Abstract
Coronaviruses can become zoonotic, as in the case of COVID-19, and hunting, sale, and consumption of wild animals in Southeast Asia increases the risk for such incidents. We sampled and tested rodents (851) and other mammals and found betacoronavirus RNA in 12 rodents. The sequences belong to two separate genetic clusters and are closely related to those of known rodent coronaviruses detected in the region and distantly related to those of human coronaviruses OC43 and HKU1. Considering the close human-wildlife contact with many species in and beyond the region, a better understanding of virus diversity is urgently needed for the mitigation of future risks.
Electronic supplementary material
The online version of this article (10.1007/s00705-020-04683-7) contains supplementary material, which is available to authorized users.
Severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2), the causative agent of the global COVID-19 outbreak in humans, was first detected in the city of Wuhan, Hubei Province, in the People’s Republic of China in late 2019 [1]. The suspected index case probably contracted the virus at a local seafood and wildlife market in the city, yet the exact species of animal that hosted the virus remains unknown. Phylogenetic analysis of the SARS-CoV-2 genome indicates a strong likelihood that the reservoir species is a bat, as in the case of the related betacoronaviruses SARS-CoV-1 and Middle East respiratory syndrome-related coronavirus (MERS-CoV) [2]. An involvement of another intermediate host between bats and humans in the transmission of SARS-CoV-2 remains unknown. Wildlife and bushmeat markets are common across Southeast Asia, and represent a significant risk for the transfer of zoonotic pathogens between wildlife and humans. Indeed, 75% of all emerging infectious diseases in the past decades have their origin in wildlife, including highly pathogenic influenza viruses (H5N1), ebolaviruses, henipaviruses, and hantaviruses, among others [3].
The CoVs most closely related to SARS-CoV-2 were isolated from bats living in Yunnan province, in the south of China, not far from the 423-km-long border with the landlocked Lao People’s Democratic Republic (Laos) [4]. Both countries were involved in the United States Agency for International Development’s (USAID) Emerging Pandemic Threats PREDICT program, and surveillance of bats in wildlife markets in rural areas in Laos using family-level PCR assays revealed the presence of CoV RNA in 41 animals [5]. In addition to bats, rodents are recognized as significant hosts of viral zoonoses and represent an important potential host for zoonotic viral spillover in Laos through their frequent incidental and intentional interaction with humans [6, 7].
Multiple groups in Laos are at high risk of zoonotic viral spillover from wildlife, including from rodents, due to their occupation or economic or geographic circumstances. People contact rodents incidentally and intentionally in various ways. In traditional-style homes, especially in rural areas, rodents are often able to easily enter the houses in search of food and shelter. These circumstances promote incidental contact with rodent urine and feces during everyday life when household members clean their houses. Rodents also commonly raid food storage areas, including rice storage huts near paddies. In terms of more direct and intentional contact, some species of rodents, including the Indian giant flying squirrel (Petaurista philippensis), Finlayson’s squirrel (Callosciurus finlaysonii), red-cheeked flying squirrel (Hylopetes spadiceus), and others, are hunted or trapped in rural forested areas using traps, guns, sticks, or other implements. Designated for food or for medicinal purposes, depending on the species, the rodents are consumed within the hunter’s village or enter the value chain to reach markets. The value chain involves a series of intermediaries that transport animals from small villages to progressively larger populated areas. At the market, animals are sold to locals or to Lao people visiting from other areas of the country, and often to foreign visitors from neighbouring Thailand, China, and Vietnam [8, 9]. Even though the sale of these animals is illegal, Laos attracts many wildlife trade tourists simply because wildlife products are more available. Throughout this value chain, people are exposed to blood, viscera, feces, and saliva of rodents and can be further exposed to these materials during the butchering process, where butchers can accidentally cut themselves with knives, allowing efficient transmission of viruses from rodents to humans to occur [9]. Considering the significant interactions of wildlife and especially rodents with humans in Laos, we were interested in investigating the presence of CoVs in these animals, which can be primary or intermediate hosts for CoVs with zoonotic potential.
