The TGN is a central compartment for protein sorting and secretion. Hecht et al. demonstrate that Cab45, a Ca2+-dependent sorting protein in the TGN, is phosphorylated by the Golgi kinase Fam20C. Phosphorylation of Cab45 decreases its TGN retention and increases the export of the Cab45 client LyzC.
Abstract
The TGN is a key compartment for the sorting and secretion of newly synthesized proteins. At the TGN, soluble proteins are sorted based on the instructions carried in their oligosaccharide backbones or by a Ca2+-mediated process that involves the cargo-sorting protein Cab45. Here, we show that Cab45 is phosphorylated by the Golgi-specific protein kinase Fam20C. Mimicking of phosphorylation translocates Cab45 into TGN-derived vesicles, which goes along with an increased export of LyzC, a Cab45 client. Our findings demonstrate that Fam20C plays a key role in the export of Cab45 clients by fine-tuning Cab45 oligomerization and thus impacts Cab45 retention in the TGN.
Introduction
The Golgi apparatus is the main sorting hub of the protein secretory pathway within cells. Much of this activity occurs in the most distal cisternae of the Golgi, known as the TGN (Chege and Pfeffer, 1990; Gleeson et al., 2004; Klumperman, 2011; De Matteis and Luini, 2008; Munro, 2005).
Over recent decades, studies have elucidated the mechanisms by which sorting takes place at the TGN to explain the trafficking of transmembrane proteins (Fölsch et al., 1999; 2001; Fölsch, 2005, 2008; Munro, 1995; Welch and Munro, 2019) and the transport of lysosomal hydrolases to endosomes and lysosomes (Mellman and Nelson, 2008). A process fundamental to all sorting events is the congregation of cargo molecules in the TGN, where they interact with cytosolic coat complexes that initiate the formation and budding of vesicles (Ang and Fölsch, 2012; Bonifacino, 2014; Guo et al., 2014; Traub and Bonifacino, 2013). However, many soluble secreted molecules contain neither a transmembrane domain nor a recognition motif for known cargo receptors, which poses a challenge as to how these molecules are sorted and trafficked (Kienzle and von Blume, 2014; Pakdel and von Blume, 2018).
We have previously described a novel sorting mechanism that explains the sorting of certain soluble secreted molecules. In this, secretory pathway Ca2+ ATPase 1 (SPCA1), a TGN-specific calcium ion (Ca2+) ATPase, interacts with cofilin1 and F-actin at its cytosolic interface, promoting Ca2+ influx into the lumen of the TGN (von Blume et al., 2009, 2011, 2012; Kienzle et al., 2014; Pizzo et al., 2010). As a result of this local Ca2+ increase, the Ca2+-binding protein calcium-binding protein 45 kD (Cab45) oligomerizes and binds secretory cargoes (clients), such as lysozyme C (LyzC), thereby segregating them from the bulk milieu of the TGN lumen (Blank and von Blume, 2017; Crevenna et al., 2016). Cab45–client complexes are then sorted into specific sphingomyelin (SM)–rich vesicles and transported to the plasma membrane for secretion (Deng et al., 2018). Other factors that influence the sorting of the Cab45–client complexes into SM-rich vesicles remain unknown.
Family with sequence similarity 20 member C (Fam20C) is a recently discovered serine/threonine kinase found in the Golgi apparatus, which phosphorylates >100 secreted substrates within the secretory pathway (Tagliabracci et al., 2012, 2013, 2015). Interestingly, many of these are Ca2+-binding and secreted proteins (Tagliabracci et al., 2015).
This study analyzes the influence of Fam20C on the SPCA1/Cab45 sorting machinery. We show that Fam20C phosphorylates Cab45 on distinct residues and thereby decreases Cab45 retention in the TGN. In this regard, our data present evidence that phosphorylation fine-tunes the oligomerization-dependent sorting process without modulating the general Ca2+-binding ability of Cab45. Moreover, phosphorylation of Cab45 drives the sorting of Cab45-client LyzC into SM-rich vesicles, leading to enhanced secretion of the cargo. Overall we propose that Fam20C regulates Cab45-dependent client sorting by modulating its release into vesicles at the TGN.
Results
Depletion of Fam20C impairs secretion of LyzC
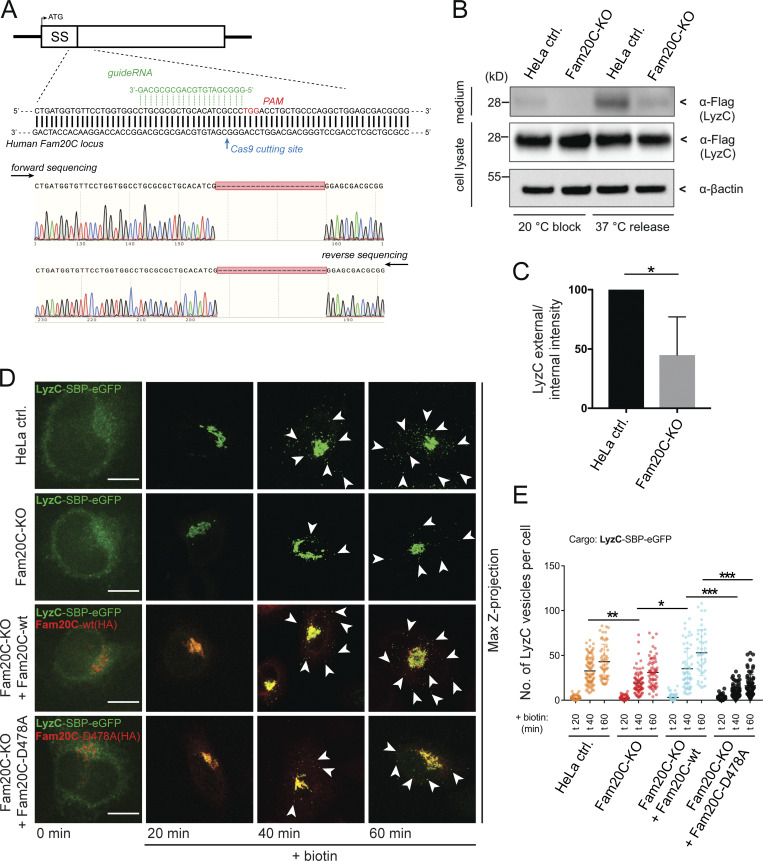
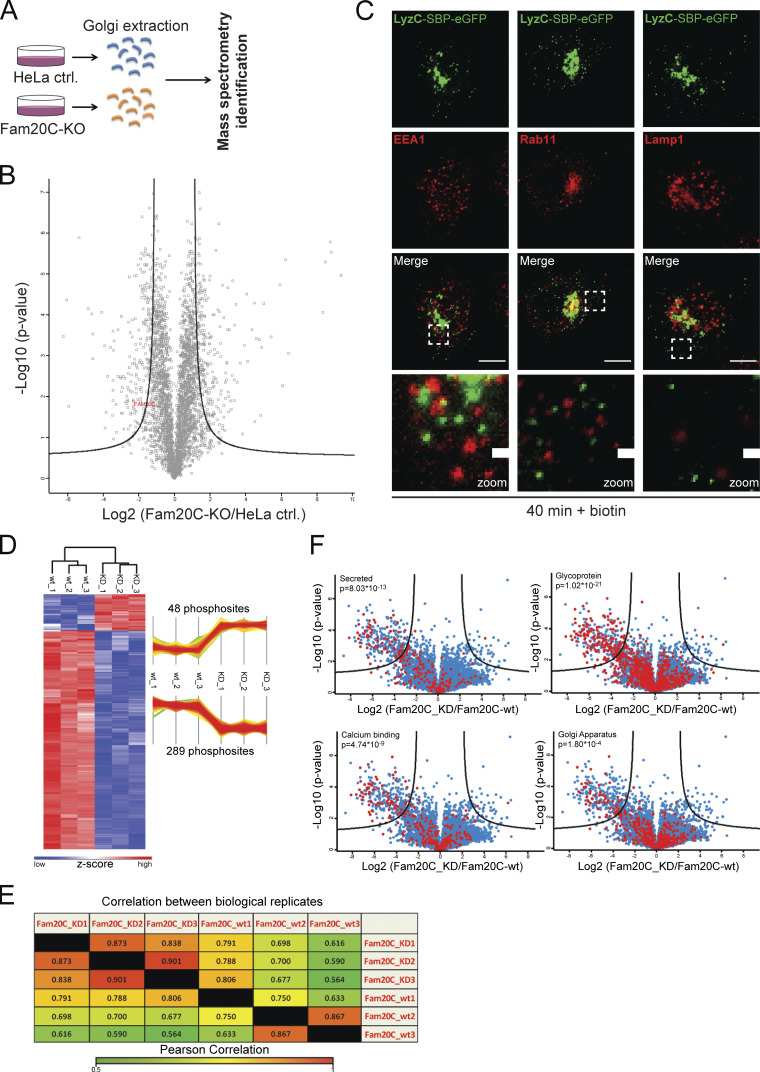
It has previously been shown that the majority of Fam20C substrates are secreted proteins (Tagliabracci et al., 2015); however, whether the kinase has a directing role in cargo secretion has not yet been investigated. To address if Fam20C plays a role in Cab45-dependent cargo sorting, a Fam20C knockout (KO) cell line was generated using CRISPR/Cas9 technology (Cong et al., 2013). The sequencing of a clone (Fig. 1 A) detected the deletion of 22 bp at the predicted Cas9 cutting site and leads to the premature termination of the protein. Additionally, we confirmed the KO of Fam20C at the protein level by mass spectrometry (MS) analysis (Fig. S1, A and B).
Figure 1.
Depletion of endogenous Fam20C impairs sorting and secretion of LyzC. (A) HeLa Fam20C KO cells were generated using the CRISPR/Cas9 technique. Fam20C was targeted downstream of the signal sequence (SS) with gRNA next to Cas9 cutting site PAM. Sequencing of a Fam20C-KO clone highlight the deletion of 22 bp. (B) Western blot analysis of the secretion of LyzC-Flag in HeLa control (ctrl.) and Fam20C-KO cells after 20°C block and 37°C release. β-Actin was used as loading control. (C) Western blots of four independent experiments (B) were quantified by densitometry with ImageJ. The bar graph represents the means ± SD of densitometric values of external LyzC-Flag, normalized to internal levels in percentage. Statistical test, Kolmogorov–Smirnov. (D) Representative immunofluorescence images of the LyzC-RUSH experiments, showing LyzC transport in different cell lines. Cells were fixed at 0, 20, 40, and 60 min after biotin addition and costained with anti-HA antibody. Arrowheads indicate post-Golgi vesicles. Scale bars, 10 µm. (E) LyzC vesicle formation was quantified from RUSH experiments (D), analyzing z-stack images (d = 0.35 µm). A scatter dot plot represents the means (± SD) of at least three independent experiments (n > 45 cells per condition). Statistical test, Kruskal–Wallis. *, P < 0.05; **, P < 0.01.; ***, P < 0.001.
Figure S1.
MS analysis confirming Fam20C protein KO and Fam20C-dependent phosphorylation sites of Cab45 (related to Figs. 1 and 3). (A) Scheme of the MS approach used to identify Fam20C protein KO. Golgi fractions of HeLa control and Fam20C-KO cells were isolated by sucrose gradient centrifugation. Equal protein amounts were separated by SDS PAGE and analyzed by MS (n = 3). (B) Volcano plot of the identified proteins of HeLa control cells versus Fam20C-KO cells. Fam20C was identified in HeLa-WT cells by MS/MS (indicated in red), but not in Fam20C-KO cells. (C) Colocalization of vesicular LyzC-SBP-EGFP against endosomal and lysosomal markers was analyzed using immunofluorescence microscopy. HeLa cells were transfected with RUSH construct LyzC-SBP-EGFP and incubated for 40 min with biotin. Cells were fixed and stained with α-EEA1 antibody (early endosomes), α-Rab11 antibody (recycling endosomes), and α-Lamp1 antibody (lysosomes). Scale bars, 10 µm. The magnification of the inset is shown in the lowest panel (scale bars, 1 µm). (D) Heatmap and profile plot illustrating hits that were significantly phosphorylated in the kinase-dead (KD) Fam20C samples (48 phosphosites) or Fam20C-WT samples (289 phosphosites). Red and blue represent the high and low z-scores, respectively. (E) Quantification of the analyzed Fam20C-WT and Fam20C-KD samples using Pearson correlation analysis. Hits from each sample were compared with hits from samples of the same group (biological replicates) as well as with hits from samples from the other group (Fam20C-WT vs. Fam20C-KD). High and low correlations between samples are indicated by red and green, respectively. (F) Categorization of the detected protein hits into subgroups (red dots). The phosphorylated proteins were analyzed for similarities (secreted, glycosylated, calcium-binding, and Golgi-localized proteins). P values describe the amount of enriched phosphorylated proteins in each subgroup.
We next examined the role of Fam20C in the secretion of the Cab45 client LyzC (von Blume et al., 2012; Crevenna et al., 2016; Deng et al., 2018). We transfected HeLa cells and Fam20C-KO cells with LyzC-Flag and analyzed the supernatants by SDS-PAGE and Western blotting. To investigate the role of Fam20C in the TGN export of LyzC, we incubated these cells at 20°C for 2 h to promote the accumulation of LyzC in the TGN followed by incubation at 37°C for 1 h to release the protein. Western blot analyses (Fig. 1 B) revealed a reduction of LyzC secretion after 37°C release in the Fam20C-KO cells of ∼50% compared with the WT cells (Fig. 1 C). This is in line with the reduced secretion in Cab45 KO-cells (Crevenna et al., 2016).
Next, we used the retention using selective hooks (RUSH) system to analyze the packaging of LyzC into secretory vesicles in Fam20C-KO cells. The RUSH system allowed us to track the cargo transport through the secretory pathway (Boncompain and Perez, 2012; Boncompain et al., 2012). Control and Fam20C-KO cells were transfected with the RUSH construct LyzC-streptavidin-binding peptide (SBP)-EGFP and fixed after various time points. We also analyzed rescued Fam20C-KO cells that stably reexpressed Fam20C-WT, as well as the kinase-dead variant Fam20C-D478A (Tagliabracci et al., 2012). The expression of Fam20C-WT and Fam20C-D478A was confirmed by staining cells with an α-HA antibody (Fig. 1 D). Without biotin, LyzC was trapped in the ER (0 min), whereas biotin addition induced LyzC transport through the secretory pathway. TGN-derived vesicles at different time points were quantified (Fig. 1 E). LyzC was observed to localize in the Golgi of the HeLa control cells 20 min after adding biotin; after 40 min, LyzC was sorted and packed into TGN-derived vesicles, with 32 ± 15 vesicles per cell (Fig. 1, D and E). To exclude that analyzed LyzC vesicles in the RUSH approach are following the endosomal/lysosomal pathway, we performed costainings in HeLa cells using specific endosomal/lysosomal markers (Fig. S1 C).
Consistent with the results of the secretion assay (Fig. 1, B and C), the Fam20C-KO cells showed a significant delay in the formation of LyzC vesicles after 40 min (with 19 ± 12 vesicles per cell). This effect was fully rescued by reexpressing Fam20C-WT in Fam20C-KO cells (resulting in 35 ± 22 vesicles per cell), but not with the kinase-dead variant (10 ± 7 vesicles per cell; Fig. 1, D and E). A comparison of the reexpression of Fam20C-WT versus Fam20C-D478A showed a significantly higher number of LyzC vesicles in the former, even at 60 min after biotin addition (with 53 ± 25 and 16 ± 12 vesicles per cell, respectively).
Together, these results indicate that Fam20C kinase activity drives the sorting and secretion of the Cab45-dependent client LyzC.
