Abstract
The immune system is a dynamic mesh of molecules, cells and tissues spanning the entire organism. Despite a wealth of knowledge about the components of the immune system, little is known about the general rules governing the organismal circuitry of immunity. Deciphering the immune system at the scale of the whole organism is crucial to understanding fundamental problems in immunobiology and physiology, and to manipulate immunity for maintaining health and preventing disease. Here I discuss the emerging principles of inter-organ communications during immune responses by focusing on three common themes that are the regulation of the (i) composition, (ii) condition and (iii) coordination of communicating organs by molecular and cellular factors. Based on these common principles, I emphasize fundamental gaps in our knowledge of organismal immune processes and the outlook to tackle immunity at the scale of the whole organism.
Introduction
Organs exchange information. For example, organs sensing food, light or stress send signals to other organ systems, allowing the organism to maintain homeostasis [1,2]. Mammalian immunity is one of the most striking examples of such inter-organ communications (Figure 1). The immune system evolved to cope with pathogens anywhere in the body – may it be a parasite residing the gut or a virus spreading to multiple organs. As a result, molecules, cells and tissues with immunological functions are ubiquitously and dynamically distributed across the organism.
Figure 1. Inter-organ communications in the mammalian immune system.
Simplified schematic of the communications events taking place between organs during organismal immunological processes. A sender organ processes input signals such as pathogens, injury or stress and releases molecular or cellular mediators. Mediators reach one or more distant, receiver organs via blood and/or lymph. The receiver organ modifies its immunological and/or physiological states, and, in cases of complex inter-organ circuits, may further propagates information to other organs.
However, while the systemic property of immunity is obvious, remarkably little is known about the general rules guiding immune processes across organs. For example, when the concentration of a cytokine varies in the blood as a result of host defense or disease, we most often lack a clear picture of the sender and receiver organs and cells that are involved. Another example is the migration dynamics of immune cells across the body which remains to be elucidated for most cell types.
Thus, a fundamental challenge in immunology today is to develop new ways to study the structure, regulation and function of the immunological events that cross organ boundaries. Deciphering the design principles of inter-organ immune signaling will yield insights into the functions and malfunctions of immunity at an unprecedented scale, that of the whole organism. Here I discuss examples of inter-organ communications in immunology by focusing on three common themes that are the regulation of the (i) composition, (ii) condition and (iii) coordination of communicating organs by molecular and cellular factors. I also emphasize fundamental questions in this emerging field and the outlook to answer them.
Molecular and cellular immune factors involved in inter-organ communications
All tissues in the body can secrete factors with local or systemic effects. A survey of 32 human tissues estimated that 10-20% of the transcripts found in any given tissue produce secreted proteins [3]. In some cases, the percentage of transcripts encoding secreted proteins can be much higher due to tissue specialization, including 70 and 40% for the pancreas and liver respectively [3]. While many factors have key roles in inter-organ communications, including metabolites, growth factors or extracellular vesicles, I focus here on cytokines which sensu stricto include interleukins, chemokines and other overlapping families of secreted immune factors. Although the concepts discussed below apply to other molecular species with immune functions, I primarily discuss cytokines for simplicity and because (1) they can be secreted by most, if not all, nucleated cells, (2) they can act as autocrine, paracrine and endocrine messengers, and (3) their primary function is the regulation of the immune system.
Many aspects of cytokine biology have been under investigation for decades, including their structural and signaling properties, their impact on cell proliferation, differentiation or death, and their association with human diseases [4-17]. In addition, recent work has begun to reveal key properties of the inter-cellular communications mediated by cytokines. For example, quantitative models helped explain the dynamics of cytokine production and consumption in cell ensembles [18], or the integration of multiple cytokine signals by T lymphocytes [19,20].
In response to local or systemic cues such as cytokines, immune cells relocate across the body as they mature and guard the host against pathogens. For example, the T cell life cycle starts in the bone marrow, continues in the thymus and, for naive T cells, throughout the body until encountering a cognate antigen [21]. However, despite this wealth of knowledge about immune cells and cytokines, we know surprisingly little about the organismal circuitry of the cytokine system and its systemic impact on cells. We also lack dynamic models that help to explain inter-organ molecular and cellular exchanges during immune processes.
Though the roles of cytokines and immune cells are seemingly countless in health and disease, I argue that common themes can be found in inter-organ signaling and can be useful as a conceptual guide for the much-needed exploration of the immune system at the scale of the whole body. To illustrate this point, I examine below examples of inter-organ crosstalk which fall into three categories based on the ability of immune cells and cytokines to regulate the (i) composition, (ii) condition and (iii) coordination of organ systems.
