In Chlamydomonas, the TRP-channel PKD2/polycystin-2 is attached to both the axonemal microtubules and extracellular glycoprotein polymers projecting from the ciliary surface. The design suggests a mechanosensory role of PKD2 in motile cilia.
Abstract
Mutations in the channel protein PKD2 cause autosomal dominant polycystic kidney disease, but the function of PKD2 in cilia remains unclear. Here, we show that PKD2 targets and anchors mastigonemes, filamentous polymers of the glycoprotein MST1, to the extracellular surface of Chlamydomonas cilia. PKD2–mastigoneme complexes physically connect to the axonemal doublets 4 and 8, positioning them perpendicular to the plane of ciliary beating. pkd2 mutant cilia lack mastigonemes, and mutant cells swim with reduced velocity, indicating a motility-related function of the PKD2–mastigoneme complex. Association with both the axoneme and extracellular structures supports a mechanosensory role of Chlamydomonas PKD2. We propose that PKD2–mastigoneme arrays, on opposing sides of the cilium, could perceive forces during ciliary beating and transfer these signals to locally regulate the response of the axoneme.
Introduction
Transient receptor potential (TRP) channel proteins are a superfamily of cation channels often with sensory functions, including members that respond to mechanical cues, such as sound and touch. Mechanical gating is also proposed for polycystic kidney disease 2 (PKD2)/polycystin-2/TRPP2, a nonselective TRP channel located in the ciliary membrane of species ranging from protists to mammals (Barr and Sternberg, 1999; Huang et al., 2007; Pazour et al., 2002; Yoder et al., 2002). Mutations in PKD2 or its binding partner PKD1, an 11-transmembrane helix protein with a large extracellular domain, cause autosomal dominant polycystic kidney disease (ADPKD), a monogenic disorder for which no cure is available (Wu and Somlo, 2000). The PKD1/2 complex is thought to function as a receptor-channel complex, with PKD1 acting as a receptor for ligands or mechanical cues, which stimulates PKD2 to open, resulting in Ca2+ influx into the cilium (Kim et al., 2016; Liu et al., 2018; Praetorius and Spring, 2001; Su et al., 2018; Yoshiba et al., 2012; Yuan et al., 2015). However, the nature of the in vivo stimulus of the PKD1/2 complex and the downstream signaling events in kidney cilia remain controversial. Similarly, it is unknown how PKD2 in nodal cilia contributes to the sensing of the nodal flow, which is required for proper left–right patterning (Pennekamp et al., 2002). Moreover, the current leading hypothesis, that primary cilia in the kidney and embryonic node function as mechanically sensitive calcium-signaling organelles, has been challenged by Delling et al. (2016). Thus, further analyses of PKD proteins within the context of the ciliary membrane are needed.
Although the role of calcium in primary cilia is unclear, calcium is known to regulate motile cilia in various ways, ranging from causing changes in the waveform to modulating behavioral responses (Collingridge et al., 2013). Several distinct calcium channels have been identified in the motile cilia of protists, and loss of such channels has been linked to specific ciliary motility changes (Dunlap, 1977; Kamiya and Witman, 1984). The voltage-gated calcium channel CAV2, for example, mediates the photophobic escape behavior of Chlamydomonas; and TRP11, a V-type TRP channel, is required for cells to avoid collision with obstacles (Fujiu et al., 2009, 2011). PKD2 is the only member of the TRP polycystin (TRPP) class of TRP channels in Chlamydomonas; it is a cargo of intraflagellar transport (IFT) and localizes to the ciliary membrane (Huang et al., 2007). However, protists lack an apparent homologue of PKD1, raising the question of whether Chlamydomonas PKD2 has different interaction partners. Partial knockdown of PKD2 in Chlamydomonas decreased gametic fusion during mating; however, a more rigorous analysis of PKD2 function was prevented by the lack of more penetrant mutants (Huang et al., 2007). Here, we used a novel Chlamydomonas pkd2 insertional mutant that lacks PKD2 expression to characterize the function, interactions, 3D structure, and large-scale organization of PKD2 in cilia. We observed that PKD2 arrays are anchored along just two of the nine doublet microtubules (DMTs) and that PKD2 organizes mastigonemes, extracellular polymers of the glycoprotein mastigoneme-like protein 1 (MST1). Mutants in PKD2 and MST1 swim with reduced velocity, indicating a motility-related role of the PKD2–mastigoneme complex in Chlamydomonas cilia.
Results
PKD2 loss reduces the swimming velocity of Chlamydomonas
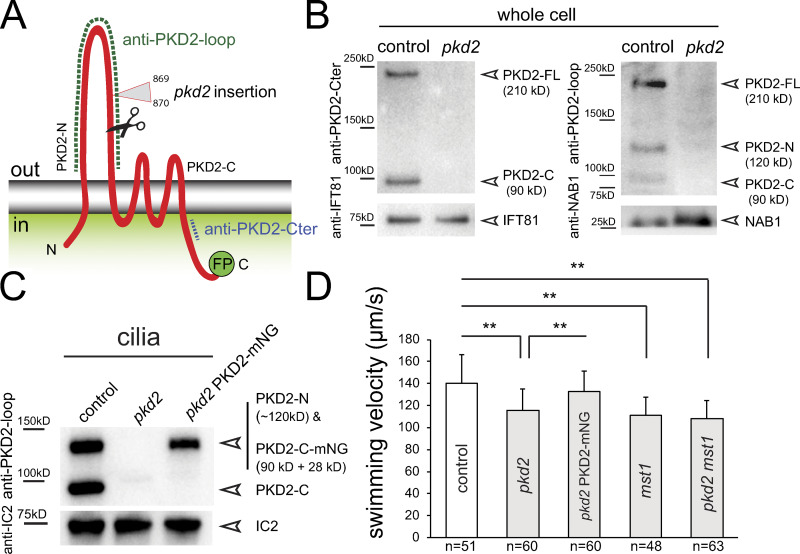
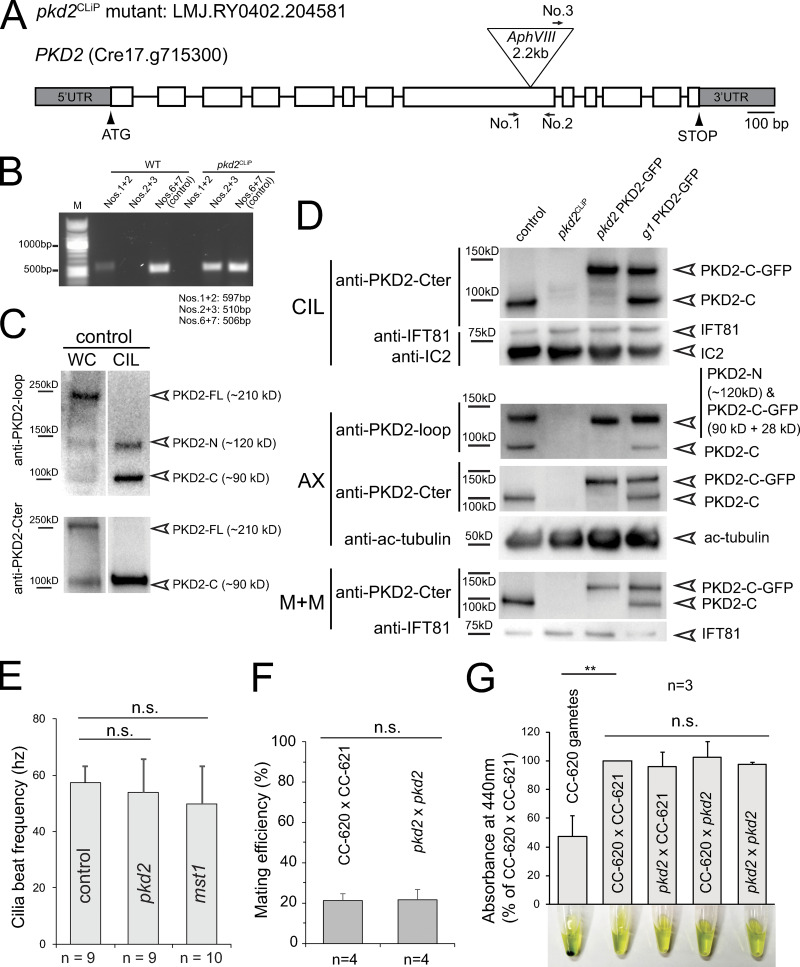
Strain LMJ.RY0402.204581 from the Chlamydomonas Library Project (CLiP) collection carries an insertion in exon 8 of the PKD2 gene (Figs. 1 A and S1, A and B; Li et al., 2016). To test for the presence of PKD2 in the mutant, we used two previously characterized antibodies, anti-PKD2-loop and anti-PKD2-Cter, raised against the extracellular loop and a peptide near the C-terminus of PKD2, respectively (Fig. 1 A; Huang et al., 2007). Huang et al. (2007) reported that full-length Chlamydomonas PKD2 (PKD2-FL; 210 kD) is cleaved into two fragments (PKD2-N and PKD2-C of 120 and 90 kD, respectively); the fragments enter cilia while PKD2-FL is excluded. As expected, PKD2 was present in control samples, whereas both antibodies failed to detect full-length PKD2 or its fragments in whole-cell and cilia samples from the CLiP mutant (Fig. S1, C and D). Thus, this strain likely lacks PKD2, and we refer to it as pkd2CLiP.
Figure 1.
Loss of PKD2 reduces Chlamydomonas swimming velocity. (A) Schematic presentation of PKD2. The relative positions of the insertion site of the AphVIII cassette in pkd2CLiP (triangle), the proteolytic cleavage site (scissors), and the antigens for the two PKD2 antibodies (dashed lines) are marked. C-terminal FP (GFP or mNG tag). (B) Western blot analysis of whole-cell samples from control and pkd2 probed with anti-PKD2-loop and anti-PKD2-Cter. Antibodies to NAB1 and IFT81 were used as loading controls. (C) Western blot of cilia isolated from control (CC-5235), pkd2, and pkd2 PKD2-mNG probed with anti-PKD2-loop and anti-IC2 as a loading control. (D) Swimming velocity of the control (CC-5235), pkd2, pkd2 PKD2-mNG rescue, mst1, and pkd2 mst1 double mutant strains. The error bars indicate SD, and the number of cells analyzed (n) are indicated. Significance based on two-tailed t test is indicated (**, P ≤ 0.01).
Figure S1.
CLiP strain LMJ.RY0402.204581 is a pkd2 null mutant. (A) Schematic representation of the PKD2 gene and of the insertion in pkd2. The positions of PCR primers used in B are indicated. (B) Agarose gel of PCR products using wild-type and pkd2CLiP genomic DNA as templates; g-β primers are used as control. (C) Western blot analysis of whole cell (WC) and isolated cilia (CIL) of control (CC-5235) cell samples using anti-PKD2-loop (top) and anti-PKD2-Cter (bottom). Note that only the two proteolytic fragments of PKD2 are present in cilia. (D) Western blot analysis of cilia (CIL), axonemes (AX), and the ciliary membrane + matrix (M+M) isolated from control (CC-5235), pkd2CLiP mutant, pkd2 PKD2-GFP, and g1 PKD2-GFP. Duplicate membranes were stained with anti-PKD2-loop and anti-PKD2-Cter as indicated; antibodies to IFT81, acetylated tubulin (ac-tubulin), and the axonemal protein IC2 were used as loading controls. (E) Average ciliary beat frequency of control (CC-5235), pkd2, and mst1 mutant cells. The SDs (error bars) and the number of cells analyzed (n) are indicated. Based on a two-tailed t test, the differences are not significant (n.s.). (F) Mating efficiency is shown by the percentage of mating cells in the mixture of two populations of gametes of the opposite mating types. The SDs (indicated by error bars)s (indicated by error bars) and the number of independent biological replicates (n) are indicated. Based on a two-tailed t test, the differences are not significant (n.s.). (G) Zygotes are initially cell-wall deficient and susceptible to cell lysis by detergent. Thus, the release of chlorophyll from detergent-treated cells can be used to assess mating efficiency in a population assay (Huang et al., 2007). The absorbance at 440 nm is displayed in percentage of that measured for the supernatant of the CC-620 plus × CC-621 minus control mating; a suspension of CC-620 gametes was used as a negative control; note green pellet of unlysed cells. The SDs (error bars) and the number of biological replicates (n) are indicated. The significance is based on two-tailed t test (**, P ≤ 0.01).
