Abstract
Plant recognition and defence against pathogens employs a two-tiered perception system. Surface-localized pattern recognition receptors (PRRs) act to recognize microbial features, whereas intracellular nucleotide-binding leucine-rich repeat receptors (NLRs) directly or indirectly recognize pathogen effectors inside host cells. Employing the tomato PRR LeEIX2/EIX model system, we explored the molecular mechanism of signalling pathways. We identified an NLR that can associate with LeEIX2, termed SlNRC4a (NB-LRR required for hypersensitive response-associated cell death-4). Co-immunoprecipitation demonstrates that SlNRC4a is able to associate with different PRRs. Physiological assays with specific elicitors revealed that SlNRC4a generally alters PRR-mediated responses. SlNRC4a overexpression enhances defence responses, whereas silencing SlNRC4 reduces plant immunity. Moreover, the coiled-coil domain of SlNRC4a is able to associate with LeEIX2 and is sufficient to enhance responses upon EIX perception. On the basis of these findings, we propose that SlNRC4a acts as a noncanonical positive regulator of immunity mediated by diverse PRRs. Thus, SlNRC4a could link both intracellular and extracellular immune perceptions.
Keywords: EIX, endocytosis, immune receptors, LeEIX2, NB-LRR, NLR, pattern triggered immunity, SlNRC4
1 |. INTRODUCTION
Plants are constantly being attacked by a wide range of pathogens that cause significant crop losses (Strange & Scott, 2005). Plant defence mechanisms depend on the capacity of each individual cell to initiate immune responses using both cell surface and intracellular receptors (Couto & Zipfel, 2016; Dodds & Rathjen, 2010). Plant immunity can be triggered by two types of molecules: microbial-associated molecular patterns (MAMPs) recognized at the cell surface and pathogen effectors recognized predominantly inside host cells (Henry, Yadeta, & Coaker, 2013; Thomma, Nurnberger, & Joosten, 2011). MAMP recognition results in pattern-triggered immunity (PTI), whereas effector recognition results in effector-triggered immunity (ETI).
Plant cell surface receptors include receptor-like kinases (RLKs) and receptor-like proteins (RLPs), which can function as pattern recognition receptors (PRRs) to perceive MAMPs (Bohm, Albert, Fan, Reinhard, & Nurnberger, 2014; Macho & Zipfel, 2014; Zipfel, 2014). RLKs possess an extracellular leucine-rich repeat (LRR) domain potentially involved in ligand binding, a single transmembrane domain and a cytoplasmic kinase domain. RLPs share the same basic structure, but lack a kinase or any other obvious signalling domain. Therefore, RLPs are assumed to rely on other proteins to initiate a signalling cascade (Gust & Felix, 2014; Tor, Lotze, & Holton, 2009). The most extensively studied RLK is FLS2, which recognizes bacterial flagellin and the flagellin-derived peptide flg22, leading to the induction of defence responses (Gomez-Gomez & Boller, 2000; Schlecht, Keske, Hierholzer, & Felix, 1999; Zipfel et al., 2004). In tomato (Solanum lycopersicum), several RLPs have been implicated in defence responses. The tomato LeEIX2 RLP recognizes and responds to the fungal MAMP-EIX (ethylene-inducing xylanase; Ron & Avni, 2004). Other extracellular RLPs can also recognize extracellular effectors, thus triggering ETI. The tomato Cf family and Ve1 receptors confer immunity upon recognition of Cladosoporium fulvum effector proteins and the Ave1 effector from Verticillium dahliae, respectively (Kawchuk et al., 2001; Ron & Avni, 2004; Takken et al., 1999; van der Hoorn et al., 2005).
Intracellular immune receptors respond to translocated effectors and activate ETI (Bonardi, Cherkis, Nishimura, & Dangl, 2012; Cui, Tsuda, & Parker, 2015). Most intracellular immune receptors belong to the nucleotide-binding leucine-rich repeat (NB-LRR or NLR) super-family. The activation of NLRs occurs either directly, by binding pathogen-secreted effectors, or indirectly, by detecting effector-induced perturbation of host immune signalling components (Win et al., 2012). NLRs consist of a central NB domain, which includes a catalytic P-loop motif responsible for the NLR activation state, and a C-terminal LRR, which is highly polymorphic and thought to confer recognition specificity (Bonardi & Dangl, 2012). NLRs are classified into two subgroups, based on their N-terminal domain: TIR-NB-LRR (TNL) proteins containing a Toll-like domain and the CC-NB-LRR (CNL) proteins characterized by a coiled-coil domain (Marone, Russo, Laido, De Leonardis, & Mastrangelo, 2013). In Arabidopsis thaliana, TNLs form the largest group of NLRs (Yu et al., 2014), whereas most solanaceous NLRs belong to the CNL class (Andolfo et al., 2014). Recently, the NLR protein required for hypersensitive response (HR)-associated cell death (NRC) family of NLRs has been demonstrated to act as helper NLRs that contribute to sensor NLR perception in solanaceous plants (Gabriels et al., 2007; Wu et al., 2017). NbNRC4 in particular has been demonstrated to be a helper NLR that contributes to immunity mediated by sensor NLRs that perceive distinct effectors from bacterial, viral, filamentous nematodes, and insect pathogens and pests (Wu et al., 2017). Within plant genomes, NLRs are organized either as isolated genes or as linked clusters that are thought to enable rapid evolution of intracellular immune receptors (McDowell & Simon, 2006).
