Abstract
PURPOSE
The availability of increasing volumes of multiomics, imaging, and clinical data in complex diseases such as cancer opens opportunities for the formulation and development of computational imaging genomics methods that can link multiomics, imaging, and clinical data.
METHODS
Here, we present the Imaging-AMARETTO algorithms and software tools to systematically interrogate regulatory networks derived from multiomics data within and across related patient studies for their relevance to radiography and histopathology imaging features predicting clinical outcomes.
RESULTS
To demonstrate its utility, we applied Imaging-AMARETTO to integrate three patient studies of brain tumors, specifically, multiomics with radiography imaging data from The Cancer Genome Atlas (TCGA) glioblastoma multiforme (GBM) and low-grade glioma (LGG) cohorts and transcriptomics with histopathology imaging data from the Ivy Glioblastoma Atlas Project (IvyGAP) GBM cohort. Our results show that Imaging-AMARETTO recapitulates known key drivers of tumor-associated microglia and macrophage mechanisms, mediated by STAT3, AHR, and CCR2, and neurodevelopmental and stemness mechanisms, mediated by OLIG2. Imaging-AMARETTO provides interpretation of their underlying molecular mechanisms in light of imaging biomarkers of clinical outcomes and uncovers novel master drivers, THBS1 and MAP2, that establish relationships across these distinct mechanisms.
CONCLUSION
Our network-based imaging genomics tools serve as hypothesis generators that facilitate the interrogation of known and uncovering of novel hypotheses for follow-up with experimental validation studies. We anticipate that our Imaging-AMARETTO imaging genomics tools will be useful to the community of biomedical researchers for applications to similar studies of cancer and other complex diseases with available multiomics, imaging, and clinical data.
INTRODUCTION
Major collaborative initiatives have unleashed a myriad of multiomics, clinical, and imaging data for large patient cohorts in studies of cancer, such as multiomics and clinical data from The Cancer Genome Atlas (TCGA)1 and the Clinical Proteomic Tumor Analysis Consortium (CPTAC)2 and radiographic and histopathology imaging data from The Cancer Imaging Archive (TCIA).3 For example, the brain tumor section of TCGA provides multiomics profiles, including RNA sequencing (RNA-seq), DNA copy number variation, and DNA methylation data for approximately 500 patients with glioblastoma multiforme (GBM)4,5 and approximately 500 patients with low-grade glioma (LGG).6 The TCIA Visually AcceSAble Rembrandt Images (VASARI)7-9 project curated a feature set of approximately 30 magnetic resonance imaging (MRI)–derived features on the basis of specialists’ review that is available for approximately 200 patients with GBM and approximately 180 patients with LGG.
CONTEXT
Key Objective
Imaging-AMARETTO provides software tools for imaging genomics through multiomics, clinical, and imaging data fusion within and across multiple patient studies of cancer, toward better diagnostic and prognostic models of cancer.
Knowledge Generated
Our network-based imaging genomics tools serve as powerful hypothesis generators that facilitate the testing of known hypotheses and uncovering of novel hypotheses for follow-up with experimental validation studies. Our case study that integrated multiple studies of brain cancer illustrates how Imaging-AMARETTO can be used for imaging diagnostics and prognostics by interrogating multimodal and multiscale networks for imaging biomarkers to identify their clinically relevant underlying molecular mechanisms.
Relevance
We anticipate that our Imaging-AMARETTO tools for network-based fusion of multiomics, clinical, and imaging data will directly lead to better diagnostic and prognostic models of cancer. In addition, our tools for network biology and medicine will open new avenues for drug discovery by integrating pharmacogenomics data into these networks, toward better therapeutics of cancer.
A trade-off exists between the number of patients included in a data set and the depth of analysis that has moved to increasing levels of refinement, ranging from studying tissues, to cell populations, to single-cell10-12 sequencing. For example, the Ivy Glioblastoma Atlas Project (IvyGAP)13-15 provides 270 transcriptomic profiles refined through histopathology imaging and annotated by a consensus of histopathologists for studying anatomic structures and cancer stem cells for a subset of approximately 30 patients from the TCGA GBM cohort.
In parallel, quantitative imaging provides tools that are capable of processing large volumes of radiography and histopathology images, such as deep convolutional neural networks. The promising field of radiogenomics is based on the idea that entities at different scales, such as molecules, cells, and tissues, are linked to one another and, therefore, may be modeled as a whole.16 Studies have shown that quantitative image features extracted from radiography imaging data are associated and predictive of gene expression patterns from tissues of matched tumors.17-20 Recent efforts further expand this work to link multiomics data with both radiography and histopathology imaging,21,22 toward developing methods for imaging genomics.
These large archives of multimodal and multiscale data sources provide complementary insights into the mechanistic basis of cancer, toward better diagnosis and treatment, and open unprecedented modeling opportunities to link multiomics data with clinical and imaging phenotypes. As a solution to imaging genomics, we introduce the Imaging-AMARETTO software tools to systematically interrogate networks derived from multiomics data for relevance to imaging biomarkers of clinical outcomes. We demonstrate the utility of these imaging genomics tools by integrating three patient brain tumor databases, including the TCGA GBM and LGG and the IvyGAP GBM cohorts. We uncover known and novel drivers of tumor-associated microglia and macrophage mechanisms, and neurodevelopmental and stemness mechanisms, with interpretation of the underlying molecular mechanisms in light of imaging biomarkers of clinical outcomes.
METHODS
The *AMARETTO Software Architecture
The *AMARETTO framework (Fig 1) provides tools for network-based fusion of multiomics, clinical, and imaging data within and across multiple patient studies of cancer. Specifically, this framework offers modular and complementary solutions to multimodal and multiscale aspects of network-based modeling within and across studies of cancer through the AMARETTO and Community-AMARETTO algorithms, respectively. In this work, we present an imaging genomics software toolbox that comprises the newly formulated Imaging-AMARETTO and Imaging-Community-AMARETTO algorithms that together facilitate interpretation of patient-derived multiomics networks for their relevance to radiography and histopathology imaging biomarkers of clinical outcomes. Resources of the *AMARETTO23 software toolbox are available through GitHub, Bioconductor, R Jupyter Notebook, GenePattern, GenomeSpace, and GenePattern Notebook.
FIG 1.
The Imaging-AMARETTO and Imaging-Community-AMARETTO software architecture. The overall framework offers modular and complementary solutions to multimodal and multiscale aspects of network-based modeling within and across multiple studies of cancer. Specifically, (1) The AMARETTO algorithm learns networks of regulatory modules or circuits (circuits of drivers and target genes) from functional genomics or multiomics data (eg, DNA copy number variation [CNV], DNA methylation [MET], RNA gene expression [RNA]) within each study of cancer (eg, within The Cancer Genome Atlas [TCGA] glioblastoma multiforme [GBM], Ivy Glioblastoma Atlas Project [IvyGAP] GBM, or TCGA low-grade glioma [LGG] cohorts separately); (2) the Community-AMARETTO algorithm learns communities or subnetworks of regulatory circuits that are shared or distinct across networks derived from multiple studies of cancer (eg, across the TCGA GBM, IvyGAP GBM, and TCGA LGG cohorts); and (3) the Imaging-AMARETTO and Imaging-Community-AMARETTO algorithms associate these circuits (AMARETTO) and subnetworks (Community-AMARETTO) to clinical, molecular, and imaging-derived biomarkers by mapping radiography and histopathology imaging data onto the networks and assessing their clinical relevance for imaging diagnostics.