To eliminate possible second-site mutations that can occur in mutants derived by random insertional mutagenesis, we outcrossed the pkd2CLiP mutant to the wild-type strain g1, resulting in strain pkd2, and used this strain for most subsequent experiments (Fig. 1, B and C). The outcrossed pkd2 swam with reduced velocity (∼116 µm/s compared with 140 µm/s for the g1 control; Fig. 1 D). The motility defect cosegregated with the pkd2 mutation in subsequent progeny, and swimming was restored to near-wild-type velocity when fluorescent protein–tagged PKD2 (PKD2-FP, i.e., PKD2-GFP or PKD2-mNeonGreen [PKD2-mNG]) was introduced into the pkd2 strain (Fig. 1, C and D). The pkd2 mutant displayed normal mating behavior, phototaxis, photophobic backward swimming, and pH shock–induced cilia shedding, most of which require calcium influx into cilia (Fig. S1, F and G; and not depicted). We conclude that the loss of PKD2 reduces the swimming velocity of Chlamydomonas.
Ciliary PKD2 is anchored in two rows along the cilium
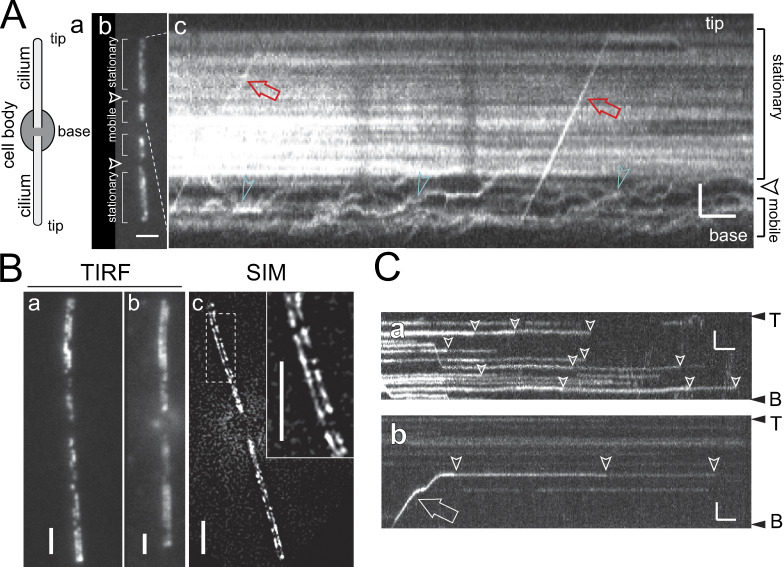
The slow-swimming phenotype of the pkd2 mutant suggests a motility-related function of PKD2 and raises the question of where within the cilium PKD2 is located. Using expression in wild-type Chlamydomonas cells, Huang et al. (2007) showed the presence of stationary and mobile PKD2-GFP in cilia, the latter moved by IFT. Total internal reflection fluorescence (TIRF) microscopy additionally identified two distinct compartments of PKD2-FP along the cilia of cells lacking endogenous PKD2: in the distal approximately two-thirds of the cilia, PKD2-FP was largely stationary (Figs. 2 A and S2 A; and Video 1). In contrast, most PKD2-FP particles in the proximal part of the cilia moved either by a slow random walk indicative of diffusion or processively by IFT (Figs. 2 A and S2 A). An exclusion zone of ∼1.5-µm length largely devoid of PKD2-FP separated the distal and proximal PKD2-FP regions, except for occasional particles passing through by diffusion or IFT (Figs. 2 A and S2 A). Notably, TIRF microscopy of favorably positioned cells indicated that stationary PKD2-FP localized along two rows in the distal cilium (Fig. 2, Ba and Bb; and Video 2). Two rows of PKD2-mNG were particularly discernable using structured illumination microscopy (SIM) of pkd2 PKD2-mNG cells (Fig. 2 Bc). In previous studies, a sizable portion of PKD2 or PKD2-GFP remained attached to the axonemes when cilia were extracted with nonionic detergents (Fig. S1 D; Huang et al., 2007; Pazour et al., 2005). Thus, stationary PKD2-GFP is likely bound to the axoneme.
Figure 2.
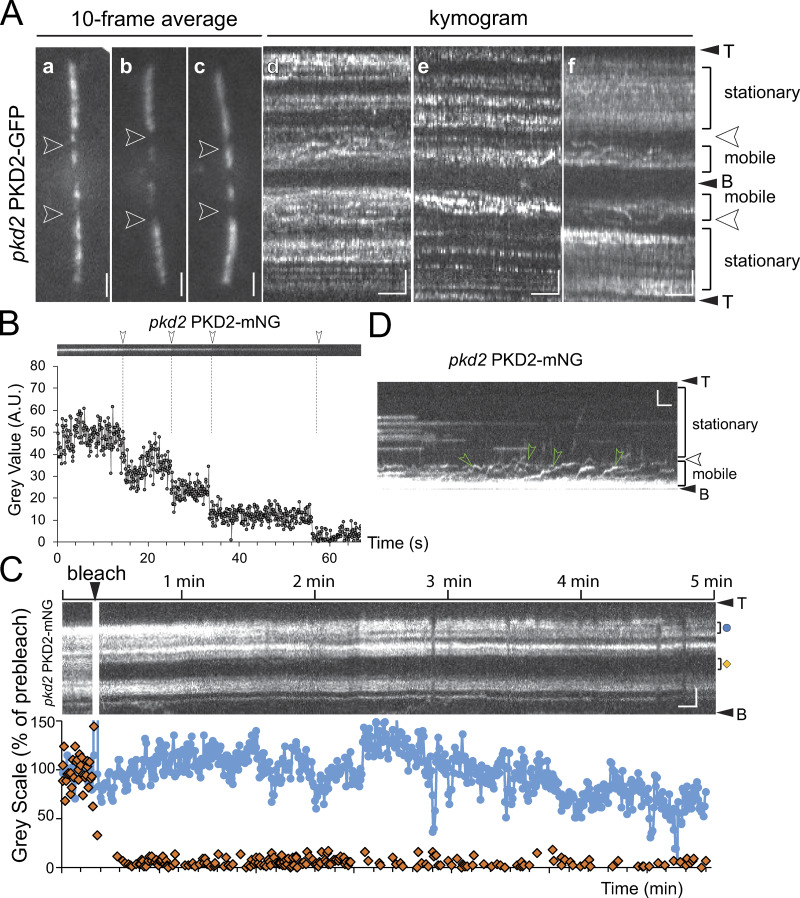
Two rows of PKD2 line the cilium. (A) Schematic representation of an adhered Chlamydomonas cell (a), TIRF image (b), and kymogram (c) of a pkd2 PKD2-GFP (b) and a pkd2 PKD2–mNG (c) cell. Arrowheads indicate the exclusion zone. The trajectories of PKD2-FP undergoing IFT (red arrows) and diffusion (green arrowheads) are marked. Horizontal trajectories indicate stationary PKD2-mNG particles. Bars = 2 s and 2 µm. (B) TIRF (a and b) and SIM (c) images of live pkd2 PKD2-mNG cells; the inset in c shows the boxed area at higher magnification. Bars = 2 µm. (C) Kymograms showing stepwise bleaching (arrowheads) of stationary PKD2-mNG particles (a and b) and of a PKD2-mNG particle freshly delivered by IFT (arrow) indicative of the presence of multiple copies of tagged PKD2 in each particle. The ciliary base (B) and tip (T) are indicated. Bars = 2 s and 2 µm.
Figure S2.
Compartmentalization of PKD2-FP in cilia. (A) Galleries of still images (a–c) and kymograms (d–f) of the cilia of pkd2 PKD2-GFP cells. The mobile and stationary regions of PKD2-GFP are indicated. Arrowheads, PKD2-GFP exclusion zone. Bars = 2 µm (a–c); 2 s and 2 µm (d–f). (B) Kymogram (top) and signal quantification (bottom) of one PKD2-mNG particle in the cilium of a pkd2 PKD2-mNG rescue cell during bleaching; arrowheads and dashed lines mark the bleaching steps. (C) FRAP analysis of ciliary PKD2-mNG in pkd2 PKD2-mNG. Top: Kymogram of the pre- and postbleach signals. The regions used to quantify the intensity in the bleached area (orange diamond) and an unbleached control area (blue dot) are marked. Bottom: Plot of signal intensity in percentage of the prebleach intensity of the bleached and the control region. Bars = 2 s and 2 µm. (D) Kymogram showing the dynamics of PKD2-mNG in the proximal cilium. The cilium was partially photobleached to better visualize PKD2-mNG moving in the mobile region. Green arrowheads indicate diffusing PKD2-mNG. Bars = 2 s and 2 µm.
Video 1.
Mobile and stationary PKD2-mNG in Chlamydomonas cilia. TIRF video and kymogram of a pkd2 PKD2-mNG cilium. PKD2-mNG is mostly stationary in the distal region of the cilium but mobile in the proximal region. Transport of PKD2-mNG by IFT is shown toward the end of the video. The cilium tip points toward the top. T, cilium top; B, cilium base. Images were acquired at 10 fps and are replayed at 4×; the video corresponds to Fig. 2 A.
Video 2.
PKD2-mNG is positioned in two rows along the distal region of Chlamydomonas cilia. TIRF video showing the organization of PKD2-mNG in two rows along the distal region of the cilium; PKD2-mNG was expressed in the pkd2 mutant. Images were acquired at 10 fps. To generate the final video, we converted the original image stack using the “walking average” command in ImageJ set to 10 frames. The video corresponds to Fig. 2 Bb and is displayed at 50 fps (5× speed).
Several studies document the ability of PKD2 to oligomerize, but the organization of the native PKD2 complex inside the ciliary membrane has not yet been examined (Shen et al., 2016; Su et al., 2018). Thus, we analyzed the photobleaching dynamics of PKD2-mNG signals in cilia at high laser intensities (Figs. 2 C and S2 B). PKD2-mNG particles in the distal cilium typically bleached in several discernable steps (average 2.8 steps, n = 23 particles), indicating that each particle contains several copies of PKD2-mNG. Because of the high density of PKD2-mNG at the beginning of the experiments, some bleaching steps were likely missed. The data reveal that PKD2 forms oligomeric particles with at least three subunits in cilia. FRAP assays showed minor to no recovery of the PKD2-FP signal in the distal cilium, indicative of stable anchoring with little protein exchange (Fig. S2 C). In contrast, fluorescent PKD2 continued to enter the proximal ciliary region when observed at high light conditions, indicating a dynamic exchange of PKD2-FP between the cell body and the proximal ciliary region (Fig. S2 D). In summary, Chlamydomonas PKD2-FP is organized into two distinct ciliary compartments, a proximal region in which PKD2 is mobile, and a distal region in which two rows of PKD2 are stably anchored.
PKD2 interacts with MST1, the main protein of mastigonemes
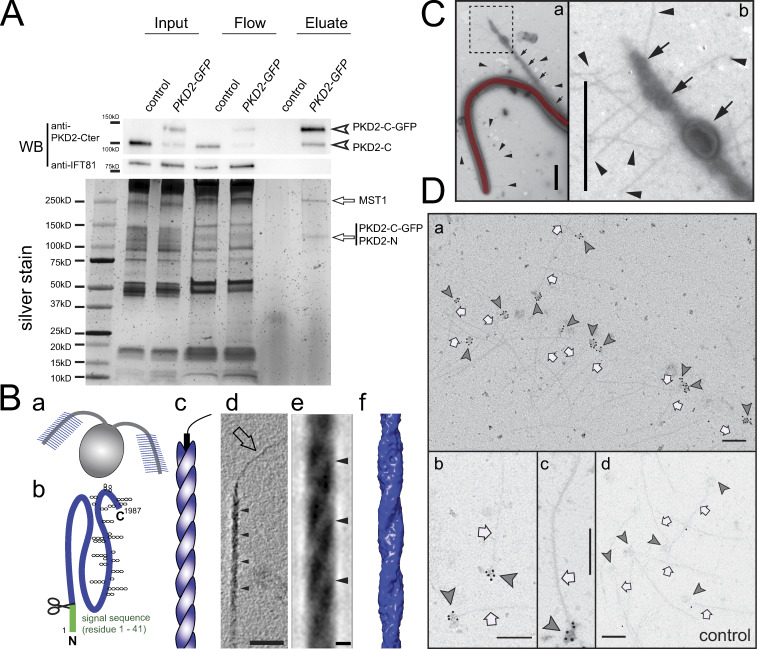
PKD1, which in metazoan cilia interacts with PKD2, is absent from Chlamydomonas, raising the question of whether Chlamydomonas PKD2 has another binding partner. Using anti-GFP nanobody beads, we captured PKD2-GFP from detergent extracts of cilia isolated from a wild-type strain (g1) transformed with PKD2-GFP (Fig. 3 A). Because this strain expresses both untagged and tagged PKD2, it allowed us to test whether PKD2 interacts with itself as implied by the imaging data; a wild-type strain (g1) was used as a control (Fig. S1 D). Western blotting using anti-PKD2-Cter identified two bands in the g1 PKD2-GFP eluates, representing the C-terminal fragments of the endogenous and of the GFP-tagged PKD2, indicating that PKD2 interacts with PKD2-GFP (Fig. 3 A, top). Silver staining showed two bands at 250 and 120 kD in the g1 PKD2-GFP eluate, which were absent in the control eluate; a band corresponding in size to the endogenous PKD2-C (90 kD) was not visible, presumably owing to its low abundance in the eluate (Fig. 3 A, bottom). Mass spectrometry of the 120-kD band identified peptides of GFP and both the N- and the C-terminal fragments of PKD2, suggesting that the two fragments remain associated within the ciliary PKD2 complex. The 250-kD band was identified as MST1.
