Summary:
Elite control of HIV replication is associated with polyfunctional lymphoid memory CD8+ T cells that lack overt cytolytic activity and home to B cell follicles.
The functional properties of circulating CD8+ T cells have been associated with immune control of HIV. However, viral replication occurs predominantly in secondary lymphoid tissues, such as lymph nodes (LNs). We used an integrated single-cell approach to characterize effective HIV-specific CD8+ T cell responses in the LNs of elite controllers (ECs), defined as individuals who suppress viral replication to extremely low levels in the absence of antiretroviral therapy (ART). Higher frequencies of total memory and follicle-homing HIV-specific CD8+ T cells were detected in the LNs of ECs compared with the LNs of chronic progressors (CPs). Moreover, HIV-specific CD8+ T cells potently suppressed viral replication without demonstrable cytolytic activity in the LNs of ECs, which harbored substantially lower amounts of CD4+ T cell-associated HIV DNA and RNA compared with the LNs of CPs. Single-cell RNA sequencing (scRNAseq) analyses further revealed a distinct transcriptional signature among HIV-specific CD8+ T cells from the LNs of ECs, typified by the downregulation of inhibitory receptors and cytolytic molecules and the upregulation of multiple cytokines, predicted secreted factors, and components of the protein translation machinery. Collectively, these results provide a mechanistic framework to expedite the identification of novel antiviral factors, highlighting a potential role for the localized deployment of non-cytolytic functions as a determinant of immune efficacy against HIV.
INTRODUCTION
AIDS is a persistent global health issue with no existing vaccine or cure. Most individuals infected with HIV experience high levels of ongoing viral replication, leading to a progressive loss of CD4+ T cells and disease onset in the absence of antiretroviral therapy (ART). However, a small subset of HIV-infected individuals (< 1%), termed elite controllers (ECs), spontaneously control viral replication below the limit of detection and generally do not progress to AIDS. It is established that virus-specific CD8+ T cells are critical determinants of the EC phenotype in humans and rhesus macaques (1, 2). In addition, HIV-specific CD8+ T cells in ECs are qualitatively distinct from HIV-specific CD8+ T cells in chronic progressors (CPs), typically displaying enhanced polyfunctionality (3, 4), cytolytic activity (5–7), and proliferative capacity (5, 8), as well as a more differentiated memory phenotype and a characteristic specificity profile (4, 9–11). These attributes have been documented primarily among circulating lymphocytes, however, whereas HIV replication occurs predominantly in lymphoid tissues (LTs) (12–15).
LTs are major reservoir sites for HIV. Recent studies have further demonstrated that almost 99% of viral RNA (vRNA)+ cells in SIV-infected rhesus macaques occur in LTs (16), reinforcing the need to understand anatomically colocalized mechanisms of immune control. It has long been known that circulating CD8+ T cells are more cytolytic than CD8+ T cells in the LTs of donors infected with HIV (17). Moreover, a state of immune privilege exists in LTs, which limits immunosurveillance by cytolytic HIV-specific CD4+ and CD8+ T cells (18, 19). In conjunction with the identification of distinct LT-resident memory CD8+ T cell subsets (20–22), these observations suggest that HIV-specific CD8+ T cells limit viral replication in LTs via effector mechanisms that differ from those employed by circulating HIV-specific CD8+ T cells (22). It also seems reasonable to propose that non-cytolytic suppression rather than cytolytic eradication dictates effective immune control of HIV, given reports of ongoing viral evolution (23, 24) and the presence of replication-competent viral strains in ECs (25). However, this proposition remains unproven to date, because previous studies have not defined the antiviral efficacy and functional characteristics of HIV-specific CD8+ T cells in the LTs of ECs.
In this study, we used a variety of methodological approaches, including polychromatic flow cytometry and single-cell RNA sequencing (scRNAseq) analyses, to compare the functional and transcriptional properties of HIV-specific CD8+ T cells in the peripheral blood and lymph nodes (LNs) of ECs and CPs. Our findings demonstrate that the maintenance of effective viral control is associated with polyfunctional HIV-specific memory CD8+ T cells with a weak cytolytic signature that preferentially home to B cell follicles in the LNs of ECs.
RESULTS
CD8+ T cells actively suppress HIV replication in the LNs of ECs
To define the nature of protective CD8+ T cell responses in LNs, where HIV replicates in vivo, we obtained tissue biopsies (cervical, iliac, inguinal, mesenteric, pelvic, or peribronchial LNs) and fine needle aspirates (inguinal LNs) from HIV+ individuals on ART and untreated HIV+ individuals categorized as acute seroconverters, ECs, or CPs (Supplementary Table 1). Extremely low levels of vRNA+ cells were detected in the LNs of ECs compared with the LNs of CPs (p = 0.0174, Fig. 1a). In line with previous findings (26), vRNA+ cells were visualized in the B cell follicles and the paracortical region (T cell zone) (Supplementary Fig. 1). Total CD4+ T cell-associated HIV DNA and RNA measurements were also lower in the LNs of ECs compared with the LNs of CPs (p = 0.0428 and p = 0.0016, respectively, Fig. 1b), and CD8+ T cells from the LNs of ECs displayed greater efficacy in a modified viral suppression assay (27) compared to CD8+ T cells from the LNs of CPs (p = 0.0224) and ART (p = 0.0323, Fig. 1c). These results suggest that CD8+ T cells maintain active control of HIV replication in the LNs of ECs.
Figure 1.
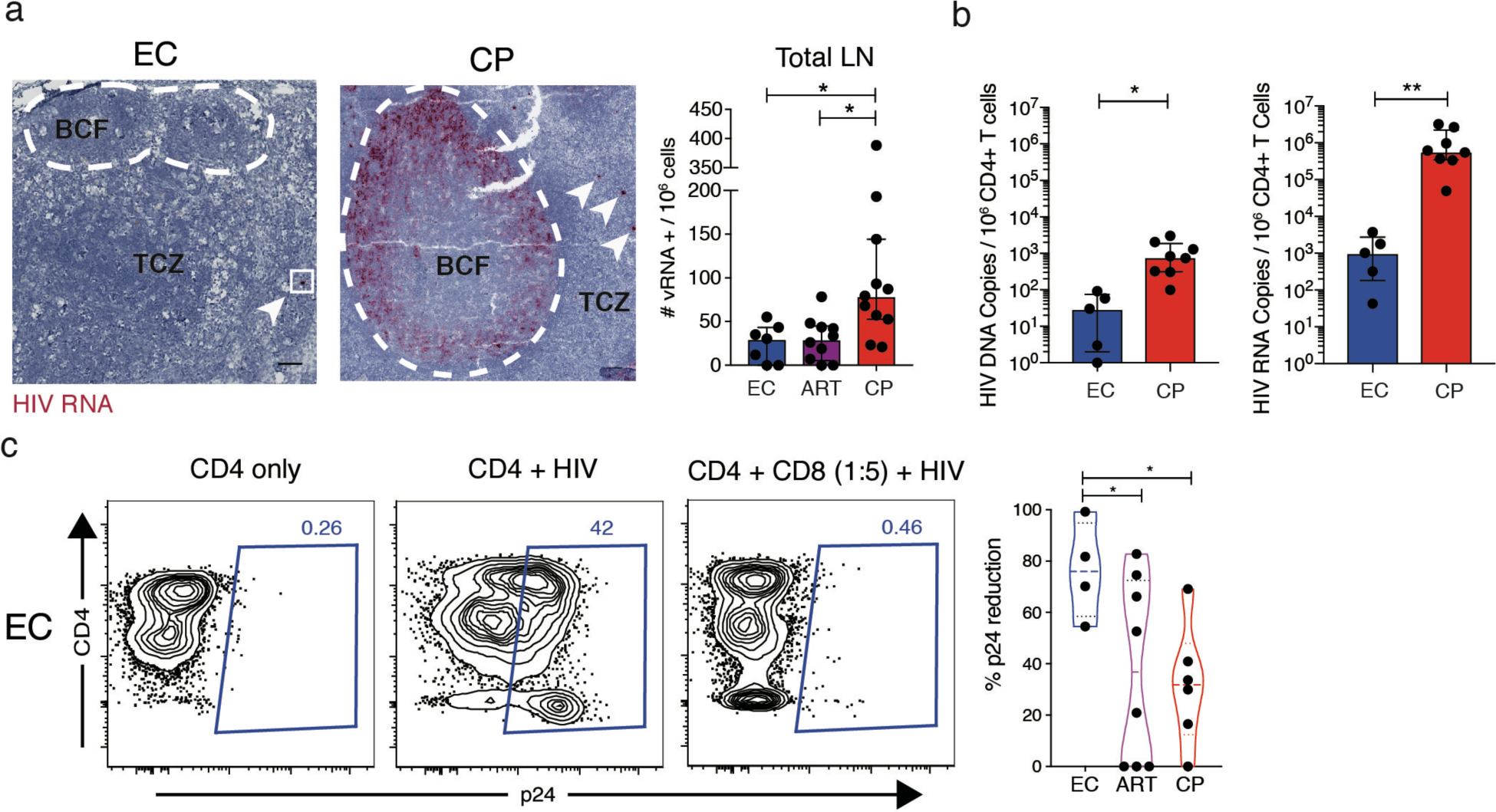
CD8+ T cells from the LNs of ECs display superior antiviral efficacy. a, Representative RNAscope images of paraffin-embedded LN biopsies from one EC and one CP (left) and data quantification across donor groups (right). TCZ: T cell zone; BCF: B cell follicle. b, Quantification of cell-associated HIV DNA and RNA from negatively-selected CD4+ T cells by qPCR. One data point on the DNA plot was adjusted from 0 to 1 for graphical purposes. c, Representative flow plots from a viral suppression assay using LN-derived CD8+ T cells (left) and data quantification across donor groups (right). Dotted lines represent median and interquartile range. Significance was determined using the Kruskal-Wallis test with Dunn’s correction (a) or unpaired t-test (b) or Welch ANOVA with Benjamini-Hochberg correction. * p < 0.05, ** p < 0.01.
