Abstract
Head and neck squamous cell carcinoma (SCCa) is primarily managed by surgical resection. Recurrence rates after surgery can be as high as 55% if residual cancer is present. In this study, hyperspectral imaging (HSI) is evaluated for detection of SCCa in ex-vivo surgical specimens. Several methods are investigated, including convolutional neural networks (CNNs) and a spectral-spatial variant of support vector machines. Quantitative results demonstrate that additional processing and unsupervised filtering can improve CNN results to achieve optimal performance. Classifying regions that include specular glare, the average AUC is increased from 0.73 [0.71, 0.75 (95% confidence interval)] to 0.81 [0.80, 0.83] through an unsupervised filtering and majority voting method described. The wavelengths of light used in HSI can penetrate different depths into biological tissue, while the cancer margin may change with depth and create uncertainty in the ground-truth. Through serial histological sectioning, the variance in cancer-margin with depth is also investigated and paired with qualitative classification heat maps using the methods proposed for the testing group SCC patients.
Keywords: Hyperspectral imaging, convolutional neural network, deep learning, optical biopsy, intraoperative imaging, head and neck surgery, head and neck cancer
1. INTRODUCTION
Head and neck cancer is the 6th most common cancer world-wide, with majority of cancers of the oral cavity and oropharynx being squamous cell carcinoma (SCC).1 Approximately two-thirds of SCC patients present with advanced disease, either stage III or IV.2 Surgical resection is the primary management SCC of the aerodigestive tract, potentially with concurrent chemo-radiation therapy depending on the extent of the disease.3
A modern advance in robotic oncologic surgical techniques of the head and neck, trans-oral robotic surgery (TORS), has been developed using the da Vinci robotic system (Intuitive Surgical, Sunnyvale, CA) and demonstrated use in resection of orophangyeal and laryngeal SCC with promising results on functional preservation.4 The da Vinci system relies on conventional optical imaging using a high resolution monitor, but studies have aimed to expand the optical imaging capabilities utilized. Fluorescence guided robotic surgeries have found a wide range of uses in gastroenterology, urology, and gynecology.5 For use in TORS, a fluorescent nuclear contrast agent can be visualized with high-resolution micro-endoscopic imaging for oropharyngeal SCC with technical feasibility in three patient case reports.6 Several label-free, optical imaging approaches have been presented for TORS.7–9 Label-free fluorescence lifetime imaging has been integrated with TORS for 25 patients with head and neck cancer.7 Additionally, early patient reports show that optical narrow-band imaging has shown success in promoting negative surgical margins in TORS for SCC resection.8,9
The local recurrence rates for SCC cases depend on the successful removal of the cancer. For surgeries with negative cancer margins, the local recurrence rate is 12–18%, compared to local recurrence rates up to 30–55% if positive cancer margins are determined.10–12 Moreover, positive cancer margins have a greatly reduced disease-free survival, with estimates ranging from 7 to 52%, compared to disease-free survival rates of 39–73% for negative margins.13,14 Disease recurrence greatly affects likelihood for additional surgeries, reduced quality of life, complications from surgery, and increased mortality rates.15
Hyperspectral imaging (HSI) is a non-contact optical imaging modality capable of acquiring a single image of potentially hundreds of discrete wavelengths. Preliminary research demonstrates that HSI has potential for providing diagnostic information for various diseases.16 Machine learning methods, including support vector machine (SVM) and convolutional neural networks (CNNs), the latter being an implementation of artificial intelligence, have demonstrated near human-level ability for image classification tasks.17,18 Preliminary studies from our group show that HSI combined with machine learning may yield diagnostic information with potential applications for surgical use.19–22 HSI uses wavelengths of light that can penetrate different depths into biological tissue, so it is possible that the cancer margin may change with depth and create uncertainty in ground truth.
This study aims to investigate the ability of HSI to detect SCC in surgical specimens from the upper aerodigestive tract using several distinct machine learning pipelines. Additionally, another objective of this work is to investigate the limiting factors of HSI-based SCC detection, including specular glare, noise and blur, and uncertainty in the ground truth due to changes in superficial cancer margin with depth, all of which must be thoroughly explored to understand the potential of HSI in the operating room. This work expands upon previous cross-validation experiments with CNN-only methods to include multiple machine learning pipelines involving CNNs and other state-of-the-art methods. Additionally, the proposed methods are tested on five HSI from five SCC patients, and the accuracy of the corresponding ground truths from these five tissues are investigated by serial histological sectioning.
2. METHODS
2.1. Experimental Design
In collaboration with the Otolaryngology Department and the Department of Pathology and Laboratory Medicine at Emory University Hospital Midtown, 203 head and neck cancer patients undergoing surgical cancer resection were recruited for our hyperspectral imaging studies. In our previous study, we have evaluated the efficacy of using HSI for optical biopsy.19,23 Excised tissue samples of head and neck squamous cell carcinoma (HNSCC) and normal tissue were collected from the upper aerodigestive tract sites, including tongue, larynx, pharynx, and mandible. Three tissue samples were collected from each patient, i.e., a sample of the tumor, a normal tissue sample, and a sample at the tumor-normal interface and were scanned with a hyperspectral imaging system.22,24 In this study, we selected 12 patients for this analysis, which were divided into two groups. The first 7 patients were used for evaluating quantitative results, expanding from previously published results.21 Five additional patients were classified using models trained from the first 7 patients, and these 5 patients results are presented qualitatively and also investigated is the variation in the cancer margin with tissue depth.
2.2. Hyperspectral Imaging
Hyperspectral images were acquired of ex-vivo surgical specimens using a previously described CRI Maestro imaging system (Perkin Elmer Inc., Waltham, Massachusetts), which capture images that are 1040 by 1,392 pixels and a spatial resolution of 25 μm per pixel.19,24–26 Each hypercube contains 91 spectral bands, ranging from 450 to 900 nm with a 5 nm spectral sampling interval. The hyperspectral data were normalized at each wavelength sampled for all pixels by subtracting the inherent dark current and dividing by a white reference disk, according to the following equation.23,26
| (1) |
2.3. Histological Imaging
To obtain a ground truth labelling for the tissue specimens imaged with HSI, tissues were inked after imaging to preserve orientation, fixed in formalin, paraffin embedded, sectioned from the top of the imaging surface, haemotoxylin and eosin (H&E) stained, and digitized. The ex-vivo tissue sections were reviewed by a board certified pathologist with expertise in H&N pathology and the cancer margins were outlined using digitized histology in Aperio ImageScope (Leica Biosystems Inc, Buffalo Grove, IL, USA).