Regulating the cellular composition of organs
The first category of inter-organ crosstalk reflects the role that cytokines and immune cells can play in regulating the cellular composition of distant organs. For example, the composition of the hematopoietic compartment of the bone marrow can be dramatically remodeled during the switch from steady state to so-called emergency hematopoiesis during systemic bacterial infection [22]. Endothelial cells from multiple tissues, including heart, liver, kidney, spleen and bone marrow, can detect lipopolysaccharide (LPS) via Toll-like receptor (TLR) 4 and release granulocyte colony-stimulating factor (G-CSF) into the blood circulation. G-CSF then acts on myeloid restricted progenitors in the bone marrow to increase granulopoiesis [23]. Interestingly, this concept of the remote regulation of hematopoiesis has been observed in other contexts, including in lung adenocarcinoma where the release of a soluble receptor in the blood triggers an increase in neutrophil maturation and recruitment to the tumor [24].
As demonstrated with hematopoiesis, the efflux and afflux of immune cells can modify the cellular composition of an organ. Such changes in composition can be temporary during an acute response or long-term as seen, for example, with the seeding of macrophages throughout the body during embryonic development [25]. Another example of this paradigm is the memory T cell compartment. Pioneering work revealed that memory T cells can distribute to most lymphoid and non-lymphoid organs in the body upon systemic challenge [26,27]. Recently, changes in immune cell composition were observed in mice during cycles of fasting or caloric restrictions as a result of inter-organ signals involving bone marrow, liver and lymphoid tissues. The numbers of monocytes and lymphocytes in blood and peripheral organs were strongly reduced, while in the bone marrow, lymphocyte numbers increased and monocyte egress decreased [28-30], which highlights the complex regulation of various immune cell compartments across organs.
Conditioning the functional state of organs
Cytokines may condition a tissue to perform a specific task related to a physiologic or defense need for the host. For instance, type I interferons (IFNs) produced in one organ can trigger an antiviral state in distant tissues. Respiratory viral infection can lead to the activation of antiviral genes in the bone marrow [31], while skin infection with a live attenuated strain of Vaccinia virus triggers a whole-body antiviral state through inter-organ IFN signaling [32]. Perhaps systemic IFN signaling evolved to arm distant tissues with antiviral defenses as a means for the host to prevent the spread of a virus across the body [32].
Similar to cytokines, immune cells modify the state of tissues as part of various inter-organ circuits. For example, neutrophils contribute to liver tissue repair prior to migrating to the lungs and subsequently the bone marrow, where they die by apoptosis [33]. Another example comes from memory T cells that reside in tissues, so called TRM cells [34-36], which have been shown to trigger organ-wide anti-microbial states [32,37]. Further, upon skin injection of a live attenuated strain of Vaccinia virus, CD8+ TRM cells have been shown to distribute broadly across distant organs, such as lung and liver, and to establish intercellular circuits that are tissue-specific and important for protection. The resulting multi-organ web of TRM cells can trigger organ-wide, antiviral states in tissues targeted by the virus as a means to limit viral spread [32].
Physiologic and immune coordination across organs
Cytokines may act by coordinating the physiological pathways of multiple organs either in parallel or serially. For example, TNF-α, IL-1β and IL-6 have been much studied for their roles in the inter-organ communications regulating metabolism [38]. Another example is TGF-β2 that is released by subcutaneous adipose tissue after exercise, leading to increased glucose uptake by muscle, heart and brown adipose tissue and beneficial metabolic effects across the body [39]. In addition, secreted factors may impact distant organs indirectly via, for example, a nervous system relay [40]. For example, IL-1β produced in the gut [41] or GDF15 in the liver or kidney [42] can act on the brain to respectively modify host anorexic behavior or hepatic triglyceride fluxes that are key for heart function during sepsis. Together, these examples highlight the power of cytokines in coordinating the activities of multiple organs either directly, through sensing of a given cytokine by multiple tissues, or indirectly, by acting on non-immune relays to communicate between organs such as neurons.
Furthermore, a variety of immune cell types migrate between organs to coordinate immunosurveillance and protective responses across the body. For example, progenitor and mature innate and adaptive immune cells share similarities in their recirculation patterns across organs. Hematopoietic progenitor cells originating in the bone marrow traffic to multiple nonlymphoid tissues where they temporarily reside prior to returning to the blood via the lymph, similarly to naive T cells [43]. Innate lymphoid cells (ILCs) were recently found to also follow inter-organ paths. Group 2 ILCs migrate from the gut to peripheral tissues such as lungs to protect the host from helminth infection [44].
Open questions about inter-organ immune crosstalk
Several fundamental questions arise from the observations reported in the case studies discussed above. Indeed, while it is clear that cytokines and immune cells cross organ boundaries to coordinate host protection and physiology, little is known about the design principles of these inter-organ circuits.
First, we lack a clear picture of the scope of these organismal communications. For cytokines, we often do not know which ones are released from which organs to impact which distant tissues, in what biological contexts do these cytokinic communications occur, and what are the temporal and spatial parameters at play during inter-tissue crosstalk? For cells, the organismal migration patterns of circulating immune cell types and subsets are not well understood and difficult to track experimentally. In addition, the full complement of the immune cell types that are involved in such inter-organ pathways is likely unknown. For example, recent work has shown that cells thought to be largely tissue resident were in fact able to recirculate and reach distant tissues in some conditions, including both innate [44] and adaptive [45,46] cells.
