Abstract
Natural killer (NK) cells are cytotoxic innate lymphoid cells (ILCs) that mediate antiviral and antitumor responses and require the transcriptional regulator Eomesodermin (Eomes) for early development. However, the role of Eomes and its molecular program in mature NK cell biology is unclear. To address this, we develop a tamoxifen-inducible, type-1-ILC-specific (Ncr1-targeted) cre mouse and combine this with Eomes-floxed mice. Eomes deletion after normal NK cell ontogeny results in a rapid loss of NK cells (but not ILC1s), with a particularly profound effect on penultimately mature stage III NK cells. Mechanisms responsible for stage III reduction include increased apoptosis and impaired maturation from stage II precursors. Induced Eomes deletion also decreases NK cell cytotoxicity and abrogates in vivo rejection of major histocompatibility complex (MHC)-class-I-deficient cells. However, other NK cell functional responses, and stage IV NK cells, are largely preserved. These data indicate that mature NK cells have distinct Eomes-dependent and -independent stages.
Keywords: NK cell, Ncr1-specific, Eomes, cytotoxicity, homeostasis, inducible-cre model, innate lymphoid cell, maturation, in vivo
Graphical Abstract

The transcription factor Eomes is important for early natural killer (NK) cell development. Wagner et al. utilize an inducible, type 1 ILC-specific cre model to demonstrate a stage-specific role for Eomes in NK cell survival and homeostasis as well as a persistent requirement for Eomes in promoting NK cell cytotoxicity.
Introduction
Natural killer (NK) cells are the founding members of a family of innate lymphoid cells (ILCs) (Colonna, 2018) that also includes helper ILC1s, ILC2s, and ILC3s (Cherrier et al., 2018). NK cells protect against infection and mediate antitumor responses via two primary effector functions: production of immunomodulatory cytokines and direct cytotoxicity (Caligiuri, 2008, Vivier et al., 2008). NK cells recognize target cells based on the integration of signals from numerous germline DNA-encoded activating and inhibitory receptors. In general, activating receptors recognize cell stress ligands, while inhibitory receptors recognize major histocompatibility complex (MHC) class I molecules (Lanier, 2005). In addition, NK cells constitutively express cytokine receptors that support their development and tune their function by ligating activating or inhibitory signals in the surrounding environment (Romee et al., 2014, Yang et al., 2010).
The developmental and transcription factor programs of NK cells are distinct from those of helper ILCs. ILC development begins in the bone marrow (BM) from the common lymphoid progenitor (CLP), which gives rise to both the NK and helper ILC lineages. Thereafter, most terminal NK cell maturation takes place in peripheral tissues (Cherrier et al., 2018, Yu et al., 2013). The NK progenitor (NKP) differentiates through several NK cell maturation stages classically characterized based on the presence or absence of the surface markers CD27 and CD11b (Chiossone et al., 2009). In addition, the activating receptor NKp46, encoded by Ncr1, is induced during stage I (CD27−CD11b−) of differentiation, followed by the integrin CD49b (DX5), and MHC-class-I-recognizing Ly49 activating and inhibitory receptors (Yu et al., 2013). Numerous transcription factors are required to regulate ILC development and promote the different lineages (Ishizuka et al., 2016, Leong et al., 2017). NK cells and ILC1s both express T-bet, but of type-1 ILCs, only NK cells express Eomesodermin (Eomes), another T-box transcription factor critical for their development (Colonna, 2018). Recent data have revealed that NK cells and ILC1s exhibit plasticity depending on their environment and specifically that NK cells can convert to ILC1-like cells in transforming growth factor β (TGF-β)-rich tumors and tissues (Berrien-Elliott et al., 2019, Cortez et al., 2017, Gao et al., 2017), which can involve downregulation of Eomes.
However, the mechanisms whereby modulation of Eomes could alter NK cellular identity and function is unclear. Eomes, like T-bet, regulates gene expression by binding T-box DNA elements. Eomes is generally required for early embryonic development, as evidenced by mice with global Eomes gene deletion exhibiting embryonic lethality (Russ et al., 2000). An important role for Eomes has been demonstrated in CD8 T cell biology; it promotes the development of central memory CD8 T cells as well as the cytotoxic T cell effector program by positively regulating interferon γ (IFN-γ), perforin, and granzyme B (Banerjee et al., 2010, Intlekofer et al., 2005, Pearce et al., 2003). Eomes is also critical for NK cell development, and mice with hematopoietic- or constitutive NK-cell-specific Eomes deletion display a severe and global decrease in conventional NK cell numbers but preserved ILC1s (Daussy et al., 2014, Gordon et al., 2012, Pikovskaya et al., 2016). Eomes is expressed by NK cells of all maturation stages, with highest expression in the relatively immature stage II and III populations (Gordon et al., 2012). NK cell terminal maturation is thought to rely on T-bet, whose expression increases in terminally mature (stage IV) NK cells, opposite to the decrease in Eomes (Daussy et al., 2014, Townsend et al., 2004). Functionally, Eomes expression correlates with NK cell IFN-γ production in vivo (Gill et al., 2012) and Prf1 transcription (Pearce et al., 2003), but T-bet has also been shown to regulate NK cell cytotoxic protein expression (Townsend et al., 2004). Thus, the importance of Eomes in mature NK cell homeostasis and function remains unclear.
Studies of Eomes in NK cell homeostasis and function have been limited by a lack of appropriate inducible genetic models. In the constitutive Ncr1-cre models available (and similarly for Vav1-cre), cre-driven Eomes deletion in immature BM NK cells results in abrogation of NK cell development and thus precludes the study of Eomes in mature NK cell biology (Gordon et al., 2012, Pikovskaya et al., 2016). Indeed, what were once thought to be Eomes-negative NK cells in such constitutive models are now better understood to have been type 1 ILCs (Gordon et al., 2012). To overcome this limitation, we developed a type-1-ILC-specific tamoxifen-inducible cre mouse model and confirmed its properties using a Rosa26-YFP reporter allele. Eomes-floxed mice were crossed to this model in order to elucidate the role of Eomes in mature NK cell biology. Here, we studied the impact of induced Eomes deletion on mature NK cell homeostasis and function. We demonstrate that there are Eomes-dependent (stage II/III) and independent (stage IV) murine NK cell subsets and that loss of Eomes does not reverse NK cell maturation, as has been previously suggested (Gordon et al., 2012). In addition, we demonstrate a requirement of ongoing Eomes expression for NK cell cytotoxicity and in vivo responses to MHC-I-deficient target cells.
Results
The Ncr1-iCreERT2 Tamoxifen-Inducible cre Model Specifically Activates cre within Type 1 ILCs
Mouse models with constitutive type 1 ILC-specific cre expression utilizing Ncr1 regulatory elements (Eckelhart et al., 2011, Narni-Mancinelli et al., 2011) have limitations. In these models, cre expression initiates with normal Ncr1 gene expression in immature BM stage I NK cells (Walzer et al., 2007). Hence, loxP-directed excision events occur early in NK cell development, precluding the study of developmentally critical genes in mature NK cells. To overcome this barrier, we developed a type-1-ILC-specific, tamoxifen-inducible cre mouse (Figure 1 A) generated by genetic targeting of a tamoxifen-responsive iCreERT2 cassette into the Ncr1 locus. This cassette is linked to NKp46 C-terminal translation via a P2A ribosomal skip site. This Ncr1-iCreERT2 model was crossed to a reporter allele consisting of a lox-stop-lox (LSL)-flanked YFP cassette genetically targeted to the Rosa26 locus in order to track cre nuclear activity (Srinivas et al., 2001). To test the timing of cre expression in this model, mice underwent oral gavage with 3 mg tamoxifen for 3 consecutive days (Heger et al., 2014, Herold et al., 2014), and 3 days later, YFP expression was analyzed in various tissues (Figure 1B). YFP expression was observed in NKp46+ cells of the blood, spleen, and liver (≥90% YFP+) as well as BM and lymph node (LN) (≥80% YFP+). YFP expression was restricted to NKp46+ cells and not expressed by other hematopoietic lineages, including T cells (Figure 1B; data not shown). Similar to other iCreERT2 models (Kristianto et al., 2017, Maurel et al., 2019), mature (8- to 12-week-old) Ncr1-iCreERT2 mice had small background levels of cre nuclear localization (∼5%–10%) in NKp46+ cells in the absence of tamoxifen that increased slowly over time (Figures 1B and S1). Therefore, in this report, experiments were performed in 8- to 12-week-old mice unless otherwise noted.
Figure 1.
Tamoxifen Induces Robust and Type-1-ILC-Specific cre Activity in Mice Harboring the Ncr1-iCreERT2 Knockin Locus
(A) Schematic depicting the Ncr1-iCreERT2 locus and mouse model.
(B) Mean ± SEM type 1 ILC (NK1.1+CD3-) and T cell (CD3+) percent YFP+ in the blood (Bl), spleen (Sp), bone marrow (BM), lymph node (LN), and liver (Liv) of Ncr1-iCreERT2 × Rosa26YFPLSL mice 2 days following three consecutive daily doses of 3 mg tamoxifen in corn oil (or no tamoxifen [No Tam]). Representative flow cytometry data from the spleen of a tamoxifen-treated mouse shown to the right. n = 2–3 mice per group, two independent experiments.
(C) Ncr1-iCreERT2 × Rosa26YFPLSL mice were treated with three consecutive daily doses of tamoxifen. Summary data show mean ± SEM. YFP positivity among splenic NK cells, liver NK cells (CD49a− DX5+) and liver ILC1 cells (CD49a+ DX5−) weekly for 5 weeks thereafter. n = 2 mice per group, one experiment.
(D) Schematic of tamoxifen regimens.
(E) PCR was performed for the presence of Eomes floxed (fl; 730 bp) or WT (620 bp) alleles on heterozygous Eomesfl/WT control genomic DNA (lanes 1 and 2) and YFP+ splenic NK cells (NK1.1+ CD3−, lane 3) or T cells (CD3+, lane 4) sorted from ILC-EomesΔ/Δ mice treated with the Tam-3d regimen. Empty lanes were cropped as shown by the border.