Imaging-AMARETTO From a User’s Perspective
The workflow for multiomics, clinical, and imaging data fusion includes utilities to learn networks from individual patient cohorts using Imaging-AMARETTO and to link networks across multiple related patient cohorts using Imaging-Community-AMARETTO. Together, these workflows allow users to integrate patient tumor-derived multiomics or transcriptional profiles with clinical and molecular characteristics and radiography and histopathology imaging features within and across related patient cohorts. The Imaging-AMARETTO source code in R is available from GitHub.24 An R Jupyter Notebook that provides stepwise guidelines for running the source code directly from GitHub for its application to brain cancer is available from Google Colaboratory.25
Imaging-AMARETTO supports multiple workflows: a patient cohort with only transcriptional profiles and a patient cohort with multiomics profiles. When only RNA-seq data are available for a cohort, a predefined list of candidate driver genes is required, which can be selected or uploaded by the user. Predefined lists of known drivers are available for collections of transcription factors (TFutils26,27 and Molecular Signatures Database [MSigDB]28,29 C3) and cancer driver genes (COSMIC30 Cancer Gene Census, MSigDB Hallmark). When genetic or epigenetic data are also available, they can help to guide the selection of candidate driver genes. Potential cancer drivers are identified as somatic recurrent cancer aberrations from genetic and epigenetic data sources using GISTIC31 and MethylMix.32,33 The GISTIC algorithm is used to identify copy number amplifications and deletions from DNA copy number variation data. The MethylMix algorithm is used to identify hyper- and hypomethylated sites from DNA methylation data. The user also specifies the number of regulatory modules to be learned from the data and the percentage of most varying genes to be included in the analysis. Networks of regulatory modules are inferred from RNA-seq data using an iterative optimization procedure. The algorithm34-37 starts with an initialization step that clusters the genes into modules of co-expressed target genes. For each of these modules, we learn the regulatory programs as a linear combination of candidate driver genes that best predict their target genes’ expression using Elastic Net–regularized regression.38 Target genes are then reassigned to the regulatory programs that best explain their gene expression levels as estimated by the predictive power of the regulatory programs’ respective regularized regression models when predicting the target genes’ expression.34-37 The algorithm iterates over these two steps until convergence. This analysis generates a network of regulatory modules, defined as a group of target genes collectively activated or repressed by their associated drivers.
Imaging-Community-AMARETTO: Linking Imaging-AMARETTO Networks Across Cohorts
To compare and integrate networks of regulatory modules across multiple cohorts, the user can submit two or more Imaging-AMARETTO networks and optionally add known networks as collections of signatures to guide subnetwork learning and interpretation, such as immune cell (CIBERSORT39,40) and stemness41-43 signatures. The algorithm34 creates a module map of all pairwise comparisons between modules across multiple networks to assess the extent of overlapping genes between all pairs of modules (−log10 P value, hypergeometric test). This module map is partitioned using an edge betweenness community detection algorithm (Girvan-Newman44) that groups the modules into subnetworks or communities across the multiple networks. These communities represent shared behavior across two or more cohorts, and modules not assigned to communities are reported as distinct behavior specific for each cohort. This analysis generates subnetworks or communities of regulatory modules that are shared or distinct across multiple Imaging-AMARETTO networks derived from multiple cohorts and further refines shared and distinct behavior of modules with respect to their specific drivers.
Downstream Utility for Interpreting Clinical and Experimental Outcomes
We developed several downstream utilities that facilitate interpretation of the Imaging-AMARETTO networks, including functional characterization, driver validation, clinical correlation, and imaging association. To functionally characterize modules and communities, we provide signatures from known gene sets databases (MSigDB) that can be augmented with user-defined signatures, such as immune cell,39,40 stromal cell,45 and stemness41-43 signatures. Regulatory modules and communities are assessed for enrichment in these known functional categories (hypergeometric test).
To validate the predicted drivers as regulators of their targets in modules and communities, we assess whether activator or repressor drivers have a direct or indirect impact on their targets using experimental genetic perturbation data. The user can test signatures derived from genetic perturbation experiments, such as signatures of target genes bound to transcription factors measured in protein-DNA–binding chromatin immunoprecipitation sequencing (ChIP-Seq) experiments (Encyclopedia of DNA Elements,46 ChIP-X Enrichment Analysis,47 Harmonizome48) or defined by motif binding (MSigDB C3), or signatures of genes induced or repressed in response to genetic knockdown or overexpression experiments of drivers (Library of Integrated Network-Based Cellular Signatures [LINCS]/Connectivity Map [CMAP],49,50 Harmonizome).
To characterize regulatory modules for clinical outcomes and molecular biomarkers, the user can submit phenotypes known for all or subsets of samples and specify the statistical hypothesis tests to use for each phenotype. Examples of clinical and molecular phenotypes include survival data, molecular subclasses (eg, mesenchymal, proneural, or classical GBM,51 astrocytoma or oligodendroglioma LGG) and biomarkers (eg, IDH mutation, EGFR amplification, MGMT methylation status). Our implementation supports survival analysis using Cox proportional hazards regression, nominal two-class and multiclass analysis using the Wilcoxon rank sum and Kruskal-Wallis tests, and continuous or ordinal analysis using the Pearson linear and Spearman rank correlation tests. These clinical and molecular phenotype associations are assessed for each of the regulatory modules in individual cohorts and combined for communities across cohorts.
Finally, to interpret regulatory modules for relevance to radiography or histopathology imaging features, associations with these imaging features can be assessed. Examples of radiography and histopathology phenotypes include the 30 TCIA VASARI MRI features defined by expert consensus for the TCGA GBM and LGG cohorts, the IvyGAP13-15 histopathology imaging features characterizing RNA-seq samples refined for anatomic structures and cancer stem cells defined by expert consensus for the IvyGAP GBM cohort, and radiography and histopathology imaging features derived using quantitative imaging methods.20,52-54
Users are provided with all results in the form of hypertext markup language (HTML) reports that are generated in an automated manner for individual cohorts using Imaging-AMARETTO and multiple cohorts using Imaging-Community-AMARETTO. These reports include searchable tables within and across modules and communities, including statistics (ie, coefficients, P values, false discovery rate [FDR] values) for functional enrichment, driver validation, clinical and molecular biomarkers, and radiography and histopathology imaging features. These reports also include heat map visualizations for modules (Figs 2-6) and graph visualizations for communities (Appendix Fig A1). Source code is also provided to convert Imaging-AMARETTO and Imaging-Community-AMARETTO networks for depositing networks in the NDEx55 network database, taking advantage of its interactive features.