Figure 3.
PKD2 binds to the mastigoneme protein MST1. (A) PKD2-GFP pull-down assay using a wild-type control strain (g1) and a strain expressing both endogenous and GFP-tagged PKD2. Shown are the detergent extracts of isolated cilia (Input), the flow-throughs, and the eluates from the anti-GFP nanobody trap. Top: Western blot (WB) showing pull-down of both the GFP-tagged and the endogenous C-terminal fragment of PKD2 indicating PKD2 oligomers. Antibodies to IFT81 were used as loading control. Bottom: Silver-stained SDS-PAGE. Mass spectrometry identified the bands marked by arrows as MST1 and a mix of the PKD2-C-GFP and PKD2-N. (Ba–Bc) Schematic representation of Chlamydomonas with mastigonemes in blue (a), MST1 with the predicted signal sequence in green (b; trapezoids indicate glycosylation), and a single mastigoneme (c). (Bd) 20-nm-thick tomographic slice of a mastigoneme showing the periodic substructure (arrowheads) of the main filament and the thinner extended tip (arrow). Bar = 50 nm. (Be and Bf) 1-nm-thick tomographic slice (e) and a full-volume 3D isosurface rendering representation (f) of the subtomogram average based on 1,977 repeat units from 73 mastigonemes after applying fourfold symmetry. Bar = 5 nm. (C) Whole-mount negative stain of an intact Chlamydomonas wild-type cilium. Mastigonemes (arrowheads), released vesicle-like structures (arrows), and cilium (colored in red) are marked in panel a. Enlarged image of the boxed area in panel a showing the vesicle-like structures and mastigonemes (b). Bar = 500 nm. (D) Anti-GFP whole-mount immunogold staining of mastigonemes released from pkd2 PKD2-GFP cells (a–c) and control cilia (d). 10-nm gold complexes decorate the base (arrowheads) of many mastigonemes (arrows) of the PKD2-GFP strain. Bars = 200 nm.
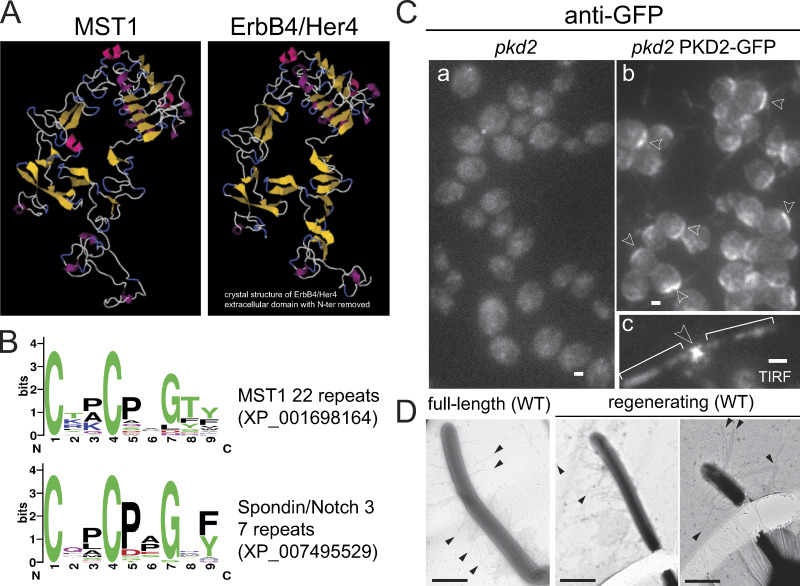
MST1 is the main component of mastigonemes, thin hairs present in two rows in the distal approximately two-thirds of Chlamydomonas cilia (Fig. 3 Ba; Witman et al., 1972; Bergman et al., 1975; Nakamura et al., 1996). The protein is predicted to possess a cleavable N-terminal ER signal sequence (SignalP5.0) and to encompass a cysteine-rich EGFR-like fold (Figs. 3 Bb and S3, A and B). MST1 lacks a transmembrane domain, suggesting that the protein is targeted for secretion (Figs. 3 Bb). The cryo-electron tomography (cryo-ET) average of mastigonemes indicates a helical substructure formed by two protofilaments twisting around each other with a pitch of ∼40 nm; the mastigonemes terminate with a thin tip (Fig. 3, Bc–f; and Video 3; Goodenough et al., 1985). To summarize, mastigonemes are helical extracellular polymers predominantly consisting of MST1.
Figure S3.
MST1 has an EGF-like extracellular fold. (A) Phyre2 homology-based structures of the C-terminal domain of MST1 and the extracellular domain of the receptor tyrosine-protein kinase ErbB4/Her4 showing the conservation of the EGFR extracellular domain fold (Kelley et al., 2015). (B) Weblogo of the 22 cysteine-rich repeats from MST1 and 7 similar motifs of the extracellular domain of Spondin/Notch3 (Crooks et al., 2004). (C) Staining of methanol-fixed pkd2 (a) and pkd2 PKD2-GFP (b) cells with anti-GFP. Arrowheads in b indicate PKD2-GFP signals near the basal bodies. TIRF live image of a pkd2 PKD2-GFP cell (c). Arrowhead in c indicates x-shaped localization of PKD2-GFP near the basal bodies. Brackets, cilia. Bar = 2 µm. (D) Whole-mount negative staining of wild-type cells showing mastigonemes of similar length on full-length and regenerating cilia. Arrowheads, mastigonemes. Bars = 1 µm.
Video 3.
3D visualization of Chlamydomonas mastigonemes. At the beginning of the video, cross-sectional and longitudinal tomographic slices of a mastigoneme average show the helical substructure of the mastigoneme filament; a 3D isosurface rendering representation of the averaged mastigoneme filament is shown at the end. The video plays at 20 fps and corresponds to Fig. 3, Bd–Bf.
To test if PKD2 is attached to the mastigonemes in vivo, we used whole-mount immunogold labeling EM. Cells gliding on a smooth surface typically in a straight line will release vesicular material with attached mastigonemes from the cilia, resulting in a straight trail of membranous material (Fig. 3 C; Bergman et al., 1975). We allowed cells to glide on EM grids, which, after washing, detergent treatment, fixation, and blocking, were incubated with anti-GFP followed by anti-IgG conjugated to 10-nm gold particles. Most mastigonemes released from the pkd2 PKD2-GFP strain were densely decorated with gold particles at one end (Fig. 3, Da–Dc). The labeled end encompasses an amorphous bulb and represents where mastigonemes were attached to the cilia. Only a few randomly distributed gold particles were detected in the wild-type control samples (Fig. 3 Dd). Therefore, PKD2 is attached to the base of the mastigonemes, and the two are often released together from cilia under stress, supporting the notion that PKD2 and mastigonemes form a stable complex.
PKD2 anchors mastigonemes on the ciliary surface
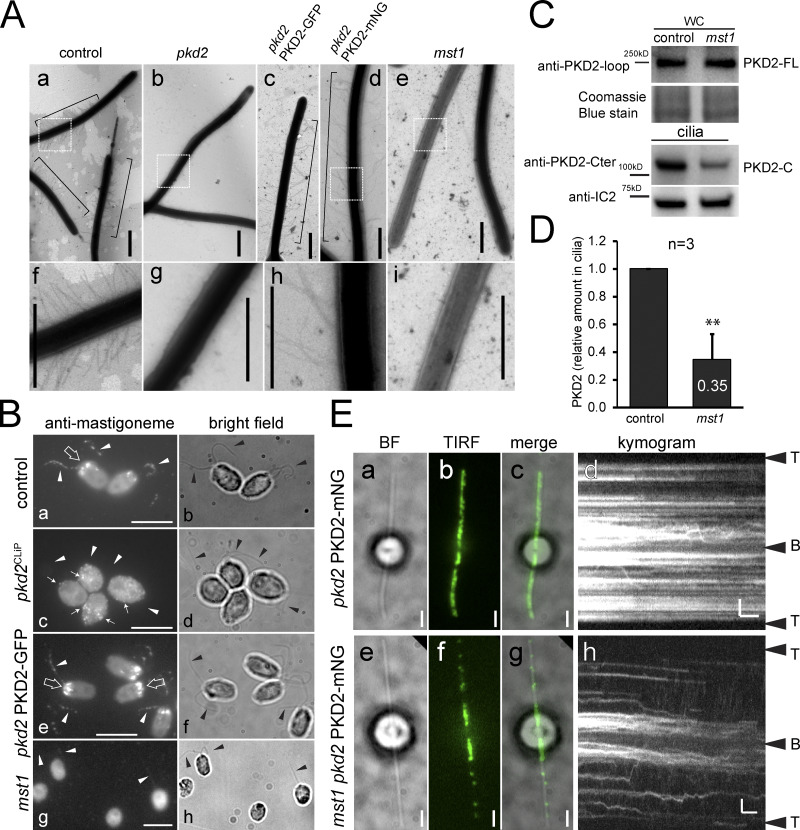
Because PKD2 was found at the base of mastigonemes, we wondered if Chlamydomonas PKD2 is required to anchor mastigonemes on the ciliary membrane. Mastigonemes were readily detected on cilia of control cells by whole-mount EM but were not observed on the cilia of the pkd2 mutant (Fig. 4, Aa, Af, Ab, and Ag). Introduction of PKD2-FP into pkd2 by mating (Fig. 4 Ac) or transformation (Fig. 4, Ad and Ah) restored mastigonemes on cilia. Thus, PKD2 is required for the assembly of mastigonemes onto the cilia surface. Immunofluorescence staining using a previously characterized antibody to MST1 confirmed the absence of mastigonemes from pkd2 cilia (Fig. 4, Bc and Bd; Nakamura et al., 1996). In addition to the signal along the cilia, anti-MST1 stained a cellular pool of MST1 near the basal bodies in control and pkd2 PKD2-GFP rescue cells (Fig. 4, Ba, Bb, Be, and Bf; Nakamura et al., 1996). In pkd2, the pool was absent; instead, small puncta of MST1 were dispersed throughout the cell, indicating that targeting of MST1 to the ciliary base was affected (Fig. 4 Bc). PKD2-FP was also present near the basal bodies, where it could colocalize with MST1 (Fig. S3 C). While not observed for the thin mastigonemes of Chlamydomonas, the large and complex mastigonemes of Ochromonas are present inside secretory vesicles near the ciliary base (Bouck, 1971). Moreover, Chlamydomonas PKD2 is present in intracellular vesicles (Wood and Rosenbaum, 2014). We propose that MST1 polymerizes inside such vesicles into mastigonemes onto PKD2 located in the vesicular membrane. The cytoplasmic portions of PKD2 direct such vesicles to the ciliary base for fusion, which will expose the mastigonemes to the extracellular environment; PKD2–mastigoneme complexes then move laterally from the plasma membrane into the cilium. Thus, PKD2 is required to recruit MST1/mastigonemes to the ciliary base and to mount mastigonemes onto the ciliary membrane. The data indicate a novel role for PKD2 in targeting extracellular components to the ciliary surface.
Figure 4.
PKD2 organizes ciliary mastigonemes. (A) Whole-mount EM of control (CC-5235; a and f), pkd2 (b and g), pkd2 PKD2-GFP (c), and pkd2 PKD2-mNG (d and h) rescue strains and mst1 (e and i). Mastigonemes are indicated by brackets. f, g, h, and i are enlarged images of the boxed areas in a, b, d, and e. Bars = 800 nm. (B) Immunofluorescence staining with anti-MST1 and the corresponding bright-field images of formaldehyde-fixed control (CC-5235; a and b), pkd2CLIP (c and d), pkd2 PKD2-GFP (e and f), and mst1 (g and h) mutant cells. Note that a pool of MST1 is present near the basal bodies (large arrows) of control cells, but absent in pkd2 and restored in pkd2 PKD2-GFP. In pkd2, anti-MST1 signals were dispersed throughout the cell body (c, small arrows); these were absent in mst1 (g). Arrowheads indicate the position of the cilia. Bar = 2 µm. (C) Western blot analysis of control (CC-5235) and mst1 whole-cell (WC) samples and isolated cilia. Anti-IC2 and Coomassie staining were used as loading controls. (D) Relative amount of PKD2 in mst1 cilia. Ciliary PKD2 protein levels were normalized for anti-IC2 staining. The SD (error bar) and the number of independent biological replicates (n) are indicated. Significance based on two-tailed t test is indicated (**, P ≤ 0.01). (E) PKD2-mNG distribution in pkd2 PKD2-mNG (a–d) and mst1 pkd2 PKD2-mNG (e–h) cells. Bright-field (a and e), TIRF (b and f), merged images (c and g), and the corresponding kymograms (d and h) are shown. Bars = 2 s and 2 µm.