2.4. Histological Ground-Truths
Binary masks are constructed from histological images and matched to the HSI in two methods. The first method is to investigate only ideal quality pixels, which is constructed by avoiding HSI regions with a large amount of specular glare. The second method is to investigate the degrading effect of specular glare on classification. For the ground-truth mask avoiding glare, only regions that contain sufficient area to extract 25×25 spatial patches without any specular glare are included. For the ground-truth mask including glare regions, the entire tissue is included for patch-making, but the top 1% of glare pixels are identified by fitting a gamma distribution to pixel intensities and no patch centered on a glare pixel is included. Figure 1 shows an example of the two masks generated from two different patients. Both masks will be used for evaluating quantitative testing results and are referred to as ‘ground-truth mask avoiding glare’, and ‘ground-truth mask including glare’, respectively.
Figure 1:
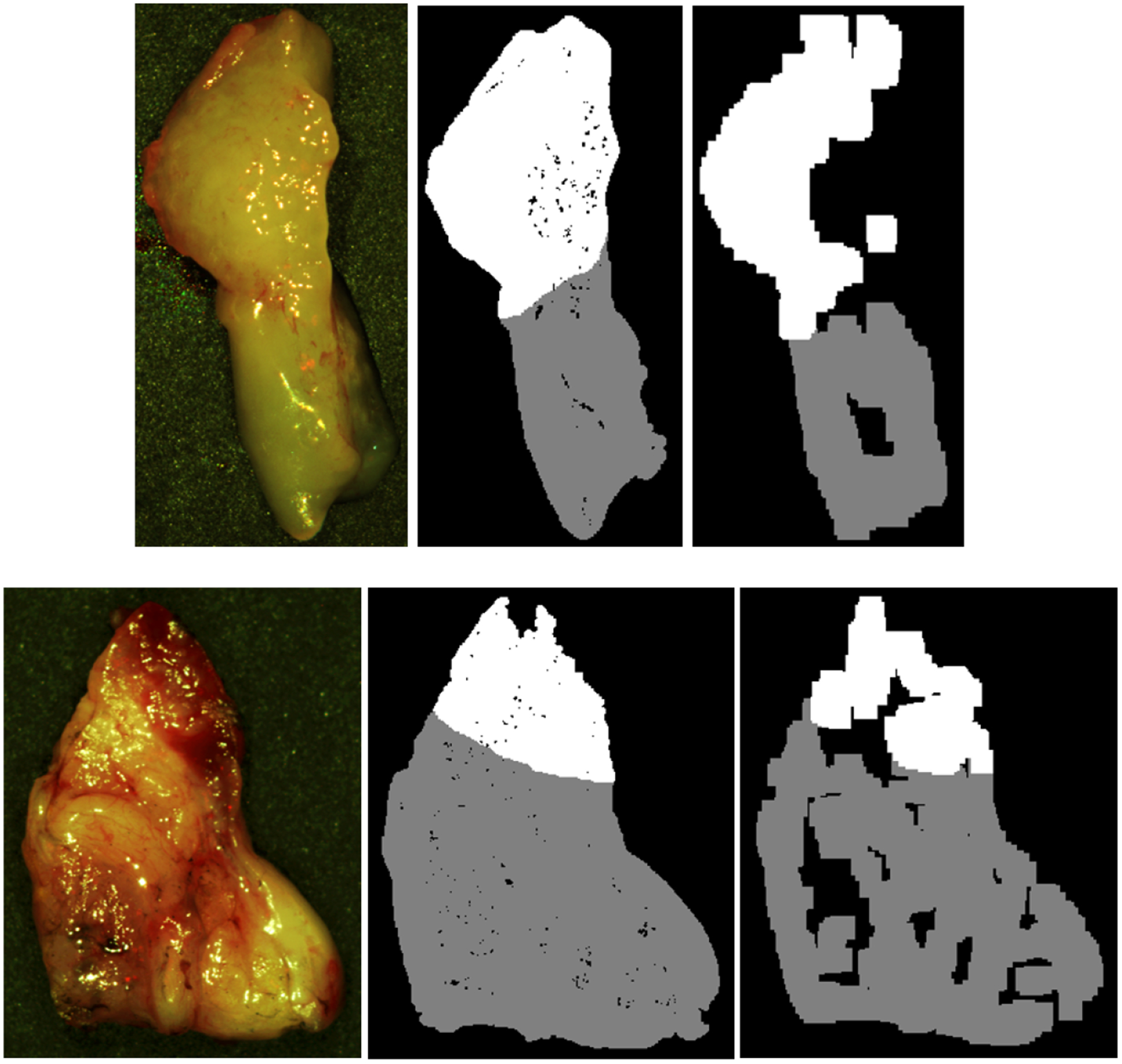
Two representative tissue specimens from different patients. Left to right: HSI-RGB composite image; binary ground-truth mask including glare regions, generated by only removing patches centered on specular glare; binary ground-truth mask avoiding glare regions, generated by sufficient area to extract 25×25 patches and avoiding specular glare. SCC in shown in white, and normal tissue shown in grey.
2.5. Effect of Sectioning Depth on Cancer Margin Ground-Truth
HSI uses wavelengths of light that can penetrate different depths into biological tissue. It possible that the cancer margin may change with depth and create uncertainty in the classification results because of the variable penetration of the HSI spectrum. To investigate this, additional histological sectioning was performed on 5 tumor-margin tissues (see Figure 2). First, the thickness of the remaining paraffin embedded tissue was estimated. Next, from the surface already sectioned, which is the top of the tissue that was optically imaged, five more sections were obtained, up to 300 microns additionally sectioned into the tissue. Therefore, the depth of sectioning was performed down to approximately 0.3 to 0.5 mm from the HS imaged surface.
