. S1, A and B). Embryos expressing sGFP::MEI-2 were fully viable (100%, n > 200, n = 4; Fig. S1 C), indicating that the sGFP tag does not interfere with the essential function of Katanin. Western blot experiments using GFP antibodies confirmed the production of sGFP::MEI-2 at the expected size (Fig. S1 B). Using a similar approach, we tried to generate sGFP knock-in mei-1 allele, but we did not recover any viable homozygous strains, indicating that the sGFP tag alters MEI-1 function, consistent with previous observations (Beard et al., 2016; Joly et al., 2016).
Figure S1.
Construction and characterization of a C. elegans line expressing endogenously tagged MEI-2 with sGFP. (A) Schematics of the mei-2 locus showing insertion of sGFP before the ATG initiation codon by CRISPR/Cas9-based genome editing. The nucleotide and protein sequence of the end of the sGFP (green) and the linker (purple) are indicated. (B) Western blot analysis of embryonic extracts from N2 or sGFP::MEI-2 strain exposed to mock (control), mei-2, or mei-1 RNAi using GFP and MEI-1 antibodies (upper panel) and tubulin (middle panel) antibodies. The lower panel shows Ponceau staining of the membrane. (C) Graphs showing the percentage of viability of N2 or sGFP::MEI-2 strain exposed to mock (Ctrl), mei-2, or mei-1 RNAi. Error bars represent SEM. n = 4 independent experiments performed in triplicate with >100 embryos each. (D) Spinning disk confocal micrographs of adult worm expressing sGFP::MEI-2 (in green) and mCherry::HIS-11 (in red) exposed to control RNAi. Arrowheads point to mitotic spindles. Sp, spermathecal; ooc, oocytes. The asterisk (*) indicates the –1 oocytes arrested in prophase of meiosis I. Note that in this worm, the oocytes adjacent to the spermatheca have resumed meiosis. (E) Spinning disk confocal micrographs of early embryos expressing sGFP::MEI-2 (in green) and mCherry::HIS-11 (in red) during meiosis. The anterior of the embryo is oriented toward the left in this and other figures. Scale bar represents 5 µm. (F) Spinning disk confocal micrographs of adult worm expressing sGFP::MEI-2 (in green) and mCherry::HIS-11 (in red) exposed to mei-2(RNAi). Insets are higher magnifications of the boxed regions. Head of the worm is on the left of the picture. The full worm was reconstituted from ∼10 different views and assembled using Photoshop. The germline is delimited by dashed lines. Sp, spermatheca; Ooc, oocytes; Emb, embryos; *, –1 oocyte. Scale bar represents 50 µm.
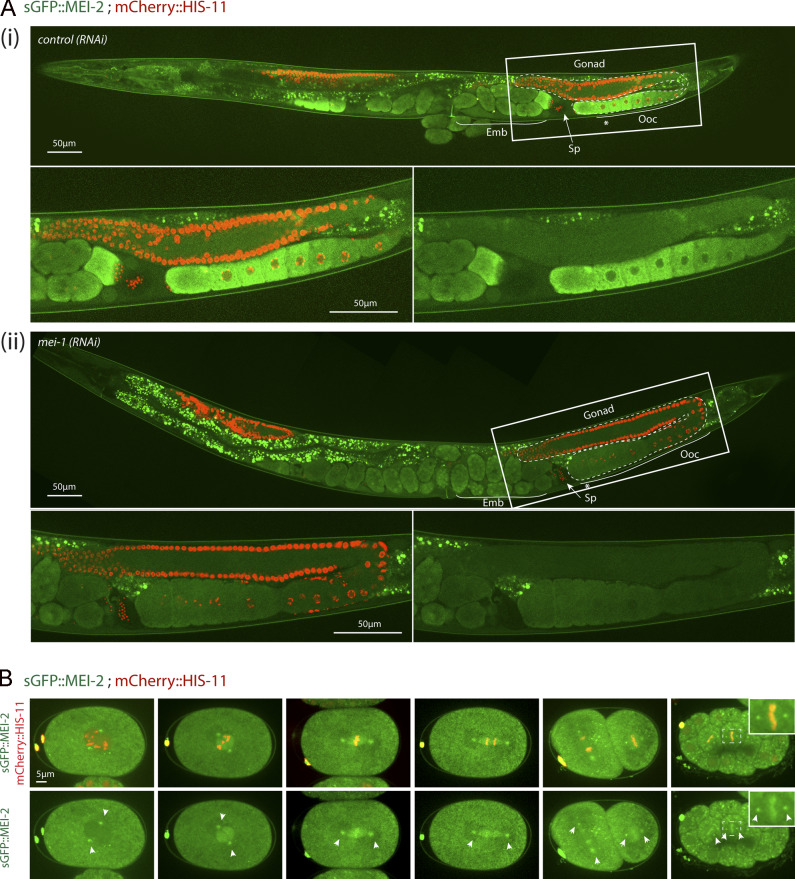
To visualize MEI-2 dynamics during C. elegans development, we used spinning disk confocal microscopy and recorded anesthetized adult worms expressing sGFP::MEI-2 and mCherry::HIS-11 as a DNA marker. In the germline, sGFP::MEI-2 was detected in the proximal gonad, where it localized to mature oocytes, consistent with previous immunolocalization studies (Srayko et al., 2000). sGFP::MEI-2 was exclusively cytoplasmic and gradually accumulated to reach a maximum level in the “–1 oocytes” (Fig. 1 A, asterisk), which are close to the spermatheca (Fig. 1 Ai). The GFP signal is specific to Katanin, because it was strongly reduced upon inactivation of mei-1 by RNAi (Fig. 1 Aii). Consistently, we noticed that MEI-2, when produced in Escherichia coli, is unstable in the absence of MEI-1. Thus, sGFP::MEI-2 is a good proxy to visualize Katanin dynamics in C. elegans. In early and late embryos, a bright GFP signal was detected on the polar bodies, but the signal in the cytoplasm was strongly reduced compared with the signal detected in oocytes (Fig. 1 Ai). Live imaging of dissected embryos revealed that sGFP::MEI-2 localizes to the meiotic spindle during meiosis I and meiosis II (Fig. S1 E), consistent with previous MEI-2 immunolocalization studies (Srayko et al., 2000). Unexpectedly, sGFP::MEI-2 was also found in mitotic embryos, not only at the one-cell stage but also in later stages. In one-cell embryos, sGFP::MEI-2 first localized to the centrosomes before nuclear envelope breakdown and then invaded the pronuclear space and accumulated on the mitotic spindle and to the chromosomes. sGFP::MEI-2 was similarly dynamically recruited to the centrosomes and chromosomes during mitosis at the two-cell stage, as well as in later stages (Fig. 1 B). Although it was believed that Katanin is eliminated after meiosis and does not participate in mitosis in C. elegans embryos, our observations indicate that a fraction of Katanin is present during embryogenesis.
Figure 1.
sGFP::MEI-2 dynamics during C. elegans development. (A) Spinning disk confocal micrographs of adult worm expressing sGFP::MEI-2 (in green) and mCherry::HIS-11 (in red) exposed to control (i) or mei-1(RNAi) (ii). Insets are higher magnifications of the boxed regions. Head of the worm is on the left of the picture. The full worm was reconstituted from ∼10 different views and assembled using Photoshop. The germline is delimited by dashed lines. Sp, spermatheca; Ooc, oocytes; Emb, embryos; *, –1 oocyte. Scale bar represents 50 µm. (B) Spinning disk confocal micrographs of early embryos expressing sGFP::MEI-2 (in green) and mCherry::HIS-11 (in red) at the one-cell (P0) and two-cell (AB and P1) stages. The anterior of the embryo is oriented toward the left in this and other figures. Scale bar represents 5 µm.
Phosphorylation of the N-terminal part of MEI-1 inhibits MT-stimulated Katanin ATPase activity
The finding that Katanin is readily detectable during embryogenesis prompted us to determine the exact molecular mechanisms regulating Katanin activity and stability in space and time. Several lines of evidence indicate that Katanin is regulated through phosphorylation: (a) MEI-1 exhibits a complex phosphorylation pattern by 2D gel electrophoresis (Gomes et al., 2013), (b) inactivation of the PP4PPFR-1 complex alters this phosphorylation pattern, (c) PP4PPFR-1 acts genetically as a Katanin activator (Gomes et al., 2013; Han et al., 2009), and (d) MBK-2 directly phosphorylates MEI-1 at the meiosis-to-mitosis transition, possibly targeting Katanin for degradation (Pang et al., 2004; Pellettieri et al., 2003; Quintin et al., 2003; Stitzel et al., 2006). As MBK-2 localizes at the centrosomes and chromosomes in mitotic embryos during embryogenesis (Pellettieri et al., 2003), similarly to Katanin, we revisited the exact role of MBK-2 in the regulation of Katanin activity and stability.
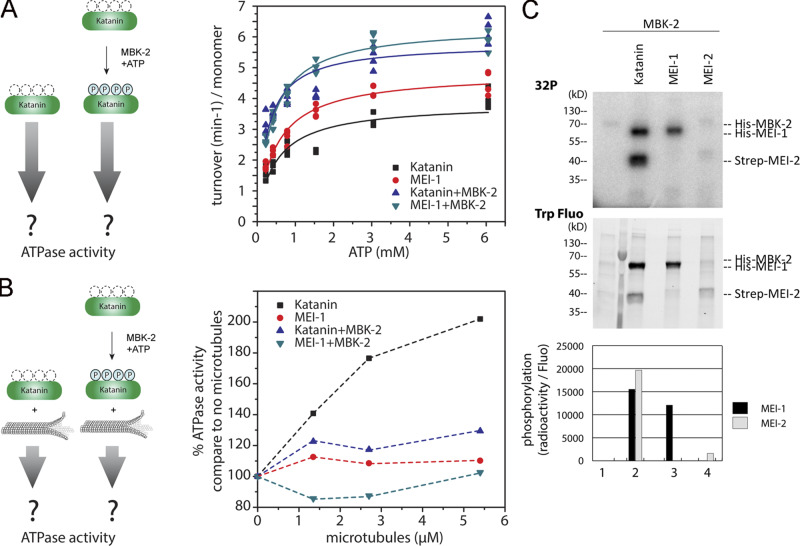
It has been shown that Katanin ATPase activity is stimulated by the presence of MTs (McNally and Vale, 1993; Hartman et al., 1998; Joly et al., 2016; Zehr et al., 2020). We thus investigated whether Katanin phosphorylation by MBK-2 could regulate Katanin ATPase activity stimulated by MTs. To this end, we prephosphorylated Katanin by MBK-2 and measured its ATPase activity in vitro, in the absence (Fig. 2 A) or presence (Fig. 2 B) of MTs. We first controlled that phosphorylation of Katanin by MBK-2 is not drastically changing the ATPase activity of the enzyme (Vmax or Km, Fig. 2 A). We then tested the effect of MTs on Katanin ATPase activity. While MTs stimulated the ATPase activity of Katanin by approximately two- to threefold, they failed to stimulate the ATPase activity of Katanin that had been prephosphorylated by MBK-2, which exhibits only a basal ATPase activity in these conditions (Fig. 2 B). These results indicate that Katanin phosphorylation by MBK-2 inhibits its MT-stimulated ATPase activity.