Mastigoneme loss affects the assembly of ciliary PKD2 rows
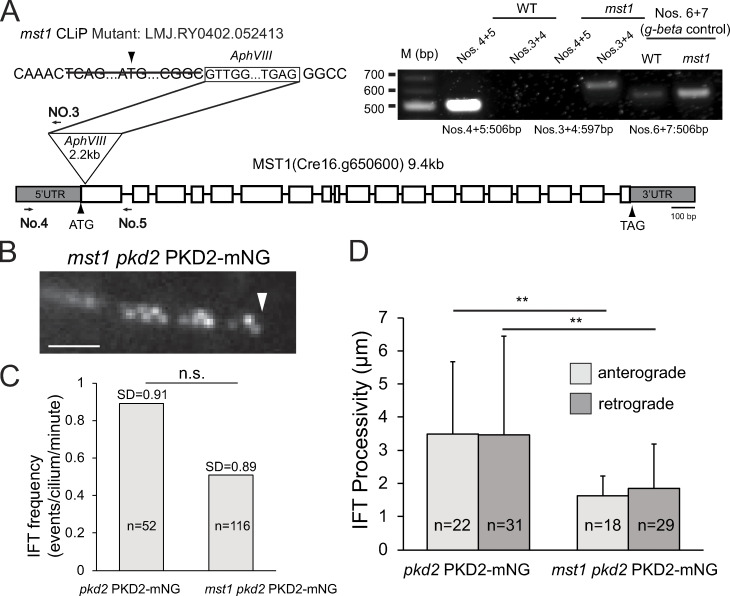
The extracellular domain between transmembrane helix 1 and 2 of mammalian PKD2 is a hotspot for disease-related mutations, likely because of its role in PKD2 oligomerization (Su et al., 2018). MST1 binding likely involves the large extracellular domain of Chlamydomonas PKD2, raising the question of whether mastigonemes contribute to the formation of PKD2 complexes (Fig. 1 A). We obtained CLiP strain LMJ.RY0402.052413, which carries an insertion in the MST1 gene that deletes 75 bp encoding the start and parts of the signal peptide, indicating that the strain may be a null mutant for MST1 (Fig. S4 A). Staining with anti-MST1 or whole-mount EM failed to detect mastigonemes on the mutant cilia; therefore, we named this strain mst1 (Fig. 4, Ae, Ai, Bg and Bh). An MST1 pool or puncta observed in control and pkd2 cell bodies, respectively, were absent in mst1 (Fig. 4 B). Nakamura et al. (1996) reported a ∼10% reduction in swimming velocity after mastigonemes were removed from wild-type cilia by treatment with anti-MST1. In agreement, the mst1 mutant also swam with a reduced velocity (112 µm/s or ∼15% slower than wild-type), resembling the swimming defect of pkd2 (Fig. 1 D). Both mutant strains had near-wild-type ciliary beat frequencies (54 and 50 Hz for pkd2 and mst1, respectively; Fig. S1 E). A similar reduction in swimming velocity was also observed for the pkd2 mst1 double mutant (Fig. 1 D), providing genetic support that PKD2 and MST1 act in the same pathway. We conclude that both MST1 and PKD2 are required to achieve a high, wild-type swimming velocity in Chlamydomonas.
Figure S4.
Loss of MST1 interferes with PKD2 assembly and transport. (A) Diagram showing the insertion in the mst1 mutant strain. The triangle marks the insertion site. Arrows indicate the positions of the primers used for PCR. Insert: Agarose gel of PCR products using wild-type and mst1 genomic DNA as templates. G-β amplified with primers 6 and 7 was used as control for DNA quality. (B) Detail of an mst1 pkd2 PKD2-mNG cilium showing the organization of the residual PKD2-mNG in two rows. Arrowhead, ciliary tip. Bar = 2 µm. (C) IFT frequency (anterograde and retrograde combined) of PKD2-mNG in pkd2 PKD2-mNG and mst1 pkd2 PKD2-mNG cilia. The SDs (error bars) and number of cilia analyzed (n) are indicated; n.s., difference is not statistically significant based on two-tailed t test. (D) IFT processivity (anterograde and retrograde, respectively) as measured by average run length of PKD2-mNG particles in pkd2 PKD2-mNG and mst1 pkd2 PKD2-mNG cilia. The SDs (error bars) and number of events analyzed (n) are indicated. **, P ≤ 0.01; significance based on two-tailed t test.
The amount of endogenous PKD2 was normal in whole-cell samples of the mst1 strain. In mutant cilia, however, PKD2 was reduced to ∼34% of the amount present in control cilia (Fig. 4, C and D). Thus, the loss of MST1 impacts the entry and/or incorporation of PKD2 into cilia. In vivo imaging of an mst1 pkd2 PKD2-mNG strain showed that the amount of PKD2-mNG in the distal ciliary segment was generally reduced but varied between individual cells (Fig. 4 E). Residual stationary PKD2-mNG in the distal region of mst1 cilia was still organized into two rows (Fig. S4 B). However, a considerable portion of PKD2-mNG remained mobile, suggesting that the presence of mastigonemes promotes axonemal anchoring of PKD2 (Fig. 4, Ee–Eh; and Video 4). IFT of PKD2-mNG in mst1 cilia was less frequent and less processive (1.7 vs. 3.5 µm run length in control cilia), suggesting that PKD2-MST1 complexes are better IFT cargoes than PKD2 alone (Fig. S4, C and D). However, PKD2-mNG remained abundant in the proximal region of mst1 cilia, which also lacks mastigonemes in wild-type cilia (Fig. 4 E; Nakamura et al., 1996). Thus, mastigonemes are not required for the entry and accumulation of PKD2 into the proximal ciliary region. On short regenerating cilia, mastigonemes were of similar length (∼700–900 nm) as those on full-length cilia, indicating that mastigonemes do not form by stepwise assembly on the ciliary surface (Fig. S3 D). Because mastigonemes are primarily absent from the proximal region of growing and full-length cilia (Fig. S3 D; Nakamura et al., 1996), we propose that full-length mastigonemes bound to PKD2 enter the cilium and then pass quickly through the proximal region to be captured in the distal region of the cilium.
Video 4.
Increased mobility of PKD2-mNG in the absence of mastigonemes. TIRF video of PKD2-mNG in cilia of a mst1 pkd2 PKD2-mNG cell and corresponding kymogram. Images were acquired at 10 fps, and the video is replayed at 2× speed. The video is related to Fig. 4, Ee–Eh.
PKD2–mastigoneme complexes are anchored perpendicular to the plane of the ciliary beating
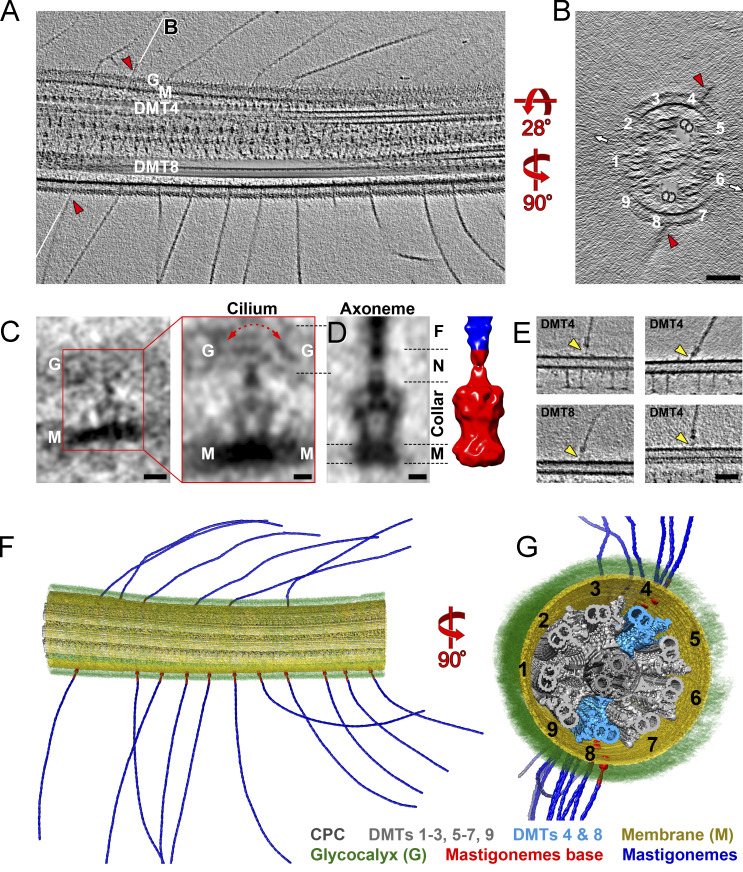
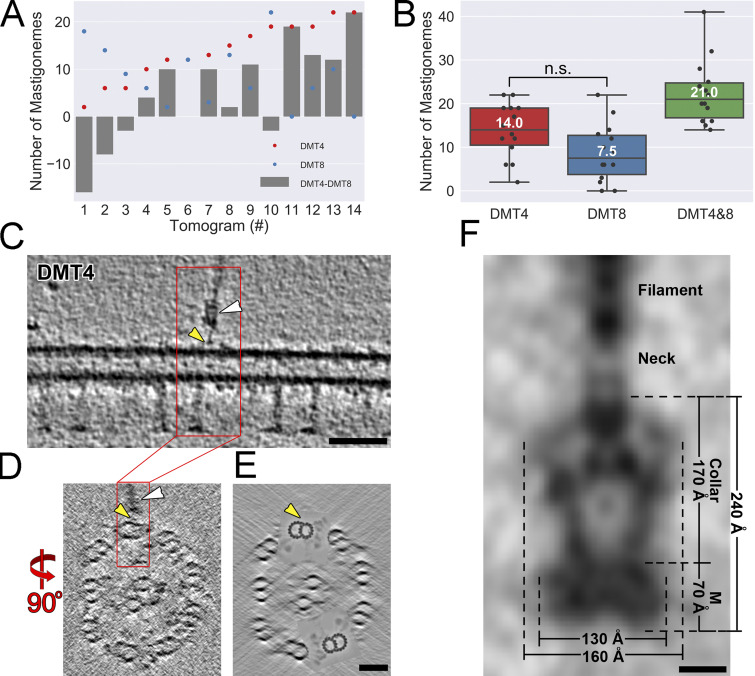
PKD2 anchors mastigonemes in two rows along the distal two-thirds of the cilium. We used cryo-ET to determine the structure of the mastigoneme base and the position of the PKD2–mastigoneme complexes with respect to the axonemal core structure in 3D at high resolution (Fig. 5 and Video 5). In Chlamydomonas, the nine axonemal DMTs are structurally and functionally different. DMT1, for example, lacks outer dynein arms and is located in the plane of the ciliary beating (Hoops and Witman, 1983; Lin et al., 2012). Using such doublet-specific structural features, we identified the nine DMTs and determined that the mastigonemes are positioned near DMTs 4 and 8 (Fig. 5, A, B, and E–G; and Fig. S5, C–E). Thus, mastigonemes project from the ciliary surface perpendicular to the plane of the ciliary beat (Fig. 5, B, F, and G). The spacing and distribution of the mastigonemes along the ciliary length were somewhat variable, and we did not detect a significant preference for either DMT4 or DMT8 (Fig. S5, A and B).
Figure 5.