In tomato, LeEIX2/EIX-mediated PTI responses include a reactive oxygen species (ROS) burst, induction of ethylene biosynthesis, expression of pathogenicity-related proteins, and the HR (Avni, Bailey, Mattoo, & Anderson, 1994; Bailey, Dean, & Anderson, 1990; Ron & Avni, 2004). EIX was shown to specifically bind to the plasma membrane of responsive cultivars of both tomato and tobacco (Hanania & Avni, 1997; Ron & Avni, 2004). After binding, EIX triggers internalization of LeEIX2 on endosomes (Bar & Avni, 2009; Ron & Avni, 2004).
Several attempts have been made to elucidate LeEIX2-mediated defence signalling by identifying LeEIX2-associated proteins (Bar, Sharfman, Schuster, & Avni, 2009; Liebrand et al., 2013). However, proteins activating downstream signalling responses have thus far remained elusive. Here, we report the isolation and identification of a tomato LeEIX2-associated NLR protein that is a member of the NRC family. We have named this protein SlNRC4a according to its close orthologs in Nicotiana benthamiana (NbNRC4a,b) (Wu et al., 2017). Here, we show an intriguing and noncanonical involvement of SlNRC4a in PTI, as a positive regulator of LeEIX2/EIX-mediated defence responses.
2 |. MATERIALS AND METHODS
2.1 |. Plant materials and growth conditions
Nicotiana tabacum cv samsun NN, N. benthamiana, and S. lycopersicum cv M82 and IL7–5 were grown from seeds in soil (Green Mix; Even-Ari, Ashdod, Israel) in a growth chamber, under long day conditions (16 hr:8 hr, light:dark) at 24°C. For stable transformation, tomato seeds were surface sterilized and germinated on Nitsch medium (Nitsch & Nitsch, 1969; Duchefa, Haarlem, The Netherlands).
2.2 |. Plasmid construction
For overexpression assays, LeEIX2 cDNA C-terminally tagged with green fluorescent protein (GFP) was cloned into the SalI site of pBINPLUS (van Engelen et al., 1995) using the following primers: LeEIX2 forward primer 5′-ATGTCGACATGGGCAAAAGAACTAATC-3′ and LeEIX2 reverse primer 5′-ATGTCGACGTTCCTTAGCTTTCCCTTCAGTC-3′. SlNRC4a cDNA C-terminally tagged with GFP, mCherry, or 2xHA was cloned into the SalI XbaI sites of pBINPLUS (van Engelen et al., 1995) using the following primers: SlNRC4a forward primer 5′-GTCGACATGGCAGATGCAGTGG-3′ and SlNRC4a reverse primer 5′-TCTAGAATTTTCAGGTGGGTATATGCTTAG-3′ between the CAM35SΩ promoter containing the translation enhancer signal and the Nos terminator. The truncated mutant was generated from the full-length plasmid using the following reverse primers and subcloned as described above: SlNRC4a-CCd reverse primer 5′-ACTCTAGAGTTACGAATGTCTTTG-3′ and SlNRC4aCRIPSR fragment reverse primer 5′-TCTAGATGTATTTGTTCCACCAAAACTTTCCACTGGCTCACTATTTG-3′.
2.3 |. Tomato stable transformation
Transgenic LeEIX2 GFP line and slnrc4a CRISPR lines were generated using IL7–5 and M82 tomato seedlings, respectively, by cotyledon transformation according to McCormick et al. (1986). Agrobacterium tumefaciens strain GV3101 harbouring the relevant constructs was used for cotyledon cocultivation. Homozygous T5 LeEIX2 GFP line and homozygous T2 slnrc4a CRISPR lines slnrc4a-2 and slnrc4a-5 were used for experimental work.
2.4 |. Transient expression by agroinfiltration
Binary vector clones were introduced by electroporation into A. tumefaciens strain GV3101. Agrobacterium cells were grown in LB medium containing 50 mg L−1 of kanamycin, 40 mg L−1 of gentamycin, and 100 mg L−1 of rifampicin overnight at 28°C, diluted into virulence (VIR) induction medium (50 mM of MES pH 5.6, 0.5% [w/v] glucose, 1.7 mM of NaH2PO4, 20 mM of NH4Cl, 1.2 mM of MgSO4, 2 mM of KCl, 17 μM of FeSO4, 70 μM of CaCl2, and 200 μM of acetosyringone), and grown for six additional hours until OD600 reached 0.4–0.6. Suspensions containing single or mixed Agrobacterium cultures were diluted to a final OD600 of 0.15–0.2 in VIR induction medium. Cultures were infiltrated with a needless syringe into leaves of N. tabacum cv samsun NN or N. benthamiana. Leaves were harvested 40 hr after injection for ethylene biosynthesis and ROS assays or confocal microscopy analysis.
2.5 |. LeEIX2-associated protein identification by immunopurification, followed by tryptic digest and mass spectrometry
Immunopurification from the transgenic tomato line expressing LeEIX2-GFP was performed as described below. Nine grams of plant tissue from Leaves 4–6 of 8-week-old TL4 or control IL7–5 tomato plants were harvested for immunopurification. As LeEIX2 possesses a transmembrane domain, fractionation methodology was applied in order to enrich the detergent soluble fraction. Proteins were extracted using three volumes of detergent-free extraction buffer (EB: 150 mM of NaCl, 50 mM of phosphate buffer pH 7.5, and 2 mM of MgCl2) and one complete protease inhibitor tablet (Roche, Germany) per 50 ml of EB. Pellets were ground with two volumes of EB containing 0.5% Triton X-100. The supernatant was diluted with three volumes of detergent-free EB in order to obtain a final concentration of 0.2% Triton X-100 before adding GFP-TrapA beads according to manufacturer’s recommendations (Chromotek, Planegg-Martinsried, Germany). Beads were incubated for 4 hr and then washed five times with detergent-free EB. Tryptic on-bead digestion was performed. Peptides were subjected to mass spectrometry as previously described (Liu, Elmore, Lin, & Coaker, 2011). Mass spectra were detected on a Q Exactive mass spectrometer (Thermo Fisher Scientific). Raw mass spectrometry data were searched with X!Tandem version Sledgehammer against the tomato proteome ITAG2.3 (SolGenomics, ftp://ftp.solgenomics.net/proteomics/Solanum_lycopersicum/) and imported into Scaffold 4.8.3 (Proteome Software) as described by Yachdav et al. (2014). Mass spectrometry was performed at the UC Davis Proteomics Core.