Figure 2.
MEI-1 phosphorylation by MBK-2 inhibits MT-stimulated Katanin ATPase activity. (A) ATPase activity of Katanin or MEI-1 alone before and after phosphorylation by MBK-2 as a function of different ATP concentrations. Turnover from four independent experiments are represented and fitted using Origin. (B) ATPase activity of Katanin or MEI-1 alone before and after its phosphorylation by MBK-2 as a function of MT concentration. (C) Radioactive in vitro kinase assay using MBK-2 kinase and Katanin, MEI-1 or MEI-2 alone as substrates. Autoradiograph of SDS-PAGE showing γ-[32P] incorporation into MEI-1 and MEI-2 (upper panel). The graph corresponds to the quantification of the radioactivity incorporated into the MEI-1 or MEI-2 divided by the total amount of MEI-1 or MEI-2 quantified using tryptophan fluorescence (Stain Free; Bio-Rad). This experiment is representative of one experiment that was reproduced three times.
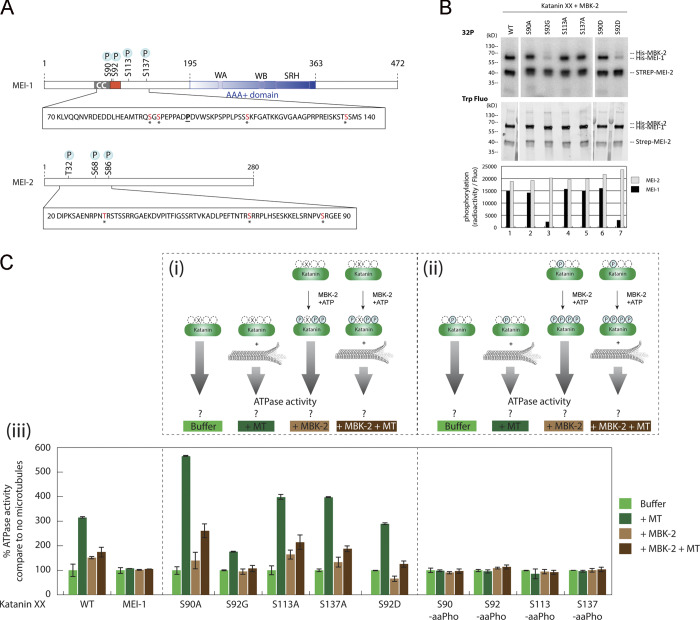
We next set out to identify the Katanin residues phosphorylated by MBK-2 responsible for this activity. To this end, we performed in vitro kinase assays using highly purified components (MBK-2 and Katanin) produced in E. coli (Fig. 2 C) and subjected the reaction product to liquid chromatography/tandem mass spectrometry (LC-MS/MS) analysis (Fig. 3 A). This analysis showed that MBK-2 phosphorylates MEI-1 at S92, consistent with a previous report (Stitzel et al., 2006), but also at S90, S113, and S137 (Fig. 3 A), consistent with the appearance of multiple phosphorylated MEI-1 isoforms in 2D gel analysis of embryonic extracts (Gomes et al., 2013). All these residues are located in the N-terminal regulatory domain of MEI-1. MBK-2 also phosphorylates MEI-2 at T32, S68, and S86 but only if MEI-1 is present, indicating that MEI-1 is essential for MBK-2–dependent MEI-2 phosphorylation (Fig. 2 C). Consistent with MBK-2 phosphorylating MEI-1 and MEI-2 at multiple sites, MBK-2 induced important mobility shifts to MEI-1 and MEI-2, with the appearance of multiple slow migrating bands on Phos-Tag SDS-PAGE that typically resolves the different phosphorylated isoforms (Fig. S2 A).
Figure 3.
Phosphorylation of the N-terminal disordered region of MEI-1 inhibits the stimulatory effect of MTs on Katanin ATPase activity. (A) Schematic representation of MEI-1 and MEI-2. The positions of phosphorylated sites identified by LC-MS/MS after in vitro phosphorylation of Katanin by MBK-2 are indicated by asterisks (*). Red box represents the PEST sequence, a predicted interacting site for MEL-26 (Fig. S5). AAA+ domain in blue contains Walker A (WA) and Walker B (WB) motifs as well as second region of homology (SRH). CC, coiled-coil. The position of the gain-of-function mutation P99L is indicated (underlined). (B) Radioactive in vitro kinase assays using MBK-2 kinase and Katanin, WT or containing serine to alanine substitutions in MEI-1 N-terminal at specific positions, as substrates. Autoradiograph of SDS-PAGE showing γ-[32P] incorporation in MEI-1 and MEI-2 (upper panel). Total Katanin present in each lane of the same SDS-PAGE using Stain Free (Bio-Rad), based on tryptophan labeling (middle panel). Graphs showing quantification of the ratio of radioactive MEI-1 and MEI-2 versus the total amount of protein (bottom panel). Gels were quantified using ImageJ, and results are represented in the histogram. The results shown are representative of one experiment that was reproduced three times. (C) Unmodified and phosphorylated Katanin ATPase activity in the absence or presence of MTs. The top panels (Ci and Cii) show the schematics of the experiment and the different Katanin versions used in the assay. Ciii, the bottom panel, shows Katanin ATPase activity presented as percentage of Katanin ATPase activity measured without MTs and without MBK-2. For each Katanin, the measured ATP activity in the absence of MT and MBK-2 was normalized to 100% (first condition, light green bars). (Ci) Katanin (green oval), WT or containing site-specific serine to alanine substitution in MEI-1, was phosphorylated or not by MBK-2, and then the ATPase activity of the enzyme was measured in the absence or presence of MTs. (Cii) Katanin containing MEI-1 phosphorylated at specific sites by genetic code expansion in E. coli was phosphorylated or not by MBK-2, and then the ATPase activity of the enzyme was measured in the absence or presence of MTs. Experiments in each condition were performed at least four times.
Figure S2.
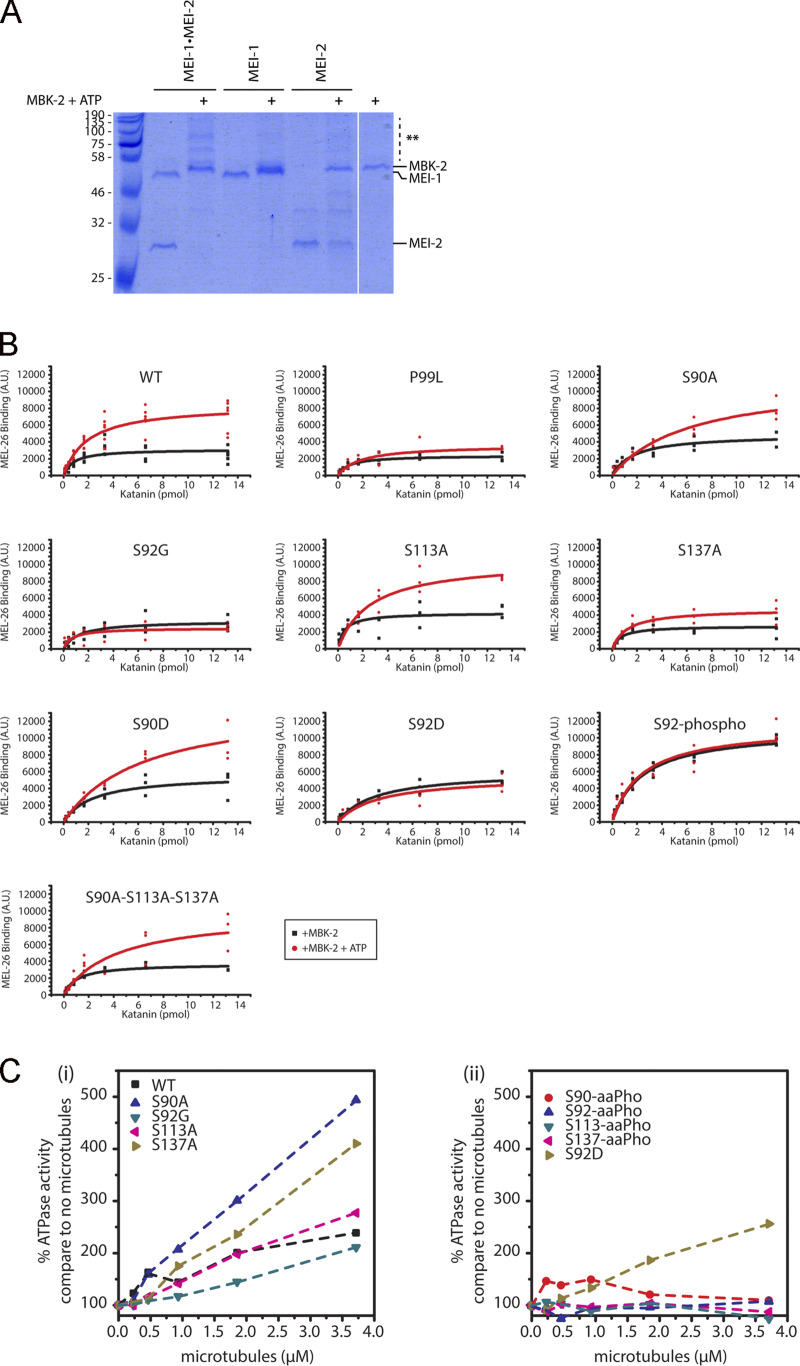
Phosphorylation of Katanin affects MEL-26 binding and its MT-dependent activity. (A) Qualitative analysis of phosphorylated Katanin on PhosTag SDS-PAGE. Katanin, MEI-1, or MEI-2 phosphorylated or not by MBK-2 ± ATP was separated on PhosTag SDS PAGE 7% to resolve the phosphorylated forms and revealed by Coomassie blue staining. (B) Quantitative analysis of MEL-26 binding to Katanin. Detailed quantification of the MEL-26 interaction detected on DotBlot membranes (presented in Fig. 4). Quantification was performed using ImageJ plugins. Raw values from at least three different experiments were plotted and analyzed using Origin. (C) Phosphorylation of the N-terminal Katanin region inhibits MT-dependent stimulation of Katanin ATPase activity. (Ci) Graphs showing the ATPase activity of Katanin WT and variants in the presence of increasing concentrations of MTs. (Cii) Graphs showing the ATPase activity of Katanin uniquely phosphorylated at specific sites (S-aaPho) compared with a Katanin variant harboring a phosphomimetic residue (S92D) in the presence of increasing concentrations of MTs.
To evaluate the relative contribution of each site of MEI-1 phosphorylation by MBK-2, we produced and purified Katanin with each “phospho-site” substituted by a nonphosphorylatable residue (alanine or glycine) and repeated the in vitro kinase assay with MBK-2 in the presence of radiolabeled [γ32P]-ATP (Fig. 3 B). We substituted S92 by a glycine because we were unable to produce a full-length version of MEI-1 with the S92 substituted by an alanine in E. coli. Substitution of S92 by glycine (S92G) decreased [γ32P]-ATP incorporation by 80%, whereas the individual substitution of the other residues (S90A or S113A or S137A) had only a modest effect. We conclude that S92 is the main residue phosphorylated by MBK-2, consistent with the previous report (Stitzel et al., 2006).