CD8+ T cells exhibit weak cytolytic activity in the LNs of ECs
In peripheral blood, the frequency of cytolytic HIV-specific CD8+ T cells, defined by expression of perforin and granzyme B, correlates inversely with plasma viral load (pVL) (5–7). We therefore hypothesized that enhanced CD8+ T cell-mediated cytolytic activity may underlie viral control in the LNs of ECs. However, lower frequencies of perforin+, granzyme B+, and perforin+ granzyme B + memory CD8+ T cells were detected in the LNs of ECs compared with the LNs of CPs (p = 0.0001, p = 0.0025, and p = 0.0003, respectively, Fig. 2a and Supplementary Fig. 2a,b). The frequency of perforin+ granzyme B+ memory CD8+ T cells also correlated positively with pVL (r = 0.8213, p < 0.0001, Fig. 2b). Importantly, perforin and granzyme B were highly expressed among circulating memory CD8+ T cells in donor-matched samples from ECs, indicating that the non-cytolytic phenotype was restricted to LNs (Supplementary Fig. 2c). Immunohistochemical analyses confirmed that fewer perforin+ CD8+ T cells were present in the LNs of ECs compared with the LNs of CPs (Fig. 2c). However, the frequency of granzyme B+ CD8+ T cells was not different between groups (Fig. 2d).
Figure 2.

Cytolytic CD8+ T cells are rare in the LNs of ECs. a, Representative flow plots showing perforin and granzyme B expression in LN-derived memory CD8+ T cells (left) and data quantification across donor groups (right). b, Correlation between the frequency of perforin+ granzyme B+ LN memory CD8+ T cells and pVL. Values for pVL below the limit of detection were plotted as 40 copies/mL. Significance was determined using Spearman’s rank correlation. c, d, Representative immunohistochemistry images of LN biopsies (left) stained for perforin (c) or granzyme B (d) and data quantification across donor groups (right). e, Representative flow plots showing HIV-specific tetramer staining of LN-derived memory CD8+ T cells (left) and data quantification across donor groups (right). Each dot represents a distinct tetramer+ population in the quantification plot. f, Representative flow plots showing HIV-specific tetramer+ cells (colored contours) overlaid on total LN-derived memory CD8+ T cells (grey background) gated to display perforin and granzyme B expression (left) and data quantification across donor groups (right). Each dot represents a distinct tetramer+ population in the quantification plot. g, Representative flow plots of redirected killing assays (left) and data quantification across donor groups (right). h, Representative flow plots showing CXCR5 expression on LN-derived memory CD8+ T cells (left) and data quantification across donor groups (right). i, Quantification of CXCR5 expression on LN-derived tetramer+ memory CD8+ T cells. Each dot represents a distinct tetramer+ population. j, Representative flow plots showing perforin and granzyme B expression in LN-derived CXCR5+ and CXCR5− memory CD8+ T cells (left) and data quantification across donor groups (right). Error bars represent median and interquartile range. Significance was determined using the Kruskal-Wallis test with Dunn’s correction (a, c, d, e, f, h, i, k) or a two-way ANOVA (g). * p < 0.05, ** p < 0.01, *** p < 0.001.
We then used human leukocyte antigen (HLA) class I-matched tetramers to examine HIV-specific CD8+ T cells. Higher frequencies of tetramer+ CD8+ T cells were present in the LNs of ECs compared with the LNs of CPs (p = 0.04) (Fig. 2e). However, these HIV-specific CD8+ T cells expressed very low levels of perforin and granzyme B, especially in the LNs of ECs (Fig. 2f). In response to cognate peptide stimulation, HIV-specific CD8+ T cells from the LNs of ECs also failed to upregulate either perforin or granzyme B, unlike CD8+ T cells paired by donor and specificity from the peripheral blood of ECs (Supplementary Fig. 3a). Moreover, CD8+ T cells from the LNs of ECs did not exhibit discernable expression of perforin or granzyme B after 3 days in culture with HIV-infected CD4+ T cells from the same LNs (Supplementary Fig. 3b).
To quantify rather than infer cytolytic activity, we performed ex vivo redirected killing assays. In contrast to circulating CD8+ T cells, donor-matched CD8+ T cells from the LNs of ECs largely failed to kill P815 mastocytoma target cells pre-coated with a CD3-specific monoclonal antibody, which mimics signals delivered via the TCR (Fig. 2g). A similar anatomical discrepancy was observed using paired samples from CPs (Fig. 2g). The addition of a live/dead dye to the redirected killing assays confirmed that the detection of active-caspase 3 captured most of killed targets as only a minor fraction of those cells was live/dead+ active-caspase 3− (Supplementary Fig. 4a,b). Furthermore, extending the redirected killing assays to 24 hours did not result in a significant increase in the killing activity of LN (p=0.183), tonsil (p=0.415), and blood CD8+ T cells (p=0.159), ruling out the involvement of temporally delayed killing mechanisms (Supplementary Fig. 4b).
Collectively, these data suggest that cytolytic activity does not contribute mechanistically to the protective attributes of CD8+ T cells in the LNs of ECs.
CD8+ T cells display increased follicle-homing potential in the LNs of ECs
B cell follicles are thought to represent immunoprivileged sites that potentiate HIV persistence as a consequence of limited immunosurveillance by CD8+ T cells (2, 28–31). Previous studies have also demonstrated an inverse correlation between the frequency of follicle-homing (CXCR5+) CD8+ T cells and pVL (18, 32, 33). In line with these findings and the relative absence of vRNA+ cells in the B cell follicles of ECs (Fig. 1b), we detected higher frequencies of CXCR5+ memory CD8+ T cells in the LNs of ECs compared with the LNs of CPs (p = 0.0288, Fig. 2h). A similar trend was observed for HIV-specific CXCR5+ CD8+ T cells (p = 0.124, Fig. 2i). However, perforin and granzyme B were typically expressed at higher frequencies among CXCR5− CD8+ T cells compared with CXCR5+ CD8+ T cells, reaching significance in the ART cohort (p = 0.0026, Fig. 2j).
HIV-specific CD8+ T cells have a distinct transcriptional profile in the LNs of ECs
To identify potential correlates of viral control in LTs, we analyzed the transcriptomes of resting HIV-specific CD8+ T cells from the LNs of ECs and CPs (Fig. 3a). The scRNAseq data were initially processed using an unsupervised kernel-based algorithm to determine cellular similarity in a global and unbiased manner (see Materials and Methods). Clusters of transcriptionally distinct cells were present in each donor group (Fig. 3b). Importantly, normalized expression levels of PRF1 and GZMB were lower in HIV-specific CD8+ T cells from the LNs of ECs compared with HIV-specific CD8+ T cells from the LNs of CPs (Fig. 3c), and normalized expression levels of transcripts encoding other cytolytic molecules, including GZMA, GZMH, GZMK, GZMM, FASL, and TNFSF10 (34, 35), were either comparable between groups or enriched in HIV-specific CD8+ T cells from the LNs of CPs compared with HIV-specific CD8+ T cells from the LNs of ECs (Fig. 3c).
Figure 3.
