BRCA1/2 help maintain genomic integrity by stabilizing stalled forks for repair and thus act as tumor suppressors. Duan et al. identify the E3 ligase RFWD3 as a novel modulator of stalled fork stability in BRCA2-deficient cells and show that blocking fork degradation by depleting MRE11 is not sufficient for full fork recovery.
Abstract
BRCA1/2 help maintain genomic integrity by stabilizing stalled forks. Here, we identify the E3 ligase RFWD3 as an essential modulator of stalled fork stability in BRCA2-deficient cells and show that codepletion of RFWD3 rescues fork degradation, collapse, and cell sensitivity upon replication stress. Stalled forks in BRCA2-deficient cells accumulate phosphorylated and ubiquitinated replication protein A (ubq-pRPA), the latter of which is mediated by RFWD3. Generation of this intermediate requires SMARCAL1, suggesting that it depends on stalled fork reversal. We show that in BRCA2-deficient cells, rescuing fork degradation might not be sufficient to ensure fork repair. Depleting MRE11 in BRCA2-deficient cells does block fork degradation, but it does not prevent fork collapse and cell sensitivity in the presence of replication stress. No such ubq-pRPA intermediate is formed in BRCA1-deficient cells, and our results suggest that BRCA1 may function upstream of BRCA2 in the stalled fork repair pathway. Collectively, our data uncover a novel mechanism by which RFWD3 destabilizes forks in BRCA2-deficient cells.
Introduction
Germline mutations in the tumor suppressors BRCA1 and BRCA2 are most commonly associated with an exceptionally high risk of breast and ovarian cancer (Kuchenbaecker et al., 2017; Lord and Ashworth, 2016; Narod and Foulkes, 2004; Rebbeck and Domchek, 2008; Welcsh and King, 2001). Mutations in BRCA2 also confer a predisposition to pancreatic and prostate cancers and melanoma (Castro et al., 2013; Mocci et al., 2013). Furthermore, individuals carrying homozygous or compound heterozygous missense mutations in BRCA1 or BRCA2 also show symptoms classically present in Fanconi anemia (FANC) families; hence, they are also classified as members of the FANC family (FANCS and FANCD1, respectively; Sawyer et al., 2015; Stewart and Elledge, 2002).
BRCA1 and BRCA2 act as tumor suppressors, primarily because of their role in maintaining genomic stability (O’Donovan and Livingston, 2010; Roy et al., 2011; Kolinjivadi et al., 2017a). They are important players in homologous recombination (HR)–driven double-strand break repair (DSBR; Moynahan et al., 2001), interstrand cross-link repair (Bunting et al., 2012; Cipak et al., 2006), and stalled replication fork repair (Kolinjivadi et al., 2017a; Long et al., 2014; Pathania et al., 2011, 2014; Schlacher et al., 2011, 2012; Willis et al., 2014). Defective DSBR and its association with inherited cancer is well established (Walsh and King, 2007); however, recent studies also suggest a strong correlation between defective stalled fork repair and predisposition to hereditary cancer. A majority of the hereditary breast cancer–predisposing genes such as BRCA1, BRCA2, PALB2, RAD51C, and BRIP1, which are known members of DSBR machinery, also play a critical role in the repair of stalled replication forks (Murphy et al., 2014; Pathania et al., 2011; Schlacher et al., 2011, 2012; Somyajit et al., 2015). Stalled forks, when not repaired, lead to increased replication stress, a prime driver of tumorigenesis (Gaillard et al., 2015; Macheret and Halazonetis, 2015). Given the importance of maintaining genomic stability via efficient repair of stalled forks and the clinical relevance of this phenomenon, it is important to understand how these proteins function at stalled replication forks.
In response to endogenous and exogenous DNA-damaging agents, replication forks tend to stall and generate intermediates, which include three-way junctions (Y structures) and four-way junctions (Michel et al., 2004; Neelsen and Lopes, 2015), the latter commonly referred to as reversed fork and/or “chicken foot” structures (Neelsen and Lopes, 2015; Quinet et al., 2017). Three-way junctions mostly mark sites that have stretches of single-stranded DNA (ssDNA) as a result of uncoupling of the replicative minichromosome maintenance protein complex helicase complex from the DNA polymerase (Byun et al., 2005). Reversed forks or chicken foot structures are formed upon remodeling of three-way junctions to generate four-way junctions that resemble the branched structure of Holliday junctions. These intermediates likely stabilize stalled forks and help in their efficient repair (Branzei and Foiani, 2010; Quinet et al., 2017).
Recent studies have shown that loss of BRCA1/2 increases stalled fork degradation (Kolinjivadi et al., 2017b; Lemaçon et al., 2017; Taglialatela et al., 2017), in part because of excessive resection of the reversed forks by MRE11 nuclease (Schlacher et al., 2011). It has also been shown that BRCA1 helps generate phosphorylated replication protein A (pRPA)–coated ssDNA at stalled forks (Pathania et al., 2011; Tian et al., 2013), which recruits repair factors like ATRIP to these structures (Pathania et al., 2011). This pRPA-coated ssDNA intermediate serves as a docking and activation site for many repair and checkpoint proteins, including ATR, ATRIP, and CHK1 (Ciccia and Elledge, 2010; Zou and Elledge, 2003).
Given that fork rescue is emerging as one of the ways that BRCA2 tumor cells acquire resistance, it is critical that we uncover all the players and events that are involved in this process. It is not yet clear whether rescuing degradation of reversed forks in BRCA2-deficient cells is enough for efficient resolution and full recovery of stalled fork to allow cell survival. It is also not clear whether BRCA1 and BRCA2 work independently of one another and/or whether there is a hierarchy among their respective functions at the stalled fork.
Here, we show that upon replication stalling, BRCA2-deficient cells, but not BRCA1-deficient cells, accumulate hyperubiquitinated RPA-coated ssDNA (ubq-RPA) at stalled forks. We find that this RPA ubiquitination is performed by RING finger–type E3 ubiquitin ligase RFWD3, which has recently been identified as a FANC protein (FANCW; Knies et al., 2017). RFWD3 has been shown to ubiquitylate RPA to promote HR-dependent fork repair (Elia et al., 2015), and during interstrand cross-link repair, it has been reported to ubiquitylate RPA and RAD51 to promote their removal from DNA (Feeney et al., 2017; Inano et al., 2017).
We find that RFWD3 contributes to increased fork destabilization and sensitivity to fork-stalling agents in BRCA2-deficient cells. Generation of this hyperubiquitinated RPA-bound ssDNA intermediate is dependent on SMARCAL1, implying that it is dependent on fork reversal. Interestingly, although codepletion of MRE11 in BRCA2-deficient cells does rescue fork degradation as shown before (Schlacher et al., 2011, 2012), these rescued reversed forks are not conducive to efficient repair. We find that rescue of fork degradation in BRCA2-depleted cells does not guarantee fork repair and cell survival. However, blocking RPA ubiquitination in BRCA2-depleted cells by codepletion of RFWD3 alleviates fork instability, reverses fork degradation, protects these cells against fork collapse, and rescues their sensitivity to fork-stalling agents. These data suggest that BRCA2-deficient cells acquire resistance to replication-stalling agents in part by down-regulating RFWD3. We also provide evidence of a hierarchal relationship between BRCA1 and BRCA2 in the stalled fork repair pathway. Together, this study provides insight into new players and events that drive tumorigenesis and chemoresistance in BRCA2-deficient cells.
Results
Stalled fork intermediates formed in BRCA1- and BRCA2-deficient cells are different
Both BRCA1 and BRCA2 are required for repair of stalled replication forks (Kolinjivadi et al., 2017a; Long et al., 2014; Pathania et al., 2011, 2014; Schlacher et al., 2011, 2012; Willis et al., 2014). To address whether there is a separation of function between these two proteins as they stabilize stalled forks, we first asked whether the stalled fork intermediates formed in absence of either BRCA1 and/or BRCA2 are similar in nature.
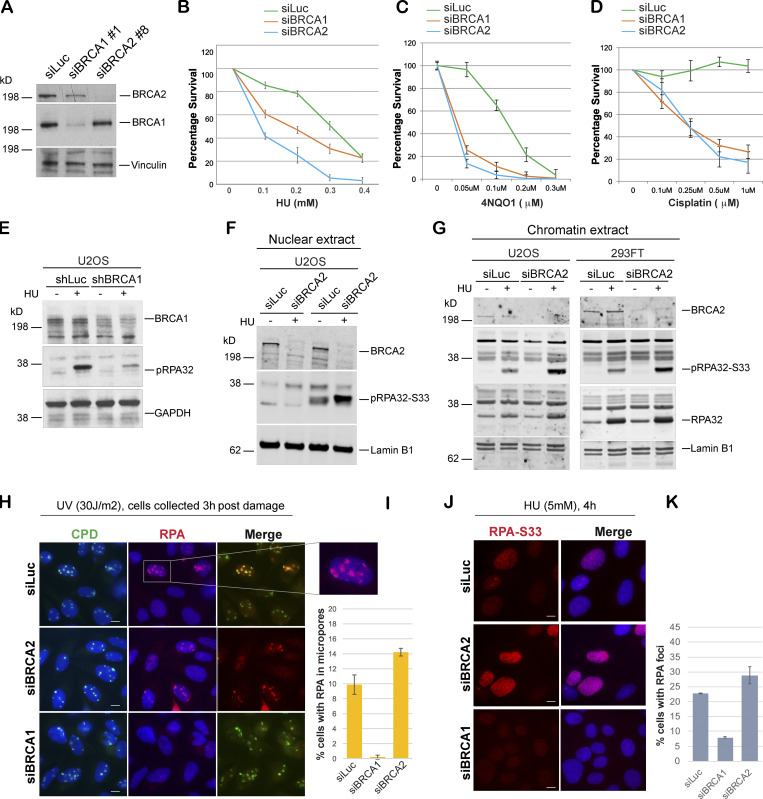
Depletion of either BRCA1 or BRCA2 with siRNA led to a marked sensitivity to stalled fork–inducing agents like hydroxyurea (HU), 4NQO1 (4-nitroquinoline-1-oxide), or cisplatin in U2OS cells (Fig. 1, A–D; and Fig. S1, A and B). These agents stall the progression of replication forks by either depleting deoxynucleoside triphosphates (e.g., HU), or by causing cross-links and/or DNA adducts (cisplatin and 4-NQO1, respectively). We confirmed that loss of BRCA2 leads to increased sensitivity to HU in multiple cell lines (RPE1 and HeLa; Fig. S1, C and D).
Figure 1.
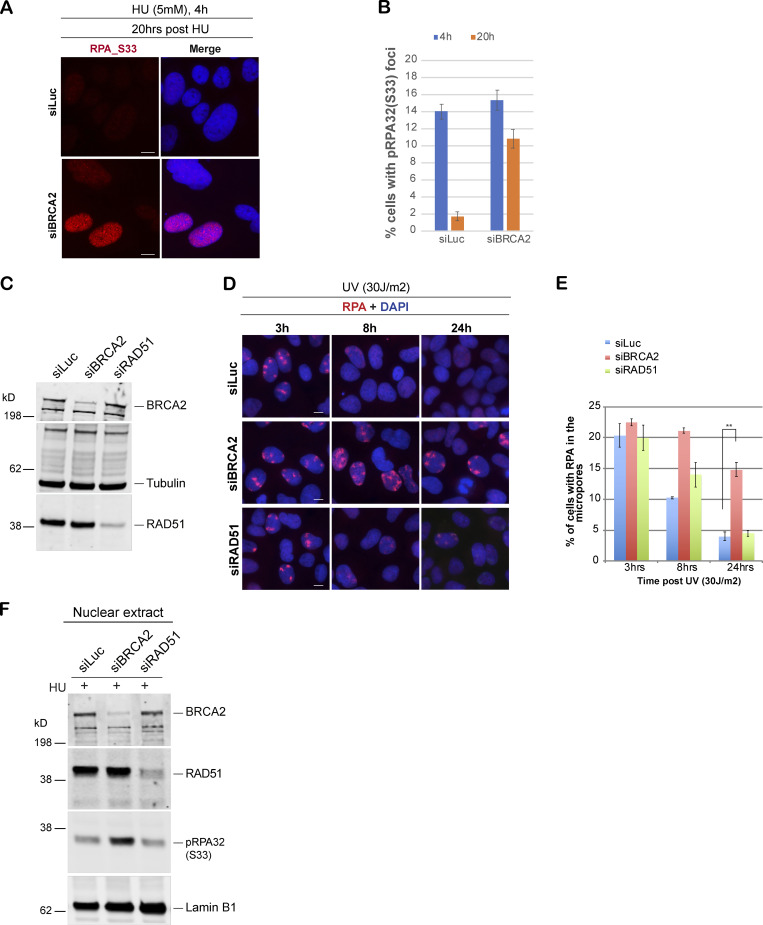
BRCA1- and BRCA2-depleted cells form different stalled fork intermediates. (A) Western blot analysis of total BRCA1 and BRCA2 protein levels in U2OS cells transfected with siLuc (control), siBRCA1, and siBRCA2. (B–D) CellTiter-Glo–based cell survival assay was used to determine the sensitivity of BRCA1- or BRCA2-deficient cells to various DNA damage–inducing agents. U2OS cells transfected with indicated siRNAs were treated with HU (4 d), 4NQO1 (5 h), or cisplatin (1 d) with indicated doses. Cells were harvested 6 d after the start of the drug treatment, and cell viabilities were tested by detecting the generation of luminescent signal, which is directly proportional to the number of cells present in the culture. Error bars represent SD between triplicates. (E–G) Western blot analysis of RPA32 and pRPA32 (S33) accumulation in control cells and BRCA1- or BRCA2-depleted cells. Cells were treated with 5 mM HU and harvested 3 h after treatment. Whole cell (E), nuclear (F), and chromatin (G) extracts were prepared. (H and I) IF and graph of RPA32 recruitment in U2OS control (siLuc) cells and BRCA1- or BRCA2-depleted cells. Cells were irradiated with 30 J/m2 of UV through a micropore membrane to generate localized sites of DNA damage and harvested 3 h after damage. Cells were costained with RPA32 and CPD. CPD served as marker of the sites of UV damage. Scale bars indicate 10 µm. (J and K) IF and graph of pRPA32-S33 recruitment in U2OS control cells and BRCA1- or BRCA2-depleted cells. Cells were treated with 5 mM HU and harvested 4 h after damage. Cells were stained with pRPA32-S33. Scale bars in J indicate 10 µM. Error bars indicate SD between triplicates.
Figure S1.
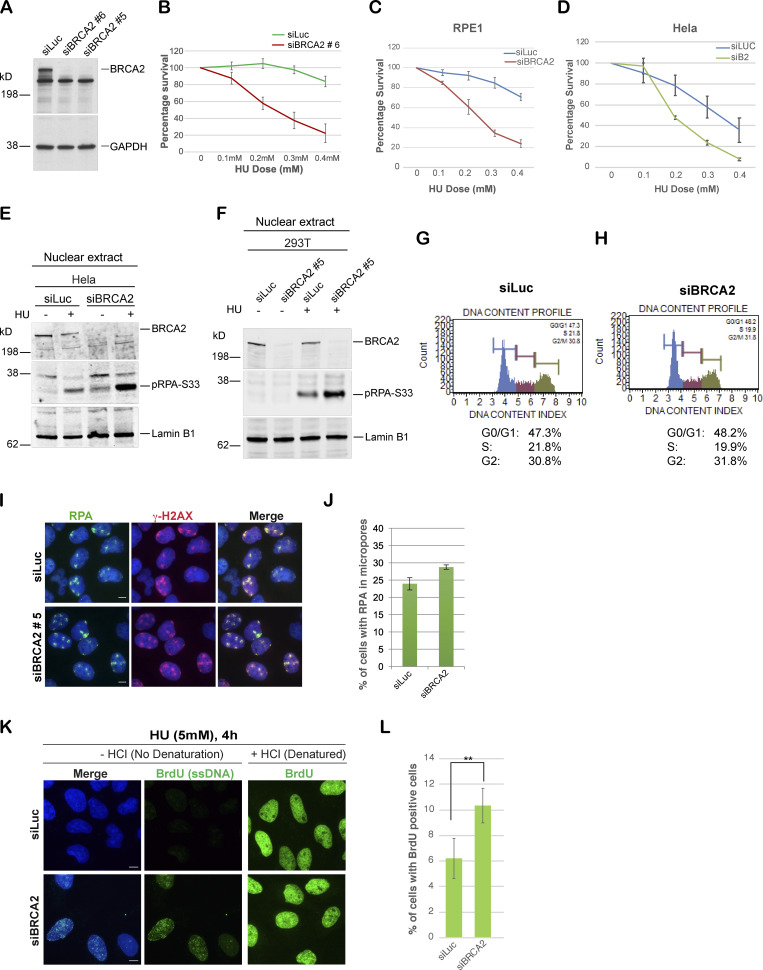
BRCA2-deficient cells are sensitive to stalled fork–inducing agents and show increased accumulation of pRPA32 and ssDNA upon stalled fork–inducing DNA damage. (A) Western blot analysis to detect knockdown efficiencies of different BRCA2-specific siRNAs. (B) CellTiter-Glo–based cell survival assay to determine the sensitivity of U2OS cells transfected with a different BRCA2-specific siRNA (siBRCA2#6) to the stalled folk–inducing agent HU. (C and D) CellTiter-Glo–based cell HU survival assay to determine the sensitivity of RPE1 cells (C) and HeLa cells (D) transfected with BRCA2-specific siRNA. (E) Western blot analysis of RPA32 accumulation in HeLa cells transfected with indicated siRNAs. Cells were treated with 5 mM HU and harvested 3 h after treatment. Nuclear extracts were prepared. (F) Western blot analysis of nuclear extract. RPA32 accumulation in HEK293T cells transfected with a different BRCA2-specific siRNA (siBRCA2#5) was analyzed. (G and H) Cell cycle analysis of U2OS control and BRCA2-depleted cells by the MUSE system (details in Materials and methods). (I and J) IF analysis and graph of RPA32 recruitment in U2OS cells transfected with siLuc or a different BRCA2-specific siRNA (siBRCA2#5). γ-H2AX served as a control to mark sites of DNA damage. Cells were irradiated with 30 J/m2 of UV as indicated above. Scale bars in I indicate 10 µm. (K and L) BrdU assay for detection of ssDNA generation after HU treatment. U2OS cells were fixed 4 h after 5 mM HU treatment and immunostained for BrdU with and without denaturation of DNA with HCl. Scale bars in K indicate 10 µm. **, P value < 0.05. Statistical significance was determined by the two-tailed Student’s t test and the error bars indicate SD (n = 3). Related to Fig. 1.