To test the contribution of each MEI-1 phospho-site to the inhibition of MT-dependent stimulation of the ATPase activity of Katanin, we produced and purified Katanin with MEI-1 singly phosphorylated at each position (S90, S92, S113, and S137) using phospho-serine incorporation in E. coli by genetic code expansion (Rogerson et al., 2015). Soluble Katanin complexes with MEI-1 phosphorylated at each position were produced in E. coli, indicating that phospho-serine incorporation does not drastically alter Katanin solubility or stability. Then, we measured the ATPase activity of Katanin, nonmodified or phosphorylated at each position in the presence or absence of MTs (Fig. 3 Ciii). Strikingly, while MTs greatly stimulated the ATPase activity of nonmodified Katanin (WT or variants), they failed to stimulate the ATPase activity of Katanin singly phosphorylated at any of the four serine residues, indicating that MEI-1 phosphorylation in the N-terminal regulatory domain renders Katanin insensitive to the presence of MTs (Fig. 3 Ciii). Introduction of phosphomimetic residue (aspartate), however, failed to phenocopy the effect of MEI-1 phosphorylation on the ATPase activity of Katanin (Fig. 3 C, S92D vs. S92-aaPho). Indeed, introduction of a negatively charged aspartate at position S92 did not inhibit MT-dependent ATPase activity of Katanin, whereas the phosphorylation of this variant by MBK-2 totally abrogated MT-dependent ATPase activity stimulation. Taken together, these results indicate that MEI-1 phosphorylation in its N-terminal regulatory domain renders Katanin insensitive to the presence of MTs. We conclude that MBK-2, by phosphorylating the catalytic MEI-1 Katanin subunit, inhibits Katanin ATPase activity stimulated by MTs.
Site-specific MEI-1 phosphorylation enhances its affinity for MEL-26
The finding that Katanin ATPase activity is inhibited by MEI-1 phosphorylation might explain why Katanin is tolerated during mitosis. However, a significant Katanin fraction is targeted for degradation after meiosis. Given that the MBK-2–dependent S92 phosphorylation of MEI-1 has been implicated in Katanin degradation (Stitzel et al., 2006), we investigated whether phosphorylation of that site is necessary and sufficient to target Katanin for CRL3MEL-26-mediated degradation, or whether the other MEI-1 phosphorylation sites also contribute to this process.
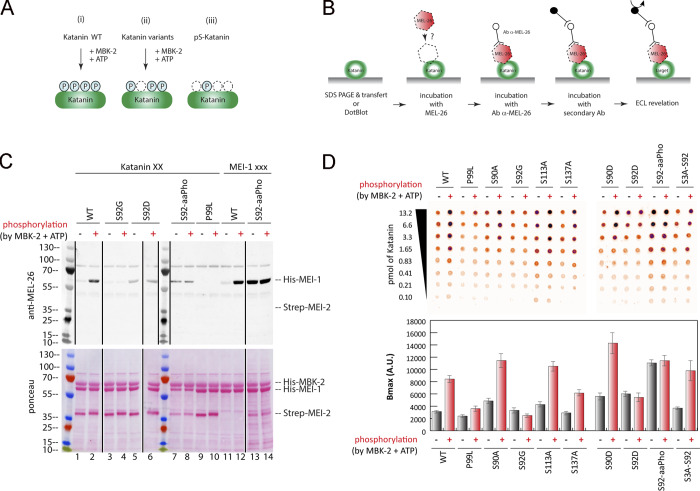
Although the interaction between MEL-26 and MEI-1 has been detected by different approaches, including yeast two-hybrid (Gomes et al., 2013; Pintard et al., 2003b), no assays have been used to detect and quantify the direct interaction between the purified proteins. We thus developed a sensitive assay (Far-Western ligand binding assay) to monitor and quantify the interaction between MEL-26 and MEI-1 (Fig. 4). Briefly, Katanin was separated on SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membrane (denaturing conditions) or directly spotted on nitrocellulose membrane (native conditions). The membrane was then incubated with MEL-26, and MEL-26–Katanin interaction was detected with anti-MEL-26 antibodies (Fig. 4).
Figure 4.
Site-specific MEI-1 phosphorylation at S92 increases MEI-1 affinity for MEL-26. (A) Schematic of the different phosphorylated forms (P, phosphate) of Katanin (green oval) used in the assay. (Ai) WT Katanin phosphorylated by MBK-2. (Aii) Katanin with site-specific substitutions replacing serine by nonphosphorylatable alanine. (Aiii) Site-specific Katanin phosphorylation by genetic code expansion in E. coli. (B) Schematics of the different steps of the Far-Western blot method used to detect the interaction between MEI-1 and MEL-26. Purified Katanin (green oval) was separated on SDS-PAGE and transferred to PVDF membrane (denatured as in C) or directly spotted on nitrocellulose membrane (native as in D). The membrane was then incubated with purified MEL-26 (red hexagon). After extensive washing of the membrane, the interaction between MEL-26 and MEI-1 was detected using MEL-26 antibodies and classic ECL revelation. (C) MEL-26–Katanin interaction detected by Far-Western ligand binding assay after separation of Katanin or MEI-1 using SDS-PAGE (denaturing conditions). Top panel corresponds to MEL-26 detection via specific antibody against MEL-26 and ECL revelation. Bottom panel shows the same membrane stained with Ponceau Red to show protein loading. Experiment was performed at least three times, and the membranes presented are representative of the results. (D) MEL-26–Katanin interaction detected by Far-Western ligand binding assay after spotting decreasing amounts (top > down) of Katanin on nitrocellulose membrane (native conditions). Histogram summary of the maximal binding (Bmax) values extracted from the fitted binding curve are presented in Fig. S2.
In this assay, MEL-26 weakly interacted with unmodified MEI-1. However, a robust binding was observed between MEL-26 and MEI-1 phosphorylated by MBK-2 (Fig. 4, C and D [lanes 1–2 and 11–12]). Consistent with previous genetic data, indicating that the product of the gain of function allele MEI-1 P99L is refractory to degradation (Clark-Maguire and Mains, 1994; Pintard et al., 2003b), this variant failed to robustly interact with MEL-26 even after phosphorylation by MBK-2 (Fig. 4, C and D [lanes 9–10]). Likewise, only weak binding was observed between the nonphosphorylatable MEI-1 S92G and MEL-26 (Fig. 4, C and D [lanes 3–4]). Individually substituting the other serines with alanines (nonphosphorylatable) did not alter MEL-26 binding to MEI-1 even after MEI-1 phosphorylation. Importantly, strong binding between MEI-1 and MEL-26 was observed using MEI-1 uniquely phosphorylated at S92 by phosphoserine incorporation in E. coli (MEI-1 S92-aaPho, Fig. 4, C and D [lanes 13–14]). The MEI-1 S92D phospho-mimetic also constitutively interacted with MEL-26, although at a reduced level, indicating that aspartate only partially mimics MEI-1 phosphorylation (Fig. 4, C and D [lanes 5–6]). Prior phosphorylation of MEI-1 S92D by MBK-2 did not enhance MEL-26 affinity for MEI-1 (compare Fig. 4 C and D, lanes 5 and 6), further indicating that MEI-1 phosphorylation at the other serines does not contribute to MEI-1 binding to MEL-26. Taken together, these observations unequivocally show that MEI-1 phosphorylation by MBK-2 at S92, but not at the other serines (S90, S113, or S131), increases MEI-1 affinity for MEL-26 in vitro.
MEI-1 phosphorylation at residue S92 is necessary and sufficient to target Katanin for degradation
To decipher whether MEI-1 phosphorylation at S92 is necessary and sufficient for MEI-1 degradation in vivo, we generated lines expressing FLAG::mei-1 RNAi-resistant transgenes (Fig. 5 A; hereafter FLAG::mei-1R) producing FLAG::MEI-1 WT, the nonphosphorylatable FLAG::MEI-1 S92A, or the phosphomimetic FLAG::MEI-1 S92D proteins using Mos1-mediated Single Copy Insertion (MosSCI; Frøkjaer-Jensen et al., 2008). We then tested the ability of the transgenes to support viability upon endogenous MEI-1 depletion by RNAi.
Figure 5.
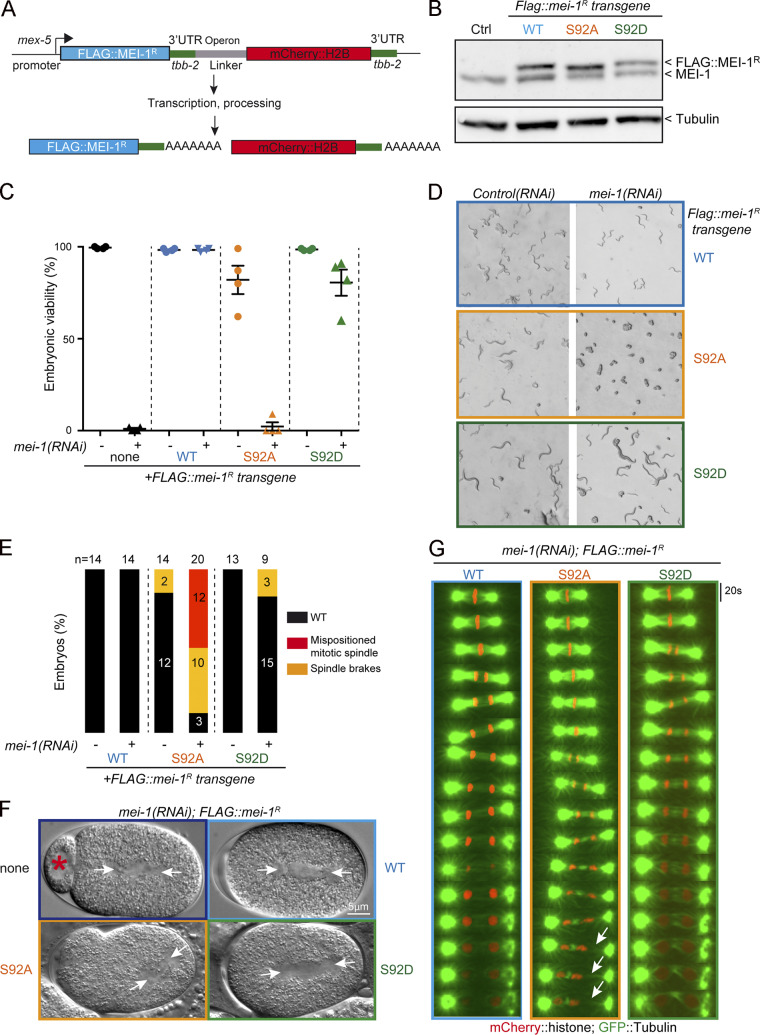
The nonphosphorylatable MEI-1 S92A version is toxic in vivo in the absence of endogenous MEI-1. (A) Schematic showing the strategy used for expressing RNAi-resistant mei-1R S92A and mei-1RS92D transgenes (mei-1R) in the C. elegans germline. The operon linker is the intercistronic region from the gpd-2/gpd-3 operon. (B) Western blot analysis of embryonic extracts of the indicated genotype using MEI-1 (upper panel) and tubulin (lower panel) antibodies. (C) Viability of embryos expressing FLAG::mei-1R transgenes WT and mutants upon RNAi-mediated inactivation of endogenous mei-1. Error bars represent SEM; n = 4 independent experiments performed in triplicate with >100 embryos each. (D) DIC images of animal progeny with the indicated genotype exposed to control or mei-1(RNAi) for 36 h at 23°C. (E) Percentage of mitotic embryos similar to WT (black bars), presenting a misaligned mitotic spindle (red bars) or a mitotic spindle that breaks during mitosis (orange bars). The number of embryos analyzed is indicated and was generated by aggregation of more than three independent experiments. (F) DIC images of one-cell-stage embryos of the indicated genotype upon inactivation of endogenous mei-1. Arrowheads indicate the mitotic spindle. (G) Live images of mitotic spindles of the indicated genotype carrying GFP::α-Tubulin (green) and Histone2B::mCherry (red) transgenes. Arrows indicate locations where the mitotic spindle breaks.