Fam20C buds with Cab45 in SM-rich vesicles
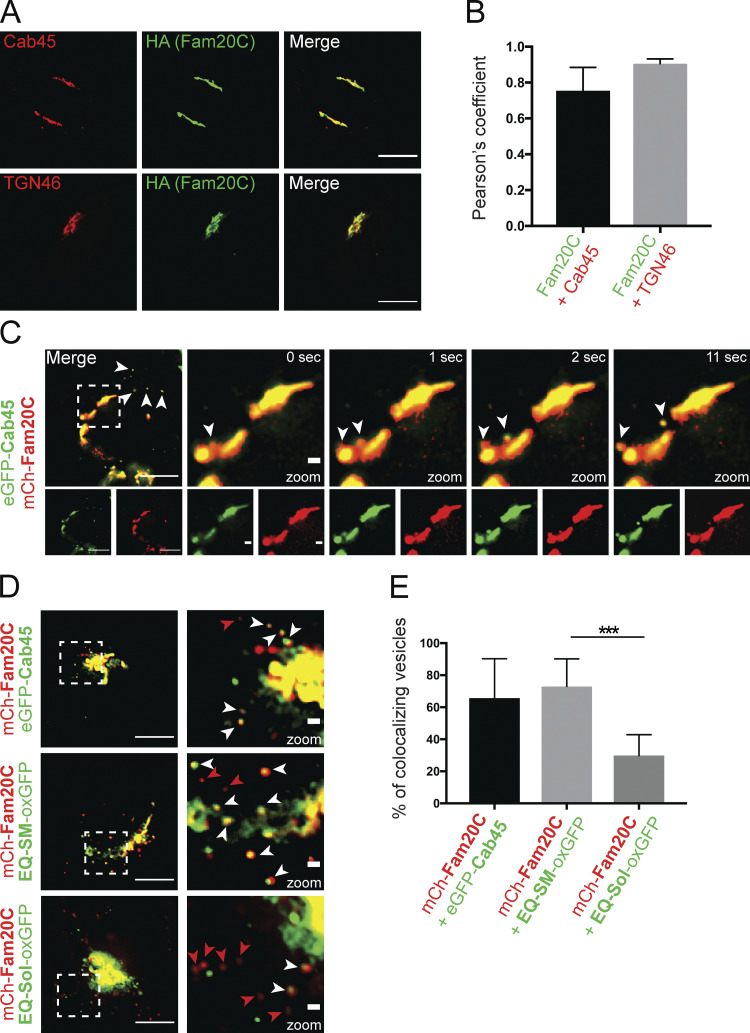
Given the finding that Fam20C depletion delayed the sorting of LyzC from the Golgi, we performed colocalization studies to investigate if there was an interconnection between the kinase and Cab45. First, we defined the localization of Fam20C and Cab45 relative to each other using immunofluorescence microscopy in fixed HeLa cells stained with antibodies against endogenous Cab45, Fam20C (HA antibody), and TGN46 (Fig. 2 A). Here, Fam20C colocalized with Cab45 (Pearson’s correlation coefficient [r] = 0.754 ± 0.071) and TGN46 (r = 0.906 ± 0.026; Fig. 2 B). To investigate this co-occurrence in living cells, we monitored Cab45 and Fam20C budding by time-lapse imaging. HeLa cells were transfected with EGFP-Cab45 and mCherry-Fam20C, and vesicle formation was monitored over time. Interestingly, both proteins were budding from the TGN in the same vesicle (Fig. 2 C).
Figure 2.
Fam20C buds with Cab45 in TGN-derived vesicles. (A) Colocalization of Fam20C-HA, Cab45, and TGN46 in stable Fam20C cell lines was demonstrated by using immunofluorescence microscopy. Scale bars, 10 µm. (B) Pearson’s correlation coefficients were determined from A using ImageJ. The bar graph represents the means ± SD (n > 8 cells per condition). (C) The budding of Fam20C vesicles from the Golgi was observed in living cells expressing mCherry-Fam20C and EGFP-Cab45. Time-lapse movies were acquired. Scale bars, 10 µm. Arrowheads indicate colocalizing vesicles. Higher magnification shows Golgi (inset; scale bars, 1 µm). (D) Example micrographs are showing the Fam20C vesicle budding in living cells that expressed mCherry-Fam20C with EGFP-Cab45, EQ-SM-oxGFP, or EQ-Sol-oxGFP. Scale bars, 10 µm. Higher magnification shows Golgi (inset; scale bars, 1 µm). White arrowheads indicate colocalizing vesicles, and red arrowheads indicate Fam20C-only vesicles. (E) The numbers of colocalizing vesicles (D) were quantified. Data were collected from three independent experiments (n > 230 vesicles per condition). The bar graph illustrates the means (± SD) of colocalizing vesicles in percentage. Statistical test, Mann–Whitney. ***, P < 0.001.
We reported recently that Cab45 is sorted into SM-rich vesicles and transported with its clients to the cell surface (Deng et al., 2018). To further verify the sorting of Fam20C, we investigated the colocalization of Fam20C with SM-rich vesicles in living cells. We used time-lapse microscopy to observe HeLa cells cotransfected with mCherry-Fam20C and EGFP-Cab45, EQ-SM-oxGFP (a nontoxic SM reporter protein derived from equinatoxin II [EQ]), or EQ-Sol-oxGFP (a nontoxic SM-binding–deficient reporter mutant). Fam20C vesicles were counted and analyzed for colocalization (Fig. 2 D). On average, out of 14 budded Fam20C vesicles, 9 were positive for Cab45 (65% ± 24%). Fam20C budded predominately in SM-positive vesicles (20 out of 26; 73% ± 17%), but not together with SM-binding–deficient EQ-Sol (13 out of 46; 30% ± 13%; Fig. 2 E).
These results suggested the close proximity of Fam20C and Cab45 at TGN exit sites before budding by going in the same transport carriers.
Fam20C phosphorylates Cab45 on distinct residues
The close proximity of Fam20C and Cab45 acts as a striking indicator that Fam20C could directly phosphorylate Cab45. This idea was also supported by the fact that many of the recently identified Fam20C substrates play roles in Ca2+ homeostasis or are actual Ca2+-binding proteins (Tagliabracci et al., 2015). So far, published screens indicated that Cab45 contains phosphorylated residues (e.g., Mertins et al., 2016; Zhou et al., 2013). However, to date, no kinase responsible for these modifications could be pinpointed.
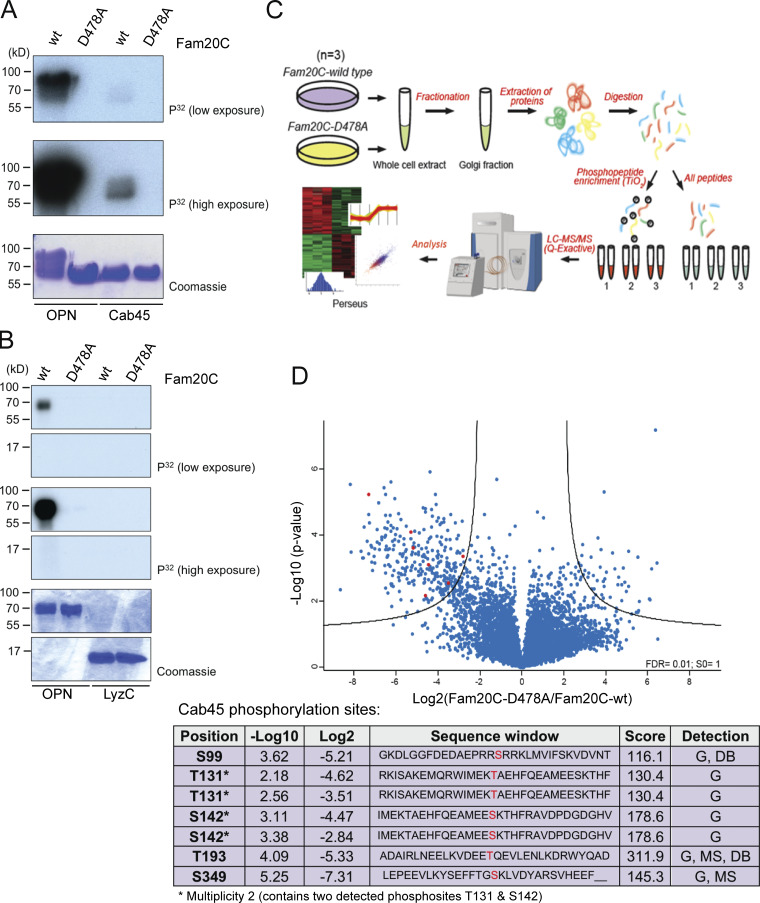
To study the interdependence between the two proteins, recombinant Cab45 was purified from HEK293 cell culture supernatants (Li et al., 2013) and subsequently subjected to in vitro kinase assays with Fam20C-WT and the kinase-dead mutant Fam20C-D478A (Fig. 3 A). Osteopontin (OPN), a highly phosphorylated Fam20C substrate was used as a positive control (Tagliabracci et al., 2015). The results illustrated in Fig. 3 A clearly prove that Fam20C, but not the inactive mutant, is indeed able to phosphorylate Cab45 in vitro. Furthermore, the lack of phosphorylation of LyzC in the same Fam20C kinase assay (Fig. 3 B) additionally indicates that phosphorylation of Cab45, but not of LyzC itself, might be responsible for the sorting defect of LyzC (Fig. 1, B–E).
Figure 3.
Fam20C phosphorylates Cab45 on five distinct residues. (A and B) In vitro Fam20C kinase assays. Recombinant Cab45-WT and LyzC were incubated with Fam20C-WT or Fam20C-D478A in the presence of [γ-32P]ATP. 32P incorporation was tested by SDS-PAGE and autoradiography. OPN was used as positive control. (C) Scheme of the MS approach used to identify Fam20C-dependent Cab45 phosphorylation in vivo. (D) Volcano plot is showing the phosphorylation change of phosphosites, detected in Golgi fractions of Fam20C-WT and D478A cells (C). Cab45 phosphorylation sites (red) are listed in Table 1. Abbreviations used: DB, database (www.phosphosites.org); G, sites detected in the Golgi fractions (C and D); MS, MS after kinase assay (A).
Table 1. Primers and oligomers used for cloning of constructs.
| Construct | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) |
|---|---|---|
| pLPCX-Cab45-5pXA | T131: GGATCATGGAGAAGGCGGCCGAGCACTTC | S99: AGCTTCCTCCGGGCCCGCCGCGGCTC |
| T142: GAGGCCATGGAGGAGGCCAAGACACACTTCCG | ||
| S193: TCAAAGTGGATGAGGAAGCACAGGAAGTCCTGGA | ||
| S349: CGAGTTCTTCACGGGCGCCAAGCTGGTGGACTAC | ||
| pLPCX-Cab45-5pXE | T131: GGATCATGGAGAAGGAAGCCGAGCACTTC | S99: AGCTTCCTCCGTTCCCGCCGCGGCTC |
| T142: GAGGCCATGGAGGAGGAAAAGACACACTTCCG | ||
| S193: TCAAAGTGGATGAGGAAGAACAGGAAGTCCTGGA | ||
| S349: CGAGTTCTTCACGGGCGAAAAGCTGGTGGACTAC | ||
| pB-T-PAF-Cab45-WT/5pXA/5pXE | Fragment1: GGCGGCCATCACAAGTTTGTACAGATGGCTACAGGCTCCCGG | Fragment1: GGTTGGCAGGCCGGCATTCACCTCC |
| Fragment2: GGAGGTGAATGCCGGCCTGCCAACC | Fragment2: CCAGCACACTGGATCAGTTATCTATGCTTAAAACTCCTCGTGCACGCTGCG | |
| pBT-PAF-GFP-Cab45-WT/5pXA/5pXE | Fragment1: GGCGGCCATCACAAGTTTGTACAGCTAGCATGGCTACAGGCTCCCG | Fragment1: CCGCTTCCACCTCCACCAGATCTGTGATGATGATGGTGATGAAGCTTTGTTC |
| Fragment2: CTGGTGGAGGTGGAAGCGGTAGCAAAGGAGAAGAACTTTTCACTGG | Fragment2: CTTTGGAAGTACAGGTTCTCGGATCCGGCCGGCCGACCTCCACCTTTGTAGAGCTC | |
| Fragment3: GAGGTCGGCCGGCCGGATCCGAGAACCTGTACTTCCAAAGTGGCGCGCCACGGCCTGCCAACCAC | Fragment3: GGATCAGTTATCTATGCGGCCGCTCTAGATTAAAACTCCTCGTGCACGC | |
| pLPCX-SS-EGFP-Cab45-5pXA/5pXE | Fragment1: CGTGACCGCCGCCCCGGAATTCCGGCCTGCCAACCACTCG | Fragment1: TTTATCGATGTTTGGCCGAGGCGACCGGTTTAAAACTCCTCGTGCACGCTGCG |
| pLPCX-Fam20C-HA | Fragment1: CGTAGATCTATGAAGATGATGCTGGTGCGCCG | Fragment1: GCACAATTGTTAAGCGTAGTCTGGGACGTCGTATGGGTACCTCGCCGAGGCGGCTCTGTG |
| pLPCX-Fam20C-D478A-HA | D478A: CATCATCCACTTAGCCAATGGAAGAGG | |
| pLPCX-mCherry-Fam20C | Fragment1: CCATAAAGCTTATACGAATTCATAGCCATGGCTACAGGCTCCCGGACGTCCCTGCTCCTGGCTTTTGGCCTGCTCTGCCTGCCCTGGCTTCAAGAGGGCAGTGCCTTCCCAACCATTCCCTTATCCTCGGGAA | Fragment1: TTCCCGAGGATAAGGGAATGGTTGGGAAGGCACTGCCCTCTTGAAGCCAGGGCAGGCAGAGCAGGCCAAAAGCCAGGAGCAGGGACGTCCGGGAGCCTGTAGCCATGG |
| Fragment2: CCCTTATCCTCGGGAACAAAGCTTATGGTGAGCAAGGGCGAGG | Fragment2: CCTCCCCGGGGGAGAGGAATTCCTTAGCGTAGTCTGGGACGTCGTATGGGTA | |
| Fragment3: TACCCATACGACGTCCCAGACTACGCTAAGGAATTCCTCTCCCCCGGGGAGG | Fragment3: TATCGATGTTTGGCCGAGGCGGCCGCTTACCTCGCCGAGGCGGCTCTGTG | |
| pIRESneo3-Str-KDEL_SS-SBP-tagRFP-Cab45 | Fragment1: CTTGCCACAACCCGGGAGGCGCGCCATGGCTACAGGCTCCCGGACGTCCC | Fragment1: CTTAATCAGCTCTTCGCCCTTAGACACACCTGCAGGTGGTTCACGTTGACCTTG |
| Fragment2: CAAGGTCAACGTGAACCACCTGCAGGTGTGTCTAAGGGCGAAGAGCTGATTAAG | Fragment2: AGTTAATTAATTGGCCCTCGAGGCCTTAAAACTCCTCGTGCACGCTGCGC |
To identify single Fam20C phosphorylation sites on Cab45 in living cells, we used sucrose gradient centrifugation to purify Golgi fractions from cells that stably express Fam20C-WT or Fam20C-D478A. These fractions were analyzed by MS and phosphoproteomics, verifying the phosphorylation status of Cab45 within the Golgi compartment (Fig. 3 C). In this regard, we predominately found Fam20C-regulated phosphorylated sites in cells expressing Fam20C-WT, but not in D478A samples (Fig. S1 D), with a strong correlation between biological replicates (Fig. S1 E).
Among the hits, we detected phosphosites of typical Fam20C substrates (Tagliabracci et al., 2012, 2015), including nucleobindin, proprotein convertase subtilisin, and amyloid β A4 protein (data not shown), as well as phosphosites of Cab45. In total, five specific Cab45 residues were identified that were phosphorylated in Golgi fractions from Fam20C-WT, but not Fam20C-D478A, cells (labeled red and listed in Fig. 3 D). Strikingly, two of these sites (T193 and S349) were also identified in the in vitro kinase assay (Fig. 3 A), when the final product of the assay was analyzed by MS (data not shown). Sites S99 and T193 have previously been reported (Mertins et al., 2016; Zhou et al., 2013), confirming the results of the present phosphoproteomics. Interestingly, many of the identified protein hits could be clustered into four subgroups to Golgi-localizing, secreted, glycosylated, and Ca2+-binding proteins (Fig. S1 F), and Cab45 matches in all of these categories.