2.6 |. Co-immunoprecipitation
Co-immunoprecipitation assays were performed as described by Leibman-Markus, Schuster, and Avni (2017). N. benthamiana leaves transiently co-expressing LeEIX2-GFP, LeEIX2, SlNRC4a-HA, or SlNRC4a-mCHERRY were harvested 40 hr after infiltration. Leaf petioles were immersed in EIX 300 μg ml−1 (or water as mock) for 7 min and then transferred to water for an additional 7 min. Five hundred milligrams of leaf tissue was used for co-immunoprecipitation, with 13 μl of GFP-TrapA beads (Chromotek, Planegg-Martinsried, Germany).
2.7 |. ROS measurement
ROS burst was measured as previously described by Leibman-Markus et al. (2017). Leaf disks (0.5 cm in diameter) were taken from either transiently expressing tobacco plants 40 hr post-infection or stable transgenic tomato lines. Disks were floated in 250 μl of ddH2O in a white 96-well plate (SPL Life Sciences, Korea) for 4–6 hr at room temperature. After incubation, the water was completely removed and ROS measurement reaction containing EIX 1 μg ml−1, flg22 1 mM, or dimethyl sulfoxide (DMSO) as mock was added and light emission was immediately measured using a microplate luminometer (Turner BioSystems Veritas, California, USA).
2.8 |. Ethylene measurement
Ethylene biosynthesis was measured as previously described by Leibman-Markus et al. (2017). Leaf disks (0.9 cm in diameter) were taken from transiently expressing tobacco plants 40-hr postinfection, and virus-induced gene silencing (VIGS) inoculated tomato plants or stable transgenic tomato lines. Five (tobacco) and six (tomato) disks were sealed in each 10-ml flask containing 1 ml of assay medium (with or without 1 μg ml−1 of EIX) and incubated with shaking for four (tobacco) and five (tomato) hours at room temperature. Ethylene production was measured by gas chromatography (Varian 3350, Varian, California, USA).
2.9 |. Fungal resistance assay
Botrytis cinerea isolates Bc-16 (Elad & Yunis, 1993) and was grown on PDA plates containing 250 mg L−1 of chloramphenicol, at 22°C. Ten days after subculturing, spores were collected from sporulated plates and suspended in water supplemented with 0.2% glucose and 0.2% KH2PO4. A 10-μl drop of concentrated spore solution (5 × 104/mL) was placed on 3–4 leaflets each of the fourth and/or fifth leaf on each tomato plant. Drops were allowed to dry for several minutes, and infected leaves were placed in closed plastic bags to maintain humidity. Lesion areas were measured using ImageJ.
2.10 |. Bioinformatics analyses
Tomato SlNRC4a (Solyc04g007070) was used to identify homologs in the tomato genome (ITAG release 2.40). The phylogenetic tree of SlNRC4a homologs was built using multiple alignment (CLUSTAL_W) and maximum likelihood (Phylogeny.fr) methods, with bootstrap values based on 1,000 iterations (Dereeper et al., 2008).
Secondary structure prediction for SlNRC4a and SlNRC4aCRISPR was performed using PROF method of Protein Predict (Yachdav et al., 2014). SlNRC4a, SlNRC4aCRISPR, and StRx (Genebank accession number CAB50786) Protein alignment was performed using CLUSTAL_W in BioEdit.
2.11 |. Virus-induced gene silencing
VIGS was performed in tomato according to Liu, Schiff, and Dinesh-Kumar (2002). A. tumefaciens strain GV3101 harbouring TRV RNA1 (pYL155) and TRV RNA2 (pYL170) was mixed in a 1:1 ratio in infiltration buffer. TRV RNA2 empty served as a control, SlNRC4a was cloned into the XhoI and BamHI sites of TRV RNA2 using the following primers: forward primer 5′-CCCTCGAGTGCAGAACATGTCAGATCTTTCTATTG-3′ and reverse primer 5′-GAGGATCCAGTCGGGGATGGCAATTTTG-3′. The resulting 358-bp fragment targets all three SlNRC4 genes.
Agrobacterium mixtures were infiltrated into cotyledons of 10-day-old tomato M82 seedlings. Six-week-old plants were used for gene level expression assessment and physiological assays. Four plants per treatment were used in each experiment.
