Highlights
-
•
BRSV and P. multocida interaction cause severe pneumonia in cattle.
-
•
ICAM1 regulated P. multocida adherence to upper respiratory epithelial cells.
-
•
BRSV infection decreased ICAM1 expression in bovine upper respiratory tract.
-
•
Upper respiratory tract as a gateway to prevent pneumonia in lower respiratory tract is disrupted by BRSV infection.
Keywords: bovine respiratory epithelial cell, BRDC, BRSV, co-infection, ICAM-1, Pasteurella multocida
Abstract
The synergistic infection of bovine respiratory syncytial virus (BRSV) and Pasteurella multocida (PM) may predispose cattle to develop severe pneumonia. Previously, we reported that BRSV infection significantly decreased PM adherence to the upper respiratory epithelial cells. It may allow bacteria to invade into the lower respiratory tract and lead to severe pneumonia. To investigate whether BRSV infection regulates the cell surface adherence receptor on bovine trachea epithelial cells (bTECs), we performed proteomic and functional analyses. BRSV infection decreased the expression of intercellular adhesion molecule-1 (ICAM1) on bTECs. Inhibition and knockdown experiments using anti-ICAM1 antibody and siRNAs targeting ICAM1 indicated that PM adherence to bTECs was dependent on ICAM1 expression. These data suggest that under normal conditions bTECs may capture PM in the upper respiratory tract, while BRSV infection reverses this mechanism. The proposed gateway function of bTECs is disrupted by BRSV infection that may facilitate bacterial invasion into the lower respiratory tract and lead to secondary or more severe respiratory infection.
1. Introduction
Bovine respiratory syncytial virus (BRSV) is a one of the major viral pathogens associated with bovine respiratory disease complex (BRDC) that has a severe economic impact on the cattle industry worldwide (Larsen et al., 2001; Härtel et al., 2004; Beaudeau et al., 2010; Timsit et al., 2016). BRSV is an enveloped, non-segmented, negative-stranded RNA virus and a member of the Orthopneumovirus genus within the Pneumoviridae family (König et al., 2004). BRSV typically causes primary infection of the respiratory tract and can predispose cattle to secondary infections by bacterial pathogens (Larsen et al., 2001; Tjønehøj et al., 2003; Agnes et al., 2013).
The respiratory epithelial cells are the first line of defence and function as a physical barrier to protect against invading pathogens (Agnes et al., 2013; Eberle et al., 2016; Cozens et al., 2019), including BRDC-causing pathogens (Liu et al., 2018; Johnston et al., 2019). Under normal conditions, the upper respiratory tract epithelial cells are responsible for inhibiting microbial invasion by trapping pathogens to adherence factors on the cellular surface (Masaki et al., 2011; Mata et al., 2012). In humans, the respiratory epithelial cells utilize the intercellular adhesion molecule-1 (ICAM1) molecule to capture Haemophilus influenzae (Novotny and Bakaletz, 2016). Respiratory viruses can modulate the expression of epithelial cell adhesion molecules, such as ICAM1, carcinoembryonic antigen-related cell adhesion molecule, vascular cell adhesion molecules, platelet-activating factor receptor, and fibronectin (Wang et al., 2000, 2009; Ishizuka et al., 2003; Golda et al., 2011; Gulraiz et al., 2015; Othumpangat et al., 2016). These studies suggest that respiratory viruses may play an important role in preconditioning the cell surface receptors thereby facilitating bacterial adherence to the adhesion molecules.
Previously, we demonstrated that the bacteria associated with BRDC, Pasteurella multocida (PM), adhered significantly higher to the bovine trachea epithelial cells (bTECs) than to lower bovine epithelial respiratory cells (BRECs), which are located in the bronchus (bovine bronchus epithelial cells; bBECs) and lung (bovine lung epithelial cells; bLECs) of the cow. The adherence of PM to the bTECs was markedly decreased by BRSV infection, which was not observed with either bBECs or bLECs. This suggests that BRSV infection may abolish the barrier function of the upper respiratory tract, thereby providing a “gateway” to bacterial pathogens (Sudaryatma et al., 2019). The adhesion molecules involved in the bovine respiratory “gateway” and their functions remain to be elucidated. In this study, we identified a cell surface receptor on the BRECs that is regulated by BRSV infection. We also investigated an interaction between this surface receptor and PM adherence to the bTECs.
2. Materials and Methods
2.1. Culture of bovine respiratory epithelial cells
Bovine respiratory epithelial cells (BRECs) were collected from freshly slaughtered adult Japanese black cattle (n = 3). BRECS were isolated from the bovine trachea (bTECs), bronchus (bBECs), and lung (bLECs) of the cattle, as described previously (Sudaryatma et al., 2019). Briefly, the organs were sectioned, and BRECs were isolated by suspension in isolation medium comprising Dulbecco’s modified Eagle’s medium/Nutrient Mixture F-12 GlutaMAX (DMEM/F12; Thermo Fisher Scientific, MA, US) supplemented with 15% heat-inactivated fetal bovine serum (FBS; Biowest, France), 200 U/ml penicillin, 200 mg/ ml streptomycin, 2.5 μg/ml amphotericin-B, 15 ng/ml epidermal growth factor, 1% insulin-transferrin-selenium, 1 μg/ml hydrocortisone, 1% non-essential amino acid, and 4 mM L-glutamine (all obtained from Wako, Japan). Tissues from each animal were confirmed free from BRDC-related viruses or bacteria by real-time PCR (Kishimoto et al., 2017). The isolated BRECs were maintained and subcultured every 5–7 days using culture medium comprising DMEM/F12, 2% FBS, 100 U/ml penicillin, 100 mg/ml streptomycin, 1 μg/ml amphotericin-B, 10 ng/ml epidermal growth factor, 1% insulin-transferrin-selenium, 1% non-essential amino acid, and 2 mM L-glutamine (Wako, Japan).