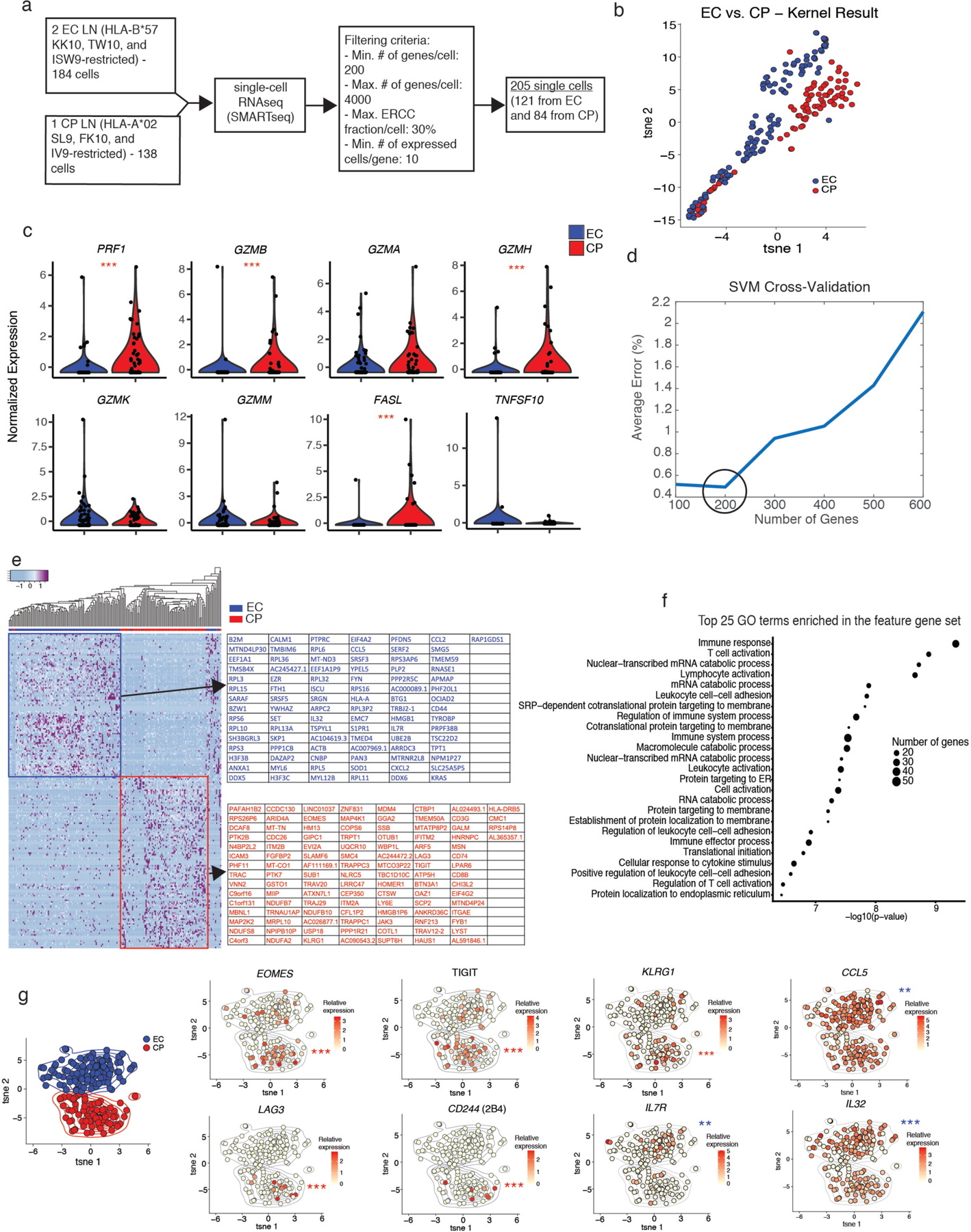
Distinct transcriptomic signatures characterize HIV-specific CD8+ T cells from the LNs of ECs and CPs. a, Set-up of the scRNAseq experiment, including data exclusion criteria. b, tSNE visualization of the unsupervised kernel-based algorithm. c, Violin plots of cytolysis-related genes showing z-normalized expression levels. d, Average percent error of k-fold cross-validation of support vector machine (SVM) models using subsets of the L0-norm-ranked gene list. Each cross-validation was reiterated 100 times. e, Heatmap showing z-normalized expression of the 200 feature genes. f, Gene ontology (GO) analysis of the feature gene list using topGO. The top 25 enriched terms were reported, and p-values were calculated using the classic Fisher method. g, tSNE feature plots showing the distribution of gene expression. tSNE coordinates were calculated using the first 50 principal components and iterated 1,000 times. The color gradient displays relative log-normalized gene expression levels. Significance was determined using scDD calculations (c, g). * p < 0.05, ** p < 0.01, *** p < 0.001.
We then applied a supervised machine-learning algorithm to determine which transcripts best characterized HIV-specific CD8+ T cells from the LNs of ECs and HIV-specific CD8+ T cells from the LNs of CPs (see Materials and Methods). A set of 200 genes reliably distinguished each donor group (average error < 0.5%) (Fig. 3d). Hierarchical clustering of these ‘feature’ genes clarified the key inter-group differences (Fig. 3e), most of which could be assigned to immune response pathways via gene ontology (GO) analysis (Fig. 3f), and t-Distributed Stochastic Neighbor Embedding (tSNE) visualization generated two distinct clusters of HIV-specific CD8+ T cells that clearly differentiated ECs from CPs (Fig. 3g). Overlaying transcript levels of specific ‘feature’ genes onto the tSNE plot further revealed that HIV-specific CD8+ T cells from the LNs of CPs preferentially expressed inhibitory receptors, such as TIGIT, LAG3, CD244, as well as the terminal differentiation marker KLRG1 and the transcription factor EOMES, collectively indicating an exhausted phenotype (36–38), whereas HIV-specific CD8+ T cells from the LNs of ECs preferentially expressed IL7R, which is crucial for homeostasis (39), and several chemokines/cytokines, such as CCL5 and IL32, collectively indicating a highly functional memory phenotype (Fig. 3g).
To confirm and extend these findings, we reanalyzed our scRNAseq data using the reproducibility-optimized test statistic (ROTS) (40), a Bayesian model of differential distribution (scDD) (41), and Seurat (42). A total of 2,264 differentially expressed genes achieved significance in at least two of these independent analytical frameworks (Supplementary Fig. 5a and Supplementary Table 2). These genes were enriched for immune-related terms, such as ‘immune response’, ‘immune system process’, and ‘defense response’, in GO analyses (Supplementary Fig. 5b). Of particular note, ingenuity pathway analysis (IPA) further identified a core cassette of 11 transcripts encoding predicted secreted factors that were selectively upregulated in HIV-specific CD8+ T cells from the LNs of ECs, including TNF, CCL5, RNASE1, and IL32, which have been shown to suppress HIV replication (43–49) (Supplementary Fig. 5c).
HIV-specific CD8+ T cells are polyfunctional and translate proteins efficiently in the LNs of ECs
In response to cognate peptide stimulation, higher frequencies of HIV-specific CD8+ T cells from the LNs of ECs produced TNF compared with HIV-specific CD8+ T cells from the LNs of CPs (p = 0.0003) and from the LNs of ARTs (p = 0.039) (Fig. 4a,b). A similar trend was observed for IFNγ, MIP-1β, and IL-2 (Fig 4b). Moreover, HIV-specific CD8+ T cells from the LNs of ECs were frequently polyfunctional, whereas HIV-specific CD8+ T cells from the LNs of CPs were predominantly monofunctional (Fig. 4c,d).
Figure 4.
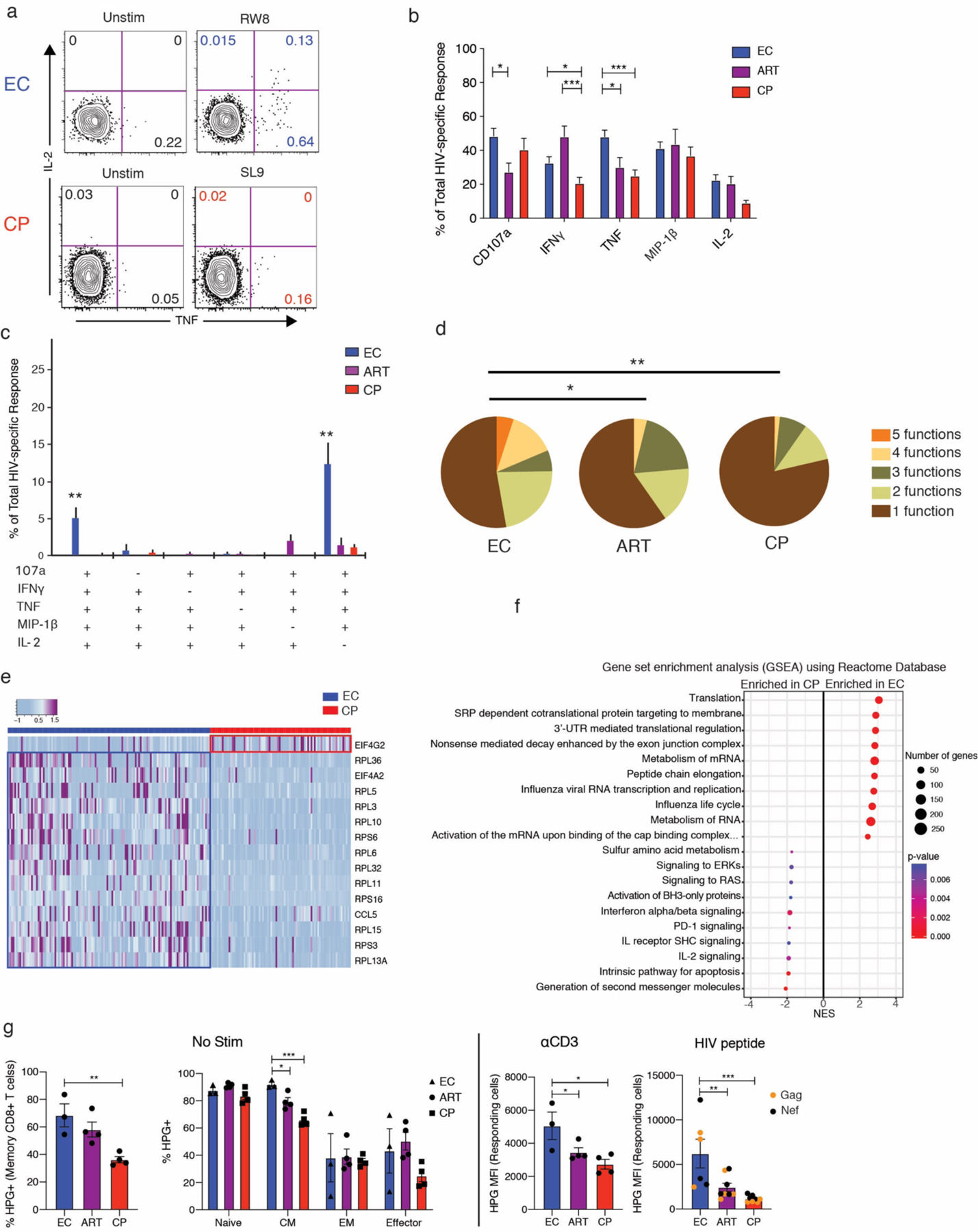
Polyfunctionality is associated with protein translation efficiency in HIV-specific CD8+ T cells from the LNs of ECs. a, Representative flow plots showing the production of TNF and IL-2 by HIV-specific CD8+ T cells from the LNs of one EC and one CP in response to cognate peptide stimulation. Plots were pre-gated on total memory CD8+ T cells. b, Quantification of cytokine production by HIV-specific CD8+ T cells from LNs. The total response for each readout was determined by summing the corresponding frequencies after background subtraction of all Boolean gates using permutations of CD107a, IFNγ, TNF, MIP-1β, and IL-2. Error bars represent median and interquartile range. c, Polyfunctionality plot of HIV-specific CD8+ T cells from LNs. Only combinations of 4 and 5 functions are shown. Error bars represent mean and SEM. d, Pie charts summarizing all combinations of functions. e, Heatmap showing z-normalized expression levels of genes from the feature list identified by GO protein translation terms. f, Gene set enrichment analysis (GSEA) of the scRNAseq data with reference to the Reactome Pathway Database, reporting the top 10 enriched pathways in ECs and CPs. g, Quantification of protein translation efficiency by LN CD8+ T cells. LNMCs were incubated with HPG in methionine-free medium for 6 hours in the presence or absence of HIV peptide pools or monoclonal antibodies specific for CD3, CD28, and CD49d. Responding cells were defined by Boolean-gating for IFN+ and/or TNF+. Error bars represent mean and SEM. Significance was determined using one-way (d, g) or two-way ANOVA (b, g). * p < 0.05, ** p < 0.01, *** p < 0.001.