Second, in the context of cytokines whose organismal effects have been documented, we often lack information about the sender and receiver cells involved and that may be hematopoietic or not. For example, cytokines such as IL-22 are secreted by hematopoietic cells and target non-hematopoietic cells [47]. One corollary to this lack of knowledge about the sender and receiver cells for any given cytokine is that, in most cases, the cytokine signaling relays that are responsible for the mobilization, migration (influx and efflux), positioning and adaptation of immune cells within a tissue remain unclear.
Third, what are the combinatorial effects of cytokines and other signals on cells and tissues? Many diseases are associated with increased levels of multiple cytokines in the blood. Presumably, each receiver cell and organ for those endocrine signals could respond to more than one cytokine at the same time. For example, at the level of T cells, the strength of a response is equal to the sum of its parts, including cytokine signals [19,20]. Whether such simplifying principle will hold true in other cases remains to be tested but it is worth considering for the study of inter-organ signaling.
Overall, addressing the questions highlighted above will help to identify the common rules governing the inter-organ circuity of immunity. Further, although I focused on cytokines, similar points can be raised for other inter-organ factors, including metabolites, hormones, antibodies, microbial components or even self-antigens, which can cross organ boundaries in type 1 diabetes [48].
Outlook on studying inter-organ communications
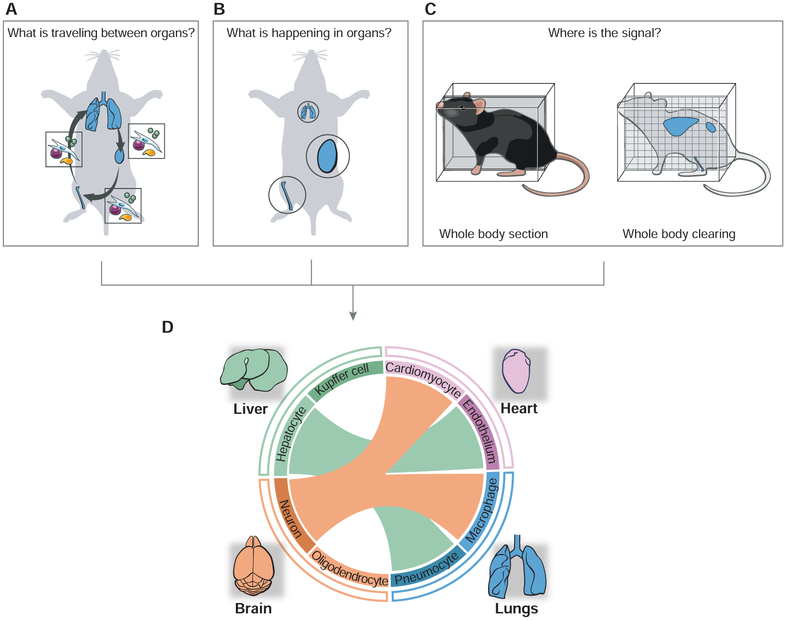
I discussed examples of inter-organ crosstalk and their impact on the composition, condition and coordination of organs during immune processes, as well as fundamental questions for the future. What is the outlook to answer those questions and tackle the challenges posed by the ubiquitous nature of immune factors across the body? The central challenge is twofold: organism-wide sampling and connecting the dynamic events involved in inter-organ signaling, which will require the development of new tools and approaches (Figure 2).
Figure 2. Studying immunological processes at the scale of the whole body.
(A-C) Schematics illustrating the fundamental challenges associated with studying inter-organ signaling. The three upper panels illustrate how to identify the mediators of inter-organ communications and their impacts on organ states. In A, the boxes illustrate the molecular and cellular mediators of inter-organ communications – using lungs, bone and kidney as a hypothetical network of communicating organs. In B, organs involved in a systemic communication circuit are shown in circles whose size is proportional to a given activity or effector mechanism. In C, organism-wide imaging is illustrated using as examples whole-body sectioning or clearing.
(D) Towards organism-level analyses of immune circuitry and its integration with host physiology. Data obtained through the approaches listed above (A-C) are integrated as a hypothetical inter-organ network, which is represented as a circular plot with links (colored lines) between communicating organs (outer circle) and cell types (inner circle) within each organ. The color of the lines linking organs depicts the sender organ.
First, to identify the mediators that carry information across organs, it is critical to improve methods to profiles cells and molecules in the circulatory systems of the body (i.e., blood, lymph). For example, sampling the influx and efflux of molecules and cells from multiple organs across the body will help to inform the identity of inter-organ signals and their fluxes in individual organs (Figure 2A). Such methods have long been employed to study the fluxes of substrates across organs and were successfully employed to measure changes in arteriovenous metabolomic profiles across most organs in pigs [49]. Sampling of immune cells in the lymph has also revealed key properties for memory T cells [50]. Thus, combining large-scale sampling of bodily fluids and measurements of various molecular and cellular entities will be a powerful means to identify inter-organ messengers.