2.2. Virus and bacteria
The BRSV strain 2205027-1 and PM strain 2368 were used as described previously (Sudaryatma et al., 2019). For infection, virus and/or bacteria were diluted in antibiotic- and serum-free DMEM/F12 to achieve an approximate multiplicity of infection (MOI). BRSV was inactivated by treatment with ultraviolet light for 1 h (UV-inactivated BRSV). The inactivation was confirmed by plaque assay for measuring infectivity (Sudaryatma et al., 2018).
2.3. BRSV infection of BRECs
BRECs were infected with BRSV as described previously (Sudaryatma et al., 2019). Briefly, respiratory epithelial cells from different organ tissues (bTECs, bBECs, and bLECs) were seeded for 24 h on a 12-well plate coated with collagen (Sumitomo Bakelite, Japan). Cells were infected with live BRSV (MOI 0.1 or MOI 1) or UV-inactivated BRSV. Cells were washed three times with phosphate buffered saline (PBS), and incubated with culture medium. Cells were maintained for 3 days after infection.
2.4. Proteomic analysis of BRSV-infected BRECs
Cell lysates of BRSV-infected and uninfected BRECs were fixed in methanol for proteomic analysis preparation. Liquid chromatography analysis was performed using an Ultimate3000 RSLCnano (Thermo Fisher Scientific, MA, USA) and a Q-Exactive (Thermo Fisher Scientific, MA, USA). Proteins were classified as positively detected if peaks were identified in 2 out of 3 treatment groups. Protein analysis was performed using the Proteome Discoverer 2.2, and Venn diagrams were designed using the Interactivenn software (www.interactivenn.net) (Heberle et al., 2015). The differentially expressed proteins were determined by a ratio <0.67 with a t-test p-value cutoff <0.05.
2.5. Blockade of ICAM1 in BRECs with anti-ICAM1 antibody
The ICAM1 blocking assay was performed using 1, 5 or 25 μg/ml mouse anti-ICAM1 antibody (ab2213; Abcam, Japan). An isotype control antibody (25 μg/ml) was used as control. Cells were seeded onto a 12-well plate for 3 days and then treated with a serial dilution series of anti-ICAM1 antibody for 1 h at 37 °C. The cells were tested for PM adherence.
2.6. Knockdown of ICAM1 in the BRECs with siRNA
Knockdown of bovine ICAM1 genes was performed by reverse-transfection of BRECs with small interfering RNA (siRNA). Two pairs of stealth RNAi designed to target the bovine ICAM1 gene were purchased from Thermo Fisher Scientific (CA, USA). The paired sequences used were ICAM1-A: 5’-UAGCUUCAAUGAACCUCACCGUAUA-3’ (sense) and 5’-UAUACGGUGAGGUUCAUUGAAGCUA-3’ (anti-sense), and ICAM1-B: 5’-CAAGGGCUGGAACUCUUCCAGAACA-3’ (sense) and 5’-UGUUCUGGAAGAGUUCCAGCCCUUG-3’ (anti-sense). The cells were transfected with a mixture of 30 pmol siRNA and 9 μl Lipofectamine RNAiMAX Reagent (Thermo Fisher Scientific, UK) in Opti-MEM (Thermo Fisher Scientific, UK). The Stealth RNAi Negative Control (Thermo Fisher Scientific, CA, USA) was used as a non-targeting control in the experiments. At 24 h post-transfection, the culture media were replaced with fresh media, and cells were maintained for 3 days for further experiments.
2.7. Adherence assay
All treatment groups (BRSV-infected (live or UV-inactivated), uninfected control, antibody-treated, siRNA-treated, or untreated cells) were exposed to PM (MOI 100) for 1 h. Dissociated cells was lysed, and the number of adherent bacteria on the cells was quantified as colony forming unit as previously described (Sudaryatma et al., 2019). For fluorescence analysis, PM was labelled with FITC as described previously (Sudaryatma et al., 2018). Antibody-treated and siRNA-treated cells were exposed to FITC-labelled PM for 1 h and observed using an Olympus DP-74 (Olympus, Japan) confocal fluorescent microscope. The nuclei of the cells were visualized with Hoechst-33342 stain (ab145597; Abcam, Japan)).
2.8. Western blot analysis
The BRSV-infected or siRNA-treated cells seeded in a 6-well plate were lysed with 300 μl lysis buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.6, 1% NP-40, protease inhibitor, and phosphatase inhibitor) and sonicated thrice for 10 secs. The protein concentrations of the samples were determined using a bicinchoninic acid protein assay reagent kit (TaKaRa Bio, Japan). Protein per sample (10 μg) was loaded into each lane of a 4–15% SDS polyacrylamide gel (Bio-Rad Laboratories, CA, USA), separated by electrophoresis, and then transferred to a PVDF membrane (Bio-Rad Laboratories, CA, USA) using a Transblot semi-dry system (Bio-Rad Laboratories, CA, USA). The membranes were blocked overnight at 4 °C with blocking buffer (25 mM Tris, pH 8.0, 125 mM NaCl, 0.1% Tween 20, and 3% bovine serum albumin). The membranes were incubated with either mouse anti-ICAM1 (ab2213; Abcam, Japan) or mouse anti-GAPDH (ab9482; Abcam, Japan) primary antibodies at room temperature for 1 h. After vigorous washing, the membranes were incubated with secondary goat anti-mouse IgG antibody conjugated with horseradish peroxidase (ab205719; Abcam, Japan) at room temperature for 1 h. The chemiluminescent signals were detected using the western blot hyper HRP substrate (TaKaRa Bio, Japan) and a ChemiDoc Touch Imaging System (Bio-Rad Laboratories, CA, USA).