All transgenes were readily expressed in embryos; FLAG::MEI-1 S92A and S92D were produced at the expected size and at levels similar to FLAG::MEI-1 WT and endogenous MEI-1 (Fig. 5 B). Embryos from animals expressing these transgenes were viable (>80% embryonic viability for FLAG::mei-1R S92A), indicating that these variants do not act as strong dominant negatives (Fig. 5, C and D). We next used RNAi to deplete endogenous mei-1 to evaluate the functional consequences of expressing FLAG::MEI-1 WT, S92A, or S92D as the sole source of MEI-1 (Fig. S3 and 6 A).
Figure S3.
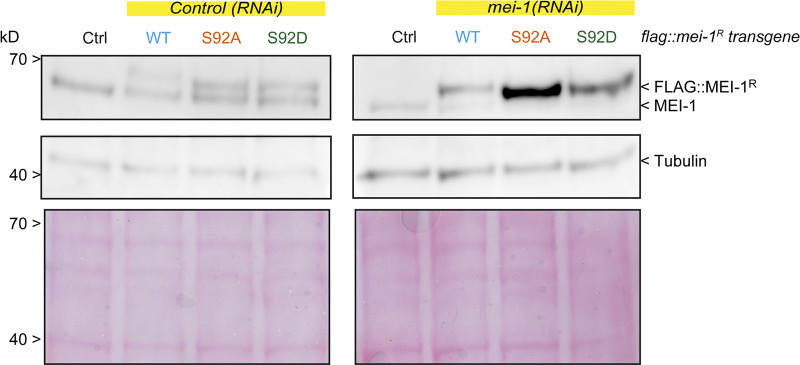
MEI-1 phosphorylation (S92) is necessary and sufficient to target MEI-1 for degradation. Western blot analysis of embryonic extracts from N2 (WT control, ctrl), FLAG::MEI-1 WT (light blue), FLAG::MEI-1 S92A (orange), and FLAG::MEI-1 S92D (green), animals exposed to mock (control) or mei-1(RNAi) using anti-MEI-1 (upper panel) and anti-tubulin (middle panel, loading control) antibodies. The lower panel shows Ponceau staining of the membrane.
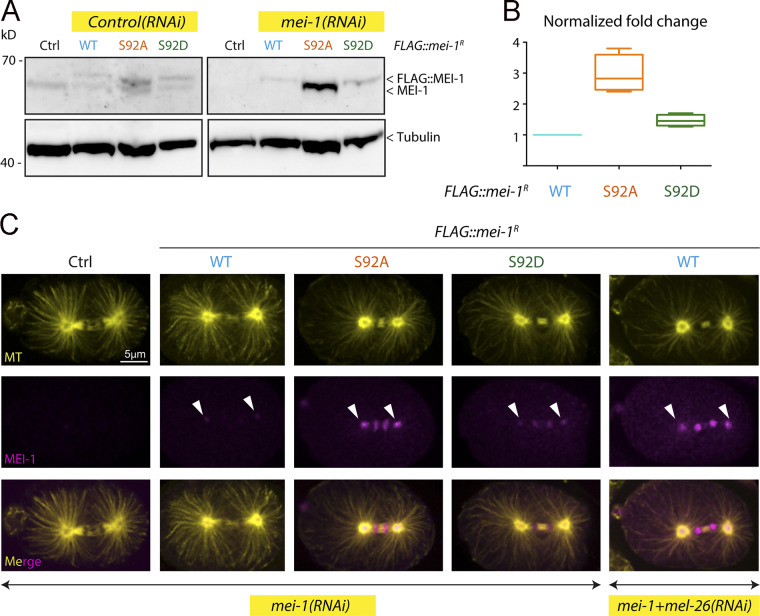
Figure 6.
Site-specific MEI-1 phosphorylation (S92) is necessary and sufficient to target MEI-1 for degradation. (A) Western blot analysis of embryonic extracts from N2 (WT control, ctrl), FLAG::MEI-1 WT (light blue), FLAG::MEI-1 S92A (orange), and FLAG::MEI-1 S92D (green), animals exposed to mock (ctrl) or mei-1(RNAi) using MEI-1 (upper panel) and tubulin (lower panel, loading control) antibodies. (B) Quantification of the ratio of MEI-1 versus tubulin signal intensity over three independent experiments revealed that MEI-1 levels accumulate by threefold in FLAG::MEI-1 S92A embryos compared with the FLAG::MEI-1 WT control or FLAG::MEI-1 S92D upon endogenous inactivation of mei-1 by RNAi. Box-and-whiskers plot presenting MEI-1 levels quantified over three experiments. The whiskers represent the min and max values. (C) Representative spinning disk confocal micrographs of fixed early embryos of the indicated genotype immunostained with MEI-1 (magenta) and MT (yellow) antibodies. White arrowheads indicate the persistence of MEI-1 in mitosis at the centrosomes and the chromosomes. Additional representative images of embryos in metaphase are presented in Fig. S4.
Consistent with previous reports, RNAi-mediated mei-1 inactivation in WT “control” N2 animals resulted in severe embryonic lethality (Fig. 5 C); mei-1(RNAi) embryos presented the large polar bodies phenotype resulting from defects in female meiotic spindle assembly (Fig. 5 and S4). Animals expressing FLAG::mei-1R S92A depleted of endogenous mei-1 also produced dead embryos. However, this transgene fully rescued the defects in meiotic spindle assembly (Fig. S4), indicating that the observed embryonic lethality is likely due to defective mitosis. We then used differential interference contrast (DIC) microscopy to film the division of one-cell embryos. Embryos expressing FLAG::mei-1R S92A displayed severe defects in MT-mediated processes (Fig. 5 F). Mitotic spindles were misoriented, ectopic cleavage furrows formed after cytokinesis, and nuclei became displaced toward the cell cortex after mitosis. We confirmed these observations using spinning disk confocal microscopy by recording tubulin and histone fused to GFP and mCherry, respectively. In embryos expressing FLAG::mei-1R S92A, the mitotic spindle was shorter, failed to elongate, and often broke during anaphase (Fig. 5, E and F). The mitotic spindle generally broke between the sister chromatids at the central spindle, resulting in the mispositioning of the midbody, but in other cases, the spindle broke at the kinetochore MTs, resulting in the segregation of all the chromosomes to one daughter cell (Fig. 5 G). All these phenotypes are typical of embryos expressing a persistent form of MEI-1 (Clark-Maguire and Mains, 1994; Kurz et al., 2002; Pintard et al., 2003b). Remarkably, embryos expressing FLAG::mei-1R S92D, which constitutively interacts with MEL-26, did not show these phenotypes and were mainly viable in the absence of endogenous MEI-1 (Fig. 5 C). Taken together, these observations strongly suggest that phosphorylation of MEI-1 at a single serine (S92) is both necessary and sufficient to target Katanin for degradation after meiosis. Consistently, Western blot experiments revealed that FLAG::MEI-1 S92A, accumulated in embryos depleted of endogenous mei-1. Quantitative analysis of multiple Western blot experiments revealed that FLAG::MEI-1 S92A accumulated threefold compared with FLAG::MEI-1 WT. Consistent with our observations that FLAG::MEI-1 S92D constitutively interacts with MEL-26, FLAG::MEI-1 S92D did not accumulate in embryos, suggesting that introduction of the phosphomimetic residue at position 92 is sufficient to target Katanin for degradation by the CRL3MEL-26 E3 ligase.
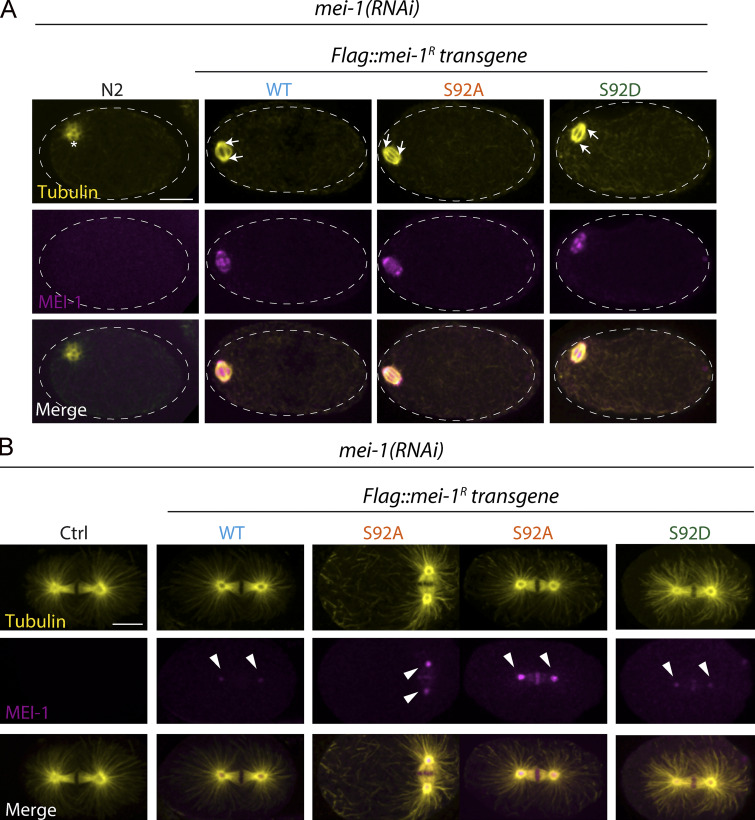
Figure S4.
Immunolocalization of Katanin during meiosis and mitosis. Additional representative spinning disk confocal micrographs of fixed early embryos of the indicated genotype immunostained with anti-MEI-1 (magenta) and anti-tubulin (yellow) antibodies. White arrowheads indicate the persistence of MEI-1 in mitosis at the centrosomes and the chromosomes. (A) Meiotic embryos. (B) Mitotic embryos.