Figure 2:
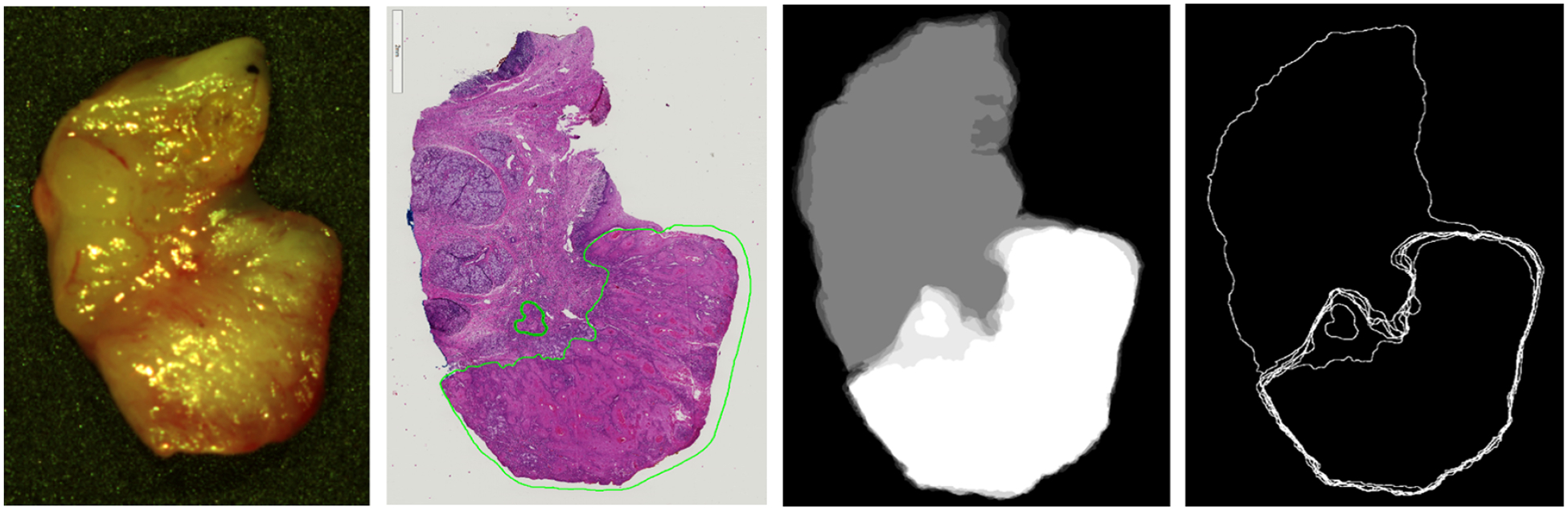
Representative tissue specimen that underwent serial histological sectioning to evaluate superificial cancer margin variation with tissue depth. Left to right: HSI-RGB image, first histological slice, binary mask overlay of the 6 histological images (white is SCC and grey is normal), topographical map of the variation with depth of the cancer margin.
2.6. Machine Learning Techniques
2.6.1. Data Pre-processing
A pre-processing chain was applied to the data mainly to reduce the noise in the spectral signatures (Figure 3). The machine learning methods detailed below were tested with and without the following pre-processing steps. The proposed pre-processing chain is based on 4 steps: image calibration, operating bandwidth selection, noise filtering, and data normalization. In the first step, the previously described method to normalize the data using white and dark references is performed. Next, the spectral bands are truncated between 490–790 nm, so the final hypercube contains only 61 bands, which will be used for classification. Spectral bands outside this range were noisy because they were too close to the limits of the HSI camera. In the third step, a smoothing filter is applied to the data in order to reduce the spectral noise. Finally, each pixels spectral signature is normalized between 0 and 1.
Figure 3:

Block diagram of the proposed pre-processing chain.
2.6.2. Convolutional Neural Network
A previously described and trained convolutional neural network was used to classify oral cavity tissues implemented using TensorFlow on a NVIDIA Titan-XP GPU.17,18,21,27 In summary, an Inception CNN was designed and trained using the leave-one-patient-out cross-validation. Validation was performed on all seven held-out patients to obtain the results previously reported and shown again in Table 1.21 The 95% confidence intervals were created using a bootstrapping method by sampling 1000 pixels from each class with replacement from each patient and calculating the AUC; the method was performed 1000 times for each patient and the 2.5 and 97.5 percentiles were reported. The saved, trained models from the patients from the previous work were used to classify the 5 testing group HSI patients that underwent serial histological sectioning for surgical margin variation with depth evaluation. The probabilities of all models were averaged per patient to obtain qualitative probability heat-maps, scaled from 0 to 1, where 0 represents normal class and 1 represents high probability that the tissue belongs in the cancer class.
Table 1:
Results of inter-patient cross-validation of SCC versus normal, obtained using the leave-one-patient-out method. Average AUCs reported with bootstrapped 95% confidence interval.
| Ground Truth | Classifier | Avg. AUC [95% Confidence Interval] |
|---|---|---|
| Avoiding Glare | spatial-SVM | 0.71 [0.68, 0.74] |
| CNN [21] | 0.86 [0.82, 0.89] | |
| spatial-SVM Processed | 0.82 [0.80, 0.84] | |
| CNN Processed | 0.84 [0.81, 0.86] | |
| HELICoiD | 0.82 [0.79, 0.84] | |
| CNN+HELICoiD | 0.82 [0.79, 0.85] | |
| Including Glare | spatial-SVM | 0.69 [0.67, 0.71] |
| CNN [21] | 0.73 [0.71, 0.76] | |
| spatial-SVM Processed | 0.76 [0.74, 0.77] | |
| CNN Processed | 0.78 [0.76, 0.81] | |
| HELICoiD | 0.79 [0.77, 0.81] | |
| CNN+HELICoiD | 0.81 [0.80, 0.83] |
2.6.3. HELICoiD Algorithm
The results of the CNN classification method and the generated probability maps were compared to the results obtained by a machine learning pipeline previously developed for intraoperative detection of brain cancer using HSI.28–32 In summary, a spatial-spectral classification algorithm, here referred to as HELICoiD (Figure 4a), was implemented using a classification map obtained by a support vector machine (SVM) classifier that is spatially homogenized by employing a combination of a one spectral band principal component analysis (PCA) decomposition through a K-nearest Neighbors (KNN) filter (Figure 4a, part A). After that, the result of the KNN filtering is merged with an unsupervised segmentation map generated by a hierarchical K-means (HKM) algorithm through a majority voting (MV) method.33 The result of this algorithm is a classification map that includes both the spatial and spectral features of the HS images. In addition, for this application, the KNN filtering is applied again to the MV probabilities and the PCA one-band-representation to homogenize the results (Figure 4a, part B). The quantitative classification results of 7 patients are reported in Table 1, obtained by using leave-one-out cross-validation. In addition, similar to the CNN method, the saved, trained models from these patients were used to classify 5 tissue specimens that were imaged with HSI and underwent serial histological sectioning for margin depth evaluation. The probabilities of all models were averaged per patient to obtain qualitative probability heat-maps, scaled from 0 to 1, where 0 represents normal class and 1 represents high probability that the tissue belongs in the cancer class. Furthermore, a pipeline that combines the CNN architecture with the HELICoiD algorithm was proposed. In this case, the spatial-spectral stage of HELICoiD (PCA+SVM+KNN) is replaced by the CNN architecture (Figure 4b).