To seek an explanation for these findings, we returned to our GO analysis of the 200 ‘feature’ genes. In addition to immune-related terms, several protein translation-related terms were also highlighted, including ‘SRP-dependent cotranslational protein targeting to membrane’, ‘protein targeting to ER’, and ‘translation initiation’ (Fig. 3f). Most of these genes encoded ribosomal protein subunits that were preferentially expressed in HIV-specific CD8+ T cells from the LNs of ECs (Fig. 4e). Gene set enrichment analysis (GSEA) similarly identified ‘translation’, ‘SRP-dependent cotranslational protein targeting to membrane’, and ‘3’ UTR-mediated translation regulation’ as the most highly enriched gene sets in the EC group (Fig. 4f).
We then performed in vitro translation experiments to measure the uptake of fluorescently labeled L-homopropargylglycine (HPG) in resting and activated CD8+ T cells from the LNs of ECs, ARTs, and CPs. Higher frequencies of resting memory CD8+ T cells from ECs incorporated HPG into newly synthesized proteins compared with resting memory CD8+ T cells from CPs (p = 0.003) (Fig. 4g). While subsets including naïve, effector memory (EM), and effector CD8+ T cells did not show any significant difference, central memory (CM) CD8+ T cells from LNs of ECs showed a higher frequency of HPG+ cells (Fig. 4g). Moreover, upon stimulation with either anti-CD3 antibody or HIV peptides, responding CD8+ T cells from LNs of ECs exhibited significantly higher HPG MFI than those from LNs of ARTs (p = 0.04 and 0.004, respectively) and CPs (p = 0.01 and 0.0008, respectively) (Fig. 4g). These results suggest that efficient protein translation can explain, at least in part, the antiviral efficacy and polyfunctional nature of HIV-specific CD8+ T cells in ECs.
DISCUSSION
CD8+ T cells are required for effective immune control of HIV infection, but it remains unknown if LT CD8+ T cells can control and suppress HIV replication in LT. Here, we show that LT CD8+ T cells in HIV+ ECs efficiently suppress viral replication despite lack of a strong cytolytic response. Our data instead demonstrate that ECs launch a robust LT CD8+ T cell response with potential to traffic to B cell follicles, a critical anatomical location of the HIV reservoir (2, 15). Furthermore, their LT CD8+ T cells downregulate inhibitory receptor expression and upregulate the production of multiple soluble factors and cytokines, which is associated with an efficient protein translation capacity. While cause and effect arguments can always be made, the failure of CD8+ T cells from ART-treated LT to achieve the characteristics of ECs indicates that the ECs’ functional profile is not simply an effect of low viral load or poor immunologic signals. Collectively, these data provide the signature of an effective HIV-specific CD8+ T cell response in LT and demonstrate that maintenance of HIV control is likely achieved through high magnitudes of non-cytolytic functional CD8+ T cell-mediated responses that can enter B cell follicles.
ECs are defined as individuals with undetectable viral load in the absence of ART. However, ultrasensitive assays have demonstrated that many ECs have low level viral load in the plasma and ongoing viral evolution(23–25), indicating that HIV might be readily detectable in LN structures. Using imaging and sensitive PCR assays, we demonstrate detectable but considerably lower HIV-RNA and -DNA levels in EC compared to CP LNs, comparable to ART-treated individuals. Although we cannot formally exclude the possibility of viral evolution within a single LN, due to technical issues of receiving enough cells from the same LN over time, it is clear that viral replication is well controlled in the LN compartment. Similar to previous analyses identifying a close association of viral control to MHC-class I restriction, we indeed found that LN CD8+ T cells from ECs efficiently suppress HIV in autologous CD4+ T cells. In contrast, LN CD8+ T cells from CPs and ARTs did not suppress HIV infection to the same degree, confirming that CD8+ T cells in LNs are likely a key component of effective HIV control in ECs.
Depletion studies in the macaque model have demonstrated that CD8+ cells are indispensable for the maintenance of SIV control even in very low viral load conditions, such as in ART-treated and EC animals (2, 50). However, these studies have not identified what type of CD8+ T cell functions are associated with viral control. We and others have demonstrated superior killing ability of blood CD8+ T cells from ECs (6, 7). Therefore, it came to our surprise that cytolytic molecule expression was lower in EC than CP CD8+ T cells, aligning with a general state of limited immunosurveillance by cytolytic T cells in HIV-infected LNs (42). We cannot definitively exclude the role of cytolytic mechanisms for viral control, particularly during early infection. However, our data at least demonstrate that a strong cytolytic response is not required for HIV-specific CD8+ T cells to maintain long-term control of HIV replication. These data are consistent with previous observations demonstrating that terminally differentiated effector memory T cells, those with the highest cytolytic capacity, are mostly present in blood and highly vascularized organs (51). Limited cytolytic activity in LT is likely an evolutionarily conserved feature of the immune system to limit tissue and antigen-presenting cell destruction under chronic antigen stimuli. In fact, in hepatitis B virus infection, CD8+ T cells clear the virus without killing infected hepatocytes via their secretion of IFNγ and TNFα (52). In other infections such as vaccinia virus, measles virus, and herpes simplex virus, non-cytolytic mechanisms of viral control are well documented (53, 54). Similar mechanisms have been described in chronic HIV and SIV infection (55–57). Our results add to this body of literature and also suggests that spatial distribution and efficient translation of antiviral proteins by a robust HIV-specific CD8+ T cell response is key to mediate an effective HIV-specific CD8+ T cell response in LTs.
Current efforts to develop a functional cure of HIV largely focus on enhancing the cytolytic function of CD8+ T cells (58). Though our study does not completely exclude a role for cytotoxicity, it argues that other types of antiviral mechanisms are involved in the maintenance of viral control in LT and worth further investigation. These data support a model where high magnitudes of functional HIV-specific CD8+ T cells can maintain control of HIV replication in LT but may not eradicate the infection due to low cytolytic activity. They also provide fresh impetus in the search for novel antiviral factors (59), together with new insights into the mechanisms that underpin immune control of HIV.
MATERIALS AND METHODS
Samples
Peripheral blood samples, tissue biopsies (cervical, iliac, or inguinal LNs), and fine needle aspirates (inguinal LNs) were obtained from HIV+ individuals on antiretroviral therapy (ART, n = 10) and untreated HIV+ individuals categorized as acute seroconverters (Fiebig stage IV–VI, n = 7), elite controllers (ECs, n = 12), or chronic progressors (CPs, n = 13). Donors were recruited at three different sites: INER-CIENI, Mexico City, Mexico; the University of California, San Francisco, USA; and the University of Pennsylvania, Philadelphia, USA. Clinical characteristics are summarized in Supplementary Table 1. For microscopy experiments, we also obtained paraffin-embedded LN slides from CPs (n = 7) via the Pathology Core at the Hospital of the University of Pennsylvania (Philadelphia). All donors provided informed consent in line with protocols approved by the INER-CIENI Ethics Committee, the Federal Commission for the Protection against Sanitary Risk (COFEPRIS), and the Institutional Review Boards of the University of California (San Francisco) and the University of Pennsylvania (Philadelphia). Sample sizes were not predetermined by power calculations, and investigators were not blinded to group identity during the course of the study.
Peripheral blood mononuclear cells (PBMCs) were isolated using standard density gradient centrifugation, and LN mononuclear cells (LNMCs) were collected from whole biopsies via mechanical homogenization. Fine needle aspirates were processed for immediate experimentation. PBMCs and LNMCs from whole biopsies were cryopreserved at −140°C.