To further confirm these observations, we used specific MEI-1 antibodies and immunofluorescence to monitor MEI-1 levels during mitosis (Fig. 6 C). We also stained MTs to visualize the mitotic spindle. WT control embryos depleted of MEI-1 presented the large polar body phenotype but assembled a normal mitotic spindle (Fig. 6 C, Ctrl). MEI-1 was undetectable in these embryos, indicating that our RNAi conditions efficiently depleted endogenous MEI-1. In embryos expressing FLAG::MEI-1 WT, the mitotic spindle was properly aligned along the anteroposterior axis of the embryos, and long astral MTs contacting the cortex were observed (Fig. 6 C, WT). MEI-1 was present on the centrosomes and chromosomes, but it was barely detectable. This staining is highly specific, confirming our live-imaging analysis using sGFP::MEI-2 and indicating that Katanin is present at low levels in mitotic embryos (Figs. S1 and 1). In embryos expressing only FLAG::MEI-1 S92A, MEI-1 readily accumulated at high levels to the centrosomes and chromosomes (Fig. 6 C, S92A and S92D). The mitotic spindle was often misoriented in these embryos (Fig. 5 F [white arrows] and Fig. S4 B). Consistent with Western blot experiments, FLAG::MEI-S92D did not accumulate in early embryos, although MEI-1 levels were slightly higher than in embryos expressing FLAG::MEI-1 WT (Fig. S4 and 6 C). Taken together, these results indicate that MEI-1 phosphorylation of the single residue S92 by MBK-2 is both necessary and sufficient to target MEI-1 for ubiquitin-mediated degradation by the CRL3MEL-26 E3 ligase. We conclude that MEI-1 phosphorylation regulates both Katanin activity and stability to deliver the appropriate level of MT-severing activity during C. elegans development (Fig. 7).
Figure 7.
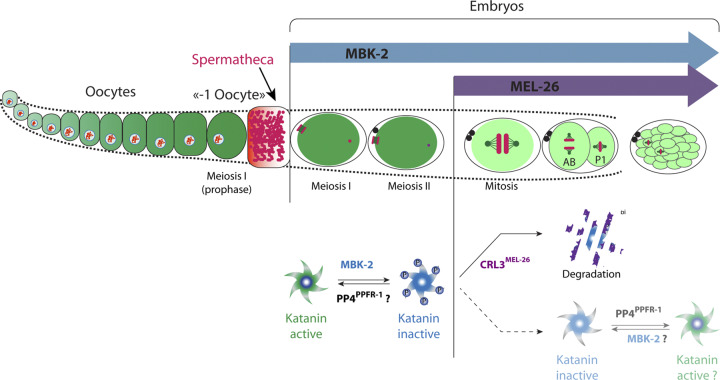
Working model of Katanin expression and regulation by cycles of phosphorylation and dephosphorylation during C. elegans development. Schematic representation of Katanin levels and regulation in C. elegans oocytes and embryos. In the top panel, the level of green color represents the amount of Katanin present in oocytes and embryos. Katanin accumulates during oogenesis to reach a maximal level in the oocyte located closed to the spermatheca (-1 oocyte). Katanin levels remain elevated during meiosis I and II but drop after meiosis as a consequence of the activation of the MBK-2 kinase and expression of the MEL-26 E3-ligase subunit. The bottom panel presents a model of the molecular mechanisms regulating Katanin levels and activity. During meiosis, Katanin might cycle between dephosphorylated (Katanin active, green) and phosphorylated (inactive, light blue) states. After meiosis, a large Katanin fraction is targeted for ubiquitin-mediated degradation by the CRL3MEL-26 E3-ligase while a fraction escaping proteasomal degradation might cycle between phosphorylated and dephosphorylated states.
Discussion
The evolutionarily conserved MT-severing AAA-ATPase enzymes are important regulators of MT dynamics, with instrumental functions in cell division and cell migration (McNally and Roll-Mecak, 2018). However, how these enzymes are regulated in space and time during development to deliver the appropriate levels of MT-severing activity is poorly understood. Using an integrated combination of genetics, live-cell imaging, and biochemical approaches, we investigated the spatiotemporal expression of Katanin during C. elegans development and dissected the molecular mechanisms regulating its activity and stability. In particular, we show that phosphorylation of the N-terminal regulatory domain of the catalytic MEI-1 subunit regulates both Katanin stability and ATPase activity. It has been shown that MTs stimulate, by an unknown mechanism, the ATPase activity of Katanin, which is essential for MT severing (McNally and Vale, 1993; Hartman and Vale, 1999; Joly et al., 2016; Zehr et al., 2020). Here we show that phosphorylation of the N-terminal regulatory domain of MEI-1 suppresses the stimulatory effect of MTs on Katanin ATPase activity, presumably inhibiting MT severing. In addition, we demonstrate unequivocally that site-specific phosphorylation of MEI-1 at S92 by MBK-2, but not at the other serines, is both necessary and sufficient to enhance the interaction between Katanin and the E3-ligase adaptor MEL-26, and to send Katanin to proteasome-dependent degradation. As the PP4PPFR-1 phosphatase likely dephosphorylates MEI-1 to stimulate Katanin activity (Gomes et al., 2013; Han et al., 2009), our results strongly suggest that cycles of phosphorylation and dephosphorylation regulate Katanin levels and activity in space and time during development, to deliver the appropriate amount of MT-severing activity.
Spatiotemporal expression and regulation of Katanin during C. elegans development
A wealth of genetic evidence indicates that Katanin is essential for meiotic spindle assembly, but it is toxic for the assembly of the mitotic spindle that forms in the same cytoplasm 20 min later. Previous work has established that Katanin is abundant in oocytes but is rapidly targeted for degradation after meiosis by the CRL3MEL-26 E3 ligase to allow mitotic spindle assembly. It was thus believed that C. elegans Katanin is entirely eliminated after meiosis and might not be produced during embryogenesis. Here, we used a strain expressing MEI-2 endogenously tagged with sGFP using CRISPR-based genome editing to monitor Katanin dynamics during C. elegans development. Consistent with immunolocalization studies, we confirmed that Katanin accumulates in oocytes to reach a maximum level in the –1 oocytes, which are located close to the spermatheca (Fig. 1 A, asterisk). The sGFP::MEI-2 signal in oocytes was undetectable upon inactivation of mei-1 by RNAi, indicating that sGFP::MEI-2 is a good proxy to visualize Katanin dynamics in worms. Unexpectedly, spinning disk confocal live-cell imaging revealed that sGFP::MEI-2 is also present during embryogenesis, although at a reduced level compared with oogenesis, where it is dynamically recruited to the mitotic spindle. Immunolocalization studies using highly specific MEI-1 antibodies also confirmed the presence of MEI-1 in mitotic embryos. Thus both subunits (MEI-1 and MEI-2) are present in embryos where Katanin may regulate aspects of chromosome segregation, similar to the situation in other systems (Jiang et al., 2017; Sharp and Ross, 2012; Zhang et al., 2007).
The machinery regulating Katanin activity and stability is also expressed during embryogenesis. Interestingly, seminal observations using the temperature-sensitive mel-26(ct61) allele and temperature shift experiments have shown that MEL-26 is continuously required from fertilization to gastrulation (26-cell stage; Mains et al., 1990). As MEI-1 is the only essential target of MEL-26 (Lu et al., 2004), this strongly suggests that MEI-1 levels are continuously regulated by the CRL3MEL-26 E3 ligase during embryogenesis. Also consistent with this hypothesis, MEL-26 is expressed throughout embryogenesis but decreases to low levels by the 64-cell stage (Johnson et al., 2009).
Similar to MEL-26 and Katanin, MBK-2 is expressed during embryogenesis, where it localizes to the centrosomes and chromosomes during mitosis (Pellettieri et al., 2003). Finally, the PP4PPFR-1 phosphatase, which activates Katanin by dephosphorylating MEI-1, is also present and active during mitosis. Indeed, down-regulation of PP4 phosphatase complex PP4PPFR-1 enhances the viability of embryos expressing a stabilized version of mei-1 (Gomes et al., 2013). PP4 may directly interact with the N-terminal part of MEI-1 to accomplish this function, as suggested by the presence of a short motif [102-WSKPSPPLPSSS-113] (bold and underlined represent the PP4-consensus binding motif WxxP) in the N-terminal part of MEI-1, presenting striking similarities with the recently identified consensus binding motif for PP4 protein phosphatases (Ueki et al., 2019). Collectively, these observations indicate that Katanin, along with the machinery regulating its levels (MBK-2 and MEL-26) and/or activity (MBK-2 and PP4), is present and active during embryogenesis. Our observations, along with the available literature, suggest the following model (Fig. 7). Katanin accumulates during oogenesis to reach a maximum level in the –1 oocytes, which are close to the spermatheca. In oocytes, MEL-26 is absent (Johnson et al., 2009), and MBK-2 is kept inactive at the cortex. During metaphase-to-anaphase transition of meiosis I, MBK-2 is activated and released into the cytoplasm (Cheng et al., 2009; Pellettieri et al., 2003; Stitzel et al., 2007). During meiosis, MEI-1 is likely subjected to cycles of phosphorylation and dephosphorylation that regulate its activity in space and time. In particular, Katanin is required to sever MTs between the second polar body and the future female pronucleus, an event stimulated by the PP4PPFR-1 phosphatase (Gomes et al., 2013). Upon mitotic entry, MEL-26 is produced (Mains et al., 1990), and MEI-1 gets phosphorylated on residue S92 and sent for degradation by the 26S proteasome. However, a fraction of MEI-1 escapes ubiquitin-mediated degradation and is dynamically recruited to the centrosomes and chromosomes during mitosis, at least until gastrulation. In embryos, levels and activity of Katanin are thus regulated by cycles of phosphorylation and dephosphorylation and ubiquitin-mediated degradation.
An evolutionarily conserved mechanism to regulate Katanin activity by phosphorylation?
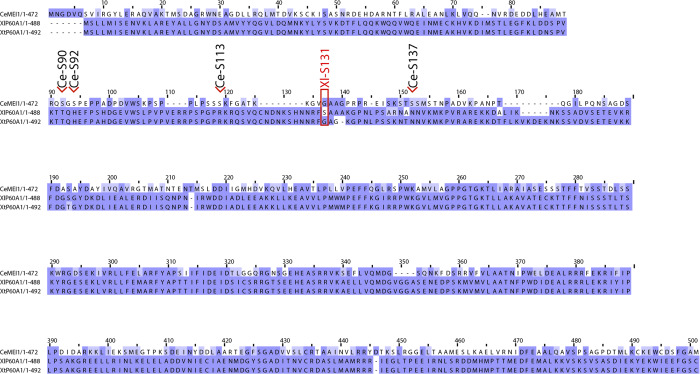
The sites modified by MBK-2, although not conserved, are all located in the N-terminal regulatory domain of MEI-1 (Fig. S5). Previous work has shown that differences in Katanin activity in the egg cytoplasm between Xenopus laevis and Xenopus tropicalis account for spindle scaling. Increased activity of Katanin scales the X. tropicalis spindle smaller compared with X. laevis (Loughlin et al., 2011). This difference in activity is regulated by the Aurora B kinase (Loughlin et al., 2011). A single phosphorylation of the N-terminal regulatory domain of X. laevis p60 subunit (S131) inhibits the MT-severing activity of the enzyme (Loughlin et al., 2011). This inhibitory phosphorylation site is not conserved in X. tropicalis Katanin, which exhibits higher Katanin-dependent MT-severing activity. How phosphorylation of X. laevis Katanin inhibits its activity, however, is poorly understood. It has been proposed that phosphorylation of serine 131 might affect the oligomeric state of the enzyme based on the observation that a phosphomimetic p60 S131E has reduced MT-stimulated ATPase activity at low concentrations but recovers WT activity at higher enzyme concentrations (Whitehead et al., 2013).