Figure 4:
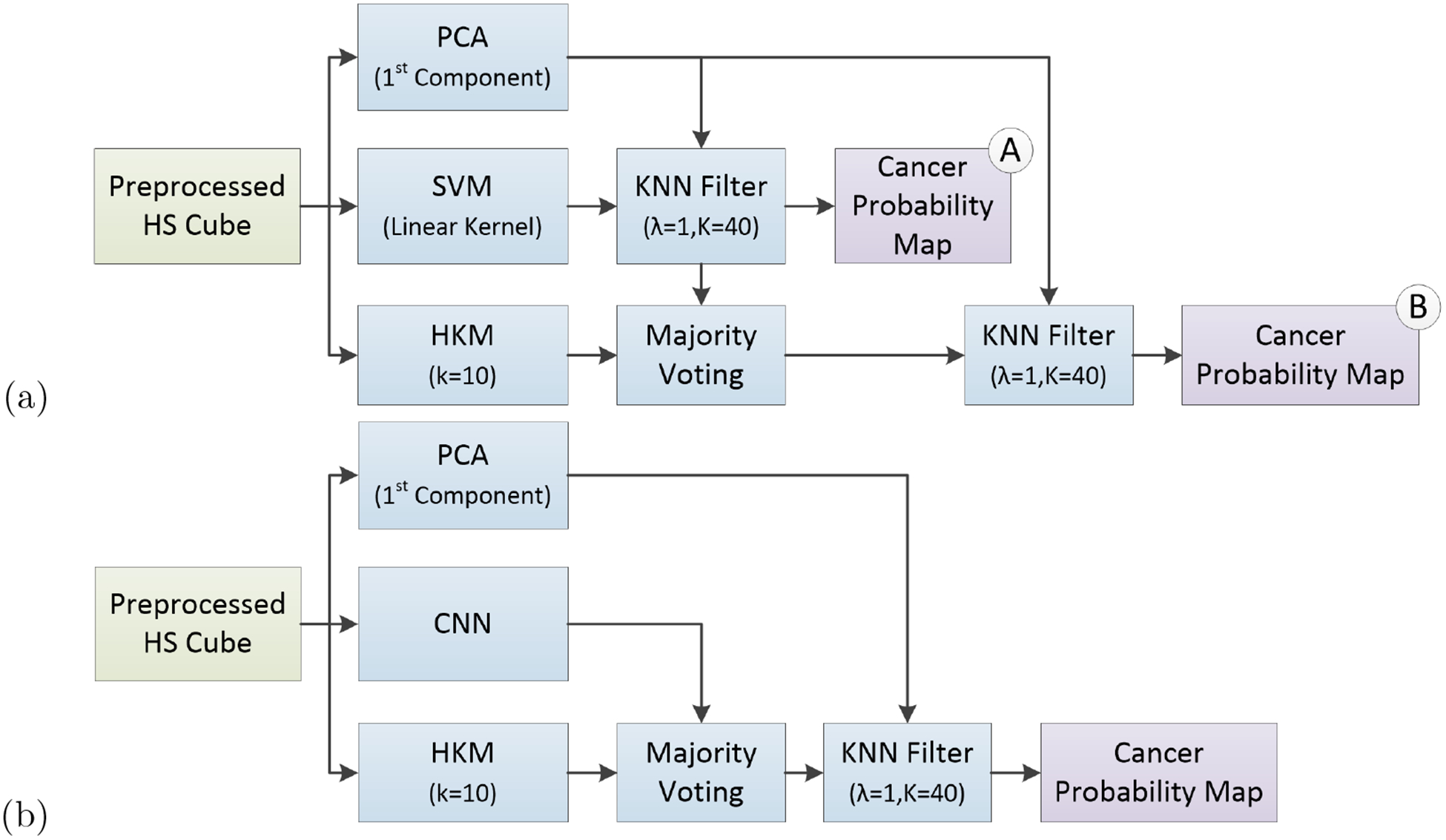
Block diagrams of the proposed classification frameworks. (a) HELICoiD algorithm with the additional KNN filter. (b) Pipeline of the mixed algorithm.
3. RESULTS
Quantitative results from the leave-one-patient-out cross-validation, using both the ground truth regions that include glare pixels and the sub-sampled masks that include only ideal quality regions that exclude glare, show that the CNN classifier group outperformed the SVM classifier group using the average area under the curve (AUC) of the receiver operator characteristic (ROC), as shown in Table 1 and Figure 5. The results are reported using seven-fold cross-validation to validate on all 7 patients.