RNAscope
Next generation in situ hybridization was performed on LN biopsies as previously described(60).
qPCR quantification of cellular HIV-1 DNA and RNA
Cell-associated DNA and RNA were purified using an AllPrep DNA/RNA Mini Kit (Qiagen), concentrated in a SpeedVac System (Thermo Fisher Scientific), and normalized to cell equivalents via quantitative PCR (qPCR) with reference to genomic telomerase reverse transcriptase for DNA and expressed ribosomal protein lateral stalk subunit P0 for RNA (Thermo Fisher Scientific). Total cell-associated HIV DNA and RNA were quantified via qPCR using a QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems) with the LTR-specific primers F522–43 (5’-GCCTCAATAAAGCTTGCCTTGA-3’; HXB2 522–543) and R626–43 (5’-GGGCGCCACTGCTAGAGA-3’; HXB2 626–643) and a FAM-BQ probe (5’-CCAGAGTCACACAACAGACGGGCACA-3’). Cell-associated HIV DNA copy number was determined in a final reaction volume of 20 μl incorporating 4 pmol of each primer, 4 pmol of probe, 5 μl of DNA, and 10 μl of 2x TaqMan Universal Master Mix II, with UNG (Thermo Fisher Scientific). Thermal parameters were as follows: 50 °C for 2 minutes (1 cycle), 95 °C for 10 minutes (1 cycle), and 95 °C for 15 seconds followed by 59 °C for 1 minute (60 cycles). Cell-associated HIV RNA copy number was determined in a final reaction volume of 20 μl incorporating 4 pmol of each primer, 4 pmol of probe, 5 μl of RNA, 0.5 μl of reverse transcriptase, and 10 μl of 2x TaqMan RNA-to-CT 1-Step (Thermo Fisher Scientific). Thermal parameters were as follows: 48 °C for 20 minutes (1 cycle), 95 °C for 10 minutes (1 cycle), and 95 °C for 15 seconds followed by 59 °C for 1 minute (60 cycles). For HIV DNA measurements, external quantification standards were prepared from the ACH-2 cell line in a background of HIV–human cellular DNA, calibrated against the Division of AIDS Virology Quality Assurance Program HIV-1 DNA Quantification Standard (National Institutes of Health AIDS Reagent Program). For HIV RNA measurements, external quantification standards were prepared from full-length NL4–3 virion RNA, and copy number was determined using a Real-Time HIV-1 Viral Load Assay (Abbott Molecular), calibrated against the Division of AIDS Virology Quality Assurance Program HIV-1 RNA Quantification Standard (National Institutes of Health AIDS Reagent Program). Copy number for all samples was determined in triplicate by extrapolation against a seven-point standard curve (1–10,000 cps).
Peptide stimulation assay
Cryopreserved LNMCs and PBMCs were thawed and rested overnight at 37 °C in RPMI medium supplemented with 10% fetal bovine serum, 1% L-glutamine, and 1% penicillin/streptomycin (R10). Cells were then washed in R10 and resuspended at 2 × 106 cells/mL. Approximately 0.5 × 106 to 1 × 106 cells were used per condition, with lower bounds defined by cell availability in each sample. All stimulation conditions included anti-CD28 and anti-CD49d (each at 1 μg/mL, BD Biosciences), GolgiStop (0.7 μL/mL, BD Biosciences), and brefeldin A (1 μg/mL, Sigma-Aldrich). Cells were stimulated for 6 hours at 37°C with peptides matching optimal epitopes derived from HIV (each at a final concentration of 1 μg/mL, New England Biolabs). Positive controls incorporated staphylococcal enterotoxin B (1 μg/mL, Sigma-Aldrich). Degranulation was detected via the addition of anti-CD107a–PE-Cy5 (eBioH4A3, eBioscience) at the start of the assay to capture surface-mobilized events in real time (61).
Flow cytometry
LNMCs and PBMCs were stained with anti-CCR7–APC-Cy7 (G043H7, BioLegend) for 10 minutes at 37 °C. Cells were then labeled with LIVE/DEAD Fixable Aqua (Thermo Fisher Scientific) for 10 minutes at room temperature to identify non-viable events, stained with a cocktail of directly-conjugated monoclonal antibodies for 20 minutes at room temperature to detect surface markers, fixed/permeabilized using a Cytofix/Cytoperm Kit (BD Biosciences), stained with another cocktail of directly-conjugated monoclonal antibodies for 1 hour at room temperature to detect intracellular markers, and fixed in 1% paraformaldehyde (Sigma-Aldrich). Data were acquired using an LSRII flow cytometer (BD Biosciences) and analyzed with FlowJo software v9.9.4 (Tree Star Inc.).
Antibodies
The following directly-conjugated reagents were used in flow cytometry experiments. Tetramer panel: anti-CCR7–APC-Cy7 (G043H7, BioLegend), anti-CD14–BV510 (M5E2, BioLegend), anti-CD19–BV510 (HIB19, BioLegend), anti-CD3–BV711 (UCHT1, BioLegend), anti-CD4–PE-Cy5.5 (S3.5, Thermo Fisher Scientific), anti-CD8–BV570 (RPA-T8, BioLegend), anti-CD27–BV785 (O323, BioLegend), anti-CD45RO–BV650 (UCHL1, BioLegend), anti-CD69–PE-Cy5 (TP1.55.3, Beckman Coulter), anti-CD103–BV605 (2E7, BioLegend), anti-CXCR5–AF488 (RF8B2, BD Biosciences), anti-perforin–PE-Cy7 (dG9, eBioscience), anti-granzyme B–AF700 (GB11, BD Biosciences), anti-Tbet–PE-Dazzle (4B10, BioLegend), and anti-Eomes–AF647 (WD1928, eBioscience). Peptide stimulation panel: anti-CD107a–PE-Cy5 (eBioH4A3, eBioscience), anti-CCR7–APC-Cy7 (G043H7, BioLegend), anti-CD14–BV510 (M5E2, BioLegend), anti-CD19–BV510 (HIB19, BioLegend), anti-CD3–BV711 (UCHT1, BioLegend), anti-CD4–PE-Cy5.5 (S3.5, Thermo Fisher Scientific), anti-CD8–BV570 (RPA-T8, BioLegend), anti-CD27–BV785 (O323, BioLegend), anti-CD45RO–BV650 (UCHL1, BioLegend), anti-CXCR5–AF647 (RF8B2, BD Biosciences), anti-IFNγ–FITC (B27, BD Biosciences), anti-TNF–BV605 (MAb11, BioLegend), anti-IL-2–APC-R700 (MQ1–17H12, BD Biosciences), anti-MIP-1β–PE-Cy7 (D21–1351, BD Biosciences), anti-perforin–BV421 (B-D48, BioLegend), anti-granzyme B–PE-TxRed (GB11, Thermo Fisher Scientific), and anti-Tbet–PE (4B10, eBioscience). Redirected killing assay panel: anti-active caspase 3–FITC (C92–605, BD Biosciences), anti-CD3–PE (SK7, BD Biosciences), and anti-perforin–PE-Cy7 (dG9, eBioscience). Suppression assay panel: anti-p24–FITC (KC57, Beckman Coulter), anti-CD14–BV510 (M5E2, BioLegend), anti-CD19–BV510 (HIB19, BioLegend), anti-CD3–BV711 (UCHT1, BioLegend), anti-CD4–PE-Cy7 (RPA-T4, BioLegend), anti-CD8–BV570 (RPA-T8, BioLegend), anti-CD25–Tricolor (CD25–3G10, Thermo Fisher Scientific), anti-perforin–BV421 (B-D48, BioLegend), and anti-granzyme B–PE-TxRed (GB11, Thermo Fisher Scientific). Translation assay panel: anti-CCR7–APC-Cy7 (G043H7, BioLegend), anti-CD14–BV510 (M5E2, BioLegend), anti-CD19–BV510 (HIB19, BioLegend), anti-CD3–BV711 (UCHT1, BioLegend), anti-CD4–APC (S3.5, Thermo Fisher Scientific), anti-CD8–BV570 (RPA-T8, BioLegend), anti-CD45RO–BV650 (UCHL1, BioLegend), and azide–AF488 (A10266, Thermo Fisher Scientific), anti-TNF-PECy7 (Mab11, Thermo Fisher Scientific), and anti-IFNγ-AF700 (B27, BD Biosciences). Viral quantification panel: anti-CCR7–APC-Cy7 (G043H7, BioLegend), anti-CD3-APC-R700 (UCHT1, BD Biosciences), anti-CD4–PE-Cy7 (RPA-T4, BioLegend), anti-CD8 PE-Cy5.5 (RPA-T8, eBiosciences), anti-CD14–BV510 (M5E2, BioLegend), anti-CD19–BV510 (HIB19, BioLegend), anti-CD45RA-PE-CF594 (HI100, BD Biosciences), anti-CXCR5–AF647 (RF8B2, BD Biosciences), and, anti-PD-1-BV421 (EH12.2H7, BioLegend).
Tetramers
HLA class I tetramers conjugated to BV421 or PE were produced as described previously (62). The following specificities were used to detect HIV-specific CD8+ T cells: A*0201-IV9 (ILKEPVHGV), A*0201-SL9 (SLYNTVATL), A*0201-FK10 (FLGKIWPSHK), A*0201-TV9 (TLNAWVKVV), A*2402-RW8 (RYPLTFGW), A*2402-KW9 (KYKLKHIVW), A*2402-RL9 (RPMTYKGAL), B*0702-GL9 (GPGHKARVL), B*0702-HI10 (HPRVSSEVHI), B*0702-SM9 (SPAIFQSSF), B*2705-KK10 (KRWIILGLNK), B*3501-VY10 (VPLDEDFRKY), B*3501-NY9 (NSSKVSQNY), B*5701-KF11 (KAFSPEVIPMF), B*5701-TW10 (TSTLQEQIGW), B*5701-ISW9 (ISPRTLNAW), and B*5701-QW9 (QASQEVKNW).
Immunohistochemistry
LN biopsy material was cut at a thickness of 5 μm and processed as described previously (63). Briefly, tissue sections were heated in 0.01% citraconic anhydride containing 0.05% Tween-20 and incubated overnight at 4 °C with monoclonal or polyclonal antibodies specific for perforin (1:100, 5B10/VP-P967, Vector Laboratories Inc.) or granzyme B (1:200, HPA003418, Sigma-Aldrich). Slides were then washed in TBS containing 0.05% Tween-20, and endogenous peroxidases were blocked using 1.5% (v/v) H2O2 in TBS, pH 7.4. Antigens were revealed using mouse Polink-1 or rabbit Polink-2 HRP in conjunction with ImmPACT DAB (Vector Laboratories Inc.) and Warp Red (Biocare Medical Inc.). Slides were then washed in H2O, counterstained with haematoxylin, mounted in Permount (Thermo Fisher Scientific), and scanned at 200x magnification using a ScanScope CS System (Aperio Technologies). Representative regions of interest (0.4 mm2) were identified visually, and high-resolution images were extracted from the whole-tissue scans. The percent area positive for CD4+ T cells was quantified using CellProfiler v3.1.5 (64).