Phosphorylation-mimetic Cab45 accumulates in TGN-derived vesicles
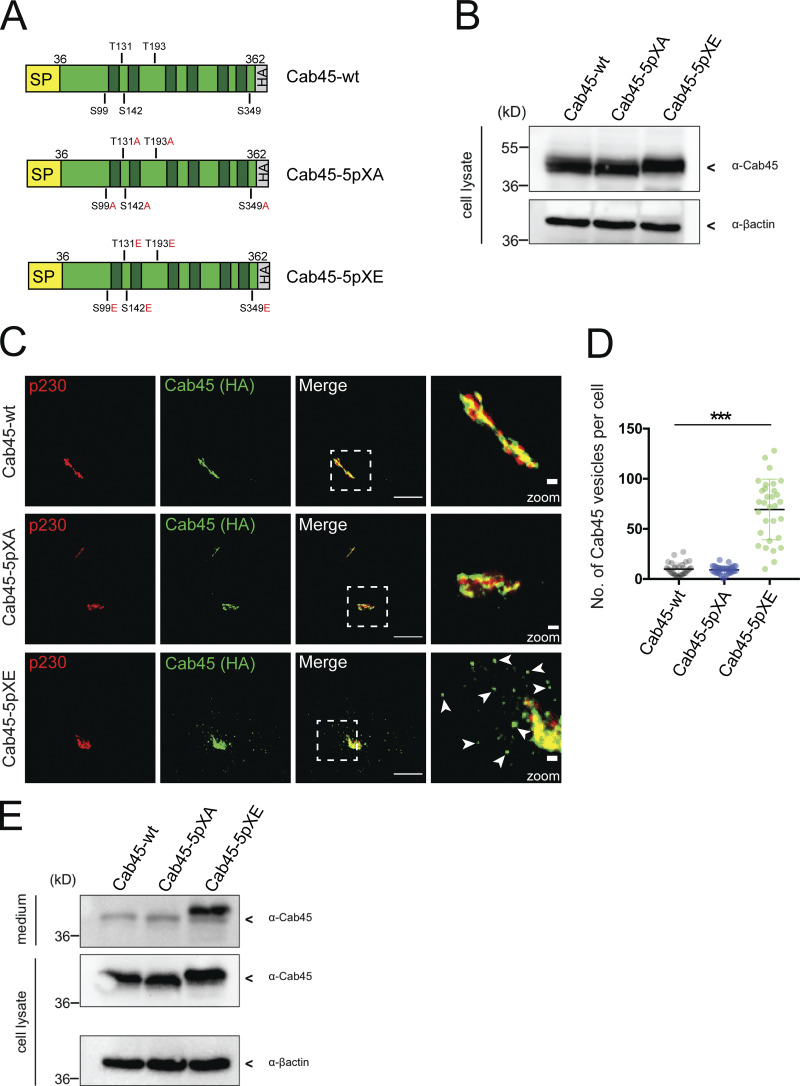
To further analyze the role of the identified Cab45 phosphorylation sites, all five sites were mutated to either alanine (Cab45-5pXA), to generate a phosphorylation-deficient mutant of Cab45, or glutamic acid, to mimic the phosphorylated protein (Cab45-5pXE; Fig. 4 A). Stable cell lines expressing these constructs were generated by retroviral transduction of HeLa Cab45-KO cells (Crevenna et al., 2016). These cells were tested for equal Cab45 expression levels by Western blot analysis (Fig. 4 B). We fixed these cells and analyzed them by immunofluorescence microscopy. Surprisingly, we observed changes in the localization of the phospho-mimetic mutant Cab45-5pXE (Fig. 4 C); Cab45-WT and Cab45-5pXA almost completely colocalized with Golgi-marker peripheral trans-Golgi membrane protein (p230), whereas Cab45-5pXE showed strong accumulation of vesicular structures in close proximity to the TGN (Fig. 4 C). The number of Cab45 vesicles per cell in cells expressing Cab45-WT, Cab-5pXA, and Cab45-5pXE were quantified from z-stack images (d = 0.35 µm; Fig. 4 D). Overall, cells expressing Cab45-5pXE showed a significantly higher number of Cab45 vesicles (69 ± 30 per cell) than cells expressing Cab45-WT (10 ± 6 vesicles per cell) and Cab45-5pXA (9 ± 4 vesicles per cell). We analyzed the origin of these vesicles by performing a block at 20°C for 2 h to inhibit protein export from the TGN. Analyses of the cell culture supernatants with SDS-PAGE and Western blotting detected Cab45-5pXE but almost no Cab45-WT or Cab45-5pXA (Fig. 4 E). In addition, we revealed no colocalization with a Sec16A marker (ER exit sites, Fig. S2 A). These results suggest that Cab45-5pXE vesicles are post-Golgi vesicles that have already left the TGN.
Figure 4.
Phospho-mimicking Cab45 localizes in post-Golgi vesicles. (A) Schemes of WT and phosphomutant Cab45-HA constructs with signal peptide (SP). Identified phosphorylation sites (serine [S] and threonine [T]) were replaced by alanine (A) or glutamic acid (E). The constructs were stably transduced in Cab45-KO cells. (B) Western blot shows Cab45 expression levels of stable Cab45-WT and phosphomutant cell lines. β-Actin was used as a loading control. (C) Steady-state localization of Cab45-WT, Cab45-5pXA, and Cab45-5pXE was analyzed by immunofluorescence microscopy. Stable cell lines were stained with α-p230 and α-HA antibody (Cab45). Scale bars, 10 µm. Higher magnification (insets) show Golgi (scale bars, 1 µm). (D) The numbers of Cab45 vesicles (C) were quantified by analyzing z-stack images (d = 0.35 µm), n > 30 cells. A scatter dot plot represents mean ± SD of counted vesicles per cell. Statistical test, Kruskal–Wallis. (E) The post-Golgi origin of Cab45 vesicles was confirmed by Western blotting after performing a 20°C block in stable cell lines. β-Actin was used as loading control. ***, P < 0.001.
Figure S2.
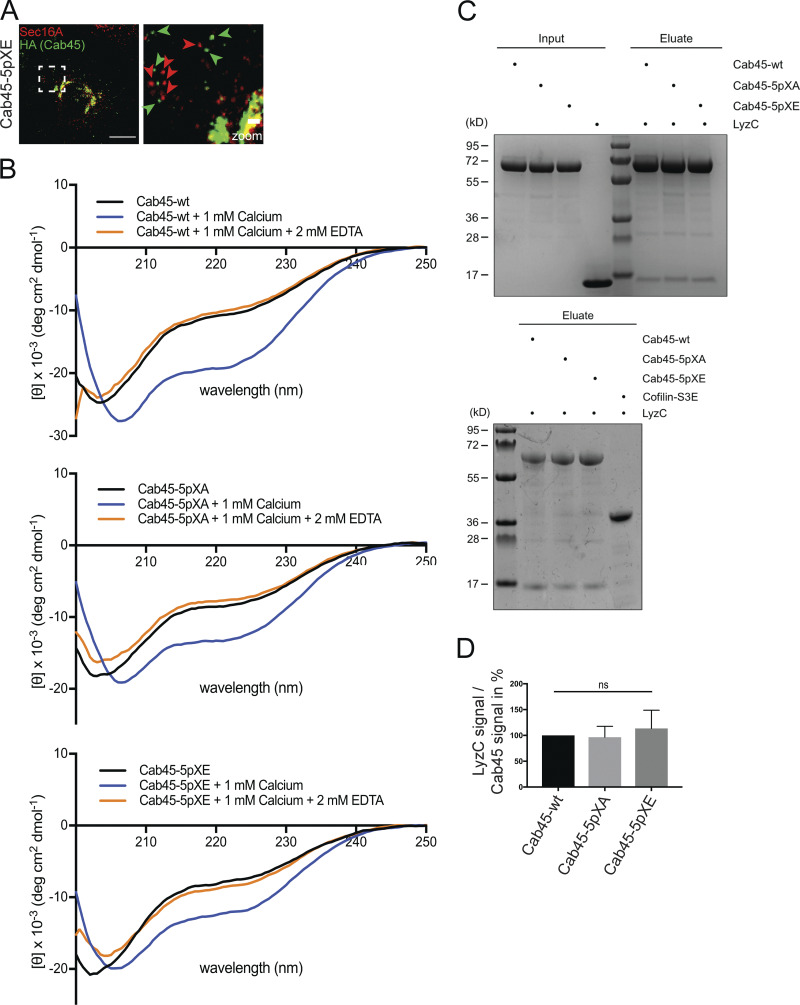
Mutation of Cab45 phosphorylation sites alters the localization, but not the secondary structure or cargo-binding ability, of the protein (related to Fig. 4). (A) Representative immunofluorescence images of cells that stably expressed Cab45-5pXE, stained against Cab45-5pXE (α-HA) and ER exit sites (α-Sec16A). Scale bar, 10 µm. The magnification of the inset is shown in the right panel (scale bar, 1 µm). The green arrowheads indicate Cab45-5pXE vesicles, and the red arrowheads indicate Sec16A-positive ER exit sites. (B) Far-ultraviolet CD spectroscopy measurements of recombinant Cab45-WT, Cab45-5pXA, and Cab45-5pXE. The secondary structures of the proteins were analyzed only in the buffer after adding 1 mM Ca2+ and followed by 2 mM EDTA. (C) Coimmunoprecipitation of recombinant LyzC with Cab45-WT and phosphomutants analyzed by Coomassie staining. Recombinant Cab45 proteins were enriched by Ni2+ beads, washed, and incubated with recombinant LyzC (upper blot). Cofilin-S3E was used as a negative control (lower blot). (D) Coomassie gels of three independent experiments described in C were quantified by densitometry with ImageJ software. The bar graph represents the means ± SD of densitometric values of LyzC signal normalized to Cab45 signal in percent. Statistical test, Kruskal–Wallis.
In a recent study, we used circular dichroism (CD) spectroscopy to examine Cab45, showing it to be rather unstructured but changing to a more α-helical secondary structure upon addition of Ca2+ (Deng et al., 2018). Adding EDTA reversed this effect completely. In the present study, we subjected recombinant Cab45-WT, Cab45-5pXA, and Cab45-5pXE to CD spectroscopy (Fig. S2 B). This showed no difference in secondary structure between Cab45-WT and the phosphomutants, either in the untreated protein sample or after adding Ca2+ and EDTA. Also, cargo binding was not changed in Cab45 phosphomutants, since we additionally performed coimmunoprecipitations to validate LyzC cargo binding of the Cab45 mutants (Fig. S2, C and D).
Together, these findings indicated that mimicking Cab45 phosphorylation accelerated export of Cab45 and promoted its occurrence in TGN-derived vesicles.
Mimicking Cab45 phosphorylation drives client secretion
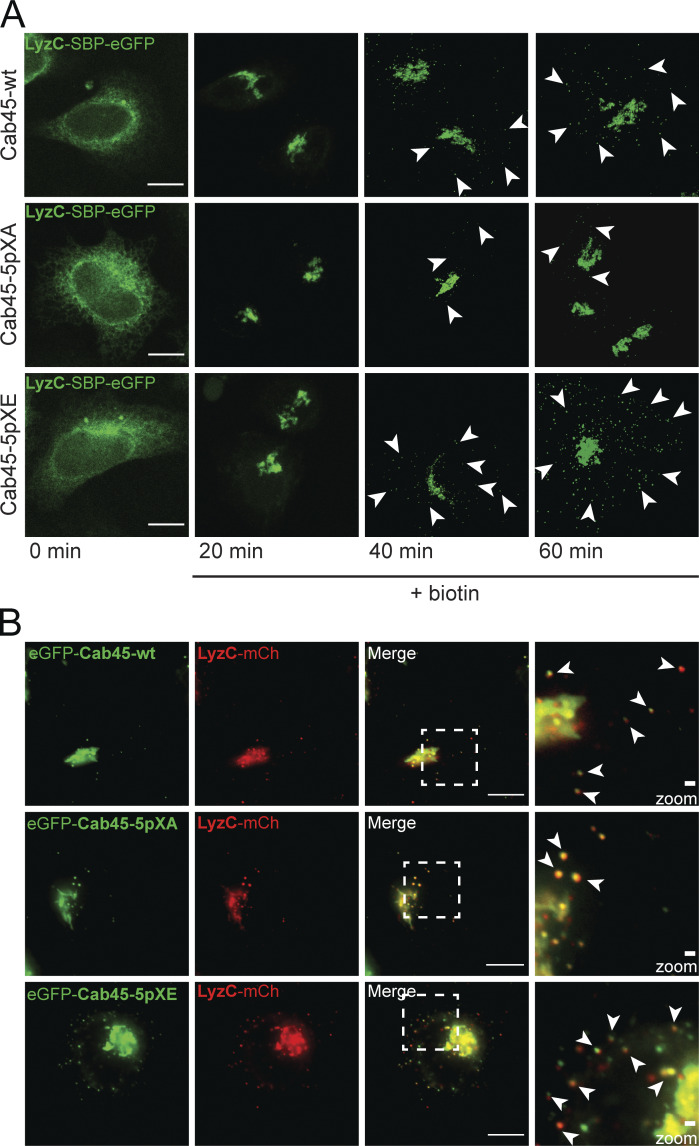
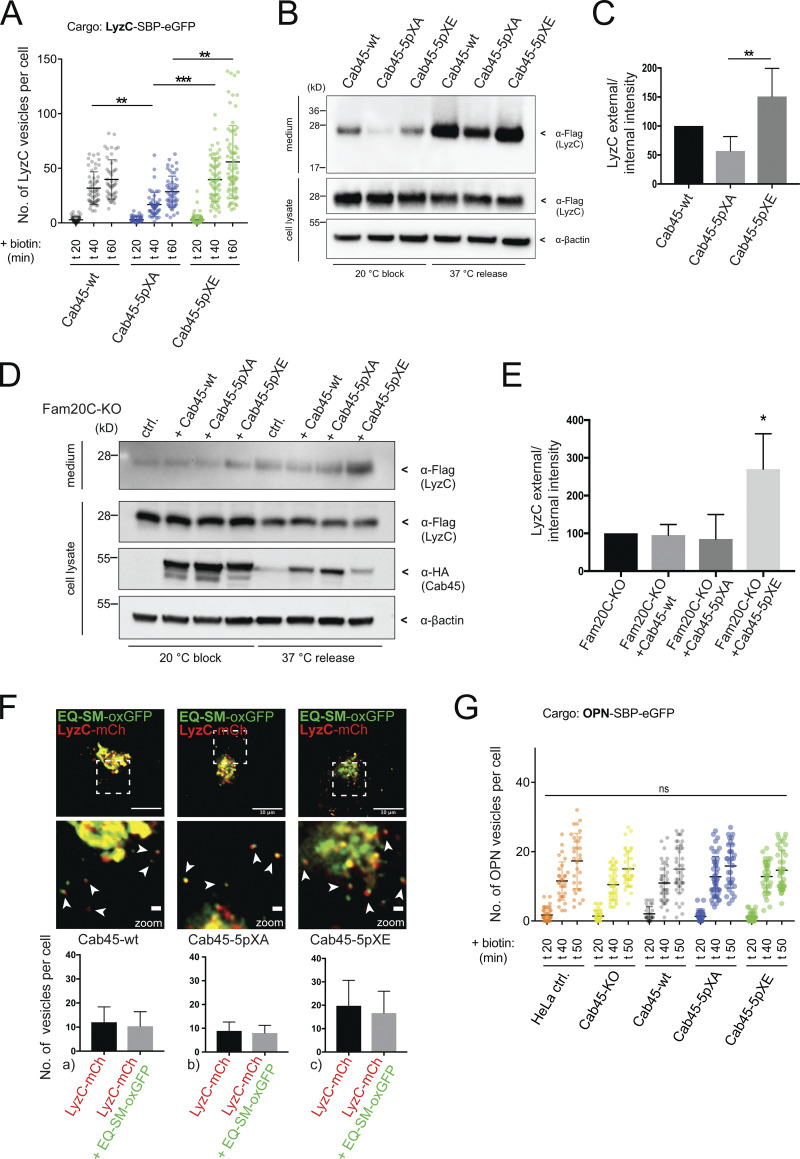
Next, we investigated whether LyzC export was influenced by the phosphorylation status of Cab45 by using the RUSH system in cells expressing Cab45-WT, Cab45-5pXA, or Cab45-5pXE. Fig. S3 A shows representative images of cells at different time points after biotin addition. Quantification of the number of LyzC vesicles (Fig. 5 A) showed a significant reduction in the number of vesicles per cell, 40 min after biotin addition, in Cab45-5pXA cells (16 ± 11) compared with Cab45-WT (31 ± 14) and Cab45-5pXE cells (39 ± 19). The same phenotype was also observed 60 min after biotin addition (Cab45-WT: 39 ± 18 vesicles per cell; Cab45-5pXA: 28 ± 14 vesicles per cell; Cab45-5pXE: 55 ± 33 vesicles per cell). Overall, Cab45-5pXE showed significantly more LyzC vesicles than Cab45-5pXA at 40 and 60 min after biotin addition.
Figure S3.
Specific LyzC sorting of Cab45 phosphomutants (related to Fig. 5). (A) Representative immunofluorescence images of RUSH experiments, showing LyzC transport in Cab45-WT and phosphomutant cell lines. The cells were transfected with LyzC-SBP-EGFP and fixed at 0, 20, 40, and 60 min after addition of biotin. Z-stack images (d = 0.35 µm) were analyzed. The arrowheads indicate cytoplasmic vesicles. Scale bars, 10 µm. (B) Cab45 phosphomutants and Cab45-WT were sorted together with LyzC into the same vesicles. HeLa WT cells were transfected with different EGFP-Cab45 constructs and LyzC-mCherry. Time-lapse movies were acquired to observe vesicle budding over time. Scale bars, 10 µm. The magnification of the inset is shown in the last panel. Bars, 1 µm. Arrowheads indicate secretory vesicles that contained both fluorescent proteins.