(F) Tam-6d-treated Eomes WT and ILC-EomesΔ/Δ mouse tissues were analyzed for Eomes expression. Summary data show mean ± SEM percent Eomes positivity in YFP+NKp46+ cells. Representative flow histograms show Eomes protein in Eomes WT (shaded blue line) and ILC-EomesΔ/Δ NK cells (empty blue line), and NK1.1− CD3− lymphocytes (background, shaded gray) in the spleen and BM. n = 5 Eomes WT mice, 9 ILC-EomesΔ/Δ mice, two independent experiments.
This model is also useful to assess cellular dynamics within the NKp46+ ILC compartment. NK cells and tissue-resident ILC1s were compared in a 3-day tamoxifen “pulse” YFP “chase” experiment. Here, we treated Ncr1-iCreERT2 × Rosa26YFPLSL mice with tamoxifen and then assessed splenic and liver NK cells as well as liver ILC1s for YFP positivity over the next 5 weeks. The proportion of YFP+ liver ILC1s remained high (≥90%), consistent with replenishment from NKp46+ tissue-resident populations and/or less cellular attrition. In contrast, splenic and liver NK cells displayed considerable turnover, and YFP positivity returned to near-baseline levels by the end of the 5-week period (Figure 1C). Thus, Ncr1-iCreERT2 x Rosa26YFPLSL mice exhibited robust and specific cre activity in NKp46+ ILCs after tamoxifen administration, which was tracked in subsequent experiments using YFP. For the remainder of the study, experiments were performed at three time points relative to tamoxifen administration: Tam-3d, Tam-6d, and Tam-9d (Figure 1D).
Tamoxifen Rapidly Eliminates Eomes in NKp46+ Cells of Ncr1-iCreERT2 × Eomesfl/fl Mice
We next crossed Ncr1-iCreERT2 × Rosa26YFPLSL mice to mice with floxed Eomes alleles (Zhu et al., 2010). Eomes allele excision was confirmed in splenocytes of Ncr1-iCreERT2 KI/WT × Rosa26YFPLSL × Eomes fl/fl (henceforth ILC-EomesΔ/Δ) mice after the Tam-3d regimen, as PCR analysis of the Eomes locus revealed excision in flow-sorted YFP+ NK cells but not YFP− T cells (Figure 1E). This was verified at the protein level after the Tam-6d regimen using flow cytometry in ILC-EomesΔ/Δ compared to control NK cells (Figure 1F). Thus, tamoxifen-induced cre efficiently translocated to the nucleus and excised Eomes in mature NK cells within 2 days.
Induced Eomes Deletion Results in a Rapid Loss of NK Cells, Most Prominently Stage III
To assess the impact of induced Eomes deletion on the NK cell compartment, we treated ILC-EomesΔ/Δ and control mice with the Tam-6d regimen and then assessed NK cell numbers and maturation. We observed a significant decrease in global YFP+ NK cell numbers in ILC-EomesΔ/Δ compared to wild-type (WT) control mice in all tissues examined (blood, spleen, BM, LN, and liver; Figure 2 A). Notably, induced Eomes deletion had a particularly profound effect on less mature stage II (CD27+CD11b−) and stage III (CD27+CD11b+) NK cells. Stage III NK cells, in particular, were significantly decreased in number and percentage in all tissues analyzed (Figure 2B). While stage IV (CD27−CD11b+) NK cell numbers were reduced in the blood, BM, and LN in ILC-EomesΔ/Δ mice, their relative proportion increased in all tissues except the liver, where it was unchanged. As expected, Eomes-dependent NK cells were decreased in both the proportion of YFP+ NKp46+ cells and absolute number in the liver, while the proportion of Eomes-independent ILC1s increased, but numbers remained unchanged (Figure 2C) (Sojka et al., 2014). Despite evidence that Eomes and T-bet negatively cross-regulate one another (Daussy et al., 2014), we did not observe increased T-bet protein levels in ILC-EomesΔ/Δ NK cells (Figure 2D). Thus, induced Eomes deletion negatively affects NK cell numbers and homeostasis, particularly those of the stage III subset.
Figure 2.
Induced Eomes Deletion Results in a Significant Decrease in Global NK Cell Numbers, with
a Particularly Profound Loss of Stage III NK Cells
(A) Mean ± SEM. YFP+ NKp46+ cell percentage of lymphocytes and number in the blood (Bl; per mL), spleen (Sp), BM (per femur), LN (per inguinal LN), or liver (Liv) of ILC-EomesΔ/Δ versus WT mice following the Tam-6d regimen.
(B) Summary data show mean ± SEM. YFP+ NK1.1+ cell stage-specific percentages and numbers in ILC-EomesΔ/Δ or Eomes WT mice in the five different tissues. ILC1s are included in liver gating. Representative flow cytometry data are shown for the spleen.
(C) Summary data show mean ± SEM. YFP+ NK cell (CD49a− DX5+) versus ILC1 (CD49a+ DX5−) percentage and numbers in the liver. n = 9 ILC-EomesD/D mice, 5 Eomes WT mice, two independent experiments.
(D) Summary data show mean ± SEM. T-bet MFI in YFP+ ILC-EomesΔ/Δ and WT NKp46+ cells across the type 1 ILC compartment, normalized to the mean T-bet MFI of ILC-EomesΔ/Δ NKp46+ cells in each tissue. n = 9 ILC-EomesΔ/Δ mice, 5 Eomes WT mice, two independent experiments. Data were compared using false-discovery-rate-corrected t tests.
*p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Loss of Stage III NK Cells in ILC-EomesΔ/Δ Mice Results from Apoptosis and Altered Cell-Cycling Dynamics
We hypothesized that the decrease in NK cell numbers observed following Eomes deletion could be the result of enhanced apoptosis or altered cell cycling. This was tested at an earlier time point after tamoxifen gavage (Tam-3d), when cells undergoing apoptosis or attempting to proliferate may still be quantifiable. At this time point, >70% of splenic ILC-EomesΔ/Δ and control NK cells were YFP+, Eomes protein was nearly absent, and reductions in NK cells had started but were more modest compared to the Tam-6d time point (Figures 3A and 3B). A significantly greater fraction of apoptotic (Annexin V+) stage III NK cells were present in the spleen and BM of ILC-EomesΔ/Δ compared to WT mice (Figure 3C). This was associated with a significant increase in caspase activation in splenic stage III NK cells of ILC-EomesΔ/Δ compared to control mice (Figure 3D). To further understand this apoptosis phenotype, we performed unbiased RNA sequencing (RNA-seq) of flow-sorted stage II, III, and IV ILC-EomesΔ/Δ and WT splenic NK cells. Gene set enrichment analysis (GSEA) of the RNA-seq data revealed significant enrichment of the Hallmark Apoptosis Pathway in ILC-EomesΔ/Δ, compared to WT, stage II NK cells (Figure 3E). This enrichment was not observed in stage III or IV NK cell comparisons (Figure S2). These data are consistent with the observed loss of stage III NK cells, as they suggest that induced Eomes deletion activates apoptotic machinery in stage II NK cells that die at stage III.
Figure 3.
Induced Eomes Deletion Results in Increased Stage III NK Cell Apoptosis and Altered Cell-Cycling Dynamics
ILC-EomesΔ/Δ and Eomes WT mice were treated with the Tam-3d regimen, and then apoptosis and cell cycling dynamics were assessed.
(A) YFP expression, percentage, and number of splenic ILC-EomesΔ/Δ and Eomes WT NK cells. n = 5–6 mice per group, two independent experiments.
(B) Mean ± SEM percentage of Eomes+ splenic NK cells. Representative flow histogram shows Eomes protein in splenic Eomes WT (shaded blue line) and ILC-EomesΔ/Δ NK cells (empty blue line), and NK1.1- CD3- lymphocytes (background, shaded gray). n = 8 ILC-EomesΔ/Δ mice, 4 Eomes WT mice, two independent experiments.
(C) Mean ± SEM. Annexin V+ ILC-EomesΔ/Δ or Eomes WT NK cells by stage in the BM and spleen. n = 12–17 mice per group, four to seven independent experiments.
(D) Mean ± SEM percent of ILC-EomesΔ/Δ or Eomes WT splenic stage II or III NK cells with activated poly-caspase expression. n = 5–6 mice per group, two independent experiments.
(E) GSEA of RNA-seq data from ILC-EomesΔ/Δ and Eomes WT stage II NK cells. An enrichment plot for the hallmark apoptosis pathway is shown to the left, with all significantly enriched or downregulated pathways shown to the right.
(F) Mean ± SEM proportion of G0, G1, and S/G2/M stage III Eomes WT and ILC-EomesΔ/Δ NK cells in the spleen and BM, with representative flow plots shown. n = 9–12 mice per group, three or four independent experiments.
(G) Mean ± SEM proportion of G0, G1, and S/G2/M stage III ILC-EomesΔ/Δ and Eomes WT NK cells in the spleen and BM one day after the Tam-3d regimen. n = 6 mice per group, two independent experiments. Data were compared using t tests with false discovery rate correction where appropriate.
*p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
ILC-EomesΔ/Δ and WT NK cells were also co-stained for Ki67 (a marker of proliferation) and DNA content to assess cell-cycle dynamics (Figures 3F, 3G, and S3A). The cell-cycle distribution of bulk, stage II, and stage IV NK cells was identical between WT and ILC-EomesΔ/Δ mice in the spleen and BM (Figure S3B). However, a significantly greater percentage of stage III NK cells were in G1 in ILC-EomesΔ/Δ compared to WT mice (Figure 3F). No cells progressed to S/G2/M, however, even when analyzed an additional day after the Tam-3d regimen (Figure 3G). This suggests that induced Eomes deletion promotes a G0 to G1 transition in stage III NK cells, yet the cell cycle is unable to progress beyond that point, possibly due to cell-cycle-arrest-related apoptosis. Eomes-deleted NK cells are capable of cell division, however, as demonstrated via transfer experiments into a homeostatic proliferation environment (Figure S4). Here, both ILC-EomesΔ/Δ and Eomes WT YFP+ splenic NK cells showed robust and comparable proliferation 14 days post-transfer into Rag2−/−γc−/− recipient mice (Figure S4B). While restrictions on our flow panel did not allow for stage-specific analyses, given the deleterious effect of Eomes on stage III NK cell numbers, we hypothesize that the proliferated ILC-EomesΔ/Δ NK cells were largely stage II and IV. Short-term proliferation of ILC-EomesΔ/Δ and WT YFP+ NK cells after 4 days in vivo was also similar but significantly reduced compared to cre-negative NK cells, suggesting that induction of cre concurrent with a strong proliferation signal may alter normal cellular proliferation (Figure S4A) (Loonstra et al., 2001). Collectively, these findings suggest that Eomes loss promotes stage III apoptosis either by directly activating apoptotic genes in precursor stage II NK cells or as a result of cell-cycle arrest.