Samples were collected from both live and freshly killed animals, either trapped in or around village homes or voluntarily provided by local hunters upon their return to the village following hunting forays. With the permission of market operators and vendors, samples were also collected from freshly killed animals for sale in markets. Oral and rectal swab specimens were collected in duplicate into individual 1.5-mL screw-top cryotubes containing either 500 µl of TRIzol Reagent (Invitrogen) or Universal Viral Transport Medium (BD), respectively. Samples were then placed directly in liquid nitrogen dry shippers and, upon arrival at the laboratory, transferred to a -80 °C freezer. Staff wore N95 masks, nitrile gloves, dedicated clothing, washable shoes or shoe covers, and protective eyewear during sampling of both live and dead animals.
RNA was extracted using a Zymo Direct-zol RNA kit and stored at -80ºC until analysis. RNA was converted into cDNA using a Maxima H Minus First Strand cDNA Synthesis Kit (Thermo Scientific) and was stored at -20 °C until analysis. Two conventional nested broad-range PCR assays, both targeting conserved regions in the RNA-dependent RNA polymerase gene (RdRp), were used to test the samples for CoV cDNA. The first PCR amplifies a product of approximately 286 nt between the primer binding sites. The first-round (CoV-FWD1: CGT TGG IAC WAA YBT VCC WYT ICA RBT RGG and CoV-RVS1: GGT CAT KAT AGC RTC AVM ASW WGC NAC ATG) and second-round (CoV-FWD2: GGC WCC WCC HGG NGA RCA ATT and CoV-RVS2: GGW AWC CCC AYT GYT GWA YRT C) primers of this PCR were specifically designed for the detection of a broad range of CoVs [10]. The second PCR was used in two modified versions, one of them specifically targeting a broad range of CoVs in bats and the other broadly targeting CoVs of other hosts. In both cases, the semi-nested PCR primers CoV-FWD3 (GGT TGG GAY TAY CCH AAR TGT GA) and CoV-RVS3 (CCA TCA TCA SWY RAA TCA TCA TA) were used for the first round. In the second round, either CoV-FWD4/Bat (GAY TAY CCH AAR TGT GAY AGA GC) or CoV-FWD4/Other (GAY TAY CCH AAR TGT GAU MGW GC) was used as the forward primer, while the reverse primer was again CoV-RVS3 [11]. Both versions amplify 387 nt between the primer binding sites. CoV-RNA-positive samples were subjected to a cytochrome b PCR assay to verify the host species. The primers CytB_F (GAG GMC AAA TAT CAT TCT GAG G) and CytB_R (TAG GGC VAG GAC TCC TCC TAG T) were used to amplify a primer-flanked 435-nt fragment of this highly conserved mitochondrial gene [12].
PCR products were subjected to electrophoresis in a 1.5% agarose gel, and products of the expected amplicon sizes were excised. DNA was extracted using a QIAGEN QIAquick Gel Extraction Kit and was sent for commercial Sanger sequencing (1st BASE). All results from sequencing were analyzed using Geneious 7.1 software and primer trimmed, and the consensus sequences were compared to the GenBank database (BLASTn, NCBI).
All sequences were deposited in the GenBank database under accession numbers MT083286, MT083287, MT083291-MT083296, MT083363-MT083365 and MT083405.
Maximum-likelihood phylogenetic trees were constructed including members of different genera (Alphacoronavirus, Betacoronavirus, and Gammacoronavirus) and species of known CoVs as well as other CoVs detected in Laos during the PREDICT project. Only a single sequence was included for isolates with more than 95% nucleotide sequence identity. Multiple sequence alignments were made in Geneious (version 11.1.3, MUSCLE alignment), and regions supported by less than 50% of the sequences were excluded. Bayesian phylogeny of the polymerase gene fragment was inferred using MrBayes (version 3.2) with the following parameters: Datatype=DNA, Nucmodel=4by4, Nst=1, Coavion=No, # States=4, Rates=Equal, 2 runs, 4 chains of 1,000,000 generations. The sequence of a whale gammacoronavirus served as outgroup to root the trees, with sampling after every 1,000 steps during the process to monitor phylogenetic convergence [13]. The average standard deviation of split frequencies was below 0.0074 for the Watanabe PCR-based analysis and below 0.0054 for the Quan PCR-based analysis (MrBayes recommended final average <0.01). The first 10% of the trees were discarded, and the remaining ones were combined using TreeAnnotator (version 2.5.1; https://beast.bio.ed.ac.uk) and displayed with FIGTREE (1.4.4; https://tree.bio.ed.ac.uk/) [14].