2.9. Quantification of mRNA ICAM-1 BRSV-infected BRECs
BRECs were infected with BRSV (MOI = 1). Cell lysates were collected at 6, 12, and 24 h post- infection (hpi). Total RNA was extracted from the cell lysates using a a RNeasy Mini Kit (Qiagen, Germany) according to the manufacturer’s protocol. Quantitative RT-PCR (qRT- PCR) was performed using a One Step TB Green PrimeScript plus RT-PCR kit (TaKaRa Bio, Japan). The primer pairs to amplified ICAM1 mRNA was used ICAM1-F: 5’-ACCAATTTCTCTTGCCGCTG and ICAM1-R: 5’-ACCGTGAGCCTACTTCCACA-3’. Data were normalized to expression of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Amplification was performed on a LightCycler 96 system (Roche, CT, USA). All experiments were performed in triplicate. Relative expression of mRNA between infected samples and uninfected controls was calculated using the 2−ΔΔCT method and expressed as a -fold change.
2.10. Statistical analysis
The data represent the mean and standard error (SEM) of at least three independent experiments in triplicate from three different animals. Statistical analyses were performed using one-way analysis of variance and Tukey’s Multiple Comparison Test. p-values <0.05 were considered statistically significant. Data analysis was performed using RStudio, version 1.0.143.
3. Results
3.1. Proteomic analysis in BRECs infected with BRSV
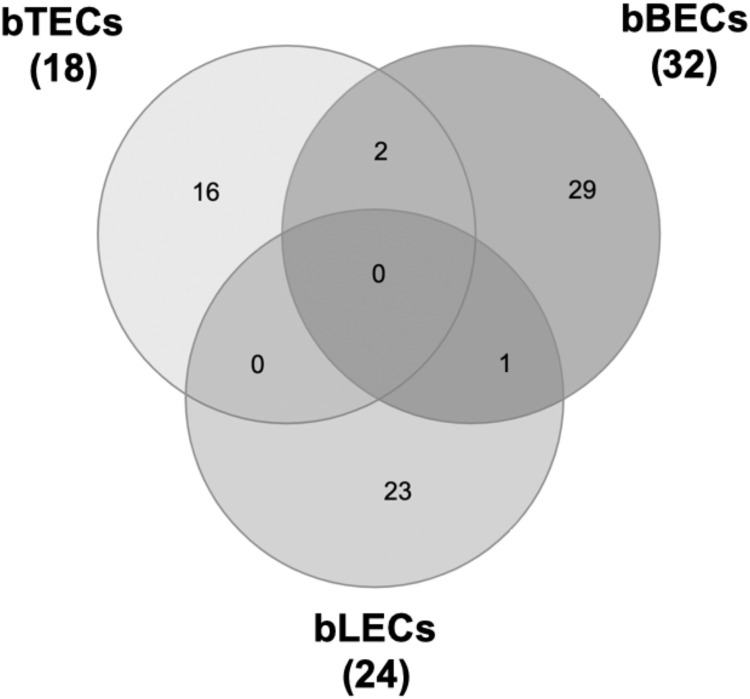
Previously, we demonstrated that the PM adherence to bTECs (but not to bBECs or bLECs) was significantly reduced by BRSV infection (Sudaryatma et al., 2019). To identify downregulated protein expression induced by BRSV infection, proteomic analysis of BRECs was performed. Proteomic analysis discovered protein changes to expression levels upon BRSV infection (bTECs: 2,523; bBECs: 2,727; and bLECs: 2,636 differentially expressed proteins). Proteins with expression levels <0.67-fold compared with uninfected control were classified as significantly downregulated (Fig. 1 ). Of these, only eighteen proteins were significantly downregulated in BRSV-infected bTECs (Supplemental Table 1). Gene ontology analysis revealed that the candidate proteins was showed to be intercellular adhesion molecule-1 (ICAM1) were involved in the membrane and protein binding pathways, according to the cellular component and molecular function annotations, respectively. Downregulated ICAM1 was showed in bTECs and bBECs infected with BRSV, but not in bLEC. Differentially expressed protein level of ICAM1 in BRSV-infected bTEC (0.118) was significantly lower (p < 0.05) than BRSV-infected bBEC (0.445).
Fig. 1.
Venn diagram illustrating downregulated candidate proteins in BRSV-infected cells. Proteomic analysis was performed comparing three different epithelial cell types isolated from the bovine respiratory tract.