Figure 5.
Fam20C-dependent Cab45 phosphorylation drives client sorting and secretion. (A) LyzC vesicle formation was quantified from LyzC-RUSH experiments by analyzing z-stack images (d = 0.35 µm). Cell lines were fixed at 0, 20, 40, and 60 min after biotin addition. A scatter dot plot represents the means ± SD of at least three independent experiments (n > 45 cells per condition). Statistical test, Kruskal–Wallis. (B) Western blot analysis of the secretion of LyzC-Flag in Cab45-WT and phosphomutant cells after 20°C block and 37°C release. β-Actin was used as loading control. (C) Western blots of four independent experiments (B) were quantified by densitometry using ImageJ. The bar graph represents the means (± SD) of densitometric values of external LyzC-Flag, normalized to the internal levels in percentage. Statistical test, Kruskal–Wallis. (D) Western blot analysis of the secretion of LyzC-Flag in Fam20C-KO cells rescued with Cab45 and phosphomutants after 20°C block and 37°C release. β-Actin was used as loading control. (E) Western blots of three independent experiments (D) were quantified by densitometry using ImageJ. The bar graph represents the means ± SD of densitometric values of external LyzC-Flag, normalized to the internal levels in percentage. Statistical test, ordinary one-way ANOVA. (F) Sorting of LyzC in EQ-SM-vesicles was controlled by performing live-cell experiments acquiring time-lapse movies. Example micrographs depict the Golgi of Cab45-WT and phosphomutants that expressed LyzC-mCherry and EQ-SM-oxGFP. Scale bars, 10 µm; magnification scale bars, 1 µm. Arrowheads indicate secretory vesicles containing both fluorescence proteins. The means ± SD of post-Golgi LyzC vesicles per cell positive for EQ-SM were quantified (n = 8 cells per condition). (G) OPN vesicle formation was quantified from OPN-RUSH experiments by analyzing z-stack images (d = 0.35 µm). Cell lines were fixed at 0, 20, 40, and 50 min after biotin addition. A scatter dot plot represents the means ± SD of at least three independent experiments (n > 28 cells per condition). Statistical test, Kruskal–Wallis. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Furthermore, we correlated the number of vesicles with the actual secreted amount of protein. To this end, we examined protein secretion from these cells by Western blotting (Fig. 5 B). Consistent with the results of the RUSH assay, LyzC secretion quantified by densitometric analysis was lower in Cab45-5pXA cells (57%) than in cells expressing Cab45-5pXE (150%) and Cab45-WT (100%; Fig. 5 C).
We continued by performing rescue experiments in Fam20C-KO cells transfected with LyzC and cotransfected Cab45-WT, Cab45-5pXA, or Cab45-5pXE, respectively (Fig. 5, D and E). By doing so, we tested the impact of the phosphomutants on the secretion of LyzC in the absence of Fam20C and, accordingly, their ability to compensate for the loss of the kinase. Strikingly, only Fam20C-KO cells rescued with Cab45-5pXE showed significantly higher LyzC secretion, which indicates that the phosphomimetic mutant is capable of compensating the loss of kinase activity in living cells.
In previous publications, we showed that Cab45-WT was packed together with LyzC in SM-rich vesicles (Deng et al., 2018). To address whether Cab45 phosphorylation changes sorting integrity into SM-rich vesicles, cell lines expressing the three Cab45 variants were cotransfected with LyzC-mCherry and EQ-SM-oxGFP and evaluated using time-lapse recordings. The LyzC vesicles per cell were counted, and colocalization with EQ-SM-oxGFP was examined (Fig. 5 F). Notably, the highest number of colocalizing vesicles per cell was monitored in cells expressing Cab45-5pXE (c, 16 ± 9) compared with Cab45-WT (a, 11 ± 6) and Cab45-5pXA (b, 8 ± 3). Of 104 LyzC vesicles observed in Cab45-WT cells, 91 were positive for EQ-SM-oxGFP (86% ± 10%). Similar ratios were determined for Cab45-5pXA cells (64 out of 71 vesicles; 91% ± 9%) and Cab45-5pXE cells (133 out of 158 vesicles; 84% ± 5%).
Using the same experimental setup, we also tested the colocalization of Cab45 and LyzC in transiently transfected HeLa cells. As expected, all three Cab45 proteins colocalized with LyzC-mCherry (Fig. S3 B). Based on these data, phosphorylation seems to promote secretion in general but does not influence sorting into correct vesicles.
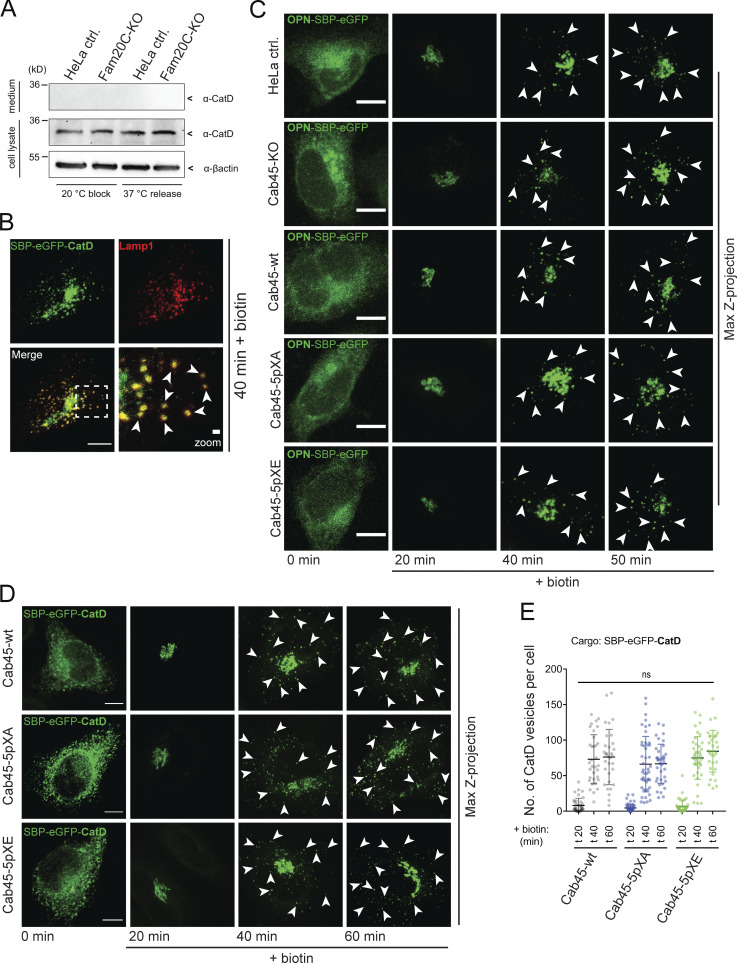
To ensure that the phosphorylations particularly influence the sorting of Cab45 clients, two control proteins were tested, the bulk flow cargo OPN and the lysosomal hydrolase cathepsin D (CatD). OPN is a secreted protein and also phosphorylated by Fam20C (Tagliabracci et al., 2012). In contrast, CatD, a nonsecretory (Fig. S4 A), lysosomal cargo (Fig. S4 B), is sorted via sortilin into clathrin-coated vesicles (Braulke and Bonifacino, 2009) and is therefore not dependent on the SPCA1/Cab45 sorting machinery (Crevenna et al., 2016; Deng et al., 2018). In summary, no differences in the formation of OPN-SBP-EGFP vesicles were observed by using the RUSH approach (Fig. 5 G and Fig. S4 C). Additionally, we verified the unaffected sorting of CatD in the Cab45 phosphomutant cell lines (Fig. S4, D and E) by the same method.
Figure S4.
Sorting of non-Cab45 cargos is not affected in Cab45 phosphomutants (related to Fig. 5). (A) Western blot analysis of the secretion of CatD in HeLa control and Fam20C-KO cells. Endogenous CatD was trapped in the Golgi at 20°C and released for 1 h at 37°C. The supernatants (upper panel) and cell lysates (lower panels) were tested for CatD by Western blotting. β-Actin was used as loading control. (B) Representative immunofluorescence images of colocalization of vesicular SBP-EGFP-CatD with the lysosomal marker Lamp1. HeLa cells were transfected with RUSH construct and incubated for 40 min with biotin. Cells were fixed and stained with α-Lamp1 antibody. Scale bar, 10 µm. The magnification of the inset is shown in the lower right pane (scale bar, 1 µm). (C) Representative immunofluorescence images of RUSH experiments showing OPN transport in HeLa control, Cab45-KO, Cab45-WT, and Cab45 phosphomutant cell lines. Cells were transfected with OPN-SBP-EGFP and fixed at 0, 20, 40, and 50 min after the addition of biotin. Z-stack images (d = 0.35 µm) were analyzed. The arrowheads indicate cytoplasmic vesicles. Scale bars, 10 µm. (D) Representative immunofluorescence images of RUSH experiments showing cathepsin D (CatD) transport in Cab45-WT and phosphomutant cell lines. Cells were transfected with SBP-EGFP-CatD and fixed at 0, 20, 40, and 60 min after the addition of biotin. Z-stack images (d = 0.35 µm) were analyzed. The arrowheads indicate cytoplasmic vesicles. Scale bars, 10 µm. (E) The numbers of CatD budding vesicles (D) were quantified. The cytoplasmic vesicles were counted at each time point by analyzing z-stack images (d = 0.35 µm). Scatter dot plot represents the means ± SD of at least three independent experiments (n > 30 cells per condition). Statistical test, Kruskal–Wallis.
These data show that Cab45 phosphorylation modulates the rate of budding of secretory vesicles containing its specific clients.
Phosphorylation of Cab45 accelerates vesicle budding at the TGN
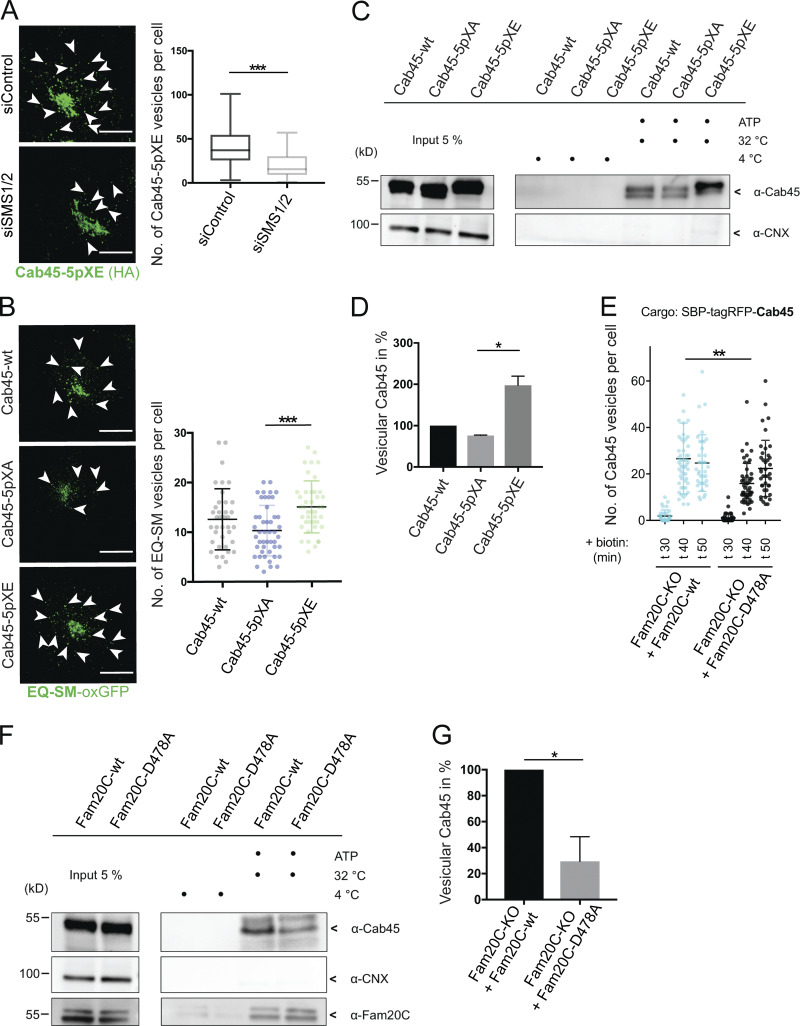
Given the finding that Cab45 sorting into SM-rich vesicles appeared not to be affected in the phosphomutant cells, we investigated the role of SM in this process. SM is synthesized by two isoforms of SM synthase (SMS) in the luminal leaflets of Golgi membranes (Barenholz and Thompson, 1980). We therefore depleted SMS1 and SMS2 in Cab45-5pXE cells with siRNA and analyzed z-stack images (d = 0.35 µm) to quantify the numbers of Cab45-5pXE vesicles stained with α-HA antibody (Fig. 6 A). We observed a significant decrease in the number of Cab45-5pXE vesicles in cells treated with siSMS1/2 siRNA (19 ± 13 vesicles per cell) compared with cells transfected with control siRNA (40 ± 20 vesicles per cell). From this, we concluded that SM is important for the TGN export of Cab45-5pXE.
Figure 6.
Mimicking Cab45 phosphorylation results in accelerated TGN export. (A) Representative immunofluorescence pictures of cells that stably expressed Cab45-5pXE, treated with control or SMS1/2 siRNA. Cells were stained against Cab45 (α-HA), and numbers of vesicles were quantified from z-stack images (d = 0.35 µm). Arrowheads indicate post-Golgi vesicles. Scale bars, 10 µm. Box and whiskers blot represents means with minimum to maximum values of at least three independent experiments (n > 57 cells per condition). Statistical test, Mann–Whitney. (B) Example micrographs of Cab45-WT and phosphomutant cell lines transfected with EQ-SM-oxGFP, after 20°C block and 37°C release. EQ-SM-oxGFP vesicles were quantified from z-stack images (d = 0.35 µm). Scale bars, 10 µm. A scatter dot plot represents means ± SD of at least three independent experiments (n > 37 cells per condition). Statistical test, Kruskal–Wallis. (C) Vesicular budding assay of Cab45 phosphomutants. TGN vesicle budding was initiated by an ATP regeneration system. Released vesicles were analyzed by Western blotting for Cab45 and CNX. (D) Western blots of three independent experiments (C) were quantified by densitometry with ImageJ. The bar graph represents the means ± SD of densitometric values of vesicular Cab45 in percentage. Statistical test, Kruskal–Wallis. (E) Cab45 vesicle formation was quantified from Cab45-RUSH experiments by analyzing z-stack images (d = 0.35 µm). Fam20C-KO cells expressing Fam20C-WT or Fam20C-D478A were fixed at 0, 30, 40, and 50 min after biotin addition. A scatter dot plot represents the means ± SD of at least three independent experiments (n > 35 cells per condition). Statistical test, Kruskal–Wallis. (F) Vesicular budding assay of endogenous Cab45. TGN vesicle budding was initiated by an ATP regeneration system in Fam20C-KO cells, which were transfected with Fam20C-WT and D478. Released vesicles were analyzed by Western blotting for Cab45, Fam20C (HA), and CNX. (G) Western blots of four independent experiments (F) were quantified by densitometry with ImageJ. The bar graph represents the means ± SD of densitometric values of vesicular Cab45 in percentage. Statistical test, Welch’s unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
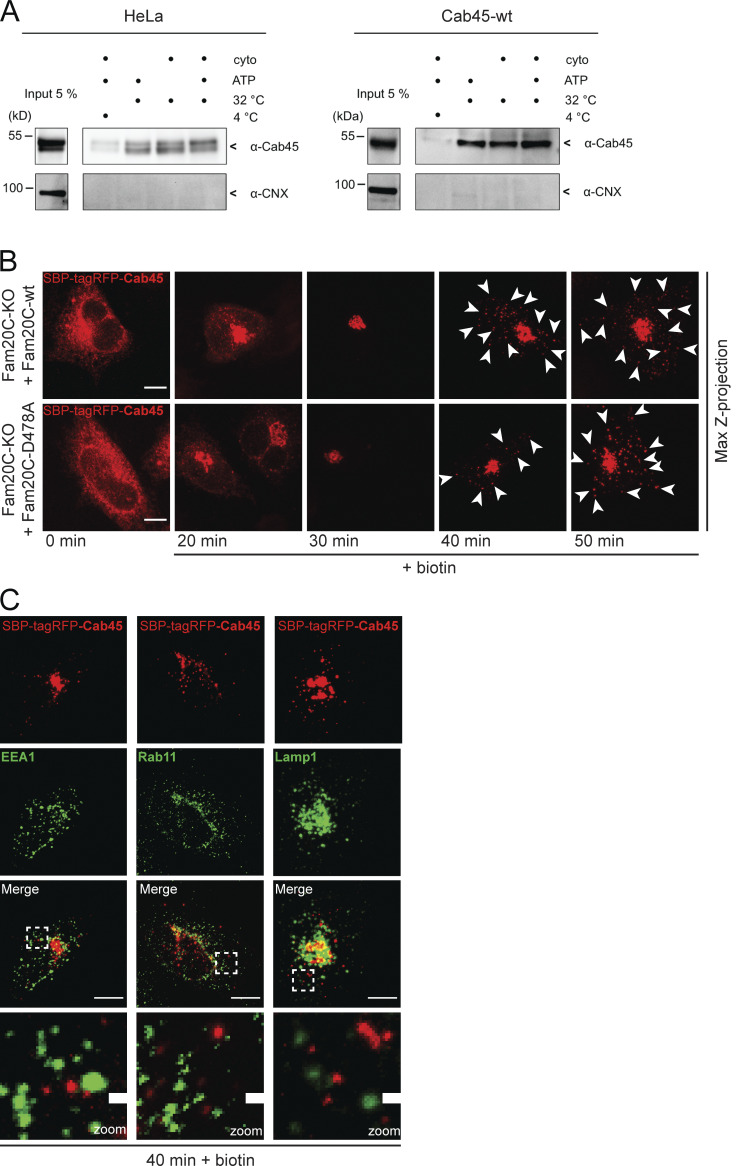
To investigate if the phosphorylation status of Cab45 also influences the export and formation of SM-containing vesicles, we transfected Cab45-WT and the phosphomutant cell lines with EQ-SM-oxGFP and performed a 20°C incubation block for 1 h followed by a release at 37°C for 1 h. An analysis of z-stack images detected significantly more EQ-SM-oxGFP vesicles in the cells expressing Cab45-5pXE (15 ± 5 vesicles per cell) than in those expressing Cab45-5pXA (10 ± 5 vesicles per cell; Fig. 6 B). We therefore conclude that mimicking the negative charges of Cab45 phosphorylation results in its increased TGN export. Next, we analyzed Cab45 vesicular budding using a semi-intact budding assay. Cab45-WT and phosphomutant cell lines were permeabilized with digitonin and incubated with an ATP regeneration system at 32°C to generate TGN-derived vesicles (Wakana et al., 2012; Deng et al., 2018). Vesicular fractions were collected by ultracentrifugation and analyzed by SDS-PAGE and Western blotting (Fig. 6 C). Beforehand, we validated budding of endogenous and overexpressed Cab45 in dependency of ATP and rat liver cytosol (Fig. S5 A). Similar to what was published in Deng et al. (2018), we could not promote budding of Cab45 with rat liver cytosol. Therefore, we performed our budding assays in the absence of rat liver cytosol.