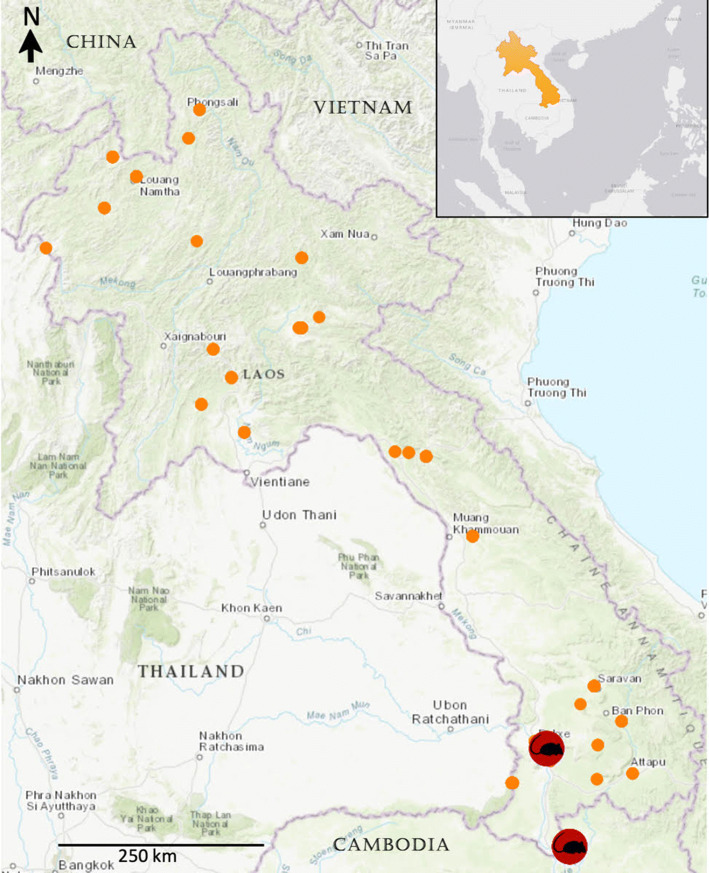
During the sampling phases of the project (2010-2013 and 2016-2018) 851 rodents, 124 carnivores, 44 primates, eight tree shrews, and one colugo (Galeopterus variegatus) were sampled and tested for the presence of CoV RNA (Fig. 1). The rodents belonged to the families Sciuridae (475), Muridae (370), Diatomyidae (5), and Hystricidae (1); the carnivores belonged to the families Viverridae (121), Mustelidae (2), and Felidae (1); the primates belonged to the families Cercopithecidae (23), Lorisidae (18), and Hylobatidae (2), with one remaining unidentified, and all of the tree shrews were of the Tupaiidae family (8) (Supplement 1).
Fig. 1.
Geographical map indicating all sampling sites within Laos (orange dots), highlighting the locations where coronaviruses were detected in rodents
CoV RNA was detected in 12 rodents, which corresponds to 1.4% of the rodents sampled. All 12 of them were sampled in the south of Laos, and all but one were oral swab samples (Fig. 1, Supplement 1). No CoV RNA was detected in any of the carnivores, primates, tree shrews, or the colugo. In all 12 positive animals, it was the Watanabe PCR that amplified CoV nucleic acid. Eleven of the isolates were identical or very similar to each other, and all 12 were similar to CoVs found in different rodents in neighboring China and Vietnam. Nine of the CoV-RNA-positive animals were caught in the same area within a time window of 5 days (Fig. 2, Table 1). Nine of the positive animals, all Rattus exulans, were found in and around human dwellings, while the other were three squirrels (one Dremomys rufigenis, two Menetes berdmorei), sampled at a wet market not far from Pakse, the capital city of Champasak province, where they were being sold for consumption. Eleven of the positive animals were sampled in the dry season (December), and one was sampled in the wet season (June).