Single-cell RNAseq
The experimental setup was described previously in Buggert et al. (22). Briefly, single HIV-tetramer+ cells were index sorted using a FACSAria II (BD Biosciences) directly into 96-well microtiter plates containing lysis buffer. Out of the 552 sorted cells ,full genome was sequenced from 221 cells. Cellular nucleic acids were recovered by processing the lysates with an RNEasy Plus Micro Kit (Qiagen) and then binding to RNAClean (SPRI) beads (Beckman Coulter). ERCC RNA control (Ambion) was added to the RNAClean beads prior to purification. Reverse transcription (RT) was done using oligo-dT primers. Whole transcriptome cDNA was then amplified by PCR for 22–24 cycles using universal priming sites primers and Kapa HiFi Hot Start ReadyMix. After post-PCR clean-up, amplified whole transcriptome cDNA was barcoded using Illumina Nextera libraries and sequenced to a depth of about 2 million 150bp paired-end reads/cell on a HiSeq 4000 (Illumina).
Single-cell RNAseq analyses
A kernel-method learning framework was used to assess the pairwise similarity between cells and compute the distance metric that best fitted the structure of the data (https://arxiv.org/pdf/1808.02061.pdf). We used a rank-based, nonparametric function to learn cell-to-cell similarity, and the kernelization of tSNE allowed us to use data from all available features. Dimensionality reduction was performed using kernel tSNE, wherein the pairwise similarity between cells was computed using the Semblance kernel as a distance measure. In determination of the cell-to-cell similarity matrix, the Gini index was used to account for the distribution of each gene and weigh it appropriately in the kernel calculation. The Gini index enabled prioritization of genes with high variance as being the genes that are more probable to be useful features for niche group detection. Single cells were then projected onto a two-dimensional space using kernel-t-distributed stochastic neighbor embedding, to enable intuitive visualization of hidden structures within the data. The supporting code is freely available online and implemented in the R Package Semblance (https://cran.rproject.org/web/packages/Semblance/index.html).
For support vector machine (SVM) analyses, genes were selected using a t-score cut-off of p < 0.01. Selected genes were then ranked using L0-norm regularization. The L0-norm-ranked gene lists were fed into the support vector machine (SVM) algorithm for cross-validation (k-fold = 10) in subsets (increment = 100 genes). The average prediction error for each cross-validation was used to determine how well each subset of genes classified the desired labels. All steps were performed in MATLAB (MathWorks). The L0-norm regularization was implemented using MATLAB Feature Selection Library.
Differentially expressed genes were identified using three approaches: ROTS (40), which optimizes the t-statistics based on the inherent characteristics of the data; scDD (41) which is a differential expression analysis method that accounts for the possibility of multimodally distributed gene expression; and Seurat (42), which uses the non-parametric Mann-Whitney U-test to assess the null hypothesis that a randomly selected mean expression value for a given gene in one group will have an equal chance of being less than or greater than a randomly selected mean expression value for the same gene in a second group. As different genes in the dataset exhibited different distribution properties, these approaches in combination allowed us to winnow the list of differentially expressed genes, specifying a cut-off of p < 0.05 in at least two out of the three outputs. The scDD p-values were reported for simplicity.
The topGO package Bioconductor v3.8 was used for GO analyses (https://bioconductor.org/packages/release/bioc/html/topGO.html). GO terms associated with each gene were obtained using the August 2017 ENSEMBL Database. Significance was determined using the classic Fisher method. GSEA analyses were performed using software developed by the Broad Institute(65, 66).
Redirected killing assay
P815 mastocytoma target cells were labeled with LIVE/DEAD Violet (Thermo Fisher Scientific) and TFL4 (OncoImmun), washed twice in PBS, and incubated for 30 minutes at room temperature with anti-CD3 (5μg/mL, UCHT1, BioRad). CD8+ T cells were negatively selected from LNMCs or PBMCs using a CD8+ T Cell Enrichment Kit (StemCell Technologies). Isolated CD8+ T cells were rested in R10 for at least 45 minutes at 37 °C and then incubated with anti-CD3-coated P815 cells at different effector-to-target ratios in a 96-well V-bottom plate for 4 hours at 37 °C. Cells were then stained as described above (see Antibodies section) and acquired using an LSRII flow cytometer (BD Biosciences).
Killing capacity was calculated by subtracting the frequency of active caspase3+ TFL4+ LIVE/DEAD− P815 cells in target-only wells from the frequency of active caspase3+ TFL4+ LIVE/DEAD− P815 cells in wells containing effector cells.
Viral suppression assay
The viral suppression assay was modified from Sáez-Cirion et al.(27). Briefly, CD4+ T cells were positively selected from LNMCs or PBMCs using a CD4+ T Cell Enrichment Kit (StemCell Technologies) and activated using a cocktail of IL-2 (100 U/mL, Chiron), anti-CD3 (1 μg/mL, UCHT1, Bio-Rad), anti-CD28 (1 μg/mL, L293, BD Biosciences), and anti-CD49d (1 μg/mL, L25, BD Biosciences). Concurrently, autologous CD8+ T cells were negatively selected from LNMCs or PBMCs using a CD8+ T Cell Enrichment Kit (StemCell Technologies) and rested in R10. After 2 days, CD4+ T cells were infected with HIV-1 BAL by spinoculation and incubated with or without autologous CD8+ T cells in the absence of exogenous IL-2. Cells were harvested after a further 3 days, stained as described above (see Antibodies section), and acquired using an LSRII flow cytometer (BD Biosciences).
Suppression capacity was calculated by dividing the frequency of p24+ CD4+ T cells in wells containing autologous CD8+ T cells by the frequency of p24+ CD4+ T cells in wells lacking autologous CD8+ T cells.
HPG translation assay
The protein translation assay was adapted from Araki et al.(67). Briefly, LNMCs were rested overnight and incubated for 30 minutes in methionine-free RPMI medium supplemented with 10% fetal bovine serum, 1% L-glutamine, and 1% penicillin/streptomycin. The cultures were then supplemented with Click-iT HPG (100 μM, Thermo Fisher Scientific). For stimulation conditions, overlapping HIV peptide pools at a final concentration of 2μg/mL/peptide (PepMix HIV (NEF) Ultra or PepMix HIV (GAG) Ultra, JPT Peptide Technologies) or anti-CD3 antibody (UCHT1, BioRad) were used in the presence of brefeldin A and monensin. Cells were harvested after a further 6 hours, stained as described above (see Antibodies), and acquired using an LSRII flow cytometer (BD Biosciences), following the instructions in the Click-iT Plus Alexa Fluor Picolyl Azide Toolkit (Thermo Fisher Scientific).
Statistics for non-sequencing data
Data were checked for normality using the Shapiro-Wilk normality test. Parametric tests were used if the data passed the normality test, and non-parametric tests were used if the data failed the normality test. Multiple corrections for two-way ANOVA were performed using the two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli. Specific tests are indicated in the relevant figure legends. Analyses were performed using R Studio or Prism v7.0 (GraphPad).
Supplementary Material
ACKNOWLEDGMENTS
We thank the Virus and Reservoirs Core at the Penn Center for AIDS Research for assistance with viral quantification, the Pathology Core at the Hospital of the University of Pennsylvania for access to biopsy materials, Kajsa Noyan-Gertler for assistance with viral suppression assays, and Mingyao Li and Yan Che for initial contributions to the scRNAseq analyses. This study was funded in part by the Oregon National Primate Research Center National Institutes of Health (NIH) grant P51OD011092 and by federal funds from the National Cancer Institute under contract HHSN261200800001E. S.D., A.R., and D.C.D. were supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases at the NIH. M.A.M., J.A.H., and M.R.B. were supported by the Penn Center for AIDS Research (P30 AI045008). M.A.M. was further supported by NIH R21 grant AI129636, the Campbell Foundation, and a grant from the W.W. Smith Charitable Trust (A1701). M.R.B. was further supported by NIH R01 grants AI076066 and AI118694 and the BEAT-HIV Delaney Collaboratory (UM1AI126620). D.A.P. was supported by a Wellcome Trust Senior Investigator Award (100326/Z/12/Z). The SCOPE cohort was supported by the University of California, San Francisco (UCSF)-Gladstone Institute of Virology and Immunology Center for AIDS Research (P30 AI027763), the Delaney AIDS Research Enterprise (AI127966), and the amfAR Institute for HIV Cure Research (109301). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services (DHHS), nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
COMPETING INTEREST STATEMENT
The authors declare no competing interests.
DATA AVAILABILITY
The sequencing data reported in this paper have been deposited in the Gene Expression Omnibus Database with the accession number GSE110684.