Figure S5.
TGN export of Cab45 (related to Fig. 6). (A) Vesicular budding assays of Cab45. The budding of TGN-derived vesicles was tested in HeLa cells (left blot) and overexpressed Cab45-WT (right blot) in dependency of ATP and rat liver cytosol at 32°C. The released vesicles were collected and analyzed by Western blotting for Cab45 and CNX. Cells incubated at 4°C were used as the negative control. (B) Representative immunofluorescence images of RUSH experiments showing Cab45 transport in Fam20C KO cells that reexpress Fam20C-WT and the kinase-dead Fam20C-D478A. The cells were transfected with SBP-tagRFP-Cab45 and fixed at 0, 20, 30, 40, and 50 min after the addition of biotin. Z-stack images (d = 0.35 µm) were analyzed. The arrowheads indicate cytoplasmic vesicles. Scale bars, 10 µm. (C) Colocalization of vesicular SBP-tagRFP-Cab45 against endosomal and lysosomal markers was checked using immunofluorescence microscopy. HeLa cells were transfected with RUSH construct SBP-tagRFP-Cab45 and incubated for 40 min with biotin. Cells were fixed and stained with α-EEA1 antibody (early endosomes), α-Rab11 antibody (recycling endosomes) and α-Lamp1 antibody (lysosomes). Scale bars, 10 µm. The magnification of the inset is shown in the lowest panel. Scale bars, 1 µm.
Consistent with the results demonstrated in Fig. 4, C–E, we detected significantly more Cab45 in the vesicular fraction of cells expressing Cab45-5pXE than in cells expressing Cab45-5pXA (Fig. 6, C and D). Western blots against calnexin (CNX) were used as a control to show that samples were not contaminated with whole-cell lysate.
We followed the dependence of the Cab45 TGN export on the Fam20C activity (Fig. 6 E). Fam20C-KO cells that stably reexpressed Fam20C-WT or Fam20C-D478A were transfected with the RUSH construct SBP-tagRFP-Cab45 and fixed at specific time points. In the absence of biotin, Cab45 localized in the ER, whereas at 0–30 min after biotin addition, it was simultaneously transported to the Golgi in both cell lines (Fig. S5 B). When Cab45 was packaged into TGN-derived vesicles, at 40 min after biotin addition, there were significantly more Cab45 vesicles detectable in the cells reexpressing Fam20C-WT (26 ± 15 vesicles per cell) than in those expressing the kinase-dead variant (15 ± 9 vesicles per cell). In line with the LyzC-RUSH (Fig. S1 C), we performed costainings to exclude colocalization of Cab45 vesicles with endosomal/lysosomal compartments (Fig. S5 C).
Finally, we analyzed the vesicle budding of endogenous Cab45 (Fig. 6 F). Fam20C-KO cell lines were transfected with Fam20C-WT or Fam20C-D478A constructs, permeabilized, and incubated with an ATP regeneration system at 32°C. Consistent with the results of the Cab45-RUSH assay, this showed a significantly higher amount of endogenous Cab45 in the vesicular fraction of the cells transfected with Fam20C-WT compared with the cells transfected with Fam20C-D478A (Fig. 6 G). It was possible to detect both Fam20C variants (WT and D478) in the same vesicular fraction, but not CNX.
Mutations of Cab45 phosphosites influence Cab45 oligomerization
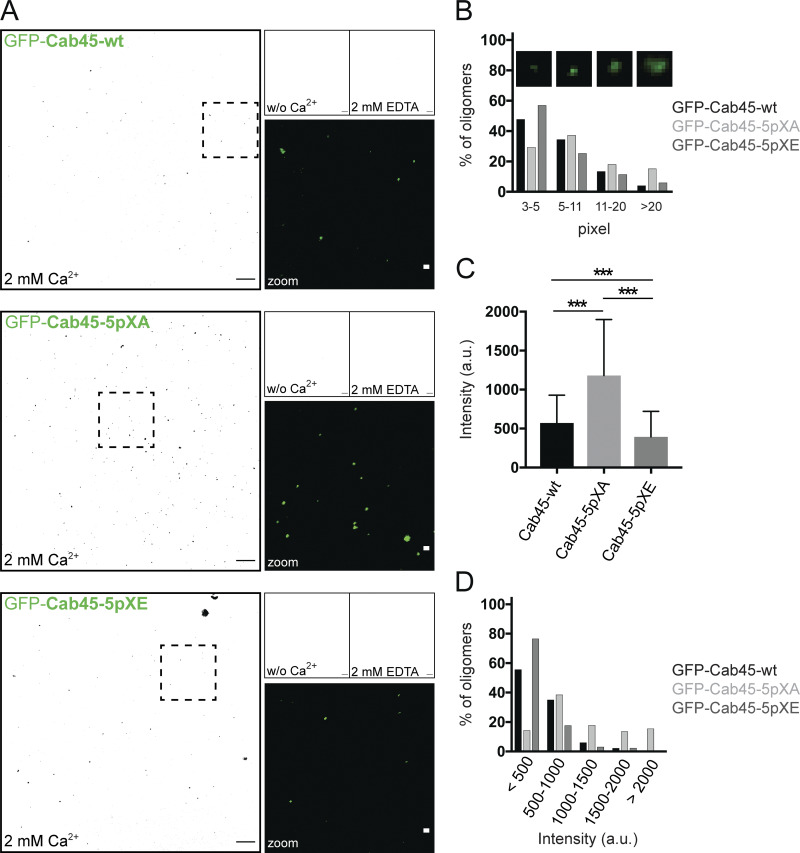
Cab45 oligomerizes in a Ca2+-dependent manner in vitro (Crevenna et al., 2016; Deng et al., 2018). To test the impact of phosphorylation on oligomerization, we investigated oligomerization behavior of the phosphomutant proteins using a confocal microscopy-based oligomerization assay (Fig. 7 A). For visualization, we purified GFP-Cab45-WT and GFP-phosphomutants. Whereas in the absence of Ca2+, no Cab45 oligomers were observed (Fig. 7 A), we clearly could detect oligomer formation of all recombinant Cab45-variants after incubation with 2 mM Ca2+. This effect was reversible by adding 2 mM EDTA.
Figure 7.
Mutations of Cab45 phosphosites influence Cab45 oligomerization. (A) In vitro oligomerization assay analyzed by confocal microscopy. Representative fluorescence pictures (black and white conversion) of recombinant GFP-Cab45WT and phosphomutants incubated without (w/o) Ca2+, together with 2 mM Ca2+ or after additional treatment with 2 mM EDTA. Scale bars, 10 µm. Insets display higher magnification of recombinant proteins treated with 2 mM Ca2+ in original colors (scale bars, 1 µm). (B) Distribution of Cab45 oligomers (2 mM Ca2+) according to their particle size in percentage. Insets show examples of oligomers with pixel units of 3–5, 5–11, 11-20, and >20. Data were collected from two independent experiments (n > 268 oligomers per condition) using ImageJ. (C) Average intensity values of oligomers formed by Cab45-WT and phosphomutants, incubated with 2 mM Ca2+. For analysis, individual background intensities were subtracted, and the pixel with the highest intensity value within the oligomer was measured using ImageJ. Data were collected from two independent experiments (n > 124 oligomers per condition). Statistical test, Kruskal–Wallis. (D) Distribution of Cab45 oligomers according to their intensity values in percentage. Oligomers analyzed in C were grouped based on their intensities (<500 a.u., 500–1,000 a.u., 1,000–1,500 a.u., 1,500–2,000 a.u., and >2,000 a.u.). ***, P < 0.001.
Furthermore, we subdivided the observed particles into four groups: very small (3–5 pixel units), small (5–11 pixel units), medium (11–20 pixel units), and large oligomers (>20 pixel units; Fig. 7 B). Interestingly, very small oligomers were mainly formed by Cab45-WT and Cab45-5pXE (47% and 57% of all analyzed oligomers), whereas the major fraction of Cab45-5pXA had a pixel size of 5–11 units (37%). In general, among larger particles (>11 pixel units), Cab45-5pXA oligomers showed the highest proportion.
In addition, we determined the intensities of counted oligomers by measuring the pixel with the highest intensity within each particle. On average, we could detect significant higher intensity levels in oligomers formed by Cab45-5pXA, followed by Cab45-WT and Cab45-5pXE (Fig. 7 C). The same datasets were also used to display the distribution of oligomers according to their intensity values (Fig. 7 D). Strikingly, the highest number of oligomers of Cab45-WT (55%) and Cab45-5pXE (76%) showed low intensity (maximum <500 a.u.); in contrast, most Cab45-5pXA oligomers (38%) reached maximum intensity values between 500 and 1,000 a.u. Also among higher intensity levels (maximum >1,000 a.u.), oligomers formed by Cab45-5pXA made up the greatest fraction.
Overall, our data suggest that upon incubation with 2 mM of Ca2+, the Cab45-5pXA mutant forms mainly larger oligomeric structures with higher intensities compared with oligomers formed by Cab45-WT and Cab45-5pXE. These findings indicate a potential oligomerization-prone behavior of the phosphorylation-deficient mutant.
Discussion
In this study, we demonstrate that Fam20C has a significant impact on protein sorting and secretion, specifically of Cab45 clients (Crevenna et al., 2016; Deng et al., 2018), by phosphorylating the sorting protein Cab45 on five distinct residues (S99, T131, S142, T193, and S349). Phosphorylation of Cab45 facilitates its exit from the TGN by reducing the size of the Cab45 oligomers, whereas Ca2+-binding seems to be unaffected. Hence, phosphorylation of Cab45 enhances the sorting and secretion of its client LyzC, without impairing the cargo binding. For the first time, we propose a sorting event of soluble secreted proteins, mediated by a TGN luminal protein complex, which is regulated by phosphorylation.
Cab45 is a Ca2+-binding protein with six EF-hand domains that localizes in the lumen of the Golgi apparatus (Blank and von Blume, 2017; von Blume and Hausser, 2019; von Blume et al., 2012; Crevenna et al., 2016; Pakdel and von Blume, 2018). In the presence of Ca2+ that has been locally pumped into the TGN by the Ca2+ ATPase SPCA1, Cab45 oligomerizes and sorts cargos (von Blume et al., 2012; Kienzle et al., 2014; Crevenna et al., 2016). This oligomerization process is required for packaging Cab45 clients into SM-rich transport vesicles at the TGN (Deng et al., 2018).
In search of other regulatory factors that contribute to the sorting of clients, we detected phosphorylation of Cab45 via Golgi kinase Fam20C. By mimicking the negative charges of the phosphorylation, Cab45-5pXE translocated in accumulated vesicles around the Golgi. A similar vesicular phenotype was observed before for Cab45-6EQ, a Ca2+-binding-deficient mutant of Cab45, that is no longer able to form large oligomeric structures. Cab45-6EQ is highly secreted and was shown to mis-sort LyzC (von Blume et al., 2012; Crevenna et al., 2016, Deng et al., 2018). In contrast, we demonstrate the correct sorting of LyzC into SM-rich vesicles in cells expressing the phosphomutants Cab45-5pXA (phosphorylation-deficient) or Cab45-5pXE (Fig. 5 F). Strikingly, in vitro analyses of these phosphomutant proteins uncovered that preventing its phosphorylation results in an increased oligomerization potential of Cab45 (Fig. 7). Since none of the predicted phosphorylation sites target EF-hand domains, we presume that Ca2+ binding is not disturbed upon Cab45 phosphorylation. Additionally, CD data presented in Fig. S2 B support this hypothesis, as protein folding and therefore the overall ability to bind Ca2+ does not seem to be affected. Noteworthy, the recombinant WT protein behaves in a similar way as the phospho-mimicking mutant Cab45-5pXE in the oligomerization assay, which might indicate that the protein purified from HEK293 cells also has phosphorylated sites. This behavior is also supported by experiments performed in cells (Fig. 5, A–C; and Fig. 6 B) where significant differences are more dominant between Cab45-5pXE and Cab45-5pXA, whereas the Cab45-WT phenotype is more similar to the Cab45-5pXE phenotype. In contrast, preventing phosphorylation of Cab45-WT by using transfected Fam20C-KO cells phenocopies the secretion phenotype of Cab45-5pXA (Fig. 5, D and E). This observation supports the idea that Cab45 is already phosphorylated under physiological conditions in the Golgi.
Is the localization of Cab45 also influenced by its oligomerization potential?