2.12 |. RNA extraction and qRT-PCR analysis
Plant total RNA was extracted using SV Total RNA Isolation System (Promega, Wisconsin, USA). Four micrograms of RNA samples were used for first strand cDNA synthesis using M-MLV reverse transcriptase (Promega, Wisconsin, USA) and oligodT15. qRT-PCR was performed according to the Fast SYBR Green Master Mix protocol (Life Technologies, Thermo Fisher, Massachusetts, USA), using a StepOnePlus machine (Thermo Fisher, Massachusetts, USA). SlNRC4a expression was examined using forward primer 5′-ACAACAATCAAGCACTTCAGGC-3′ and reverse primer 5′-TCTCCAAAGAAGGAC-CCTGAGTT-3′ and the endogenous control gene Solyc04g015210 with forward primer 5′-TTGTCCAGGAGGAACAGGGTT-3′ and reverse primer 5′-ACCAAGTCCCGGCATTC-CTA-3′. Endogenous normalizing gene Sl-cyclophilin (Solyc01g111170) was amplified using forward primer 5′-TGAGTGGCTCAACGGAAAGC-3′ and reverse primer 5′-CCA-ACAGCCTCTGCCTTCTTA-3′. qRT-PCR was performed using the following program: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 45 s. Control samples without reverse transcriptase did not generate a PCR product after 40 amplification cycles, indicating the samples were free of genomic DNA contamination.
2.13 |. Western blot
Western blot was performed on N. benthamiana leaves transiently expressing relevant constructs. One hundred milligrams of plant tissue were ground to a fine powder with liquid nitrogen, and three volumes of extraction buffer (50 mM of Tris-HCl, pH 7.5, 2 mM of MgCl2, 150 mM of NaCl, 140 mM of β-mercaptoethanol, 2 mM of phenylmethylsulfonyl fluoride (PMSF), and one complete protease inhibitor tablet, without EDTA [Roche, Germany] per 50 ml) were added. Samples were centrifuged, and supernatant cytosolic fraction was discarded (or collected if required). Pellets were ground using two volumes of EB with 1% Triton X-100 and incubated in a rotating wheel at 4°C for 20 min before centrifugation. Supernatant samples (TSM) were collected and boiled after adding sample buffer (8% sodium dodecyl sulphate, 40% glycerol, 200 mM of Tris-Cl, pH 6.8, 388 mM of dithiothreitol, and 0.1 mg ml−1 of bromophenol blue dye). Samples were run in sodium dodecyl sulphate-polyacrylamide gel electrophoresis, blotted onto nitrocellulose membranes, and incubated with antibodies as required: mouse anti-mCherry and rat anti-GFP (Chromotek, Planegg-Martinsried, Germany) and mouse anti-HA (BioLegend, California, USA).
2.14 |. Confocal microscopy
Confocal microscopy images were acquired using a Zeiss LSM780 confocal microscope system with the Objective LD SC Plan-Apochromat 20×/1.0 Corr M32 or the Objective C-Apochromat 40×/1.2 W Corr M27, as described in the relevant figures. Images were acquired using two tracks. Track 1 collected the chlorophyll fluorescence using an excitation laser wavelength of 633 nm (2% power) and an emission collection range of 652–721 nm. Track 2 used two different channels to collect GFP and dsRed or mCherry fluorescence using an excitation laser of 488 nm (5% power) and 561 nm (3% power), respectively. For GFP, the emission was collected in the range of 493–535 nm, and for mCherry, emission was collected in the range of 588–641. Images of 8 bits and 1024 × 1024 pixels were acquired using a pixel dwell time of 1.27, pixel averaging of 4, and pinhole of 1 airy unit. Z-stacks were acquired ensuring an overlap of 50% for each slice.
Image analysis was conducted with Fiji-ImageJ using the raw images (Schindelin et al., 2012). We used the Coloc2 tool for colocalization analysis, the 3D object counter tool for quantifying endosome numbers, and the measurement analysis tool for quantifying pixel intensity (Schindelin et al., 2012).
2.15 |. CRISPR-Cas9 genome editing
CRISPR-Cas9 editing was performed in tomato according to Xie, Minkenberg, and Yang (2015). An sgRNA targeting the first intron of SlNRC4a was designed to include a BstYI restriction site three nucleotides upstream of the PAM site to enable screening of editing events. The sgRNA was divided into 2 parts. Each part was amplified using an sgRNA spacer primer (sgRNA-F 5′-taggtctccAGTGAGCAGgttttagagctagaaat-3′, sgRNA-R 5′-atggtctcaCACTATTTGTGTGTCtgcaccagccgggaa-3′) and terminal specific primers containing a FokI site. After FokI digestion, the fragment was inserted into the BsaI digested modified pUC57-cloning vector containing a U6 promoter and subsequently subcloned into the binary vector pMR286 (Mily Ron unpublished plasmid collection). A. tumefaciens strain GV3101 harbouring pMR286 with SlNRC4a sgRNA was used for S. lycopersicum cv M82 transformation. Total DNA was extracted from transformed plants and used as a template for SlNRC4a sgRNA flanking-fragment amplification. PCR fragments were digested with BstYI, and completely uncut fragments were presumed to originate from plants edited at both alleles and further verified by sequencing. Relevant transgenic lines were selfed, and the resulting T1 plants were re-analysed. Leaves 4–5 of 6-week-old plants were used for physiological assays.
3 |. RESULTS
3.1 |. Tomato SlNRC4a associates with LeEIX2
In order to identify LeEIX2-associated proteins involved in innate immune signalling upon EIX elicitation, we employed the introgression line (IL) 7-5, which originates from a cross between the green-fruited species Solanum pennellii and cultivated tomato (cv M82) (Eshed & Zamir, 1995; Steinhauser et al., 2011). M82 is an EIX responsive cultivar, possessing a functional LeEIX2 receptor, whereas IL7-5 does not respond to EIX elicitation, possessing a truncated LeEIX2 receptor (Ron, 2004). We stably transformed IL7-5 with LeEIX2 possessing a C-terminal GFP tag (LeEIX2-GFP) under the 35S promoter. The resulting transgenic line (TL) TL4 was able to mount an HR upon infiltration with EIX, in a similar manner to cv M82 plants, thus demonstrating the ability of the transgene to functionally complement the introgression of the LeEIX2 native locus (Figure 1a). The LeEIX2-GFP fusion protein was detected in the complemented line TL4 using Western blotting and confocal microscopy (Figure 1b,c).