Second, inter-organ signaling impacts the states of the communicating organs. Thus, experimental methods are needed to characterize the dynamic changes occurring across communicating organs (Figure 2B). For example, multi-tissue gene expression studies have started to contribute to addressing this challenge by identifying shared and tissue-specific expression patterns that vary in health and disease [51-59]. In addition, organ-level expression can detect immunological changes driven by cell composition or direct gene regulation, even in rare cells [32,37,60,61]. Multi-tissue profiling approaches will further help to decipher interorgan circuits when combined with, for example, (1) ongoing efforts to map the cellular composition of organs at large [62-64], or by focusing on immune cells [65,66], (2) single-cell measurement tools that are becoming increasingly multiparametric [67], (3) methods to locate molecules and cells in tissue sections or whole tissues [68-75], and (4) computational deconvolution methods, whereby cellular composition and contribution are inferred from bulk expression or epigenomic measurements [76,77].
Third, and perhaps most difficult, how can we establish causal relationships between systemic mediators and the regulation of multi-organ processes (Figure 2D)? First, chemical, genetic or other perturbations will help to understand inter-organ pathways as long as such perturbations are applied to a sender organ specifically while monitoring the impact on putative receiver organs. Modifying molecular and cellular mediators through engineering can broaden the range of perturbations available to tease apart inter-organ signaling. For example, recent advances in engineering proteins [78] and cytokines [79] will be invaluable tools, with application including the targeting of a given cytokine to a tissue of interest [80]. In addition, the ability to recreate multi-tissue systems in vitro is likely to generate useful toy models that are easy to manipulate to tease apart inter-organ signaling [81-83].
Next, computational approaches that can be applied to the study inter-organ signaling are emerging. For example, a simple example comes from the mining of the expression patterns of secreted proteins and their cognate receptors across nine organs upon infection, which helped to infer putative inter-organ connectivity maps that can then be used as starting points for in depth studies [32]. In addition, methods such as systems genetics that leverages the analysis of multi-tissue datasets across individuals from various genetic backgrounds [84] and large-scale text mining of the PubMed database [85] also provide powerful tools to decipher inter-organ signaling. Complementing these approaches with quantitative models built for inter-organ timescales and processes will be crucial to understand the emerging principles of systemic communications in immunology. The development of such models will benefit from the acquisition of time series data at the scale of the whole-body, which is becoming increasingly feasible using whole-tissue gene expression [32].
Last, recent developments in imaging modalities across the organism will be valuable to observe and quantify the dynamics of molecules and cells across multiple organs in parallel, including, for example, immuno-PET [86], whole-body sectioning [27,87,88] or clearing methods [89] (Figure 2C). Combining whole-body imaging with reporters of various cellular activities or recent advances in cellular barcoding approaches for lineage tracing [90-92] is likely to yield important insights for our understanding of whole-body immunity.
Conclusions
Using examples of the regulation of organ composition, condition and coordination by the immune system, I have emphasized how little we know about organismal immunity. Deciphering the immune system at the scale of the whole organism is crucial to understanding fundamental problems in immunobiology and physiology, and to manipulate immunity for maintaining health and preventing disease. Although studying inter-organ immune signaling is a daunting challenge, I highlighted several paths forward based upon recent advances in immunology and beyond. Approaches likely to help us in this endeavor include (1) finding a minimal set of models to focus on for the comparative analysis of inter-organ circuits, (2) mapping organism-wide communications by developing molecular and cellular measurement, imaging and perturbation tools, and (3) creating synthetic assemblies of tissues and organs that mimic key features of interorgan processes.
Acknowledgments
I am grateful to many colleagues in the field and members of the lab for critical discussions. I thank anonymous reviewers for helpful comments and Sigrid Knemeyer for help with figures. This work was kindly supported by a National Institutes of Health Director’s New Innovator Award (1DP2AI145100-01) and the Elliot and Ruth Sigal Melanoma Research Alliance Young Investigator Award.