Figure 5:
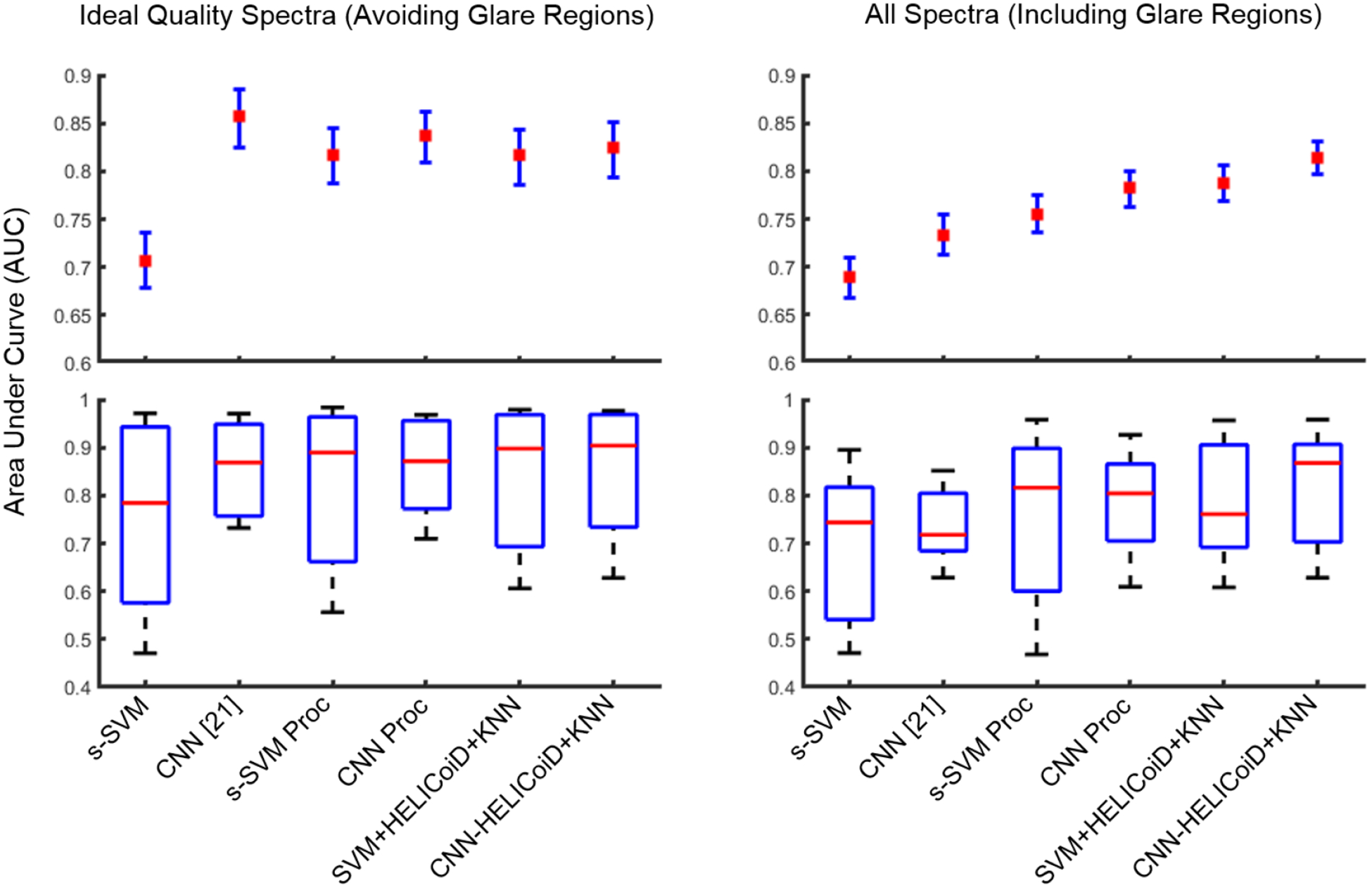
Results of inter-patient cross-validation of SCC versus normal, obtained using the leave-one-patient-out method. Top: average AUCs reported with 95% confidence interval. Bottom: box plots with the range in black, 75th and 25th percentile in blue, and median in red.
When classification is performed of only ideal quality pixels (obtained from the sub-sampled mask, see Figure 1), the results indicate that additional preprocessing of the spectral signature and addition of the HELICoiD method and KNN filtering do not significantly improve the results compared to only using the CNN, see Figure 5. The average AUCs for the CNN groups are 0.86, 0.84, and 0.82 for the CNN, CNN with preprocessed input data, and CNN+HELICoiD method. The average AUCs for the traditional ML groups are 0.71, 0.82, and 0.82 for the SVM+PCA+KNN without and with preprocessed input data, and HELICoiD method, respectively. The 95% confidence intervals overlap for all groups except for basic spatial SVM, as shown in Table 1. Additionally, all methods have a similar interquartile range and median distribution, see Figure 5.
However, when the classification is performed over the entire HSI tissue area, which includes classification of pixels that contain more noise and variability, the CNN+HELICoiD method classifies with an average AUC of 0.81, which outperforms other methods tested. The average AUC values range from 0.69 to 0.79 for the traditional ML groups, and the CNN alone with or without pre-processing has an average AUC of 0.78 and 0.73, respectively, see Table 1 for complete results with 95% confidence intervals. Moreover, this second scenario corresponds to a more realistic application of HSI. In summary, from cross-validation experiments including specular glare in the ground-truth mask, the best classification method was using the CNN with pre-processed input as the probabilty heat maps for the HELICoiD+KNN filtering method, and using SVM+PCA+KNN heat map instead of the CNN-generated heatmap yielded slightly lower results.
For generalization and application, HSI from five patients with SCC comprised the hold-out testing group and were classified using the saved models that were trained and cross-validated using the 7 patient training/validation group. Qualitative investigation of the five, testing patients was performed, classified with the both the CNN using pre-processed input alone and the method of additional HELICoiD+KNN filtering of the pre-processed CNN-generated probability maps. As shown in Figure 6, the CNN+HELICoiD+KNN technique performs better on the 5 testing patients, in agreement with the quantitative metrics from the cross-validation group. The SCC probability heat maps are shown with binary masks depicting the uncertainty and variation in the superficial cancer margin with depth. Additionally, the extremes in the superficial margin are overlaid on the cancer heat maps. Depending on the tissue, as demonstrated, the margin changes by about 1 mm depending on sectioning depth. Therefore, our qualitative results can be interpreted within the range of uncertainty of the ground truth to provide more insight to the classification potential of machine learning methods using HSI for cancer detection.