RPA-coated ssDNA is a critical intermediate in the efficient repair of stalled forks and for initiating the intra-S phase checkpoint by recruiting ATR (Zou and Elledge, 2003). We have shown previously that BRCA1 is required for the generation of this critical intermediate (Pathania et al., 2011). To address whether BRCA2, like BRCA1, plays a role in generation of this repair intermediate, we treated BRCA1/2-depleted cells and control cells (shLuc and/or siLuc; shRNA or siRNA against luciferase, respectively) with HU to induce stalled replication forks. As shown before, HU-induced DNA damage resulted in accumulation of pRPA32 in control cells, but not in BRCA1-depleted cells (Fig. 1 E). Surprisingly, BRCA2 depletion did not affect pRPA32 accumulation after HU-induced DNA damage (Fig. 1, F and G; and Fig. S1, E and F). On the contrary, after HU treatment, BRCA2-depleted cells show an increase in pRPA32 (Fig. 1, F and G). We confirmed that this accumulation of pRPA32 was on chromatin (Fig. 1 G) and was occurring in multiple different cell lines (HEK293FT, U2OS, and HeLa; Fig. 1, F and G; and Fig. S1 E) and with different BRCA2-specific siRNA (Fig. S1 F). We also confirmed that there was no cell cycle perturbation upon BRCA2 depletion that could account for this increase in pRPA32 in these cells. Control and BRCA2-depleted cells showed very similar cell cycle profiles (Fig. S1, G and H).
To further confirm the differences in RPA accumulation upon fork stalling in BRCA1- and BRCA2-depleted cells, we performed immunofluorescence (IF) staining for RPA32 in cells treated with UV or HU. UV irradiation was performed through a micropore filter to generate sites of localized stalled forks/DNA damage (Pathania et al., 2011). RPA32 was efficiently recruited to sites of UV damage, where it colocalized with cyclobutane pyrimidine dimers (CPDs) in both control and BRCA2-depleted cells (Fig. 1, H and I; and Fig. S1, I and J). This is in marked contrast to what happens in BRCA1-depleted cells (Pathania et al., 2011; Fig. 1, H and I). Likewise, after HU-induced replication fork stalling, BRCA1-depleted cells, but not BRCA2-depleted cells, show decreased phosphorylated RPA32 (S33) foci (Fig. 1, J and K), a marker associated with ssDNA accumulation at stalled forks (Sirbu et al., 2011; Zeman and Cimprich, 2014). To directly address whether there is an increase in ssDNA accumulation in BRCA2-depleted cells undergoing fork stalling, we adopted a previously established approach to study accumulation of ssDNA in cells after DNA damage (Rubbi and Milner, 2001). Despite similar BrdU incorporation in both control and BRCA2-depleted cells (+HCl panel, Fig. S1 K), a higher proportion of BRCA2-depleted cells showed BrdU-positive cell population under nondenaturing (−HCl) conditions (Fig. S1, K and L), implying an increase in ssDNA accumulation in these cells.
Together, these data indicate that stalled fork intermediates formed in BRCA1-depleted cells are different from those in BRCA2-depleted cells. Furthermore, the striking difference in their ability to generate RPA-coated ssDNA raises the possibility that these proteins function at different steps in the stalled fork repair pathway.
BRCA1 may function upstream of BRCA2 in the stalled fork repair pathway
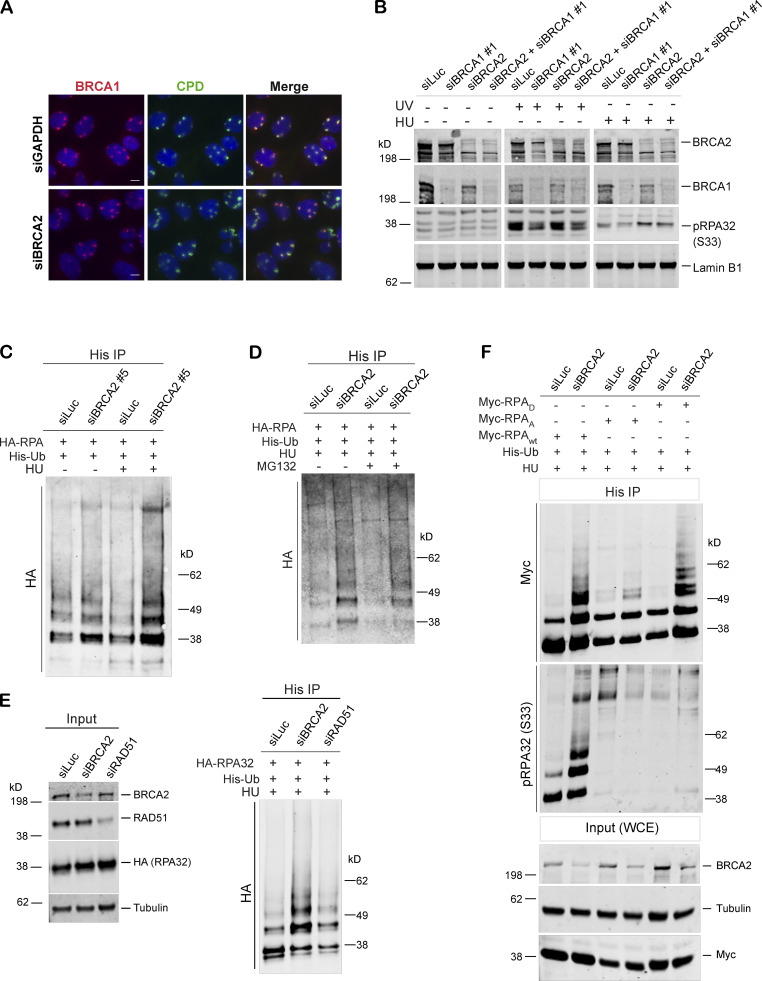
Having established that BRCA1 and BRCA2 loss leads to accumulation of different stalled fork intermediates, we next sought to determine whether depletion of BRCA2 would affect recruitment of BRCA1 to sites of stalled replication forks. We found that BRCA1 was efficiently recruited to UV-induced stalled forks in BRCA2-depleted cells (siBRCA2; Fig. S2 A), raising the possibility that BRCA1 might function independently and upstream of BRCA2.
Figure S2.
BRCA1 may function upstream of BRCA2 in the stalled fork repair pathway, and BRCA2 depletion results in hyperubiquitination of RPA after stalled fork–inducing DNA damage. (A) BRCA1 recruitment was not affected in cells depleted of BRCA2. IF analysis of BRCA1 recruitment in U2OS control cells and BRCA2-depleted cells after UV irradiation as indicated above. Cells were costained with BRCA1 and CPD. Scale bars indicate 10 µm. (B) Western blot analysis of RPA32 accumulation in U2OS cells transfected with indicated siRNAs. (C) Immunoprecipitation analysis of RPA32 ubiquitination in HEK293T cells transfected with siLuc or a different BRCA2-specific siRNA (siBRCA2#5). (D) Immunoprecipitation analysis of RPA ubiquitination in HEK293T control and BRCA2-deficient cells in the absence or presence of MG132. HEK293T cells with indicated siRNAs were transfected with His-tagged ubiquitin and HA-tagged RPA32. Cells were treated with 5mM HU for 3 h. Before harvesting the cells, 10 µM MG132 was added for 1 h. (E) Immunoprecipitation analysis of RPA ubiquitination in HEK293T cells transfected with control (siLuc) or BRCA2 orRAD51 siRNAs. Experimental conditions used are as described before. (F) Immunoprecipitation analysis to study the relationship between RPA phosphorylation and ubiquitination by expressing various RPA mutants. In RPAD mutant, both of the cyclin-cdk2 phosphorylation sites and six of the stress-dependent phosphorylation sites (S8, S11, S12, S13, T21, and S33) were replaced by aspartate. In RPAA mutant, these same sites were converted to alanine to prevent phosphorylation. HEK293T cells were transfected with indicated siRNAs, followed by transfection with His-tagged ubiquitin and Myc-tagged WT or RPAA, RPAD mutant RPA. Cells were processed for His immunoprecipitation as described above. Blot was probed with anti-Myc and anti-pRPA32 (S33) antibodies. Cells were collected after 4 h of HU treatment (5 mM). A different BRCA1-specific siRNA (siBRCA1#1) was used in B. Related to Figs. 2 and 4.
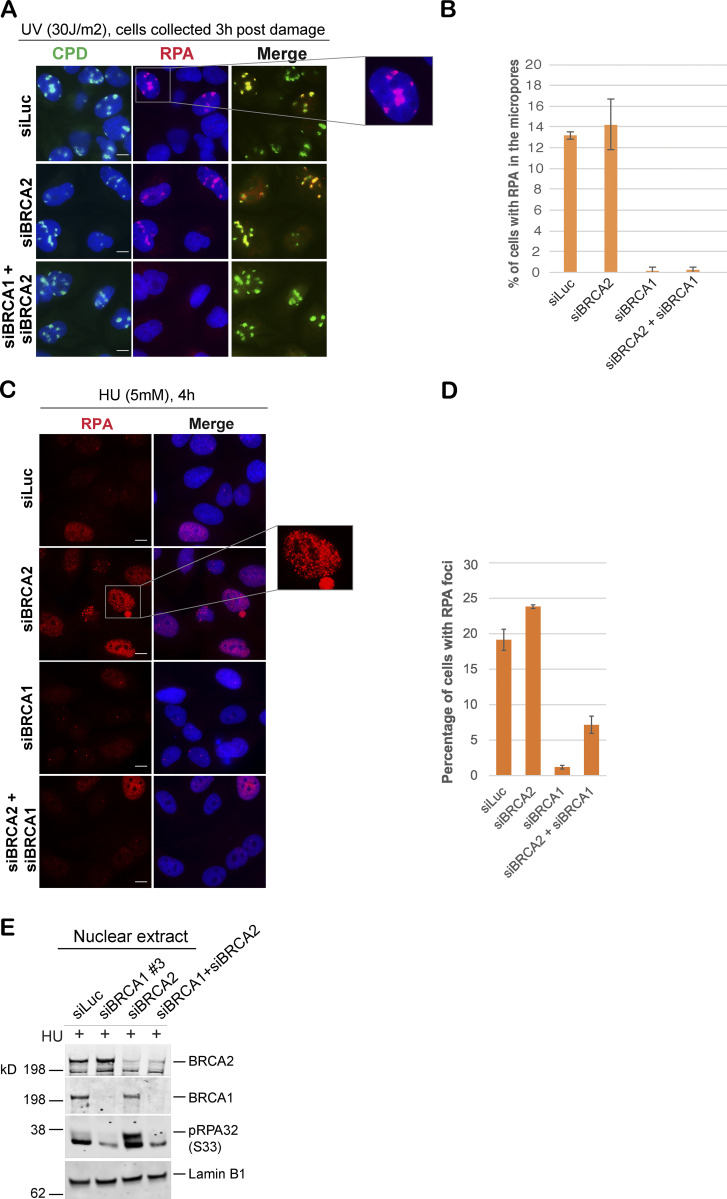
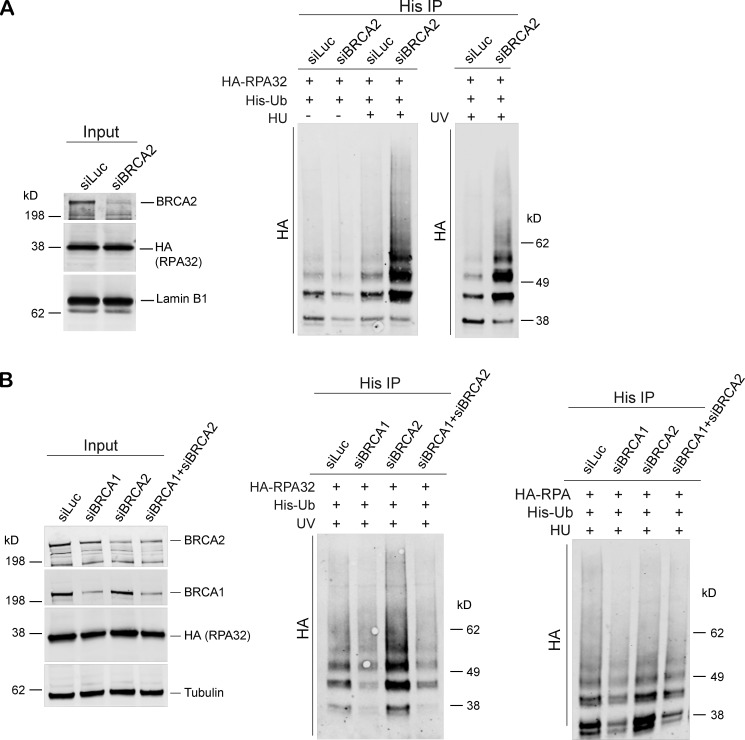
Given that depletion of BRCA1 reduced pRPA32 accumulation on chromatin in response to stalled fork–inducing damage (Pathania et al., 2011), whereas loss of BRCA2 led to excessive accumulation of pRPA32 on chromatin, we asked whether cells codepleted of both BRCA1 and BRCA2 phenocopy the pRPA32 defect, as seen in BRCA1-depleted cells. Indeed, loss of BRCA1 in BRCA2-depleted cells did suppress the accumulation of RPA32 and pRPA32 in response to UV- and HU-induced DNA damage (Fig. 2, A–E; and Fig. S2 B).
Figure 2.
BRCA1 may function upstream of BRCA2 in the stalled fork repair pathway. (A and B) IF analysis and graph of RPA32 recruitment in U2OS cells transfected with control siRNA (siLuc) or siRNA for BRCA1 or BRCA2, or both BRCA1 and BRCA2. These cells were fixed 3 h after UV damage (30 J/m2). Scale bars in A indicate 10 µm. (C and D) IF analysis and graph of RPA32 recruitment in U2OS cells transfected with control siRNA (siLuc) or siRNA for BRCA1, BRCA2, or both BRCA1 and BRCA2. Cells were collected 4 h after HU treatment (5 mM). Scale bars in C indicate 10 µm. Error bars indicate SD between triplicates. (E) Western blot analysis of nuclear extracts. RPA32 accumulation in U2OS cells transfected with indicated siRNAs was analyzed. Cells were treated with 5 mM HU and harvested 3 h after treatment. Nuclear extracts were prepared.
These data also show that stalled fork repair intermediates that accumulate in BRCA1- and BRCA2-depleted cells are different in nature. In BRCA1-depleted cells, these intermediates have little to no ssDNA, whereas BRCA2-depleted cells are enriched for stalled fork intermediates with excess accumulation of ssDNA/pRPA32. Importantly, codepletion of BRCA1 in BRCA2-depleted cells strongly inhibits excessive accumulation of pRPA32-coated stalled fork intermediates. While we cannot determine whether BRCA1 and BRCA2 are part of the same stalled fork repair pathway or different pathways, these results show that loss of each of these critical players leaves behind a different stalled fork intermediate and raises the possibility that BRCA1 functions upstream of BRCA2.
RPA is persistently associated with stalled replication forks in BRCA2-deficient cells
We next asked whether pRPA32-coated ssDNA in BRCA2-deficient cells resolved normally. Unlike control cells, where pRPA32/RPA32 is lost from a majority of the HU- (Fig. 3, A and B) and UV-induced (Fig. 3, C–E) DNA damage sites in due course (∼24 h), suggesting efficient resolution of the stalled forks, BRCA2-depleted cells revealed persistent accumulation of pRPA32/RPA32 (Fig. 3, A–E).