3.2. BRSV-regulated ICAM1 expression in the upper respiratory tract
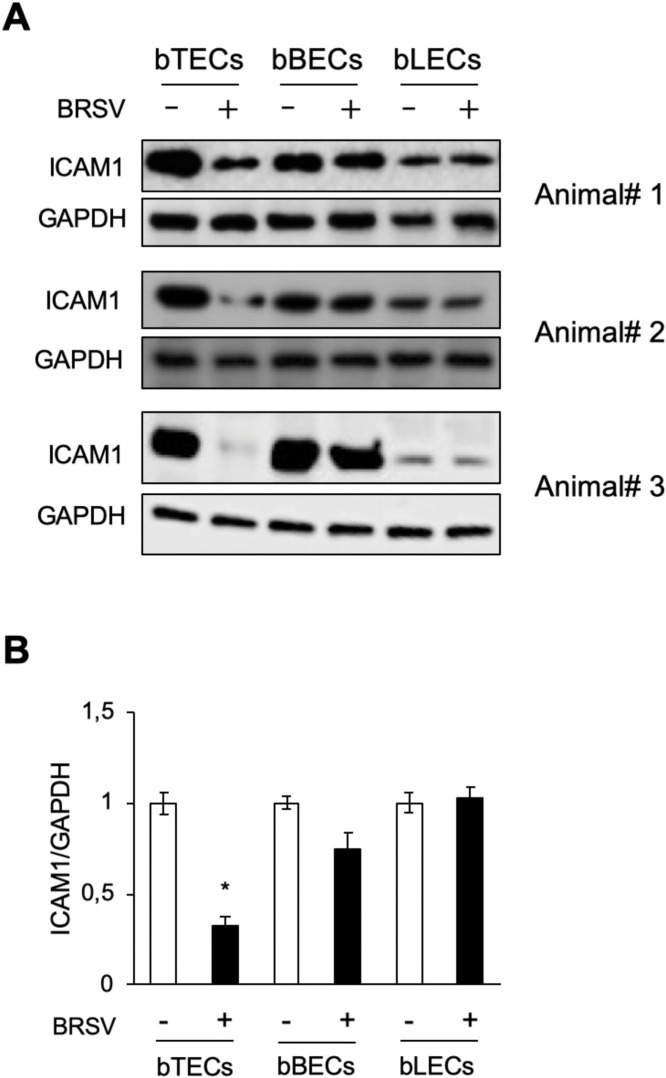
To confirm that BRSV infection reduced ICAM1 expression in the BRECs, western blot analysis was performed. As expected, ICAM1 expression decreased following BRSV treatment in both bTECs and bBECs compared with uninfected cells (Fig. 2 A). This difference was only statistically significant (p < 0.05) in bTECs (Fig. 2B). This result was supported by analysis of ICAM1 mRNA (Supplemental Fig. 1). A downregulation of ICAM1 was not observed in BRSV-infected bLECs. Interestingly, ICAM1 expression in uninfected bTECs was significantly higher than that in uninfected bBECs and bLECs (Fig. 2A). ICAM1 downregulation in bTECs was virus-dependent and correlated with the MOI used. This was based on the observation that cells infected with low MOI or UV-inactivated virus exhibited relatively normal levels of ICAM1 (Figs. 3 A and 3B).
Fig. 2.
Downregulation of ICAM1 in bTECs infected with BRSV. (A) Western blot analysis of three different epithelial cell type samples infected with BRSV (MOI 1) (+) or control (-) for 3 days. ICAMI expression was evaluated compared with positive loading control (GAPDH). (B) Quantification of relative ICAM1 expression was calculated by dividing the band density of ICAM1 by that of the GAPDH loading control. The data represent n = 3; error bars: SEM; *p < 0.05.
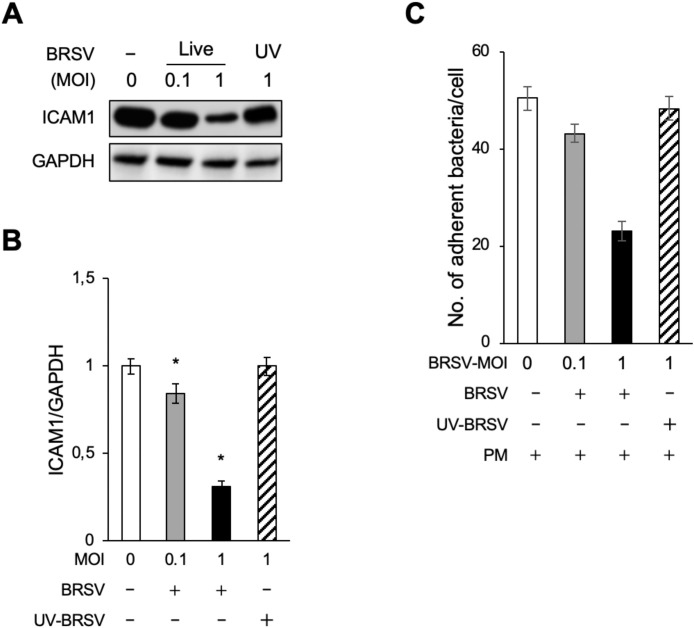
Fig. 3.
Pasteurella multocida (PM) adherence to BRSV-infected bTECs is dependent on ICAM1 expression. (A) ICAM1 expression in bTECs infected with increasing MOIs of live BRSV was evaluated by western blot. UV-inactivated virus served as a negative control. No-treatment control denoted by (-). (B) Quantification of relative ICAM1 expression level was calculated by dividing the band density of ICAM1 by that of the GAPDH loading control. The data represent n = 3; error bars: SEM. (C) PM adherence to bTECs infected with increasing MOIs of BRSV (+) was evaluated by counting PM-positive colonies. The data represent n = 3; error bars: SEM; *p < 0.05.