Figure S5.
Sequence alignment. Protein sequence alignment of p60 subunit from X. tropicalis, X. laevis, and C. elegans. Red box marks S131 X. laevis phosphorylated residue identified in Loughlin et al. (2011). Ce-Sx point to serine identified in our study. Sequence references: X. laevis (NP_001084226.1), X. tropicalis (NP_001072433.1), and C. elegans (P34808.1)
In C. elegans MEI-1, similar to the situation in X. tropicalis p60, this residue is a glycine (Fig. S5). However, introduction of phosphomimetic residue in place of the site modified by MBK-2 does not have the same effect as the S131E mutation. For instance, the ATPase activity of Katanin containing MEI-1 S92D is normally stimulated by MT in vitro, and this protein supports Katanin function during meiotic spindle assembly. Nevertheless, Katanin with MEI-1 uniquely phosphorylated at S92 is refractory to the presence of MTs. The observed difference between MEI-1 S92D and MEI-1 S92-aaPho might be due to the charge densities, distributions, and pKa of the carboxyl group of aspartic acid residue, which are quite different from a phosphate group. Whereas MEI-1 S92D mimics partially S92 phosphorylation in promoting MEI-1 binding to MEL-26, it fails to phenocopy the effect of MEI-1 phosphorylation in inhibiting MT-stimulated ATPase activity of Katanin. Similar effects were observed with Katanin variants containing S90, S113, or S137 residues singly phosphorylated at each position. These results suggest that C. elegans Katanin activity requires an optimal steric arrangement of these four residues, but introduction of a single phosphate on one of these sites is sufficient to perturb Katanin regulatory domain organization and enable MT-dependent stimulation of the ATPase activity. We speculate that a similar mechanism may operate in the case of X. laevis p60 and possibly also for Katanin in other species.
How do MTs stimulate Katanin ATPase activity?
While MEI-1 phosphorylation does not alter basal Katanin ATPase activity, phosphorylation of a single residue of the N-terminal regulatory domain is sufficient to strongly inhibit the ATPase activity stimulated by MTs. To decipher the exact role of this MEI-1 phosphorylation, it will be essential to understand how MTs stimulate Katanin ATPase activity. MTs could affect different properties of the oligomer. Similar to other AAA+ ATPases, Katanin is organized as a hexamer. The organization of the subunits within the hexamer allows the formation of the nucleotide binding and hydrolysis pocket at the interface between subunits. The binding of MTs might have an effect on (a) the formation of the oligomer or on (b) the nucleotide pocket organization, leading to more efficient ATP binding and/or hydrolysis. In both cases, upon MT binding, Katanin would undergo a structural rearrangement leading to enhanced ATPase activity. From a mechanistic point of view, controlling the activity of an enzyme directly by using substrate binding as signal is a very efficient mechanism limiting the unproductive use of ATP hydrolysis. In a similar way, maintaining a basal ATPase activity might be a selective advantage to ensure a very short delay of response when interacting with the substrate. Here we reveal an additional regulatory level: the phosphorylation of the N-terminal regulatory domain antagonizing the MT-binding signal and ensuring spatial and temporal regulation of the enzyme activity.
Materials and methods
Plasmids
The list of plasmids used in this study is provided in Table S1.
Nematode strains, culture conditions, and RNAi
C. elegans strains were cultured and maintained using standard procedures (Brenner, 1974; Stiernagle, 2006). Transgenic worms expressing FLAG::mei-1R transgenes were generated by Mos1-mediated single-copy gene insertion using the strain EG6699 (ttTi5605 II; unc-119(ed3) III; oxEx1578; Frøkjaer-Jensen et al., 2008). Correct insertion of the transgene was verified by PCR. The list of C. elegans strains used in this study is provided in Table S2.
Embryonic lethality assays
Embryonic lethality was assayed after leaving L4 larvae on IPTG plates seeded with bacteria expressing mock or mei-1 dsRNA at 22°C–25°C. After 24– 36 h, five adult worms were transferred onto a new plate. After 5 h, the worms were removed from the plate, and the progeny were counted after 24 h. Embryonic lethality was then scored as the percentage of dead embryos found among the progeny.
Protein purification
Katanin (6xHis-MEI-1 WT or variant Strep-MEI-2) were purified from BL21 (DE3) bacteria transformed with pNJ plasmids (see Table S1) as described in Joly et al. (2016). Before storage, buffer was exchanged to 50 mM Tris HCl, pH 8.0, 500 mM NaCl, and 5% glycerol using G25 columns, and the proteins were snap frozen in liquid nitrogen and stored at −80°C.
6xHis-MEI-1 and Strep-MEI-2 were purified from BL21 (DE3) bacteria transformed with pNJ420 and pNJ440, respectively, following the same protocol as previously described in Joly et al. (2016) and stored in 50 mM Tris HCl, pH 8.0, 500 mM NaCl, and 5% glycerol.
Production and purification of Katanin phosphorylated at specific sites by genetic code expansion in E. coli
Phospho-inserted Katanin was purified from BL21 (DE3) ΔSerB strain transformed with pNJ797 and derivative, allowing the production of MEI-1 WT-6xHis and Strep-MEI-2 or MEI-1 Xaa-Pho-6xHis and Strep-MEI-2 derivative, repectively (see Table S1). Briefly, 1 liter of Luria-Bertani medium supplemented with 1 mM phosphoserine was inoculated with overnight culture (1% vol/vol) and grown at 37°C to OD600nm 0.4–0.6. The protein production was induced with 0.5 mM final concentration of IPTG for 5 h at 24°C. After centrifugation, cells were resuspended in 50 mM Tris HCl, pH 8.0, 500 mM NaCl, and 5% glycerol and broken by sonication. The supernatant was loaded onto a StrepTrap HP (GE Healthcare), and after washing, the protein was eluted using 10 mM desthiobiotin. The protein eluate was directly loaded on a HiTrap Chelating HP column (GE Healthcare) precharged with nickel, and the bound proteins were eluted using imidazole in the lysis buffer after extensive washes. The buffer was exchanged to 50 mM Tris HCl, pH 8.0, 500 mM NaCl, and 5% glycerol using G25 columns, and the proteins were snap frozen in liquid nitrogen and stored at −80°C. All the phospho-inserted purifications were verified by mass spectrometry and showed 100% of phosphoserine amino acid at the chosen position.
6xHis-MBK-2 was purified from BL21 (DE3) transformed with pNJ644, allowing the production of 6xHis-MBK-2. Briefly, 1 liter of Luria-Bertani medium was inoculated with an overnight culture (1% vol/vol) and grown at 37°C to OD600nm 0.4–0.6. Protein production was induced with 0.1 mM final concentration of IPTG for 16 h at 18°C. After centrifugation, cells were resuspended in 50 mM Tris HCl, pH 8.0, 500 mM NaCl, and 5% glycerol and broken by sonication. The supernatant was loaded on a HiTrap Chelating HP column (GE Healthcare) precharged with nickel, and the bound proteins were eluted using imidazole in the lysis buffer after extensive washes. The buffer was exchanged to 50 mM Tris HCl, pH 8.0, 500 mM NaCl, and 5% glycerol using G25 columns, and the proteins were snap frozen in liquid nitrogen and stored at −80°C.
Tubulin was purified from pig brain using three rounds of polymerization/depolymerization cycles and resuspended into BRB80 buffer (80 mM Pipes-KOH, pH 6.8, 1 mM MgCl2, and 1 mM EGTA). Rhodamine-labeled tubulin from porcine brain was purchased from Cytoskeleton. Protein concentrations were estimated using the method of Lowry et al. (1951), and protein purity was estimated on SDS-PAGE stained with Coomassie blue.
Far-ligand binding assay (Far-Western blot)
Far-Western ligand binding assays were performed essentially as described (Wu et al., 2007) on NuPAGE 4–12% (Invitrogen) run in MOPS buffer and transferred to a 0.45-mm PVDF membrane. For the dot blot assay, sample (2 µl) was directly spotted onto nitrocellulose membrane and air dried. The membranes were then incubated for 6 h at 4°C in blocking solution (TBS-Tween 0.05% with 5% milk and 1% BSA). Membranes were then incubated in the blocking solution containing 2 mg of GST-MEL-26 for 14 h at 4°C. After washing, membranes were incubated with anti-MEL-26 antibody (1:2,000) for 4 h, and anti-rabbit-HRP antibody was used. Western blot was revealed using enhanced chemiluminescence (ECL) Immobilon Western (Millipore) and scanned with ImageQuant LAS4000 (GE). Blots were analyzed and quantified using ImageJ (National Institutes of Health).
ATPase activity
Steady-state ATPase assays were used to monitor Katanin ATPase activity and performed as described in Joly et al. (2016). Briefly, assays were performed at 28°C in the presence of an NADH-coupled regeneration system (Nørby, 1988) in 100 µl final volume, in buffer containing 25 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 1 mM DTT, 1 mM NADH, 10 mM phosphoenol pyruvate, 10 U/ml pyruvate kinase, 20 U/ml lactate dehydrogenase, ATP (0–20 mM), and Katanin or MEI-1 (0–10 µM). For the MT-stimulated ATPase assay, a fixed concentration of Katanin or MEI-1 (0.4 µM) was used and mixed with different concentrations of MTs (as indicated). OD340 was read using a SpectraMax2 plate reader. Experiments were performed at least five times.
In vitro kinase assay
In vitro kinase assay was performed at 30°C for 30 min in 10 µl containing 50 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 500 mM NaCl, 5% glycerol, 0–2 µM His-MBK-2, and 0–2 µM Katanin or MEI-1 or MEI-2. After 5-min incubation at 30°C, reactions were initiated by adding either 0.2 mM ATP or a mix of 0.2 mM ATP and 5 µCi γ-[32P]ATP (for radioactive experiments) for 30 min at 30°C. Reactions were stopped by adding 5 µl of Laemmli buffer and immediately boiled for 5 min. All the samples were loaded on 12% SDS-PAGE (TGX Bio-Rad) and run in 1× Tris/glycine/SDS buffer. Proteins were detected using Stain Free technology and ChemiDoc MP Imaging System (Bio-Rad), and radioactivity was measured by Amersham Typhoon Imager (GE) and analyzed using ImageJ. Mass spectrometry analysi was performed using the same method as described in Tavernier et al. (2015).