IL-15 Signaling Is Altered in Stage II and III NK Cells as a Result of Induced Eomes Deletion but Does Not Account for Stage III NK Cell Loss
Interleukin-15 (IL-15) is the major cytokine involved in NK cell development, survival, and proliferation. Three major pathways are activated downstream of the IL-15 receptor (IL-15R): JAK/STAT (JAK1/3 and STAT3/5), mitogen-activated protein kinase (MAPK), and phosphatidylinositol 3-kinase (PI3K) (Mishra et al., 2014). STAT5, in particular, is known to be critical for NK cell survival, since it promotes the transcription of anti-apoptotic molecules like Bcl-2 and Mcl-1 (Huntington et al., 2007, Ranson et al., 2003). Since Eomes is known to regulate the β chain of the IL-15R (CD122) (Intlekofer et al., 2005), we hypothesized that induced Eomes deletion resulted in decreased expression of the intermediate-affinity (β/γ) IL-15R and thus decreased IL-15 signaling and STAT5 phosphorylation in vivo, leading to stage III NK cell apoptosis. Indeed, a significant decrease in CD122 expression on both stage II/III and stage IV Tam-3d-treated ILC-EomesΔ/Δ compared to WT NK cells was observed (Figure 4 A). We used phospho-flow cytometry to interrogate activation of downstream signaling pathways in response to low-dose (10 ng/mL) and high-dose (100 ng/mL) IL-15 in NK cells from WT and ILC-EomesΔ/Δ mice treated with the Tam-3d regimen (Figure 4B). Unexpectedly, we observed no defect in pSTAT5 activation at either IL-15 concentration tested and, accordingly, no difference in Bcl-2 protein expression between ILC-EomesΔ/Δ and WT NK cells (Figure S5A). In contrast, pERK and pAkt median fluorescence intensity (MFI) was decreased in ILC-EomesΔ/Δ compared to WT stage II/III NK cells in response to low-dose, but not high-dose, IL-15. This is consistent with higher levels of IL-15 being required to activate these pathways, compared to STAT5, in murine NK cells (Marçais et al., 2014). While they are not considered to be the principal regulators of NK cell apoptosis, the PI3K/Akt/mTOR and MAPK pathways have been shown to play important roles in promoting NK cell survival (Huntington et al., 2007). However, stage III NK cells were not rescued in mice that received exogenous IL-15 concomitantly with tamoxifen (Figures S5B–S5D). Thus, the negative effect of Eomes deletion on stage III NK cell survival is likely not the result of impaired IL-15 signaling.
Figure 4.
Induced Eomes Deletion Results in Modest Differences in IL-15 Signaling
(A) Mean ± SEM. CD122 MFI on stage II/III (CD27+) versus stage IV (CD27−CD11b+) splenic NK cells from Eomes WT and ILC-EomesΔ/Δ mice treated with the Tam-3d regimen. Representative flow histograms also shown. n = 6 mice per group, two independent experiments.
(B) Mean ± SEM pSTAT5, pERK, and pAkt MFI fold change over baseline (unstimulated) in splenic NK cells from Eomes WT and ILC-EomesΔ/Δ mice treated with the Tam-3d regimen. Cells were stimulated with 10 or 100 ng/mL IL-15 for 15 min (pSTAT5) or 1 h (pERK, pAkt). n = 7–10 mice per group, three or four independent experiments. Data were compared using t tests.
*p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Induced Eomes Deletion Impairs NK Cell Maturation In Vivo
The PI3K and MAPK pathways are involved in other aspects of NK cell homeostasis, including maturation (Tassi et al., 2007). We hypothesized that the stage III NK cell defect might also be due to impaired maturation from precursor stage II cells. To test this hypothesis, we treated ILC-EomesΔ/Δ and WT mice with the Tam-3d regimen then sorted YFP+ CD27+ (stage II/III) or YFP+ CD27− CD11b+ (stage IV) NK cells, transferred them into congenic recipients, and allowed them to mature for 2 weeks in vivo (Figure 5 A) (Sullivan et al., 2015). Stage IV NK cells from both ILC-EomesΔ/Δ and WT control mice were stable and remained stage IV after 2 weeks (Figure 5B), contrary to previous results utilizing ex vivo TAT-cre-induced Eomes deletion (Gordon et al., 2012). However, the majority of ILC-EomesΔ/Δ NK cells that were stage II/III at the time of transfer were stage II after 2 weeks in vivo, with only a small number of stage IV NK cells developing. This was in contrast to WT cells that showed expected in vivo maturation (Figures 5B and 5C). To account for any bias introduced by sorting on YFP+ cells, this experiment was repeated by sorting stage II/III or stage IV cell trace violet (CTV)-labeled NK cells from ILC-EomesΔ/Δ or CD45.1 WT mice prior to tamoxifen, and co-transferring cells of the same stage into CD45.2 recipients. The recipient mice were then treated with 3 days of tamoxifen, and in vivo maturation was assessed 2 weeks later with similar results (Figure S6). These in vivo maturation experiments indicate that Eomes is required for normal stage II to III NK cell maturation. Thus, the particularly profound defect in stage III NK cells following Eomes deletion also arises from a stage II to III maturation block.
Figure 5.
Eomes Is Required for Normal NK Cell Maturation
(A) ILC-EomesΔ/Δ and Eomes WT mice were treated with the Tam-3d regimen. Splenic NK cells were sorted to YFP+CD27+ (stage II/III) or YFP+CD27−CD11b+ (stage IV) and transferred into CD45.2 WT recipients. 2 weeks later, YFP+ cell maturation was assessed.
(B) Stage distribution of ILC-EomesΔ/Δ or Eomes WT sorted stage II/III or IV NK cells before (light blue) or 2 weeks after transfer (dark blue).
(C) Representative flow cytometry data showing maturation of stage II/III ILC-EomesΔ/Δ or Eomes WT NK cells after 2 weeks in vivo. N ≥ 3 pooled mice per group, three independent experiments. Data were compared using t tests.
*p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
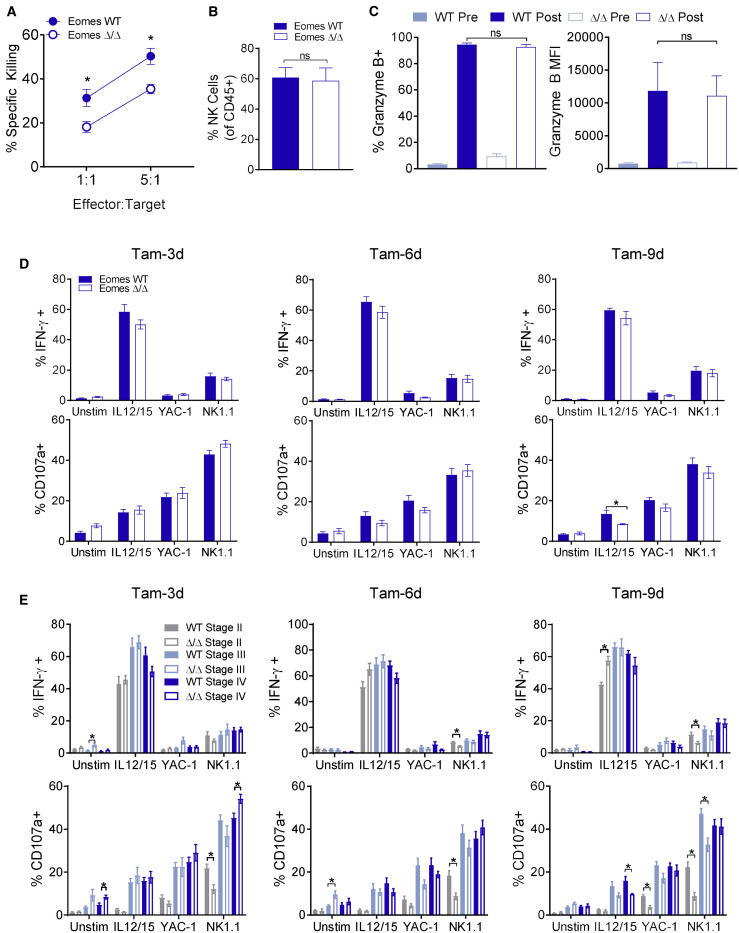
Eomes Deletion Impairs NK Cell Cytotoxicity, but Not Cytokine Production
ILC-EomesΔ/Δ mice were next used to assess the impact of Eomes deletion on mature NK cell function. To assess ex vivo cytotoxic responses, we treated ILC-EomesΔ/Δ and WT mice with the Tam-3d regimen, and on the 2nd day of tamoxifen also administered poly(I:C), a synthetic Toll-like receptor 3 (TLR3) agonist that activates NK cells (Fehniger et al., 2007). 24 h later, splenocytes were harvested, and enriched NK cells were used as effectors in flow-based killing assays against MHC-class I-deficient RMA-S leukemia targets. We observed a significant decrease in RMA-S killing by ILC-EomesΔ/Δ NK cells at both effector-to-target ratios examined (Figure 6 A). NK cells were equally enriched in the two groups (Figure 6B), demonstrating that the killing defect was not due to different numbers of ILC-EomesΔ/Δ versus control NK cells, but rather was cell-intrinsic. NK cell killing of target cells involves the targeted release of cytotoxic-protein-containing granules. Granzyme B protein levels were not affected by induced Eomes deletion (Figure 6C), consistent with prior reports showing robust granzyme B protein levels in Eomes-deficient ILCs (Gordon et al., 2012). However, RNA-seq analysis revealed a ≥2-fold decrease in perforin mRNA in stage II and III ILC-EomesΔ/Δ relative to WT NK cells (Figure S7A), consistent with prior reports of NK cell perforin regulation by Eomes (Intlekofer et al., 2005). Indeed, we performed Eomes chromatin immunoprecipitation sequencing (ChIP-seq) on primary, IL-15-expanded murine NK cells and confirmed the Prf1 transcription start site to be a direct target of Eomes (Table S1). In addition, since inducible Eomes deletion might impact the NK cell granular exocytosis pathway more globally (Galandrini et al., 2013), we performed GSEA of our RNA-seq data for the Kyoto Encyclopedia of Genes and Genomes (KEGG) NK-cell-mediated cytotoxicity pathway and observed significant downregulation of this pathway in ILC-EomesΔ/Δ compared to WT NK cells of all stages (Figure S7B). Thus, induced Eomes deletion appears to affect several components of the NK cell cytotoxic program, including perforin.