Figure 3.
BRCA2 depletion results in increased accumulation of pRPA32 after stalled fork–inducing DNA damage. (A and B) IF analysis and graph of RPA32 recruitment in U2OS cells transfected with control siRNA (siLuc) and siRNA for BRCA2. Cells were treated with 5 mM HU for 4 h and harvested right after or 20 h after treatment. Scale bars in A indicate 10 µm. (C) Western blot analysis of whole-cell lysate served as input for IF. (D and E) IF analysis and graph of RPA32 recruitment in U2OS cells transfected with control siRNA (siLuc) or siRNA for BRCA1, BRCA2, or RAD51. Cells were fixed 3, 8, and 24 h after UV damage (30 J/m2). **, P value < 0.05. Statistical significance was determined by the two-tailed Student’s t test and the error bars indicate SD (n = 3). Scale bars in D indicate 10 µm. (F) Western blot analysis of RPA32 accumulation in U2OS cells transfected with control siRNA (siLuc) or siRNA for RAD51. Cells were treated with 5 mM HU and harvested 3 h after treatment. Nuclear extracts were prepared.
We next sought to determine factors that contribute to excessive accumulation of RPA in BRCA2-depleted cells. BRCA2 has been shown to displace RPA and load RAD51 at DSBs (Jensen et al., 2010), and though BRCA2 is not required for RAD51-driven stalled fork reversal (Zellweger et al., 2015), it has been shown to help establish stable association of RAD51 with the regressed arms of a stalled fork (Mijic et al., 2017). We wondered if the accumulation of RPA at stalled forks in BRCA2-depleted cells was a result of inefficient RAD51 loading. If true, one might predict that RAD51 depletion would result in a similar phenotype.
RAD51-depleted cells were irradiated through micropore filters to generate sites of stalled replication forks, and the kinetics of RPA32 recruitment and its loss from the sites of stalled replication forks was studied in these cells. Unlike BRCA2-depleted cells, RAD51-depleted cells do not show persistent RPA32 accumulation at sites of stalled forks and/or accumulation of pRPA32 in the nuclear extracts prepared from cells undergoing replication fork stalling (Fig. 3, C–F). These data indicate that pRPA32 accumulation in BRCA2-depleted cells is not due to inability of BRCA2-depleted cells to replace RPA32 with RAD51 at stalled forks.
BRCA2 depletion results in hyperubiquitination of RPA at stalled replication forks
We next sought to determine whether pRPA32 accumulation in BRCA2-depleted cells (Fig. 1, F–K; and Fig. S1, E and F) was associated with changes in RPA modification, especially ubiquitination. RPA ubiquitination is known to occur during DNA replication and after DNA damage induced by agents like HU, UV, and mitomycin C (Elia et al., 2015; Feeney et al., 2017; Inano et al., 2017; Lin et al., 2018; Maréchal and Zou, 2015). As shown in Fig. 4 A, BRCA2 depletion led to increased RPA ubiquitination (hyperubiquitination) after exposure to stalled fork–inducing agents like HU and UV (Fig. 4 A and Fig. S2 C). Furthermore, incubation with MG132 did not change ubiquitination levels (Fig. S2 D), suggesting that the RPA hyperubiquitination observed upon BRCA2 depletion does not lead to proteasomal degradation.
Figure 4.
BRCA2 depletion results in hyperubiquitination of RPA after HU-induced replication stress. (A) Immunoprecipitation (IP) analysis of RPA ubiquitination in HEK293T cells transfected with control siRNA (siLuc) or siRNA for BRCA1. These cells were transfected with His-tagged ubiquitin (His-Ub) and HA-tagged RPA32 (HA-RPA32) and treated with 5 mM HU or irradiated with 30 J/m2 of UV. Cells were harvested 3 h after treatment. Cell extracts were immunoprecipitated with Ni-resin and immunoblotted. Blot was probed with anti-HA Ab. (B) Immunoprecipitation analysis of RPA ubiquitination in HEK293T cells transfected with control siRNA (siLuc) or siRNA for BRCA1, BRCA2, or both BRCA1 and BRCA2. Experimental conditions used are as described above.
We next investigated whether BRCA1 depletion also led to similar hyperubiquitination of RPA32 in cells undergoing replication stress. Cells were depleted of BRCA1 and then tested for RPA32 ubiquitination after HU- and/or UV-induced DNA damage. Unlike BRCA2, BRCA1 depletion did not lead to ubiquitination of RPA32 (Fig. 4 B). Furthermore, codepletion of BRCA1 in BRCA2-depleted cells suppressed RPA32 hyperubiquitination (Fig. 4 B). This is consistent with our hypothesis that BRCA1 functions upstream of BRCA2 in the stalled fork repair pathway.
In keeping with our observation that RAD51 depletion does not lead to increased accumulation of pRPA32-coated ssDNA at stalled forks, we find that depletion of RAD51 does not lead to increased ubiquitination of RPA32 after HU-induced DNA damage (Fig. S2 E).
To further characterize the factors that drive RPA ubiquitination in BRCA2-depleted cells, we next asked whether RPA phosphorylation affects RPA ubiquitination in these cells. We worked with two constructs (RPAA and RPAD) that harbor alanine or aspartate mutations in eight DNA damage– and cell cycle–responsive phosphorylation sites (S8, S11, S12, S13, T21, S23, S29, and S33), with the aspartate mutations serving as a mimic of phosphorylation. These mutants have been shown previously to be competent in getting recruited to sites of DNA damage like that induced by HU (Vassin et al., 2004). We found that relative to nontargeting controls, BRCA2 depletion induced ubiquitination of RPAwt construct more effectively than the RPAA mutant (Fig. S2 F). These results are in keeping with a prior report using a similar RPA mutant in WT cells (Dubois et al., 2017).
Interestingly, we see an increase in RPA ubiquitination of the RPAD mutant in BRCA2-depleted cells relative to control cells (Fig. S2 F). We also noticed that majority of ubiquitinated RPA in BRCA2-depleted cells was phosphorylated at S33 residue (Fig. S2 F), indicating that phosphorylated RPA is a strong target for ubiquitination. As expected, RPAA and RPAD mutants do not show S33 phosphorylation, because they lack S33.
Together, these results show that hyper-RPA ubiquitination occurs in BRCA2-depleted cells and that loss of BRCA1 suppresses accumulation of this hyperubiquitinated RPA intermediate at the stalled forks. These results also suggest that phosphorylation of RPA is conducive and probably a necessary step toward its ubiquitination by an E3 ligase.
Hyperubiquitination of RPA after BRCA2 depletion is dependent on RFWD3 E3 ligase
Having established that RPA is hyperubiquitinated in BRCA2-depleted cells undergoing replication stress, we next addressed the E3 ligase that carries out RPA ubiquitination in BRCA2-depleted cells. RFWD3 is an E3 ligase that is required for the ubiquitination of RPA after DNA damage and has been shown to assist in stabilizing stalled fork for HR-dependent repair (Elia et al., 2015) as well as for timely removal of RPA after mitomycin C-induced DNA damage (Feeney et al., 2017; Inano et al., 2017).
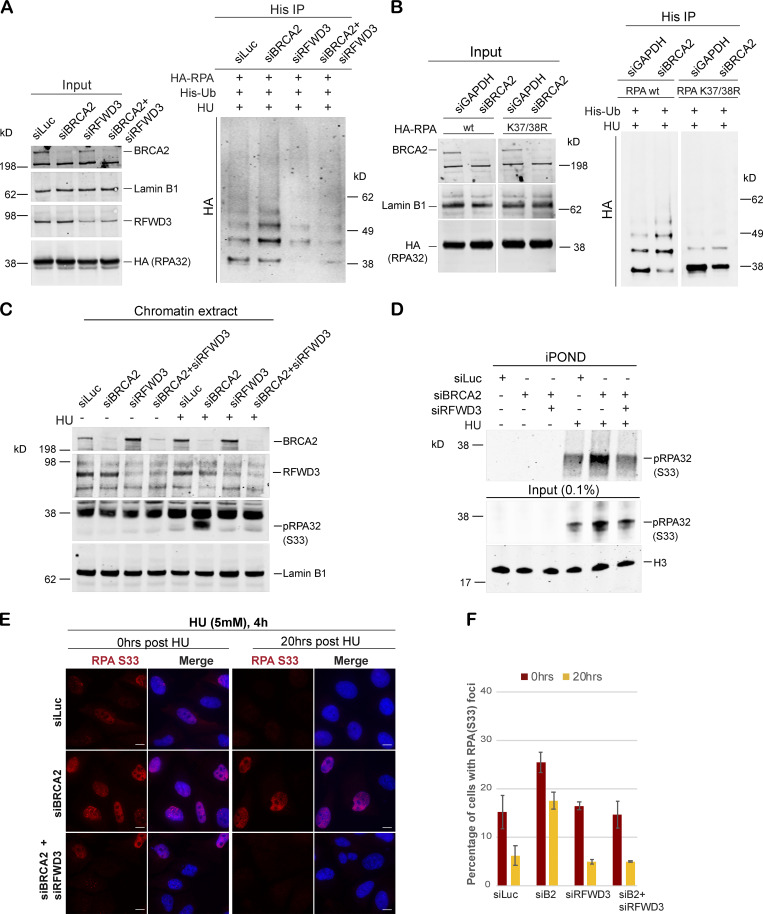
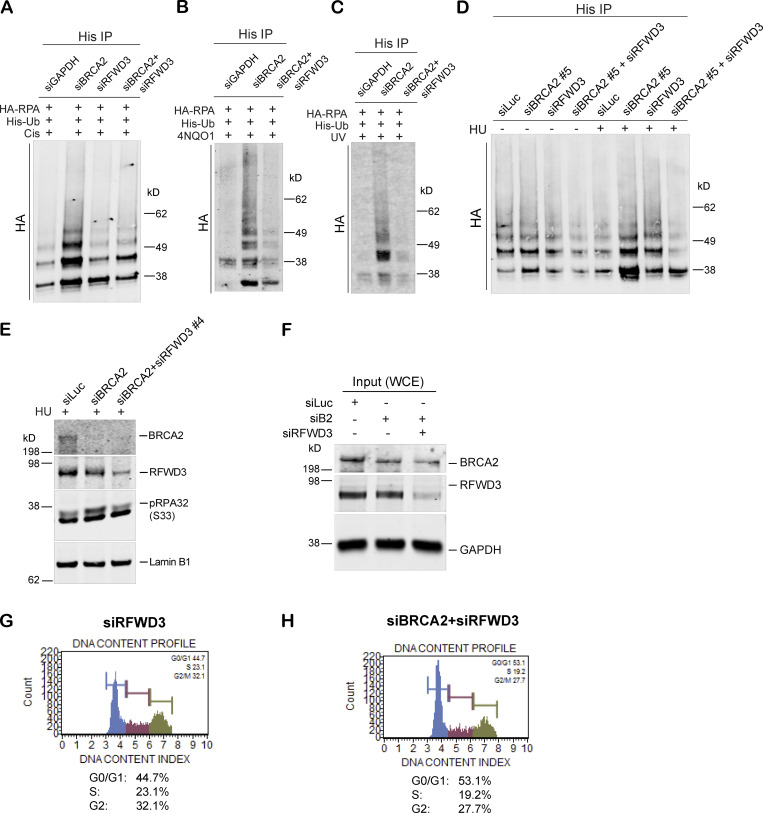
We asked whether RFWD3 is the E3 ligase that was responsible for hyperubiquitination of RPA in BRCA2-depleted cells. Codepletion of RFWD3 resulted in loss of RPA hyperubiquitination in BRCA2-depleted cells treated with different replication-stalling agents (Fig. 5 A and Fig. S3, A–D). This suggested that hyperubiquitination of RPA32 in BRCA2-depleted cells is driven by the E3 ligase RFWD3. Previous studies have identified lysine residues K37 and K38 on RPA32 as sites of RFWD3-dependent ubiquitination (Elia et al., 2015). To investigate whether these are the same lysine residues that are ubiquitinated by RFWD3 in response to stalled fork formation in BRCA2-depleted cells, we transfected siBRCA2- and siLuc (control)-treated cells with WT RPA32 and/or RPA32 with K37 and K38 mutated to arginine (K37R/K38R_RPA32), thus rendering these two lysine residues unavailable for ubiquitination. As shown in Fig. 5 B, the K37/38R RPA mutant is severely compromised for ubiquitination in BRCA2-depleted cells. This confirms that these RPA residues are prime targets for hyperubiquitination by RFWD3 in BRCA2-depleted cells undergoing replication stress.
Figure 5.
Hyperubiquitination of RPA in BRCA2-deficient cells undergoing replication stress is performed by the E3 ligase RFWD3. (A) Immunoprecipitation analysis of RPA ubiquitination in HEK293T cells transfected with control siRNA (siLuc) or siRNA for BRCA2, RFWD3, or both BRCA2 and RFWD3. HEK293T cells with indicated siRNAs were transfected with His-tagged ubiquitin and HA-tagged RPA32. Cells were treated with 5mM HU and harvested 3 h after treatment. His immunoprecipitation was done as indicated above, and blots were immunoblotted with HA. (B) Lysine K37 and K38 amino acid residues in RPA get ubiquitinated by RFWD3 in BRCA2-depleted cells. Cells were transfected with indicated siRNAs, followed by transfection with HA-tagged WT or K37/38R RPA mutant. Cells were processed for His-immunoprecipitation as described above. (C) Western blot analysis of RPA32 accumulation on chromatin after disrupting RPA ubiquitination by codepletion of RFWD3 in BRCA2-depleted cells. U2OS cells transfected with indicated siRNAs were treated with 5 mM HU and harvested 3 h after treatment. Chromatin extracts were prepared, and relevant Western blot was probed with pRPA32 (S33). (D) Western blot analysis of cells with indicated siRNA for input and captured proteins isolated by iPOND. Cells were pulse labeled with EdU for 10 min followed by treatment with 5 mM HU and harvested 3 h after treatment. (E and F) IF analysis to study the effect of RPA ubiquitination on RPA32 accumulation in BRCA2-deficient cells. Scale bars in E indicate 10 µm. (E) U2OS cells transfected with indicated siRNAs were treated with 5 mM HU for 4 h and fixed right after (0 h) or 20 h after treatment. Cells were stained with pRPA32-S33. (F) Graph indicates percentage of cells with pRPA-S33 foci after HU induced DNA damage. Error bars indicate SD (n = 3).
Figure S3.
Hyperubiquitination of RPA after BRCA2 depletion is performed by the E3 ligase RFWD3. (A–C) Immunoprecipitation analysis of RPA ubiquitination in HEK293T cells transfected with indicated siRNAs. Cells were treated with the following drugs: 2 µM cisplatin for 5 h (A), 1 mg/ml 4NQO1 for 3 h (B), or 30 J/m2 of UV for 3 h (C). (D) Immunoprecipitation analysis of RPA ubiquitination in response to HU in HEK293T cells treated with indicated siRNAs. A different BRCA2-specific siRNA (siBRCA2#5) was used for this experiment. (E) Western blot analysis of RPA32 accumulation after disrupting RPA ubiquitination by codepletion of RFWD3 in BRCA2-depleted cells. A different RFWD3-specific siRNA (siRFWD3#4) was used for this experiment. (F) Western blot analysis of whole-cell lysate served as input for iPOND. (G and H) Cell cycle analysis of RFWD3-depleted and BRCA2/RFWD3–codepleted cells by MUSE-based cell cycle assay. Related to Fig. 5.
Accumulation of phosphorylated RPA in BRCA2-depleted cells is dependent on RFWD3
To determine whether the increase in pRPA32 in BRCA2-depleted cells was dependent on its hyperubiquitination by RFWD3, we codepleted RFWD3 from BRCA2-depleted cells and tested these cells for pRPA32 accumulation. Codepletion of RFWD3 almost completely blocked the increased accumulation of pRPA32 on chromatin in BRCA2-depleted cells (Fig. 5 C and Fig. S3 E). To further confirm that these changes in pRPA32 accumulation were indeed occurring on nascent DNA at stalled replication forks, we performed iPOND (isolation of proteins on nascent DNA) analysis (Sirbu et al., 2012). Just as we had observed in nuclear and the chromatin extracts, loss of BRCA2 resulted in increased pRPA32 accumulation (Fig. 5 D; input in Fig. S3 F), and codepletion of RFWD3 in BRCA2-depleted cells led to rescue of this phenotype (Fig. 5 D). Histone H3 served as a positive loading control in these experiments. As before, we also confirmed that there were no cell cycle perturbations due to RFWD3 depletion and that this did not account for the rescue we observed in BRCA2-depleted cells (Fig. S3, G and H).
To further confirm that pRPA32 accumulation in BRCA2-depleted cells can be rescued by codepleting these cells of RFWD3, we performed IF-based analysis. Here too, cells depleted of both BRCA2 and RFWD3 showed faster resolution of pRPA32 foci upon HU treatment (compare siB2 vs siB2+siRFWD3, for 0 h and 20 h after HU treatment; Fig. 5, E and F).