Western blotting and antibodies
Western blot analysis was performed using standard procedures (Sambrook et al., 1989). Antibodies include rabbit anti-MEI-1 (Pintard et al., 2003b), mouse anti-tubulin (DM1A, Sigma-Aldrich), and goat anti-GFP (polyclonal; Rockland, 039600-101-215). HRP-conjugated anti-mouse, anti-rabbit, and anti-goat antibodies (Sigma-Aldrich) were used at 1:3,000, and the signal was detected with chemiluminescence (Millipore).
The anti-MEI-1 antibody purified on strip was further purified on acetone powder prepared from adult worms depleted of MEI-1 by RNA interference.
Live imaging and image analysis
Live imaging of full worms
Imaging of full worms was performed using a spinning disk confocal microscope. Adult worms were anesthetized using levamisole and transferred onto an agarose pad on a 24 × 60-mm coverslip mounted on a metal holder. A coverslip (18 × 18 mm) was placed on top. Live imaging was performed at 23°C using a spinning disc confocal head (CSU-W1; Yokogawa Corporation of America) mounted on a DMI8 inverted microscope (Leica) equipped with 491- and 561-nm lasers (491-nm 150 mW; DPSS Laser 561 nm 150 mW) and a complementary metal-oxide-semiconductor (Orca-Flash 4 V2+, Hamamatsu). Acquisition parameters were controlled by MetaMorph software (Molecular Devices). In all cases, a 40×, HC PL APO 40×/1.30 Oil CS2 (Leica 506358) lens was used, and ∼10 z-sections were collected at 0.5-µm intervals. Each “reconstituted worm” was assembled in Adobe Photoshop from 8–10 fields of view. Captured images were processed using ImageJ and Photoshop.
Liveimaging of embryos
Imaging of dissected embryos were performed using a spinning disk confocal microscope. Adult worms were dissected in 4 µl of L-15 blastomere culture medium on a 24 × 60-mm coverslip mounted on a metal holder. The drop of medium was surrounded by a ring of petroleum jelly that served as a spacer, preventing compression of the embryos. A coverslip (18 × 18 mm) was placed on top to seal the chamber and prevent evaporation during filming. Live imaging was performed at 23°C using a spinning disc confocal head (CSU-X1; Yokogawa Corporation of America) mounted on an Axio Observer.Z1 inverted microscope (Zeiss) equipped with 491- and 561-nm lasers (OXXIUS 488 nm 150 mW, OXXIUS Laser 561 nm 150 mW) and an electron-multiplying charge coupled device (QuantEM 512SC, Photometrics). Acquisition parameters were controlled by MetaMorph software (Molecular Devices). In all cases a 63×, Plan-Apochromat 63×/1.4 Oil (Zeiss) lens was used, and approximately four z-sections were collected at 1-µm and 10-s intervals. Captured images were processed using ImageJ and Photoshop.
Live imaging was performed at 23°C using a spinning disc confocal head (CSU-X1; Yokogawa Corp. of America) mounted on a Ti-E inverted microscope (Nikon) equipped with 491- and 561-nm lasers (Roper Scientific) and a charge-coupled device camera (Coolsnap HQ2; Photometrics). Acquisition parameters were controlled by MetaMorph software (Molecular Devices). In all cases a 60×, 1.4 NA PlanApochromat lens with 2 × 2 binning was used, and four z-sections were collected at 2-µm intervals every 20 s.
Live imaging of mitotic divisions by DIC
For the visualization of early embryonic development in live specimens by DIC (Fig. 5), embryos were obtained by cutting open gravid hermaphrodites using two 21-gauge needles. Embryos were handled individually and mounted on a coverslip in 7 µl of egg buffer (Shelton and Bowerman, 1996). The coverslip was placed on a 2% agarose pad. Time-lapse DIC images were acquired by an Axiocam Hamamatsu ICcI camera (Hamamatsu Photonics) mounted on a Zeiss AxioImager A1 microscope equipped with a Plan Neofluar 100×/1.3-NA objective (Zeiss), and the acquisition system was controlled by Axiovision software (Zeiss). Images were acquired at 10-s intervals.
Immunofluorescence and imaging
10–15 worms were dissected in 7 µl of blastomere culture solution on subbing solution–coated slides using a 21-gauge needle to release embryos. An 18 × 18-mm coverslip was placed onto the drop, and the slide was frozen on a block of metal precooled on packed dry ice. After 20 min, the coverslip was flicked off, and the slide was plunged into −20°C methanol for 20 min. Slides were rehydrated in PBS for 5 min and incubated overnight at 4°C with 30 µl of primary antibodies in 3% BSA in PBS in a wet chamber. Affinity-purified rabbit anti-MEI-1 antibody was used at a dilution of 1:500, mouse monoclonal anti-α-tubulin antibody (DM1A; Sigma-Aldrich) was used at 1:400, and the secondary antibodies were coupled to the fluorophores Alexa Fluor 488 or 543 (Molecular Probes) used at 1:1,000. Embryos were mounted in Vectashield mounting medium with DAPI. Fixed embryos were imaged using a spinning disk confocal X1 microscope with 63× objective. Captured images were processed using ImageJ and Photoshop.
Graphs and statistical analysis
Prism 6 (GraphPad Software) was used to generate graphs and proceed to statistical analysis. The tests used are mentioned in the figure legends.
Online supplemental material
Fig. S1 shows the construction and characterization of a C. elegans line expressing MEI-2 endogenously tagged with sGFP. Fig. S2 shows the qualitative analysis of phosphorylated Katanin on PhosTag SDS-PAGE, after Coomassie staining (A); the quantitative analysis of the interaction between MEL-26 and Katanin (B); and that phosphorylation of the N-terminal Katanin region inhibits MT-dependent stimulation of Katanin ATPase activity (C). Fig. S3 shows that MEI-1 phosphorylation (S92) is necessary and sufficient to target MEI-1 for degradation. Fig. S4 shows the localization of Katanin during meiosis and mitosis on a fixed sample using immunostaining. Fig. S5 shows multiple protein sequence alignment of p60 subunits. Table S1 is the list of plasmids used in this study. Table S2 is the list of worm lines used in this study.
Supplementary Material
lists the plasmids used in this study.
lists the transgenic worm lines used in this study.
Acknowledgments
We thank Ana Beatriz Diaz for early strain constructions. We thank P. Moussounda and R. Servouze for help with media preparation. We acknowledge the ImagoSeine core facility of the Institut Jacques Monod, member of IBiSA and the France-BioImaging (ANR-10-INBS-04) infrastructure and the Institut Jacques Monod ‘Structural and Fonctional proteomic platform.” We thank J. Dumont for sharing microscopy equipment. We thank B. Bowerman, A. Guichet, R. Karess, J. Dumont, B. Lacroix, and B. Ossareh-Nazari for critical reading of the manuscript. We also thank WormBase.
Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). E. Beaumale is supported by a PhD fellowship from the Ministry of Research. N. Joly was supported by an Association pour la Recherche sur le Cancer Project Fellowship and is supported by funding “Dynamic Research” from Agence Nationale de la Recherche (ANR-18-IDEX-0001), Université de Paris excellency initiative (IdEx). Work in the laboratory of L. Pintard is supported by the French Agence Nationale de la Recherche under grant no. ANR-17-CE13-0011-01/01, by Ligue Contre le Cancer (Programme Equipes Labellisées). L. Pintard’s team is also supported by Agence Nationale de la Recherche, laboratory excellence (LabEx) “Who Am I?” ANR-11-LABX-0071 and the Université de Paris IdEx ANR-18-IDEX-0001, through the “Investments for the Future” program.
The authors declare no competing financial interests.
Author contributions: N. Joly and L. Pintard designed the study. N. Joly and E. Beaumale performed GFP::MEI-2 localization in worms. L. Van Hove and N. Joly generated DNA constructs. E. Beaumale, L. Van Hove, L. Martino, and L. Pintard generated C. elegans lines and performed experiments in worms. N. Joly performed all remaining experiments and data analysis with E. Beaumale and L. Pintard. N. Joly, E. Beaumale, and L. Pintard assembled the figures. N. Joly and L. Pintard wrote the manuscript with input from all co-authors.