Figure 6.
Induced Eomes Deletion Does Not Significantly Alter NK Cell Functional Responses but Impairs Ex Vivo Cytotoxicity
(A and B) ILC-EomesΔ/Δ or Eomes WT mice were treated with the Tam-3d regimen. On day 2 of tamoxifen, mice were also administered 300 μg poly(I:C) intraperitoneally (i.p.) 24 h later, NK cells were enriched from splenocytes pooled from 2-3 mice per group and used in a flow-based killing assay against RMA-S targets. (A) Percent specific killing of RMA-S cells at the indicated effector/target ratios and (B) mean ± SEM. NK cell percentage of CD45+ cells from splenocytes used in the killing assay. n = 3 sets of pooled mice per group, three independent experiments.
(C) Mean ± SEM granzyme B percent positive and MFI of NK cells immediately before (in the blood) and 24 h after (in the spleen) poly(I:C). n = 7–11 mice per group, two or three independent experiments.
(D and E) Splenocytes from ILC-EomesΔ/Δ or Eomes WT mice treated with the Tam-3d, Tam-6d, or Tam-9d regimens were stimulated with plate-bound anti-NK1.1, IL-12 (10 ng/mL) + IL-15 (10 ng/mL) or YAC-1 lymphoma targets (10:1 effector:target [E:T]) in a 6-h functional assay. Summary data show (D) overall mean ± SEM percent IFN-γ or CD107a positive NK cells and (E) IFN-γ and CD107a positivity by stage in response to the different stimuli. n = 8–9 mice per group, three independent experiments. Data were compared using false-discovery-rate-corrected t tests.
*p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
To complement cytotoxicity assessments, we examined in vitro NK cell cytokine and degranulation (surface CD107a) responses after stimulation. A time course of ILC-EomesΔ/Δ versus WT NK cell function following the Tam-3d, Tam-6d, or Tam-9d regimens was utilized (Figures 6D and 6E). At each time point, we observed no differences in ILC-EomesΔ/Δ versus WT NK cell degranulation or IFN-γ production in response to cytokine (IL-12/15) stimulation, YAC-1 lymphoma, or activating receptor NK1.1 ligation (Figure 6D). We also investigated stage-specific ILC-EomesΔ/Δ versus WT NK cell function, and similarly, there were few substantial differences. The most consistent was a modest deficit in ILC-EomesΔ/Δ stage II NK cell functional responses to anti-NK1.1 ligation (Figure 6E). Thus, in vitro NK cell cytokine and degranulation responses were largely preserved upon induced Eomes deletion, despite reduced ex vivo cytotoxicity.
Induced Eomes Deletion Impairs Rejection of MHC-Class-I-Deficient Splenocytes In Vivo
To investigate the importance of induced Eomes deletion on NK cell function in vivo, we assessed the ability of NK cells to reject MHC-class I-deficient targets (Bix et al., 1991). ILC-EomesΔ/Δ and WT mice were treated with the Tam-3d regimen and injected intravenously (i.v.) on day 3 with a 50:50 mix of WT and β2 m−/− (MHC-class-I-deficient) splenocytes. 18–20 h later, mice were sacrificed and the WT:β2 m−/− ratio of transferred cells in the spleen assessed (Figure 7 ). β2 m−/− cells were efficiently rejected in WT mice, but no rejection occurred in ILC-EomesΔ/Δ mice (Figures 7A–7C). Indeed, the WT:β2 m−/− ratio of transferred cells in the spleen after 18–20 h was the same for ILC-EomesΔ/Δ mice as it was for negative control mice that had been NK cell depleted. Alterations in the NK cell Ly49 repertoire could underlie this finding, and prior publications have demonstrated that Eomes is required for the generation of a diverse Ly49 repertoire, but not its maintenance (Gordon et al., 2012). However, there were no substantial differences in Ly49 receptor expression between ILC-EomesΔ/Δ and WT NK cells (Figure 7D), suggesting that Ly49 alterations were unlikely to account for the impaired in vivo rejection phenotype. Thus, the impaired ex vivo cytotoxicity of ILC-EomesΔ/Δ NK cells was manifest in vivo, as the cells showed no ability to reject MHC-class-I-deficient targets.
Figure 7.
Inducible Eomes Deletion Impairs In Vivo NK Cell Rejection of β2 m−/− Splenocytes
(A–C) Eomes WT and ILC-EomesΔ/Δ mice were treated with the Tam-3d regimen. On day 3, mice received tamoxifen as well as 2 × 106 WT and 2 × 106 β2 m−/− splenocytes via tail vein injection. Transferred splenocytes were CFSE+ and differentially labeled with CTV. 18–20 h later, mice were sacrificed, and the WT versus β2 m−/− content of CFSE+ splenocyte events was assessed. As a control for NK-independent killing, some mice received 200 μg anti-NK1.1 antibody i.p. 2 days before splenocyte transfer. (A) β2 m−/− versus WT composition of transferred (CFSE+) splenocytes 18–20 h post-transfer with representative flow in (B). (C) Mean ± SEM percent rejection by ILC-EomesΔ/Δ versus WT mice. Percent rejection = (1 − [(ratio WT:β2 m −/−)experimental/(ratio WT:β2 m−/−)NK depleted]) × 100. n = 10 Eomes WT mice, 10 ILC-EomesΔ/Δ KO mice, 6 NK-depleted mice, two independent experiments.
(D) Ly49 repertoire of Eomes WT and ILC-EomesΔ/Δ YFP+ NK cells following the Tam-3d regimen. n = 8–11 mice, three or four independent experiments. Data were compared using t tests with false discovery rate correction where appropriate.
*p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Discussion
The role of Eomes in mature NK cell homeostasis and function has been incompletely understood, in large part due to a lack of inducible, conditional genetic models. Constitutive Ncr1-cre and Vav-cre models have conclusively demonstrated that DX5+ NK cells fail to develop in the absence of Eomes (Gordon et al., 2012, Pikovskaya et al., 2016). While some studies attempted to investigate the functional and phenotypic properties of cells that did develop in these models, particularly in the liver, these results are challenging to interpret in light of our current understanding that these were ILC1s and not Eomes-deficient NK cells. Since constitutive Ncr1-cre and Vav-cre models delete genes early in NK cell development (stage I or earlier), they preclude the study of genes required for normal NK cell ontogeny (like Eomes). In addition, global inducible models are complicated by cell-extrinsic effects and the requirement for adoptive transfer experiments (Madera et al., 2018). To overcome these limitations, we developed a highly penetrant, tamoxifen-inducible type-1-ILC-specific cre mouse. A transgenic inducible model that expressed cre in a small minority of NKp46+ cells was previously reported but lacked highly penetrant type 1 ILC-specific gene targeting (Nabekura and Lanier, 2016). The Ncr1-iCreERT2 reported here results in near-complete Eomes protein loss throughout the NK cell compartment in 2 days and permitted the demonstration of a persistent requirement for Eomes in mature NK cell homeostasis and function. Like most cre-loxP systems, our Ncr1-iCreERT2 model has limitations that must be considered during experimental design, including small background levels of cre expression in older mice and potentially altered proliferation kinetics. These limitations reinforce the importance of having a reporter for cre activity in experimental mice.
Eomes deletion negatively affected NK cell homeostasis, with a particularly profound effect on stage III NK cells. We determined that the stage III deficit arose from several mechanisms, including increased apoptosis (likely caspase dependent), altered cell cycling, and impaired maturation from stage II NK cells. IL-15-induced activation of PI3K/Akt/mTOR and MAPK was impaired in response to low-dose IL-15 in ILC-EomesΔ/Δ ΝΚ cells, yet exogenous IL-15 failed to rescue stage III NK cells in vivo. Thus, Eomes appears to regulate NK cell survival via mechanisms that are not rescued by exogenous IL-15. Indeed, we observed enrichment of apoptosis-related genes specifically in ILC-EomesΔ/Δ compared to WT stage II NK cells, which likely undergo cell death as they transition to stage III. These data reveal a role for Eomes in regulating hematopoietic cell apoptosis, a greater understanding of which could potentially lead to novel ways of supporting NK cells in the setting of adoptive transfer. The deficit in PI3K and MAPK activation by IL-15 at physiologic cytokine levels could, however, contribute to the maturation defect observed in stage II ILC-EomesΔ/Δ ΝΚ cells (Tassi et al., 2007). Importantly, and in contrast to prior models that induced Eomes deletion ex vivo in mature NK cells (Gordon et al., 2012), we demonstrate that mature (stage IV) NK cells are largely Eomes independent and do not revert to an immature state if Eomes is lost. In addition, although we saw no overall deficits in NK cell homeostatic proliferation upon induced Eomes deletion, stage-specific analyses as well as investigations of activation or inflammation-driven proliferation were not performed and will be important to fully appreciate the impact of Eomes deletion on NK cell proliferation. For the last decade, NK cell maturation has largely been classified based on CD27 and CD11b expression (Chiossone et al., 2009). Our findings suggest that an alternate way of characterizing NK cell maturation is based on Eomes dependence. As such, stage II/III NK cells are Eomes dependent, while stage IV NK cells are predominantly Eomes independent. Terminally mature NK cells downregulate Eomes and upregulate T-bet, and so we hypothesize that stage IV NK cell homeostasis is T-bet dependent. Additional studies utilizing the inducible cre model with both T-bet and Eomes floxed alleles will clarify these T-box transcription factor requirements.
