Abstract
Root-knot nematodes from the genus Meloidogyne are polyphagous plant endoparasites and agricultural pests of global importance. Here, we report the high-quality genome sequence of Meloidogyne luci population SI-Smartno V13. The resulting genome assembly of M. luci SI-Smartno V13 consists of 327 contigs, with an N50 contig length of 1,711,905 bp and a total assembly length of 209.16 Mb.
Keywords: Genome, Genomics, Illumina, Meloidogyne luci, PacBio, Root-knot nematode.
Root-knot nematodes (RKN) from the genus Meloidogyne parasitize a wide range of host plants and have a global distribution. They are considered the most important group of plant-parasitic nematodes (Jones et al., 2013). Field infestations result in economic damage due to reduction or loss of crop yield with estimated global annual losses of $110bn (Danchin et al., 2013; Bebber et al., 2014). Among RKN, the tropical species belonging to Meloidogyne Clade I reproduce asexually by mitotic parthenogenesis (except M. floridensis) and parasitize a broader range of hosts than their sexual relatives (Castagnone-Sereno and Danchin, 2014). Several genomes of Clade I tropical Meloidogyne spp. have been sequenced (Abad et al., 2008; Lunt et al., 2014; Blanc-Mathieu et al., 2017; Szitenberg et al., 2017) and have revealed them to be complex allopolyploids with heterozygous duplicated genome regions and abundant transposable elements (Blanc-Mathieu et al., 2017; Szitenberg et al., 2017). Previous genome assemblies largely relied on short-read next-generation sequencing which limited the contiguity of the assemblies. Sato et al. (2018) found that applying long-read sequencing technologies such as Pacific Biosciences single-molecule real-time (SMRT) significantly improved the contiguity of their Meloidogyne arenaria assembly.
The species belonging to the Meloidogyne ethiopica group include the closely related species M. ethiopica, M. inornata and M. luci (Gerič Stare et al., 2019). The phylogenetic positions of different populations of M. ethiopica group species within Clade I Meloidogyne are incompletely resolved. Isolated specimens of M. luci in Europe were previously misidentified as M. ethiopica due to their high similarity (Gerič Stare et al., 2017). We used long-read Pacific Biosciences Sequel and short-read Illumina HiSeqX sequencing data to produce a high-quality Meloidogyne luci genome assembly. The M. luci population SI-Smartno was isolated from tomato plants grown in a commercial production greenhouse in Šmartno, Slovenia (Gerič Stare et al., 2018). A line (V13) was reared from the progeny of a single female and multiplied on tomato (Solanum lycopersicum “Val”). Nematode eggs were obtained by hypochlorite extraction (Hussey and Barker, 1973) and cleaned by sucrose flotation (McClure et al., 1973). Genomic DNA (gDNA) was obtained by phenol-chloroform extraction from the nematode eggs ground in liquid nitrogen. Following fluorometric quantification (Qubit; Thermo Fisher Scientific), a total of 6.64 μg of gDNA was used for Illumina whole-genome sequencing (WGS) on HiSeqX platform. 150 bp paired-end reads were generated from 350 bp insert TruSeq DNA PCR-Free libraries, yielding 206,071,630 reads (30.9 Gb). Reads were quality checked with FastQC v0.11.8 (Andrews, 2018) and trimmed with Trimmomatic v0.36 (Bolger et al., 2014) using the Phred quality score cutoff at 20. Prior to Pacific Biosciences SMRT sequencing on the Sequel, gDNA was assessed using the Femto Pulse system (Agilent) and a total of 10 μg of gDNA was used. The SMRTbell Express template prep kit (Pacific Biosciences) was used to prepare PacBio library >20 kb using standard protocol without shearing (Procedure & Checklist – Preparing >15 kb Libraries Using SMRTbell® Express Template Preparation Kit). Blue Pippin (Sage Science, MA, USA) was used for size selection (25 kb cutoff). Libraries were sequenced on the Sequel using v2.1 Sequencing and Binding kits generating 3,617,847 reads (42.4 Gb). We generated approximately 150-fold and 200-fold genome coverage using Illumina and PacBio data, respectively. Adapter and barcode sequences were filtered out within the Sequel instrument and assembled with HGAP4 pipeline (SMRT Link suite v5.1.0.26412, Pacific Biosciences) and polished with Pilon (Walker et al., 2014) using trimmed Illumina data.