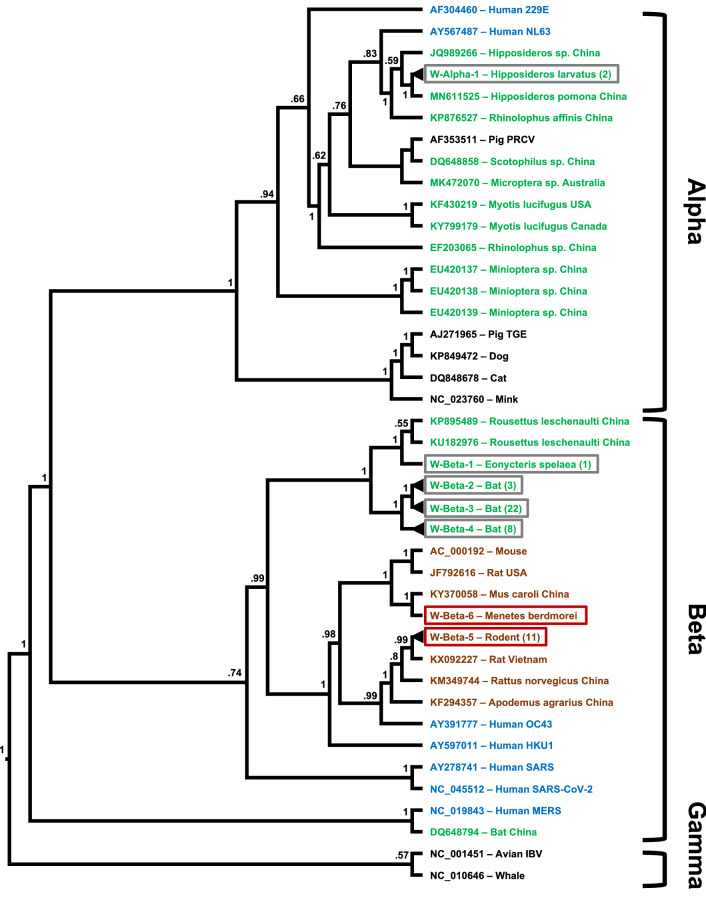
Fig. 2.
Maximum-likelihood phylogenetic tree of coronaviruses presented as a proportional cladogram, based on the 387-nt RdRp region targeted by the PCR by Watanabe et al. [11]. The tree includes the sequences detected here (red boxes) and those described previously in Laos (grey boxes) and indicates the number of isolates with less than 5% difference in brackets for isolates. GenBank accession numbers are listed for published sequences from outside of Laos, while sequences obtained during the PREDICT project are identified by cluster names (compare Table 1 and Supplemental 3). Green font indicates coronavirus sequences obtained from bats; brown font indicates rodents; blue, humans; and black, other hosts. The host species and country of sequence origin are indicated for bats and rodents if applicable. No species is indicated for isolates if detected in more than 1 species (compare Table one and Supplement 3). Numbers at nodes indicate bootstrap support
Table 1.
List of CoV RNA-positive rodent samples
| Group | Isolate & GenBank no | Host | BLASTn 2020-02-25 |
Collection date & PCR |
|---|---|---|---|---|
| W-Beta-5 | LAAR0140 MT083291 |
Rodent, Rattus exulans |
97% Coronaviridae sp. isolate 7565L07R6RATCoV (KX092227) | 2016-12-09 Watanabe |
| W-Beta-5 | LAAR0156 MT083292 |
Rodent, Rattus exulans |
97% Coronaviridae sp. isolate 7565L07R6RATCoV (KX092227) | 2016-12-09 Watanabe |
| W-Beta-5 | LAAR0174 MT083293 |
Rodent, Rattus exulans |
97% Coronaviridae sp. isolate 7565L07R6RATCoV (KX092227) | 2016-12-10 Watanabe |
| W-Beta-5 | LAAR0198 MT083294 |
Rodent, Rattus exulans |
97% Coronaviridae sp. isolate 7565L07R6RATCoV (KX092227) | 2016-12-12 Watanabe |
| W-Beta-5 | LAAR0202 MT083295 |
Rodent, Rattus exulans |
97% Coronaviridae sp. isolate 7565L07R6RATCoV (KX092227) | 2016-12-12 Watanabe |
| W-Beta-5 | LAAR0206 MT083363 |
Rodent, Rattus exulans |
97% Coronaviridae sp. isolate 7565L07R6RATCoV (KX092227) | 2016-12-12 Watanabe |
| W-Beta-5 | LAAR0208 MT083296 |
Rodent, Rattus exulans |
97% Coronaviridae sp. isolate 7565L07R6RATCoV (KX092227) | 2016-12-12 Watanabe |
| W-Beta-5 | LAAR0209 MT083286 |
Rodent, Rattus exulans |
97% Coronaviridae sp. isolate 7565L07R6RATCoV (KX092227) | 2016-12-13 Watanabe |
| W-Beta-5 | LAAR0210 MT083287 |
Rodent, Rattus exulans |
97% Coronaviridae sp. isolate 7565L07R6RATCoV (KX092227) | 2016-12-13 Watanabe |
| W-Beta-5 | LAAR0235 MT083364 |
Rodent, Dremomys rufigenis |
97% Coronaviridae sp. isolate 7565L07R6RATCoV (KX092227) | 2016-12-18 Watanabe |
| W-Beta-5 | LAAR0239 MT083365 |
Rodent, Menetes berdmorei |
97% Coronaviridae sp. isolate 7565L07R6RATCoV (KX092227) | 2016-12-19 Watanabe |