Functionally, we observed that induced Eomes deletion impaired NK cell killing, as evidenced by both in vivo rejection of MHC-class-I-deficient splenocytes and ex vivo killing of tumor target cells. While the decreased NK cell number arising from induced Eomes deletion could be a contributing factor, the killing defect in vivo was disproportionate to the NK cell deficit, and the ex vivo killing assay was performed with equal NK cell numbers. Thus, Eomes deletion impairs NK cell killing in vivo and ex vivo via NK-cell-intrinsic mechanisms. NK cells kill via exocytosis of cytotoxic-protein-containing granules and via the ligation of death receptors on target cells. Ongoing Eomes-regulated transcription is not required for short-term granzyme B protein expression, as we observed no reduction in granzyme B protein levels in ILC-EomesΔ/Δ compared to WT NK cells. In contrast, RNA-seq revealed a significant reduction in perforin mRNA in ILC-EomesΔ/Δ stage II and III NK cells compared to WT, and Eomes ChIP-seq confirmed Prf1 to be an NK cell target of Eomes. While we were unable to quantify perforin protein due to technical limitations of available flow-based antibodies, decreases in perforin protein are likely to have occurred given the perforin sensitivity of RMA-S cells (Wallin et al., 2003). However, perforin mRNA was unchanged by induced Eomes deletion in stage IV NK cells, and since these cells are major NK cytotoxic mediators, this suggested that other components of the NK cell cytotoxic machinery may have been altered by induced Eomes deletion. Indeed, we observed downregulation of the KEGG NK-cell-mediated cytotoxicity pathway in ILC-EomesΔ/Δ NK cells of all stages, demonstrating a broader role for Eomes in regulating NK cell cytotoxicity. It is also possible that alterations in death receptor ligand expression contributed to the killing defects observed. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), in particular, is expressed on tissue-resident, Eomes-negative ILC1s, so it is possible that it is upregulated upon Eomes deletion. However, RMA-S cells are relatively TRAIL and FasL resistant in short-term killing assays (Screpanti et al., 2001, Wallin et al., 2003). Thus, while it will be important to investigate death receptor ligand expression and function on ILC-EomesΔ/Δ NK cells in the future, they likely did not contribute substantially to our cytotoxicity phenotype.
Unlike the cytotoxicity results, short-term NK cell degranulation and IFN-γ production were largely unaltered by induced Eomes deletion. It has been suggested that Eomes may be important for sustained NK cell functional responses and that Eomes downregulation, for example in cells infiltrating a tumor, might be a sign of NK cell exhaustion (Gill et al., 2012). As such, it will be informative to perform studies of sustained NK cell function (such as infection with murine cytomegalovirus [MCMV] or tumors) using our inducible cre system to assess the role of Eomes in these types of NK cell responses. Intriguingly, our ChIP-seq data, performed on IL-15-expanded NK cells, revealed a weak binding peak near the granzyme B promoter (Table S1). We speculate that in settings of sustained activation, Eomes may play a role in regulating the expression of this cytotoxic protein as well and that this may be a mechanism through which Eomes loss contributes to NK cell exhaustion. A greater understanding of how Eomes deletion affects the entire NK-cell-activating (e.g., NKG2D and NKp30) and inhibitory (e.g., NKG2A) repertoire will also be important to fully understand virus and tumor model findings.
This conditional, inducible cre model has many potential applications beyond the study of developmentally critical genes, which will make it important for addressing other outstanding questions in the type 1 ILC field. For example, it could be used to study NK cell populations that arise only after specific physiologic triggers, such as MCMV-specific innate memory or cytokine-induced memory-like NK cells. In addition, since we demonstrated that ILC1s regenerate at a much slower rate than NK cells, or from NKp46+ precursors, this model may be useful to selectively ablate genes within ILC1s, thereby permitting studies of the in vivo functions of these distinct but related cell types. Thus, this study provides a critical tool for the field to address new questions and avenues of investigation in type 1 ILC biology.
Limitations
Submission of this manuscript occurred during the SARS-CoV-2 pandemic, limiting our ability to perform experiments to address all issues raised during peer review. Some experiments, while of great interest, were beyond the feasible scope of the manuscript in light of restrictions imposed to contain the pandemic. These issues will be addressed in future studies, including (1) whether death receptor ligands (TRAIL and FasL) are differentially expressed on ILC-EomesΔ/Δ versus control NK cells and whether they contribute significantly to ILC-EomesΔ/Δ NK cell cytotoxicity; (2) whether induced Eomes deletion alters sustained NK cell function, as determined by an in vivo virus or tumor model; (3) if the expression of other activating and inhibitory receptors (e.g., NKG2D, NKp30, and NKG2A) differs between ILC-EomesΔ/Δ and control NK cells; (4) evaluation of perforin protein in addition to decreased mRNA in stage II and III ILC-EomesΔ/Δ NK cells; and (5) a comprehensive assessment of the impact of Eomes deletion on NK cell proliferation.
In summary, we developed a type-1-ILC-specific inducible cre mouse to identify a stage-specific requirement for Eomes in regulating NK cell homeostasis and cytotoxicity. In addition to elucidating the role of Eomes in mature NK cell biology, this study highlights the importance of complementing studies of constitutive cre with inducible models in order to elucidate the biological roles of genes at all stages of cellular development. As such, we anticipate that the Ncr1-iCreERT2 model will provide important insights into the nascent field of type 1 ILC biology.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PE-eFluor 610 CD3e anti-mouse mAb (145-2C11) | Thermo Fischer | Cat# 61-0031-82; RRID:AB_2574514 |
| APC CD11b anti-mouse mAb (M1/70) | Thermo Fischer | Cat# 17-0112-82; RRID:AB_469343 |
| PE-Cyanine7 CD27 anti-mouse mAb (LG.7F9) | Thermo Fischer | Cat# 25-0271-82; RRID:AB_1724035 |
| APC CD49b anti-mouse mAb (DX5) | Thermo Fischer | Cat# 17-5971-82; RRID:AB_469485 |
| eFluor660 CD107a anti-mouse mAb (1D4B) | Thermo Fischer | Cat# 50-1071-82; RRID:AB_11149501 |
| eFluor450 CD122 anti-mouse mAb (TM-b1) | Thermo Fischer | Cat# 48-1222-82; RRID:AB_2016697 |
| PerCP-eFluor710 EOMES anti-mouse mAb (Dan11mag) | Thermo Fischer | Cat# 46-4875-82; RRID:AB_10597455 |
| APC Granzyme B anti-human mAb (GB12) | Thermo Fischer | Cat# MHGB05; RRID:AB_10373420 |
| PE IFN gamma anti-mouse mAb (XMG1.2) | Thermo Fischer | Cat# 12-7311-82; RRID:AB_466193 |
| APC Ki-67 anti-mouse mAb (SolA15) | Thermo Fischer | Cat# 17-5698-82; RRID:AB_2688057 |
| APC Ly-49H anti-mouse mAb (3D10) | Thermo Fischer | Cat# 17-5886-82; RRID:AB_2688057 |
| PerCP-eFluor710 Ly-49G2 anti-mouse mAb (4D11) | Thermo Fischer | Cat# 46-5781-82; RRID:AB_1834437 |
| PE-Cyanine7 NKp46 anti-mouse mAb (29A1.4) | Thermo Fischer | Cat# 25-3351-82; RRID:AB_2573442 |
| APC Anti-Mouse CD3e mAb (145-2C11) | BD Biosciences | Cat# 561826; RRID:AB_10896663 |
| APC CD11b mAb (M1/70) | BD Biosciences | Cat# 561690; RRID:AB_10897015 |
| PE anti-mouse CD27 mAb (LG.3A10) | BD Biosciences | Cat# 561785; RRID:AB_10896150 |
| PE anti-rat/mouse CD49a mAb (Ha31/8) | BD Biosciences | Cat# 562115; RRID:AB_562115 |
| PE anti-mouse Ly-49C and Ly-49I mAb (5E6) | BD Biosciences | Cat# 553277; RRID:AB_394751 |
| PerCP-Cy5.5 anti-mouse NK-1.1 mAb (PK136) | BD Biosciences | Cat# 561111; RRID:AB_10564092 |
| BV421 Akt (pS473) mAb (M89-61) | BD Biosciences | Cat# 562599; RRID:AB_2737674 |
| PE ERK1/2 (pT202/pY204) mAb (20A) | BD Biosciences | Cat# 612566; RRID:AB_399857 |
| Alexa Fluor 647 Stat5 (pY694) mAb (47/Stat5(pY694)) | BD Biosciences | Cat# 562076; RRID:AB_11154412 |
| Brilliant Violet 605 anti-mouse CD45 mAb (30-F11) | BioLegend | Cat# 103139; RRID:AB_2562341 |
| Brilliant Violet 605 anti-mouse CD45.1 mAb (A20) | BioLegend | Cat# 110737; RRID:AB_11204076 |
| PE anti-mouse Ly-49AB6 mAb (A1/Ly49A) | BioLegend | Cat# 138703; RRID:AB_2134787 |