FIG 2.
Imaging-AMARETTO predicts STAT3 and AHR as known drivers of tumor-associated microglia and macrophage mechanisms in low-grade glioma (LGG). These heat maps present module 125 from The Cancer Genome Atlas (TCGA) LGG cohort that is a member of community 5 shared across the three cohorts (TCGA glioblastoma multiforme [GBM], Ivy Glioblastoma Atlas Project GBM, and TCGA LGG). For all patient-derived samples (rows), the heat maps show driver genes’ multiomics profiles (columns), including DNA methylation and RNA gene expression data (left panels); target genes’ (columns) RNA gene expression levels (middle panel); and relevant biomarkers (columns), including clinical and molecular and imaging phenotypes (right panels). This module includes eight driver genes (FNDC3B, IQGAP1, ANO6, ELK3, STAT3, TMOD3, CASP8, and ITPRIPL2) that jointly act as activator drivers of the 114 target genes in this module, including AHR and STAT3. Six driver genes are methylation driven (hypomethylation of ANO6, CASP8, ITPRIPL2, STAT3, and TMOD3 and hypermethylation of ANO6 and ELK3, inversely associated with their gene expression levels). Survival analysis reveals that increased expression of the genes in this module is associated with shorter survival (P = 7.3e-11; false discovery rate [FDR] = 1.0e-9). This module distinguishes between the histological subtypes (P = 1.6e-15; FDR = 2.1e-14), with lower expression representing the oligodendroglioma subtype (P = 1.0e-14; FDR = 1.1e-13) and higher expression reflecting the astrocytoma subtype (P = 1.5e-11; FDR = 2.8e-10). IDH mutation (mut) status and 1p19q subtypes are associated with the module expression (P = 3.9e-22; FDR = 1.5e-21), with higher expression levels representing the wild-type (wt) status. Association of Visually AcceSAble Rembrandt Images magnetic resonance imaging features with this module shows that the proportion of enhancing tumor (f5; P = 8.96e-7; FDR = 0.0000134) and enhancement intensity (f4; P = .0000395; FDR = 0.000494) are correlated with gene expression, while the proportions of nonenhancing tumor (f6; P = .0000452; FDR = 0.00036) and cortical involvement (f20; P = .0175; FDR = 0.118) are inversely correlated with expression. Module gene expression levels also distinguish between the thicknesses of enhancing margin (f11; P = .00137; FDR = 0.02).71 CNV, copy number variation.
FIG 6.
Imaging-AMARETTO predicts CCR2 and OLIG2 as co-acting known activator and repressor drivers and MAP2 as novel repressor driver linking tumor-associated microglia and macrophage mechanisms with neurodevelopmental and stemness mechanisms in glioblastoma multiforme (GBM). These heat maps present module 98 from The Cancer Genome Atlas (TCGA) GBM cohort that is a member of community 2 shared across the three cohorts (TCGA GBM, Ivy Glioblastoma Atlas Project GBM, and TCGA low-grade glioma). For all patient-derived samples (rows), the heat maps show driver genes’ multiomics profiles (columns), including DNA methylation and RNA gene expression data (left panels); target genes’ (columns) RNA gene expression levels (middle panel); and relevant biomarkers (columns), including clinical and molecular and imaging phenotypes (right panels). This module includes 10 driver genes (TEC, SPINT1, CCR2, MAN1A1, OLIG2, NPAS3, MAP2, RUFY3, NTRK3, and CSPG5) that jointly act as activator (TEC, SPINT1, CCR2, MAN1A1) and repressor (OLIG2, NPAS3, MAP2, RUFY3, NTRK3, CSPG5) drivers of the 98 target genes in this module. Two driver genes, MAP2 and RUFY3, are methylation driven (hypomethylation of MAP2 and hyper- and hypomethylation of RUFY3, inversely associated with their gene expression levels). Higher expression of the genes in this module represents the mesenchymal molecular subclass (P = 5.1e-35; false discovery rate [FDR] = 5.9e-34), while lower expression of genes in this module comprises the classical (P = 6.1e-15; FDR = 3.8e-14), G-CIMP (P = .000012; FDR = 0.000030), and proneural (P = .00034; FDR = 0.00062) molecular subclasses. IDH1 mutation status is associated with the module expression (P = .00069; FDR = 0.0019), with higher expression levels representing the wild-type status. Association of Visually AcceSAble Rembrandt Images magnetic resonance imaging features with this module shows that the proportion of nonenhancing tumor is inversely correlated (f6; P = .045; FDR = 0.13) with nonenhancing proportion < 33% (P = .0086; FDR = 0.067) and the proportion of enhancing tumor is correlated (f5; P = .020; FDR = 0.28) with the module gene expression.75
RESULTS
To demonstrate its utility, we applied the Imaging-AMARETTO workflow to three studies of brain tumors: multiomics profiles from approximately 500 patients and approximately 30 radiography MRI features for approximately 200 patients from the TCGA GBM cohort,56 multiomics profiles from approximately 500 patients and approximately 30 radiography MRI features for approximately 180 patients from the TCGA LGG cohort,56 and for a subset of approximately 30 patients from the TCGA GBM patient cohort 270 transcriptomic profiles refined through histopathology imaging and annotated with imaging features that characterize anatomic structures for 122 samples and cancer stem cells for 148 samples were used from the IvyGAP GBM project.57
Disease progression in glioma is characterized by infiltration of resident microglia and peripheral macrophages in the tumor microenvironment and by pervasive infiltration of tumor cells in the healthy surroundings of the tumor.58 Understanding microglia and macrophage physiology and its complex interactions with tumor cells can elucidate their roles in glioma progression and uncover potentially interesting druggable targets.
Our results show that Imaging-AMARETTO captures these hallmarks of glioma, for example, key drivers of tumor-associated microglia and macrophage mechanisms59 mediated by STAT3, AHR, and CCR2, and neurodevelopmental and stemness mechanisms60 that involve OLIG2. Our findings recapitulate recent discoveries59,60 and provide interpretation of the molecular mechanisms in light of imaging biomarkers of clinical outcomes. Of note, Imaging-Community-AMARETTO also uncovers novel key master drivers that are shared by these distinct key mechanisms.
Imaging-AMARETTO Deciphers Clinical Relevance of Multiomics Modules of Key Driver Mechanisms
Of clinical relevance, higher expression levels of STAT3, AHR, and CCR2 modules (Figs 2, 3, and 6) are associated with shorter survival in GBM and LGG, and these modules also distinguish between molecular subclasses of GBM and LGG. In GBM, the mesenchymal subclass is represented by higher expression of these modules compared with the classical and proneural subclasses. In LGG, the astrocytoma subtype is characterized by higher expression of these modules compared with the oligodendroglioma subtype.
FIG 3.