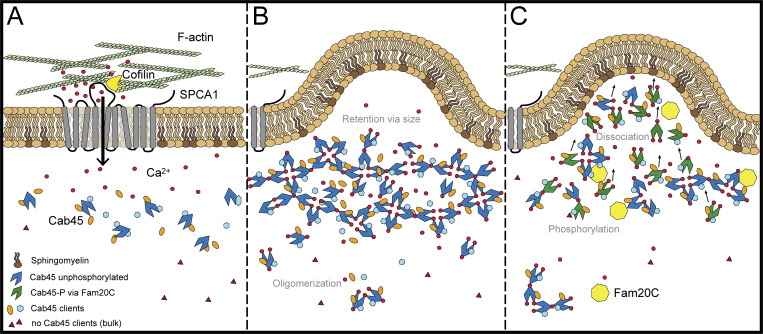
The oligomerization of Cab45 is a Ca2+-dependent process and can be dissolved upon the incubation of recombinant protein with Ca2+ chelators like EGTA (Crevenna et al., 2016). Interestingly, endogenous Golgi-localized Cab45 was secreted in higher amounts when cells were treated with the Ca2+ ionophore A23187 (von Blume et al., 2012), indicating the requirement of Ca2+ for its retention in the TGN. Similar behavior was also suggested earlier for ER-located Ca2+-binding proteins (e.g., reticuloplasmins) that are released from the ER when cells are treated with Ca2+ ionophores (Booth and Koch, 1989). The authors further suggest that perturbation of Ca2+ directly influences the retention of the proteins. So far, Cab45 was thought to reside in the Golgi (Scherer et al., 1996); however, not much is known about the retention mechanisms of Golgi-resident proteins, especially of soluble proteins. Nucleobindin 1, another soluble protein, has (similar to Cab45) two EF-hand domains and localizes mostly in the cis-Golgi before it is partly secreted (Miura et al., 1994; Lin et al., 1998, 1999). Interestingly, Nucleobindin 1 is a known Fam20C substrate (Tagliabracci et al., 2015), but how the protein regulates its localization and if it is able to oligomerize is not established. Since our data show that Cab45 localizes in the TGN but also goes together with its clients into SM-rich vesicles (Deng et al., 2018), we assume that the release of Cab45 into secretory vesicles is essential for the sorting. We therefore state that phosphorylation of Cab45 fine-tunes the size of the oligomers, which drives the export of the protein from the TGN in the following way (Fig. 8). Upon Ca2+ influx into the TGN by SPCA1, Cab45 oligomerizes, binds secretory clients, and segregates them away from the bulk of proteins (A). As a consequence, Cab45 forms large oligomeric structures, which are too big to be packaged into secretory vesicles and so are retained together with the clients in the TGN (B). This size according retention was hypothesized earlier for Golgi-resident transmembrane proteins that aggregate and so are excluded from transport vesicles (Nilsson et al., 1993). As a way to actively regulate the secretion of cargo molecules, we propose that Fam20C phosphorylates Cab45, which causes disassembly of the Cab45 multimers (C). This was also investigated for other proteins, such as Sae2, a protein involved in double-strand break repair (Fu et al., 2014). Sae2 disassembles into active monomers in a stepwise manner upon phosphorylation, which in addition enhances Sae2’s solubility. Similarly, phosphorylation of the heat shock protein αB-crystallin reduces the size of oligomers (Ito et al., 2001). We furthermore assume that oligomer dissociation decreases the retention of Cab45 within the TGN. In support of this idea, we found more phosphorylation-mimicking Cab45 in vesicular fractions by performing semi-intact budding and RUSH assays (Fig. 6, C–G). Consequently, cargo that is bound to Cab45 is packed along with disassembled multimers into SM-rich vesicles for further transport. In this regard, secretion of LyzC was significantly reduced in Fam20C-KO cells (Fig. 1, B and C), even though LyzC itself is not phosphorylated by Fam20C (Fig. 3 B; and Fig. 5, D and E). Defects in LyzC secretion are phenocopied in Cab45-KO cells (Crevenna et al., 2016), further emphasizing the importance of Fam20C in Cab45-mediated cargo sorting. Since phosphorylation and dissociation of oligomers are occurring close to “TGN exit sites” (Fig. 2 C), this would in addition explain why Fam20C and Cab45 bud together in the same SM-rich vesicles. Overall, this process enables Fam20C to regulate the exact time point of cargo release by modifying Cab45, but it could also trigger its own secretion.
Figure 8.
Fam20C phosphorylates Cab45 and regulates client secretion by oligomer disassembly. The model depicts the newly investigated role of Fam20C in Cab45 client sorting. A subset of soluble secretory proteins is sorted via the SPCA1/Cab45 sorting machinery. (A) SPCA1 interacts with cofilin1 on its cytosolic interface, recruits F-actin and promotes Ca2+ influx into the lumen of the TGN. (B) In the presence of Ca2+, Cab45 binds secretory clients and forms large oligomeric structures, which are, because of their size, excluded from packaging into vesicles. (C) Fam20C phosphorylates Cab45 on specific residues, whereby Cab45 multimers disassemble and TGN retention is abolished. This allows clients together with Cab45 to be packed into SM-rich vesicles.
However, internal or external stimuli that are necessary to initiate Cab45 phosphorylation and control this process require further investigations. It is noteworthy that SPCA1 pumps Mn2+ along with Ca2+ into the Golgi, and Mn2+ was shown to be a crucial cofactor for Fam20C activity (Tagliabracci et al., 2012). This fact might explain how both processes, Ca2+-binding and phosphorylation, are coordinated on time. Another question that has to be addressed is how the budding of SM-rich vesicles is regulated. Similar to Cab45, glycophosphatidylinositol-anchored proteins do not contain cytosolic tails to interact with coat components (Mayor and Riezman, 2004). In HeLa cells, these proteins are also sorted into SM-rich vesicles at the TGN (Deng et al., 2016). It is assumed that upon oligomerization of glycophosphatidylinositol-anchored proteins, tightly packed lipid platforms are combined that facilitate its sorting to the apical plasma membrane (Paladino et al., 2004). Potentially, Cab45 binds to transmembrane proteins or lipid components, whereas oligomerization creates similarly “SM-rich sorting domains.” The Cab45–client complex might in addition have curvature-active properties analogous to the endocytosis of toxins such as Shiga toxin, which favors and stabilizes vesicle formation (Cummings et al., 1998; Johannes and Römer, 2010; Pezeshkian et al., 2017; Römer et al., 2007). Subsequently, the dissociation of large oligomeric complexes upon Cab45 phosphorylation might promote its internalization into SM-rich vesicles. However, further investigations are necessary to elucidate this. So far, no interaction between Cab45 and transmembrane proteins or lipids has been described. From our semi-intact budding assays, we assume that budding may not require a cytosolic coat, such as clathrin.
In summary, we were able to demonstrate that phosphorylation of Cab45 by Fam20C regulates Cab45 retention within the TGN by changing its oligomerization potential and thus enhances secretion of Cab45 clients. As phosphorylation is a fast and controllable modification, the predicted mechanism not only contributes to the overall sorting capacity of the cell but also allows cells to dynamically adjust secretion to environmental changes. These results provide unique mechanistic insight into how secreted proteins are exported from the TGN, which is essential for the understanding of cellular homeostasis and communication.
Materials and methods
Cloning and constructs
All clonings were conducted using Phusion High-Fidelity Polymerase and T4 ligase (Thermo Fisher Scientific) according to the manufacturer’s advice. Sequences of all plasmids were verified using the SmartSeq Kit from Eurofins Genomics or KECK sequencing (Yale University). All restriction enzymes were purchased from NEB and also used as recommended.
The cloning of the pLPCX-Cab45 constructs was previously described (Crevenna et al., 2016). Using site-directed mutagenesis, point mutations were introduced to generate missense mutations substituting five phosphorylation sites (S99, T131, S142, T193, and S349) of Cab45 to alanine (A) or glutamic acid (E). The resulting constructs were named pLPCX-Cab45-5pXA (for alanine substitutions) and pLPCX-Cab45-5pXE (for glutamic acid substitutions). All three constructs contained a C-terminal HA tag.
For protein purification from mammalian HEK293 cells, Cab45-WT, Cab45-5pXA, and Cab45-5pXE were inserted into the piggyBac system as described previously (Li et al., 2013). The used piggyBac backbone vector (PB-T-PAF) and also PB-RN and PBase were a gift of James Rini, University of Toronto, Ontario, Canada (Li et al., 2013).
To generate a PB-T-PAF-His-SUMO-Cab45 construct, Gibson assembly with two fragments was used. The first fragment containing a SS-His-SUMO tag was amplified from the pI-secSUMOstar-Cab45 construct as described previously (Crevenna et al., 2016). The second fragment containing Cab45 or the phosphorylation mutants was amplified from the corresponding pLPCX constructs described above. The PB-T-PAF vector was linearized with NheI and NotI-HF restriction enzymes.
PB-T-PAF-GFP-Cab45 variants were cloned in a similar way. PB-T-PAF was linearized with NheI and NotI-HF restriction enzymes. Insert was integrated using three-fragment Gibson Assembly. Fragment 1 (SS-His-tag) and fragment 3 (Cab45-WT/variants) were amplified from PB-T-PAF-His-SUMO-Cab45 constructs above. Superfolder GFP (fragment 2) was amplified from sfGFP-N1 (Addgene; #54737). All fragments contained proper overlaps for Gibson Assembly approach.
For live-cell microscopy, pLPCX-Cab45 constructs were fused with EGFP. The cloning strategy of the EGFP-Cab45 construct was described before (Deng et al., 2018). For a one-fragment Gibson assembly, this vector was linearized with EcoRI-HF and NotI-HF. The Cab45 5pXA or 5pXE inserts were amplified with suitable overhangs from the corresponding pLPCX constructs described above.
Full-length Fam20C was amplified from a pCMV-Sports6 vector obtained from Vincent Tagliabracci’s laboratory (UT Southwestern Medical Center, Dallas, TX) by PCR. This fragment was inserted with EcoRI/BglII restrictions sites into the pLPCX empty vector (Addgene). To generate an inactive kinase as a control, D478 was mutated to alanine by site-directed mutagenesis. For easy Western blot, an HA tag was attached at the C-terminus of Fam20C.
Cloning of pLPCX-LyzC-mCherry was previously described (Deng et al., 2018). For the mCherry-Fam20C fusion construct, three-fragment Gibson assembly was used. Fragment 1 (forward and reverse sequence) includes the signal peptide with overhangs to pLPCX vector and mCherry (fragment 2) and was directly ordered from IDT. mCherry was amplified from pLPCX-LyzC-mCherry (see above). Fam20C (fragment 3) was amplified from the full-length pLPCX plasmid described above. For this purpose, the backbone was linearized with the restriction enzymes BamHI-HF and NotI-HF.
The pIRESneo3-Str-SBP-EGFP vector was a gift from Franck Perez, Institut Curie Centre de Recherche, Paris, France (Addgene; plasmid #65264). The other RUSH constructs pIRESneo3-Str-KDEL-SBP-EGFP-CatD and pIRESneo3_Str-LyzC-KDEL-SBP-EGFP were published previously (Deng et al., 2018). Cab45 RUSH construct pIRESneo3-Str-SBP-tagRFP-Cab45 was generated via two-fragment Gibson approach. Creating overlapping parts, Fragment 1 (SS-SBP) was amplified from CatD-RUSH construct (see Deng et al., 2018); tagRFP-Cab45 (fragment 2) was amplified from pLPCX-SP-tagRFP-Cab45 (not published). For this construct, tag-RFP was amplified from pCAG-tagRFP-NLS-HA-Bxb1 (Addgene; plasmid #65625). Fragments were reinserted into CatD-RUSH vector linearized with AscI and XhoI. The osteopontin (OPN) RUSH construct pIRESneo3-Str-OPN-SBP-EGFP was cloned via one-fragment Gibson assembly approach. OPN-V5 was amplified from cDNA4-OPN-V5 (a gift from Vincent Tagliabracci, UT Southwestern Medical Center, Dallas, TX) and inserted into pIRESneo3-Str-SBP-EGFP that was linearized with AscI and XhoI.
The pcDNA3.1_Flag-LyzC plasmid was a gift from V. Malhotra (Center of Genomic Regulation, Barcelona, Spain; von Blume et al., 2012).
The plasmids Eq-SM-oxGFP and Eq-sol-oxGFP were are gift from C. Burd (Yale University, New Haven, CT).
Cell culture and stable transfection
All cell lines were maintained in DMEM supplemented with 10% FCS (Gibco) and Penicillin/Streptomycin (P/S) at 37°C and 5% CO2. To create stable expressing HeLa cell lines, plasmids of Cab45-WT, Cab45-5pXA, Cab45-5pXE, Fam20C-WT, and Fam20C-KD were introduced via VSV-G pseudotyped retroviral vectors as described previously (Crevenna et al., 2016).
Doxycycline-inducible stable HEK293 cell lines for protein expression were generated using the transposon-based piggyBac system (Li et al., 2013). Therefore, HEK293 cells were transfected with PB-T-PAF (gene of interest introduced), PB-RN, and PBase (total DNA 1.5 µg; ratio 8:1:1) using polyethylenimine (PEI; Alfa Aesar). Cell culture medium was removed the next day. 48 h after transfection, selection of cells was performed in supplemented DMEM containing 10 µg/ml puromycin dihydrochloride (Sigma-Aldrich) and 500 µg/ml G 418 disulfate salt (Sigma-Aldrich).
Transient transfection and siRNA knockdown
Cells were transiently transfected with 1.25 mg/ml PEI (Alfa Aesar) or Lipofectamine LTX PLUS (Thermo Fisher Scientific) following manufacturer protocols.
For siRNA knockdown, the following oligos were used, as well as scrambled negative control from Invitrogen: SMS1, 5′-GACGGCAGCUUCAGCAUCAAGAUUA-3′; SMS2: 5′-UCAAUAGUGGGACGCAGAUUCUGUU-3′.
20 nM siRNA was added to 100 µl Opti-MEM reduced serum medium (Gibco by Life Technologies) together with 12 µl HiPerFect Transfection Reagent (Qiagen). The siRNA transfection mix was incubated at room temperature for 15 min and added dropwise to the seeded cells. Transfection was performed 24–48 h before further analysis.
CRISPR/Cas9 KO cell lines
HeLa Fam20C KO cells were generated using the CRISPR/Cas9 technique. Fam20C was targeted downstream of the signal sequence with gRNA next to the Cas9 recognition site protospacer adjacent motif (PAM). 20 nt gRNA oligos targeting Fam20C (DMP4) were designed using www.genome-engineering.org/crispr (Ran et al., 2013).
Fam20C-gRNA sequence: 5′-GGGCGATGTGCAGCGCGCAG-3′
gRNA sequences were synthesized by Metabion. Oligos (forward and reverse sequence) were cloned into mammalian expression vector pX459 (Addgene; plasmid #62988) using BbsI cutting sites. HeLa parental cells were transfected using PEI as described above. 24 h after transfection, cells were selected in 2 µg/ml puromycin for 48 h. Single clones were isolated, expanded, and checked for deletion by sequencing with following sequencing primers: forward, 5′-CACCGATGGACCTTGACCC-3′; reverse, 5′-AGTGGGACGAGAGGTTGGAG-3′.
In addition, KO was checked using MS analysis. Golgi fractions of HeLa WT and Fam20C-KO cells were isolated as described previously (Crevenna et al., 2016) and separated via SDS-PAGE. Samples from gel lanes were digested with trypsin by an in-gel digestion protocol, and peptides were extracted and purified via C18 StageTips. Peptides were analyzed in a Q Exactive HF machine using data dependent acquisition scheme using Higher Energy Collisional Dissocation (HCD) fragmentation. Raw data were processed using the Maxquant computational platform, and the peak lists were searched against human reference proteome database from UniProt. All identifications were filtered at 1% false discovery rate (FDR) and label-free quantification.
Protein expression and purification of His-Sumo-Cab45 variants
Doxycycline-inducible HEK293-Cab45 cells were grown in p15 culture dishes until they reached full confluence. Cells were carefully washed three times with DPBS (Gibco) before starting protein expression. Therefore, cells were incubated for 20 h with DMEM supplemented with 1 µg/ml doxycycline monohydrate (LKT Laboratories) and 1 µg/ml Aprotinin (Sigma-Aldrich). Cell culture supernatant was collected, centrifuged (5 min; 1,000 rpm, 4°C) to remove dead cells, and filtered via 0.2-µm syringe filters. His-SUMO/GFP-Cab45 variants were purified using nickel-based affinity chromatography as described previously (Crevenna et al., 2016). Protein was stored at −80°C with 10% vol/vol after snap freezing in liquid nitrogen. His-Cofilin-S3E was purified as described previously (Kienzle et al., 2014).
Antisera
The antisera used were rat monoclonal anti-HA (Roche; catalog #11867423001), rabbit monoclonal anti-KIAA0310 (Thermo Fisher Scientific; catalog #A300-648A-M), mouse monoclonal anti-cis-Golgi matrix protein 130 (BD Biosciences; catalog #610823), mouse monoclonal anti-CNX (Sigma-Aldrich; catalog #C7617), mouse monoclonal anti-p230 (BD Biosciences; catalog #611281), sheep polyclonal anti-TGN46 (AbD Serotec; catalog #AHP500G), mouse monoclonal anti-β-actin (Sigma-Aldrich; catalog #A5441), mouse monoclonal ANTI-FLAG M2-peroxidase (HRP; Sigma-Aldrich; catalog #A8592), mouse monoclonal anti-EEA1 antibody (BD Biosciences; catalog #610457), rabbit polyclonal anti-Rab11 antibody (gift from the De Camilli laboratory, Yale School of Medicine, New Haven, CT, self-made), rabbit monoclonal anti-Lamp1 antibody (Cell Signaling; catalog #9091S), and mouse monoclonal anti-CatD antibody (BD Biosciences; catalog #610801).