REFERENCES
- 1.Deeks SG, Walker BD, Human Immunodeficiency Virus Controllers: Mechanisms of Durable Virus Control in the Absence of Antiretroviral Therapy, Immunity 27, 406–416 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Fukazawa Y, Lum R, Okoye AA, Park H, Matsuda K, Bae JY, Hagen SI, Shoemaker R, Deleage C, Lucero C, Morcock D, Swanson T, Legasse AW, Axthelm MK, Hesselgesser J, Geleziunas R, Hirsch VM, Edlefsen PT, Piatak M, Estes JD, Lifson JD, Picker LJ, B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers, Nat. Med 21, 132–139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, Roederer M, Koup RA, HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells, Blood 107, 4781–4789 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Almeida JR, Price DA, Papagno L, Arkoub ZA, Sauce D, Bornstein E, Asher TE, Samri A, Schnuriger A, Theodorou I, Costagliola D, Rouzioux C, Agut H, Marcelin A-G, Douek D, Autran B, Appay V, Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover, Journal of Experimental Medicine 204, 2473–2485 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Migueles SA, Laborico AC, Shupert WL, Sabbaghian MS, Rabin R, Hallahan CW, Van Baarle D, Kostense S, Miedema F, McLaughlin M, Ehler L, Metcalf J, Liu S, Connors M, HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors, Nat Immunol 3, 1061–1068 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Migueles SA, Osborne CM, Royce C, Compton AA, Joshi RP, Weeks KA, Rood JE, Berkley AM, Sacha JB, Cogliano-Shutta NA, Lloyd M, Roby G, Kwan R, McLaughlin M, Stallings S, Rehm C, O’Shea MA, Mican J, Packard BZ, Komoriya A, Palmer S, Wiegand AP, Maldarelli F, Coffin JM, Mellors JW, Hallahan CW, Follman DA, Connors M, Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control, Immunity 29, 1009–1021 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hersperger AR, Pereyra F, Nason M, Demers K, Sheth P, Shin LY, Kovacs CM, Rodriguez B, Sieg SF, Teixeira-Johnson L, Gudonis D, Goepfert PA, Lederman MM, Frank I, Makedonas G, Kaul R, Walker BD, Betts MR, Blankson J, Ed. Perforin expression directly ex vivo by HIV-specific CD8 T-cells is a correlate of HIV elite control, PLoS Pathog 6, e1000917 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKinnon LR, Kaul R, Kimani J, Nagelkerke NJ, Wachihi C, Fowke KR, Ball TB, Plummer FA, HIV-specific CD8+ T-cell proliferation is prospectively associated with delayed disease progression, Immunology and Cell Biology 90, 346–351 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Addo MM, Draenert R, Rathod A, Verrill CL, Davis BT, Gandhi RT, Robbins GK, Basgoz NO, Stone DR, Cohen DE, Johnston MN, Flynn T, Wurcel AG, Rosenberg ES, Altfeld M, Walker BD, Zhang L, Ed. Fully differentiated HIV-1 specific CD8+ T effector cells are more frequently detectable in controlled than in progressive HIV-1 infection, PLoS ONE 2, e321 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goulder PJ, Phillips RE, Colbert RA, McAdam S, Ogg G, Nowak MA, Giangrande P, Luzzi G, Morgan B, Edwards A, McMichael AJ, Rowland-Jones S, Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS, Nat. Med 3, 212–217 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Migueles SA, Sabbaghian MS, Shupert WL, Bettinotti MP, Marincola FM, Martino L, Hallahan CW, Selig SM, Schwartz D, Sullivan J, Connors M, HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors, PNAS 97, 2709–2714 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pantaleo G, Graziosi C, Butini L, Pizzo PA, Schnittman SM, Kotler DP, Fauci AS, Lymphoid organs function as major reservoirs for human immunodeficiency virus, PNAS 88, 9838–9842 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pantaleo G, Graziosi C, Demarest JF, Butini L, Montroni M, Fox CH, Orenstein JM, Kotler DP, Fauci AS, HIV infection is active and progressive in lymphoid tissue during the clinically latent stage of disease, Nature 362, 355–358 (1993). [DOI] [PubMed] [Google Scholar]
- 14.Pantaleo G, Graziosi C, Demarest JF, Cohen OJ, Vaccarezza M, Gantt K, Muro Cacho C, Fauci AS, Role of Lymphoid Organs in the Pathogenesis of Human Immunodeficiency Virus (HIV) Infection, Immunological Reviews 140, 105–130 (1994). [DOI] [PubMed] [Google Scholar]
- 15.Perreau M, Savoye A-L, De Crignis E, Corpataux J-M, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G, Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production, J. Exp. Med 210, 143–156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estes JD, Kityo C, Ssali F, Swainson L, Makamdop KN, Del Prete GQ, Deeks SG, Luciw PA, Chipman JG, Beilman GJ, Hoskuldsson T, Khoruts A, Anderson J, Deleage C, Jasurda J, Schmidt TE, Hafertepe M, Callisto SP, Pearson H, Reimann T, Schuster J, Schoephoerster J, Southern P, Perkey K, Shang L, Wietgrefe SW, Fletcher CV, Lifson JD, Douek DC, McCune JM, Haase AT, Schacker TW, Defining total-body AIDS-virus burden with implications for curative strategies, Nat. Med (2017), doi: 10.1038/nm.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersson J, Behbahani H, Lieberman J, Connick E, Landay A, Patterson B, Sönnerborg A, Loré K, Uccini S, Fehniger TE, Perforin is not co-expressed with granzyme A within cytotoxic granules in CD8 T lymphocytes present in lymphoid tissue during chronic HIV infection, AIDS 13, 1295–1303 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Reuter MA, Del Río Estrada PM, Buggert M, Petrovas C, Ferrando-Martinez S, Nguyen S, Sada Japp A, Ablanedo-Terrazas Y, Rivero-Arrieta A, Kuri-Cervantes L, Gunzelman HM, Gostick E, Price DA, Koup RA, Naji A, Canaday DH, Reyes-Terán G, Betts MR, HIV-Specific CD8+ T Cells Exhibit Reduced and Differentially Regulated Cytolytic Activity in Lymphoid Tissue, Cell Reports 21, 3458–3470 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buggert M, Nguyen S, McLane LM, Steblyanko M, Anikeeva N, Paquin-Proulx D, Del Río Estrada PM, Ablanedo-Terrazas Y, Noyan K, Reuter MA, Demers K, Sandberg JK, Eller MA, Streeck H, Jansson M, Nowak P, Sönnerborg A, Canaday DH, Naji A, Wherry EJ, Robb ML, Deeks SG, Reyes-Terán G, Sykulev Y, Karlsson AC, Betts MR, Douek DC, Ed. Limited immune surveillance in lymphoid tissue by cytolytic CD4+ T cells during health and HIV disease, PLoS Pathog 14, e1006973 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Woon HG, Braun A, Li J, Smith C, Edwards J, Sierro F, Feng CG, Khanna R, Elliot M, Bell A, Hislop AD, Tangye SG, Rickinson AB, Gebhardt T, Britton WJ, Palendira U, Munz C, Ed. Compartmentalization of Total and Virus-Specific Tissue-Resident Memory CD8+ T Cells in Human Lymphoid Organs, PLoS Pathog 12, e1005799 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar BV, Ma W, Miron M, Granot T, Guyer RS, Carpenter DJ, Senda T, Sun X, Ho S-H, Lerner H, Friedman AL, Shen Y, Farber DL, Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites, Cell Reports 20, 2921–2934 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buggert M, Nguyen S, Salgado-Montes de Oca G, Bengsch B, Darko S, Ransier A, Roberts ER, Del Alcazar D, Brody IB, Vella LA, Beura L, Wijeyesinghe S, Herati RS, Del Río Estrada PM, Ablanedo-Terrazas Y, Kuri-Cervantes L, Sada Japp A, Manne S, Vartanian S, Huffman A, Sandberg JK, Gostick E, Nadolski G, Silvestri G, Canaday DH, Price DA, Petrovas C, Su LF, Vahedi G, Dori Y, Frank I, Itkin MG, Wherry EJ, Deeks SG, Naji A, Reyes-Terán G, Masopust D, Douek DC, Betts MR, Identification and characterization of HIV-specific resident memory CD8+ T cells in human lymphoid tissue, Science Immunology 3, eaar4526 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mens H, Kearney M, Wiegand A, Shao W, Schonning K, Gerstoft J, Obel N, Maldarelli F, Mellors JW, Benfield T, Coffin JM, HIV-1 Continues To Replicate and Evolve in Patients with Natural Control of HIV Infection, Journal of Virology 84, 12971–12981 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Connell KA, Brennan TP, Bailey JR, Ray SC, Siliciano RF, Blankson JN, Control of HIV-1 in elite suppressors despite ongoing replication and evolution in plasma virus, Journal of Virology 84, 7018–7028 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blankson JN, Bailey JR, Thayil S, Yang H-C, Lassen K, Lai J, Gandhi SK, Siliciano JD, Williams TM, Siliciano RF, Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors, Journal of Virology 81, 2508–2518 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boritz EA, Darko S, Swaszek L, Wolf G, Wells D, Wu X, Henry AR, Laboune F, Hu J, Ambrozak D, Hughes MS, Hoh R, Casazza JP, Vostal A, Bunis D, Nganou-Makamdop K, Lee JS, Migueles SA, Koup RA, Connors M, Moir S, Schacker T, Maldarelli F, Hughes SH, Deeks SG, Douek DC, Multiple Origins of Virus Persistence during Natural Control of HIV Infection, Cell (2016), doi: 10.1016/j.cell.2016.