RFWD3 depletion rescues fork degradation in BRCA2-depleted, but not BRCA1-depleted, cells
We next addressed whether excessive ubiquitination of RPA32 at stalled forks in BRCA2-depleted cells could impair fork stability. It has been shown previously (Schlacher et al., 2011) that stalled replication forks in BRCA2-depleted cells are degraded by MRE11 nuclease. Given that RFWD3 loss rescued excess RPA accumulation phenotype of BRCA2-depleted cells, we wondered whether its depletion also affects degradation of the stalled forks in BRCA2-depleted cells.
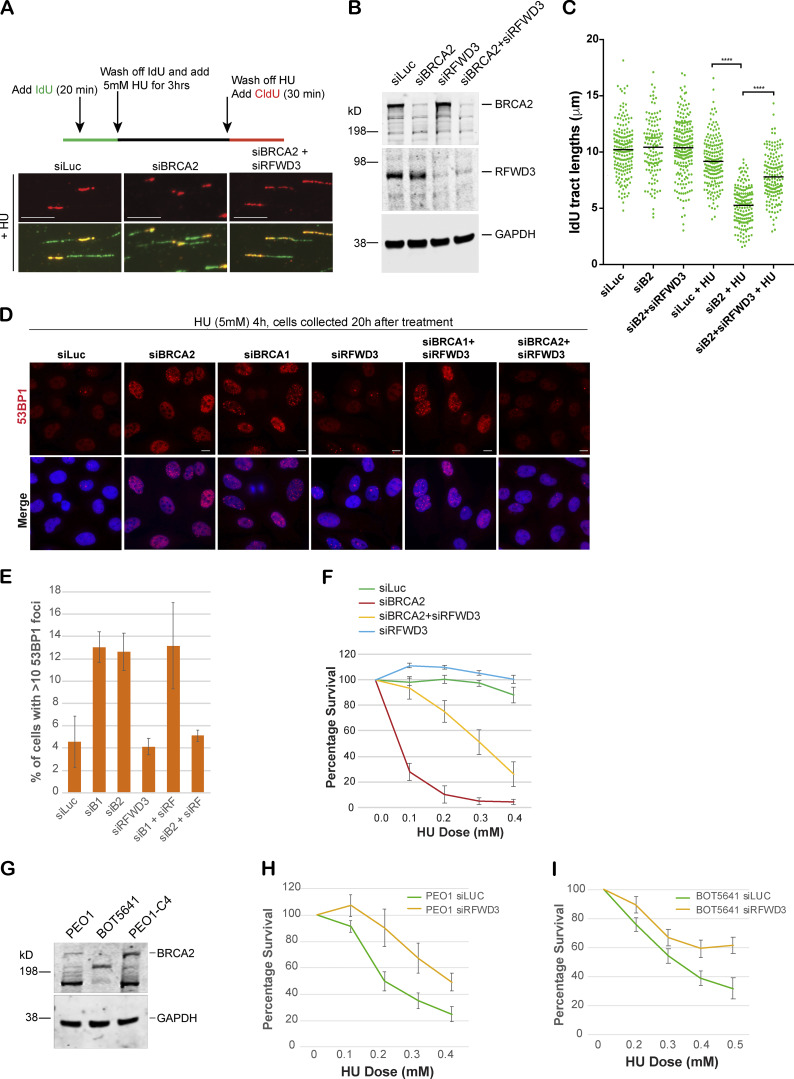
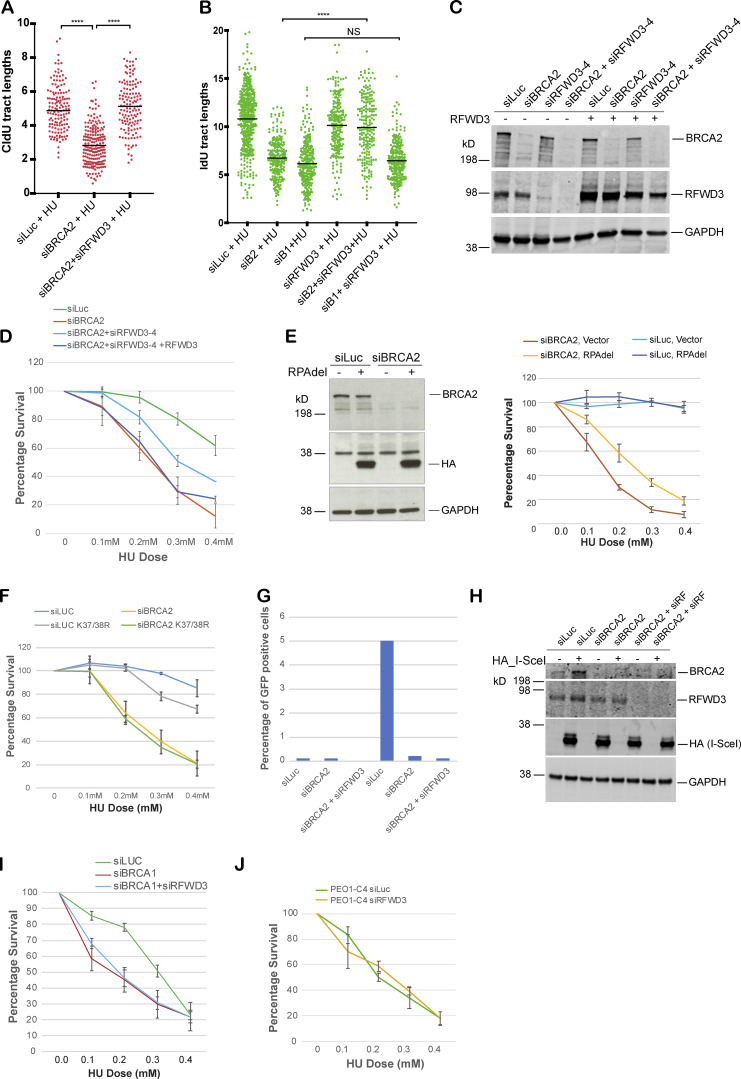
Control (siLuc) and BRCA2-depleted (siBRCA2) U2OS cells were exposed to HU, and DNA fiber analysis was performed on these cells as described before (Schlacher et al., 2011). 5-iodo-2′-deoxyuridine (IdU) was added to cells for 20 min and then washed off, followed by HU treatment for 3 h. This was followed by 5-chloro-2′-deoxyuridine (CldU) treatment (Fig. 6 A). In the untreated samples, CldU was added right after IdU treatment. The length of the IdU-labeled strand was measured in untreated and HU-treated samples by ImageJ software. As shown before, any shortening of the IdU tract upon HU treatment served as a measure of the fork instability (Schlacher et al., 2011, 2012; Somyajit et al., 2015). As expected, in control cells (siLuc), there was little change in IdU tract length after HU treatment; however, in BRCA2-depleted cells, IdU tract length shortened from ∼10.2 µm to ∼5 µm after HU treatment, revealing increased fork degradation (Fig. 6, B and C). We found that loss of RFWD3 significantly rescued the fork degradation phenotype of BRCA2-depleted cells (Fig. 6 C). We also noticed that although the restart frequency of these forks was not altered significantly in BRCA2-depleted cells compared with controls (Schlacher et al., 2011; Ying et al., 2012), the tract length of restarted forks was significantly shorter in BRCA2-depleted cells (Fig. S4 A). Thus, although fork restart occurred at the same frequency in BRCA2-depleted cells as in control cells, these data suggest that there is either a delay in the restart of the stalled forks in BRCA2-depleted cells and/or the restarted forks are not as processive as those in the control cells, as observed previously (Lemaçon et al., 2017). Interestingly, codepletion of RFWD3 in BRCA2-depleted cells fully rescued this defect in these cells (Fig. S4 A). Thus, RFWD3 destabilizes and affects restart of the stalled forks in BRCA2-depleted cells.
Figure 6.
RFWD3 depletion rescues fork degradation, fork collapse, and cell sensitivity to stalled fork–inducing agents in BRCA2-depleted cells and BRCA2 mutant tumor cells. (A) Top: Schematic of how DNA fiber experiment was performed. Bottom: Representative tracts from DNA fiber experiments with U2OS cells transfected with indicated siRNAs. Green and red tracts correspond to IdU and CldU incorporation, respectively. Scale bars indicate 10 µm. (B) Western blot analysis of whole-cell lysate to confirm knockdown of BRCA2 and RFWD3 after transfecting cells with indicated siRNAs. These extracts are from the cells used for Fiber assay. (C) Scatterplots compare the tract lengths of IdU-labeled fibers between different siRNA conditions and in the presence or absence of HU, with black lines indicating the median. ****, P < 0.0005. (D and E) IF and graph of 53BP1 recruitment in U2OS cells transfected with indicated siRNAs. Cells were treated with 5 mM HU for 4 h and then collected 20 h after treatment. Graph indicates the percentage of cells with more than 10 53BP1 foci per cell. Scale bars in D indicate 10 µm. (F) CellTiter-Glo–based cell survival assay was used to determine the sensitivity of U2OS cells transfected with indicated siRNAs to HU. HU was added for 4 d, and cells were allowed to recover for 2 d before harvesting them for CellTiter-Glo–based analysis. (G) Western blot analysis of total BRCA2 protein levels in different BRCA2 tumor lines. PEO1 is an ovarian cancer cell line that has a BRCA2 homozygous mutation 5193C>G, which would normally result in a stop codon at amino acid 1655. BOT5641 is a BRCA2 breast tumor line that has a c.6486_6489del mutation. (H and I) CellTiter-Glo–based cell survival assay was used to determine the sensitivity of BRCA2 mutant tumor lines PEO1 (H) and BOT5641 (I) to HU after depletion of RFWD3. Error bars indicate SD between triplicates.
Figure S4.
RFWD3 depletion rescues fork degradation, fork collapse, and cell sensitivity to stalled fork–inducing agents in BRCA2-depleted cells and BRCA2 mutant tumor cells. (A) Fork restart analysis. Scatterplots compare the tract lengths of CldU in cells treated with different siRNAs and in the presence of HU, with black horizontal lines indicating the median of all the tracts counted (>200). ****, P < 0.0005. (B) Scatterplots compare the tract lengths of IdU in U2OS cells transfected with different siRNAs and in the presence of HU, with black horizontal lines indicating the median of all the tracts counted (>200). ****, P < 0.0005. (C) Western blot shows expression level of indicated proteins in control (siLuc), BRCA2-depleted (siBRCA2), and BRCA2- and RFWD3-codepleted (siBRCA2+siRFWD3) cells. (D) CellTiter-Glo–based cell survival assay was used to determine the sensitivity of BRCA2- and RFWD3-codepleted cells to HU after reconstituting these cells with WT RFWD3. (E and F) CellTiter-Glo–based cell survival assay was used to determine the sensitivity of BRCA2-depleted cells expressing truncated (lacking residues 243–262) “Del” HA-RPA32 (E) or K37/38R mutant of the RPA32 (F) to HU. (G and H) HR assay to determine HR efficiency. U2OS cell line with a stably integrated DRGFP reporter was used for this assay. Cells were transfected with HA-tagged I-SceI–expressing plasmid to induce DSBs. Graph indicates percentage of GFP-positive cells transfected with control (siLuc), BRCA2, RFWD3, or BRCA2 and RFWD3 together. GFP was detected by flow cytometry. (I) Loss of RFWD3 does not rescue sensitivity of BRCA1-depleted cells to HU. CellTiter-Glo–based cell survival assay was used to determine the sensitivity of BRCA1- and RFWD3-codepleted cells to HU. (J) CellTiter-Glo–based cell survival assay was used to determine the sensitivity of PEO1-C4 to HU after depletion of RFWD3. PEO1-C4 is a BRCA2-proficient ovarian cancer cell line. Error bars indicate SD between triplicates. Related to Fig. 6.
Given that loss of RFWD3 rescues fork degradation in BRCA2-depleted cells, we next asked whether loss of RFWD3 can also reduce the increased fork collapse that is observed in these cells. 53BP1 nuclear foci were used to mark the presence of collapsed forks (assayed ∼20 h after HU-induced DNA damage; Harding and Bristow, 2012; Sotiriou et al., 2016). We treated cells with 5 mM HU for 4 h and then allowed the cells to grow for another 20 h before fixing the cells for analysis of collapsed forks. While control cells resolved the stalled forks formed in the presence of HU, BRCA2 depletion led to an increase in the number of cells with 53BP1 foci, reflecting an increase in the number of cells undergoing fork collapse (Fig. 6, D and E). Importantly, codepletion of RFWD3 in these cells fully rescued this phenotype (Fig. 6, D and E). We also observed that although both BRCA1- and BRCA2-depleted cells undergo fork degradation and fork collapse, RFWD3 depletion rescues these defects only in BRCA2-depleted cells and not in BRCA1-depleted cells (Fig. 6, D and E; and Fig. S4 B). These results highlight the difference between stalled fork intermediates formed in cells depleted of BRCA1 or BRCA2. It remains to be seen whether the difference in nature of stalled fork intermediates formed in absence of each of these proteins also dictates factors that can rescue fork defects in them.
Collectively, these results indicate that RFWD3 contributes to increased fork degradation and fork collapse in BRCA2-depleted, but not BRCA1-depleted, cells.
RFWD3 contributes to increased sensitivity of BRCA2-deficient cells to stalled fork–inducing agents
As shown above, BRCA2 depletion leads to increased accumulation of RFWD3-dependent hyperubiquitinated pRPA-coated ssDNA at stalled forks. Depletion of the E3 ligase RFWD3 reverses pRPA accumulation and fork degradation in BRCA2-deficient cells. We next addressed whether loss of RFWD3 can rescue increased replication stress in BRCA2-depleted cells and provide them with a survival advantage. Codepletion of RFWD3 in BRCA2-depleted cells partially rescued the sensitivity of BRCA2-depleted cells to HU (Fig. 6 F). We confirmed this by using two different RFWD3-specific siRNAs (Fig. 6 F and Fig. S4, C and D). We also confirmed the specificity of the RFWD3 siRNAs by adding back siRFWD3-resistant RFWD3 and reversing the effect of siRFWD3 (Fig. S4, C and D). This result suggests that RFWD3-dependent RPA ubiquitination, along with increased fork degradation, contributes to increased cell sensitivity of BRCA2-deficient cells to replication-stalling agents.
We further investigated the effect of suppressing RPA32 ubiquitination by expressing ubiquitination-defective RPA32 in BRCA2-depleted cells. The ubiquitination-defective RPA32s we used in this study are (1) the K37/38R mutant of the RPA32 and (2) the RPAdel mutant in which the RFWD3 interaction domain (20 amino acids) is deleted (Elia et al., 2015). Both RPA mutants exhibit reduced ubiquitination in the presence of replication-stalling agents (Elia et al., 2015). We find that expression of RPAdel partially rescued the sensitivity of BRCA2-depleted cells to HU (Fig. S4 E). However, expression of K37/38R mutant of RPA32 was not able to consistently rescue the sensitivity of BRCA2-depleted cells to HU (Fig. S4 F). This difference in the ability of the two different RPA32 ubiquitination mutants might be due to the differences in the extent of their ubiquitination defect. The K37/38R mutant of RPA32 completely abrogates the ability of RPA32 to become ubiquitinated but does not affect the ubiquitination of other RPA subunits (RPA70 and RPA14). The RPA32-del mutant, on the other hand, can inhibit ubiquitination of all three RPA subunits, as it disrupts RPA32’s interaction with RFWD3 (Gong and Chen, 2011), and can affect RFWD3 recruitment to chromatin (Liu et al., 2011).
We next asked whether loss of RFWD3 can also rescue defective HR DSBR function in BRCA2-depleted cells. We did not see a similar rescue of HR defect in these cells. To test the HR efficiency, we used a U2OS cell line with a stably integrated direct-repeat GFP reporter (Moynahan et al., 2001). In this cell line, the DSB is induced by expression of I-SceI, HR-dependent repair of the DSB produces a functional copy of GFP, and these GFP-positive cells are scored by FACS-based analysis. As expected, BRCA2-depleted cells (siBRCA2) show fewer GFP-positive cells compared to control siLuc-treated cells (Fig. S4 G). However, codepletion of RFWD3 in BRCA2-depleted cells did not rescue HR deficiency in these cells (Fig. S4 G). Equal expression of I-SceI in these cells was confirmed by checking for levels of HA-tagged I-SceI in all the samples (Fig. S4 H).
Given that RFWD3 depletion did not rescue fork degradation and/or fork collapse in BRCA1-depleted cells, we hypothesized that loss of RFWD3 might not rescue sensitivity of BRCA1-depleted cells to stalled fork–inducing agent like HU. We find that this is indeed true and loss of RFWD3 does not rescue sensitivity of BRCA1-deficient cells (Fig. S4 I). These results suggest that increased RPA32 ubiquitination by RFWD3 contributes to increased sensitivity to replication-stalling agents in BRCA2-depleted, but not BRCA1-depleted, cells. This further suggests that stalled fork intermediates formed in absence of BRCA1 and BRCA2 are modulated by different protein factors and that RFWD3 plays a role in fork stability in BRCA2-deficient cells, but not in BRCA1-deficient cells.