| W-Beta-6 | LAAR0561 MT083405 |
Rodent, Menetes berdmorei |
94% Rodent coronavirus isolate RtMc-CoV-2/YN2013 (KY370058) | 2018-06-12 Watanabe |
Since there are abundant contact opportunities for wildlife pathogens and humans in Laos, and considering that coronavirus-zoonotic events can involve intermediate hosts, as in the cases of SARS and MERS, we focused our screening on non-bat species potentially capable of playing that role. We found CoV RNA in swab samples from several of the rodents examined in the study. Earlier, we noted a relatively high number of diverse CoVs detected in various species of bats all over Laos (Supplements 2 and 3) [5]. This corresponds to similar findings in other countries with a tropical climate, and a hypothesis has been suggested that bats may serve as a seeding host for zoonotic CoV infections [15, 16]. The 1.4% prevalence of CoV RNA in rodents was much lower than what had been detected in bats in Laos; however, such observations have been made repeatedly, re-emphasizing the role of bats as a primary CoV source [5, 15–18]. It is worth noting that we targeted rodents most likely to be in contact with humans and transmit virus and found fewer CoV-RNA-positive animals than in other studies of rodents in the region or elsewhere. A variety of factors may explain the lower incidence of coronavirus-positive animals in this study, including the sample types tested. Studies in which CoV RNA was more frequently detected in rodents have used intestine or fecal matter for their studies, while we tested oral and rectal swab samples [18–20]. We employed swab sampling to minimize harm to live animals and to avoid any damage to rodents possessed by hunters or market vendors during sampling. Organ collection was rarely feasible, even from dead rodents in market settings, since the size and mass of the animals are used to determine their selling price. While potentially underestimating the actual CoV circulation, swab sampling has the advantage of being minimally invasive, quick, and applicable to all species.
The 12 rodent CoV sequences we found fall into two clusters, with 11 of them differing by only one nucleotide. Therefore, these 11 likely belong to the same strain that may have been circulating at that time, since they were obtained from rodents in the same southern region of Laos during December 2016 (Table 1). Both CoV strains detected here cluster with other betacoronaviruses previously detected in rodents in the region (Fig. 2, Table 1). This suggests that the viruses had a longer evolutionary history within rodent hosts and probably did not originate from a recent cross-species transmission event.