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sáez-Cirión A, Shin SY, Versmisse P, Barré-Sinoussi F, Pancino G, Ex vivo T cell–based HIV suppression assay to evaluate HIV-specific CD8+ T-cell responses, Nat Protoc 5, 1033–1041 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Folkvord JM, Anderson DM, Arya J, MaWhinney S, Connick E, Microanatomic relationships between CD8+ cells and HIV-1-producing cells in human lymphoid tissue in vivo, J. Acquir. Immune Defic. Syndr 32, 469–476 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Folkvord JM, Armon C, Connick E, Lymphoid follicles are sites of heightened human immunodeficiency virus type 1 (HIV-1) replication and reduced antiretroviral effector mechanisms, AIDS Res. Hum. Retroviruses 21, 363–370 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Connick E, Mattila T, Folkvord JM, Schlichtemeier R, Meditz AL, Ray MG, McCarter MD, MaWhinney S, Hage A, White C, Skinner PJ, CTL fail to accumulate at sites of HIV-1 replication in lymphoid tissue, J. Immunol 178, 6975–6983 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Connick E, Folkvord JM, Lind KT, Rakasz EG, Miles B, Wilson NA, Santiago ML, Schmitt K, Stephens EB, Kim HO, Wagstaff R, Li S, Abdelaal HM, Kemp N, Watkins DI, MaWhinney S, Skinner PJ, Compartmentalization of simian immunodeficiency virus replication within secondary lymphoid tissues of rhesus macaques is linked to disease stage and inversely related to localization of virus-specific CTL, J. Immunol 193, 5613–5625 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li S, Folkvord JM, Rakasz EG, Abdelaal HM, Wagstaff RK, Kovacs KJ, Kim HO, Sawahata R, MaWhinney S, Masopust D, Connick E, Skinner PJ, Silvestri G,Ed. Simian Immunodeficiency Virus-Producing Cells in Follicles Are Partially Suppressed by CD8+ Cells In Vivo, Journal of Virology 90, 11168–11180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rahman MA, McKinnon KM, Karpova TS, Ball DA, Venzon DJ, Fan W, Kang G, Li Q, Robert-Guroff M, Associations of Simian Immunodeficiency Virus (SIV)-Specific Follicular CD8+T Cells with Other Follicular T Cells Suggest Complex Contributions to SIV Viremia Control, J. Immunol, ji1701403 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russell JH, Ley TJ, Lymphocyte-mediated cytotoxicity, Annu. Rev. Immunol 20, 323–370 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Voskoboinik I, Whisstock JC, Trapani JA, Perforin and granzymes: function, dysfunction and human pathology, Nature Reviews Immunology 15, 388–400 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Buggert M, Tauriainen J, Yamamoto T, Frederiksen J, Ivarsson MA, Michaëlsson J, Lund O, Hejdeman B, Jansson M, Sönnerborg A, Koup RA, Betts MR, Karlsson AC, Luban J, Ed. T-bet and Eomes are differentially linked to the exhausted phenotype of CD8+ T cells in HIV infection, PLoS Pathog 10, e1004251 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wherry EJ, Kurachi M, Molecular and cellular insights into T cell exhaustion, Nature Reviews Immunology 15, 486–499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tauriainen J, Scharf L, Frederiksen J, Naji A, Ljunggren H-G, Sönnerborg A, Lund O, Reyes-Terán G, Hecht FM, Deeks SG, Betts MR, Buggert M, Karlsson AC, Perturbed CD8(+) T cell TIGIT/CD226/PVR axis despite early initiation of antiretroviral treatment in HIV infected individuals, Sci Rep 7, 40354 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carrette F, Surh CD, IL-7 signaling and CD127 receptor regulation in the control of T cell homeostasis, Semin. Immunol 24, 209–217 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suomi T, Seyednasrollah F, Jaakkola MK, Faux T, Elo LL, Poisot T, Ed. ROTS: An R package for reproducibility-optimized statistical testing, PLoS Comput. Biol 13, e1005562 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korthauer KD, Chu L-F, Newton MA, Li Y, Thomson J, Stewart R, Kendziorski C, A statistical approach for identifying differential distributions in single-cell RNA-seq experiments, Genome Biol. 17, 222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R, Integrating single-cell transcriptomic data across different conditions, technologies, and species, Nat. Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lane BR, Markovitz DM, Woodford NL, Rochford R, Strieter RM, Coffey MJ, TNF-α Inhibits HIV-1 Replication in Peripheral Blood Monocytes and Alveolar Macrophages by Inducing the Production of RANTES and Decreasing C-C Chemokine Receptor 5 (CCR5) Expression, The Journal of Immunology 163, 3653–3661 (1999). [PubMed] [Google Scholar]
- 44.Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P, Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells, Science 270, 1811–1815 (1995). [DOI] [PubMed] [Google Scholar]
- 45.Appay V, Rowland-Jones SL, RANTES: a versatile and controversial chemokine, Trends in Immunology 22, 83–87 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Bedoya VI, Boasso A, Hardy AW, Rybak S, Shearer GM, Rugeles MT, Ribonucleases in HIV Type 1 Inhibition: Effect of Recombinant RNases on Infection of Primary T Cells and Immune Activation-Induced RNase Gene and Protein Expression, https://home-liebertpub-com.proxy.library.upenn.edu/aid 22, 897–907 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Zapata W, Aguilar-Jiménez W, Feng Z, Weinberg A, Russo A, Potenza N, Estrada H, Rugeles MT, Identification of innate immune antiretroviral factors during in vivo and in vitro exposure to HIV-1, Microbes and Infection 18, 211–219 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Monteleone K, Di Maio P, Cacciotti G, Falasca F, Fraulo M, Falciano M, Mezzaroma I, D’Ettorre G, Turriziani O, Scagnolari C, Interleukin-32 isoforms: expression, interaction with interferon-regulated genes and clinical significance in chronically HIV-1-infected patients, Med. Microbiol. Immunol 203, 207–216 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Ribeiro-Dias F, Saar Gomes R, de Lima Silva LL, Dos Santos JC, Joosten LAB, Interleukin 32: a novel player in the control of infectious diseases, Journal of Leukocyte Biology 101, 39–52 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Cartwright EK, Spicer L, Smith SA, Lee D, Fast R, Paganini S, Lawson BO, Nega M, Easley K, Schmitz JE, Bosinger SE, Paiardini M, Chahroudi A, Vanderford TH, Estes JD, Lifson JD, Derdeyn CA, Silvestri G, CD8+ Lymphocytes Are Required for Maintaining Viral Suppression in SIV-Infected Macaques Treated with Short-Term Antiretroviral Therapy, Immunity 45, 656–668 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thome JJC, Yudanin N, Ohmura Y, Kubota M, Grinshpun B, Sathaliyawala T, Kato T, Lerner H, Shen Y, Farber DL, Spatial map of human T cell compartmentalization and maintenance over decades of life, Cell 159, 814–828 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guidotti LG, Ishikawa T, Hobbs MV, Matzke B, Schreiber R, Chisari FV, Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes, Immunity 4, 25–36 (1996). [DOI] [PubMed] [Google Scholar]
- 53.Guidotti LG, Chisari FV, To kill or to cure: options in host defense against viral infection, Curr. Opin. Immunol 8, 478–483 (1996). [DOI] [PubMed] [Google Scholar]
- 54.Guidotti LG, Chisari FV, Noncytolytic control of viral infections by the innate and adaptive immune response, Annu. Rev. Immunol 19, 65–91 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Walker CM, Moody DJ, Stites DP, Levy JA, CD8+ lymphocytes can control HIV infection in vitro by suppressing virus replication, Science 234, 1563–1566 (1986). [DOI] [PubMed] [Google Scholar]
- 56.Klatt NR, Shudo E, Ortiz AM, Engram JC, Paiardini M, Lawson B, Miller MD, Else J, Pandrea I, Estes JD, Apetrei C, Schmitz JE, Ribeiro RM, Perelson AS, Silvestri G, Douek DC, Ed. CD8+ Lymphocytes Control Viral Replication in SIVmac239-Infected Rhesus Macaques without Decreasing the Lifespan of Productively Infected Cells, PLoS Pathog 6, e1000747 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wong JK, Strain MC, Porrata R, Reay E, Sankaran-Walters S, Ignacio CC, Russell T, Pillai SK, Looney DJ, Dandekar S, Douek DC, Ed. In Vivo CD8+ T-Cell Suppression of SIV Viremia Is Not Mediated by CTL Clearance of Productively Infected Cells, PLoS Pathog 6, e1000748 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trautmann L, Kill: boosting HIV-specific immune responses, Curr Opin HIV AIDS 11, 409–416 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levy JA, The search for the CD8+ cell anti-HIV factor (CAF), Trends in Immunology 24, 628–632 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Deleage C, Wietgrefe SW, Del Prete G, Morcock DR, Hao XP, Piatak M, Bess J, Anderson JL, Perkey KE, Reilly C, McCune JM, Haase AT, Lifson JD, Schacker TW, Estes JD, Defining HIV and SIV Reservoirs in Lymphoid Tissues, Pathog Immun 1, 68–106 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Betts MR, Brenchley JM, Price DA, De Rosa SC, Douek DC, Roederer M, Koup RA, Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation, J. Immunol. Methods 281, 65–78 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Price DA, Brenchley JM, Ruff LE, Betts MR, Hill BJ, Roederer M, Koup RA, Migueles SA, Gostick E, Wooldridge L, Sewell AK, Connors M, Douek DC, Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses, Journal of Experimental Medicine 202, 1349–1361 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schuetz A, Deleage C, Sereti I, Rerknimitr R, Phanuphak N, Phuang-Ngern Y, Estes JD, Sandler NG, Sukhumvittaya S, Marovich M, Jongrakthaitae S, Akapirat S, Fletscher JLK, Kroon E, Dewar R, Trichavaroj R, Chomchey N, Douek DC, O Connell RJ, Ngauy V, Robb ML, Phanuphak P, Michael NL, Excler J-L, Kim JH, de Souza MS, Ananworanich J, RV254/SEARCH 010 and RV304/SEARCH 013 Study Groups, Desrosiers RC, Ed. Initiation of ART during early acute HIV infection preserves mucosal Th17 function and reverses HIV-related immune activation, PLoS Pathog 10, e1004543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM, CellProfiler: image analysis software for identifying and quantifying cell phenotypes, Genome Biol. 7, R100 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC, PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes, Nat. Genet 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 66.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles, PNAS 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Araki K, Morita M, Bederman AG, Konieczny BT, Kissick HT, Sonenberg N, Ahmed R, Translation is actively regulated during the differentiation of CD8(+) effector T cells, Nat Immunol 18, 1046–1057 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