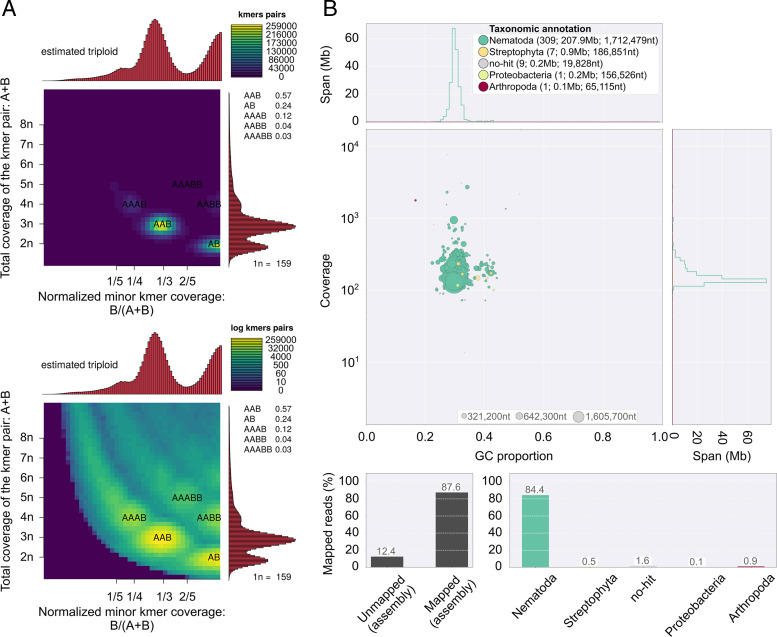
The assembled M. luci SI-Smartno genome consists of 327 contigs with a minimum contig length of 10,147 bp and N50 contig length of 1,711,905 bp. The total length of assembly is 209.16 Mb. Smudgeplot v0.1.3 (Ranallo-Benavidez et al., in press) and Jellyfish v1.0 (Marçais and Kingsford, 2011) were used to estimate genome ploidy based on the counting of k-mers (k = 21) on short-read data. The genome is estimated to be triploid (AAB). Blobtools (Laetsch and Blaxter, 2017) was used to assess contaminant DNA presence (Fig. 1B). The assembly is currently the most contiguous RKN assembly (Table 1) available with an estimated coverage of 95.16% of the coding space based on Core Eukaryotic Genes Mapping Approach (CEGMA) analysis (Parra et al., 2007) and the average number of CEGs at 2.88 supports the triploid genome model (Fig. 1A). The polished assembly was 88.1% complete based on the eukaryote set (n = 303) of Benchmarking Universal Single-Copy Orthologs (Simão et al., 2015). The assembly of M. luci SI-Smartno can now be used to determine the correct phylogenetic position of the clade, identification of genetic changes related to the origins of virulence, and in the study of evolutionary history of this organism.
Figure 1:
Genome ploidy estimation and contaminant analysis of the Meloidogyne luci SI-Smartno genome assembly. (A) Smudgeplots showing the coverage and distribution of k-mer pairs that fit to triploid genome model. (B) Blobplot showing the lack of contamination of assembly by foreign (non-Nematoda) genetic material.
Table 1.
Summary statistics of the Meloidogyne luci genome assembly compared to the genome assemblies of other Meloidogyne spp. currently available in DDBJ/ENA/GenBank.
| Species | Strain/isolate desig-nation | Accession (DDBJ/ENA/GenBank) | Assembly size (Mb) | Genome coverage | Number of contigs/scaffolds | N50 | GC content (%) | Number of pre-dicted genes | CEGMA score (% complete) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| M. luci | SI-Smartno V13 | ERS3574357 | 209.16 | 200 | 327 | 1,711,905 | 30.2 | n/a | 95.2 | This study |
| M. incognita | Morelos | GCA_000180415.1 | 82.10 | 5 | 9,538 | 12,786 | 31.4 | 19,212 | 77 | Abad et al. (2008) |
| M. incognita | W1 | GCA_003693645.1 | 121.96 | 100 | 33,351 | 16,520 | 30.6 | 24,714 | 83 | Szitenberg et al. (2017) |
| M. incognita | V3 | GCA_900182535.1 | 183.53 | 100 | 12,091 | 38,588 | 29.8 | 45,351 | 97 | Blanc-Mathieu et al. (2017) |
| M. javanica | VW4 | GCA_003693625.1 | 150.35 | 300 | 34,316 | 14,128 | 30.2 | 26,917 | 90 | Szitenberg et al. (2017) |