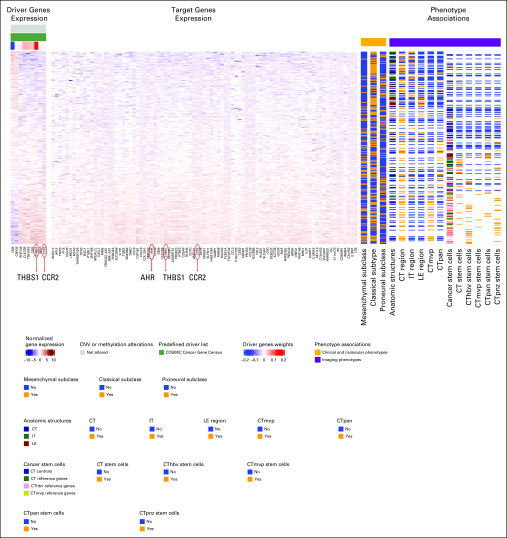
Imaging-AMARETTO predicts AHR and CCR2 as known drivers and identifies TBHS1 as a novel driver of tumor-associated microglia and macrophage mechanisms in glioblastoma multiforme (GBM). These heat maps present module 64 from the Ivy Glioblastoma Atlas Project (IvyGAP) GBM cohort that is a member of community 5 shared across the three cohorts (The Cancer Genome Atlas [TCGA] GBM, IvyGAP GBM, and TCGA low-grade glioma). For all patient-derived samples (rows), the heat maps show driver genes’ functional genomics profiles (columns), specifically RNA gene expression profiles (left panel); target genes’ (columns) RNA gene expression levels (middle panel); and relevant biomarkers (columns), including clinical and molecular and imaging phenotypes (right panels). This module includes nine driver genes (THBS1, CLEC2B, TNFAIP3, DSE, RNF149, MGP, CCR2, CSPG5, and CKB) that jointly act as activators (eg, CCR2, a squamous cell carcinoma tumor-rejection antigen recognized by T lymphocytes and candidate for specific immunotherapy) and repressors (CSPG5 and CKB) that drive the 87 target genes in this module, including AHR, CCR2, and THBS1. Association analyses confirm that higher expression of module genes reflects samples derived from patients in the mesenchymal subclass (P = .000047; false discovery rate [FDR] = 0.00015), while lower expression represents the classical subclass (P = .032; FDR = 0.067). Association of histopathology imaging features that study anatomic structures reveals that genes in this module distinguish between the samples derived from distinct anatomic structures (P = 1.8e-15; FDR = 4.3e-15), (continued on following page)where cellular tumor (CT; P = .0000078; FDR = 0.000013) samples and, in particular, with substructure microvascular proliferation (CTmvp; P = 8.7e-14; FDR = 6.2e-13) and pseudopalisading cells around necrosis (CTpan; P = .018; FDR = 0.027) show elevated expression of module genes, while leading edge (LE; P = .018; FDR = 0.023) and infiltrating tumor (IT; P = .0014; FDR = 0.0045) samples show lower expression. Association of histopathology imaging features targeting cancer stem cells reveals that samples derived from cancer stem cells are generally associated with higher module gene expression compared with nonstem cells (P = 4.0e-16; FDR = 8.6e-15) and, specifically, elevated expression in stem-cell v control samples from substructures of the CT (P = .0000059; FDR = 0.00018), including hyperplastic blood vessels (CThbv; P = 1.2e-11; FDR = 5.2e-10), perinecrotic zone (CTpnz; P = 3.4e-9; FDR = 3.4e-8), CTpan (P = .000087; FDR = 0.00032), and CTmvp (P = .0039; FDR = 0.029).72 CNV, copy number variation.
Diametrically opposed, higher expression levels of OLIG2 modules (Figs 4-6) are associated with better survival in GBM and LGG, and these modules also distinguish between molecular subclasses of GBM and LGG but in the opposite direction. In GBM the classical and proneural subclasses are represented by higher expression of these modules compared with the mesenchymal subclass. In LGG, the oligodendroglioma subtype is characterized by higher expression of these modules compared with the astrocytoma subtype.
FIG 4.
Imaging-AMARETTO predicts OLIG2 as known driver of neurodevelopmental and stemness mechanisms in low-grade glioma (LGG). These heat maps present module 91 from The Cancer Genome Atlas (TCGA) LGG cohort that is a member of community 2 shared across the three cohorts (TCGA glioblastoma multiforme [GBM], Ivy Glioblastoma Atlas Project GBM, and TCGA LGG). For all patient-derived samples (rows), the heat maps show driver genes’ multiomics profiles (columns), including DNA methylation and RNA gene expression data (left panels); target genes’ (columns) RNA gene expression levels (middle panel); and relevant biomarkers (columns), including clinical and molecular and imaging phenotypes (right panels). This module includes nine driver genes (SOX8, OLIG2, EBF4, NLGN2, SHD, RANBP17, FERMT1, LOC254559, and C11orf63) that jointly act as activator and repressor (C11orf63) drivers of the 94 target genes in this module, including OLIG2. Six driver genes are methylation driven (hypomethylation of FERMT1 and OLIG2 and hypermethylation of C11orf63, NLGN2, RANBP17, and SOX8, inversely associated with their gene expression levels). Survival analysis reveals that increased expression of the genes in this module is associated with better survival (P = 7.1e-11; false discovery rate [FDR] = 1.0e-9). This module distinguishes between the histological subtypes (P = .00047; FDR = 0.00084), with lower expression representing the astrocytoma subtype (P = .00016; FDR = 0.00039) and higher expression reflecting the oligodendroglioma subtype (P = .0027; FDR = 0.0047). IDH mutation (mut) status and 1p19q subtypes are associated with the module expression (P = 2.0e-26; FDR = 1.1e-25), with lower expression levels representing the wild-type (wt) status. Association of Visually AcceSAble Rembrandt Images magnetic resonance imaging features with this module shows that the proportion of enhancing tumor (f5; P = .016; FDR = 0.032) is inversely correlated with gene expression, while the proportion of nonenhancing tumor (f6; P = .032; FDR = 0.066), cortical involvement (f20; P = .018; FDR = 0.12), and the presence of cysts (f8; P = .0095; FDR = 0.14) are correlated with expression.73 CNV, copy number variation.
Imaging-AMARETTO Deciphers Histopathology Imaging Biomarkers of Key Driver Mechanisms
Histopathology imaging features of anatomic structures show that higher expression of STAT3, AHR, and CCR2 modules (Fig 3) distinguishes between samples derived from the cellular tumor compared with those from leading edge and infiltrating tumor regions. Higher expression of OLIG2 modules distinguishes infiltrating tumor from cellular tumor samples.
Features representative of cancer stem cells show that higher expression of STAT3, AHR, and CCR2 modules (Fig 3) distinguishes cancer stem-cell samples from their non–stem cancer cell counterparts. This observation is consistent across the distinct substructures of the cellular tumor, including hyperplastic blood vessels, microvascular proliferation, perinecrotic zone, and pseudopalisading cells around necrosis. Diametrically opposed, higher expression of OLIG2 modules distinguishes non–stem cancer cells from cancer stem cells consistently across these microvascular and necrosis substructures.