FIGURE 1.
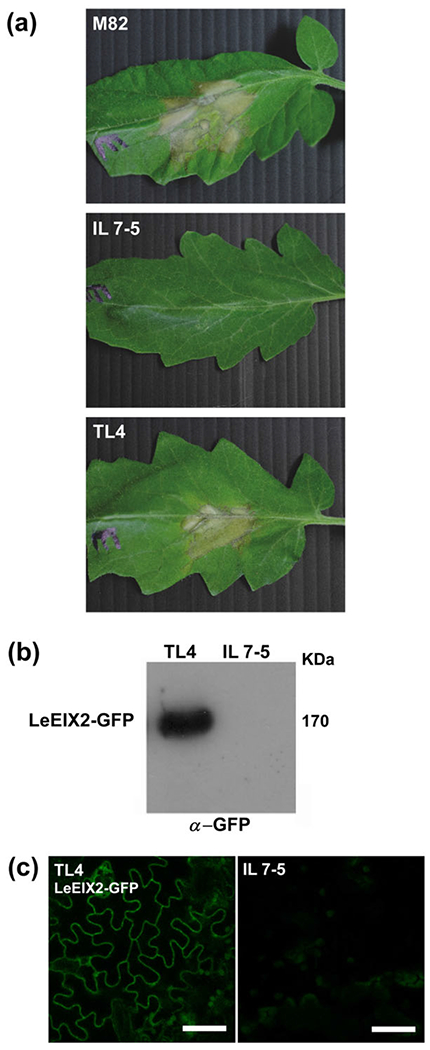
Functional complementation of IL7-5 with LeEIX2-GFP. Different tomato genotypes were compared and analysed: M82—A Solanum lycopersicum EIX responsive cultivar. IL7-5—An EIX nonresponsive Solanum pennellii × Solanum lycopersicum introgression line. TL4—A transgenic IL7–5 expressing LeEIX2-GFP. (a) Hypersensitive response. Leaves were injected with 2 μg ml−1 of EIX. Hypersensitive response development was photographed 72-hr postinjection. (b) Western blot. Total proteins were extracted fromTL4 and IL7-5, and TSM fractions were separated on a sodium dodecyl sulphate-polyacrylamide gel electrophoresis gel and LeEIX2-GFP detected by anti-GFP Western blot. (c) Confocal imaging. TL4 as well as IL7-5 (untransformed control) were analysed for GFP expression by confocal microscopy. Images were acquired using a Zeiss LSM 780 confocal microscope (20× objective, scale bar 50 μm)
To identify LeEIX2-GFP copurifying proteins, TL4 plants were pretreated with either EIX or water (mock treatment), and the Triton X-100 soluble membranes (TSM) protein fraction was subjected to immunoprecipitation using GFP-TrapA (Leibman-Markus et al., 2017). Purified proteins were subjected to tryptic on-bead digestion, and tryptic peptides were analysed by mass spectrometry. Peptides predicted to be encoded by more than 700 loci were detected. In both samples originating from TL4 (with EIX or mock), but not from IL7-5 (Control), peptides from the pulled-down LeEIX2-GFP transgene were detected, as well as previously described LeEIX2 interactors, SlSOBIRl and SlSOBIRl-like (Table S1; Liebrand et al., 2013). In addition, peptides matching the predicted protein encoded by Solyc04g007070 were also detected (Table S1). Based on sequence analysis of conserved domains, the protein is an NLR of the CC class (CNLs). Solyc04g007070 resides in an NLR cluster with three additional CNLs (Solyc04g007030, Solyc04g007050, and Solyc04g007060) and two TNLs (Solyc04g007075 and Solyc04g007090; Figure S1). Both Solyc04g007070 and Solyc04g007060 belong to clade CNL-14 of tomato, which contains 240 NLRs (Andolfo et al., 2014). Three members of this clade have been previously studied and reported as SlNRC1-3 (for NLR protein required for HR-associated cell death) (Gabriels et al., 2007; Wu et al., 2017). Recently, two NRC members from N. benthamiana, NbNRC4a and NbNRC4b, were found to define a distinct NbNRC4 subfamily within the Solanaceae CNL/NRC family (Wu et al., 2017). NbNRC4s were shown to act as a helper NLR, working in conjunction with other NLRs to confer resistance to diverse effectors (Wu et al., 2017). All three tomato CNLs in the SlNRC4 subfamily (Solyc04g007070, Solyc04g007060, and Solyc04g007030) belong to the gene cluster mentioned above. We have termed them SlNRC4a, SlNRC4b, and SlNRC4c, respectively. In this work, we focus mainly on SlNRC4a.
3.2 |. Tomato SlNRC4a associates with the PRR LeEIX2
SlNRC4a, like all NLRs, is an intracellular protein and according to Plant-mPLoc predicted to be cytoplasmic (Chou & Shen, 2010). LeEIX2 is a transmembrane domain containing protein, with an extracellular domain and a short cytoplasmic tail (Ron & Avni, 2004). Nevertheless, fractionation analysis coupled with immunoblotting revealed that SlNRC4a’s protein distribution favours the TSM fraction over the cytosolic one. This pattern resembles that of LeEIX2, in contrast to the distribution of the cytosolic protein—Pelota homolog (Solyc04g009810, Figure S2a; Lapidot et al., 2015). Moreover, SlNRC4a and LeEIX2 colocalize as observed by confocal microscopy, having a Pearson correlation coefficient of 0.63 ± 0.09 (Figure S2b, c). Both biochemical and confocal microscopy evidence point to their possible physical interaction at a common subcellular location, enabling a role for SlNRC4a in the LeEIX2 signalling network.