None of the CoVs detected in Laos wildlife, neither the ones described earlier in bats nor the ones described here in rodents, have a very close relationship to CoVs currently known to cause human disease. Nevertheless, the rodent CoVs do fall into the same cluster as human CoVs OC43 and HKU1, and these two human CoVs probably originated from direct or indirect spillover events of rodent viruses sometime in the past [21]. However, we still do not know enough about the molecular mechanisms and drivers of zoonotic events to determine the level of risk with certainty. We conclude that Laos’ wildlife harbors diverse CoVs and that a potential for interspecies transmission of viruses and novel diseases exists. Human contact with wildlife such as bats and rodents is common throughout the country, with many rural households consuming bushmeat as a main source of protein and utilizing it as a trade commodity, and this risk potential is particularly relevant. Therefore, behavioral risk reduction, vigilance, ongoing surveillance and research are important to help mitigate the risks of coronavirus zoonotic disease emergence and transmission in the region, especially in the aftermath of COVID-19.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplement 2 Maximum likelihood phylogenetic tree of coronaviruses presented as a proportional cladogram, based on the RdRp region targeted by the PCR by Quan et. al. [10]. The tree includes sequences of bat coronaviruses detected in Laos (grey boxes) and indicates the number of isolates with less than 5% difference in brackets [5]. GenBank accession numbers are listed for previously published sequences, while sequences obtained during the project are identified by cluster names (Supplemental 3). Green font indicates coronavirus sequences obtained from bats, brown font indicates rodents, blue humans and black other hosts. The host species and country of sequence origin are indicated for bats and rodents if applicable, no species is indicated for novel isolates if detected in more than 1 species (Supplemental 3). Numbers at nodes indicate bootstrap support. (PDF 35 kb)
Acknowledgements
The authors would like to thank the government of Laos for the permission to conduct this study, the staff of the National Animal Health Laboratory in Vientiane, the Wildlife Conservation Society, and Metabiota, who assisted in sample collection and testing, as well as Vibol Hul and Philippe Dussart from the Institut Pasteur du Cambodge and any other involved members of the PREDICT-1 and PREDICT-2 consortium (https://ohi.vetmed.ucdavis.edu/programs-projects/predict-project/authorship). This study was made possible by the generous support of the American people through the United States Agency for International Development (USAID) Emerging Pandemic Threats PREDICT program (cooperative agreement numbers GHN-A-OO-09-00010-00 and AID-OAA-A-14-00102). The contents are the responsibility of the authors and do not necessarily reflect the views of USAID or the United States Government.
Compliance with ethical standards
Conflict of interest
Philippe Buchy is currently an employee of GSK vaccines. All other authors have no conflict of interest.
Ethical approval
Animal capture and specimen collection were approved by the Institutional Animal Care and Use Committee (IACUC, UC Davis) and the Ministry of Agriculture and Forestry of Lao PDR.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with Pneumonia in China, 2019. N Engl J Med. 2020;2020:NEJMao2001017. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu R, Zhao X, Li J, et al. Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet. 2020;2020:S0140673620302518. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu B, Zeng LP, Yang XL, Ge XY, Zhang W, Li B, Xie JZ, Shen XR, Zhang YZ, Wang N, Luo DS, Zheng XS, Wang MN, Daszak P, Wang LF, Cui J, Shi ZL. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13:e1006698. doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lacroix A, Duong V, Hul V, San S, Davun H, Omaliss K, Chea S, Hassanin A, Theppangna W, Silithammavong S, Khammavong K, Singhalath S, Greatorex Z, Fine AE, Goldstein T, Olson S, Joly DO, Keatts L, Dussart P, Afelt A, Frutos R, Buchy P. Genetic diversity of coronaviruses in bats in Lao PDR and Cambodia. Infect Genet Evol. 2017;48:10–18. doi: 10.1016/j.meegid.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han BA, Schmidt JP, Bowden SE, Drake JM. Rodent reservoirs of future zoonotic diseases. Proc Natl Acad Sci USA. 2015;112:7039–7044. doi: 10.1073/pnas.1501598112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olival KJ, Hosseini PR, Zambrana-Torrelio C, Ross N, Bogich TL, Daszak P. Host and viral traits predict zoonotic spillover from mammals. Nature. 