The PKD2–mastigoneme complexes associate with DMT4 and DMT8. (A and B) Tomographic slices of an intact Chlamydomonas wild-type cilium viewed in longitudinal section (A, thickness = 25 nm) and cross section (B, thickness = 5 nm). Mastigonemes (red arrowheads) are positioned closest to DMT4 and DMT8. The white lines labeled B in A indicate the position of the cross section shown in B. The cross section (B) shows the relative positions of the two rows of mastigonemes to the ciliary beating direction (small white arrows). For clarity, DMTs 4 and 8 are highlighted by cloning the subtomogram average of the 96-nm DMT repeat back into the raw tomogram for all axonemal repeats along DMT4 and 8. Bar = 100 nm. (C) Tomographic slices of the averaged mastigoneme base (left, thickness = 2 nm; bar = 10 nm) and after applying threefold symmetry (right, thickness = 2 nm; bar = 5 nm) from intact Chlamydomonas cilia. The averaged structure is based on 53 particles extracted from six tomograms. The ciliary membrane (M) and glycocalyx (G) are indicated. The red arrow indicates variable positions of the mastigonemes with respect to the base in our samples. (D) Tomographic slice (left, thickness = 2 nm; bar = 5 nm) and 3D isosurface rendering representation (right) of the averaged mastigoneme base from demembranated cilia (axonemes) after applying threefold symmetry. The dashed lines on the left indicate the levels of the membrane (M) and glycocalyx (G); the dashed lines on the right mark the trans-membrane (M), collar, and neck (N) subregions of the mastigoneme base and the attached mastigoneme filament (F). The dimensions of the averaged base complex are shown in Fig. S5 F. (E) Tomographic slices (thickness = 5 nm; bar = 50 nm) of demembranated cilia showing thin linkers (arrowheads) between the mastigoneme bases and DMTs; the DMT number is indicated. See Fig. S5, C–E for further details of contact site between the base complex and the DMTs. (F and G) 3D isosurface rendering of a representative region of an intact Chlamydomonas wild-type cilium (same as shown in A and B). Subtomogram averages of the mastigoneme filaments, 96-nm axonemal DMT repeats, and the mastigoneme base complexes were cloned back into the positions of individual repeat units in the raw reconstruction; selected substructures were segmented and colored as indicated for better visualization.
Video 5.
Tomographic slices and 3D visualization of a representative region of an intact wild-type Chlamydomonas cilium. The video starts with cross-sectional and longitudinal tomographic slices through a 3D reconstructed wild-type Chlamydomonas cilium. This is followed by a 3D isosurface rendering visualization of the cilium, in which selected structures are highlighted by different colors, such as the central pair complex (CPC), DMTs 1–9, membrane, glycocalyx, mastigonemes base, and the mastigonemes. The video plays at 20 fps and corresponds to Fig. 5, F and G.
Figure S5.
Mastigonemes are linked to the outer A-B-tubule junction. (A) The numbers of mastigonemes associated with DMTs 4 and 8, respectively (red and blue dots), and their difference (gray bars) in each of 14 tomograms of intact wild-type Chlamydomonas cilia. (B) The distribution of mastigoneme numbers associated with DMT4, DMT8, and their sum in tomograms from intact wild-type Chlamydomonas cilia. n.s., not significant; n = 14 cilia; two-tailed t test. (C–E) Tomographic slices of a representative demembranated wild-type Chlamydomonas cilium, as viewed in longitudinal section (C, which is an enlarged version of an image shown in Fig. 5 E; thickness: 5 nm) and cross sections (D, thickness: 0.5 nm; and E, thickness: 50 nm). A mastigoneme with its base (white arrowheads) is clearly visible, and the anchor site of the thin linker that connects the mastigoneme base to DMT4 is highlighted by yellow arrowheads. For clarity of the DMT structure, DMTs 4 and 8 are highlighted in E by cloning the subtomogram average of the 96-nm DMT repeat back into the raw tomogram for all axonemal repeats along DMT4 and 8. Bars = 50 nm. (F) Dimensions and named regions of the averaged mastigoneme base (also shown in Fig. 5 E) complex in demembranated cilia. The transmembrane disk (M), collar, neck, and mastigoneme filament are marked. Bar = 5 nm.
Tomograms of intact Chlamydomonas cilia show that each mastigoneme is attached to a membrane-embedded base structure (Fig. 5 C), which likely encompasses several copies of PKD2, as indicated by the immunogold labeling and photobleaching experiments. In agreement with the biochemical data, some mastigonemes remained attached to the axonemes after the membrane was removed with detergent. Subtomogram averages of 53 base complexes from cilia (Fig. 5 C) and 17 from axonemes (Fig. 5 D) show that the mastigoneme base has a trapezoid-shaped structure with a maximum diameter of ∼160 Å and a total length of ∼240 Å (Fig. S5 F). The base structure consists of a membrane-embedded disc and a ∼170-Å-long collar. The membrane-embedded portion of the complex has dimensions of ∼130 by 70 by 130 Å, which is similar in size to the human PKD1-PKD2 complex but larger than the PKD2 homotetramer (Shen et al., 2016; Su et al., 2018). The collar encloses a central vestibule and holds the mastigoneme via a neck region at the beginning of the glycocalyx (Fig. 5, C and D). The absence of the ciliary membrane and glycocalyx allowed us to visualize additional structural details of the averaged mastigoneme base, including a thin linker that connected each mastigoneme base to the axonemal DMT4 and DMT8 near their outer junction between the A- and B-tubules (Figs. 5 E and S5, C–E). Our biochemical, genetic, and microscopic data strongly indicate that the trapezoid particle revealed by cryo-ET encompasses several copies of PKD2, providing the first structural details of a native PKD2 channel complex within the ciliary membrane.
Discussion
Chlamydomonas PKD2 links extracellular structures to the cytoskeleton
Our data show two major interactions of the PKD2 complex in Chlamydomonas cilia: it binds to the axonemal DMT4 and DMT8, and it anchors mastigonemes, large extracellular polymers of the glycoprotein MST1, to the ciliary membrane. Association with the cytoskeleton and the ECM is characteristic from many mechanosensory channels, raising the question of whether the PKD2 complex in other systems also functions as a link between the cytoskeleton and extracellular components. Ciliary mastigonemes, including the structurally more complex tripartite mastigonemes, are present in various groups of protists, but homologues of MST1 are limited in distribution to green algae (Bouck, 1971; Bouck et al., 1978). However, several lines of evidence suggest a relationship between cilia, PKD2, and the ECM (Drummond, 2011; Seeger-Nukpezah and Golemis, 2012). For example, the ciliated endings of mechanosensory neurons in Caenorhabditis elegans and the modified cilia in mechanosensory insect sensilla, the sense organs for hearing and tactile sensation, are embedded in the ECM (Garcia-Anoveros and Corey, 1997; Keil, 2012). In mammals, the tips of certain motile cilia carry a crown of bristles that is not well characterized (Dirksen and Satir, 1972). Furthermore, ADPKD includes extrarenal phenotypes such as abdominal hernias, suggesting a role of the PKD1-PKD2 complex in ECM secretion and cell adhesion (Mangos et al., 2010). Similar to MST1, many transmembrane and ECM proteins encompass EGFR-like domains, and mammalian PKD2 displays EGF-induced conductance, which is likely because of its possible interaction with the EGF receptor (Ma et al., 2005). In C. elegans, the localization and function of PKD2 are affected by mutations in several mec genes encoding ECM proteins with EGFP repeats (De Vore et al., 2018). Thus, a yet-to-be-identified extracellular protein or polymer chain could transiently or permanently binds to PKD2 in primary cilia. Alternatively, PKD1, which is absent from protists, possesses a large extracellular domain that could perform a mechanotransductory role in the mammalian PKD1-PKD2 complex similar to that of mastigonemes in the PKD2-MST1 complex of Chlamydomonas.
Chlamydomonas PKD2 forms linear arrays in cilia
The PKD2–mastigoneme complex is physically linked to just two of the nine axonemal DMTs. An organization into two rows and association with the axoneme is also typical for mastigonemes in other protists (Markey and Bouck, 1977). PKD2 could bind directly or indirectly to the DMTs. In C. elegans and mammalian cells, the level of tubulin glutamylation regulates the abundance of PKD2 in cilia, but direct binding of PKD2 to microtubules has not been demonstrated (He et al., 2018; O’Hagan et al., 2011). In Chlamydomonas, several other circumferential asymmetries in axonemal structure exist, including the absence of outer dynein arms from DMT1, doublet specific linkers, and microtubule inner proteins; the basis for this remarkably specific targeting of protein complexes in cilia remains unknown (Hoops and Witman, 1983). Anchoring of PKD2 likely involves the thin linker observed between the Chlamydomonas PKD2 complex and the DMTs. In mechanosensory sensilla, a linker composed of both channel and nonchannel components connects the TRPN channel NOMPC to microtubules (Sun et al., 2019). In addition to positioning channels, linkers that connect channel proteins to the cytoskeleton or ECM can also function as gating springs to open the channels (Markin and Hudspeth, 1995).
The DMTs are not only structurally distinct but also contribute differentially to axonemal bending (Lin and Nicastro, 2018). Thus, the anchoring of channels to specific doublets could ensure a spatially defined axonemal response to channel activity. Although not directly linked to the axoneme, the voltage-gated calcium channel CatSper, which is required for the hyperactivation of sperm flagella, is arranged in four rows along the principal piece of human and mouse sperm flagella. Mutations in several CatSper subunits disrupt this order and cause male infertility (Chung et al., 2014, 2017; Hwang et al., 2019). Additionally, specific positioning of TRP channels has also been found for the Drosophila PKD2 homologue Almost there (AMO), which localizes to the tip of sperm flagella. Mutant amo males have motile sperm but remain infertile because the spermatozoa fail to hyperactivate, which is required for them to exit from the female spermatheca (Köttgen et al., 2011). Distinct patterns of TRP channels in the ciliary membrane could reflect species- and cell-specific adaptations to regulate the diverse bending patterns of motile cilia (Fujiu et al., 2011).
The Chlamydomonas PKD2–mastigoneme complex functions in cilia motility
Mutants in pkd2 and mst1 share a slow-swimming phenotype, indicating a motility-related role of the PKD2–mastigoneme complex in cilia. Mastigonemes, which fan out from the ciliary surface, likely increase the efficiency of the ciliary beat (Jahn et al., 1964). Thus, one interpretation is that PKD2’s role is to anchor the mastigonemes, and the loss of mastigonemes in the pkd2 mutant is responsible for the reduction of swimming velocity. However, an alternate or additional function of PKD2 is probable because mammalian PKD2 shows ion conductivity, and, based on sequence conservation, Chlamydomonas PKD2 is likely also a functional channel (Huang et al., 2007; Liu et al., 2018; Shen et al., 2016). It has been hypothesized that mechanical gating of mammalian PKD2 as a result of flow-induced passive bending of primary cilia is an important feedback mechanism in kidneys, and that when it goes amiss it results in PKD disease (Nauli et al., 2003; Praetorius and Spring, 2001). However, the change in membrane tension during passive ciliary bending is likely too small to be detected by mechanosensitive channels (Ferreira et al., 2019). Instead, if such a channel is linked to extracellular structures (such as the ECM), the axoneme, or both, the tension produced by mechanical forces acting on these associated structures is more likely sufficient to activate the channel. Ferreira et al. (2019) suggested that even flows that are too weak to bend a primary cilium could still generate sufficient drag on long extracellular polymer chains to activate an associated mechanosensitive channel. Indeed, ECM association, cytoskeletal anchoring, or both are hallmarks of many mechanosensitive channels such as that used for hearing (Gillespie and Müller, 2009). Thus, mastigonemes could mediate mechanical opening of PKD2 in Chlamydomonas cilia.
As Chlamydomonas cilia oscillate with ∼60-Hz frequency, mastigonemes will pull on PKD2, which is secured to the axoneme. The forces acting on the ciliary surface will vary in strength and direction depending on the stage of the beat cycle and the position on the cilium. The stereotypical orientation of PKD2–mastigoneme complexes on cilia could specify the timing of PKD2 opening, which in turn could locally regulate motor protein activity. In nonmotile cilia, microtubule anchoring and ECM association could underlie mechanical gating of PKD2 in response to flow-induced shear forces. Therefore, our observation that PKD2 is attached to mastigonemes and the axoneme suggests a mechanosensory role of PKD2 in motile cilia and points at a possible general functional mechanism that could also play a role in primary cilia.
Materials and methods
Strains, culture conditions, and genotyping
Chlamydomonas strains used in this study are listed in Table 1. The pkd2 (LMJ.RY0402.204581) and mst1 (LMJ.RY0402.052413) mutants are available from the Chlamydomonas Library Project (CLiP; https://www.chlamylibrary.org/allMutants). Cells were grown in modified minimal medium (M medium; https://www.chlamycollection.org/methods/media-recipes/minimal-or-m-medium-and-derivatives-sager-granick/) and maintained at 22°C with a light/dark cycle of 14:10 h; large cultures used for cilia isolation were aerated with air enriched with 0.5% CO2.