Figure 6:
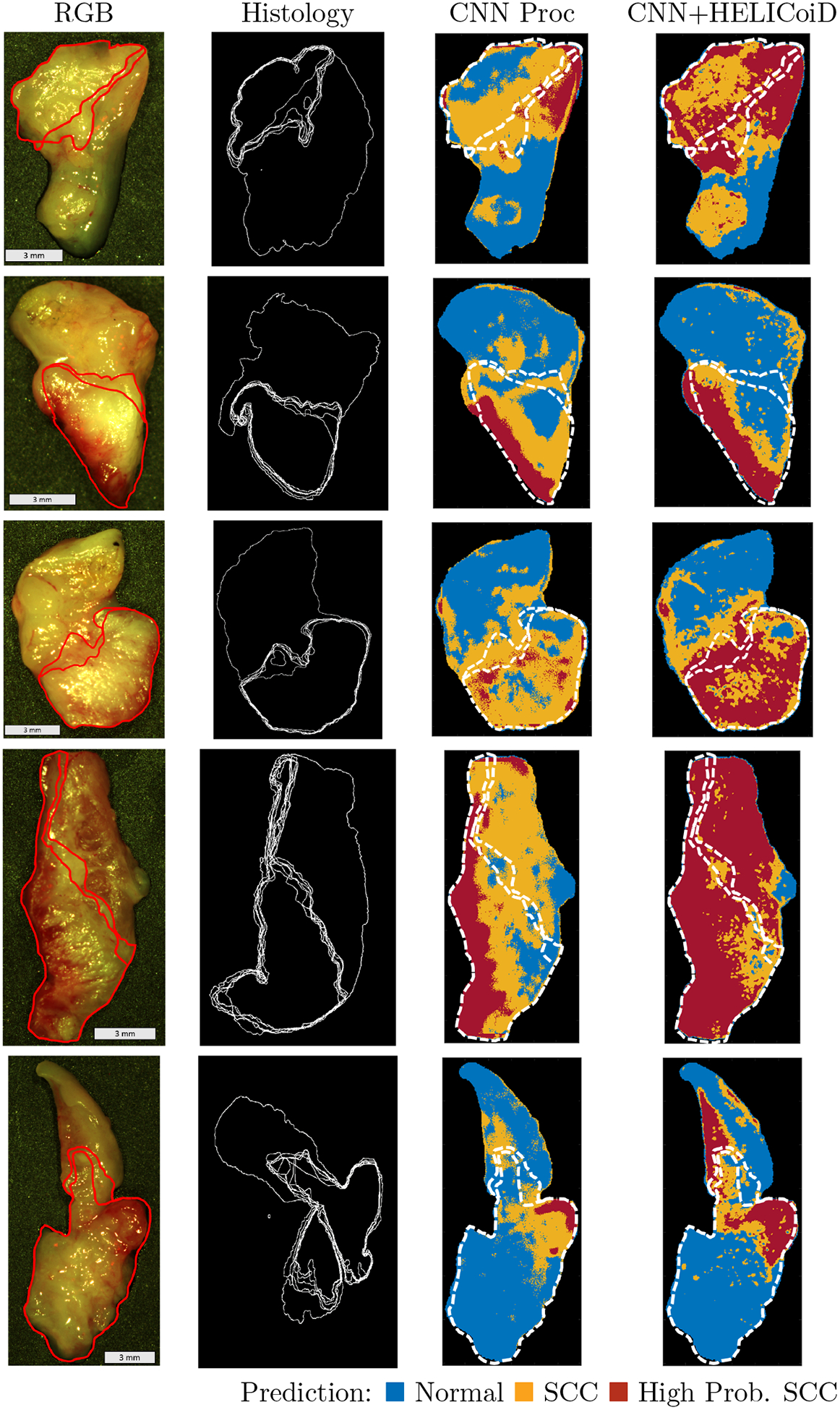
Representative results of binary cancer classification of the 5 testing patients. From left to right: HSI-RGB composite; histological ground truth showing variation in cancer margins with cancer area outlined; heat maps for cancer probability generated in two ways: 1. the CNN with pre-processed input alone, and 2. the method of additional HELICoiD+KNN filtering of the pre-processed CNN-generated probability maps. The extremes in the superficial cancer margin are overlaid on to the heat maps.
4. CONCLUSION
In this work, we presented and quantified the combination of two state-of-the-art machine learning-based classification methods for HSI of ex-vivo head and neck SCC surgical specimens. In summary, there are two methods for generating SCC prediction probability maps: the first uses a CNN, and the second uses a combination of SVM+PCA+KNN. After generation of the predicted cancer probabilities on a pixel level, the probability map is fed into a KNN filtering layer and majority voting determines the class belonging of the region of interest. Therefore, two distinct methods are compared using a 7-fold cross-validation group to obtain quantitative evaluation metrics. Additionally, the methods are tested on a group of HSI obtained from 5 SCC patients that underwent serial histological sectioning to evaluate the variation in the cancer margin with penetration depth of the light wavelengths.
The quantitative results of this paper suggest that when working with ideal quality pixels, such as the spectral signatures generated from flat surface tissue surfaces with no glare, the CNN techniques and traditional, regression-based spatial-spectral machine learning algorithms will perform with no significant difference. The average AUCs for these methods using the pre-processed input data range from 0.82 to 0.86 with overlapping confidence intervals. However, when the pixels classified contain noise, for example due to sloping of the tissue edges or specular glare from completely reflected incident light, additional spectral smoothing and additional HELICoiD+KNN filtering of the classifier improve classification results of the CNN. The top performing method was CNN+HELICoiD+KNN with an average AUC of 0.81. These methods tested outperform the traditional spectral-spatial machine learning methods employed in this study on the 7 patient validation group. Therefore, the 5 patient testing group was classified with the HELICoiD+KNN method using the CNN-generated probability map and compared to the results of the CNN-only method using pre-processed input data.
In order to test the general application of the proposed methods, HSI from five testing group patients with SCC were classified using the models that were trained and cross-validated using the five patient group. To qualitatively investigate these results, histological images of the 5 ex-vivo tissue specimens were obtained down to about 0.3 mm to determine how the superficial cancer margin may change with depth. As shown in Figure 6, the CNN+HELICoiD+KNN technique performs best on the 5 patient SCC testing group, which was the same result obtained from the 7-fold quantitative cross-validation experiment. Additionally, it can also be seen that the superficial cancer margin of the ex-vivo tissue specimens changes by about 1 millimeter. These results allow interpretation of the cancer prediction probability maps with observed variation in the ground truth. However, additional possible uncertainty may exist in the histological ground truth, for example if the angle of sectioning is skewed from the tissue plane then the ground truth could be warped which could lead to errors that cannot be corrected by deformable registration.