References
- 1.Droujinine IA, Perrimon N: Interorgan Communication Pathways in Physiology: Focus on Drosophila. Annu Rev Genet 2016, 50:539–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Owusu-Ansah E, Perrimon N: Stress signaling between organs in metazoa. Annu Rev Cell Dev Biol 2015, 31:497–522. [DOI] [PubMed] [Google Scholar]
- 3.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. : Proteomics. Tissue-based map of the human proteome. Science 2015, 347:1260419. [DOI] [PubMed] [Google Scholar]
- 4.Batlle E, Massague J: Transforming Growth Factor-beta Signaling in Immunity and Cancer. Immunity 2019, 50:924–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dougan M, Dranoff G, Dougan SK: GM-CSF, IL-3, and IL-5 Family of Cytokines: Regulators of Inflammation. Immunity 2019, 50:796–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedrich M, Pohin M, Powrie F: Cytokine Networks in the Pathophysiology of Inflammatory Bowel Disease. Immunity 2019, 50:992–1006. [DOI] [PubMed] [Google Scholar]
- 7.Hammond TR, Marsh SE, Stevens B: Immune Signaling in Neurodegeneration. Immunity 2019, 50:955–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang S, Tanaka T, Narazaki M, Kishimoto T: Targeting Interleukin-6 Signaling in Clinic. Immunity 2019, 50:1007–1023. [DOI] [PubMed] [Google Scholar]
- 9.Lambrecht BN, Hammad H, Fahy JV: The Cytokines of Asthma. Immunity 2019, 50:975–991. [DOI] [PubMed] [Google Scholar]
- 10.Lazear HM, Schoggins JW, Diamond MS: Shared and Distinct Functions of Type I and Type III Interferons. Immunity 2019, 50:907–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leonard WJ, Lin JX, O'Shea JJ: The gammac Family of Cytokines: Basic Biology to Therapeutic Ramifications. Immunity 2019, 50:832–850. [DOI] [PubMed] [Google Scholar]
- 12.Mantovani A, Dinarello CA, Molgora M, Garlanda C: Interleukin-1 and Related Cytokines in the Regulation of Inflammation and Immunity. Immunity 2019, 50:778–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGeachy MJ, Cua DJ, Gaffen SL: The IL-17 Family of Cytokines in Health and Disease. Immunity 2019, 50:892–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murakami M, Kamimura D, Hirano T: Pleiotropy and Specificity: Insights from the Interleukin 6 Family of Cytokines. Immunity 2019, 50:812–831. [DOI] [PubMed] [Google Scholar]
- 15.Ouyang W, O'Garra A: IL-10 Family Cytokines IL-10 and IL-22: from Basic Science to Clinical Translation. Immunity 2019, 50:871–891. [DOI] [PubMed] [Google Scholar]
- 16.Tait Wojno ED, Hunter CA, Stumhofer JS: The Immunobiology of the Interleukin-12 Family: Room for Discovery. Immunity 2019, 50:851–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams JW, Huang LH, Randolph GJ: Cytokine Circuits in Cardiovascular Disease. Immunity 2019, 50:941–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altan-Bonnet G, Mukherjee R: Cytokine-mediated communication: a quantitative appraisal of immune complexity. Nat Rev Immunol 2019, 19:205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gett AV, Hodgkin PD: A cellular calculus for signal integration by T cells. Nat Immunol 2000, 1:239–244. [DOI] [PubMed] [Google Scholar]
- 20.Marchingo JM, Kan A, Sutherland RM, Duffy KR, Wellard CJ, Belz GT, Lew AM, Dowling MR, Heinzel S, Hodgkin PD: T cell signaling. Antigen affinity, costimulation, and cytokine inputs sum linearly to amplify T cell expansion. Science 2014, 346:1123–1127. [DOI] [PubMed] [Google Scholar]
- 21.von Andrian UH, Mackay CR: T-cell function and migration. Two sides of the same coin. N Engl J Med 2000, 343:1020–1034. [DOI] [PubMed] [Google Scholar]
- 22.Boettcher S, Manz MG: Regulation of Inflammation- and Infection-Driven Hematopoiesis. Trends Immunol 2017, 38:345–357. [DOI] [PubMed] [Google Scholar]
- 23.Boettcher S, Gerosa RC, Radpour R, Bauer J, Ampenberger F, Heikenwalder M, Kopf M, Manz MG: Endothelial cells translate pathogen signals into G-CSF-driven emergency granulopoiesis. Blood 2014, 124:1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engblom C, Pfirschke C, Zilionis R, Da Silva Martins J, Bos SA, Courties G, Rickelt S, Severe N, Baryawno N, Faget J, et al. : Osteoblasts remotely supply lung tumors with cancer-promoting SiglecF(high) neutrophils. Science 2017, 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ginhoux F, Guilliams M: Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity 2016, 44:439–449. [DOI] [PubMed] [Google Scholar]
- 26.Masopust D, Vezys V, Marzo AL, Lefrancois L: Preferential localization of effector memory cells in nonlymphoid tissue. Science 2001, 291:2413–2417. [DOI] [PubMed] [Google Scholar]
- 27.Reinhardt RL, Khoruts A, Merica R, Zell T, Jenkins MK: Visualizing the generation of memory CD4 T cells in the whole body. Nature 2001, 410:101–105. [DOI] [PubMed] [Google Scholar]