Table 1. List of strains.
| Name | Genotype | Reference or source |
|---|---|---|
| CC-621 (wild-type) | nit1, nit2, mt− | Chlamydomonas Genetics Center |
| CC-620 (wild-type) | nit1, nit2, mt+ | Chlamydomonas Genetics Center |
| g1 (wild-type) | nit1, agg1, mt+ | Pazour et al., 1995 |
| CC-5235 and CC-4533 | cw15, mt− | CLiP |
| pkd2CLiP (LMJ.RY0402.204581) | cw15, pkd2, mt− | CLiP |
| pkd2 (progeny selected from pkd2CLiP backcrossed twice with g1) | pkd2 (both mating types) | This study |
| mst1 (LMJ.RY0402.052413) | cw15, mst1, mt− | CLiP |
| PKD2-GFP | mt+ | Huang et al., 2007 |
| pkd2 PKD2-mNG | pkd2, mt− | This study |
| pkd2 PKD2-GFP | pkd2, mt− | This study |
| mst1 pkd2 | mst1, pkd2 | This study |
| mst1 pkd2 PKD2-mNG | mst1, pkd2 | This study |
PCR was used to verify the insertions in the CLiP mutants and track the insertional alleles in mating progeny. In brief, genomic DNA was isolated from control, pkd2 (LMJ.RY0402.204581), and mst1 (LMJ.RY0402.052413). To identify the insertional alleles, primers 1–3 were used for pkd2, and primers 3–5 for mst1 (Table 2). To control for DNA quality, primers 6 and 7 were used to amplify parts of the g-β gene, and primers 8–11 were used to determine the mating type (Zamora et al., 2004).
Table 2. List of primers.
| No. | Sequence (5′ to 3′) |
|---|---|
| 1 | ACCACCAACGAGGAGATTGC |
| 2 | GATGACACGGTTGGTGAG |
| 3 | TGTCGCTGAAAGTGGAGGTC |
| 4 | GGACGTCTCCTTGGCCTTAT |
| 5 | AAGGCAGCTCCACACTCACT |
| 6 | CAAGCTGAAGAACAACCTGGTG |
| 7 | CTTGCTGGTGATGTTGAACTCG |
| 8 | TCCAACGCATAGCCATCAAC |
| 9 | TGTTTGCTAGGGGTGCAATG |
| 10 | ACCGGTGTTTACCGTCGAGT |
| 11 | CCTTTCTGTAGGGCCACCTG |
| 12 | GGACTAGTCTTTCTTGCGCTATGACACTTC |
| 13 | GCTCTAGACCATGGGATGACGGGCCCG |
| 14 | CGCCTCGAGGGCGCCTCCGGCCAGGGCGCCTCCGGCGCGATGCGAACCTTCGGGCCCA |
| 15 | CGCGGATCCCGCGCCGGAGGCGCCCTGGCCGGAGGCGCCCTGGGGCGGGGTCTCATTC |
| 16 | GCGTATTTGAAGCGGGTACCATCGATACCGTCGACCTCG |
| 17 | TATAGGGCGAATTGGGTACCCAAATACGCCCAGCCCGC |
Transgenic strain generation
To express PKD2-GFP in the pkd2 mutant, the aph7" gene conferring resistance to hygromycin was amplified by PCR with primers 12 and 13 and inserted into pHK52, a plasmid encompassing PKD2 fused at its C-terminus to GFP; the final construct was named pHK52-Hyg (Huang, et al., 2007). SpeI-linearized pHK52-Hyg was used to transform into the pkd2 mutant. To replace the GFP with mNG, the PKD2 genomic sequence was amplified by PCR with primers 14 and 15 and cloned into the pBR25-mNG plasmid linearized with HindIII and BamH1 using In-Fusion (Clontech). Similarly, the bleomycin resistant gene was PCR amplified with primers 16 and 17 and inserted into the unique Kpn1 site of the pBR25-PKD2-mNG plasmid. The resulting plasmid pPL-PKD2-mNG was linearized with PsiI and transformed into the pkd2 and mst1 mutants by electroporation (NEPA21 Super Electroporator Type II). Transformants were selected on TAP plates containing 5 µg/ml zeocin (Invitrogen) or, in the case of the pHK52-Hyg transformants, 20 µg/ml hygromycin (Bio Basic). Clones expressing FP-tagged PKD2 were identified by TIRF microscopy. Typically, 1 in ∼150 transgenic clones expressed tagged PKD2.
Mating experiments
For mating experiments, 100 ml of the vegetative cells were aerated in M medium at a light/dark cycle of 14:10 h for 4–5 d to reach a concentration of 2 × 106 cells/ml. The evening before the mating experiment, cells were transferred to 15 ml M-N medium (M medium without nitrogen; https://www.chlamycollection.org/methods/media-recipes/minimal-or-m-medium-and-derivatives-sager-granick/) and aerated overnight in constant light. In the morning, cells were transferred to 2 ml of 1/5 M-N supplemented with 10 mM Hepes and incubated for an additional 30 min. Then, gametes of the opposite mating type were mixed, incubated for 4–6 h in light, transferred to M medium 4% agar or 1.8% Phytogel (Sigma-Aldrich) plates, air-dried, and incubated overnight in light and then for ≥10 d in the dark. Plates were transferred to −20°C for 2 d and defrosted, and zygotes were allowed to germinate and grow in constant light for ∼10 d. After streaking for single cells, progeny were identified by PCR, TIRF imaging, and Western blotting.
To quantify the mating efficiency, we counted biflagellate and quadriflagellate cells in samples fixed 30 min after mixing of equal numbers of mt+ and mt− gametes and applied the following formula: % fusion = (2 quadriflagellate × 100)/(2 quadriflagellate + biflagellate). Mating efficiency was also tested in a population assay based on the feature that zygotes initially lack a cell wall and are easily lysed by detergent treatment. After mixing of the gametes, cells were stored in the dark for 45 min and treated with 0.2% NP-40 Alternative (Calbiochem, 492016) to release the chlorophyll. After 10 min, the cell suspension was centrifuged (tabletop, top speed, 5 min, RT), and the absorbance of the supernatant was measured at 440 nm in a spectrophotometer (BioTek SynergyII). For each strain tested, the absorbance was normalized using the mating between the wild-type control strains CC-620 and CC-621 (100%).
Isolation of cilia
To isolate cilia, cells were concentrated and washed with 10 mM Hepes (pH 7.4), resuspended in 10 ml of Hepes-Magnesium-Sucrose (HMS; 10 mM Hepes, pH 7.4, 5 mM MgSO4, and 4% sucrose wt/vol), and deciliated by addition of 2.5 ml of dibucaine-HCl (25 mM in H2O; Sigma-Aldrich) and vigorous pipetting (King, 1995). Next, 20 ml of 0.7 mM EGTA in HMS was added, and the cell bodies were removed by centrifugation (1,150 g, 3 min, 4°C; Sorvall Legend XTR, Thermo Fisher Scientific). The supernatant was underlain with a sucrose cushion (10 ml of 25% sucrose in HMS) and centrifuged (1,700 g, 4°C, 10 min) to remove the remaining cell bodies. The cilia in the upper phase were sedimented by centrifugation (27,000 g, 4°C, 20 min; Beckman Coulter, Avanti JXN-26) and resuspended in Hepes-Magnesium-EGTA-Potassium (HMEK; 30 mM Hepes, 5 mM MgSO4, 0.5 mM EGTA, and 25 mM KCl) supplemented with 1% protease inhibitor cocktail (Sigma-Aldrich, P9599), and extracted for 20 min on ice with NP-40 Alternative (1% final concentration). The axonemal and membrane + matrix fractions were separated by centrifugation (27,000 g, 4°C, 15 min).
For cryo-electron tomography, cilia of strain CC-4533 were detached from the cells using the pH-shock method (Witman, 1986) and purified by two centrifugations (2,400 g for 10 min) over sucrose cushions (20% sucrose, 1 mM SrCl2, 10 mM Hepes, and 1 mM DTT). The cilia-containing layer was collected and supplemented with IGEPAL CA-630 (Sigma-Aldrich; final convention 1%) to remove the ciliary membrane by a 20-min incubation at 4°C with mild agitation. The axonemes were collected by centrifugation (10,000 g for 10 min) and resuspended in 30 mM Hepes, pH 7.4, 5 mM MgSO4, 1 mM EGTA, 0.1 mM EDTA, and 25 mM KCl.
Antibodies and Western blotting
Isolated whole cells, cell bodies, and cilia samples were incubated for 10 min at 95°C in Laemmli SDS sample buffer. Protein samples were separated by SDS-PAGE using precast gels (Bio-Rad TGX) and transferred to PVDF membrane. Membranes were blocked in 4% milk (4% BSA for anti-PKD2-Cter) and stained according to standard protocols, then incubated in the diluted primary antibodies overnight at 4°C with agitation and the secondary antibodies for ∼60 min at RT. The following antibodies were used in this study: rabbit polyclonal anti-PKD2-Cter (1:2,000), anti-PKD2-loop (1:2,000; Huang et al., 2007), mouse monoclonal anti-IFT81 (1:500; Cole et al., 1998), anti-acetylated tubulin (1:10,000, Sigma-Aldrich 6-11-B), anti-IC2 (1:100; King and Witman, 1990), and anti-NAB1 (1:5,000; Agrisera). Secondary antibodies (anti-mouse or anti-rabbit IgG conjugated to horseradish peroxidase; Invitrogen) were diluted 1:2,000 and visualized using Femto Glow Western PLUS (Michigan Diagnostics) as a substrate. A ChemiDoc MP imaging system and Image Lab software (Bio-Rad Laboratories) were used for documentation and quantification of signals.
Immunoprecipitation
Cilia isolated from a wild-type expressing PKD2-GFP and an untransformed control strain were resuspended in HMEK + protease inhibitor cocktail, lysed by addition of 1% NP-40 (final concentration), and adjusted to 100 mM NaCl (final concentration) using a 2-M NaCl stock. The axonemes were removed by centrifugation (27,000 g, 4°C, 15 min) and the supernatants were incubated with anti-GFP nanobody agarose beads (Allele Biotechnology) for 1 h at 4°C using a rotisserie. The beads were washed twice with 150 mM NaCl, twice with 200 mM NaCl, and once with HMEK. Proteins were eluted using 200 mM glycine, pH 2.5. The eluate, input, and flow-through were then analyzed by Western blotting and SDS-PAGE; bands of interest were cut from silver-stained gels (Silver Stain Plus Kit, Bio-Rad Laboratories) and subjected to mass spectroscopy using an Orbitrap Elite system at the Proteomics and Mass Spectrometry Core Facility at the University of Georgia.
Whole-mount negative stain and immunogold EM
For whole-mount immunogold labeling EM, 10 µl of concentrated cells (∼2 × 107 cells/ml in M medium) were applied to formvar-coated EM grids and allowed to settle for 3 min. The specimens were then inverted onto a drop of HMEK supplemented with 0.3% NP-40 for 20 s, washed twice with HMEK, and fixed for 3 min with 1% formaldehyde in HMEK. After washing in PBS (2 × 5 min) and PBS-T (PBS + 0.05% Tween-20; 2 × 5 min), specimens were blocked for 20 min with blocking buffer (PBS-T + 1% BSA [Akron, AK8909] + 0.1% cold water fish gelatin [Sigma-Aldrich, G7765]). After blocking, grids were incubated overnight at 4°C in rabbit polyclonal anti-GFP antibody (Abcam, 6556; diluted 1:100 in blocking buffer). After washing (4× 5 min in PBS-T), grids were incubated in 10 nm gold conjugated to goat anti-rabbit IgG (Pelco, 1:20 in blocking buffer) for 1.5 h at RT. Then, grids were washed in PBS-T (3× 5 min), PBS (2× 5 min), and H2O (2× 5 min). For negative staining of immunolabeled specimens or whole cells, grids were blotted near dry and stained with 2% aqueous uranyl acetate for 90 s. Images were collected using a JEOL JEM1011 electron microscope.
Immunofluorescence
For immunofluorescence microscopy of mastigonemes, cells were fixed for 30 min at RT with freshly prepared 3.7% formaldehyde, allowed to settle onto poly-l-lysine multiwell slides for 5 min, washed three times with PBS for 10 min each, and air dried. The specimens were treated with 0.5% Triton X-100 in PBS for 5 min, blocked in PBS with 1% BSA and 0.05% Tween20, and incubated in anti-MST1 (1:500; Nakamura et al., 1996) overnight at 4°C in blocking buffer. After incubation in the secondary antibodies (anti-mouse IgG conjugated to Alexa Fluor 488 or 568; 1:1,000; Invitrogen; 90–120 min; RT) specimens were mounted with ProlongGold (Invitrogen). An Axio Imager.Z1 equipped with a 100×/1.3-NA EC Plan-Neofluar objective, an ApoTome, an axiocam MRm camera, and Axiovision (Zeiss) were used for imaging and image acquisition. To detect PKD2-GFP, we used anti-GFP 1:1,000 (mouse; Sigma-Aldrich; G 6539). Image brightness and contrast were adjusted using Photoshop (CC2018; Adobe), and figures were assembled using Illustrator (CC2018; Adobe). Acquisition conditions and adjustments of brightness and contrast were similar for images mounted together.