Imaging-AMARETTO Deciphers Radiography Imaging Biomarkers of Key Driver Mechanisms
Radiographic image features of STAT3, AHR, and CCR2 modules (Figs 2 and 6) are highly consistent across GBM and LGG. Higher expression is associated with a higher proportion of enhancing tumor, lower proportion of nonenhancing tumor, and less cortical involvement. These modules also distinguish between measures of thickness of enhancing margin in both GBM and LGG. In GBM these STAT3, AHR, and CCR2 modules show higher expression in association with eloquent cortex, while in LGG, they show higher expression in association with enhancement intensity.
Features of OLIG2 modules (Figs 4-6) are also consistent across GBM and LGG and diametrically opposed to those of STAT3, AHR, and CCR2. In both GBM and LGG, higher expression is associated with a higher proportion of nonenhancing tumor and lower proportion of enhancing tumor. In GBM, higher expression is also associated within speech receptive eloquent cortex, while in LGG, higher expression is associated with cortical involvement and the presence of cysts.
Imaging-Community-AMARETTO Uncovers Known Key and Novel Master Drivers Linking Mechanisms
Recent discoveries of STAT3, AHR, and CCR2 as drivers of tumor-associated microglia and macrophage mechanisms59 are captured by modules in communities 1 and 5: TCGA LGG module 125 (Fig 2) shows hypomethylation of STAT3 as activator driver of AHR, with higher expression associated with shorter survival and astrocytoma LGG, and IvyGAP GBM module 64 (Fig 3) shows that higher expression of AHR and CCR2 is associated with the presence of cancer stem cells and microvascular substructures and suggests as novel activator driver, THBS1, that plays important roles in macrophage infiltration and angiogenesis,61 vascularization,62 and tumorigenesis62 in glioma. OLIG2 as a driver of neurodevelopmental and stemness mechanisms60,63,64 is captured by modules in community 2: (1) TCGA LGG module 91 (Fig 4) shows hypomethylation of OLIG2 as activator driver of this module, with higher expression associated with better survival, oligodendrocyte LGG, and IDH1 wild-type status; (2) TCGA GBM module 75 (Fig 5) is driven by OLIG2, with higher expression associated with proneural and classical versus mesenchymal GBM, and suggests as novel repressor driver THBS1 and as novel activator driver hypomethylation of neuronal marker MAP2 that plays important roles in microtubule-associated neurogenesis65,66 and reduces invasiveness67 and stemness68 in glioma; and (3) TCGA GBM module 98 (Fig 6) shows CCR2 and OLIG2 co-acting as activator and repressor drivers, respectively, highlighting their diametrically opposed behavior, with higher CCR2 and lower OLIG2 expression associated with mesenchymal versus proneural and classical GBM and suggesting as novel repressor driver hypomethylation of MAP2, consistent with observations in TCGA GBM module 75 (Fig 5). Using knockdown experiments of THBS1 from LINCS/CMAP, we confirmed that THBS1 acts as activator and repressor of its targets in IvyGAP GBM module 64 (Fig 3) and TCGA GBM module 75 (Fig 5), respectively.
FIG 5.
Imaging-AMARETTO predicts OLIG2 as known driver and identifies MAP2 and THBS1 as novel drivers of neurodevelopmental and stemness mechanisms in glioblastoma multiforme (GBM). These heat maps present module 75 from The Cancer Genome Atlas (TCGA) GBM cohort that is a member of community 2 shared across the three cohorts (TCGA GBM, Ivy Glioblastoma Atlas Project GBM, and TCGA low-grade glioma). For all patient-derived samples (rows), the heat maps show driver genes’ multiomics profiles (columns), including DNA copy number variation (CNV), DNA methylation, and RNA genes’ expression data (left panels); target genes’ (columns) RNA gene expression levels (middle panel); and relevant biomarkers (columns), including clinical and molecular and imaging phenotypes (right panels). This module includes 10 driver genes (CSPG5, MAP2, OLIG2, CKB, GCSH, PPP2R2B, HEPACAM, CDH10, CTNND2, and THBS1) that jointly act as activator and repressor (THBS1) drivers of the 84 target genes in this module. One driver gene, GCSH, is copy number driven (somatic recurrent copy number deletions and amplifications associated with its gene expression), and three driver genes are methylation driven (hypomethylation of CDH10, MAP2, and PPP2R2B, inversely associated with their gene expression). Higher expression of genes in this module comprises the classical (P = 9.9e-13; false discovery rate [FDR] = 5.1e-12) and proneural (P = .0000024; FDR = 0.0000056) molecular subclasses, while lower expression of genes in this module represents the mesenchymal molecular subclass (P = 6.1e-40; FDR = 2.3e-38). Association of Visually AcceSAble Rembrandt Images (VASARI) magnetic resonance imaging features with this module shows that the proportion of nonenhancing tumor (f6; P = .013; FDR = 0.060, with nonenhancing proportion < 33%, P = .0014; FDR = 0.027, and nonenhancing proportion < 5%, P = .032; FDR = 0.17) and speech receptive eloquent cortex (f3; P = .046; FDR = 0.75) are correlated with the module gene expression.74
Thus, Imaging-Community-AMARETTO (Appendix Fig A1) identified THBS1 and MAP2 as novel master drivers across the three STAT3, AHR, CCR2, and OLIG2 communities that provide new insights into how these distinct key mechanisms are linked in glioma. Interesting avenues for further exploration with experimental validation studies include testing novel hypotheses of THBS1 and MAP2 as master regulators of shared mechanisms that involve macrophage infiltration, vascularization, tumorigenesis, invasion, stemness, and neurogenesis in glioma.
DISCUSSION
We developed the Imaging-AMARETTO algorithms and software tools for imaging genomics to facilitate systematic interrogation of regulatory networks derived from multiomics data within and across related patient studies for their relevance to radiography and histopathology imaging features that predict clinical outcomes. We demonstrated its utility through application to three patient studies of brain tumors, including multiomics and radiography imaging data from the TCGA GBM and LGG studies and transcriptional and histopathology imaging data from the IvyGAP GBM study.
Our results show that Imaging-AMARETTO recapitulates known key drivers of tumor-associated microglia and macrophage mechanisms (STAT3, AHR, and CCR2) and neurodevelopmental and stemness mechanisms (OLIG2). Imaging-AMARETTO provides interpretation of the underlying molecular mechanisms in light of imaging biomarkers of clinical outcomes, and Imaging-Community-AMARETTO also uncovered novel master drivers THBS1 and MAP2 that establish relationships across these distinct mechanisms.
Of note, the querying of the Imaging-AMARETTO networks for modules whose elevated expression is inversely associated with proportions of enhancing tumor and cancer stem cells on radiography and histopathology imaging, respectively, shows that these modules are putatively coregulated by activator drivers OLIG2 and MAP2 and repressor drivers STAT3, AHR, CCR2, and THBS1. Thus, we hypothesize that restoration of the function of OLIG2 and MAP2 and attenuation of the expression of STAT3, AHR, CCR2, and THBS1 potentially shift their target genes’ expression to more benign functional states associated with better survival in GBM and LGG.