In order to confirm the association between LeEIX2 and SlNRC4a, co-immunoprecipitation (co-IP) experiments were performed. We transiently co-expressed tomato SlNRC4a-mCherry, LeEIX2-GFP, or LeEIX2 in N. benthamiana (an EIX nonresponsive plant) and pulled down LeEIX2-GFP by anti-GFP IP. LeEIX2-GFP is weakly detected in the inputs and is highly concentrated in IPs, indicating a highly efficient pull down. SlNRC4a-mCherry was successfully pulled down in the presence of LeEIX2-GFP but not when using untagged LeEIX2 as a control (Figure 2a) or the transmembrane-intrinsic protein AtPIP2A tagged with GFP (Figure S3a), verifying the assay specificity. In order to further confirm the association, the reciprocal co-IP experiments were performed, pulling down SlNRC4a-mCherry by anti-mCherry IP. LeEIX2-GFP was successfully pulled down in the presence of SlNRC4a-mCherry but not when using free mCherry as a control (Figure S3b). The association between the two proteins was enhanced twofold in the presence of EIX, suggesting an EIX-dependent association (Figure 2a,b).
FIGURE 2.
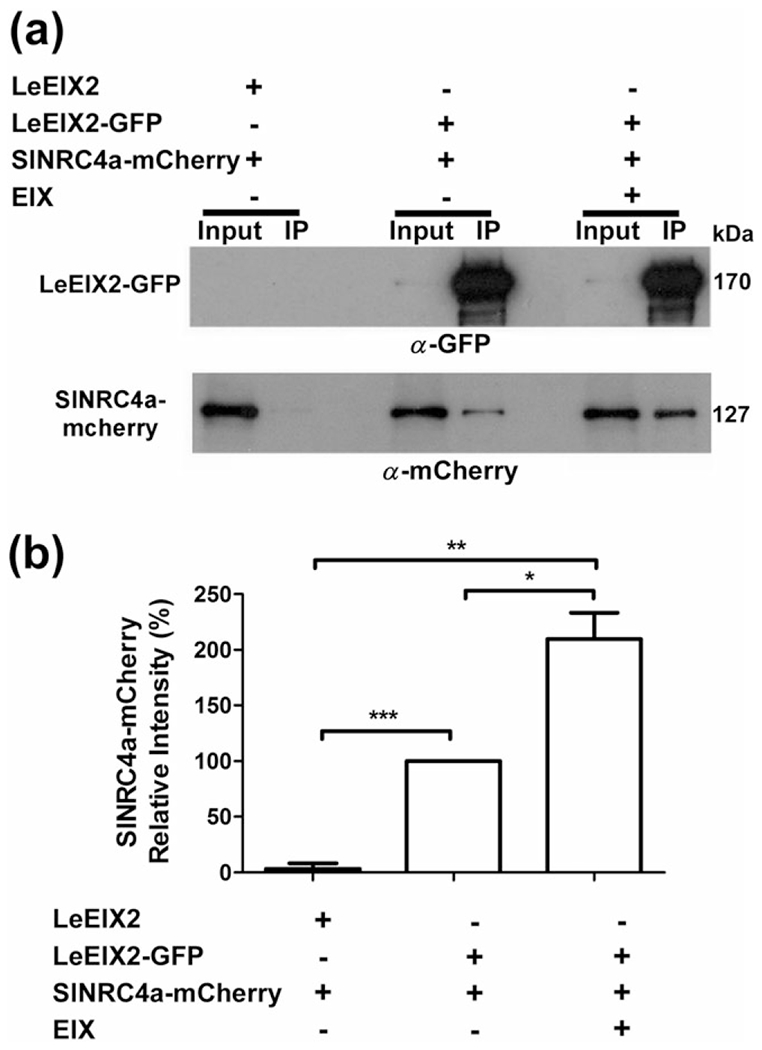
SlNRC4a and LeEIX2 associate in planta when expressed in Nicotiana benthamiana. (a) Plants were transiently cotransformed with LeEIX2-GFP or LeEIX2 and SlNRC4a-mCherry. Leaves were harvested and treated with either EIX or water (mock) at the petiole for 15 min. Triton X-100 soluble membrane (TSM) protein fractions were immunoprecipitated (IPed) using GFP affinity beads. Input IP samples were subjected to sodium dodecyl sulphate-polyacrylamide gel electrophoresis and immunoblot analyses. Membranes were probed with anti-GFP antibodies to detect LeEIX2-GFP and anti-mCherry antibodies to detect SlNRC4a-mCherry. (b) SlNRC4a-mCherry signal intensity was determined using Coloc2 from FIJI-ImageJ, by dividing co-IP intensity by input intensity. SlNRC4a-mCherry signal intensity, without EIX treatment, was defined as 100%. Error bars represent the average ± SD of three independent experiments. Asterisks indicate significant differences (t test, *P < 0.05, **P < 0.005, ***P < 0.001)
3.3 |. SlNRC4a enhances EIX-mediated defence responses
The association of SlNRC4a with LeEIX2 suggests a possible role for SlNRC4a in PTI signal transduction. We examined the effect of SlNRC4a overexpression on the ROS burst and ethylene production elicited by EIX. We transiently expressed SlNRC4a in N. tabacum cv samsun NN (an EIX responsive cultivar) followed by EIX application. SlNRC4a expression enhanced EIX-elicited defence responses when compared with free GFP as control, leading to a 73% increase in total ROS burst, with more than a 50% increase at peak levels and a 30% increase in ethylene induction (Figure 3a,b).