As BRSV infection had the greatest effect on bTEC expression of ICAM1 (Fig. 2), this cell type was used for further experiments. The bTECs, pre-infected with different MOIs of BRSV or UV-inactivated BRSV control, were exposed to PM. PM adherence to bTECs was significantly decreased (p < 0.05) in BRSV-infected cells compared with uninfected cells (Fig. 3C). This decrease was virus-dependent because minimal changes to PM adherence were observed in cells infected with low MOI or UV-inactivated virus. These results suggest that decreased PM adherence to BRSV-infected bTECs is correlated with a downregulation of ICAM1 expression.
3.3. PM adherence to upper respiratory cells is ICAM1 dependent
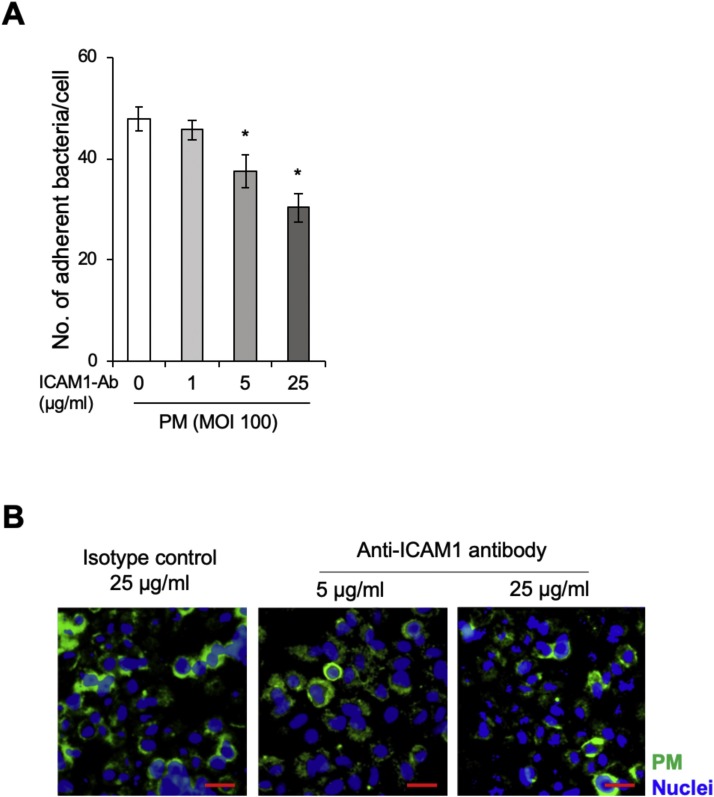
The aforementioned results suggest a link between ICAM1 expression and PM adherence to respiratory epithelial cells. To elucidate the role of ICAM1 for PM adherence, bTECs were treated with an ICAM1-specific antibody and exposed to PM. Antibody treatment was observed to decrease PM adherence to bTECs in a dose-dependent manner (Fig. 4 A). Furthermore, fluorescence microscopy analysis demonstrated that ICAM1 inhibition reduced the signal intensity of FITC-labelled PM in bTECs (Fig. 4B).
Fig. 4.
Specific inhibition of ICAM1 decreased Pasteurella multocida (PM) adherence to bTECs. (A) Bar graph representing bTEC treatment with increasing concentrations of anti-ICAM1 antibody and incubation with PM. The degree of PM adherence to the cells was evaluated by counting PM-positive colonies. The data represent n = 3; error bars: SEM; *p < 0.05. (B) FITC-labeled PM (green) adherence to the antibody-treated cells was observed with a fluorescence microscope. An isotype control antibody was used as control. The nuclei were stained with Hoechst-33342 (blue) (scale bar: 30 μm).
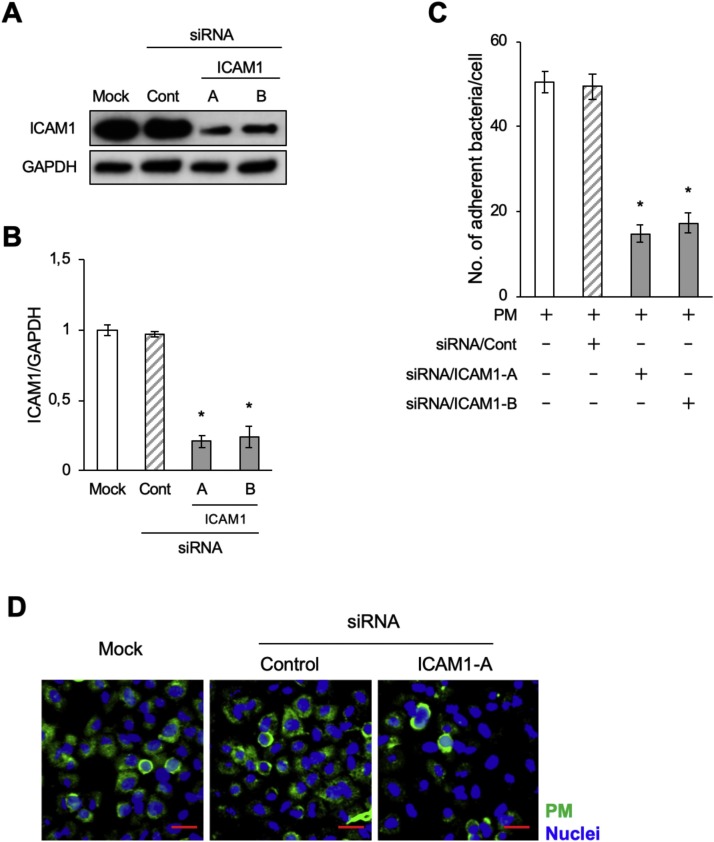
We further performed ICAM1 knockdown experiments in bTECs to examine a link between ICAM1 expression and PM adherence. The bTECs were transfected with two pairs of siRNAs (siICAM-A and siICAM-B) targeting the bovine ICAM1 gene. Significantly low levels (p < 0.05) of ICAM1 were observed in siRNA-treated cells compared with untreated cells (Figs. 5 A and 5B). Consistent with the ICAM1 inhibition experiment, a significant decrease in PM adherence to bTECs was observed by ICAM1 siRNA knockdown (p < 0.05) (Fig. 5C). Together, these results support the hypothesis that PM adherence to bTECs is ICAM1-dependent.