- 28.Collins N, Han SJ, Enamorado M, Link VM, Huang B, Moseman EA, Kishton RJ, Shannon JP, Dixit D, Schwab SR, et al. : The Bone Marrow Protects and Optimizes Immunological Memory during Dietary Restriction. Cell 2019, 178:1088–1101 e1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagai M, Noguchi R, Takahashi D, Morikawa T, Koshida K, Komiyama S, Ishihara N, Yamada T, Kawamura YI, Muroi K, et al. : Fasting-Refeeding Impacts Immune Cell Dynamics and Mucosal Immune Responses. Cell 2019, 178:1072–1087 e1014. [DOI] [PubMed] [Google Scholar]
- 30.Jordan S, Tung N, Casanova-Acebes M, Chang C, Cantoni C, Zhang D, Wirtz TH, Naik S, Rose SA, Brocker CN, et al. : Dietary Intake Regulates the Circulating Inflammatory Monocyte Pool. Cell 2019, 178:1102–1114 e1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hermesh T, Moltedo B, Moran TM, Lopez CB: Antiviral instruction of bone marrow leukocytes during respiratory viral infections. Cell Host Microbe 2010, 7:343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kadoki M, Patil A, Thaiss CC, Brooks DJ, Pandey S, Deep D, Alvarez D, von Andrian UH, Wagers AJ, Nakai K, et al. : Organism-Level Analysis of Vaccination Reveals Networks of Protection across Tissues. Cell 2017, 171:398–413 e321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Hossain M, Thanabalasuriar A, Gunzer M, Meininger C, Kubes P: Visualizing the function and fate of neutrophils in sterile injury and repair. Science 2017, 358:111–116. [DOI] [PubMed] [Google Scholar]
- 34.Masopust D, Soerens AG: Tissue-Resident T Cells and Other Resident Leukocytes. Annu Rev Immunol 2019, 37:521–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gebhardt T, Palendira U, Tscharke DC, Bedoui S: Tissue-resident memory T cells in tissue homeostasis, persistent infection, and cancer surveillance. Immunol Rev 2018, 283:54–76. [DOI] [PubMed] [Google Scholar]
- 36.Park SL, Gebhardt T, Mackay LK: Tissue-Resident Memory T Cells in Cancer Immunosurveillance. Trends Immunol 2019, 40:735–747. [DOI] [PubMed] [Google Scholar]
- 37.Ariotti S, Hogenbirk MA, Dijkgraaf FE, Visser LL, Hoekstra ME, Song JY, Jacobs H, Haanen JB, Schumacher TN: T cell memory. Skin-resident memory CD8(+) T cells trigger a state of tissue-wide pathogen alert. Science 2014, 346:101–105. [DOI] [PubMed] [Google Scholar]
- 38.Lee YS, Wollam J, Olefsky JM: An Integrated View of Immunometabolism. Cell 2018, 172:22–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takahashi H, Alves CRR, Stanford KI, Middelbeek RJW, Pasquale N, Ryan RE, Xue R, Sakaguchi M, Lynes MD, So K, et al. : TGF-beta2 is an exercise-induced adipokine that regulates glucose and fatty acid metabolism. Nat Metab 2019, 1:291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Godinho-Silva C, Cardoso F, Veiga-Fernandes H: Neuro-Immune Cell Units: A New Paradigm in Physiology. Annu Rev Immunol 2019, 37:19–46. [DOI] [PubMed] [Google Scholar]
- 41.Rao S, Schieber AMP, O'Connor CP, Leblanc M, Michel D, Ayres JS: Pathogen-Mediated Inhibition of Anorexia Promotes Host Survival and Transmission. Cell 2017, 168:503–516 e512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luan HH, Wang A, Hilliard BK, Carvalho F, Rosen CE, Ahasic AM, Herzog EL, Kang I, Pisani MA, Yu S, et al. : GDF15 Is an Inflammation-Induced Central Mediator of Tissue Tolerance. Cell 2019, 178:1231–1244 e1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Massberg S, Schaerli P, Knezevic-Maramica I, Kollnberger M, Tubo N, Moseman EA, Huff IV, Junt T, Wagers AJ, Mazo IB, et al. : Immunosurveillance by hematopoietic progenitor cells trafficking through blood, lymph, and peripheral tissues. Cell 2007, 131:994–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang Y, Mao K, Chen X, Sun MA, Kawabe T, Li W, Usher N, Zhu J, Urban JF, Paul WE Jr., et al. : S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science 2018, 359:114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beura LK, Fares-Frederickson NJ, Steinert EM, Scott MC, Thompson EA, Fraser KA, Schenkel JM, Vezys V, Masopust D: CD4(+) resident memory T cells dominate immunosurveillance and orchestrate local recall responses. J Exp Med 2019, 216:1214–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klicznik MM, Morawski PA, Hollbacher B, Varkhande SR, Motley SJ, Kuri-Cervantes L, Goodwin E, Rosenblum MD, Long SA, Brachtl G, et al. : Human CD4(+)CD103(+) cutaneous resident memory T cells are found in the circulation of healthy individuals. Sci Immunol 2019, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dudakov JA, Hanash AM, van den Brink MR: Interleukin-22: immunobiology and pathology. Annu Rev Immunol 2015, 33:747–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wan X, Zinselmeyer BH, Zakharov PN, Vomund AN, Taniguchi R, Santambrogio L, Anderson MS, Lichti CF, Unanue ER: Pancreatic islets communicate with lymphoid tissues via exocytosis of insulin peptides. Nature 2018, 560:107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jang C, Hui