This case study illustrates how Imaging-AMARETTO can be used for imaging diagnostics and prognostics by interrogating multimodal and multiscale networks for imaging biomarkers to identify their clinically relevant underlying molecular mechanisms. Our network-based imaging genomics tools are powerful hypothesis generators that facilitate the testing of known hypotheses and uncovering of novel hypotheses for follow-up with experimental validation studies. We anticipate that our tools for network-based fusion of multiomics, clinical, and imaging data will lead to better diagnostic and prognostic models of cancer and will open new avenues for drug discovery by integrating pharmacogenomic data into these networks, toward better therapeutics of cancer.69,70
ACKNOWLEDGMENT
We thank Aviv Regev, PhD, for helpful discussions on developing network biology and medicine approaches for studying complex human diseases. We also thank Howard Weiner, MD, and Vijay Kuchroo, DVM, PhD, for helpful insights into deciphering the mechanistic basis of gliomas.
APPENDIX
FIG A1.
Imaging-Community-AMARETTO identifies known drivers AHR, STAT3, CCR2, and OLIG2 and uncovers novel master drivers THBS1 and MAP2 that link distinct key mechanisms that underlie glioma. In this Imaging-Community-AMARETTO graph, the nodes represent the regulatory modules or circuits that are learned from the three studies of brain cancer (The Cancer Genome Atlas [TCGA] glioblastoma multiforme [GBM], Ivy Glioblastoma Project [IvyGAP] GBM, and TCGA low-grade glioma [LGG]; node sizes are scaled by the number of driver and target genes in the modules), the edges represent the extent of overlapping genes between the modules across the three cohorts (edge thickness is scaled with the significance of the overlapping genes between modules), and the clouds represent how the modules across the three cohorts are grouped into the communities or subnetworks that are learned using the Girvan-Newman edge betweenness community detection algorithm. Imaging-Community-AMARETTO organized modules regulated by known drivers of tumor-associated microglia and macrophage mechanisms STAT3, AHR, and CCR2 and neurodevelopmental and stemness mechanism OLIG2 into three communities. Community 5 (C5) links STAT3, AHR, and CCR2, and C1 links AHR and CCR2 as activators of shared modules. Modules regulated by OLIG2 are represented in C2 with OLIG2 as activator of its modules. Of note, C2 also contains a module that links OLIG2 and CCR2 co-acting as repressor and activator, respectively (Fig 6). Imaging-Community-AMARETTO also uncovered THBS1 and MAP2 as novel master drivers across the three STAT3, AHR, CCR2, and OLIG2 communities that provide new insights into how these distinct key mechanisms are linked in glioma. THBS1 is an activator driver of three modules in C1 and C5, and a repressor driver of three modules in C2. MAP2 is a repressor driver of three modules in C1 and C5 and an activator driver of six modules in C2 (except repressor of TCGA GBM module 98). Taken together, C1 links AHR and THBS1 (TCGA GBM module 79). C2 links OLIG2, MAP2, and THBS1 (TCGA GBM module 75; Fig 5); OLIG2 and MAP2 (IvyGAP GBM module 38, TCGA GBM module 61); CCR2, OLIG2, and MAP2 (TCGA GBM module 98; Fig 6). C5 links CCR2, AHR, and THBS1 (IvyGAP GBM module 64; Fig 3) and AHR and STAT3 (TCGA LGG module 125; Fig 2). Using genetic knockdown experiments of THBS1 from LINCS/Connectivity Map, we confirmed that THBS1 acts as activator and repressor drivers of its targets in IvyGAP GBM module 64 (Fig 3) and TCGA GBM module 75 (Fig 5), respectively (http://portals.broadinstitute.org/pochetlab/JCO_CCI_Imaging-AMARETTO/Imaging-AMARETTO_HTML_Report_TCGA-GBM_IVYGAP-GBM_TCGA-LGG/; www.ndexbio.org/#/network/16820740-d7ea-11e9-bb65-0ac135e8bacf).
SUPPORT
Supported by the National Cancer Institute (NCI) Informatics Technology for Cancer Research (R21CA209940 [O.G., T.F.B., J.P.M., N.P.], U01CA214846 [V.C.], U01CA214846 Collaborative Set-aside [O.G., A.M.K., V.C., N.P.], U24CA194107 [J.P.M.], U24CA220341 [J.P.M.], U24CA180922 [B.J.H., N.P., A. Regev]), NCI (R01CA215072 [A.M.K.], U01CA217851 [O.G.], U01CA199241 [O.G.], Stanford CTD2 [O.G.]), National Institute of Allergy and Infectious Diseases (R03AI131066 [T.F.B., N.P.]), and National Institute of Biomedical Imaging and Bioengineering (R01EB020527 [O.G.]). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
SOFTWARE AND RESOURCES
The source code of Imaging-AMARETTO is available from GitHub, and a Notebook for its application to the three brain cancer cohorts is available from Google Colaboratory, including links to interactive HTML reports and NDEx networks: http://portals.broadinstitute.org/pochetlab/JCO_CCI_Imaging-AMARETTO/Imaging-AMARETTO_Software_Resources.html.
AUTHOR CONTRIBUTIONS
Conception and design: Olivier Gevaert, Mohsen Nabian, Shaimaa Bakr, Artur Manukyan, Erik J. Uhlmann, Jill P. Mesirov, Vincent Carey, Nathalie Pochet
Financial support: Olivier Gevaert, Anna M. Krichevsky, Jill P. Mesirov, Vincent Carey, Nathalie Pochet
Collection and assembly of data: Olivier Gevaert, Mohsen Nabian, Artur Manukyan, Jishu Xu, Nathalie Pochet
Data analysis and interpretation: Olivier Gevaert, Mohsen Nabian, Celine Everaert, Jayendra Shinde, Artur Manukyan, Ted Liefeld, Thorin Tabor, Joachim Lupberger, Brian J. Haas, Thomas F. Baumert, Mikel Hernaez, Michael Reich, Francisco J. Quintana, Anna M. Krichevsky, Vincent Carey, Nathalie Pochet
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Olivier Gevaert
Research Funding: Paragon Development Systems (Inst), Lucence Diagnostics (Inst)
Brian J. Haas
Consulting or Advisory Role: Immuneering, Diamond Age Data Science
Thomas F. Baumert
Stock and Other Ownership Interests: Alentis Therapeutics, Alentis Therapeutics (Inst)
Research Funding: Janssen Research & Development (Inst), Alentis Therapeutics (Inst)
Patents, Royalties, Other Intellectual Property: Patent applications on Claudin-1 targeting monoclonal antibodies for liver disease, fibrosis, and cancer (Inst); patent applications on liver disease drug discovery systems (Inst)
Vincent Carey
Employment: CleanSlate (I)
Honoraria: Gilead Sciences (I)
Research Funding: Bayer AG
No other potential conflicts of interest were reported.