Fig. 5.
Knockdown of ICAM1 decreased Pasteurella multocida (PM) adherence to bTECs. (A) bTECs were transfected with siRNA targeting ICAM1 (ICAM1-A and ICAM1-B), and ICAM1 expression was evaluated by western blot. (B) The relative ICAM1 expression level was calculated by dividing the band density of ICAM1 by that of the GAPDH loading control. The data represent n = 3; error bars: SEM; *p < 0.05. (C) The degree of PM adherence to bTECs was evaluated by counting PM-positive colonies. The data represent n = 3; error bars: SEM; *p < 0.05. (D) FITC-labeled PM (green) adherence to the siRNA-transfected cells (ICAM1-A) was observed with a fluorescence microscope. The nuclei were stained with Hoechst-33342 (blue) (scale bar: 30 μm).
4. Discussion
This study provides evidence that the cell surface receptor ICAM1 may play a role in PM adherence to the epithelium of the bovine upper respiratory tract. Infection with BRSV was shown to decrease ICAM1 expression on bTECs, but not on bBECs and bLECs, of the lower respiratory tract. Further, BRSV infection caused a reduced adherence of PM to bTECs. Together, these results suggest that downregulation of ICAM1, induced by BRSV infection, leads to dysfunctional adherence of PM to the upper respiratory tract. This then facilitates PM invasion into the lower respiratory tract that, in turn, can trigger pneumonia in cattle.
The tissue tropic effects of BRSV infection (e.g., virus replication, PM adherence, cytokine induction, and innate immune activity) are known to differ between the upper and lower respiratory epithelial cells (Masaki et al., 2011; Gulraiz et al., 2015; Sudaryatma et al., 2019). In this study, proteomic analysis revealed a bTEC-specific downregulation of ICAM1 by BRSV infection compared with infected BRECs (Fig. 1, Fig. 2). ICAM1 downregulation by BRSV infection was not observed in either the bBECs or bLECs. Our quantitative RT-PCR analysis demonstrated that BRSV infection decreased the level of ICAM1 mRNA in bTECs, suggesting that BRSV infection blocked the step of transcription of mRNA. ICAM1 is expressed on a variety of cells including epithelial cells, endothelial cells, fibroblasts, and leukocytes (Avadhanula et al., 2006; Gulraiz et al., 2015). Human adenovirus, parainfluenza virus-3 and respiratory syncytial virus (HRSV) modulate ICAM1 in epithelial cells (Avadhanula et al., 2006; Novotny and Bakaletz, 2016). Interestingly, other human respiratory viruses, such as rhinovirus, coronavirus, metapneumovirus, and influenza virus, are reported also to regulate cellular surface receptors including carcinoembryonic antigen-related cell adhesion molecules-1 and platelet-activating factor, which are associated with bacterial adherence (Staphylococcus aureus, Streptococcus pneumoniae, and H. influenza) (Ishizuka et al., 2003; Golda et al., 2011; Iverson et al., 2011; Lai et al., 2016). From this point of view, it would be tempting to test whether other bovine viruses can show such effect in bTECs in the future study. In case of HRSV infection, viral NS1 and NS2 proteins decreased the expression of nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) (Liu et al., 2013), a host factor modulating ICAM1 expression (Othumpangat et al., 2016). It would be intriguing to test whether NS1 and NS2 proteins of BRSV have similar activity on NF-κB in the future study. Previously, it was reported that ICAM1 facilitates adherence of H. influenza to human respiratory epithelial cells (Avadhanula et al., 2006; Novotny and Bakaletz, 2016). Mycobacterium tuberculosis binds to ICAM1 receptors with M5 ligands, allowing it to invade THP-1 cells (Bhalla et al., 2015). To the best of our knowledge, this study is the first report examining the interaction between respiratory viruses, cell surface receptors, and bacterial adherence to the bovine respiratory tract.
In our previous study examining bovine semi-primary respiratory epithelial cells, the adherence of PM to bTECs was significantly higher than that to bBECs and bLECs under normal conditions. PM adherence to bTECs, but not to bBECs and bLECs, was significantly decreased by BRSV infection (Sudaryatma et al., 2019). The present study demonstrated that the normal ICAM1 expression level was significantly higher in uninfected bTECs than in uninfected bBECs and bLECs (Fig. 2A). Furthermore, ICAM1 knockdown by anti-ICAM1 antibody treatment or ICAM1 siRNAs resulted in decreased PM adherence to bTECs (Fig. 4, Fig. 5). These results indicate that ICAM1 may be required to capture PM on the surface of bTECs in the upper respiratory tract of cattle.
Therefore, we speculate that under normal conditions, bTECs may prevent bacterial infiltration into the lower respiratory tract by maintaining the function of ICAM1. BRSV infection might destroy this “gateway” function of the bTECs by downregulating ICAM1 expression.