RFWD3 loss provides survival advantage to BRCA2 mutant tumor cells
Finally, to determine whether RFWD3 was also driving sensitivity of BRCA2 mutant tumor cells to stalled fork–inducing agents, we worked with two different BRCA2 tumor lines: PEO1, an established BRCA2 mutant ovarian line (Stordal et al., 2013), and BOT5641, a BRCA2 breast tumor line derived by us from a breast tumor section collected during surgery from a BRCA2 mutation carrier (c.6486_6489del; Fig. 6 G). These two BRCA2 tumor lines, with little to no expression of full-length BRCA2, are sensitive to stalled fork inducing agents like HU. We asked whether sensitivity of these tumor lines could be rescued in part by depleting these tumor cells of RFWD3. Sensitivity of both PEO1 and BOT5641 was partially rescued by loss of RFWD3 in these cells (Fig. 6, H and I), further confirming the role of RFWD3 in increasing replication stress in BRCA2-deficient cells and/or BRCA2 tumor cells. No such rescue was observed in a revertant BRCA2 tumor line (PE01-C4), which expresses BRCA2 (Fig. 6 G and Fig. S4 J). Given the high endogenous replication stress in tumor cells, these data would imply an increased dependence of BRCA2 tumor cells on losing RFWD3 for better survival. In keeping with this hypothesis, we find that BRCA2 mutant tumors tend to harbor RFWD3 loss. Analysis of breast, ovarian, and prostate cancer data from the cBioPortal (Cerami et al., 2012; Gao et al., 2013) shows that both breast and prostate tumors harboring BRCA2 loss tend to co-occur with RFWD3 loss (prostate cancer, n = 3,212, odds ratio [OR] = 4.18 [1.8–8.6], P = 0.0004, breast cancer, n = 3,367, OR = 6.40 [0.7–27.8], P = 0.047). In ovarian cancer, a nonsignificant trend toward co-occurrence was observed (n = 316, OR = 2.43 [0.05–25.6], P = 0.395). No significant association was found between the co-occurrence of BRCA1 and RFWD3 loss across breast, prostate, and ovarian cancer, with both breast and ovarian trending toward mutual exclusivity (OR = 0, 2.08, and 0, and P = 1, 0.39, and 1 for breast, prostate, and ovarian cancer, respectively). While supportive of the mechanism presented in this work, these data must nevertheless be interpreted with caution given the overall rarity of RFWD3 events (affecting 0.5%, 1.8%, and 1.6% of breast, prostate, and ovarian cancer cases, respectively).
Generation of the ubq-pRPA–coated ssDNA intermediate in BRCA2-deficient cells is not dependent on MRE11-driven fork resection
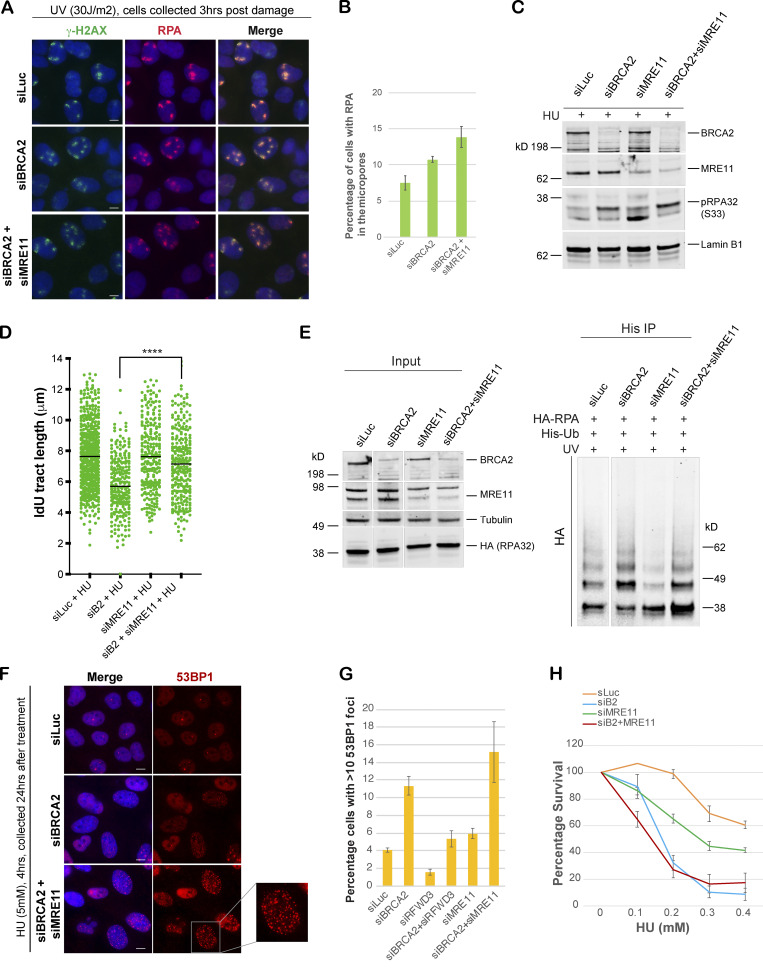
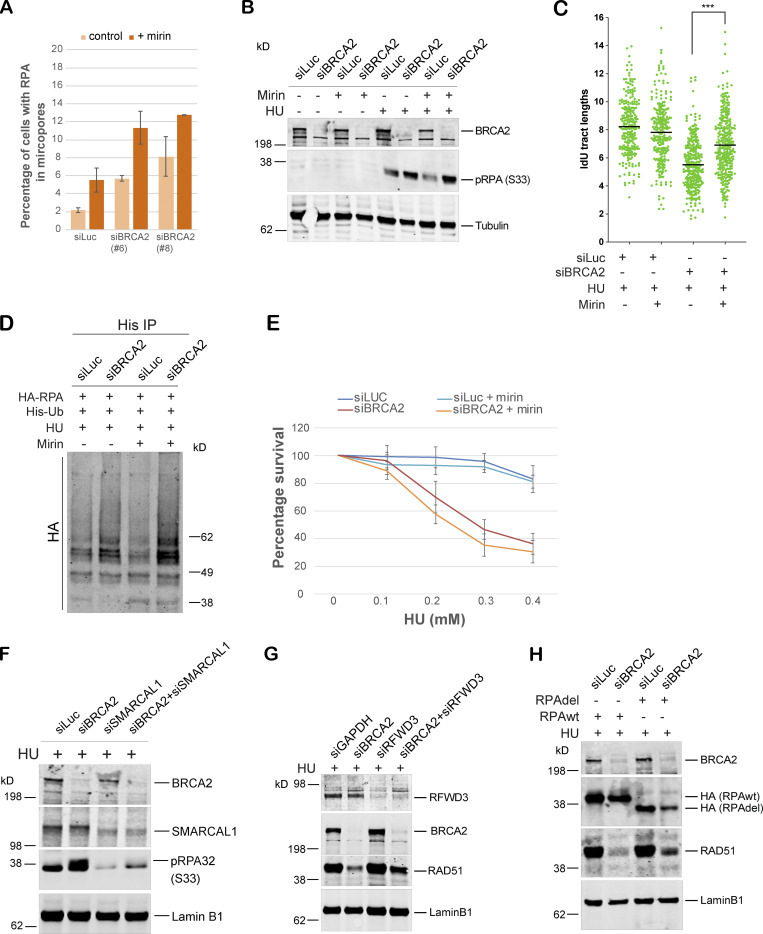
To investigate the source of excessive ssDNA that we find accumulating after replication fork stalling in BRCA2-depleted cells, we asked whether MRE11, a nuclease well documented for its role in fork degradation in BRCA2-depleted cells, was responsible. Surprisingly, we find that codepleting MRE11 in BRCA2-deficient cells and/or blocking MRE11 activity by mirin (Dupré et al., 2008) does not reduce the excessive accumulation of RPA/p-RPA32 in BRCA2-depleted cells (Fig. 7, A–C; and Fig. S5, A and B). This was surprising given that MRE11 loss helps rescue fork degradation in BRCA2-depleted cells. To address this discrepancy, we next used the same conditions (loss of MRE11 and/or addition of mirin) to study fork degradation by fiber assay. As shown before (Lemaçon et al., 2017; Schlacher et al., 2011), we too find that loss of MRE11 and/or loss of MRE11 activity does robustly reduce fork degradation in BRCA2-depleted cells (Fig. 7 D and Fig. S5 C); however, under those same conditions, we also find increased RPA accumulation. We checked the ubiquitination status of RPA in MRE11-depleted BRCA2-deficient cells (and also in cells treated with mirin) and find that the RPA is indeed hyperubiquitinated in these cells (Fig. 7 E and Fig. S5 D).
Figure 7.
Generation of the ubq-RPA–coated ssDNA intermediate in BRCA2-deficient cells is not dependent on MRE11-driven fork resection. (A and B) IF analysis, and graph of RPA32 recruitment in U2OS cells transfected with indicated siRNAs. Cells were fixed 3 h after UV damage (30 J/m2). Scale bars in A indicate 10 µm. (C) Western blot analysis of RPA32 accumulation in U2OS cells transfected with indicated siRNAs. Cells were treated with 5 mM HU and harvested 3 h after treatment. Nuclear extracts were prepared for analysis. (D) Scatterplots compare the tract lengths of IdU-labeled fibers between different siRNA conditions and in the presence of HU, with black lines indicating the median. ****, P < 0.0005. (E) Immunoprecipitation analysis of RPA ubiquitination in HEK293T cells transfected with indicated siRNAs. Experimental conditions used are as described above. (F and G) IF analysis and graph of 53BP1 recruitment in U2OS cells transfected with indicated siRNAs. Cells were treated with 5 mM HU for 4 h and then collected 20 h after treatment. Graph indicates the percentage of cells with >10 of 53BP1 foci per cell. Scale bars in G indicate 10 µm. Error bars indicate SD (n = 3). (H) CellTiter-Glo–based cell survival assay was used to determine the sensitivity of U2OS cells to HU after codepletion of MRE11 in BRCA2-depleted cells. Error bars indicate SD between triplicates.
Figure S5.
Generation of the ubq-pRPA intermediate in BRCA2-deficient cells is not dependent on MRE11-driven fork resection. Mirin was used in these experiments to suppress MRE11 nuclease activity. SMARCAL1-mediated fork reversal is required for accumulation of hyperubiquitinated RPA accumulation in BRCA2-deficient cells. (A) Quantification of IF-based analysis of RPA32 recruitment in U2OS cells transfected with control (siLuc) or BRCA2 siRNA in the absence or presence of mirin. Mirin was used to block exonuclease activity of MRE11. U2OS cells transfected with indicated siRNAs were irradiated with UV as indicated above, followed by 40 µM mirin treatment for 5 h. (B) Western blot analysis of RPA32 accumulation on chromatin in U2OS control (siLuc) or BRCA2-depleted cells (siBRCA2) in the absence or presence of mirin. U2OS cells transfected with indicated siRNAs were cotreated with 40 µM mirin and 5 mM HU for 3 h. (C) DNA fiber scatterplots compare the tract lengths of IdU under different conditions. U2OS cells transfected with indicated siRNAs were cotreated with 40 µM mirin and 5 mM HU for 3 h. More than 200 tracts were counted in each experiment. The black horizontal bar represents the median of all the tracts counted in that experiment. ***, P < 0.005. (D) RPA ubiquitination in HEK293T cells transfected with indicated siRNAs in the absence or presence of mirin. HEK293T cells transfected with indicated siRNAs were cotreated with 40 µM mirin and 5 mM HU for 3 h before collecting them for ubiquitination analysis. (E) CellTiter-Glo–based cell survival assay was used to determine the sensitivity of BRCA2-depleted cells to HU in the absence or presence of mirin. Error bars indicate SD (n = 3). (F) Western blot–based analysis of RPA32 accumulation after disrupting reversed fork formation by codepletion of SMARCAL1 in BRCA2-deficient cells. Experimental conditions used are as described above. Nuclear extracts were prepared. Blot was probed with anti-pRPA32 (S33). (G) Western blot analysis of RAD51 accumulation in the nuclear extracts after disrupting RPA ubiquitination by codepletion of RFWD3 in BRCA2-depleted cells. U2OS cells transfected with indicated siRNAs were treated with 5 mM HU and harvested 3 h after treatment. Nuclear extracts were prepared, and relevant Western blot was probed with RAD51. (H) Western blot analysis of RAD51 recruitment after disrupting RPA ubiquitination by expressing truncated (lacking residues 243–262) “Del” HA-RPA32 in BRCA2-depleted cells. Cells were treated with 5 mM HU and harvested 3 h after treatment. Nuclear extracts were prepared, and relevant Western blot was probed with RAD51. Related to Figs. 7 and 8.
Given that our data show that stalled forks with hyperubiquitinated pRPA-coated ssDNA in BRCA2-deficient cells are not conducive to fork repair, we wondered whether the reversed forks, now protected by loss of MRE11, are still resistant to efficient repair and would collapse over time. In keeping with this hypothesis, we find that depletion of MRE11 in BRCA2-depleted cells did not indeed reduce the number of cells undergoing fork collapse (Fig. 7, F and G). This is in marked contrast to what we observe upon RFWD3 depletion in BRCA2-depleted cells. In keeping with no rescue of fork collapse in BRCA2-depleted cells, loss of MRE11 and/or blocking its activity with mirin did not rescue the sensitivity of BRCA2-depleted cells to HU (Fig. 7 H, and Fig. S5 E). These data further suggest that rescue of stalled fork degradation is not necessarily the same as repairing the stalled forks and that the reverse forks might not be conducive to repair in the BRCA2-deficient setting, especially if the ssDNA at the reversed forks is coated with hyperubiquitinated RPA.
SMARCAL1-mediated fork reversal is required for accumulation of hyperubiquitinated RPA-coated ssDNA in BRCA2-deficient cells
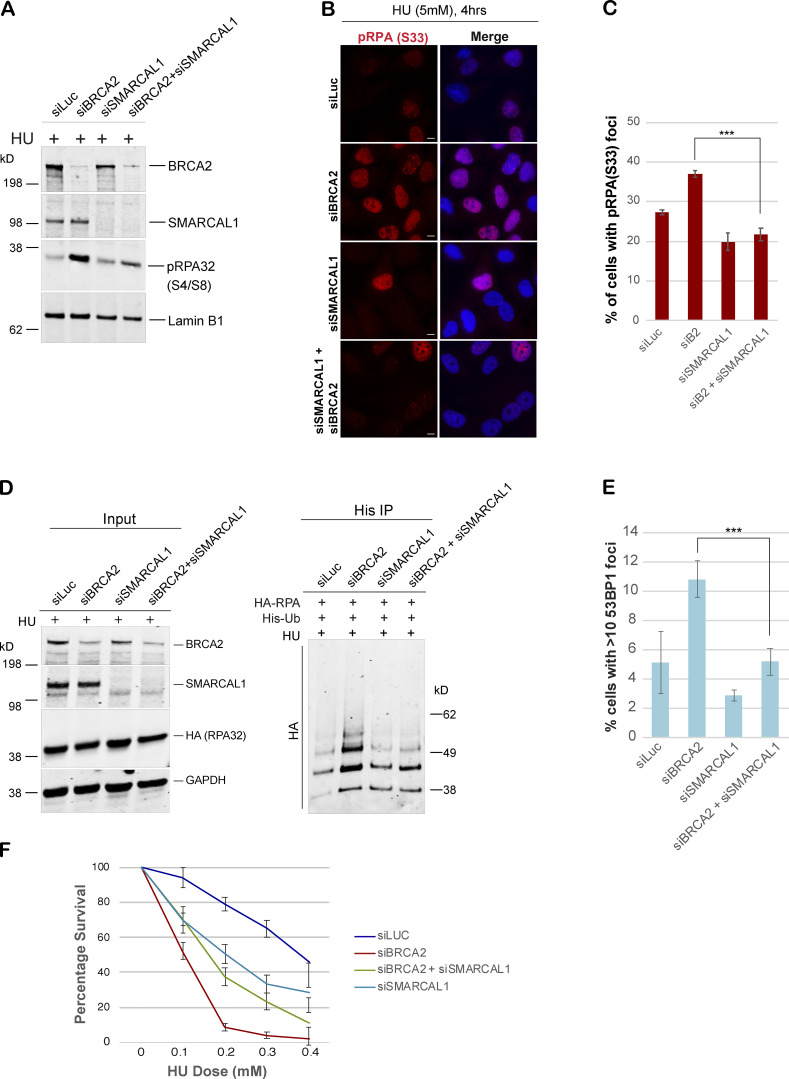
Having established that MRE11-dependent fork resection does not contribute to accumulation of excessive RPA in BRCA2-deficient cells, we next asked whether fork reversal was required for this phenomenon. We hypothesized that if fork reversal was necessary in order to generate pRPA-coated ssDNA in BRCA2-deficient cells, then blocking fork reversal by depleting cells of SMARCAL1, a fork remodeler (Kolinjivadi et al., 2017b; Quinet et al., 2017; Taglialatela et al., 2017), would inhibit pRPA accumulation after stalled fork–inducing DNA damage in these cells. Indeed, codepleting SMARCA1 in BRCA2-depleted cells strongly inhibited accumulation of pRPA32 after HU-induced fork stalling, which was confirmed by Western blot of nuclear extracts as well as IF-based assays (Fig. 8, A–C; and Fig. S5 F). We next asked whether excess RPA ubiquitination observed in BRCA2-depleted cells could also be rescued by codepletion of SMARCAL1 in these cells. Indeed, codepleting SMARCAL1 led to marked decrease in accumulation of ubiquitinated RPA32 in BRCA2-depleted cells (Fig. 8 D). In keeping with our previous observation that suppressing excessive accumulation of ubq-RPA rescues fork collapse in BRCA2-depleted cells, we find that loss of SMARCAL1 also rescues forks from collapsing (Fig. 8 E). This is in keeping with previous reports that fork degradation in BRCA2-depleted cells can be rescued by blocking fork reversal (Mijic et al., 2017; Taglialatela et al., 2017). Rescue of fork collapse by codepleting SMARCAL1 in BRCA2-depleted cells also translates into partial rescue of sensitivity of these cells to HU (Fig. 8 F).