REFERENCES
- 1. National Cancer Institute: The Cancer Genome Atlas Program, 2018. https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga.
- 2. National Cancer Institute Office of Cancer Clinical Proteomics Research: OCCPR: A leader in cancer proteomics and proteogenomics, 2019. https://proteomics.cancer.gov.
- 3. The Cancer Imaging Archive: Welcome to The Cancer Imaging Archive, 2019. https://www.cancerimagingarchive.net.
- 4. doi: 10.1038/nature07385. Cancer Genome Atlas Research Network: Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455:1061-1068, 2008 [Erratum: Nature 494:506, 2013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. doi: 10.1016/j.cell.2013.09.034. Brennan CW, Verhaak RGW, McKenna A, et al: The somatic genomic landscape of glioblastoma. Cell 155:462-477, 2013 [Erratum: Cell 157:753, 2014] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brat DJ, Verhaak RGW, Aldape KD, et al: Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 372:2481-2498, 2015. [DOI] [PMC free article] [PubMed]
- 7. Bakas S, Akbari H, Sotiras A, et al: Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data 4:170117, 2017. [DOI] [PMC free article] [PubMed]
- 8. Cancer Imaging Archive: VASARI Research Project. https://wiki.cancerimagingarchive.net/display/Public/VASARI+Research+Project.
- 9. Cancer Imaging Archive: TCGA Glioma Phenotype Research Group. https://wiki.cancerimagingarchive.net/display/Public/TCGA+Glioma+Phenotype+Research+Group.
- 10.Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tirosh I, Venteicher AS, Hebert C, et al: Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539:309-313, 2016. [DOI] [PMC free article] [PubMed]
- 12. Filbin MG, Tirosh I, Hovestadt V, et al: Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science 360:331-335, 2018. [DOI] [PMC free article] [PubMed]
- 13. Puchalski RB, Shah N, Miller J, et al: An anatomic transcriptional atlas of human glioblastoma. Science 360:660-663, 2018. [DOI] [PMC free article] [PubMed]
- 14. IvyGAP: Ivy Glioblastoma Atlas Project, 2019. https://glioblastoma.alleninstitute.org.
- 15. IvyGAP: Data download. https://glioblastoma.alleninstitute.org/static/download.html.
- 16.Rutman AM, Kuo MD. Radiogenomics: Creating a link between molecular diagnostics and diagnostic imaging. Eur J Radiol. 2009;70:232–241. doi: 10.1016/j.ejrad.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 17.Segal E, Sirlin CB, Ooi C, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25:675–680. doi: 10.1038/nbt1306. [DOI] [PubMed] [Google Scholar]
- 18.Diehn M, Nardini C, Wang DS, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A. 2008;105:5213–5218. doi: 10.1073/pnas.0801279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gevaert O, Xu J, Hoang CD, et al. Non-small cell lung cancer: Identifying prognostic imaging biomarkers by leveraging public gene expression microarray data--methods and preliminary results. Radiology. 2012;264:387–396. doi: 10.1148/radiol.12111607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Itakura H, Achrol AS, Mitchell LA, et al. Magnetic resonance image features identify glioblastoma phenotypic subtypes with distinct molecular pathway activities. Sci Transl Med. 2015;7:303ra138. doi: 10.1126/scitranslmed.aaa7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheerla A, Gevaert O. Deep learning with multimodal representation for pancancer prognosis prediction. Bioinformatics. 2019;35:i446–i454. doi: 10.1093/bioinformatics/btz342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Momeni A, Thibault M, Gevaert O: Dropout-enabled ensemble learning for multi-scale biomedical data. in Crimi A, Bakas S, Kuijf H, et al (eds): Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Cham, Switzerland, Springer International Publishing, 2019, pp 407-415. [Google Scholar]
- 23. Broad Institute: The *AMARETTO framework for network biology and medicine: Linking disease, drivers, targets and drugs via graph-based fusion of multi-omics, clinical, imaging, and perturbation data. http://portals.broadinstitute.org/pochetlab/amaretto.html.
- 24. GitHub: broadinstitute/ImagingAMARETTO. https://github.com/broadinstitute/ImagingAMARETTO.
- 25. Google Colaboratory: Imaging-AMARETTO: An imaging genomics software tool to systematically interrogate multi-omics networks for relevance to radiography and histopathology imaging biomarkers of clinical outcomes with application to studies of brain tumors. https://colab.research.google.com/drive/14u1KZJ3Gf-9qjDycyBKzBiN5VzzOa2xU#scrollTo=LujO14znmO0J.
- 26. Stubbs BJ, Gopaulakrishnan S, Glass K, et al: TFutils: Data structures for transcription factor bioinformatics. F1000Res 8:152, 2019. [DOI] [PMC free article] [PubMed]
- 27. Carey V, Gopaulakrishnan S: TFutils: Bioconductor version release (3.10). https://bioconductor.org/packages/TFutils.
- 28.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liberzon A, Birger C, Thorvaldsdóttir H, et al. The Molecular Signatures Database (MSigDB) Hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. doi: 10.1093/nar/gky1015. COSMIC: COSMIC, the Catalogue of Somatic Mutations in Cancer. https://cancer.sanger.ac.uk/cosmic. [DOI] [PMC free article] [PubMed]
- 31.Mermel CH, Schumacher SE, Hill B, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cedoz PL, Prunello M, Brennan K, et al: MethylMix 2.0: An R package for identifying DNA methylation genes. Bioinformatics 34:3044-3046, 2018. [DOI] [PMC free article] [PubMed]
- 33.Gevaert O, Tibshirani R, Plevritis SK. Pancancer analysis of DNA methylation-driven genes using MethylMix. Genome Biol. 2015;16:17. doi: 10.1186/s13059-014-0579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Champion M, Brennan K, Croonenborghs T, et al. Module analysis captures pancancer genetically and epigenetically deregulated cancer driver genes for smoking and antiviral response. EBioMedicine. 2018;27:156–166. doi: 10.1016/j.ebiom.2017.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shinde J, Everaert C, Bakr S, et al: AMARETTO: Regulatory network inference and driver gene evaluation using integrative multi-omics analysis and penalized regression: Bioconductor version release 3.10, 2019. https://bioconductor.org/packages/AMARETTO.
- 36. doi: 10.1098/rsfs.2013.0013. Gevaert O, Villalobos V, Sikic BI, et al: Identification of ovarian cancer driver genes by using module network integration of multi-omics data. Interface Focus 3:20130013, 2013 [Erratum Interface Focus 4:20140023, 2014] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gevaert O, Plevritis S. Identifying master regulators of cancer and their downstream targets by integrating genomic and epigenomic features. Pac Symp Biocomput. 2013:123–134. [PMC free article] [PubMed] [Google Scholar]
- 38.Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc Series B Stat Methodol. 2005;67:301–320. [Google Scholar]
- 39.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stanford University: CIBERSORT. https://cibersort.stanford.edu.