Swimming velocity
To determine the swimming velocity, cells were resuspended in fresh M medium, placed in a chambered plastic slide (Fisherbrand, 14-377-259) and observed using an inverted light microscope (TMS, Nikon). Images were recorded using a MU500 camera (Amscope) and Topview software at a fixed exposure time of 1 or 2 s. The length of the swimming trajectories was manually analyzed in ImageJ (National Institutes of Health) and converted into μm/s. The cilia beating frequency was determined as follows: swimming cells were recorded at 500 fps, and kymograms generated in ImageJ were used to analyze the displacement of the cell body. To measure the cilia length, cells were fixed in ∼4% formaldehyde, bright-field images were recorded using the Nikon Eclipse Ti-U microscope, and images were analyzed in ImageJ. Excel was used for statistical analysis of the data.
In vivo TIRF and SIM
Samples for in vivo imaging were prepared as follows: at RT, 10 µl of cells were placed inside of a ring of petroleum jelly or vacuum grease onto a 24 × 60-mm No. 1.5 coverslip and allowed to settle for ∼1–3 min. Then, a 22 × 22-mm No. 1.5 coverslip with 5 µl of 10 mM Hepes and 5 mM EGTA, pH 7.4, was inverted onto the larger cover glass to form a sealed observation chamber. For TIRF imaging, we used a Nikon Eclipse Ti-U inverted light microscope equipped with a 60×/1.49-NA objective lens and a 40-mW 488-nm diode laser (Spectraphysics; Lechtreck, 2013). A focused 488-nm laser beam or high laser intensities were used for photobleaching. If not mentioned otherwise, images were recorded at 10 fps using the iXon X3 DU897 EMCCD camera (Andor) and the Elements software package (Nikon). Still images mostly represent 10 frame walking averages. ImageJ with the Multiple Kymogram plug-in was used to analyze the recordings and generate kymograms. Kymograms, individual frames, and videos were cropped and adjusted for brightness and contrast in ImageJ and Photoshop CC 2018 (Adobe); Illustrator CC 2018 (Adobe) was used to assemble the figures.
For FRAP analysis, TIRF videos were opened in ImageJ FIJI, and the region of interest (ROI) was selected using the rectangle or line tool. The fluorescence intensity along the ROI was determined using the Plot Z-axis tool, and the data were exported into Excel. The fluorescence intensity in the ROI was corrected for the background fluorescence using ROI of the same size place next to the cell in SIM. The highest intensity value before the bleaching event was set to 100%, and the recovery of fluorescence in percentage of the prebleached value was calculated.
An Elyra S1 (Zeiss) microscope equipped with a 488-nm Argon laser and a 100×/1.46-NA oil-immersion objective was used for SIM imaging of pkd2 PKD2-mNG cells. Image stacks consisting of 110-nm optical sections were obtained at three different rotation angles, and SIM was processed using Zen software. The SIM image in Fig. 2 B represents a maximum-intensity projection of the entire Z-stack.
Cryo-ET and image processing
Cryosamples were prepared, imaged by cryo-ET, and processed as previously described (Nicastro et al., 2011). In brief, Quantifoil holey carbon grids (R2/2, 200 mesh; Quantifoil Micro Tools) were glow discharged and loaded onto a homemade plunge-freezing device. 3 µl of Chlamydomonas cells or isolated axonemes and 1 µl of fivefold concentrated BSA-coated 10-nm colloidal gold solution were applied to the grid (Lin and Nicastro, 2018). After brief mixing, grids were blotted from the back with filter paper for 1.5–3 s and immediately plunge frozen in liquid ethane. Vitrified specimens were transferred into a Titan Krios transmission electron microscope (Thermo Fisher Scientific) operated at 300 kV. Intact cilia or axonemes that appeared well preserved by EM inspection were imaged under low-dose conditions and using an energy filter in zero-loss mode (Gatan, Pleasanton, CA; 20-eV slit width) with −0.5 µm defocus and a Volta-Phase-Plate (Danev et al., 2014) at a magnification of 26,000× corresponding to a pixel size of ∼5.5 Å. Tilt series were collected using SerialEM with continuous series of tilt images or a dose-symmetric tilting scheme (Hagen et al., 2017) and recorded on a K2 camera (Gatan) with an electron dose rate of eight electrons/pixel/second. At each tilt angle, a video stack of 15 frames was collected with a total exposure time of 6 s. The cumulative electron dose per tilt series was limited to ∼100 e/Å2.
Frames of each video stack were aligned using a script extracted from IMOD (Kremer et al., 1996). The resulting images of a tilt series were assembled into a motion-corrected tilt series stack, which was further reconstructed into a 3D tomogram using fiducial alignment and weighted back projection in the IMOD software package (Kremer et al., 1996). To enhance the signal-to-noise ratio and improve the resolution, subtomograms that contained the mastigoneme filament repeats and mastigoneme base complexes, respectively, were extracted from the raw tomograms, aligned, and averaged with missing wedge compensation using the Particle Estimation for Electron Tomography (PEET) program to obtain subtomogram averages (Nicastro et al., 2006). Approximately 2,000 mastigoneme filament repeats and 53 mastigoneme base particles were selected from intact cilia tomograms, and 17 mastigoneme base particles were selected from demembranated cilia tomograms. After subtomogram averaging, a threefold symmetry was applied to the mastigoneme base, and a helical symmetry of four subunits per turn with a 400-Å pitch was applied to the mastigoneme averages. Subtomogram averaging was also performed of the 96-nm axonemal repeats from individual DMTs, and the nine DMTs of individual cilia were identified based on DMT-specific features (Bui et al., 2012; Lin et al., 2012). The structures were visualized as 2D tomographic slices and 3D isosurface renderings using the IMOD package (Kremer et al., 1996) and UCSF Chimera (Pettersen et al., 2004). To improve clarity of the 3D visualization in Fig. 5 (F and G) and Video 5, the raw tomogram was first denoised using a smoothing Kernel filter and a modulation transfer function filter, and then structural features were segmented and colored to generate the 3D isosurface rendering representation.
Online supplemental material
Fig. S1 shows PCR and Western blot analyses of the pkd2CLiP strain and mating assays. Fig. S2 shows in vivo imaging and photobleaching experiments of PKD2-FP. Fig. S3 shows the Phyre2 structure and repeat analysis of the EGFR-like domain of MST1 and mastigonemes on regrowing cilia. Fig. S4 shows a map and PCR analysis of the mst1 mutant locus and compares IFT transport of PKD2 in control and mst1 cells. Fig. S5 shows the distribution of mastigonemes on cilia and the dimensions of the PKD2-encompassing base complex based on cryo-EM tomography. Video 1 shows the distribution of PKD2-mNG in cilia. Video 2 shows the organization of ciliary PKD2-mNG into two rows. Video 3 shows the 3D structure of the mastigonemes. Video 4 shows the behavior of PKD2-mNG in mst1 mutant cilia. Video 5 shows a tomogram and the 3D representation of a ciliary segment with attached mastigonemes.
Acknowledgments
We are grateful to Kaiyao Huang, Dennis Diener, and Joel Rosenbaum (Yale University) for providing the PKD2-GFP vector and antibodies to PKD2, and to Gang Fu and Kai Cai (University of Texas Southwestern Medical Center) for providing tomographic data for reanalysis. We thank our colleagues Robert Bloodgood (University of Virginia), William Dentler (University of Kansas), and Juan Wang (Rutgers University) for discussion and critical reading of the manuscript. We acknowledge expert assistance by the University of Georgia’s Proteomics and Mass Spectrometry Facility, which is funded in part by the National Institutes of Health (S10RR028859) and the Biomedical Microscopy Core.
We thank D. Stoddard for management of the University of Texas Southwestern Medical Center cryo-electron microscope facility, which is funded in part by a Cancer Prevention and Research Institute of Texas Core Facility Award (RP170644). This study was supported by the National Institutes of Health (GM110413 to K. Lechtreck and GM083122 to D. Nicastro), and the Cancer Prevention and Research Institute of Texas (grant RR140082 to D. Nicastro). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare no competing financial interests.
Author contributions: P. Liu performed genetic experiments, acquired data, interpreted data, designed the experiments, and wrote the manuscript. X. Lou prepared the samples, acquired data at the microscope, performed image processing, interpreted the data, and wrote the manuscript. J.L. Wingfield acquired data at the microscope and wrote the manuscript. J. Lin prepared the samples, acquired data at the microscope, performed image processing, and wrote the manuscript. D. Nicastro and K. Lechtreck designed the experiments, analyzed and interpreted the data, and wrote the manuscript.
References
- Barr M.M., and Sternberg P.W.. 1999. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature. 401:386–389. 10.1038/43913 [DOI] [PubMed] [Google Scholar]
- Bergman K., Goodenough U.W., Goodenough D.A., Jawitz J., and Martin H.. 1975. Gametic differentiation in Chlamydomonas reinhardtii. II. Flagellar membranes and the agglutination reaction. J. Cell Biol. 67:606–622. 10.1083/jcb.67.3.606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck G.B. 1971. The structure, origin, isolation, and composition of the tubular mastigonemes of the Ochromas flagellum. J. Cell Biol. 50:362–384. 10.1083/jcb.50.2.362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck G.B., Rogalski A., and Valaitis A.. 1978. Surface organization and composition of Euglena. II. Flagellar mastigonemes. J. Cell Biol. 77:805–826. 10.1083/jcb.77.3.805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui K.H., Yagi T., Yamamoto R., Kamiya R., and Ishikawa T.. 2012. Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J. Cell Biol. 198:913–925. 10.1083/jcb.201201120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J.J., Miki K., Kim D., Shim S.H., Shi H.F., Hwang J.Y., Cai X., Iseri Y., Zhuang X., and Clapham D.E.. 2017. CatSperzeta regulates the structural continuity of sperm Ca(2+) signaling domains and is required for normal fertility. eLife. 6 e23082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J.J., Shim S.H., Everley R.A., Gygi S.P., Zhuang X., and Clapham D.E.. 2014. Structurally distinct Ca(2+) signaling domains of sperm flagella orchestrate tyrosine phosphorylation and motility. Cell. 157:808–822. 10.1016/j.cell.2014.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole D.G., Diener D.R., Himelblau A.L., Beech P.L., Fuster J.C., and Rosenbaum J.L.. 1998. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141:993–1008. 10.1083/jcb.141.4.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingridge P., Brownlee C., and Wheeler G.L.. 2013. Compartmentalized calcium signaling in cilia regulates intraflagellar transport. Curr. Biol. 23:2311–2318. 10.1016/j.cub.2013.09.059 [DOI] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.-M., and Brenner S.E.. 2004. WebLogo: a sequence logo generator. Genome Res. 14(6):1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danev R., Buijsse B., Khoshouei M., Plitzko J.M., and Baumeister W.. 2014. Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc. Natl. Acad. Sci. USA. 111:15635–15640. 10.1073/pnas.1418377111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vore D.M., Knobel K.M., Nguyen K.C.Q., Hall D.H., and Barr M.M.. 2018. Extracellular matrix regulates morphogenesis and function of ciliated sensory organs in Caenorhabditis elegans. BioRxiv. (Preprint posted July 24, 2018).