| Alexa Fluor 647 anti-mouse Ly49D mAb (4E5) | BioLegend | Cat# 138306; RRID:AB_10574955 |
| Anti-Eomes antibody - ChIP Grade | abcam | Cat# ab23345; RRID:AB_778267 |
| Anti-NK1.1 (pk136) | WashU Antibody Production Core | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tamoxifen | Sigma Aldrich | Cat# T5648 |
| Corn oil | Sigma Aldrich | Cat# C8267 |
| HyClone Classical Liquid Media: RPMI 1640 | Thermo Fisher | Cat# SH30027LS |
| Fetal Bovine Serum | Sigma Aldrich | Cat# F0926 |
| Fixation/Permeabilization Concentrate | Invitrogen | Cat# 00-5123-43 |
| Fixation/Permeabilization Diluent | Invitrogen | Cat# 00-5223-56 |
| Permeabilization Buffer (10X) | Invitrogen | Cat#00-8333-56 |
| Cytofix/Cytoperm Fixation and Permeabilization Solutionl | BD Bioscience | Cat# BDB554722 |
| murine IL-15 | Peprotech | Cat#210-15 |
| Trizol Reagent | Invitrogen | Cat#15596026 |
| Poly (I:C) LMW | Invivogen | Cat#tlrl-picw |
| 7-AAD | Sigma Aldrich | Cat#A9400 |
| Murine IL-12 | Peprotech | Cat#210-12 |
| Protein Transport Inhibitor (Containing Brefeldin A) | BD Bioscience | Cat# 555029 |
| Protein Transport Inhibitor (Containing Monensin) | BD Bioscience | Cat# 554724 |
| FxCycle Violet Stain | Invitrogen | Cat#F10347 |
| Hyclone L-Glutamine | Thermo Fisher | Cat# SH3003402 |
| Hyclone HEPES Solution | Thermo Fisher | Cat# SH3023701 |
| HyClone Non Essential Amino Acids | Thermo Fisher | Cat# SH3023801 |
| HyClone Sodium Pyruvate Solution | Thermo Fisher | Cat# SH3023901 |
| HyClone Penicillin Streptomycin 100X Solution | Thermo Fisher | Cat# SV30010 |
| Critical Commercial Assays | ||
| FAM-FLICA® Poly Caspase Assay Kit | Immunochemistry Technologies | Cat# 9120 |
| PE Annexin V Apoptosis Detection Kit | BD Bioscience | Cat# 559763 |
| CellTrace Violet Cell Proliferation Kit | Thermo Fisher | Cat# C34557 |
| Direct-zol RNA Microprep | Zymo Research | Cat#R2061 |
| EasySep Mouse NK Cell Isolation Kit | Stem Cell Technologies | Catalog # 19855 |
| Deposited Data | ||
| RNA Seq Data | This paper | GEO: GSE132942 |
| Chip Seq Data | This paper | GEO: GSE133048 |
| Experimental Models: Cell Lines | ||
| Mouse: RMA-S | Yokoyama Lab, Wash U | RRID: CVCL_2180 |
| Mouse: YAC-1 | Yokoyama Lab, Wash U | RRID: CVCL_2244 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Ncr1-ERT2-iCre | Fehniger Lab, Wash U (This paper) | N/A |
| Mouse: ROSA26 YFPlox/stop/lox | The Jackson Laboratory | Cat# JAX:006148; RRID:IMSR_JAX:006148 |
| Mouse: Eomesloxp/loxp | The Jackson Laboratory | Cat# JAX: 017293; RRID:IMSR_JAX:017293 |
| Mouse: Eomesfl/fl x Ncr1-ERT2-iCreKI/WT x Rosa-YFPKI/KI | Fehniger Lab, Wash U (This Paper) | N/A |
| Mouse: Ncr1-ERT2-iCreKI/WT x Rosa-YFPKI/KI x EomesWT/WT | Fehniger Lab, Wash U (This Paper) | N/A |
| Mouse: CD45.2 C57BL/6J | The Jackson Laboratory | Cat# JAX: 000664; RRID:IMSR_JAX:000664 |
| Mouse: CD45.1 C57BL/6J | The Jackson Laboratory | Cat# JAX: 002014; RRID:IMSR_JAX:002014 |
| Mouse: B2m−/− | Yokoyama Lab, Wash U | N/A |
| Software and Algorithms | ||
| FlowJo | Treestar Inc | https://www.flowjo.com/; RRID:SCR_008520 |
| Novoalign | Novocraft | http://www.novocraft.com/products/novoalign/; RRID:SCR_014818 |
| MACS | (Zhang et al., 2008) | https://taoliu.github.io/MACS/; RRID:SCR_013291 |
| STAR version 2.0.4b | (Dobin et al., 2013) | https://code.google.com/p/rna-star/; RRID:SCR_015899 |
| Gene Set Enrichment Analysis Software | (Subramanian et al., 2005) and (Mootha et al., 2003) | https://www.gsea-msigdb.org/gsea/index.jsp; RRID:SCR_003199 |
| Subread:feature Count version 1.4.5 | (Liao et al., 2014) | http://bioinf.wehi.edu.au/featureCounts/; RRID:SCR_009803 |
| GraphPad Prism 7 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/; RRID:SCR_002798 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Todd A. Fehniger (tfehnige@wustl.edu).
Materials Availability
The Ncr1-iCreERT2 mouse generated in this study will be made available to qualified investigators by the Lead Contact, Todd A. Fehniger (tfehnige@wustl.edu) upon request.
Data and Code Availability
The accession numbers for the RNaseq and CHIPseq data reported in this paper are GEO:GSE132942 (RNaseq) and GEO:GSE133048 (ChIPseq).
Experimental Model and Subject Details
Mice
Ncr1-iCreERT2 mice were developed in the Fehniger lab on the C57BL/6J background. Mice were bred to Rosa26 YFPLSL knock-in mice (The Jackson Laboratory, stock no. 006148) as well as Eomes fl/fl mice (The Jackson Laboratory, stock no. 017293). For all experiments, ILC-EomesΔ/Δ refers to Ncr1-iCreERT2 KI/WT x Rosa26YFPLSL x Eomes fl/fl mice. Ncr1-iCreERT2 KI/WT x Rosa26YFPLSL x Eomes WT/WT mice were used as WT controls in Figures 1E, 2, 3A, 3B, 3D, 3F, 3G, 5, 6A, and 6B, as well as Figures S1 and S3–S5. In the remaining figures, if these mice were limiting, Ncr1-iCreERT2 WT/WT x Rosa26YFPLSL x Eomes fl/fl mice as well as CD45.1 and CD45.2 C57BL/6J mice (The Jackson Laboratory, nos. 002014 and 000664) were also utilized. CD45.1 C57BL/6J mice were utilized as WT controls in Figure S6 and as cre-negative controls in Figure S4. Rag2−/−γc−/− mice (Taconic, no. 4111) were used as recipients for in vivo homeostatic proliferation assays (Figure S4). All mice (Eomes WT and ILC-EomesΔ/Δ) were treated with identical tamoxifen regimens for all experiments. Mice were orally gavaged with 3mg tamoxifen (Sigma) in corn oil (Sigma) for 2-3 days as described. Mice were bred and maintained in specific pathogen-free housing, and experiments were conducted in accordance with the guidelines of and with the approval of the Washington University Animal Studies Committee. Experiments were performed on 8-12 week old male and female mice.
Cell Lines
RMA-S and YAC-1 cell lines were maintained at 37°C in 5% CO2 in RPMI-1640 plus 10% FBS and supplements(10mM HEPES, 1% NEAA, 1% sodium pyruvate, 1% L-glutamine, 1X penicillin and streptomycin) following ATCC guidelines and kept in continuous culture for < 2 months.
Method Details
Organ isolation and processing
Mice were sacrificed by CO2 asphyxiation and organs harvested immediately. Blood was harvested by cardiac puncture prior to removal of other organs. Spleens and inguinal nodes were isolated and crushed through a 70 μm filter to generate a single cell suspension. Bone marrow was harvested by flushing femurs with a 23-gauge needle. Whole livers were desiccated with a tissue grinder then lymphocytes were isolated using a Percoll gradient. All tissues were ammonium-chloride-potassium lysed. Cell counts were obtained using propium iodide exclusion with a Cellometer counter (Nexcelcom).
Flow cytometry and cell sorting
Surface antibody staining was performed for 15 minutes at 4°C. Intracellular staining was performed in permeabilization buffer following fixation/permeabilization with kits from BD Biosciences (for IFN-γ staining) or Invitrogen (for Eomes, Granzyme B, Bcl2, and Ki67 staining). EomesΔ/Δ NK cells (and cre-positive WT cells) were gated on YFP+ events whenever possible. Since Invitrogen fixation/permeabilization quenches YFP, cells were pre-fixed in 1% PFA for 2 minutes at room temperature prior to fixation/permeabilization in order to preserve YFP when intracellular proteins were assessed after the Tam-6d regimen (Figures 1E and 2). However, pre-fixation was unable to preserve YFP after the Tam-3d regimen, so intracellular analyses were performed on bulk NK cells (which were nearly 80% YFP+, Figure 3A) at this time point. For phosphoflow cytometry assays, splenocytes were stimulated with recombinant mouse IL-15 (Peprotech) at 10ng/mL or 100ng/mL for 15 minutes (pSTAT5) or 1 hour (pERK, pAkt). Cells were then fixed with 1% PFA for 10 minutes at room temperature. Next, cells were permeabilized in ice cold methanol for 30 minutes at 4°C, then washed and stained overnight with phosphoflow antibodies. Flow cytometry data were collected on a Gallios flow cytometer (Beckman Coulter) and analyzed using FlowJo (Treestar) software. Cell sorting was performed on a BD FACS Aria II to greater than 95% purity.
Cell cycle and viability assays
Cell cycle was assessed using Ki67 and FxCycle Violet (Invitrogen) staining following Invitrogen Fixation/Permeabilization. For viability assays, freshly-isolated splenocytes were incubated at 37°C for 2-3 hours then assayed for poly-caspase activation (Immunochemistry Technologies) or surface Annexin V (performed in Annexin V buffer, both BD Biosciences).