- 41.Ben-Porath I, Thomson MW, Carey VJ, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marson A, Levine SS, Cole MF, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim J, Woo AJ, Chu J, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newman MEJ, Girvan M. Finding and evaluating community structure in networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;69:026113. doi: 10.1103/PhysRevE.69.026113. [DOI] [PubMed] [Google Scholar]
- 45.Baryawno N, Przybylski D, Kowalczyk MS, et al. A cellular taxonomy of the bone marrow stroma in homeostasis and leukemia. Cell. 2019;177:1915–1932.e16. doi: 10.1016/j.cell.2019.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.ENCODE Project Consortium The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 47.Lachmann A, Xu H, Krishnan J, et al. ChEA: Transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rouillard AD, Gundersen GW, Fernandez NF, et al: The Harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) 2016:baw100, 2016. [DOI] [PMC free article] [PubMed]
- 49.Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 50.Subramanian A, Narayan R, Corsello SM, et al. A next generation Connectivity Map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171:1437–1452.e17. doi: 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verhaak RGW, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gevaert O, Mitchell LA, Achrol AS, et al. Glioblastoma multiforme: Exploratory radiogenomic analysis by using quantitative image features. Radiology. 2014;273:168–174. doi: 10.1148/radiol.14131731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu TT, Achrol AS, Mitchell LA, et al. Magnetic resonance perfusion image features uncover an angiogenic subgroup of glioblastoma patients with poor survival and better response to antiangiogenic treatment. Neuro-oncol. 2017;19:997–1007. doi: 10.1093/neuonc/now270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nicolasjilwan M, Hu Y, Yan C, et al. Addition of MR imaging features and genetic biomarkers strengthens glioblastoma survival prediction in TCGA patients. J Neuroradiol. 2015;42:212–221. doi: 10.1016/j.neurad.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. NDEx: Welcome to the Network Data Exchange. https://home.ndexbio.org.
- 56. GenePattern: AMARETTO supporting data files. https://datasets.genepattern.org/?prefix=data/module_support_files/Amaretto.
- 57. Carey V: ivygapSE: A SummarizedExperiment for Ivy-GAP data: Bioconductor version release 3.10, 2019. https://bioconductor.org/packages/ivygapSE.
- 58.Sevenich L. Brain-resident microglia and blood-borne macrophages orchestrate central nervous system inflammation in neurodegenerative disorders and brain cancer. Front Immunol. 2018;9:697. doi: 10.3389/fimmu.2018.00697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. doi: 10.1038/s41593-019-0370-y. Takenaka MC, Gabriely G, Rothhammer V, et al: Control of tumor-associated macrophages and T cells in glioblastoma via AHR and CD39. Nat Neurosci 22:729-740, 2019 [Erratum: Nat Neurosci 22:1533, 2019] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krichevsky AM, Uhlmann EJ. Oligonucleotide therapeutics as a new class of drugs for malignant brain tumors: Targeting mRNAs, regulatory RNAs, mutations, combinations, and beyond. Neurotherapeutics. 2019;16:319–347. doi: 10.1007/s13311-018-00702-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Offer S, Menard JA, Pérez JE, et al. Extracellular lipid loading augments hypoxic paracrine signaling and promotes glioma angiogenesis and macrophage infiltration. J Exp Clin Cancer Res. 2019;38:241. doi: 10.1186/s13046-019-1228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Daubon T, Léon C, Clarke K, et al: Deciphering the complex role of thrombospondin-1 in glioblastoma development. Nat Commun 10:1146, 2019. [DOI] [PMC free article] [PubMed]
- 63.Suvà ML, Rheinbay E, Gillespie SM, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ceccarelli M, Barthel FP, Malta TM, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. doi: 10.1038/s41598-019-39852-5. Gao L, Huang S, Zhang H, et al: Suppression of glioblastoma by a drug cocktail reprogramming tumor cells into neuronal like cells. Sci Rep 9:3462, 2019 [Erratum: Sci Rep 10:2971, 2020] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yuan J, Zhang F, Hallahan D, et al. Reprogramming glioblastoma multiforme cells into neurons by protein kinase inhibitors. J Exp Clin Cancer Res. 2018;37:181. doi: 10.1186/s13046-018-0857-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Zhou Y, Wu S, Liang C, et al. Transcriptional upregulation of microtubule-associated protein 2 is involved in the protein kinase A-induced decrease in the invasiveness of glioma cells. Neuro-oncol. 2015;17:1578–1588. doi: 10.1093/neuonc/nov060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yi R, Feng J, Yang S, et al. miR-484/MAP2/c-Myc-positive regulatory loop in glioma promotes tumor-initiating properties through ERK1/2 signaling. J Mol Histol. 2018;49:209–218. doi: 10.1007/s10735-018-9760-9. [DOI] [PubMed] [Google Scholar]
- 69.Emmert-Streib F, Dehmer M, Haibe-Kains B. Gene regulatory networks and their applications: Understanding biological and medical problems in terms of networks. Front Cell Dev Biol. 2014;2:38. doi: 10.3389/fcell.2014.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wooden B, Goossens N, Hoshida Y, et al. Using big data to discover diagnostics and therapeutics for gastrointestinal and liver diseases. Gastroenterology. 2017;152:53–67.e3. doi: 10.1053/j.gastro.2016.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Imaging-AMARETTO HTML report of module 125 from the TCGA LGG cohort. http://portals.broadinstitute.org/pochetlab/JCO_CCI_Imaging-AMARETTO/Imaging-AMARETTO_HTML_Report_TCGA-GBM_IVYGAP-GBM_TCGA-LGG/TCGA_LGG/AMARETTOhtmls/modules/module125.html.
- 72. Imaging-AMARETTO HTML report of module 64 from the IvyGAP GBM cohort. http://portals.broadinstitute.org/pochetlab/JCO_CCI_Imaging-AMARETTO/Imaging-AMARETTO_HTML_Report_TCGA-GBM_IVYGAP-GBM_TCGA-LGG/Ivygap_GBM/AMARETTOhtmls/modules/module64.html.
- 73. Imaging-AMARETTO HTML report of module 91 from the TCGA LGG cohort. http://portals.broadinstitute.org/pochetlab/JCO_CCI_Imaging-AMARETTO/Imaging-AMARETTO_HTML_Report_TCGA-GBM_IVYGAP-GBM_TCGA-LGG/TCGA_LGG/AMARETTOhtmls/modules/module91.html.
- 74. Imaging-AMARETTO HTML report of module 75 from the TCGA GBM cohort. http://portals.broadinstitute.org/pochetlab/JCO_CCI_Imaging-AMARETTO/Imaging-AMARETTO_HTML_Report_TCGA-GBM_IVYGAP-GBM_TCGA-LGG/TCGA_GBM/AMARETTOhtmls/modules/module75.html.
- 75. Imaging-AMARETTO HTML report of module 98 from the TCGA GBM cohort. http://portals.broadinstitute.org/pochetlab/JCO_CCI_Imaging-AMARETTO/Imaging-AMARETTO_HTML_Report_TCGA-GBM_IVYGAP-GBM_TCGA-LGG/TCGA_GBM/AMARETTOhtmls/modules/module98.html.