Figure 8.
SMARCAL1-mediated fork reversal is required for accumulation of hyperubiquitinated RPA in BRCA2-deficient cells. (A) Western blot-based analysis of pRPA32 accumulation after disrupting reversed fork formation by codepletion of SMARCAL1 in BRCA2 deficient cells. Experimental conditions used are as described above. Nuclear extracts were prepared. Blot was probed with anti-pRPA32 (S4/S8). (B and C) IF and graphs of pRPA32 (S33) recruitment in U2OS cells transfected with the indicated siRNAs. Cells were treated with 5 mM HU and harvested 4 h after damage. ***, P < 0.005. Statistical significance was determined by the two-tailed Student’s t test and the error bars indicate SD (n = 3). Scale bars in B indicate 10 µm. (D) Immunoprecipitation analysis of RPA ubiquitination in HEK293T cells transfected with indicated siRNAs. Experimental conditions used are as described above. (E) Quantification of IF-based analysis of 53BP1 foci in U2OS cells transfected with indicated siRNAs. Experimental conditions used are as described above. Graph indicates the percentage of cells with >10 53BP1 foci per cell. ***, P < 0.005. (F) CellTiter-Glo–based cell survival assay was used to determine the sensitivity of U2OS cells to HU in cells depleted of BRCA2, SMARCAL1, and/or BRCA2+SMARCAL1. Error bars indicate SD between triplicates. Experimental conditions used are as described above.
RFWD3-dependent RPA ubiquitination affects RAD51 accumulation in BRCA2-depleted cells undergoing replication stress
RAD51 loading after fork stalling is one of the critical steps toward full repair and resolution of the stalled fork (Hashimoto et al., 2010; Kolinjivadi et al., 2017b; Lemaçon et al., 2017; Schlacher et al., 2011; Zellweger et al., 2015). It is not only required for efficient fork reversal (Lemaçon et al., 2017; Zellweger et al., 2015) but is also implicated in blocking MRE11-dependent fork degradation (Hashimoto et al., 2010; Kolinjivadi et al., 2017b; Schlacher et al., 2011). It has been shown that although BRCA2 is not required for RAD51-dependent fork remodeling (Lemaçon et al., 2017; Mijic et al., 2017), it assists in loading RAD51 on the regressed arms of the stalled fork (Mijic et al., 2017).
The results described above raise the question whether codepletion of RFWD3 in BRCA2-depleted cells allowed more efficient RAD51 loading in BRCA2-depleted cells undergoing replication stress. We addressed this question in BRCA2-depleted cells codepleted for RFWD3 and/or overexpressing RPAdel mutant. We find that depletion of RFWD3 or overexpression of RPAdel mutant both led to small but consistent increase in RAD51 accumulation in BRCA2-depleted cells (Fig. S5, G and H). We cannot at this point say that these changes in RAD51 accumulation are indeed happening at the stalled forks; however, these results do raise the possibility that reduced RPA ubiquitination in BRCA2 and RFWD3 codepleted cells could contribute to more efficient removal of RPA from stalled forks and its replacement with RAD51. Results with RPAdel mutant that cannot interact with RFWD3 point to a more direct relationship between RPA ubiquitination and RAD51 loading.
Discussion
This study provides insight into multiple important aspects of stalled fork repair pathway in BRCA1- and BRCA2-deficient cells. First, we identify E3 ligase RFWD3 as a novel modulator of stalled fork stability in BRCA2-deficient cells and propose that forks coated with ubiquitinated RPA are resistant to repair. Second, we propose that rescuing fork degradation is not always sufficient to ensure full repair and recovery in BRCA2-deficient cells and that the extent of ubq-pRPA in the protected forks influences complete fork recovery. We also provide evidence for the hierarchy that exists between BRCA1- and BRCA2-dependent function at stalled replication forks.
We show that BRCA2 loss leads to accumulation of excessive ssDNA coated by phosphorylated and hyperubiquitinated RPA in response to replication stress. RPA ubiquitination in BRCA2-deficient cells is driven by RFWD3. This intermediate impairs fork stability and is in part responsible for increased sensitivity of BRCA2-depleted cells to a fork-stalling agent like HU. We confirm this by showing that codepleting RFWD3 in BRCA2-deficient cells reduces RPA ubiquitination and its accumulation on nascent DNA at stalled forks. Notably, RFWD3 depletion rescues fork degradation, defective fork restart, and fork collapse in these cells.
RFWD3 is clearly an important player in the repair of DNA damage and has roles to play in both HR-driven stalled fork repair and cross-link repair (Elia et al., 2015; Feeney et al., 2017; Inano et al., 2017). However, in this study we made the unexpected finding that RFWD3 can also negatively affect repair under certain circumstances, one of them being BRCA2 deficient cells undergoing replication stress. We propose that in these cells, unchecked RFWD3 activity contributes to fork instability, collapse, and cell death.
We also find that RFWD3-dependent ubiquitination of RPA is in part driven by RPA phosphorylation in BRCA2-depleted cells. These results raise the possibility that RPA phosphorylation could contribute to its increased ubiquitination by RFWD3. RPA and RFWD3 interaction has been shown previously in the context of DNA replication and repair (Feeney et al., 2017; Lin et al., 2018), and this interaction is increased in response to DNA damage (Feeney et al., 2017). It remains to be seen whether HU-induced RPA phosphorylation in BRCA2-depleted cells leads to stronger interaction of RFWD3 with RPA, leading to its increased ubiquitination.
Stalled replication forks frequently undergo fork reversal, which allows forks to be stabilized while the lesion is resolved (Neelsen and Lopes, 2015; Quinet et al., 2017; Sidorova, 2017). It has been shown previously that reversed fork degradation in BRCA2-depleted cells can be rescued by codepletion of SMARCAL1, a fork remodeler, in these cells (Kolinjivadi et al., 2017b; Taglialatela et al., 2017). This suggests that reversed forks are entry points for nucleases like MRE11/CTIP/EXO1 (Lemaçon et al., 2017; Przetocka et al., 2018; Schlacher et al., 2011) to initiate fork degradation in BRCA2-depleted cells. Our data add to this observation and suggest that one of the important fork-destabilizing events in BRCA2-depleted cells could be accumulation of ubiquitinated RPA on ssDNA stretches associated with the reversed forks.
Blocking reversed fork degradation in these cells is emerging as an important means of acquiring chemoresistance (Liao et al., 2018; Mijic et al., 2017; Sidorova, 2017), making it critical for us to understand the mechanisms that drive this resistance. Interestingly, our study suggests that upon loss of MRE11, the stalled forks in BRCA2-depleted cells, although protected, are not conducive to repair and do eventually collapse. This is evident from (1) no change in RPA accumulation at stalled forks upon MRE11 depletion and/or by blocking its nuclease activity by mirin, suggesting no change in aberrant ssDNA accumulation at stalled forks in BRCA2-depleted cells, (2) increased 53BP1 foci formation (i.e., increased fork collapse), and (3) no rescue of sensitivity of BRCA2-depleted cells upon codepletion of MRE11. However, under these conditions (siMRE11 and/or use of mirin to block MRE11 nuclease activity), we do see a near-complete rescue of fork stability as shown before (Lemaçon et al., 2017; Schlacher et al., 2011). These data strongly suggest that MRE11 loss–driven fork protection does not necessarily translate into efficient fork repair and that these forks are still susceptible to fork collapse in BRCA2-depleted cells.
Based on previously published work, there are four possible sources of ssDNA. One possibility (option A) is that the ssDNA we see in BRCA2-depleted cells is present at the reversed fork, specifically the overhangs on the regressed arm. ssDNA at the ends of regressed arms and/or in the reversed forks has been shown before through images captured by electron microscopy. More specifically, Sogo et al. (2002) show that in Saccharomyces cerevisiae rad53 mutant cells, HU treatment leads to accumulation of replication intermediates, including reversed forks with ssDNA arms. Furthermore, Lemaçon et al. (2017) show that blocking MUS81 in BRCA2-deficient cells increases accumulation of ssDNA in the reversed forks.
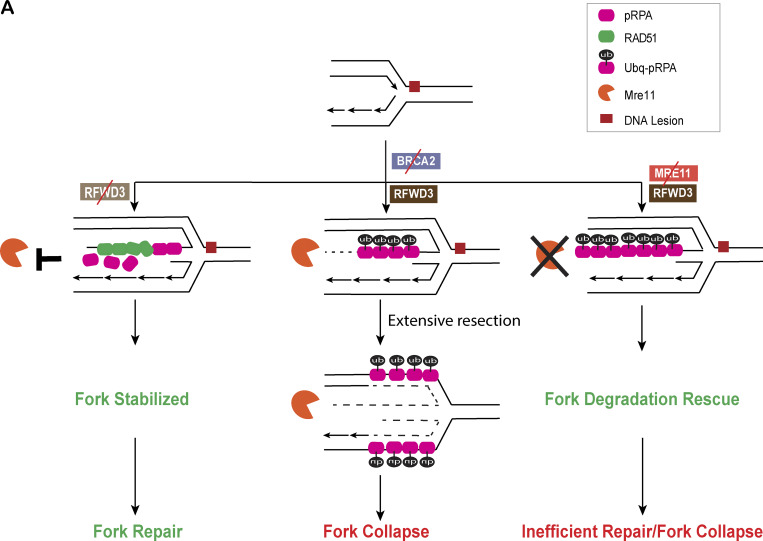
Second possibility (option B) is that ssDNA is present on the three-way junction after fork resection, (Mijic et al., 2017; Bhat et al., 2018). A third possibility (option C) is that it is present at the internal gaps as seen by EM analysis of BRCA2- and RAD51-depleted Xenopus laevis extracts (Hashimoto et al., 2010; Kolinjivadi et al., 2017b). A fourth possibility (option D) is that the source of ssDNA is fork uncoupling wherein the helicase complex uncouples from the polymerase, resulting in ssDNA at a three-way junction (Byun et al., 2005; Cortez, 2005). Given that we see near-complete suppression of pRPA accumulation upon SMARCAL1 codepletion in BRCA2-deficient cells, we propose that the source of ssDNA being coated by pRPA in BRCA2-deficient cells is a reversed fork (option A and/or B). Loss of MRE11 in BRCA2-depleted cells does allow fork stabilization; however, it is possible that this is at the expense of leaving ubiquitinated pRPA32–coated regressed arms of the fork, which might be resistant to repair (Fig. 9 A).
Figure 9.
RFWD3 is a novel modulator of stalled fork stability in BRCA2-deficient cells. (A) Model.
This study raises certain intriguing questions about RPA displacement from the forks and how the reversed forks are protected upon RFWD3 loss. One possibility, which we indicate in our model (Fig. 9 A), is that loss of RFWD3-dependent RPA ubiquitination could lead to better displacement of ssDNA-bound RPA by RAD51, leading to more effective coating of the reversed fork by RAD51. There have been conflicting reports, with one suggesting that RAD51 loading on chromatin after replication stress is BRCA2 dependent (Mijic et al., 2017), while others have shown that it is independent of BRCA2 (Ray Chaudhuri et al., 2016; Tarsounas et al., 2003). Either way, we speculate that loss of RFWD3 and the subsequent loss of ubiquitination of RPA at the reversed forks could assist in more effective loading of RAD51 (presumably in BRCA2-independent manner). In keeping with this model, we do see increased RAD51 loading in BRCA2-deficient cells either codepleted of RFWD3 and/or expressing RPA mutant that cannot get ubiquitinated by RFWD3 (RPAdel). Given that RAD51 loading on reversed forks is known to block MRE11-dependent degradation of reversed forks (Bhat et al., 2018; Kolinjivadi et al., 2017b), such loading of RAD51 at reversed forks would also help ensure that MRE11-dependent degradation of forks is reduced in BRCA2-deficient cells, thus protecting the forks even further.
Finally, we saw that stalled fork intermediates in BRCA1- and BRCA2-deficient cells were different. There are multiple studies that have described differences between BRCA1 and BRCA2 loss–associated phenotypes. While BRCA1 loss leads to an increase in tandem duplications, a form of genomic rearrangement in response to defective stalled fork repair, BRCA2 loss does not (Menghi et al., 2018; Willis et al., 2017). It has also been shown that CTIP-driven (C-terminal binding protein interacting protein) protection of reversed forks is different in BRCA1- and BRCA2-deficient cells (Przetocka et al., 2018). There is also evidence that MUS81-dependent fork rescue is specific to BRCA2-depleted cells and does not occur in BRCA1-depleted cells (Lemaçon et al., 2017). It is not yet clear what drives these differences. Our study is the first one to point to a difference in stalled fork intermediates that accumulate upon depletion of each of these proteins and has looked at cells that are codepleted for both the proteins to get an insight into any hierarchy that might exist in their roles during stalled fork repair.
We cannot rule out that BRCA1 and BRCA2 function independently in different stalled fork repair pathways; however, our results do indicate that BRCA1- and BRCA2-codepleted cells align more closely with phenotypes observed in BRCA1-depleted cells (Fig. 2, A–E; and Fig. 4 B). This raises the possibility that BRCA1 functions upstream of BRCA2. Whether it does so in a linear singular stalled fork repair pathway that both BRCA1 and BRCA2 share or in a common step in two separate pathways that BRCA1 and BRCA2 are a part of remains to be seen.
Interestingly, such a hierarchy between BRCA1 and BRCA2 is reflected in the clinical data that show that in BRCA1/BRCA2 transheterozygotes (defined as a state of heterozygosity at two different loci, which in this case are BRCA1 and BRCA2), it is the BRCA1 heterozygosity that drives the clinical phenotypes (Rebbeck et al., 2016) and not BRCA2 heterozygosity in women who have mutations in both BRCA1 and BRCA2. For example, like BRCA1 mutation carriers, transheterozygotes are more likely to be diagnosed with ovarian cancer, develop cancer at a younger age, and have estrogen receptor–negative breast cancer, different from the clinical phenotype observed in BRCA2 mutation carriers.
This study also raises the interesting possibility that RFWD3 loss will give BRCA2-deficient cells a survival advantage, especially during tumorigenesis. If this were true, there might be a significant co-occurrence of mutations in BRCA2 and RFWD3 in tumor samples. Similarly, given that BRCA1-depleted cells do not benefit from codepletion of RFWD3, one might not detect a similar correlation between RFWD3 and BRCA1. We addressed this question by analyzing datasets in publicly accessible cBioPortal for Cancer Genomics (http://www.cbioportal.org; Cerami et al., 2012; Gao et al., 2013). We looked for evidence of tumor samples that had both BRCA2 and RFWD3 mutations and/or deletions. Supporting our hypothesis, there was a significantly increased chance of co-occurrence of BRCA2 and RFWD3 mutations/deletions in prostate cancer and breast cancer. No such significant co-occurrence of somatic mutations/deletions of BRCA1 and RFWD3 was observed.
In summary, we provide new insights into both BRCA1- and BRCA2-dependent function at the stalled forks, the hierarchy that exists between them, and identify FANC protein RFWD3 (FANCW) as a critical modulator of fork stability in BRCA2-deficient cells. Such mechanistic insights provide us the framework to understand events that drive BRCA2 mutant tumorigenesis and help design effective therapeutic and preventive strategies for individuals carrying BRCA2 mutations.
Materials and methods
Cell lines and cell culture
U2OS and HeLa cells were mainly used for CellTiter-Glo, IF assays, and Western blot. HEK293T cells were mainly used for immunoprecipitation analysis. PEO1 cells were used for cell sensitivity assay. All four cell lines were cultured in DMEM supplemented with 10% of FBS. BOT5641 cells were cultured in RPMI supplemented with 10% FBS.