In vivo maturation experiments
In Figure 5, ILC-EomesΔ/Δ and Eomes WT mice were treated with the Tam-3d regimen. Next, NK cells were purified via negative selection (StemCell Technologies, Inc.) from pooled ILC-EomesΔ/Δ or Eomes WT spleens and sorted to YFP+ stage II/III (CD27+) or stage IV (CD27- CD11b+). Cells were injected into separate male CD45.2 WT recipients. 2 weeks later, mice were sacrificed and the stage distribution of YFP+ splenocytes analyzed via flow cytometry. In Figure S6, NK cells were purified from pooled ILC-EomesΔ/Δ or CD45.1 WT spleens, labeled with CTV (Invitrogen) and sorted to stage II/III (CD27+) or stage IV (CD27- CD11b+) prior to tamoxifen administration. ILC-EomesΔ/Δ and Eomes WT cells of the same stage were pooled and injected into male CD45.2 WT mice, who then received 3 daily doses of 3mg tamoxifen in corn oil via oral gavage. 2 weeks later, recipient mice were sacrificed and the stage of CTV+YFP+ (ILC-EomesΔ/Δ) and CTV+CD45.1+ (WT) cells analyzed by flow cytometry.
In vivo homeostatic proliferation
ILC-EomesΔ/Δ and Eomes WT mice along with Cre- CD45.1 WT controls were treated with the Tam-3d regimen. Splenocytes were CTV-labeled, then NK cells were purified from pooled splenocytes via negative selection (StemCell Technologies, Inc.) and sorted to NK1.1+CD3-YFP+ (ILC-EomesΔ/Δ and WT) or NK1.1+CD3- (CD45.1WT). Sorted cells were injected in CD45.2 Rag2−/−γc−/− recipients and CTV dilution of YFP+ (ILC-EomesΔ/Δ and WT) or CD45.1+ cells (CD45.1 WT) was assessed 4 or 14 days post-transfer via flow cytometry.
ChIP sequencing
In 3 separate experiments, purified NK cells from pooled CD45.2 Eomes WT splenocytes were expanded in vitro for 4 days with 100ng/mL IL-15. Cells were then harvested and approximately 10 × 106 cells were used per ChIP experiment. Chromatin immunoprecipiation was performed using the MAGnify Chromatin Immunoprecipitation System (Thermo Fisher) as per manufacturer’s instructions with a ChIP-grade anti-Eomes antibody from Abcam (ab23345). Non-immunoprecipitated DNA was used as an input control. ChIP DNA was blunt ended, had addition of “A” base to 3′ end, and had sequencing adapters ligated to the ends. The fragments were size selected to 200-600 base pairs, and underwent amplification for 15 cycles with primers incorporating p5 and p7 sequences and a unique index tag for multiplexing. The resulting libraries were sequenced using the Illumina HiSeq3000 as single reads extending 50 bases. The raw data was demultiplexed and aligned to the reference genome using Novoalign. MACS was used to call peaks. For downstream analysis, gene-associated peaks that had greater than 5-fold enrichment over input and were present in all three replicates (±500 base pairs from each other) were identified (Table S1).
RNA sequencing and Gene Set Enrichment Analysis
In 3 separate experiments, ILC-EomesΔ/Δ or CD45.2 Eomes WT mice were treated with the Tam-3d regimen. Next, NK cells were purified from pooled ILC-EomesΔ/Δ or Eomes WT splenocytes and sorted to stage II (CD27+CD11b-), stage III (CD27+CD11b+), or stage IV (CD27-CD11b+). KO cells were also YFP+. Cells were stored in Trizol at −80°C until RNA isolation using the Direct-zol RNA MicroPrep kit (Zymo Research). NextGen RNA sequencing was performed using an Illumina HiSeq 2500 sequencer. RNA-Seq reads were aligned to the Ensembl release 76 top-level assembly with STAR version 2.0.4b. Gene counts were derived from the number of uniquely aligned unambiguous reads by Subread:featureCount version 1.4.5. Differential expression analysis was performed to analyze for differences between conditions and the results were filtered for only those genes with Benjamini-Hochberg false-discovery rate adjusted p values less than or equal to 0.05.
We utilized Gene Set Enrichment Analysis (GSEA) with the Molecular Signatures Database (MSigDB) (Subramanian et al., 2005, Mootha et al., 2003) developed by the Broad Institute and UC San Diego on the RNA sequencing results of sorted stage II, III, and IV ILC-EomesΔ/Δ and Eomes WT NK cells to identify biological pathways that were enriched or downregulated in ILC-EomesΔ/Δ versus Eomes WT NK cells in respective maturation stages. Pathways were considered significantly enriched or downregulated if p < 0.05 and FDR < 0.25.
In vivo β2 m−/− rejection
ILC-EomesΔ/Δ and WT mice were orally gavaged with 3mg tamoxifen in corn oil for 3 consecutive days. On the third day, mice also received 4e6 splenocytes that were a 50/50 mix of CD45.2 WT and β2 m−/− (a gift from Wayne Yokoyama). Transferred splenocytes were differentially labeled with CTV for discrimination. In addition, on the first day of tamoxifen administration, some control mice received 200 μg anti-NK1.1 intraperitoneally (i.p.) to control for non NK-specific killing. 18-20 hours after splenocyte transfer, mice were sacrificed and the ratio of CTVhi (WT) to CTVlow (β2 m−/−) splenocytes was assessed. Percent rejection was calculated as: (1-[(Ratio WT:β2 m−/−)experimental/(Ratio WT:β2 m−/−)NK depleted])∗100 (Parikh et al., 2015).
In vitro cytotoxicity (FLoKA) assays
ILC-EomesΔ/Δ and WT mice were administered 3mg tamoxifen in corn oil via oral gavage for 2 consecutive days. On the second day, mice were also administered 300 μg of poly I:C (Invivogen) i.p. 20-24 hours post-poly I:C, splenocytes were pooled from 2-3 mice per group and NK cells were enriched via negative selection (Stem Cell Technologies). NK-enriched splenocytes were used in killing assays against CTV-labeled RMA-S targets at the indicated E:T ratios for 4 hours. Dead cells were identified via 7-AAD staining and specific killing calculated as: % 7-AAD+effector+target - % 7-AAD+target alone. Mice were cheek bled immediately prior to poly I:C administration for a baseline NK cell granzyme B readout, and splenic NK cell granzyme B was also assessed at the time of the killing assay.
In vitro functional assays
ILC-EomesΔ/Δ and Eomes WT mice were treated with the Tam-3d, Tam-6d, and Tam-9d regimens. At each time point, functional assays were performed on splenocytes. Splenocytes were either unstimulated (media alone) or stimulated with cytokines (10ng/mL IL-12 + 10ng/mL IL-15, both Peprotech), YAC-1 lymphoma targets (E:T = 10:1), or plate-bound purified anti-NK1.1 for 6 hours. Anti-CD107a (LAMP-1) antibody was added at the start of the assay to detect degranulation, and brefeldin A and monensin (BD Biosciences) were added for the final 5 hours. Cells were then surface stained, fixed/permeabilized (BD Biosciences), and intracellularly stained for IFN-γ, follow by flow cytometry analysis.
Quantification and Statistical Analysis
Statistical analyses were performed using GraphPad Prism 7 software. Tests utilized are indicated in figure legends. ∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001.
Acknowledgments
This work was supported by grants from the Howard Hughes Medical Institute (Medical Fellow Award to J.A.W.), the NIH (T32 HL007088 to J.A.W., P.W., and J.A.F.), and the NIH/NCI (F32 CA200253 to M.M.B.-E.; K12 5K12CA167540 to M.M.B.-E.; R01AI127752 to M.A.C.; and R01 AI102924, R01 CA205239, and P50 CA171963 to T.A.F.). We acknowledge the use of the Siteman Flow Core and Genome Technology Access center (NIH grant P30 CA91842) for this study. We thank Drs. Wayne Yokoyama, Daniel Link, Timothy Ley, and Anthony R. French for insightful discussion.
Author Contributions
J.A.W., P.W., and T.A.F. conceived and designed the study. All authors collected, analyzed, or assembled the data. J.A.W., P.W., and T.A.F. wrote the manuscript. All authors reviewed the data and edited and approved the final version of the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: June 2, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107720.