Secondary antibodies were purchased from Thermo Fisher Scientific (catalog numbers A-11034, A-11029, A-21209, A-21208, A-21203, 32260, 31470, and 32230).
Anti-Cab45 antibody was generated by the animal facility (immunization service) of the Max Planck Institute of Biochemistry as described previously (Crevenna et al., 2016). Full-length recombinant Cab45 (His-Sumo tagged) was used for rabbit immunization.
Protein detection by immunoblotting, Coomassie staining, and immunofluorescence
For detection of intracellular proteins, cells were washed three times with PBS and lysed with 1% TritonX-100 in PBS. Secreted proteins in supernatants were concentrated using Centrifugal Filters (Amicon Ultra). Proteins were separated according to size via SDS-PAGE.
For Coomassie staining, SDS gel was rinsed with PBS and prefixed in 40% vol/vol MeOH, 10% vol/vol acetic acid (HOAc) for 10 min. Gel was incubated in 40% vol/vol MeOH, 10% vol/vol HOAc with 0.1% weight/vol Coomassie Brilliant Blue R overnight and destained using 40% vol/vol MeOH, 10% vol/vol HOAc until single bands were visible. For storage, gel was put in 7% HOAc at 4°C.
For immunoblotting, proteins were transferred to nitrocellulose membranes using a wet blot system (Bio-Rad). Membranes were incubated in 5% BSA in TBS for at least 1 h and incubated with specific primary and corresponding secondary HRP-coupled antibody with additional washing steps with TBS-Tween (0.1%) in between. Proteins were detected via chemiluminescence (Thermo Fisher Scientific), acquired with ChemiDoc Imaging System (Bio-Rad).
For immunostaining, cells were cultured in six wells on glass slides and fixed for 10 min with 4% paraformaldehyde. After washing with PBS, cells were permeabilized for 5 min in 0.2% Triton-X 100 and 0.5% SDS in 4% BSA or saponin for at least 30 min. After washing with PBS, cells permeabilized with Triton-X 100 were blocked with 4% BSA for 1 h. Cells were incubated with primary followed by corresponding secondary antibody for 1 h at room temperature in blocking buffer in the dark. Slides were washed three times with PBS after incubation with antibody. Glass slides were mounted with ProLong Gold (Thermo Fisher Scientific). Acquisition was done using confocal laser-scanning microscope (Carl Zeiss; 40×, LSM 780, software ZEN 2010). If contrast was changed for better visualization of e.g., vesicles, this was done equally for all conditions.
Pearson’s correlation
Pearson’s correlation was performed using ImageJ 1.52c.
Secretion assay after 20°C block
For the secretion assay, cells were washed three times with PBS and incubated for 2 h at 20°C in DMEM supplemented with 100 µg/ml cycloheximide. Supernatant of control cells was taken directly after 20°C incubation to check basal secretion during block and concentrated for 5 min at 16,000 ×g at 4°C in Centrifugal Filters (Amicon Ultra; Ultracel 10K); cells were lysed with 1% TritonX-100 in PBS. Experimental cells were shifted to 37°C and incubated for 1 h to start protein release from the Golgi. Supernatants were taken and concentrated like before. Cells were lysed as well. Protein secretion was checked by Western blot analysis as described above. Protein levels (densitometry) were quantified from at least three independent experiments using Fiji 1.0 software. Supernatant signals were normalized to corresponding protein signals in cell lysates. Actin signals were used as loading controls. Values of controls (20°C block only) were subtracted from values of cells passed through 37°C release. Final values were normalized to WT cells (100%) and plotted as a bar graph.
RUSH cargo sorting assay using confocal microscopy
RUSH assay was performed as described previously (Deng et al., 2018). Different cells lines were cultured in six wells on glass slides and transfected with RUSH constructs. 16 h after transfection, cells were incubated with 40 µM d-Biotin (SUPELCO) in DMEM for different time points (20, 30, 40, 50, and 60 min or without d-Biotin (control). Cells were washed once with PBS, fixed in 4% PFA in PBS for 10 min, and mounted on coverslips using ProLong Gold (Thermo Fisher Scientific). If required, costaining was done (see above). Acquisition of either GFP or RFP was done at 20°C using a confocal laser-scanning microscope (Carl Zeiss; LSM 780 or LSM 880; 40×/1.4, 63×/1.4, 100×/1.46 NA Plan-Apochromat oil objectives, software ZEN 2010) by imaging z-stacks with a step size of 0.35 µm.
For quantification of vesicles, we empirically measured the sizes of objects between 4 and 20 pixels using the Analyze Particles function in ImageJ, which detects vesicular structures but omits larger structures such as the Golgi. While small-fragmented and isolated Golgi structures could be detected in error, such structures are rare. Furthermore, only vesicles of cells expressing the RUSH construct were counted. The Fiji macro count_fixed_vesicles_V1.3 (M. Pakdel) including the Particle Analyzer plug-in by Fiji was used to determine the number of vesicles.
Golgi vesicle budding assays
Microscopy budding assay
Live-cell experiments were done as described previously (Deng et al., 2018) at 37°C. HeLa cells were seeded in live cell μ-dishes and transfected with Fam20C-mCherry and EGFP-Cab45/Eq-SM-oxGFP/Eq-Sol-oxGFP. Stable HeLa Cab45 WT/mutant cells were transfected with LyzC-mCherry and Eq-SM-oxGFP. Medium was exchanged to live-cell imaging solution (Molecular Probes) supplemented with 10 mM glucose. Imaging was done by live-cell wide-field microscopy. Dual-channel acquisition for EGFP and mCherry was performed at 1-s intervals for 100 frames on a GE Healthcare DeltaVision Elite system based on an OLYMPUS IX-71 inverted microscope, an OLYMPUS 60×/1.42 PLAPON oil objective and a PCO sCMOS 5.5 camera. For deconvolution software softWoRx v.6.0 was used. Budding events and colocalizaton were scored manually from time-lapse series choosing one image out of frame (ImageJ).
The time-lapse movies were analyzed by identifying budding events from the Golgi. Fam20C budding events were scored whether they are EGFP-Cab45, Eq-SM-oxGFP, or Eq-Sol-oxGFP positive. For LyzC budding, presence of Eq-SM-oxGFP in Cab45-WT and mutant cells was checked.
The ratios of Cab45 budding with or without EQ-SM were calculated to total budding events. The mean and ±SD of the ratios were plotted as a bar graph.
Budding assay in semi-intact cells
Cells were incubated for 2 h at 20°C with DMEM, supplemented with FCS, Penicillin/Streptomycin, and 100 µg/ml cyclohexamide. Cells were washed 1x with PBS and trypsinized. Cells were counted, same amount of cells per condition was washed 2× with buffer A (20 mM Hepes, 250 mM D-sorbitol, 150 mM potassium acetate). Cells were permeabilized for 5 min with 40 µg/ml digitonin on ice and washed three times with buffer A. Cells were divided (control and experimental condition). Control cells were incubated for 45 min at 4°C in buffer A. Experimental cells were incubated for 45 min at 32°C with an ATP regenerating system (1 mM ATP, 40 mM creatine phosphate, 0.2 mg/ml creatine phosphokinase and 0.1 mM GTP). The reactions were centrifuged for 10 min at 10,000 g, supernatant was collected and centrifuged for 100,000 g for 1 h. Vesicular fraction (pellet) was dissolved in SDS-loading buffer and analyzed by Western blotting. Protein levels (densitometry) were quantified from at least three independent experiments using Fiji 1.0 software. Values of controls (4°C incubation) were subtracted from values of cells incubated with ATP regeneration system at 32°C. Final values were normalized to WT cells (100%) and plotted as a bar graph.
CD spectroscopy
CD spectroscopy measurements were performed in a 1-mm (path length) cuvette at 10°C on a JASCO J-715 spectrometer. Protein samples (0.2 mg/ml) were dissolved in CD buffer (20 mM Tris, pH 6.8, and 500 mM NaCl), and the indicated amounts of Ca2+ were added before spectra were recorded. An average of three (± Ca2+ analysis) independent spectra (from 195 to 250 nm with 0.1-nm spacing) were documented. Data were normalized to molecular ellipticity of protein, and a fast Fourier transform (FFT) filter was applied.
LyzC immunoprecipitation
Recombinant His-SUMO-Cab45 variants (purified as described above) and controls (His-Cofilin-S3E) were incubated with Ni2+-agarose beads (Thermo Fisher Scientific; catalog #25214) for 2 h at 4°C on turning wheel, in immunoprecipitation buffer (50 mM Tris, 100 mM NaCl, and 0.1% TritonX-100 with protease inhibitors; Roche; catalog #11836170001) containing 100 µM Ca2+. Beads were washed three times with immunoprecipitation buffer with Ca2+ and incubated for 2 h at 4°C with LyzC (Sigma-Aldrich) on a turning wheel. Beads were washed again three times and boiled in 1× Laemmli buffer. Coomassie staining was performed as described above.
MS for phosphoproteomic analysis
HeLa S3 suspensions cells stably expressing Fam20C or Fam20C-D478A (4 liter roller culture per condition) were harvested and pelleted. Pellets were then washed once in Breaking Buffer (10 mM Tris, pH 7.4, and 250 mM sucrose), diluted 1:5 in Breaking Buffer supplemented with cOmplete Tablets Mini EDTA-free (Roche) and homogenized with an EMBL cell cracker (ball size, 8.002 mm; 9-µm gap). After addition of 1 mM EDTA, the sucrose concentration of the homogenate was adjusted to 37% (weight/vol) and overlayed with 35% and 29% sucrose in 10 mM Tris, pH 7.4. Cellular components were separated by ultracentrifugation for 3 h at 133,000 g. The Golgi membrane fraction was extracted, adjusted to Breaking Buffer conditions, and snap-frozen in liquid nitrogen for storage at −80°C.
For MS analysis, all samples were lysed in MS lysis buffer (10 mM Tris, pH 7.5, 4% SDS, and 10 mM DTT), boiled and sonicated, and precipitated overnight using ice-cold acetone (vol/vol 80%). After centrifugation (4,000 g), the pellet was washed at least twice with 80% ice-cold acetone before air drying and resuspension (sonication) in TFE buffer (10% 2–2-2-trifluorethanol and 100 mM ammonium bicarbonate). Proteins were digested using LyzC and trypsin (1:100) overnight at 37°C and phosphopeptides enriched as described previously (Humphrey et al., 2015). Samples were prepared in triplets.
For liquid chromatography–tandem mass spectrometry (LC-MS/MS) sample preparation peptides were purified using in-house prepared stage tips (Rappsilber et al., 2003) Empor SPE disks SDB-RPS (Sigma-Aldrich) before LC-MS/MS analysis as described previously (Kulak et al., 2014). Briefly, stage tips were prepared by inserting two layers of SDB-RPS matrix into a 200 µl pipette tip using an in-house–prepared syringe device. Stage tips were first activated with 100 µl MS buffer C (30% MeOH and 1% trifluoroacetic acid) and then washed with 100 µl MS buffer D (2% acetonitrile and 0.2% trifluoroacetic acid) before loading of the acidified peptides (1% trifluoroacetic acid). After centrifugation, the stage tips were washed three times (200 µl each) with MS buffer D. Elution was performed using 60 µl MS buffer E (60% acetonitrile and 1.25% ammonium hydroxide). Eluates were collected in 200-µl PCR tubes and dried using a Concentrator plus SpeedVac centrifuge (Eppendorf) at 60°C. Peptides were resuspended in MS buffer F (2% acetonitrile and 0.1% trifluoroacetic acid) and briefly sonicated (Branson Ultrasonics) before LC/MS-MS analysis.
For LC-MS/MS measurements, peptides were loaded on a 20- or 50-cm reversed phase column (75-µm inner diameter, packed in-house with ReproSil-Pur C18-AQ 1.9 µm resin [Dr. Maisch]). Column temperature was maintained at 55°C using a homemade column oven. An EASY-nLC 1200 system (Thermo Fisher Scientific) was directly coupled online with the mass spectrometer (Q Exactive) via a nanoelectrospray source, and peptides were separated with a binary buffer system of MS buffer A (0.1% formic acid) and MS buffer G (80% acetonitrile and 0.1% formic acid) at a flow rate of 250 or 350 nl/min. Peptides were eluted with a nonlinear 270-min gradient of 5–60% MS buffer G. After each gradient, the column was washed with 95% MS buffer G for 5 min. The mass spectrometer was programmed to acquire in a data-dependent mode (Top10) using a fixed ion injection time strategy. Full scans were acquired in the Orbitrap mass analyzer with resolution 60,000 at 200 m/z (3E6 ions were accumulated with a maximum injection time of 25 ms). The top intense ions (N for TopN) with charge states ≥2 were sequentially isolated to a target value of 1E5 (maximum injection time of 120 ms, 20% underfill), fragmented by Higher Energy Collisional Dissocation (NCE 25, Q Exactive) and detected in the Orbitrap (Q Exactive, R = 15,000 at m/z 200).
Raw MS data were processed using MaxQuant version 1.5.3.15 (Cox and Mann, 2008) with an FDR <0.01 at the level of proteins, peptides, and modifications. Searches were performed against the Mouse or Human UniProt FASTA database (September 2015). Enzyme specificity was set to trypsin. The search included cysteine carbamidomethylation as a fixed modification and N-acetylation of protein; oxidation of methionine; and/or phosphorylation of serine, threonine, or tyrosine residue (PhosphoSTY) as variable modifications. Up to two missed cleavages were allowed for protease digestion. “Match between runs” was enabled, with a matching time window of 0.5–0.7 min. Bioinformatic analyses were performed with Perseus (www.perseus-framework.org) and Microsoft Excel and data visualized using Graph Prism (GraphPad Software) or Perseus (Tyanova et al., 2016). Significance was assessed using a one-sample t test, two-sample Student’s t test, and ANOVA analysis, for which replicates were grouped, and statistical tests were performed with permutation-based FDR correction for multiple hypothesis testing. Were indicated, missing data points were replaced by data imputation after filtering for valid values (all valid values in at least one experimental group). Error bars represent mean ± SEM or mean ± SD.
In vitro kinase assays
In vitro kinase assays were performed by the Tagliabracci laboratory as described previously (Tagliabracci et al., 2012). Fam20C was purified from insect cells as described previously (Tagliabracci et al., 2012). Incorporation of 32P into Cab45 or LyzC by Fam20C was tested by incubating 0.1 mg/ml recombinant Cab45 or OPN (control) with 0.5 mg/ml recombinant Fam20C-WT or Fam20C-KD for 2 h (reaction mixture: 50 mM Tris-HCl, pH 7.5, 10 mM MnCl2, and 1 mM [γ32P]ATP [Specific Activity = 500 cpm/pmol]). Reaction was stopped by adding 15 mM EDTA and SDS loading buffer. Products were separated by SDS-PAGE followed by Coomassie blue staining and autoradiography. Recombinant Cab45-WT was purified as described above. LyzC was purchased from Sigma-Aldrich.
Cab45 oligomerization assay and analysis
For oligomerization assays, recombinant GFP-Cab45 proteins were purified as described above. Proteins were centrifuged for 10 min at 10,000 g at 4°C to remove aggregates. 0.2 mg/ml protein in 20 mM Tris, pH 7.4, was analyzed in a Lab-Tek 8 Chamber #1.0 borosilicate coverglass system (Nunc) with confocal microscopy (Carl Zeiss; 63×/1.4 NA Plan-Apochromat oil objective, LSM 880, software ZEN 2010) at 20°C. Proteins were incubated 10 min in buffer only, with 2 mM Ca2+ and 2 mM EDTA (after Ca2+ treatment), and z-stacks (0.35 µm) were taken at the bottom area of the well.
For analysis, image showing bottom of the well in focus was used. Individual background intensity for each picture was subtracted (ImageJ macro: https://imagej.nih.gov/ij/macros/SubractMeasuredBackground.txt). Size distribution of oligomers was analyzed using ImageJ software (function: Analyze Particles; 0–1 Circularity). Oligomers were counted by the program according to their pixel size of (>3, >5, >11, and >20 pixel units). Intensity of vesicles was analyzed by measuring the intensity of the pixel with the highest intensity within a particle. According to the amount of oligomers formed by Cab45-5pXA, only a representative section of the image was analyzed.