Plasmids
The His-Ubq, HA-tagged RPAwt, RPAdel, and RPA_K37/38R, RFWD3 plasmid (resistant to siRNA #4) are described previously (Elia et al., 2015). Myc-tagged RPAwt, RPAA, and RPAD have been described previously (Murphy et al., 2014).
IF and antibodies
Cells on coverslips were washed with PBS and fixed in 4% PFA/2% sucrose solution for 15 min. The coverslips were washed again with PBS and then Triton extracted (0.5% Triton X-100 in PBS) for 4 min. Cells were incubated with their respective antibodies for 30 min at 37°C followed by incubation with secondary antibodies (FITC or Rhodamine) for 30 min at 37°C. Primary antibodies used in IF studies were RPA34 (Cal Biochem; NA18; 1:100), 53BP1 (Bethyl; A300-272A; 1:2,000), g-H2AX (Millipore; 05–636; 1:5,000), RPA32 (Thermo Fisher Scientific; PA5-22256; 1:400), pRPAS33 (Sigma; PLA0210-100 μl; 1:1,500), and BRCA1 (Upstate; 07–434; 1:400). Coverslips were mounted using mounting medium (DAPI). Images were acquired with an Axio Imager.M2 (Carl Zeiss) equipped with an Axiocam 506 color camera, controlled by Zen software.
UV irradiation with micropore filters
Cells were irradiated at 30 J/m2 by using a 254-nm UV lamp. The cells were irradiated through a 3-µM micropore membrane (Millipore; TSTP04700) and allowed to recover after irradiation for indicated times at 37°C before being fixed and stained.
CellTiter-Glo
2,000 cells were plated into each well of a transparent 96-well plate in triplicate. After 24 h, cells were treated with different drugs. CellTiter-Glo–based analysis was conducted 7 d after the drug treatment. Each well was washed twice with PBS, and then 60 μl 1:1 CellTiter-Glo Reagents (Promega; G7572)/DMEM was added to each well. Cells were incubated at 37°C for 20 min, and supernatant was then transferred to an opaque 96-well plate. Luminescence was read using a BMG Labtech luminometer.
Fiber assay
Cells were labeled with 25 µM IdU for 20 min, washed five times with PBS, and then treated with 5 mM HU for 3 h. Cells were then labeled with 250 µM CldU for 30 min. For cells that did not undergo HU treatment, CldU was added right after washing off IdU. Cells were harvested and mixed with unlabeled cells at a ratio of 1:3. Mixed cells were lysed and spread on to the slides followed by fixation with acetic acid/methanol (1:3) for 20 min. After denaturation and blocking, DNA tracts were stained with rat anti-CldU (Abcam; ab6326) and mouse anti-IdU (BD Biosciences; 555627) for 2 h at room temperature. DNA tracts were then stained with the secondary antibodies Alexa Fluor 555 goat anti-rat and Alexa Fluor 488 goat anti-mouse for 1 h.
Immunoprecipitation
Cells were transfected with His-tagged ubiquitin and HA-tagged RPA32 or Myc-tagged RPA32 using Lipofectamine 2000. 24 h after transfection, cells were treated with different DNA damage–inducing agents and then harvested. Cell pellets were lysed in Guanidine HCl buffer (6 M Guanidine HCl, 20 mM Tris HCl, pH 8, 0.5 M NaCl, 5% Glycerol, 25 mM Imidazole, pH 8, and dH2O) supplemented with protease inhibitor and phosphatase inhibitor. Cell lysates were then sonicated for 20 s at 30% amplitude twice. Sonicated lysates with 600 μg protein were incubated with Ni-NTA agarose for 3 h at room temperature. Bound complexes were then washed once in Guanidine HCl buffer, supplemented with 0.1% Tween 20, twice with Buffer B (1:4 Guanidine HCl buffer/Buffer C) supplemented with 10 mM N-ethylmaleimide, twice with Buffer C (25 mM Tris HCl, pH 6.8, 150 mM NaCl, 25 mM Imidazole, pH 6.8, 5% glycerol, 0.1% Tween 20, and dH2O) supplemented with 10 mM N-ethylmaleimide. Finally, beads were eluted in 100 μl of 1:1 Buffer C/SDS-sample buffer and then boiled.
iPOND
The iPOND experiment was performed based on a protocol described in Sirbu et al. (2012). Briefly, at 48 h after siRNA transfection, HEK293T cells were incubated with 10 µM EdU (5-ethynyl-2´-deoxyuridine) for 10 min at 37°C and harvested immediately or after 3 h treatment with 5 mM HU. Immediately fixed the cells with 1% formaldehyde in PBS for 20 min at room temperature. Quench of the cross-linking reaction by adding 1.25 M glycine. Cells were then collected and washed three times with PBS. The samples were flash frozen and stored at −80°C. The next day, cell pellets were resuspended in permeabilization buffer (0.25% Triton X-100 in PBS) at a concentration of 107 cells/ml and incubated at room temperature for 30 min. Cells were then washed at 4°C with 0.5% BSA/PBS followed by a one-time wash with PBS. Cells were then incubated in the click or no-click reaction cocktail (based on Sirbu et al.’s protocol [Sirbu et al., 2012]) for 2 h at room temperature. After washing once with 0.5% BSA/PBS and PBS alone, cells were resuspended in lysis buffer containing aprotinin and leupeptin. The cell lysates were sonicated by using Bioruptor (company and catalog number) with 25 cycles, on high, of 30 s on and 30 s off. The supernatant was then diluted 1:1 (vol/vol) in PBS containing aprotinin and leupeptin. 15 µl of lysate was saved as input sample. 15 µl of 2× SDS laemmli sample buffer (SB) was added to the input sample and stored at −80°C. The remaining lysate was incubated with magnetic streptavidin Dynabeads (Thermo Fisher Scientific; #65305) overnight at 4°C. After washing for 5 min each with cold lysis buffer,1 M NaCl, and twice more with lysis buffer, the beads were supplemented with 1:1 (vol/vol) 2× SB. The input and captured samples were incubated at 95°C for 25 min before Western blot analysis.
Cell cycle analysis
Cell cycle analysis was done using the Muse Cell Cycle Kit (#MCH100106). After 48 h of siRNA transfection, cells were fixed in 1 ml of 70% ethanol at 4°C overnight. The next day, ethanol was removed by centrifugation, and cells were washed once with PBS. Then cell pellets were incubated with 200 µl of Muse Cell Cycle Reagent at room temperature for 30 min. The samples were analyzed on the Muse Cell Analyzer (#0500-3115). Muse Cell Cycle Kit uses the nuclear DNA stain propidium iodide to discriminate cells at different stage of the cell cycle.
BrdU assay for ssDNA detection
After 24 h of siRNA transfection, cells were loaded onto coverslips and allowed to attach for 24 h. 50 µM BrdU (BD Biosciences; #517581KZ) was then added for 24 h. After incubating cells for 4 h with 5 mM HU, cells were fixed with cold 100% methanol for 30 min at −20°C and then quickly rinsed with cold acetone. After washing four times with PBS, cells were immunostained with BrdU antibody (BD Biosciences; # BDB347580) at 37°C for 1 h. Control samples were included for quantification of cells that incorporated BrdU by treating cells with 1 M HCl for 10 min before blocking and processing them for staining as the −HCl samples.
HR assay
U2OS cells with a stably integrated direct-repeat GFP reporter (Moynahan et al., 2001) were transfected with indicated siRNA and then were transfected with HA-tagged I-SceI–expressing plasmid. 48 h later, cells were collected, and the percentage of GFP-positive cells was then detected by flow cytometry.
Analysis of co-occurrence of BRCA2 and RFWD3 mutations
Co-occurrence of BRCA2 and RFWD3 was investigated in prostate, breast, and ovarian cancer using the cBioPortal (Cerami et al., 2012; Gao et al., 2013), accessed 2019/08/12. Datasets were queried using the keywords: “BRCA2: MUT_DRIVER HOMDEL” and “RFWD3: MUT_DRIVER HOMDEL”. Prostate datasets included “Metastatic Prostate Adenocarcinoma (MCTP, Nature 2012)”, “Metastatic Prostate Adenocarcinoma (SU2C/PCF Dream Team, PNAS 2019)”, “Metastatic Prostate Cancer (SU2C/PCF Dream Team, Cell 2015)”, “Prostate Adenocarcinoma (Broad/Cornell, Cell 2013)”, “Prostate Adenocarcinoma (Broad/Cornell, Nat Genet 2012)”, “Prostate Adenocarcinoma (CPC-GENE, Nature 2017)”, “Prostate Adenocarcinoma (Fred Hutchinson CRC, Nat Med 2016)”, “Prostate Adenocarcinoma (MSKCC, Cancer Cell 2010)”, “Prostate Adenocarcinoma (MSKCC, PNAS 2014)”, “Prostate Adenocarcinoma (MSKCC/DFCI, Nature Genetics 2018)”, “Prostate Adenocarcinoma (SMMU, Eur Urol 2017)”, “Prostate Cancer (MSK, 2019)”, “Prostate Cancer (MSKCC, JCO Precis Oncol 2017)”, “The Metastatic Prostate Cancer Project (Provisional, December 2018)”, and “Prostate Adenocarcinoma (TCGA, PanCancer Atlas)”. Breast cancer datasets included “Breast Cancer (METABRIC, Nature 2012 & Nat Commun 2016)”, “Breast Cancer (MSK, Cancer Cell 2018)”, “Breast Invasive Carcinoma (British Columbia, Nature 2012)”, “Breast Invasive Carcinoma (Broad, Nature 2012)”, “Breast Invasive Carcinoma (Sanger, Nature 2012)”, “Metastatic Breast Cancer (INSERM, PLoS Med 2016)”, “The Metastatic Breast Cancer Project (Provisional, October 2018)”, and “Breast Invasive Carcinoma (TCGA, PanCancer Atlas)”. Ovarian cancer datasets included “Ovarian Serous Cystadenocarcinoma (TCGA, Nature 2011)”. The number of prostate cancer samples with respectively no events, BRCA2, RFWD3, and both: 3,010, 141, 51, 10. The number of breast cancer samples with respectively no events, BRCA2, RFWD3, and both was 3,290, 57, 18, and 2. The number of ovarian cancer samples with respectively no events, BRCA2, RFWD3, and both was 282, 29, 4, and 1. Co-occurrence was assessed with a two-sided Fisher’s exact test. A similar analysis was done to determine the co-occurrence of BRCA1 and RFWD3 events as well.
siRNA
For siRNA experiments, cells were seeded in a 6-well plate and transfected with 60 pmol siRNA with RNAiMAX (Invitrogen), followed by changing medium the next day. siRNA was purchased from Dharmacon, and the siRNA sequences were as follows: siLuc, 5′-CGUACGCGGAAUACUUCGAUU-3′; siBRCA1#1, 5′-CAACAUGCCCACAGAUCAAUU-3′; siBRCA1#3, 5′-CAGCUACCCUUCCAUCAUAUU-3′; siBRCA2#8, 5′-UAAGGAACGUCAAGAGAUAUU-3′; siBRCA2#5, 5′-GAAACGGACUUGCUAUUUAUU-3′; siBRCA2#6, 5′-GGUAUCAGAUGCUUCAUUA-3′; siRFWD3#2, 5′-GGAAACAGGCCGAGUUAGAUU-3′; siRFWD3#4, 5′-GGACCUACUUGCAAACUAUUU-3′; siMRE11, 5′-GCUAAUGACUCUGAUGAUAUU-3′; siRAD51, 5′-GAGCUUGACAAACUACUUCUU-3′; siSMARCAL1, 5′-GAAUCUCACUUCCUCAAAAUU-3′; and siGAPDH, 5′-UGGUUUACAUGUUCCAAUA-3′; siBRCA2#8, siBRCA1#1, and siRFWD3#2 were used if not indicated in the figure.
Immunoblotting and antibodies
Whole-cell extracts were prepared by lysing cells in NETN450 lysis buffer (450 mM NaCl, 20 mM Tris-HCl, pH 7.8, 0.5% NP-40, 1 mM EDTA, and dH2O). For nuclear extracts, cells were lysed in Protein Extraction (PEB; 0.5% Triton X, 20 mM Hepes, pH 7, 100 mM NaCl, 3 mM MgCl2, 300 mM sucrose, and dH2O) on ice for 20 min followed by spinning at 5,000 rpm for 10 min to remove the cytoplasmic extract. Cell pellets were washed once with PBS followed by lysing in NETN 400 lysis buffer (400 mM NaCl, 20 mM Tris-HCl, pH 7.8, 0.5% NP-40, 1 mM EDTA, and dH2O) for 1 h at 4°C to generate the nuclear extract. All lysis buffers were supplemented with protease inhibitor and phosphatase inhibitor. Antibodies used for Western blot were RFWD3 (Bethyl; A301-397A; 1:2,500), BRCA2 (Bethyl; A300-005A; 1:3,000), SD118 (Calbiochem; OP107; 1:2,500), GAPDH (Santa Cruz; SC-25778; 1:4,000), GAPDH (BioLegend; 919501; 1:4,000), RAD51 (Santa Cruz; SC-8349; 1:2,500), HA (BioLegend; 901514; 1:3,000), LaminB1 (Cell Signaling; 12596; 1:3,000), pRPA32 S4/S8 (Bethyl; A300-245A; 1:2,500), pRPA S33 (Sigma; PLA0210; 1:2,500), RPA34 (Calbiochem; NA18; 1:3,000), α-Tubulin (Santa Cruz; SC-5286; 1:3,000), Mre11 (Genetex; GTX70212; 1:3,000), and SMARCAL1 (Bethyl; A301-616A; 1:2,500).
Online supplemental material
Fig. S1 shows increased sensitivity of BRCA2-depleted cells to HU in support of data presented in Fig. 1 and provides evidence for increased ssDNA in BRCA2-depleted cells. Fig. S2 shows that BRCA1 may function upstream of BRCA2 in the stalled fork repair pathway and addresses the relationship between RPA phosphorylation and ubiquitination by studying the ubiquitination status of various RPA mutants. Fig. S3 shows that hyperubiquitination of RPA after BRCA2 depletion is performed by E3 ligase RFWD3 and shows input samples from iPOND-based analysis in Fig. 5 and cell cycle analysis of RFWD3-depleted and BRCA2/RFWD3 codepleted cells. Fig. S4 shows that RFWD3 depletion rescues fork degradation, fork collapse, and cell sensitivity to stalled fork–inducing agents in BRCA2-depleted cells and shows that there is no rescue of sensitivity to HU upon codepletion of RFWD3 in BRCA1-deficient cells. Fig. S5 shows that generation of ubq-pRPA–coated ssDNA intermediate in BRCA2-deficient cells is not dependent on MRE11-driven fork resection and also shows that SMARCAL1-mediated fork reversal is required for accumulation of hyperubiquitinated RPA coated ssDNA in BRCA2-deficient cells.
Acknowledgments
We thank Dr. David Livingston, Dr. Jill Macoska, and Dr. Stephen Godin for helpful discussion and comments on the manuscript. We also thank Tiego Da’ Silva for help with making and purifying plasmid DNA and Kimberly Toomire for help with setting up Western blot experiments. We thank Dr. James Borowiec (NYU Langone Medical Center, New York, NY) for sharing the RPAwt, RPAA, and RPAD plasmids with us.
This work was supported by the Breast Cancer Research Foundation (to J.E. Garber) and a U54 DF/HCC (Dana Farber/Harvard Cancer Center) pilot grant (to S. Pathania).
The authors declare no competing financial interests.
Author contributions: H. Duan and S. Pathania conceived the study and designed the experiments. H. Duan carried out all the experiments and analyzed the data along with S. Pathania. Some Western blots, IF, and cell culture–based experiments were carried out by S. Mansour, R. Reed, M.K. Gillis, and B. Parent under the supervision of H. Duan. B. Liu carried out the initial his-ubiquitination assays to confirm increased RPA ubiquitination in BRCA2-deficient cells. cBioPortal-based statistical analysis was carried out by N. Birkbak, Z. Sztupinskzki, and Z. Szallasi. S. Pathania and H. Duan discussed and interpreted the data with A.E.H. Elia and J.E. Garber. J.E. Garber also provided the patient samples to derive BRCA2 mutant breast tumor lines. The manuscript was written by S. Pathania, and H. Duan contributed to manuscript preparation.