In conclusion, this investigation provides more information on the potential of hyperspectral imaging and machine learning for the detection of head and neck cancer. The main objective of this study was to investigate the limitations of HSI-based SCC detection, such as specular glare, noise, blurring, and tissue-edge sloping artifacts. Additionally, another objective was to evaluate the general efficacy on example test cases with uncertainty in the ground truth as the superficial cancer margin varies with penetration depth. All of the above factors are necessary to explore to understand the potential of HSI in the operating room. The proposed deep learning and machine learning methods employed to study these objectives require sufficiently large patient datasets for training, validation, and testing. Therefore, the preliminary results of this study encourage inclusion of more data and further exploration into the ability of HSI for cancer detection.
ACKNOWLEDGMENTS
This research was supported in part by the U.S. National Institutes of Health (NIH) grants (R21CA176684, R01CA156775, R01CA204254, and R01HL140325). The authors would like to thank the surgical pathology team at Emory University Hospital Midtown for their help in collecting fresh tissue specimens.
Footnotes
DISCLOSURES
The authors have no relevant financial interests in this article and no potential conflicts of interest to disclose. Informed consent was obtained from all patients in accordance with Emory Institutional Review Board policies under the Head and Neck Satellite Tissue Bank protocol.
REFERENCES
- [1].Joseph LJ, Goodman M, Higgins K, Pilai R, Ramalingam SS, Magliocca K, Patel MR, El-Deiry M, Wadsworth JT, Owonikoko TK, Beitler JJ, Khuri FR, Shin DM, and Saba NF, “Racial disparities in squamous cell carcinoma of the oral tongue among women: A SEER data analysis,” Oral Oncology 51(6), 586–592 (2015). [DOI] [PubMed] [Google Scholar]
- [2].Yao M, Epstein JB, Modi BJ, Pytynia KB, Mundt AJ, and Feldman LE, “Current surgical treatment of squamous cell carcinoma of the head and neck,” Oral Oncology 43(3), 213–223 (2007). [DOI] [PubMed] [Google Scholar]
- [3].Kim BY, Choi JE, Lee E, Son YI, Baek CH, Kim SW, and Chung MK, “Prognostic factors for recurrence of locally advanced differentiated thyroid cancer,” Journal of Surgical Oncology (2017). [DOI] [PubMed] [Google Scholar]
- [4].Weinstein GS, O’Malley BW, Magnuson JS, Carroll WR, Olsen KD, Daio L, Moore EJ, and Holsinger FC, “Transoral robotic surgery: A multicenter study to assess feasibility, safety, and surgical margins,” The Laryngoscope 122(8), 1701–1707 (2012). [DOI] [PubMed] [Google Scholar]
- [5].Landau MJ, Gould DJ, and Patel KM, “Advances in fluorescent-image guided surgery,” Annals of Translational Medicine 4(20) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Patsias A, Giraldez-Rodriguez L, Polydorides AD, Richards-Kortum R, Anandasabapathy S, Quang T, Sikora AG, and Miles B, “Feasibility of transoral robotic-assisted high-resolution microendoscopic imaging of oropharyngeal squamous cell carcinoma,” Head & Neck 37(8), 99–102 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Phipps JE, Unger J, Gandour-Edwards R, Moore MG, Beweley A, Farwell G, and Marcu L, “Head and neck cancer evaluation via transoral robotic surgery with augmented fluorescence lifetime imaging,” in [Biophotonics Congress: Biomedical Optics Congress 2018 (Microscopy/Translational/Brain/OTS)], Optical Society of America; (2018). [Google Scholar]
- [8].Vicini C, Montevecchi F, D’agostino G, De Vito A, and Meccariello G, “A novel approach emphasising intra-operative superficial margin enhancement of head-neck tumours with narrow-band imaging in transoral robotic surgery,” Acta Otorhinolaryngol Ital. 35(3) (2015). [PMC free article] [PubMed] [Google Scholar]
- [9].Tateya I, Ishikawa S, Morita S, Ito H, Sakamoto T, Murayama T, Kishimoto Y, Hayashi T, Funakoshi M, Hirano S, Kitamura M, Morita M, Muto M, and Ito1 J, “Magnifying endoscopy with narrow band imaging to determine the extent of resection in transoral robotic surgery of oropharyngeal cancer.,” Case Reports in Otolaryngology. 2014(604737) (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chen TY, Emrich LJ, and Driscoll DL, “The clinical significance of pathological findings in surgically resected margins of the primary tumor in head and neck carcinoma,” International Journal of Radiation Oncology. 13(6) (1987). [DOI] [PubMed] [Google Scholar]
- [11].Loree T and Strong E, “Significance of positive margins in oral cavity squamous carcinoma,” American Journal of Surgery 160(4) (1990). [DOI] [PubMed] [Google Scholar]
- [12].D.N Sutton DN, Brown JS, Rogers SN, Vaughan ED, and Woolgar JA, “The prognostic implications of the surgical margin in oral squamous cell carcinoma,” International Journal of Oral and Maxillofacial Surgery. 32(1) (2003). [DOI] [PubMed] [Google Scholar]
- [13].Amaral TMP, da Silva Freire AR, Carvalho AL, Pinto CL, and Kowalski LP, “Predictive factors of occult metastasis and prognosis of clinical stages i and ii squamous cell carcinoma of the tongue and floor of the mouth,” Oral Oncology 40(8), 780–786 (2004). [DOI] [PubMed] [Google Scholar]
- [14].Garzino-Demo P, Acqua AD, Dalmasso P, Fasolis M, LaTerra-Maggiore GM, Ramieri G, Berrone S, Rampino M, and Schena M, “Clinicopathological parameters and outcome of 245 patients operated for oral squamous cell carcinoma,” Journal of Cranio-Maxillofacial Surgery 34(6), 344–350 (2006). [DOI] [PubMed] [Google Scholar]
- [15].Rathod S, Livergant J, Klein J, Witterick I, and Ringash J, “A systematic review of quality of life in head and neck cancer treated with surgery with or without adjuvant treatment,” Oral Oncology 51(10), 888–900 (2015). [DOI] [PubMed] [Google Scholar]