FIGURE 3.
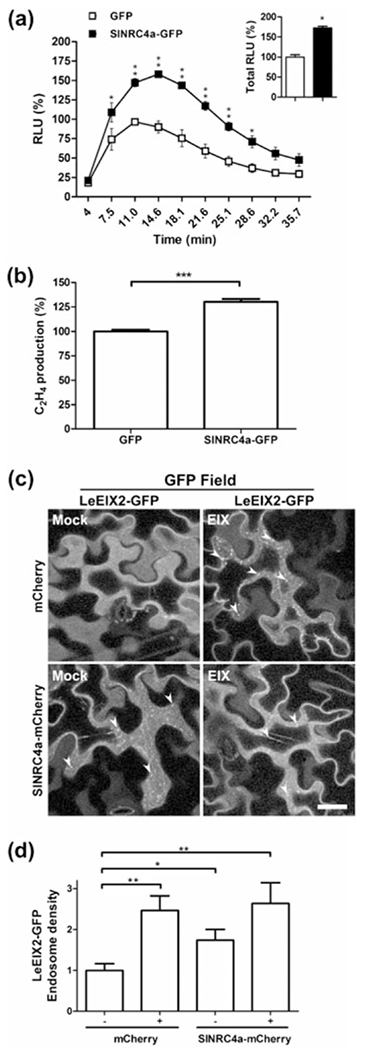
Overexpressing SlNRC4a enhances EIX-mediated defence responses in Nicotiana tabacum. (a) Leaf disks of transiently transformed N. tabacum leaves with SlNRC4a-GFP or control (free GFP) as indicated (24 hr after transformation) were floated on water for ~24 hr and then replaced with luminescence solution containing 1 μg ml−1 of EIX. Luminescence (RLU) was immediately measured to track the ROS burst. Average ± SEM values of three independent experiments, n = 12 each. Asterisks indicate significant differences with control treatment (two-way analysis of variance, *P < 0.05, **P < 0.001). (b) Leaf disks of transiently transformed N. tabacum leaves with SlNRC4a-GFP or control (free GFP) as indicated (48 hr after transformation) were floated on a solution with 250 mM of sorbitol containing 1 μg ml−1 of EIX. Ethylene biosynthesis was measured after 4 hr. Ethylene induction was defined as the ΔEthylene (Ethylene+EIX – Ethylene−EIX). The control induction average value was defined as 100%. Error bars represent the average ± SEM of three independent experiments, with asterisks denoting significant differences with control treatment (N = 17, t test, P < 0.0001). Effect of SlNRC4a overexpression on LeEIX2 endosomes in N. benthamiana: (c) Leaves transiently expressing LeEIX2-GFP and free mCherry (Control) or SlNRC4a-mCherry as indicated were treated with EIX (1 μg g−1 tissue) or water (mock) at the petiole 40 hr after transformation. LeEIX2-GFP endosomes were visualized by confocal microscopy 15-min post-EIX treatment. Representative Z-projections were acquired using Zeiss LSM 780 confocal microscope (40× objective, arrowheads indicate endosomes. Scale bar 30 μm). (d) LeEIX2-GFP endosome density was quantified using 3D object counter (Fiji-ImageJ). Error bars represent the average ± SEM of three independent replicates, four images each. Asterisks indicate significant differences from the control (N = 12, t test, *P < 0.05, **P < 0.001)
LeEIX2 has been shown to undergo endocytosis, which is highly intensified upon EIX recognition and binding. Inhibiting endosome formation reduces EIX-mediated responses, whereas arresting early endosomal trafficking increases the responses (Sharfman et al., 2011). These results suggest that LeEIX2 endosomes play an active role in signal transduction, leading us to study the effect of SlNRC4a on this process. When transiently co-expressing SlNRC4a together with LeEIX2-GFP in N. benthamiana, we observed an increase in both number and density of GFP-tagged endosomes compared with the control in the absence of EIX perception. After EIX treatment, expression of SlNRC4a did not further increase endosome accumulation (Figures 3c,d and S4). This result suggests overexpression of SlNRC4a induces an early onset of LeEIX2 endosome induction prior to elicitation—a steady-state increase independent of EIX presence. Notably, although EIX treatment and SlNRC4a co-expression are increasing LeEIX2 endosomal localization, neither are altering overall protein levels (Figure S4b). Intriguingly, EIX treatment leads to an increase of SlNRC4a overall protein levels (Figure S4c).
3.4 |. Tomato SlNRC4a associates with AtFLS2 and enhances flagellin-mediated defence response
We have demonstrated that SlNRC4a is able to associate with the RLP LeEIX2 primarily in a ligand-dependent manner and acts as a positive regulator of EIX-elicited responses (Figures 2 and 3). In order to examine the extent of SlNRC4a involvement in PTI signalling, we focused on the well-characterized RLK—Flagellin Sensing 2 (FLS2). FLS2 perceives a 22 amino acid epitope of bacterial flagellin (Chinchilla, Bauer, Regenass, Boller, & Felix, 2006). We tested whether SlNRC4a can also associate with the FLS2 receptor using co-immunoprecipitation. SlNRC4a-HA and AtFLS2-GFP were transiently co-expressed in N. benthamiana followed by GFP IP. SlNRC4a was successfully pulled down when using AtFLS2-GFP, whereas no co-IP was detected when using free GFP (Figure 4a) or the membrane-localized control AtPIP2A-GFP (Figure S3c), verifying the assay specificity. Low levels of AtFLS2-GFP are detected in input fractions and high ones in IPs, indicating a high capacity pull down. SlNRC4a was pulled down both in the absence and presence of flg22 (Figure 4a).