| M. javanica | – | GCA_900003945.1 | 235.80 | 100 | 31,341 | 10,388 | 29.9 | 98,578 | 96 | Blanc-Mathieu et al. (2017) |
| M. floridensis | – | GCA_000751915.1 | 96.67 | 200 | 58,696 | 3,698 | 30.0 | n/a | 58.1 | Lunt et al. (2014) |
| M. floridensis | SJF1 | GCA_003693605.1 | 74.85 | 100 | 8,887 | 13,261 | 30.2 | 14,144 | 84 | Szitenberg et al. (2017) |
| M. arenaria | HarA | GCA_003693565.1 | 163.75 | 100 | 46,436 | 10,504 | 30.3 | 30,308 | 91 | Szitenberg et al. (2017) |
| M. arenaria | – | GCA_900003985.1 | 258.07 | 100 | 26,196 | 16,462 | 29.8 | 103,001 | 95 | Blanc-Mathieu et al. (2017) |
| M. arenaria | A2-O | GCA_003133805.1 | 284.05 | 60 | 2,224 | 204,551 | 30.0 | n/a | 94.8 | Sato et al. (2018) |
| M. enterolobii | L30 | GCA_003693675.1 | 162.97 | 200 | 42,008 | 10,552 | 30.2 | 31,051 | 81 | Szitenberg et al. (2017) |
| M. graminicola | IARI | GCA_002778205.1 | 38.19 | 180 | 4,304 | 20,482 | 23.1 | 10,196 | 84.3 | Somvanshi et al. (2018) |
| M. hapla | VW9 | GCA_000172435.1 | 53.01 | 10 | 3,450 | 37,608 | 27.4 | 14,420 | 94.8 | Opperman et al. (2008) |
Note: n/a, not assessed.
Data availability and accession number(s)
Procedural information concerning the genome assembly and analysis presented in this paper can be found at the GitHub repository at https://github.com/CristianRiccio/mluci. The sequences have been deposited in DDBJ/ENA/GenBank under the accession number ERS3574357.
Acknowledgments
The sequencing was performed at Edinburgh Genomics and the Wellcome Sanger Institute. This research was partially funded by Slovenian Research Agency (ARRS Grants No. MR 38128, P4–0072).
References
- Abad P., Gouzy J., Aury J.-M., Castagnone-Sereno P., Danchin E. G. J., Deleury E., Perfus-Barbeoch L., Anthouard V., Artiguenave F., Blok V. C., Caillaud M.-C., Coutinho P. M., Dasilva C., De Luca F., Deau F., Esquibet M., Flutre T., Goldstone J. V., Hamamouch N., Hewezi T., Jaillon O., Jubin C., Leonetti P., Magliano M., Maier T. R., Markov G. V., McVeigh P., Pesole G., Poulain J., Robinson-Rechavi M., Sallet E., Ségurens B., Steinbach D., Tytgat T., Ugarte E., van Ghelder C., Veronico P., Baum T. J., Blaxter M., Bleve-Zacheo T., Davis E. L., Ewbank J. J., Favery B., Grenier E., Henrissat B., Jones J. T., Laudet V., Maule A. G., Quesneville H., Rosso M.-N., Schiex T., Smant G., Weissenbach J. and Wincker P.. 2008. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita . Nature Biotechnology 26:909–915. [DOI] [PubMed] [Google Scholar]
- Andrews S. 2018. FastQC – a quality control tool for high throughput sequence data. Babraham Bioinformatics, The Babraham Institute, Cambridge, available at: www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed January 15, 2019).