Supplemental Information
References
- Banerjee A., Gordon S.M., Intlekofer A.M., Paley M.A., Mooney E.C., Lindsten T., Wherry E.J., Reiner S.L. Cutting edge: the transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J. Immunol. 2010;185:4988–4992. doi: 10.4049/jimmunol.1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrien-Elliott M.M., Sun Y., Neal C., Ireland A., Trissal M.C., Sullivan R.P., Wagner J.A., Leong J.W., Wong P., Mah-Som A.Y., et al. MicroRNA-142 is critical for the homeostasis and function of type 1 innate lymphoid cells. Immunity. 2019;51:479–490.e6. doi: 10.1016/j.immuni.2019.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bix M., Liao N.S., Zijlstra M., Loring J., Jaenisch R., Raulet D. Rejection of class I MHC-deficient haemopoietic cells by irradiated MHC-matched mice. Nature. 1991;349:329–331. doi: 10.1038/349329a0. [DOI] [PubMed] [Google Scholar]
- Caligiuri M.A. Human natural killer cells. Blood. 2008;112:461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrier D.E., Serafini N., Di Santo J.P. Innate lymphoid cell development: a T cell perspective. Immunity. 2018;48:1091–1103. doi: 10.1016/j.immuni.2018.05.010. [DOI] [PubMed] [Google Scholar]
- Chiossone L., Chaix J., Fuseri N., Roth C., Vivier E., Walzer T. Maturation of mouse NK cells is a 4-stage developmental program. Blood. 2009;113:5488–5496. doi: 10.1182/blood-2008-10-187179. [DOI] [PubMed] [Google Scholar]
- Colonna M. Innate lymphoid cells: diversity, plasticity, and unique functions in immunity. Immunity. 2018;48:1104–1117. doi: 10.1016/j.immuni.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez V.S., Ulland T.K., Cervantes-Barragan L., Bando J.K., Robinette M.L., Wang Q., White A.J., Gilfillan S., Cella M., Colonna M. SMAD4 impedes the conversion of NK cells into ILC1-like cells by curtailing non-canonical TGF-β signaling. Nat. Immunol. 2017;18:995–1003. doi: 10.1038/ni.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daussy C., Faure F., Mayol K., Viel S., Gasteiger G., Charrier E., Bienvenu J., Henry T., Debien E., Hasan U.A., et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J. Exp. Med. 2014;211:563–577. doi: 10.1084/jem.20131560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckelhart E., Warsch W., Zebedin E., Simma O., Stoiber D., Kolbe T., Rülicke T., Mueller M., Casanova E., Sexl V. A novel Ncr1-Cre mouse reveals the essential role of STAT5 for NK-cell survival and development. Blood. 2011;117:1565–1573. doi: 10.1182/blood-2010-06-291633. [DOI] [PubMed] [Google Scholar]
- Fehniger T.A., Cai S.F., Cao X., Bredemeyer A.J., Presti R.M., French A.R., Ley T.J. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity. 2007;26:798–811. doi: 10.1016/j.immuni.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Galandrini R., Capuano C., Santoni A. Activation of lymphocyte cytolytic machinery: Where are we? Front. Immunol. 2013;4:390. doi: 10.3389/fimmu.2013.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Souza-Fonseca-Guimaraes F., Bald T., Ng S.S., Young A., Ngiow S.F., Rautela J., Straube J., Waddell N., Blake S.J., et al. Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat. Immunol. 2017;18:1004–1015. doi: 10.1038/ni.3800. [DOI] [PubMed] [Google Scholar]
- Gill S., Vasey A.E., De Souza A., Baker J., Smith A.T., Kohrt H.E., Florek M., Gibbs K.D., Jr., Tate K., Ritchie D.S., Negrin R.S. Rapid development of exhaustion and down-regulation of eomesodermin limit the antitumor activity of adoptively transferred murine natural killer cells. Blood. 2012;119:5758–5768. doi: 10.1182/blood-2012-03-415364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S.M., Chaix J., Rupp L.J., Wu J., Madera S., Sun J.C., Lindsten T., Reiner S.L. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;36:55–67. doi: 10.1016/j.immuni.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger K., Seidler B., Vahl J.C., Schwartz C., Kober M., Klein S., Voehringer D., Saur D., Schmidt-Supprian M. CreERT2 expression from within the c-Kit gene locus allows efficient inducible gene targeting in and ablation of mast cells. Eur. J. Immunol. 2014;44:296–306. doi: 10.1002/eji.201343731. [DOI] [PubMed] [Google Scholar]
- Herold M.J., Stuchbery R., Mérino D., Willson T., Strasser A., Hildeman D., Bouillet P. Impact of conditional deletion of the pro-apoptotic BCL-2 family member BIM in mice. Cell Death Dis. 2014;5:e1446. doi: 10.1038/cddis.2014.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntington N.D., Puthalakath H., Gunn P., Naik E., Michalak E.M., Smyth M.J., Tabarias H., Degli-Esposti M.A., Dewson G., Willis S.N., et al. Interleukin 15-mediated survival of natural killer cells is determined by interactions among Bim, Noxa and Mcl-1. Nat. Immunol. 2007;8:856–863. doi: 10.1038/ni1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer A.M., Takemoto N., Wherry E.J., Longworth S.A., Northrup J.T., Palanivel V.R., Mullen A.C., Gasink C.R., Kaech S.M., Miller J.D., et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- Ishizuka I.E., Constantinides M.G., Gudjonson H., Bendelac A. The innate lymphoid cell precursor. Annu. Rev. Immunol. 2016;34:299–316. doi: 10.1146/annurev-immunol-041015-055549. [DOI] [PubMed] [Google Scholar]
- Kristianto J., Johnson M.G., Zastrow R.K., Radcliff A.B., Blank R.D. Spontaneous recombinase activity of Cre-ERT2 in vivo. Transgenic Res. 2017;26:411–417. doi: 10.1007/s11248-017-0018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier L.L. NK cell recognition. Annu. Rev. Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- Leong J.W., Wagner J.A., Ireland A.R., Fehniger T.A. Transcriptional and post-transcriptional regulation of NK cell development and function. Clin. Immunol. 2017;177:60–69. doi: 10.1016/j.clim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Loonstra A., Vooijs M., Beverloo H.B., Allak B.A., van Drunen E., Kanaar R., Berns A., Jonkers J. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. USA. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madera S., Geary C.D., Lau C.M., Pikovskaya O., Reiner S.L., Sun J.C. Cutting edge: divergent requirement of T-Box transcription factors in effector and memory NK cells. J. Immunol. 2018;200:1977–1981. doi: 10.4049/jimmunol.1700416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais A., Cherfils-Vicini J., Viant C., Degouve S., Viel S., Fenis A., Rabilloud J., Mayol K., Tavares A., Bienvenu J., et al. The metabolic checkpoint kinase mTOR is essential for IL-15 signaling during the development and activation of NK cells. Nat. Immunol. 2014;15:749–757. doi: 10.1038/ni.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel D.B., Matsumoto T., Vallejo J.A., Johnson M.L., Dallas S.L., Kitase Y., Brotto M., Wacker M.J., Harris M.A., Harris S.E., et al. Characterization of a novel murine Sost ER T2 Cre model targeting osteocytes. Bone Res. 2019;7:6. doi: 10.1038/s41413-018-0037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A., Sullivan L., Caligiuri M.A. Molecular pathways: interleukin-15 signaling in health and in cancer. Clin. Cancer Res. 2014;20:2044–2050. doi: 10.1158/1078-0432.CCR-12-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Nabekura T., Lanier L.L. Tracking the fate of antigen-specific versus cytokine-activated natural killer cells after cytomegalovirus infection. J. Exp. Med. 2016;213:2745–2758. doi: 10.1084/jem.20160726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narni-Mancinelli E., Chaix J., Fenis A., Kerdiles Y.M., Yessaad N., Reynders A., Gregoire C., Luche H., Ugolini S., Tomasello E., et al. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc. Natl. Acad. Sci. USA. 2011;108:18324–18329. doi: 10.1073/pnas.1112064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh B.A., Piersma S.J., Pak-Wittel M.A., Yang L., Schreiber R.D., Yokoyama W.M. Dual requirement of cytokine and activation receptor triggering for cytotoxic control of murine cytomegalovirus by NK cells. PLoS Pathog. 2015;11:e1005323. doi: 10.1371/journal.ppat.1005323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce E.L., Mullen A.C., Martins G.A., Krawczyk C.M., Hutchins A.S., Zediak V.P., Banica M., DiCioccio C.B., Gross D.A., Mao C.A., et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- Pikovskaya O., Chaix J., Rothman N.J., Collins A., Chen Y.-H., Scipioni A.M., Vivier E., Reiner S.L. Cutting edge: Eomesodermin is sufficient to direct type 1 innate lymphocyte development into the conventional NK lineage. J. Immunol. 2016;196:1449–1454. doi: 10.4049/jimmunol.1502396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranson T., Vosshenrich C.A.J., Corcuff E., Richard O., Müller W., Di Santo J.P. IL-15 is an essential mediator of peripheral NK-cell homeostasis. Blood. 2003;101:4887–4893. doi: 10.1182/blood-2002-11-3392. [DOI] [PubMed] [Google Scholar]
- Romee R., Leong J.W., Fehniger T.A. Utilizing cytokines to function-enable human NK cells for the immunotherapy of cancer. Scientifica (Cairo) 2014;2014:205796. doi: 10.1155/2014/205796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ A.P., Wattler S., Colledge W.H., Aparicio S.A.J.R., Carlton M.B.L., Pearce J.J., Barton S.C., Surani M.A., Ryan K., Nehls M.C., et al. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–99. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- Screpanti V., Wallin R.P., Ljunggren H.G., Grandien A. A central role for death receptor-mediated apoptosis in the rejection of tumors by NK cells. J. Immunol. 2001;167:2068–2073. doi: 10.4049/jimmunol.167.4.2068. [DOI] [PubMed] [Google Scholar]
- Sojka D.K., Plougastel-Douglas B., Yang L., Pak-Wittel M.A., Artyomov M.N., Ivanova Y., Zhong C., Chase J.M., Rothman P.B., Yu J., et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife. 2014;3:e01659. doi: 10.7554/eLife.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan R.P., Leong J.W., Schneider S.E., Ireland A.R., Berrien-Elliott M.M., Singh A., Schappe T., Jewell B.A., Sexl V., Fehniger T.A. MicroRNA-15/16 antagonizes Myb to control NK cell maturation. J. Immunol. 2015;195:2806–2817. doi: 10.4049/jimmunol.1500949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassi I., Cella M., Gilfillan S., Turnbull I., Diacovo T.G., Penninger J.M., Colonna M. p110gamma and p110delta phosphoinositide 3-kinase signaling pathways synergize to control development and functions of murine NK cells. Immunity. 2007;27:214–227. doi: 10.1016/j.immuni.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Townsend M.J., Weinmann A.S., Matsuda J.L., Salomon R., Farnham P.J., Biron C.A., Gapin L., Glimcher L.H. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- Vivier E., Tomasello E., Baratin M., Walzer T., Ugolini S. Functions of natural killer cells. Nat. Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- Wallin R.P.A., Screpanti V., Michaëlsson J., Grandien A., Ljunggren H.G. Regulation of perforin-independent NK cell-mediated cytotoxicity. Eur. J. Immunol. 2003;33:2727–2735. doi: 10.1002/eji.200324070. [DOI] [PubMed] [Google Scholar]
- Walzer T., Bléry M., Chaix J., Fuseri N., Chasson L., Robbins S.H., Jaeger S., André P., Gauthier L., Daniel L., et al. Identification, activation, and selective in vivo ablation of mouse NK cells via NKp46. Proc. Natl. Acad. Sci. USA. 2007;104:3384–3389. doi: 10.1073/pnas.0609692104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Pang Y., Moses H.L. TGF-beta and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol. 2010;31:220–227. doi: 10.1016/j.it.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Freud A.G., Caligiuri M.A. Location and cellular stages of natural killer cell development. Trends Immunol. 2013;34:573–582. doi: 10.1016/j.it.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Ju S., Chen E., Dai S., Li C., Morel P., Liu L., Zhang X., Lu B. T-bet and eomesodermin are required for T cell-mediated antitumor immune responses. J. Immunol. 2010;185:3174–3183. doi: 10.4049/jimmunol.1000749. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers for the RNaseq and CHIPseq data reported in this paper are GEO:GSE132942 (RNaseq) and GEO:GSE133048 (ChIPseq).