Quantification and statistical analysis
For statistical evaluation, GraphPad Prism version 7.0b for Mac OS X (GraphPad Software) was used. Means ± SD are plotted in all analyzed graphs. Statistical details can be found in figure legends. Normality testing was performed using normality tests of D’Agositino–Pearson, Shapiro–Wilk, and Kolmogorov–Smirnov. A Kruskal–Wallis test followed by Dunn’s multiple comparisons test was used for RUSH cargo sorting assays, Cab45 vesicle counting, LyzC secretion of Cab45 phosphomutants, Eq-SM vesicle counting, vesicle budding phosphomutants, LyzC immunoprecipitation, and intensity analysis of oligomers. An unpaired t test was used for vesicular budding assay of endogenous Cab45. A Mann–Whitney test was used for colocalization of Fam20C with Eq-SM and Eq-Sol vesicles as well as vesicle counting of Cab45-5pXE with SMS1/2 siRNA knockdown. A Kolmogorov–Smirnov test was used for LyzC secretion of Fam20C-KO. An ordinary one-way ANOVA test was used for LyzC secretion in Fam20C-KO cells rescued with Cab45 constructs. The following P value style was used: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Online supplemental material
Fig. S1 shows experiments related to Fig. 1 and Fig. 3 in which Fam20C-KO and Cab45 phosphorylation sites were identified using MS analysis. Fig. S2 shows additional results related to Fig. 4 in which Cab45 phosphomutants were analyzed for localization, secondary structure, and cargo binding. Fig. S3 shows experiments related to Fig. 5 in which LyzC sorting was further analyzed in Cab45 phosphomutants. Fig. S4 shows experiments related to Fig. 5 in which phosphomutants of Cab45 were additionally tested for specificity in cargo sorting. Fig. S5 shows experiments related to Fig. 6 in which TGN export of Cab45 was further characterized.
Supplementary Material
Acknowledgments
J. von Blume was funded by the Deutsche Forschungsgemeinschaft Perspective Program (Boehringer Ingelheim Foundation; project grant BL 1186/4-1) and the National Institutes of Health National Institute of General Medical Sciences (award number GM134083-01). V. Tagliabracci was funded by the Welch Foundation (grant I-1911).
The authors declare no competing financial interests.
Author contributions: Conceptualization, T.K-H. Hecht, B. Blank, M. Steger, and J. von Blume; Methodology, T.K-H. Hecht, B. Blank, and J. von Blume; Investigation, T.K-H. Heinz Hecht, B. Blank, M. Steger, and V. Lopez; Writing – Original Draft, T.K-H. Hecht, B. Blank, and J. von Blume; Writing – Review & Editing, all authors; Funding Acquisition, J. von Blume; Resources, M. Mann, V. Tagliabracci, and J. von Blume; Supervision, J. von Blume.
References
- Ang S.F., and Fölsch H.. 2012. The role of secretory and endocytic pathways in the maintenance of cell polarity. Essays Biochem. 53:29–39. 10.1042/bse0530029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenholz Y., and Thompson T.E.. 1980. Sphingomyelins in bilayers and biological membranes. Biochimica Et Biophysica Acta Bba - Rev Biomembr. 604:129–158. [DOI] [PubMed]
- Blank B., and von Blume J.. 2017. Cab45-Unraveling key features of a novel secretory cargo sorter at the trans-Golgi network. Eur. J. Cell Biol. 96:383–390. 10.1016/j.ejcb.2017.03.001 [DOI] [PubMed] [Google Scholar]
- Boncompain G., and Perez F.. 2012. Synchronising protein transport in the secretory pathway. Curr Protoc Cell Biol. 15.19.1-15.19.16. [DOI] [PubMed]
- Boncompain G., Divoux S., Gareil N., de Forges H., Lescure A., Latreche L., Mercanti V., Jollivet F., Raposo G., and Perez F.. 2012. Synchronization of secretory protein traffic in populations of cells. Nat. Methods. 9:493–498. 10.1038/nmeth.1928 [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S. 2014. Vesicular transport adds a Nobel. Trends Cell Biol. 24(1):3–5. 10.1016/j.tcb.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth C., and Koch G.L.. 1989. Perturbation of cellular calcium induces secretion of luminal ER proteins. Cell. 59:729–737. 10.1016/0092-8674(89)90019-6 [DOI] [PubMed] [Google Scholar]
- Braulke T., and Bonifacino J.S.. 2009. Sorting of lysosomal proteins. Biochimica Et Biophysica Acta Bba - Mol. Cell Res. 1793:605–614. [DOI] [PubMed] [Google Scholar]
- Chege N.W., and Pfeffer S.R.. 1990. Compartmentation of the Golgi complex: brefeldin-A distinguishes trans-Golgi cisternae from the trans-Golgi network. J. Cell Biol. 111:893–899. 10.1083/jcb.111.3.893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., et al. . 2013. Multiplex genome engineering using CRISPR/Cas systems. Science. 339:819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., and Mann M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:nbt.1511. [DOI] [PubMed] [Google Scholar]
- Crevenna A.H., Blank B., Maiser A., Emin D., Prescher J., Beck G., Kienzle C., Bartnik K., Habermann B., Pakdel M., et al. . 2016. Secretory cargo sorting by Ca2+-dependent Cab45 oligomerization at the trans-Golgi network. J. Cell Biol. 213:305–314. 10.1083/jcb.201601089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings M.D., Ling H., Armstrong G.D., Brunton J.L., and Read R.J.. 1998. Modeling the carbohydrate-binding specificity of pig edema toxin. Biochemistry. 37:1789–1799. 10.1021/bi971807f [DOI] [PubMed] [Google Scholar]
- De Matteis M.A., and Luini A.. 2008. Exiting the Golgi complex. Nat. Rev. Mol. Cell Biol. 9:273–284. 10.1038/nrm2378 [DOI] [PubMed] [Google Scholar]
- Deng Y., Rivera-Molina F.E., Toomre D.K., and Burd C.G.. 2016. Sphingomyelin is sorted at the trans Golgi network into a distinct class of secretory vesicle. Proc. Natl. Acad. Sci. USA. 113:6677–6682. 10.1073/pnas.1602875113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Pakdel M., Blank B., Sundberg E.L., Burd C.G., and von Blume J.. 2018. Activity of the SPCA1 Calcium Pump Couples Sphingomyelin Synthesis to Sorting of Secretory Proteins in the Trans-Golgi Network. Dev. Cell. 47:464–478.e8. 10.1016/j.devcel.2018.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fölsch H. 2005. The building blocks for basolateral vesicles in polarized epithelial cells. Trends Cell Biol. 15:222–228. 10.1016/j.tcb.2005.02.006 [DOI] [PubMed] [Google Scholar]
- Fölsch H. 2008. Regulation of membrane trafficking in polarized epithelial cells. Curr. Opin. Cell Biol. 20:208–213. 10.1016/j.ceb.2008.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fölsch H., Ohno H., Bonifacino J.S., and Mellman I.. 1999. A novel clathrin adaptor complex mediates basolateral targeting in polarized epithelial cells. Cell. 99:189–198. 10.1016/S0092-8674(00)81650-5 [DOI] [PubMed] [Google Scholar]
- Fölsch H., Pypaert M., Schu P., and Mellman I.. 2001. Distribution and function of AP-1 clathrin adaptor complexes in polarized epithelial cells. J. Cell Biol. 152:595–606. 10.1083/jcb.152.3.595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q., Chow J., Bernstein K.A., Makharashvili N., Arora S., Lee C.F., Person M.D., Rothstein R., and Paull T.T.. 2014. Phosphorylation-regulated transitions in an oligomeric state control the activity of the Sae2 DNA repair enzyme. Mol. Cell. Biol. 34:778–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson P.A., Lock J.G., Luke M.R., and Stow J.L.. 2004. Domains of the TGN: coats, tethers and G proteins. Traffic. 5:315–326. 10.1111/j.1398-9219.2004.00182.x [DOI] [PubMed] [Google Scholar]
- Guo Y., Sirkis D.W., and Schekman R.. 2014. Protein sorting at the trans-Golgi network. Annu. Rev. Cell Dev. Biol. 30:169–206. 10.1146/annurev-cellbio-100913-013012 [DOI] [PubMed] [Google Scholar]
- Humphrey S.J., Azimifar B.S., and Mann M.. 2015. High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat. Biotechnol. 33:nbt.3327. [DOI] [PubMed]
- Ito H., Kamei K., Iwamoto I., Inaguma Y., Nohara D., and Kato K.. 2001. Phosphorylation-induced change of the oligomerization state of alpha B-crystallin. J. Biol. Chem. 276:5346–5352. [DOI] [PubMed] [Google Scholar]
- Johannes L., and Römer W.. 2010. Shiga toxins--from cell biology to biomedical applications. Nat. Rev. Microbiol. 8:105–116. 10.1038/nrmicro2279 [DOI] [PubMed] [Google Scholar]
- Kienzle C., and von Blume J.. 2014. Secretory cargo sorting at the trans-Golgi network. Trends Cell Biol. 24:584–593. 10.1016/j.tcb.2014.04.007 [DOI] [PubMed] [Google Scholar]
- Kienzle C., Basnet N., Crevenna A.H., Beck G., Habermann B., Mizuno N., and von Blume J.. 2014. Cofilin recruits F-actin to SPCA1 and promotes Ca2+-mediated secretory cargo sorting. J. Cell Biol. 206:635–654. 10.1083/jcb.201311052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman J. 2011. Architecture of the mammalian Golgi. Cold Spring Harb. Perspect. Biol. 3 a005181 10.1101/cshperspect.a005181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak N.A., Pichler G., Paron I., Nagaraj N., and Mann M.. 2014. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 11:319–324. 10.1038/nmeth.2834 [DOI] [PubMed] [Google Scholar]
- Li Z., Michael I.P., Zhou D., Nagy A., and Rini J.M.. 2013. Simple piggyBac transposon-based mammalian cell expression system for inducible protein production. Proc. Natl. Acad. Sci. USA. 110:5004–5009. 10.1073/pnas.1218620110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P., Le-Niculescu H., Hofmeister R., McCaffery J.M., Jin M., Hennemann H., McQuistan T., De Vries L., and Farquhar M.G.. 1998. The mammalian calcium-binding protein, nucleobindin (CALNUC), is a Golgi resident protein. J. Cell Biol. 141:1515–1527. 10.1083/jcb.141.7.1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P., Yao Y., Hofmeister R., Tsien R.Y., and Farquhar M.G.. 1999. Overexpression of CALNUC (nucleobindin) increases agonist and thapsigargin releasable Ca2+ storage in the Golgi. J. Cell Biol. 145:279–289. 10.1083/jcb.145.2.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor S., and Riezman H.. 2004. Sorting GPI-anchored proteins. Nat. Rev. Mol. Cell Biol. 5:110–120. 10.1038/nrm1309 [DOI] [PubMed] [Google Scholar]
- Mellman I., and Nelson W.J.. 2008. Coordinated protein sorting, targeting and distribution in polarized cells. Nat. Rev. Mol. Cell Biol. 9:833–845. 10.1038/nrm2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P., Mani D.R., Ruggles K.V., Gillette M.A., Clauser K.R., Wang P., Wang X., Qiao J.W., Cao S., Petralia F., et al. ; NCI CPTAC . 2016. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 534:55–62. 10.1038/nature18003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Kurosawa Y., and Kanai Y.. 1994. Calcium-binding activity of nucleobindin mediated by an EF hand moiety. Biochem. Biophys. Res. Commun. 199:1388–1393. 10.1006/bbrc.1994.1384 [DOI] [PubMed] [Google Scholar]
- Munro S. 1995. An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 14:4695–4704. 10.1002/j.1460-2075.1995.tb00151.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. 2005. The Golgi apparatus: defining the identity of Golgi membranes. Curr. Opin. Cell Biol. 17:395–401. 10.1016/j.ceb.2005.06.013 [DOI] [PubMed] [Google Scholar]
- Nilsson T., Slusarewicz P., Hoe M.H., and Warren G.. 1993. Kin recognition. A model for the retention of Golgi enzymes. FEBS Lett. 330:1–4. 10.1016/0014-5793(93)80906-B [DOI] [PubMed] [Google Scholar]
- Pakdel M., and von Blume J.. 2018. Exploring new routes for secretory protein export from the trans-Golgi network. Mol. Biol. Cell. 29:235–240. 10.1091/mbc.E17-02-0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paladino S., Sarnataro D., Pillich R., Tivodar S., Nitsch L., and Zurzolo C.. 2004. Protein oligomerization modulates raft partitioning and apical sorting of GPI-anchored proteins. J. Cell Biol. 167:699–709. 10.1083/jcb.200407094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezeshkian W., Gao H., Arumugam S., Becken U., Bassereau P., Florent J.-C., Ipsen J.H., Johannes L., and Shillcock J.C.. 2017. Mechanism of Shiga Toxin Clustering on Membranes. ACS Nano. 11:314–324. 10.1021/acsnano.6b05706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzo P., Lissandron V., and Pozzan T.. 2010. The trans-golgi compartment: A new distinct intracellular Ca store. Commun. Integr. Biol. 3:462–464. 10.4161/cib.3.5.12473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 8(11):2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Ishihama Y., and Mann M.. 2003. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75:663–670. 10.1021/ac026117i [DOI] [PubMed] [Google Scholar]
- Römer W., Berland L., Chambon V., Gaus K., Windschiegl B., Tenza D., Aly M.R., Fraisier V., Florent J.-C., Perrais D., et al. . 2007. Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature. 450:670–675. 10.1038/nature05996 [DOI] [PubMed] [Google Scholar]
- Scherer P.E., Lederkremer G.Z., Williams S., Fogliano M., Baldini G., and Lodish H.F.. 1996. Cab45, a novel (Ca2+)-binding protein localized to the Golgi lumen. J Cell Biol. 133(2):257–268. 10.1083/jcb.133.2.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliabracci V.S., Engel J.L., Wen J., Wiley S.E., Worby C.A., Kinch L.N., Xiao J., Grishin N.V., and Dixon J.E.. 2012. Secreted kinase phosphorylates extracellular proteins that regulate biomineralization. Science. 336:1150–1153. 10.1126/science.1217817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliabracci V.S., Pinna L.A., and Dixon J.E.. 2013. Secreted protein kinases. Trends Biochem. Sci. 38:121–130. 10.1016/j.tibs.2012.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliabracci V.S., Wiley S.E., Guo X., Kinch L.N., Durrant E., Wen J., Xiao J., Cui J., Nguyen K.B., Engel J.L., et al. . 2015. A Single Kinase Generates the Majority of the Secreted Phosphoproteome. Cell. 161:1619–1632. 10.1016/j.cell.2015.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub L.M., and Bonifacino J.S.. 2013. Cargo recognition in clathrin-mediated endocytosis. Cold Spring Harb. Perspect. Biol. 5 a016790 10.1101/cshperspect.a016790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., and Cox J.. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 13:731–740. 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- von Blume J., and Hausser A.. 2019. Lipid-dependent coupling of secretory cargo sorting and trafficking at the trans-Golgi network. FEBS Lett. 593:2412–2427. 10.1002/1873-3468.13552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Blume J., Duran J.M., Forlanelli E., Alleaume A.-M., Egorov M., Polishchuk R., Molina H., and Malhotra V.. 2009. Actin remodeling by ADF/cofilin is required for cargo sorting at the trans-Golgi network. J. Cell Biol. 187:1055–1069. 10.1083/jcb.200908040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Blume J., Alleaume A.-M., Cantero-Recasens G., Curwin A., Carreras-Sureda A., Zimmermann T., van Galen J., Wakana Y., Valverde M.A., and Malhotra V.. 2011. ADF/cofilin regulates secretory cargo sorting at the TGN via the Ca2+ ATPase SPCA1. Dev. Cell. 20:652–662. 10.1016/j.devcel.2011.03.014 [DOI] [PubMed] [Google Scholar]
- von Blume J., Alleaume A.-M., Kienzle C., Carreras-Sureda A., Valverde M., and Malhotra V.. 2012. Cab45 is required for Ca(2+)-dependent secretory cargo sorting at the trans-Golgi network. J. Cell Biol. 199:1057–1066. 10.1083/jcb.201207180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakana Y., van Galen J., Meissner F., Scarpa M., Polishchuk R.S., Mann M., and Malhotra V.. 2012. A new class of carriers that transport selective cargo from the trans Golgi network to the cell surface. EMBO J. 31:3976–3990. 10.1038/emboj.2012.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch L.G., and Munro S.. 2019. A tale of short tails, through thick and thin: investigating the sorting mechanisms of Golgi enzymes. FEBS Lett. 593:2452–2465. 10.1002/1873-3468.13553 [DOI] [PubMed] [Google Scholar]
- Zhou H., Di Palma S., Preisinger C., Peng M., Polat A.N., Heck A.J., and Mohammed S.. 2013. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J. Proteome Res. 12:260–271. 10.1021/pr300630k [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.