References
- Beard S.M., Smit R.B., Chan B.G., and Mains P.E.. 2016. Regulation of the MEI-1/MEI-2 Microtubule-Severing Katanin Complex in Early Caenorhabditis elegans Development. G3 (Bethesda). 6:3257–3268. 10.1534/g3.116.031666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman B., and Kurz T.. 2006. Degrade to create: developmental requirements for ubiquitin-mediated proteolysis during early C. elegans embryogenesis. Development. 133:773–784. 10.1242/dev.02276 [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K.C., Klancer R., Singson A., and Seydoux G.. 2009. Regulation of MBK-2/DYRK by CDK-1 and the pseudophosphatases EGG-4 and EGG-5 during the oocyte-to-embryo transition. Cell. 139:560–572. 10.1016/j.cell.2009.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Maguire S., and Mains P.E.. 1994. Localization of the mei-1 gene product of Caenorhaditis elegans, a meiotic-specific spindle component. J. Cell Biol. 126:199–209. 10.1083/jcb.126.1.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRenzo C., and Seydoux G.. 2004. A clean start: degradation of maternal proteins at the oocyte-to-embryo transition. Trends Cell Biol. 14:420–426. 10.1016/j.tcb.2004.07.005 [DOI] [PubMed] [Google Scholar]
- Dow M.R., and Mains P.E.. 1998. Genetic and molecular characterization of the caenorhabditis elegans gene, mel-26, a postmeiotic negative regulator of mei-1, a meiotic-specific spindle component. Genetics. 150:119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C., Davis M.W., Hopkins C.E., Newman B.J., Thummel J.M., Olesen S.P., Grunnet M., and Jorgensen E.M.. 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40:1375–1383. 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa M., He Y.J., Borchers C., and Xiong Y.. 2003. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat. Cell Biol. 5:1001–1007. 10.1038/ncb1056 [DOI] [PubMed] [Google Scholar]
- Gomes J.E., Tavernier N., Richaudeau B., Formstecher E., Boulin T., Mains P.E., Dumont J., and Pintard L.. 2013. Microtubule severing by the katanin complex is activated by PPFR-1-dependent MEI-1 dephosphorylation. J. Cell Biol. 202:431–439. 10.1083/jcb.201304174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Gomes J.E., Birmingham C.L., Pintard L., Sugimoto A., and Mains P.E.. 2009. The role of protein phosphatase 4 in regulating microtubule severing in the Caenorhabditis elegans embryo. Genetics. 181:933–943. 10.1534/genetics.108.096016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman J.J., and Vale R.D.. 1999. Microtubule disassembly by ATP-dependent oligomerization of the AAA enzyme katanin. Science. 286:782–785. 10.1126/science.286.5440.782 [DOI] [PubMed] [Google Scholar]
- Hartman J.J., Mahr J., McNally K., Okawa K., Iwamatsu A., Thomas S., Cheesman S., Heuser J., Vale R.D., and McNally F.J.. 1998. Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell. 93:277–287. 10.1016/S0092-8674(00)81578-0 [DOI] [PubMed] [Google Scholar]
- Jiang K., Rezabkova L., Hua S., Liu Q., Capitani G., Altelaar A.F.M., Heck A.J.R., Kammerer R.A., Steinmetz M.O., and Akhmanova A.. 2017. Microtubule minus-end regulation at spindle poles by an ASPM-katanin complex. Nat. Cell Biol. 19:480–492. 10.1038/ncb3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.L., Lu C., Raharjo E., McNally K., McNally F.J., and Mains P.E.. 2009. Levels of the ubiquitin ligase substrate adaptor MEL-26 are inversely correlated with MEI-1/katanin microtubule-severing activity during both meiosis and mitosis. Dev. Biol. 330:349–357. 10.1016/j.ydbio.2009.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly N., Martino L., Gigant E., Dumont J., and Pintard L.. 2016. Microtubule-severing activity of the AAA+ ATPase Katanin is essential for female meiotic spindle assembly. Development. 143:3604–3614. 10.1242/dev.140830 [DOI] [PubMed] [Google Scholar]
- Kuo Y.W., Trottier O., Mahamdeh M., and Howard J.. 2019. Spastin is a dual-function enzyme that severs microtubules and promotes their regrowth to increase the number and mass of microtubules. Proc. Natl. Acad. Sci. USA. 116:5533–5541. 10.1073/pnas.1818824116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz T., Pintard L., Willis J.H., Hamill D.R., Gönczy P., Peter M., and Bowerman B.. 2002. Cytoskeletal regulation by the Nedd8 ubiquitin-like protein modification pathway. Science. 295:1294–1298. 10.1126/science.1067765 [DOI] [PubMed] [Google Scholar]
- Li W., DeBella L.R., Guven-Ozkan T., Lin R., and Rose L.S.. 2009. An eIF4E-binding protein regulates katanin protein levels in C. elegans embryos. J. Cell Biol. 187:33–42. 10.1083/jcb.200903003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughlin R., Wilbur J.D., McNally F.J., Nédélec F.J., and Heald R.. 2011. Katanin contributes to interspecies spindle length scaling in Xenopus. Cell. 147:1397–1407. 10.1016/j.cell.2011.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry O.H., Rosebrough N.J., Farr A.L., and Randall R.J.. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265–275. [PubMed] [Google Scholar]
- Lu C., and Mains P.E.. 2007. The C. elegans anaphase promoting complex and MBK-2/DYRK kinase act redundantly with CUL-3/MEL-26 ubiquitin ligase to degrade MEI-1 microtubule-severing activity after meiosis. Dev. Biol. 302:438–447. 10.1016/j.ydbio.2006.09.053 [DOI] [PubMed] [Google Scholar]
- Lu C., Srayko M., and Mains P.E.. 2004. The Caenorhabditis elegans microtubule-severing complex MEI-1/MEI-2 katanin interacts differently with two superficially redundant beta-tubulin isotypes. Mol. Biol. Cell. 15:142–150. 10.1091/mbc.e03-06-0418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mains P.E., Kemphues K.J., Sprunger S.A., Sulston I.A., and Wood W.B.. 1990. Mutations affecting the meiotic and mitotic divisions of the early Caenorhabditis elegans embryo. Genetics. 126:593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally F.J., and Roll-Mecak A.. 2018. Microtubule-severing enzymes: From cellular functions to molecular mechanism. J. Cell Biol. 217:4057–4069. 10.1083/jcb.201612104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally F.J., and Vale R.D.. 1993. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 75:419–429. 10.1016/0092-8674(93)90377-3 [DOI] [PubMed] [Google Scholar]
- McNally K., Audhya A., Oegema K., and McNally F.J.. 2006. Katanin controls mitotic and meiotic spindle length. J. Cell Biol. 175:881–891. 10.1083/jcb.200608117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Reichert T., Greenan G., O’Toole E., and Srayko M.. 2010. The elegans of spindle assembly. Cell. Mol. Life Sci. 67:2195–2213. 10.1007/s00018-010-0324-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nørby J.G. 1988. Coupled assay of Na+,K+-ATPase activity. Methods Enzymol. 156:116–119. 10.1016/0076-6879(88)56014-7 [DOI] [PubMed] [Google Scholar]
- Pang K.M., Ishidate T., Nakamura K., Shirayama M., Trzepacz C., Schubert C.M., Priess J.R., and Mello C.C.. 2004. The minibrain kinase homolog, mbk-2, is required for spindle positioning and asymmetric cell division in early C. elegans embryos. Dev. Biol. 265:127–139. 10.1016/j.ydbio.2003.09.024 [DOI] [PubMed] [Google Scholar]
- Pellettieri J., Reinke V., Kim S.K., and Seydoux G.. 2003. Coordinate activation of maternal protein degradation during the egg-to-embryo transition in C. elegans. Dev. Cell. 5:451–462. 10.1016/S1534-5807(03)00231-4 [DOI] [PubMed] [Google Scholar]
- Pintard L., and Bowerman B.. 2019. Mitotic Cell Division in Caenorhabditis elegans. Genetics. 211:35–73. 10.1534/genetics.118.301367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard L., Kurz T., Glaser S., Willis J.H., Peter M., and Bowerman B.. 2003a Neddylation and deneddylation of CUL-3 is required to target MEI-1/Katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr. Biol. 13:911–921. 10.1016/S0960-9822(03)00336-1 [DOI] [PubMed] [Google Scholar]
- Pintard L., Willis J.H., Willems A., Johnson J.L., Srayko M., Kurz T., Glaser S., Mains P.E., Tyers M., Bowerman B., and Peter M.. 2003b The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature. 425:311–316. 10.1038/nature01959 [DOI] [PubMed] [Google Scholar]
- Quintin S., Mains P.E., Zinke A., and Hyman A.A.. 2003. The mbk-2 kinase is required for inactivation of MEI-1/katanin in the one-cell Caenorhabditis elegans embryo. EMBO Rep. 4:1175–1181. 10.1038/sj.embor.7400029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogerson D.T., Sachdeva A., Wang K., Haq T., Kazlauskaite A., Hancock S.M., Huguenin-Dezot N., Muqit M.M., Fry A.M., Bayliss R., et al. 2015. Efficient genetic encoding of phosphoserine and its nonhydrolyzable analog. Nat. Chem. Biol. 11:496–503. 10.1038/nchembio.1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A., and McNally F.J.. 2010. Microtubule-severing enzymes. Curr. Opin. Cell Biol. 22:96–103. 10.1016/j.ceb.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A., and Vale R.D.. 2006. Making more microtubules by severing: a common theme of noncentrosomal microtubule arrays? J. Cell Biol. 175:849–851. 10.1083/jcb.200611149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., and Maniatis T.. 1989. Molecular Cloning. A Laboratory Manual. Second edition Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sharp D.J., and Ross J.L.. 2012. Microtubule-severing enzymes at the cutting edge. J. Cell Sci. 125:2561–2569. 10.1242/jcs.101139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton C.A., and Bowerman B.. 1996. Time-dependent responses to glp-1-mediated inductions in early C. elegans embryos. Development. 122:2043–2050. [DOI] [PubMed] [Google Scholar]
- Srayko M., Buster D.W., Bazirgan O.A., McNally F.J., and Mains P.E.. 2000. MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev. 14:1072–1084. [PMC free article] [PubMed] [Google Scholar]
- Srayko M., O’Toole E.T., Hyman A.A., and Müller-Reichert T.. 2006. Katanin disrupts the microtubule lattice and increases polymer number in C. elegans meiosis. Curr. Biol. 16:1944–1949. 10.1016/j.cub.2006.08.029 [DOI] [PubMed] [Google Scholar]
- Stiernagle T. 2006. Maintenance of C. elegans. WormBook. doi/10.1895/wormbook.1.7.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitzel M.L., Cheng K.C., and Seydoux G.. 2007. Regulation of MBK-2/Dyrk kinase by dynamic cortical anchoring during the oocyte-to-zygote transition. Curr. Biol. 17:1545–1554. 10.1016/j.cub.2007.08.049 [DOI] [PubMed] [Google Scholar]
- Stitzel M.L., Pellettieri J., and Seydoux G.. 2006. The C. elegans DYRK Kinase MBK-2 Marks Oocyte Proteins for Degradation in Response to Meiotic Maturation. Curr. Biol. 16:56–62. 10.1016/j.cub.2005.11.063 [DOI] [PubMed] [Google Scholar]
- Tavernier N., Noatynska A., Panbianco C., Martino L., Van Hove L., Schwager F., Léger T., Gotta M., and Pintard L.. 2015. Cdk1 phosphorylates SPAT-1/Bora to trigger PLK-1 activation and drive mitotic entry in C. elegans embryos. Journal of Cell Biology. 208(6):661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki Y., Kruse T., Weisser M.B., Sundell G.N., Larsen M.S.Y., Mendez B.L., Jenkins N.P., Garvanska D.H., Cressey L., Zhang G., et al. 2019. A Consensus Binding Motif for the PP4 Protein Phosphatase. Mol. Cell. 76:953–964.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemu A., Szczesna E., Zehr E.A., Spector J.O., Grigorieff N., Deaconescu A.M., and Roll-Mecak A.. 2018. Severing enzymes amplify microtubule arrays through lattice GTP-tubulin incorporation. Science. 361 eaau1504 10.1126/science.aau1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlhac M.H., Terret M.E., and Pintard L.. 2010. Control of the oocyte-to-embryo transition by the ubiquitin-proteolytic system in mouse and C. elegans. Curr. Opin. Cell Biol. 22:758–763. 10.1016/j.ceb.2010.09.003 [DOI] [PubMed] [Google Scholar]
- Whitehead E., Heald R., and Wilbur J.D.. 2013. N-terminal phosphorylation of p60 katanin directly regulates microtubule severing. J. Mol. Biol. 425:214–221. 10.1016/j.jmb.2012.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Li Q., and Chen X.-Z.. 2007. Detecting protein-protein interactions by Far western blotting. Nat. Protoc. 2:3278–3284. 10.1038/nprot.2007.459 [DOI] [PubMed] [Google Scholar]
- Xu L., Wei Y., Reboul J., Vaglio P., Shin T.H., Vidal M., Elledge S.J., and Harper J.W.. 2003. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 425:316–321. 10.1038/nature01985 [DOI] [PubMed] [Google Scholar]
- Zehr E.A., Szyk A., Szczesna E., and Roll-Mecak A.. 2020. Katanin Grips the beta-Tubulin Tail through an Electropositive Double Spiral to Sever Microtubules. Dev. Cell. 52:118–131.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Rogers G.C., Buster D.W., and Sharp D.J.. 2007. Three microtubule severing enzymes contribute to the “Pacman-flux” machinery that moves chromosomes. J. Cell Biol. 177:231–242. 10.1083/jcb.200612011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists the plasmids used in this study.
lists the transgenic worm lines used in this study.