2017;546:646–650. doi: 10.1038/nature22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greatorex ZF, Olson SH, Singhalath S, Silithammavong S, Khammavong K, Fine AE, Weisman W, Douangngeun B, Theppangna W, Keatts L, Gilbert M, Karesh WB, Hansel T, Zimicki S, O'Rourke K, Joly DO, Mazet JA. Wildlife trade and human health in Lao PDR: an assessment of the Zoonotic disease risk in markets. PLoS ONE. 2016;11:e0150666. doi: 10.1371/journal.pone.0150666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pruvot M, Khammavong K, Milavong P, Philavong C, Reinharz D, Mayxay M, Rattanavong S, Horwood P, Dussart P, Douangngeun B, Theppangna W, Fine AE, Olson SH, Robinson M, Newton P. Toward a quantification of risks at the nexus of conservation and health: the case of bushmeat markets in Lao PDR. Sci Total Environ. 2019;676:732–745. doi: 10.1016/j.scitotenv.2019.04.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quan PL, Firth C, Street C, Henriquez JA, Petrosov A, Tashmukhamedova A, Hutchison SK, Egholm M, Osinubi MO, Niezgoda M, Ogunkoya AB, Briese T, Rupprecht CE, Lipkin WI. Identification of a severe acute respiratory syndrome coronavirus-like virus in a leaf-nosed bat in Nigeria. mBio. 2010;1:e00208-10. doi: 10.1128/mBio.00208-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe S, Masangkay JS, Nagata N, Morikawa S, Mizutani T, Fukushi S, Alviola P, Omatsu T, Ueda N, Iha K, Taniguchi S, Fujii H, Tsuda S, Endoh M, Kato K, Tohya Y, Kyuwa S, Yoshikawa Y, Akashi H. Bat coronaviruses and experimental infection of bats, the Philippines (2010) Emerg Infect Dis. 2010;16:1217–1223. doi: 10.3201/eid1608.100208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Townzen JS, Brower AV, Judd DD. Identification of mosquito bloodmeals using mitochondrial cytochrome oxidase subunit I and cytochrome b gene sequences. Med Vet Entomol. 2008;22:386–393. doi: 10.1111/j.1365-2915.2008.00760.x. [DOI] [PubMed] [Google Scholar]
- 13.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MRBAYES 3.2: efficient Bayesian phylogenetic inference and model selection across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouckaert R, Vaughan TG, Barido-Sottani J, Duchêne S, Fourment M, Gavryushkina A, Heled J, Jones G, Kühnert D, De Maio N, Matschiner M, Mendes FK, Müller NF, Ogilvie HA, du Plessis L, Popinga A, Rambaut A, Rasmussen D, Siveroni I, Suchard MA, Wu CH, Xie D, Zhang C, Stadler T, Drummond AJ. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2019;15:e1006650. doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuen KY, Lau SK, Woo PC. Wild animal surveillance for coronavirus HKU1 and potential variants of other coronaviruses. Hong Kong Med J. 2012;18(Suppl 2):25–26. [PubMed] [Google Scholar]
- 16.Anthony SJ, Johnson CK, Greig DJ, Kramer S, Che X, Wells H, Hicks AL, Joly DO, Wolfe ND, Daszak P, Karesh W, Lipkin WI, Morse SS, PREDICT Consortium. Mazet JAK, Goldstein T. Global patterns in coronavirus diversity. Virus Evol. 2017;3:vex012. doi: 10.1093/ve/vex012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W, Lin XD, Guo WP, Zhou RH, Wang MR, Wang CQ, Ge S, Mei SH, Li MH, Shi M, Holmes EC, Zhang YZ. Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China. Virology. 2015;474:19–27. doi: 10.1016/j.virol.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berto A, Anh PH, Carrique-Mas JJ, Simmonds P, Van Cuong N, Tue NT, Van Dung N, Woolhouse ME, Smith I, Marsh GA, Bryant JE, Thwaites GE, Baker S, Rabaa MA, VIZIONS consortium Detection of potentially novel paramyxovirus and coronavirus viral RNA in bats and rats in the Mekong Delta region of southern Viet Nam. Zoonoses Public Health. 2019;65:30–42. doi: 10.1111/zph.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ge XY, Yang WH, Zhou JH, Li B, Zhang W, Shi ZL, Zhang YZ. Detection of alpha- and betacoronaviruses in rodents from Yunnan, China. Virol J. 2017;14:98. doi: 10.1186/s12985-017-0766-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monchatre-Leroy E, Boué F, Boucher JM, Renault C, Moutou F, Ar Gouilh M, Umhang G. Identification of alpha and beta coronavirus in wildlife species in France: bats, rodents, rabbits, and hedgehogs. Viruses. 2017;9:364. doi: 10.3390/v9120364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement 2 Maximum likelihood phylogenetic tree of coronaviruses presented as a proportional cladogram, based on the RdRp region targeted by the PCR by Quan et. al. [10]. The tree includes sequences of bat coronaviruses detected in Laos (grey boxes) and indicates the number of isolates with less than 5% difference in brackets [5]. GenBank accession numbers are listed for previously published sequences, while sequences obtained during the project are identified by cluster names (Supplemental 3). Green font indicates coronavirus sequences obtained from bats, brown font indicates rodents, blue humans and black other hosts. The host species and country of sequence origin are indicated for bats and rodents if applicable, no species is indicated for novel isolates if detected in more than 1 species (Supplemental 3). Numbers at nodes indicate bootstrap support. (PDF 35 kb)