S, Zeng X, Cowan AJ, Wang L, Chen L, Morscher RJ, Reyes J, Frezza C, Hwang HY, et al. : Metabolite Exchange between Mammalian Organs Quantified in Pigs. Cell Metab 2019, 30:594–606 e593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerlach C, Moseman EA, Loughhead SM, Alvarez D, Zwijnenburg AJ, Waanders L, Garg R, de la Torre JC, von Andrian UH: The Chemokine Receptor CX3CR1 Defines Three Antigen-Experienced CD8 T Cell Subsets with Distinct Roles in Immune Surveillance and Homeostasis. Immunity 2016, 45:1270–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mele M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, et al. : Human genomics. The human transcriptome across tissues and individuals. Science 2015, 348:660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baruch K, Deczkowska A, David E, Castellano JM, Miller O, Kertser A, Berkutzki T, Barnett-Itzhaki Z, Bezalel D, Wyss-Coray T, et al. : Aging. Aging-induced type I interferon response at the choroid plexus negatively affects brain function. Science 2014, 346:89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dobrin R, Zhu J, Molony C, Argman C, Parrish ML, Carlson S, Allan MF, Pomp D, Schadt EE: Multi-tissue coexpression networks reveal unexpected subnetworks associated with disease. Genome Biol 2009, 10:R55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang T, Zhang J, Xie L, Dong X, Zhang L, Cai YD, Li YX: Crosstissue coexpression network of aging. OMICS 2011, 15:665–671. [DOI] [PubMed] [Google Scholar]
- 55.Keller MP, Choi Y, Wang P, Davis DB, Rabaglia ME, Oler AT, Stapleton DS, Argmann C, Schueler KL, Edwards S, et al. : A gene expression network model of type 2 diabetes links cell cycle regulation in islets with diabetes susceptibility. Genome Res 2008, 18:706–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boekel J, Kallskog O, Ryden-Aulin M, Rhen M, Richter-Dahlfors A: Comparative tissue transcriptomics reveal prompt inter-organ communication in response to local bacterial kidney infection. BMC Genomics 2011, 12:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kozawa S, Ueda R, Urayama K, Sagawa F, Endo S, Shiizaki K, Kurosu H, Maria de Almeida G, Hasan SM, Nakazato K, et al. : The Body-wide Transcriptome Landscape of Disease Models. iScience 2018, 2:238–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang IM, Zhang B, Yang X, Zhu J, Stepaniants S, Zhang C, Meng Q, Peters M, He Y, Ni C, et al. : Systems analysis of eleven rodent disease models reveals an inflammatome signature and key drivers. Mol Syst Biol 2012, 8:594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Samdani P, Singhal M, Sinha N, Tripathi P, Sharma S, Tikoo K, Rao KV, Kumar D: A Comprehensive Inter-Tissue Crosstalk Analysis Underlying Progression and Control of Obesity and Diabetes. Sci Rep 2015, 5:12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McKay PF, Cizmeci D, Aldon Y, Maertzdorf J, Weiner J, Kaufmann SH, Lewis DJ, van den Berg RA, Del Giudice G, Shattock RJ: Identification of potential biomarkers of vaccine inflammation in mice. Elife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brandes M, Klauschen F, Kuchen S, Germain RN: A systems analysis identifies a feedforward inflammatory circuit leading to lethal influenza infection. Cell 2013, 154:197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tabula Muris C, Overall c, Logistical c, Organ c, processing, Library p, sequencing, Computational data a, Cell type a, Writing g, et al. : Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 2018, 562:367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, Saadatpour A, Zhou Z, Chen H, Ye F, et al. : Mapping the Mouse Cell Atlas by Microwell-Seq. Cell 2018, 172:1091–1107 e1017. [DOI] [PubMed] [Google Scholar]
- 64.Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell P, Carninci P, Clatworthy M, et al. : The Human Cell Atlas. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spitzer MH, Gherardini PF, Fragiadakis GK, Bhattacharya N, Yuan RT, Hotson AN, Finck R, Carmi Y, Zunder ER, Fantl WJ, et al. : IMMUNOLOGY. An interactive reference framework for modeling a dynamic immune system. Science 2015, 349:1259425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spitzer MH, Carmi Y, Reticker-Flynn NE, Kwek SS, Madhireddy D, Martins MM, Gherardini PF, Prestwood TR, Chabon J, Bendall SC, et al. : Systemic Immunity Is Required for Effective Cancer Immunotherapy. Cell 2017, 168:487–502 e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stuart T, Satija R: Integrative single-cell analysis. Nat Rev Genet 2019, 20:257–272. [DOI] [PubMed] [Google Scholar]