- Delling M., Indzhykulian A.A., Liu X., Li Y., Xie T., Corey D.P., and Clapham D.E.. 2016. Primary cilia are not calcium-responsive mechanosensors. Nature. 531:656–660. 10.1038/nature17426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen E.R., and Satir P.. 1972. Ciliary activity in the mouse oviduct as studied by transmission and scanning electron microscopy. Tissue Cell. 4:389–403. 10.1016/S0040-8166(72)80017-X [DOI] [PubMed] [Google Scholar]
- Drummond I.A. 2011. Polycystins, focal adhesions and extracellular matrix interactions. Biochim. Biophys. Acta. 1812:1322–1326. 10.1016/j.bbadis.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap K. 1977. Localization of calcium channels in Paramecium caudatum. J. Physiol. 271:119–133. 10.1113/jphysiol.1977.sp011993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R., Fukui H., Chow R., Vilfan A., and Vermot J.. 2019. The cilium as a force sensor-myth versus reality. J. Cell Sci. 132 jcs213496 10.1242/jcs.213496 [DOI] [PubMed] [Google Scholar]
- Fujiu K., Nakayama Y., Yanagisawa A., Sokabe M., and Yoshimura K.. 2009. Chlamydomonas CAV2 encodes a voltage- dependent calcium channel required for the flagellar waveform conversion. Curr. Biol. 19:133–139. 10.1016/j.cub.2008.11.068 [DOI] [PubMed] [Google Scholar]
- Fujiu K., Nakayama Y., Iida H., Sokabe M., and Yoshimura K.. 2011. Mechanoreception in motile flagella of Chlamydomonas. Nat. Cell Biol. 13:630–632. 10.1038/ncb2214 [DOI] [PubMed] [Google Scholar]
- Garcia-Anoveros J., and Corey D.P.. 1997. The molecules of mechanosensation. Annu. Rev. Neurosci. 20:567–594. 10.1146/annurev.neuro.20.1.567 [DOI] [PubMed] [Google Scholar]
- Gillespie P.G., and Müller U.. 2009. Mechanotransduction by hair cells: models, molecules, and mechanisms. Cell. 139:33–44. 10.1016/j.cell.2009.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U.W., Adair W.S., Collin-Osdoby P., and Heuser J.E.. 1985. Structure of the Chlamydomonas agglutinin and related flagellar surface proteins in vitro and in situ. J. Cell Biol. 101(3):924–941. 10.1083/jcb.101.3.924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen W.J.H., Wan W., and Briggs J.A.G.. 2017. Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J. Struct. Biol. 197:191–198. 10.1016/j.jsb.2016.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K., Ma X., Xu T., Li Y., Hodge A., Zhang Q., Torline J., Huang Y., Zhao J., Ling K., et al. 2018. Axoneme polyglutamylation regulated by Joubert syndrome protein ARL13B controls ciliary targeting of signaling molecules. Nat. Commun. 9:3310 10.1038/s41467-018-05867-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoops H.J., and Witman G.B.. 1983. Outer doublet heterogeneity reveals structural polarity related to beat direction in Chlamydomonas flagella. J. Cell Biol. 97:902–908. 10.1083/jcb.97.3.902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Diener D.R., Mitchell A., Pazour G.J., Witman G.B., and Rosenbaum J.L.. 2007. Function and dynamics of PKD2 in Chlamydomonas reinhardtii flagella. J. Cell Biol. 179:501–514. 10.1083/jcb.200704069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J.Y., Mannowetz N., Zhang Y., Everley R.A., Gygi S.P., Bewersdorf J., Lishko P.V., and Chung J.J.. 2019. Dual Sensing of Physiologic pH and Calcium by EFCAB9 Regulates Sperm Motility. Cell. 177:1480–1494.e1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn T.L., Landman M.D., and Fonseca J.R.. 1964. Mechanism of Locomotion of Flagellates. 2. Function of Mastigonemes of Ochromonas. J. Protozool. 11:291–296. 10.1111/j.1550-7408.1964.tb01756.x [DOI] [Google Scholar]
- Kamiya R., and Witman G.B.. 1984. Submicromolar levels of calcium control the balance of beating between the two flagella in demembranated models of Chlamydomonas. J. Cell Biol. 98:97–107. 10.1083/jcb.98.1.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil T.A. 2012. Sensory cilia in arthropods. Arthropod Struct. Dev. 41:515–534. 10.1016/j.asd.2012.07.001 [DOI] [PubMed] [Google Scholar]
- Kelley L.A., Mezulis S., Yates C.M., Wass M.N., and Sternberg M.J.E.. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 10(6):845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Nie H., Nesin V., Tran U., Outeda P., Bai C.X., Keeling J., Maskey D., Watnick T., Wessely O., et al. 2016. The polycystin complex mediates Wnt/Ca(2+) signalling. Nat. Cell Biol. 18:752–764. 10.1038/ncb3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S.M. 1995. Large-scale isolation of Chlamydomonas flagella. Methods Cell Biol. 47:9–12. 10.1016/S0091-679X(08)60783-9 [DOI] [PubMed] [Google Scholar]
- King S.M., and Witman G.B.. 1990. Localization of an intermediate chain of outer arm dynein by immunoelectron microscopy. J. Biol. Chem. 265:19807–19811. [PubMed] [Google Scholar]
- Köttgen M., Hofherr A., Li W., Chu K., Cook S., Montell C., and Watnick T.. 2011. Drosophila sperm swim backwards in the female reproductive tract and are activated via TRPP2 ion channels. PLoS One. 6 e20031 10.1371/journal.pone.0020031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., and McIntosh J.R.. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76. 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Lechtreck K.F. 2013. In vivo imaging of IFT in Chlamydomonas flagella. Methods Enzymol. 524:265–284. 10.1016/B978-0-12-397945-2.00015-9 [DOI] [PubMed] [Google Scholar]
- Li X., Zhang R., Patena W., Gang S.S., Blum S.R., Ivanova N., Yue R., Robertson J.M., Lefebvre P.A., Fitz-Gibbon S.T., et al. 2016. An Indexed, Mapped Mutant Library Enables Reverse Genetics Studies of Biological Processes in Chlamydomonas reinhardtii. Plant Cell. 28:367–387. 10.1105/tpc.15.00465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., and Nicastro D.. 2018. Asymmetric distribution and spatial switching of dynein activity generates ciliary motility. Science. 360 eaar1968 10.1126/science.aar1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Heuser T., Song K., Fu X., and Nicastro D.. 2012. One of the nine doublet microtubules of eukaryotic flagella exhibits unique and partially conserved structures. PLoS One. 7 e46494 10.1371/journal.pone.0046494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Vien T., Duan J., Sheu S.H., DeCaen P.G., and Clapham D.E.. 2018. Polycystin-2 is an essential ion channel subunit in the primary cilium of the renal collecting duct epithelium. eLife. 7 e33183 10.7554/eLife.33183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma R., Li W.P., Rundle D., Kong J., Akbarali H.I., and Tsiokas L.. 2005. PKD2 functions as an epidermal growth factor-activated plasma membrane channel. Mol. Cell. Biol. 25:8285–8298. 10.1128/MCB.25.18.8285-8298.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangos S., Lam P.Y., Zhao A., Liu Y., Mudumana S., Vasilyev A., Liu A., and Drummond I.A.. 2010. The ADPKD genes pkd1a/b and pkd2 regulate extracellular matrix formation. Dis. Model. Mech. 3:354–365. 10.1242/dmm.003194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markey D.R., and Bouck G.B.. 1977. Mastigoneme attachment in Ochromonas. J. Ultrastruct. Res. 59:173–177. 10.1016/S0022-5320(77)80077-4 [DOI] [PubMed] [Google Scholar]
- Markin V.S., and Hudspeth A.J.. 1995. Gating-spring models of mechanoelectrical transduction by hair cells of the internal ear. Annu. Rev. Biophys. Biomol. Struct. 24:59–83. 10.1146/annurev.bb.24.060195.000423 [DOI] [PubMed] [Google Scholar]
- Nakamura S., Tanaka G., Maeda T., Kamiya R., Matsunaga T., and Nikaido O.. 1996. Assembly and function of Chlamydomonas flagellar mastigonemes as probed with a monoclonal antibody. J. Cell Sci. 109:57–62. [DOI] [PubMed] [Google Scholar]
- Nauli S.M., Alenghat F.J., Luo Y., Williams E., Vassilev P., Li X., Elia A.E., Lu W., Brown E.M., Quinn S.J., et al. 2003. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat. Genet. 33:129–137. 10.1038/ng1076 [DOI] [PubMed] [Google Scholar]
- Nicastro D., Fu X., Heuser T., Tso A., Porter M.E., and Linck R.W.. 2011. Cryo-electron tomography reveals conserved features of doublet microtubules in flagella. Proc. Natl. Acad. Sci. USA. 108:E845–E853. 10.1073/pnas.1106178108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D., Schwartz C., Pierson J., Gaudette R., Porter M.E., and McIntosh J.R.. 2006. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 313:944–948. 10.1126/science.1128618 [DOI] [PubMed] [Google Scholar]
- O’Hagan R., Piasecki B.P., Silva M., Phirke P., Nguyen K.C., Hall D.H., Swoboda P., and Barr M.M.. 2011. The tubulin deglutamylase CCPP-1 regulates the function and stability of sensory cilia in C. elegans. Curr. Biol. 21:1685–1694. 10.1016/j.cub.2011.08.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., Agrin N., Leszyk J., and Witman G.B.. 2005. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170:103–113. 10.1083/jcb.200504008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., Sineshchekov O.A., and Witman G.B.. 1995. Mutational analysis of the phototransduction pathway of Chlamydomonas reinhardtii. J. Cell Biol. 131(2):427–440. 10.1083/jcb.131.2.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., San Agustin J.T., Follit J.A., Rosenbaum J.L., and Witman G.B.. 2002. Polycystin-2 localizes to kidney cilia and the ciliary level is elevated in orpk mice with polycystic kidney disease. Curr. Biol. 12:R378–R380. 10.1016/S0960-9822(02)00877-1 [DOI] [PubMed] [Google Scholar]
- Pennekamp P., Karcher C., Fischer A., Schweickert A., Skryabin B., Horst J., Blum M., and Dworniczak B.. 2002. The ion channel polycystin-2 is required for left-right axis determination in mice. Curr. Biol. 12:938–943. 10.1016/S0960-9822(02)00869-2 [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., and Ferrin T.E.. 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Praetorius H.A., and Spring K.R.. 2001. Bending the MDCK cell primary cilium increases intracellular calcium. J. Membr. Biol. 184:71–79. 10.1007/s00232-001-0075-4 [DOI] [PubMed] [Google Scholar]
- Seeger-Nukpezah T., and Golemis E.A.. 2012. The extracellular matrix and ciliary signaling. Curr. Opin. Cell Biol. 24:652–661. 10.1016/j.ceb.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P.S., Yang X., DeCaen P.G., Liu X., Bulkley D., Clapham D.E., and Cao E.. 2016. The Structure of the Polycystic Kidney Disease Channel PKD2 in Lipid Nanodiscs. Cell. 167:763–773.e711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q., Hu F., Ge X., Lei J., Yu S., Wang T., Zhou Q., Mei C., and Shi Y.. 2018. Structure of the human PKD1-PKD2 complex. Science. 361 eaat9819 10.1126/science.aat9819 [DOI] [PubMed] [Google Scholar]
- Sun L., Gao Y., He J., Cui L., Meissner J., Verbavatz J.M., Li B., Feng X., and Liang X.. 2019. Ultrastructural organization of NompC in the mechanoreceptive organelle of Drosophila campaniform mechanoreceptors. Proc. Natl. Acad. Sci. USA. 116:7343–7352. 10.1073/pnas.1819371116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman G.B. 1986. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 134:280–290. 10.1016/0076-6879(86)34096-5 [DOI] [PubMed] [Google Scholar]
- Witman G.B., Carlson K., Berliner J., and Rosenbaum J.L.. 1972. Chlamydomonas flagella. I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J. Cell Biol. 54(3):507–539. 10.1083/jcb.54.3.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood C.R., and Rosenbaum J.L.. 2014. Proteins of the ciliary axoneme are found on cytoplasmic membrane vesicles during growth of cilia. Curr. Biol. 24:1114–1120. 10.1016/j.cub.2014.03.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., and Somlo S.. 2000. Molecular genetics and mechanism of autosomal dominant polycystic kidney disease. Mol. Genet. Metab. 69:1–15. 10.1006/mgme.1999.2943 [DOI] [PubMed] [Google Scholar]
- Yoder B.K., Hou X., and Guay-Woodford L.M.. 2002. The polycystic kidney disease proteins, polycystin-1, polycystin-2, polaris, and cystin, are co-localized in renal cilia. J. Am. Soc. Nephrol. 13:2508–2516. 10.1097/01.ASN.0000029587.47950.25 [DOI] [PubMed] [Google Scholar]
- Yoshiba S., Shiratori H., Kuo I.Y., Kawasumi A., Shinohara K., Nonaka S., Asai Y., Sasaki G., Belo J.A., Sasaki H., et al. 2012. Cilia at the node of mouse embryos sense fluid flow for left-right determination via Pkd2. Science. 338:226–231. 10.1126/science.1222538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Zhao L., Brueckner M., and Sun Z.. 2015. Intraciliary calcium oscillations initiate vertebrate left-right asymmetry. Curr. Biol. 25:556–567. 10.1016/j.cub.2014.12.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora I., Feldman J.L., and Marshall W.F.. 2004. PCR-based assay for mating type and diploidy in Chlamydomonas. Biotechniques. 37:534–536. 10.2144/04374BM01 [DOI] [PubMed] [Google Scholar]