- Bebber D. P., Holmes T. and Gurr S. J.. 2014. The global spread of crop pests and pathogens. Global Ecology and Biogeography 23:1398–1407. [Google Scholar]
- Blanc-Mathieu R., Perfus-Barbeoch L., Aury J.-M., Da Rocha M., Gouzy J., Sallet E., Martin-Jimenez C., Bailly-Bechet M., Castagnone-Sereno P., Flot J.-F., Kozlowski D. K., Cazareth J., Couloux A., Da Silva C., Guy J., Kim-Jo Y.-J., Rancurel C., Schiex T., Abad P., Wincker P. and Danchin E. G. J.. 2017. Hybridization and polyploidy enable genomic plasticity without sex in the most devastating plant-parasitic nematodes. PLoS Genetics 13:1–36, available at: 10.1371/journal.pgen.1006777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M. and Usadel B.. 2014. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castagnone-Sereno P. and Danchin E. G. J.. 2014. Parasitic success without sex – the nematode experience. Journal of Evolutionary Biology 27:1323–1333. [DOI] [PubMed] [Google Scholar]
- Danchin E. G. J., Arguel M.-J., Campan-Fournier A., Perfus-Barbeoch L., Magliano M., Rosso M.-N., Da Rocha M., Da Silva C., Nottet N., Labadie K., Guy J., Artiguenave F. and Abad P.. 2013. Identification of novel target genes for safer and more specific control of root-knot nematodes from a pan-genome mining. PLoS Pathogens 9:1–15, available at: 10.1371/journal.ppat.1003745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerič Stare B., Strajnar P., Širca S., Susič N. and Urek G.. 2018. Record of a new location for tropical root knot nematode Meloidogyne luci in Slovenia. EPPO Buletin 48:135–137. [Google Scholar]
- Gerič Stare B., Strajnar P., Susič N., Urek G. and Širca S.. 2017. Reported populations of Meloidogyne ethiopica in Europe identified as Meloidogyne luci . Plant Disease 101:1627–1632. [DOI] [PubMed] [Google Scholar]
- Gerič Stare B., Aydınlı G., Devran Z., Mennan S., Strajnar P, Urek G. and Širca S.. 2019. Recognition of species belonging to Meloidogyne ethiopica group and development of a diagnostic method for its detection. European Journal of Plant Pathology 154:621–633, available at: 10.1007/s10658-019-01686-2 [DOI] [Google Scholar]
- Hussey R. S. and Barker K. R.. 1973. Comparison of methods of collecting inocula for Meloidogyne spp., including a new technique. Plant Disease Reporter 57:1025–1028. [Google Scholar]
- Jones J.T., Haegeman A., Danchin E. G. J., Gaur H. S., Helder J., Jones M. G. K., Kikuchi T., Manzanilla-López R., Palomares-Rius J. E., Wesemael W. M. L. and Perry R. N.. 2013. Top 10 plant-parasitic nematodes in molecular plant pathology. Molecular Plant Pathology 14:946–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laetsch D. R. and Blaxter M. L.. 2017. BlobTools: interrogation of genome assemblies. F1000Research 6:1287. [Google Scholar]
- Lunt D. H., Kumar S., Koutsovoulos G. and Blaxter M. L.. 2014. The complex hybrid origins of the root knot nematodes revealed through comparative genomics. PeerJ 2:1, available at: 10.7717/peerj.356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure M. A., Kruk T. H. and Misaghi I.. 1973. A method for obtaining quantities of clean Meloidogyne eggs. Journal of Nematology 5:230. [PMC free article] [PubMed] [Google Scholar]
- Marçais G. and Kingsford C.. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27:764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman C. H., Bird D. M., Williamson V. M., Rokhsar D. S., Burke M., Cohn J., Cromer J., Diener S., Gajan J., Graham S., Houfek T. D., Liu Q., Mitros T., Schaff J., Schaffer R., Scholl E., Sosinski B. R., Thomas V. P. and Windham E.. 2008. Sequence and genetic map of Meloidogyne hapla: a compact nematode genome for plant parasitism. Proceedings of the National Academy of Sciences of the United States of America 105:14802–14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G., Bradnam K. and Korf I.. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23:1061–1067. [DOI] [PubMed] [Google Scholar]
- Ranallo-Benavidez T. R., Jaron K. S. and Schatz M. C.. (in press). GenomeScope 2.0 and Smudgeplots: reference-free profiling of polyploid genomes. Nature Communications. [DOI] [PMC free article] [PubMed]
- Sato K., Kadota Y., Gan P., Bino T., Uehara T., Yamaguchi K., Ichihashi Y., Maki N., Iwahori H., Suzuki T., Shigenobu S. and Shirasu K.. 2018. High-quality genome sequence of the root-knot nematode Meloidogyne arenaria genotype A2-O. Genome Announcements 6:e00519–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V. and Zdobnov E. M.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Somvanshi V. S., Tathode M., Shukla R. N. and Rao U.. 2018. Nematode genome announcement: a draft genome for rice root-knot nematode, Meloidogyne graminicola . Journal of Nematology 50:111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szitenberg A., Salazar-Jaramillo L., Blok V. C., Laetsch D. R., Joseph S., Williamson V. M., Blaxter M. L. and Lunt D. H.. 2017. Comparative genomics of apomictic root-knot nematodes: hybridization, ploidy, and dynamic genome change. Genome Biology and Evolution 9:2844–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B.J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C. A., Zeng Q., Wortman J., Young S. K. and Earl A. M.. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PloS One 9:1–14, available at: 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Procedural information concerning the genome assembly and analysis presented in this paper can be found at the GitHub repository at https://github.com/CristianRiccio/mluci. The sequences have been deposited in DDBJ/ENA/GenBank under the accession number ERS3574357.