- [16].Lu G and Fei B, “Medical hyperspectral imaging: a review,” Journal of Biomedical Optics 19(1) (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Krizhevsky A, Sutskever I, and Hinton GE, “Imagenet classification with deep convolutional neural networks,” Proceedings of the 25th International Conference on Neural Information Processing Systems 1, 1097–1105 (2012). [Google Scholar]
- [18].Szegedy C, Liu W, Jia Y, Sermanet P, Reed SE, Anguelov D, Erhan D, Vanhoucke V, and Rabinovich A, “Going deeper with convolutions,” 2015 IEEE Conference on Computer Vision and Pattern Recognition (2015). [Google Scholar]
- [19].Fei B, Lu G, Halicek MT, Wang X, Zhang H, Little JV, Magliocca KR, Patel M, Griffith CC, El-Deiry MW, and Chen AY, “Label-free hyperspectral imaging and quantification methods for surgical margin assessment of tissue specimens of cancer patients,” in [2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC)], 4041–4045 (July 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Halicek M, Little JV, Wang X, Patel M, Griffith CC, Chen AY, and Fei B, “Optical biopsy of head and neck cancer using hyperspectral imaging and convolutional neural networks,” SPIE Proceedings, Photonics West BiOS 2018: Optical Imaging, Therapeutics, and Advanced Technology in Head and Neck Surgery and Otolaryngology 10469, 104690X (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Halicek M, Little JV, Wang X, Patel M, Griffith CC, Chen AY, and Fei B, “Tumor margin classification of head and neck cancer using hyperspectral imaging and convolutional neural networks,” SPIE Proceedings, Medical Imaging 2018: Image-Guided Procedures, Robotic Interventions, and Modeling 10576, 1057605 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lu G, Little JV, Wang X, Zhang H, Patel M, Griffith CC, El-Deiry M, Chen AY, and Fei B, “Detection of head and neck cancer in surgical specimens using quantitative hyperspectral imaging,” Clinical Cancer Research (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fei B, Lu G, Wang X, Zhang H, Little JV, Patel MR, Griffith CC, El-Diery MW, and Chen AY, “Label-free reflectance hyperspectral imaging for tumor margin assessment: a pilot study on surgical specimens of cancer patients,” Journal of Biomedical Optics 22(8) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Halicek M, Lu G, Little JV, Wang X, Patel M, Griffith CC, Chen AY, and Fei B, “Deep convolutional neural networks for classifying head and neck cancer using hyperspectral imaging,” Journal of Biomedical Optics 22(6) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lu G, Wang D, Qin X, Muller S, Wang X, Chen AY, Chen ZG, and Fei B, “Detection and delineation of squamous neoplasia with hyperspectral imaging in a mouse model of tongue carcinogenesis,” Journal of Biophotonics (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lu G, Wang D, Qin X, Halig L, Muller S, Zhang H, Chen A, Pogue BW, Chen Z, and Fei B, “Framework for hyperspectral image processing and quantification for cancer detection during animal tumor surgery,” Journal of Biomedical Optics 20(12) (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS, Davis A, Dean J, Devin M, Ghemawat S, Goodfellow I, Harp A, Irving G, Isard M, Jia Y, Jozefowicz R, Kaiser L, Kudlur M, Levenberg J, Mané D, Monga R, Moore S, Murray D, Olah C, Schuster M, Shlens J, Steiner B, Sutskever I, Talwar K, Tucker P, Vanhoucke V, Vasudevan V, Viégas F, Vinyals O, Warden P, Wattenberg M, Wicke M, Yu Y, and Zheng X, “TensorFlow: Large-scale machine learning on heterogeneous systems,” (2015). Software available from tensorflow.org. [Google Scholar]
- [28].Fabelo H, Ortega S, Lazcano R, Madroal D, Callico G, Juarez E, Salvador R, Bulters D, Bulstrode H, Szolna A, Piaeiro JF, Sosa C,A, O’Shanahan J, Bisshopp S, Hernandez M, Morera J, Ravi D, Kiran BR, Vega A, Bez-Quevedo A, Yang G, Stanciulescu B, and Sarmiento R, “An intraoperative visualization system using hyperspectral imaging to aid in brain tumor delineation,” Sensors 18(2) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Florimbi G, Fabelo H, Torti E, Lazcano R, Madroal D, Ortega S, Salvador R, Leporati F, Danese G, Baez-Quevedo A, Callico GM, Juarez E, Sanz C, and Sarmiento R, “Accelerating the k-nearest neighbors filtering algorithm to optimize the real-time classification of human brain tumor in hyperspectral images,” Sensors 18(7) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ravi D, Fabelo H, Callico GM, and Yang G, “Manifold embedding and semantic segmentation for intraoperative guidance with hyperspectral brain imaging,” IEEE Trans on Medical Imaging 36, 1845–1857 (September 2017). [DOI] [PubMed] [Google Scholar]
- [31].Lazcano R, Madronal D, Salvador R, Desnos K, Pelcat M, Guerra R, Fabelo H, Ortega S, Lopez S, Callico G, Juarez E, and Sanz C, “Porting a PCA-based hyperspectral image dimensionality reduction algorithm for brain cancer detection on a manycore architecture,” Journal of Systems Architecture 77 (2017). [Google Scholar]
- [32].Madroal D, Lazcano R, Salvador R, Fabelo H, Ortega S, Callico G, Juarez E, and Sanz C, “SVM-based real-time hyperspectral image classifier on a manycore architecture,” J Syst Archit 80(C), 30–40 (2017). [Google Scholar]
- [33].Fabelo H, Ortega S, Ravi D, Kiran BR, Sosa C, Bulters D, Callico GM, Bulstrode H, Szolna A, Pineiro JF, Kabwama S, Madronal D, Lazcano R, O’Shanahan A, Bisshopp S, Hernandez M, Baez A, Yang G-Z, Stanciulescu B, Salvador R, Juarez E, and Sarmiento R, “Spatio-spectral classification of hyperspectral images for brain cancer detection during surgical operations,” PLOS ONE 13, 1–27 (March 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]