References
- Bhat K.P., Krishnamoorthy A., Dungrawala H., Garcin E.B., Modesti M., and Cortez D.. 2018. RADX Modulates RAD51 Activity to Control Replication Fork Protection. Cell Rep. 24:538–545. 10.1016/j.celrep.2018.06.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., and Foiani M.. 2010. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11:208–219. 10.1038/nrm2852 [DOI] [PubMed] [Google Scholar]
- Bunting S.F., Callén E., Kozak M.L., Kim J.M., Wong N., López-Contreras A.J., Ludwig T., Baer R., Faryabi R.B., Malhowski A., et al. 2012. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol. Cell. 46:125–135. 10.1016/j.molcel.2012.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun T.S., Pacek M., Yee M.-C., Walter J.C., and Cimprich K.A.. 2005. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19:1040–1052. 10.1101/gad.1301205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro E., Goh C., Olmos D., Saunders E., Leongamornlert D., Tymrakiewicz M., Mahmud N., Dadaev T., Govindasami K., Guy M., et al. 2013. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J. Clin. Oncol. 31:1748–1757. 10.1200/JCO.2012.43.1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2:401–404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A., and Elledge S.J.. 2010. The DNA damage response: making it safe to play with knives. Mol. Cell. 40:179–204. 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipak L., Watanabe N., and Bessho T.. 2006. The role of BRCA2 in replication-coupled DNA interstrand cross-link repair in vitro. Nat. Struct. Mol. Biol. 13:729–733. 10.1038/nsmb1120 [DOI] [PubMed] [Google Scholar]
- Cortez D. 2005. Unwind and slow down: checkpoint activation by helicase and polymerase uncoupling. Genes Dev. 19:1007–1012. 10.1101/gad.1316905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois J.C., Yates M., Gaudreau-Lapierre A., Clément G., Cappadocia L., Gaudreau L., Zou L., and Maréchal A.. 2017. A phosphorylation-and-ubiquitylation circuitry driving ATR activation and homologous recombination. Nucleic Acids Res. 45:8859–8872. 10.1093/nar/gkx571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupré A., Boyer-Chatenet L., Sattler R.M., Modi A.P., Lee J.H., Nicolette M.L., Kopelovich L., Jasin M., Baer R., Paull T.T., et al. 2008. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat. Chem. Biol. 4:119–125. 10.1038/nchembio.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia A.E.H., Wang D.C., Willis N.A., Boardman A.P., Hajdu I., Adeyemi R.O., Lowry E., Gygi S.P., Scully R., and Elledge S.J.. 2015. RFWD3-Dependent Ubiquitination of RPA Regulates Repair at Stalled Replication Forks. Mol. Cell. 60:280–293. 10.1016/j.molcel.2015.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeney L., Muñoz I.M., Lachaud C., Toth R., Appleton P.L., Schindler D., and Rouse J.. 2017. RPA-Mediated Recruitment of the E3 Ligase RFWD3 Is Vital for Interstrand Crosslink Repair and Human Health. Mol. Cell. 66:610–621.e4. 10.1016/j.molcel.2017.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard H., García-Muse T., and Aguilera A.. 2015. Replication stress and cancer. Nat. Rev. Cancer. 15:276–289. 10.1038/nrc3916 [DOI] [PubMed] [Google Scholar]
- Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6:pl1 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., and Chen J.. 2011. E3 ligase RFWD3 participates in replication checkpoint control. J. Biol. Chem. 286:22308–22313. 10.1074/jbc.M111.222869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding S.M., and Bristow R.G.. 2012. Discordance between phosphorylation and recruitment of 53BP1 in response to DNA double-strand breaks. Cell Cycle. 11:1432–1444. 10.4161/cc.19824 [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Ray Chaudhuri A., Lopes M., and Costanzo V.. 2010. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 17:1305–1311. 10.1038/nsmb.1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inano S., Sato K., Katsuki Y., Kobayashi W., Tanaka H., Nakajima K., Nakada S., Miyoshi H., Knies K., Takaori-Kondo A., et al. 2017. RFWD3-Mediated Ubiquitination Promotes Timely Removal of Both RPA and RAD51 from DNA Damage Sites to Facilitate Homologous Recombination. Mol. Cell. 66:622–634.e8. 10.1016/j.molcel.2017.04.022 [DOI] [PubMed] [Google Scholar]
- Jensen R.B., Carreira A., and Kowalczykowski S.C.. 2010. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 467:678–683. 10.1038/nature09399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knies K., Inano S., Ramírez M.J., Ishiai M., Surrallés J., Takata M., and Schindler D.. 2017. Biallelic mutations in the ubiquitin ligase RFWD3 cause Fanconi anemia. J. Clin. Invest. 127:3013–3027. 10.1172/JCI92069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolinjivadi A.M., Sannino V., de Antoni A., Técher H., Baldi G., and Costanzo V.. 2017a Moonlighting at replication forks - a new life for homologous recombination proteins BRCA1, BRCA2 and RAD51. FEBS Lett. 591:1083–1100. 10.1002/1873-3468.12556 [DOI] [PubMed] [Google Scholar]
- Kolinjivadi A.M., Sannino V., De Antoni A., Zadorozhny K., Kilkenny M., Técher H., Baldi G., Shen R., Ciccia A., Pellegrini L., et al. 2017b Smarcal1-Mediated Fork Reversal Triggers Mre11-Dependent Degradation of Nascent DNA in the Absence of Brca2 and Stable Rad51 Nucleofilaments. Mol. Cell. 67:867–881.e7. 10.1016/j.molcel.2017.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchenbaecker K.B., Hopper J.L., Barnes D.R., Phillips K.A., Mooij T.M., Roos-Blom M.J., Jervis S., van Leeuwen F.E., Milne R.L., Andrieu N., et al. ; BRCA1 and BRCA2 Cohort Consortium . 2017. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA. 317:2402–2416. 10.1001/jama.2017.7112 [DOI] [PubMed] [Google Scholar]
- Lemaçon D., Jackson J., Quinet A., Brickner J.R., Li S., Yazinski S., You Z., Ira G., Zou L., Mosammaparast N., et al. 2017. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat. Commun. 8:860 10.1038/s41467-017-01180-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H., Ji F., Helleday T., and Ying S.. 2018. Mechanisms for stalled replication fork stabilization: new targets for synthetic lethality strategies in cancer treatments. EMBO Rep. 19:19 10.15252/embr.201846263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.C., Wang Y., Hsu R., Giri S., Wopat S., Arif M.K., Chakraborty A., Prasanth K.V., and Prasanth S.G.. 2018. PCNA-mediated stabilization of E3 ligase RFWD3 at the replication fork is essential for DNA replication. Proc. Natl. Acad. Sci. USA. 115:13282–13287. 10.1073/pnas.1814521115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Chu J., Yucer N., Leng M., Wang S.-Y., Chen B.P.C., Hittelman W.N., and Wang Y.. 2011. RING finger and WD repeat domain 3 (RFWD3) associates with replication protein A (RPA) and facilitates RPA-mediated DNA damage response. J. Biol. Chem. 286:22314–22322. 10.1074/jbc.M111.222802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long D.T., Joukov V., Budzowska M., and Walter J.C.. 2014. BRCA1 promotes unloading of the CMG helicase from a stalled DNA replication fork. Mol. Cell. 56:174–185. 10.1016/j.molcel.2014.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C.J., and Ashworth A.. 2016. BRCAness revisited. Nat. Rev. Cancer. 16:110–120. 10.1038/nrc.2015.21 [DOI] [PubMed] [Google Scholar]
- Macheret M., and Halazonetis T.D.. 2015. DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol. 10:425–448. 10.1146/annurev-pathol-012414-040424 [DOI] [PubMed] [Google Scholar]
- Maréchal A., and Zou L.. 2015. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 25:9–23. 10.1038/cr.2014.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menghi F., Barthel F.P., Yadav V., Tang M., Ji B., Tang Z., Carter G.W., Ruan Y., Scully R., Verhaak R.G.W., et al. 2018. The Tandem Duplicator Phenotype Is a Prevalent Genome-Wide Cancer Configuration Driven by Distinct Gene Mutations. Cancer Cell. 34:197–210.e5. 10.1016/j.ccell.2018.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., Grompone G., Florès M.-J., and Bidnenko V.. 2004. Multiple pathways process stalled replication forks. Proc. Natl. Acad. Sci. USA. 101:12783–12788. 10.1073/pnas.0401586101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijic S., Zellweger R., Chappidi N., Berti M., Jacobs K., Mutreja K., Ursich S., Ray Chaudhuri A., Nussenzweig A., Janscak P., et al. 2017. Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat. Commun. 8:859 10.1038/s41467-017-01164-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocci E., Milne R.L., Méndez-Villamil E.Y., Hopper J.L., John E.M., Andrulis I.L., Chung W.K., Daly M., Buys S.S., Malats N., et al. 2013. Risk of pancreatic cancer in breast cancer families from the breast cancer family registry. Cancer Epidemiol. Biomarkers Prev. 22:803–811. 10.1158/1055-9965.EPI-12-0195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan M.E., Pierce A.J., and Jasin M.. 2001. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell. 7:263–272. 10.1016/S1097-2765(01)00174-5 [DOI] [PubMed] [Google Scholar]
- Murphy A.K., Fitzgerald M., Ro T., Kim J.H., Rabinowitsch A.I., Chowdhury D., Schildkraut C.L., and Borowiec J.A.. 2014. Phosphorylated RPA recruits PALB2 to stalled DNA replication forks to facilitate fork recovery. J. Cell Biol. 206:493–507. 10.1083/jcb.201404111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narod S.A., and Foulkes W.D.. 2004. BRCA1 and BRCA2: 1994 and beyond. Nat. Rev. Cancer. 4:665–676. 10.1038/nrc1431 [DOI] [PubMed] [Google Scholar]
- Neelsen K.J., and Lopes M.. 2015. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat. Rev. Mol. Cell Biol. 16:207–220. 10.1038/nrm3935 [DOI] [PubMed] [Google Scholar]
- O’Donovan P.J., and Livingston D.M.. 2010. BRCA1 and BRCA2: breast/ovarian cancer susceptibility gene products and participants in DNA double-strand break repair. Carcinogenesis. 31:961–967. 10.1093/carcin/bgq069 [DOI] [PubMed] [Google Scholar]
- Pathania S., Nguyen J., Hill S.J., Scully R., Adelmant G.O., Marto J.A., Feunteun J., and Livingston D.M.. 2011. BRCA1 is required for postreplication repair after UV-induced DNA damage. Mol. Cell. 44:235–251. 10.1016/j.molcel.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathania S., Bade S., Le Guillou M., Burke K., Reed R., Bowman-Colin C., Su Y., Ting D.T., Polyak K., Richardson A.L., et al. 2014. BRCA1 haploinsufficiency for replication stress suppression in primary cells. Nat. Commun. 5:5496 10.1038/ncomms6496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przetocka S., Porro A., Bolck H.A., Walker C., Lezaja A., Trenner A., von Aesch C., Himmels S.F., D’Andrea A.D., Ceccaldi R., et al. 2018. CtIP-Mediated Fork Protection Synergizes with BRCA1 to Suppress Genomic Instability upon DNA Replication Stress. Mol. Cell. 72:568–582.e6. 10.1016/j.molcel.2018.09.014 [DOI] [PubMed] [Google Scholar]
- Quinet A., Lemaçon D., and Vindigni A.. 2017. Replication Fork Reversal: Players and Guardians. Mol. Cell. 68:830–833. 10.1016/j.molcel.2017.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A., Callen E., Ding X., Gogola E., Duarte A.A., Lee J.E., Wong N., Lafarga V., Calvo J.A., Panzarino N.J., et al. 2016. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 535:382–387. 10.1038/nature18325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbeck T.R., and Domchek S.M.. 2008. Variation in breast cancer risk in BRCA1 and BRCA2 mutation carriers. Breast Cancer Res. 10:108 10.1186/bcr2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbeck T.R., Friebel T.M., Mitra N., Wan F., Chen S., Andrulis I.L., Apostolou P., Arnold N., Arun B.K., Barrowdale D., et al. ; HEBON . 2016. Inheritance of deleterious mutations at both BRCA1 and BRCA2 in an international sample of 32,295 women. Breast Cancer Res. 18:112 10.1186/s13058-016-0768-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R., Chun J., and Powell S.N.. 2011. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer. 12:68–78. 10.1038/nrc3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbi C.P., and Milner J.. 2001. Analysis of nucleotide excision repair by detection of single-stranded DNA transients. Carcinogenesis. 22:1789–1796. 10.1093/carcin/22.11.1789 [DOI] [PubMed] [Google Scholar]
- Sawyer S.L., Tian L., Kähkönen M., Schwartzentruber J., Kircher M., Majewski J., Dyment D.A., Innes A.M., Boycott K.M., Moreau L.A., et al. ; FORGE Canada Consortium . 2015. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov. 5:135–142. 10.1158/2159-8290.CD-14-1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K., Christ N., Siaud N., Egashira A., Wu H., and Jasin M.. 2011. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 145:529–542. 10.1016/j.cell.2011.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K., Wu H., and Jasin M.. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 22:106–116. 10.1016/j.ccr.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova J. 2017. A game of substrates: replication fork remodeling and its roles in genome stability and chemo-resistance. Cell Stress. 1:115–133. 10.15698/cst2017.12.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu B.M., Couch F.B., Feigerle J.T., Bhaskara S., Hiebert S.W., and Cortez D.. 2011. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 25:1320–1327. 10.1101/gad.2053211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu B.M., Couch F.B., and Cortez D.. 2012. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat. Protoc. 7:594–605. 10.1038/nprot.2012.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo J.M., Lopes M., and Foiani M.. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 297:599–602. 10.1126/science.1074023 [DOI] [PubMed] [Google Scholar]
- Somyajit K., Saxena S., Babu S., Mishra A., and Nagaraju G.. 2015. Mammalian RAD51 paralogs protect nascent DNA at stalled forks and mediate replication restart. Nucleic Acids Res. 43:9835–9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou S.K., Kamileri I., Lugli N., Evangelou K., Da-Ré C., Huber F., Padayachy L., Tardy S., Nicati N.L., Barriot S., et al. 2016. Mammalian RAD52 Functions in Break-Induced Replication Repair of Collapsed DNA Replication Forks. Mol. Cell. 64:1127–1134. 10.1016/j.molcel.2016.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G., and Elledge S.J.. 2002. The two faces of BRCA2, a FANCtastic discovery. Mol. Cell. 10:2–4. 10.1016/S1097-2765(02)00580-4 [DOI] [PubMed] [Google Scholar]
- Stordal B., Timms K., Farrelly A., Gallagher D., Busschots S., Renaud M., Thery J., Williams D., Potter J., Tran T., et al. 2013. BRCA1/2 mutation analysis in 41 ovarian cell lines reveals only one functionally deleterious BRCA1 mutation. Mol. Oncol. 7:567–579. 10.1016/j.molonc.2012.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taglialatela A., Alvarez S., Leuzzi G., Sannino V., Ranjha L., Huang J.W., Madubata C., Anand R., Levy B., Rabadan R., et al. 2017. Restoration of Replication Fork Stability in BRCA1- and BRCA2-Deficient Cells by Inactivation of SNF2-Family Fork Remodelers. Mol. Cell. 68:414–430.e8. 10.1016/j.molcel.2017.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarsounas M., Davies D., and West S.C.. 2003. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene. 22:1115–1123. 10.1038/sj.onc.1206263 [DOI] [PubMed] [Google Scholar]
- Tian F., Sharma S., Zou J., Lin S.Y., Wang B., Rezvani K., Wang H., Parvin J.D., Ludwig T., Canman C.E., et al. 2013. BRCA1 promotes the ubiquitination of PCNA and recruitment of translesion polymerases in response to replication blockade. Proc. Natl. Acad. Sci. USA. 110:13558–13563. 10.1073/pnas.1306534110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassin V.M., Wold M.S., and Borowiec J.A.. 2004. Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol. Cell. Biol. 24:1930–1943. 10.1128/MCB.24.5.1930-1943.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T., and King M.-C.. 2007. Ten genes for inherited breast cancer. Cancer Cell. 11:103–105. 10.1016/j.ccr.2007.01.010 [DOI] [PubMed] [Google Scholar]
- Welcsh P.L., and King M.-C.. 2001. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum. Mol. Genet. 10:705–713. 10.1093/hmg/10.7.705 [DOI] [PubMed] [Google Scholar]
- Willis N.A., Chandramouly G., Huang B., Kwok A., Follonier C., Deng C., and Scully R.. 2014. BRCA1 controls homologous recombination at Tus/Ter-stalled mammalian replication forks. Nature. 510:556–559. 10.1038/nature13295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis N.A., Frock R.L., Menghi F., Duffey E.E., Panday A., Camacho V., Hasty E.P., Liu E.T., Alt F.W., and Scully R.. 2017. Mechanism of tandem duplication formation in BRCA1-mutant cells. Nature. 551:590–595. 10.1038/nature24477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying S., Hamdy F.C., and Helleday T.. 2012. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res. 72:2814–2821. 10.1158/0008-5472.CAN-11-3417 [DOI] [PubMed] [Google Scholar]
- Zellweger R., Dalcher D., Mutreja K., Berti M., Schmid J.A., Herrador R., Vindigni A., and Lopes M.. 2015. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 208:563–579. 10.1083/jcb.201406099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman M.K., and Cimprich K.A.. 2014. Causes and consequences of replication stress. Nat. Cell Biol. 16:2–9. 10.1038/ncb2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., and Elledge S.J.. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 300:1542–1548. 10.1126/science.1083430 [DOI] [PubMed] [Google Scholar]