In conclusion, this study demonstrated that PM adherence was regulated by the ICAM1 adhesion molecule on the surface of bTECs. BRSV infection diminished ICAM1 expression and decreased the PM adherence to bTECs. We propose that BRSV infection causes ICAM1 dysfunction in the upper respiratory tract, which may permit invasion of bacteria into the lower respiratory tract. This synergistic viral–bacterial interaction in the respiratory tract may increase the risk of pneumonia in the BRDC of cattle.
Funding
This work was supported by JSPS KAKENHI (Grant Number 17K08080) and JSPS Core-to-core Program, B. Asia-Africa Science Platform. We declare that the funding body did not contribute to any aspect of the study, including experimental design, sample collection, analysis, and interpretation of data. In addition, they were not involved in writing the report or in the decision to submit the article for publication.
Acknowledgments
The authors are grateful to Dr. Nakahata and Dr. Nakatake (Faculty of Medicine, University of Miyazaki) who performed the proteomic analysis of BRSV-infected BRECs.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.vetmic.2020.108748.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Agnes J.T., Zekarias B., Shao M., Anderson M.L., Gershwin L.J., Corbeil L.B. Bovine respiratory syncytial virus and Histophilus somni interaction at the alveolar barrier. Infect. Immun. 2013;81:2592–2597. doi: 10.1128/IAI.00108-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avadhanula V., Rodriguez C.A., Ulett G.C., Bakaletz L.O., Adderson E.E. Nontypeable Haemophilus influenzae Adheres to Intercellular Adhesion Molecule 1 (ICAM-1) on Respiratory Epithelial Cells and Upregulates ICAM-1 Expression. Infect. Immun. 2006;74:830–838. doi: 10.1128/IAI.74.2.830-838.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudeau F., Ohlson A., Emanuelson U. Associations between bovine coronavirus and bovine respiratory syncytial virus infections and animal performance in Swedish dairy herds. J. Dairy Sci. 2010;93:1523–1533. doi: 10.3168/jds.2009-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla K., Chugh M., Mehrotra S., Rathore S., Tousif S., Prakash Dwivedi V., Prakash P., Kumar Samuchiwal S., Kumar S., Kumar Singh D., Ghanwat S., Kumar D., Das G., Mohmmed A., Malhotra P., Ranganathan A. Host ICAMs play a role in cell invasion by Mycobacterium tuberculosis and Plasmodium falciparum. Nat. Commun. 2015;6:6049. doi: 10.1038/ncomms7049. [DOI] [PubMed] [Google Scholar]
- Cozens D., Sutherland E., Lauder M., Taylor G., Berry C.C., Davies R.L. Pathogenic Mannheimia haemolytica Invades Differentiated Bovine Airway Epithelial Cells. Infect. Immun. 2019:87. doi: 10.1128/IAI.00078-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle K.C., McGill J.L., Reinhardt T.A., Sacco R.E. Parainfluenza Virus 3 Blocks Antiviral Mediators Downstream of the Interferon Lambda Receptor by Modulating Stat1 Phosphorylation. J. Virol. 2016;90:2948–2958. doi: 10.1128/JVI.02502-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golda A., Malek N., Dudek B., Zeglen S., Wojarski J., Ochman M., Kucewicz E., Zembala M., Potempa J., Pyrc K. Infection with human coronavirus NL63 enhances streptococcal adherence to epithelial cells. J. Gen. Virol. 2011;92:1358–1368. doi: 10.1099/vir.0.028381-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulraiz F., Bellinghausen C., Bruggeman C.A., Stassen F.R. Haemophilus influenzae increases the susceptibility and inflammatory response of airway epithelial cells to viral infections. FASEB J. 2015;29:849–858. doi: 10.1096/fj.14-254359. [DOI] [PubMed] [Google Scholar]
- Härtel H., Nikunen S., Neuvonen E., Tanskanen R., Kivelä S.-L., Aho P., Soveri T., Saloniemi H. Viral and Bacterial Pathogens in Bovine Respiratory Disease in Finland. Acta Vet. Scand. 2004;45:2. doi: 10.1186/1751-0147-45-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberle H., Meirelles V.G., da Silva F.R., Telles G.P., Minghim R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015 doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka S., Yamaya M., Suzuki T., Takahashi H., Ida S., Sasaki T., Inoue D., Sekizawa K., Nishimura H., Sasaki H. Effects of Rhinovirus Infection on the Adherence of Streptococcus pneumoniae to Cultured Human Airway Epithelial Cells. J. Infect. Dis. 2003;188:1928–1939. doi: 10.1086/379833. [DOI] [PubMed] [Google Scholar]
- Iverson A.R., Boyd K.L., McAuley J.L., Plano L.R., Hart M.E., McCullers J.A. Influenza virus primes mice for pneumonia from Staphylococcus aureus. J. Infect. Dis. 2011;203:880–888. doi: 10.1093/infdis/jiq113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston D., Earley B., McCabe M.S., Lemon K., Duffy C., McMenamy M., Cosby S.L., Kim J., Blackshields G., Taylor J.F., Waters S.M. Experimental challenge with bovine respiratory syncytial virus in dairy calves: bronchial lymph node transcriptome response. Sci. Rep. 2019;9:14736. doi: 10.1038/s41598-019-51094-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto M., Tsuchiaka S., Rahpaya S.S., Hasebe A., Otsu K., Sugimura S., Kobayashi S., Komatsu N., Nagai M., Omatsu T., Naoi Y., Sano K., Okazaki-Terashima S., Oba M., Katayama Y., Sato R., Asai T., Mizutani T. Development of a one-run real-time PCR detection system for pathogens associated with bovine respiratory disease complex. J. Vet. Med. Sci. 2017;79:517–523. doi: 10.1292/jvms.16-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König P., Giesow K., Schuldt K., Buchholz U.J., Keil G.M. A novel protein expression strategy using recombinant bovine respiratory syncytial virus (BRSV): Modifications of the peptide sequence between the two furin cleavage sites of the BRSV fusion protein yield secreted proteins, but affect processing and funct. J. Gen. Virol. 2004;85:1815–1824. doi: 10.1099/vir.0.80010-0. [DOI] [PubMed] [Google Scholar]