- 68.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X: RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 2015, 348:aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eng CL, Lawson M, Zhu Q, Dries R, Koulena N, Takei Y, Yun J, Cronin C, Karp C, Yuan GC, et al. : Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 2019, 568:235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Keren L, Bosse M, Marquez D, Angoshtari R, Jain S, Varma S, Yang SR, Kurian A, Van Valen D, West R, et al. : A Structured Tumor-Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging. Cell 2018, 174:1373–1387 e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, Evans K, Liu C, Ramakrishnan C, Liu J, et al. : Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 2018, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goltsev Y, Samusik N, Kennedy-Darling J, Bhate S, Hale M, Vazquez G, Black S, Nolan GP: Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell 2018, 174:968–981 e915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Ferrante TC, Terry R, Turczyk BM, Yang JL, Lee HS, Aach J, et al. : Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc 2015, 10:442–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stahl PL, Salmen F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, Giacomello S, Asp M, Westholm JO, Huss M, et al. : Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016, 353:78–82. [DOI] [PubMed] [Google Scholar]
- 75.Rodriques SG, Stickels RR, Goeva A, Martin CA, Murray E, Vanderburg CR, Welch J, Chen LM, Chen F, Macosko EZ: Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science 2019, 363:1463–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Avila Cobos F, Vandesompele J, Mestdagh P, De Preter K: Computational deconvolution of transcriptomics data from mixed cell populations. Bioinformatics 2018, 34:1969–1979. [DOI] [PubMed] [Google Scholar]
- 77.Titus AJ, Gallimore RM, Salas LA, Christensen BC: Cell-type deconvolution from DNA methylation: a review of recent applications. Hum Mol Genet 2017, 26:R216–R224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang PS, Boyken SE, Baker D: The coming of age of de novo protein design. Nature 2016, 537:320–327. [DOI] [PubMed] [Google Scholar]
- 79.Spangler JB, Moraga I, Mendoza JL, Garcia KC: Insights into cytokine-receptor interactions from cytokine engineering. Annu Rev Immunol 2015, 33:139–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ishihara J, Ishihara A, Sasaki K, Lee SS, Williford JM, Yasui M, Abe H, Potin L, Hosseinchi P, Fukunaga K, et al. : Targeted antibody and cytokine cancer immunotherapies through collagen affinity. Sci Transl Med 2019, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shim S, Belanger MC, Harris AR, Munson JM, Pompano RR: Two-way communication between ex vivo tissues on a microfluidic chip: application to tumor-lymph node interaction. Lab Chip 2019, 19:1013–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen WLK, Edington C, Suter E, Yu J, Velazquez JJ, Velazquez JG, Shockley M, Large EM, Venkataramanan R, Hughes DJ, et al. : Integrated gut/liver microphysiological systems elucidates inflammatory inter-tissue crosstalk. Biotechnol Bioeng 2017, 114:2648–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van Midwoud PM, Merema MT, Verpoorte E, Groothuis GM: A microfluidic approach for in vitro assessment of interorgan interactions in drug metabolism using intestinal and liver slices. Lab Chip 2010, 10:2778–2786. [DOI] [PubMed] [Google Scholar]
- 84.Seldin MM, Lusis AJ: Systems-based approaches for investigation of inter-tissue communication. J Lipid Res 2019, 60:450–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kveler K, Starosvetsky E, Ziv-Kenet A, Kalugny Y, Gorelik Y, Shalev-Malul G, Aizenbud-Reshef N, Dubovik T, Briller M, Campbell J, et al. : Immune-centric network of cytokines and cells in disease context identified by computational mining of PubMed. Nat Biotechnol 2018, 36:651–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wei W, Jiang D, Ehlerding EB, Luo Q, Cai W: Noninvasive PET Imaging of T cells. Trends Cancer 2018, 4:359–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Reinhardt RL, Jenkins MK: Whole-body analysis of T cell responses. Curr Opin Immunol 2003, 15:366–371. [DOI] [PubMed] [Google Scholar]
- 88.Southern PJ, Blount P, Oldstone MB: Analysis of persistent virus infections by in situ hybridization to whole-mouse sections. Nature 1984, 312:555–558. [DOI] [PubMed] [Google Scholar]
- 89.Susaki EA, Ueda HR: Whole-body and Whole-Organ Clearing and Imaging Techniques with Single-Cell Resolution: Toward Organism-Level Systems Biology in Mammals. Cell Chem Biol 2016, 23:137–157. [DOI] [PubMed] [Google Scholar]
- 90.Buchholz VR, Schumacher TN, Busch DH: T Cell Fate at the Single-Cell Level. Annu Rev Immunol 2016, 34:65–92. [DOI] [PubMed] [Google Scholar]
- 91.Schumacher TN, Gerlach C, van Heijst JW: Mapping the life histories of T cells. Nat Rev Immunol 2010, 10:621–631. [DOI] [PubMed] [Google Scholar]
- 92.Kester L, van Oudenaarden A: Single-Cell Transcriptomics Meets Lineage Tracing. Cell Stem Cell 2018, 23:166–179. [DOI] [PubMed] [Google Scholar]