- Lai S.H., Liao S.L., Wong K.S., Lin T.Y. Preceding human metapneumovirus infection increases adherence of Streptococcus pneumoniae and severity of murine pneumococcal pneumonia. J. Microbiol. Immunol. Infect. 2016;49:216–224. doi: 10.1016/j.jmii.2014.04.008. [DOI] [PubMed] [Google Scholar]
- Larsen L.E., Tegtmeier C., Pedersen E. Bovine respiratory syncytial virus (BRSV) pneumonia in beef calf herds despite vaccination. Acta Vet. Scand. 2001;42:113–121. doi: 10.1186/1751-0147-42-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.W., Lee T.L., Chen Y.C., Liang C.J., Wang S.H., Lue J.H., Tsai J.S., Lee S.W., Chen S.H., Yang Y.F., Chuang T.Y., Chen Y.L. PM2.5-induced oxidative stress increases intercellular adhesion molecule-1 expression in lung epithelial cells through the IL-6/AKT/STAT3/NF-κB-dependent pathway. Part. Fibre Toxicol. 2018;15:4. doi: 10.1186/s12989-018-0240-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Qin X., Xiang Y., Liu H., Gao G., Qin L., Liu C., Qu X. Progressive Changes in Inflammatory and Matrix Adherence of Bronchial Epithelial Cells with Persistent Respiratory Syncytial Virus (RSV) Infection (Progressive Changes in RSV Infection) Int. J. Mol. Sci. 2013;14(9):18024–18040. doi: 10.3390/ijms140918024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki T., Kojima T., Okabayashi T., Ogasawara N., Ohkuni T., Obata K., Takasawa A., Murata M., Tanaka S., Hirakawa S., Fuchimoto J., Ninomiya T., Fujii N., Tsutsumi H., Himi T., Sawada N. A nuclear factor-κB signaling pathway via protein kinase C δ regulates replication of respiratory syncytial virus in polarized normal human nasal epithelial cells. Mol. Biol. Cell. 2011;22:2144–2156. doi: 10.1091/mbc.e10-11-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata M., Sarrion I., Armengot M., Carda C., Martinez I., Melero J.A., Cortijo J. Respiratory Syncytial Virus Inhibits Ciliagenesis in Differentiated Normal Human Bronchial Epithelial Cells: Effectiveness of N-Acetylcysteine. PLoS One. 2012;7 doi: 10.1371/journal.pone.0048037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny L.A., Bakaletz L.O. Intercellular adhesion molecule 1 serves as a primary cognate receptor for the Type IV pilus of nontypeable Haemophilus influenzae. Cell. Microbiol. 2016;18:1043–1055. doi: 10.1111/cmi.12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othumpangat S., Noti J.D., McMillen C.M., Beezhold D.H. ICAM-1 regulates the survival of influenza virus in lung epithelial cells during the early stages of infection. Virology. 2016;487:85–94. doi: 10.1016/j.virol.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudaryatma P.E., Mekata H., Kubo M., Subangkit M., Goto Y., Okabayashi T. Co-infection of epithelial cells established from the upper and lower bovine respiratory tract with bovine respiratory syncytial virus and bacteria. Vet. Microbiol. 2019;235:80–85. doi: 10.1016/j.vetmic.2019.06.010. [DOI] [PubMed] [Google Scholar]
- Sudaryatma P.E., Nakamura K., Mekata H., Sekiguchi S., Kubo M., Kobayashi I., Subangkit M., Goto Y., Okabayashi T. Bovine respiratory syncytial virus infection enhances Pasteurella multocida adherence on respiratory epithelial cells. Vet. Microbiol. 2018;220:33–38. doi: 10.1016/j.vetmic.2018.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timsit E., Workentine M., Schryvers A.B., Holman D.B., van der Meer F., Alexander T.W. Evolution of the nasopharyngeal microbiota of beef cattle from weaning to 40 days after arrival at a feedlot. Vet. Microbiol. 2016;187:75–81. doi: 10.1016/j.vetmic.2016.03.020. [DOI] [PubMed] [Google Scholar]
- Tjønehøj K., Uttenthal Å., Viuff B., Larsen L.E., Røntved C., Rønsholt L. An experimental infection model for reproduction of calf pneumonia with bovine respiratory syncytial virus (BRSV) based on one combined exposure of calves. Res. Vet. Sci. 2003 doi: 10.1016/S0034-5288(02)00154-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.H., Kwon H.J., Jang Y.J. Rhinovirus enhances various bacterial adhesions to nasal epithelial cells simultaneously. Laryngoscope. 2009;119:1406–1411. doi: 10.1002/lary.20498. [DOI] [PubMed] [Google Scholar]
- Wang S.Z., Hallsworth P.G., Dowling K.D., Alpers J.H., Bowden J.J., Forsyth K.D. Adhesion molecule expression on epithelial cells infected with respiratory syncytial virus. Eur. Respir. J. 2000;15:358–366. doi: 10.1034/j.1399-3003.2000.15b23.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.