FIGURE 4.
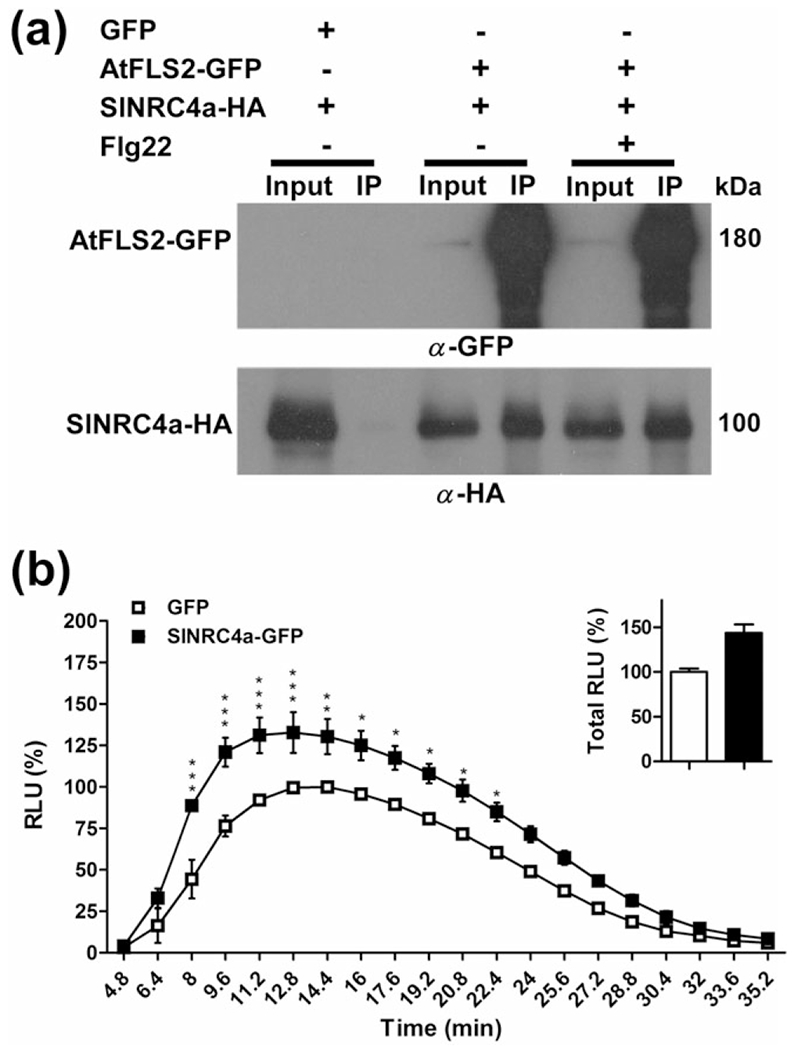
(a) SlNRC4a and FLS2 associate in planta when expressed in Nicotiana benthamiana. Plants were transiently cotransformed with pAtFLS2::AtFLS2-GFP or NOS::GFP and 35S::SlNRC4a-2xHA using Agrobacterium infiltration. Forty-eight hours after infiltration, leaves were harvested and treated with either 1 mM of flg22 or water (mock) at the petiole for 15 min; 0.5 g of leaves per treatment was used for protein extraction. Triton X-100 soluble membrane protein fractions were subjected to anti-GFP IP. Input and IPed proteins were subjected to sodium dodecyl sulphate-polyacrylamide gel electrophoresis and immunoblot analyses with anti-GFP and anti-HA. (b) Effect of overexpression of SlNRC4a in N. benthamiana on flagellin-mediated ROS burst. Leaf disks of transiently transformed N. benthamiana leaves with SlNRC4a-GFP or control (free GFP) as indicated (24 hr after transformation) were floated on water for ~24 hr and then replaced with luminescence solution containing 1 μM of flg22. Luminescence was immediately measured to track the ROS burst. Average ± SEM values of three independent experiments, n = 12 each. Asterisks indicate significant differences with control treatment (two-way analysis of variance, *P < 0.05, **P < 0.01, ***P < 0.001)
SlNRC4a associates with LeEIX2 and positively regulates downstream immune signalling outputs. In addition, SlNRC4a also associates with the PRR FLS2. This association raises the possibility of SlNRC4a also participating in FLS2-PTI signal transduction. Therefore, we tested a possible role of SlNRC4a on flagellin-mediated defence responses by measuring the ROS burst after flg22 elicitation after transiently expressing SlNRC4a in N. benthamiana (a flagellin responsive plant). SlNRC4a enhances the total amount of flg22-triggered ROS burst by 44%, with more than 30% increase at peak levels (Figure 4b). No response was detected without flg22 elicitation.
3.5 |. Silencing SINRC4 in tomato abolishes EIX-mediated defence response
The observation that SlNRC4a overexpression enhances EIX-elicited defence responses led us to employ VIGS to silence SlNRC4a, SlNRC4b, and SlNRC4c simultaneously based on their conserved nucleotide sequence (Figure 5a). The wild-type tomato cv M82 was subjected to VIGS. SlNRC4a,b transcript levels were reduced by close to 80% and SlNRC4c by 64%, whereas Solyc04g015210, an NRC family member most closely related to the SlNRC4 clade, remained unaffected as confirmed by qPCR (Figure 5b). Thus, these experiments demonstrate the efficiency and specificity of the VIGS assay.
FIGURE 5.
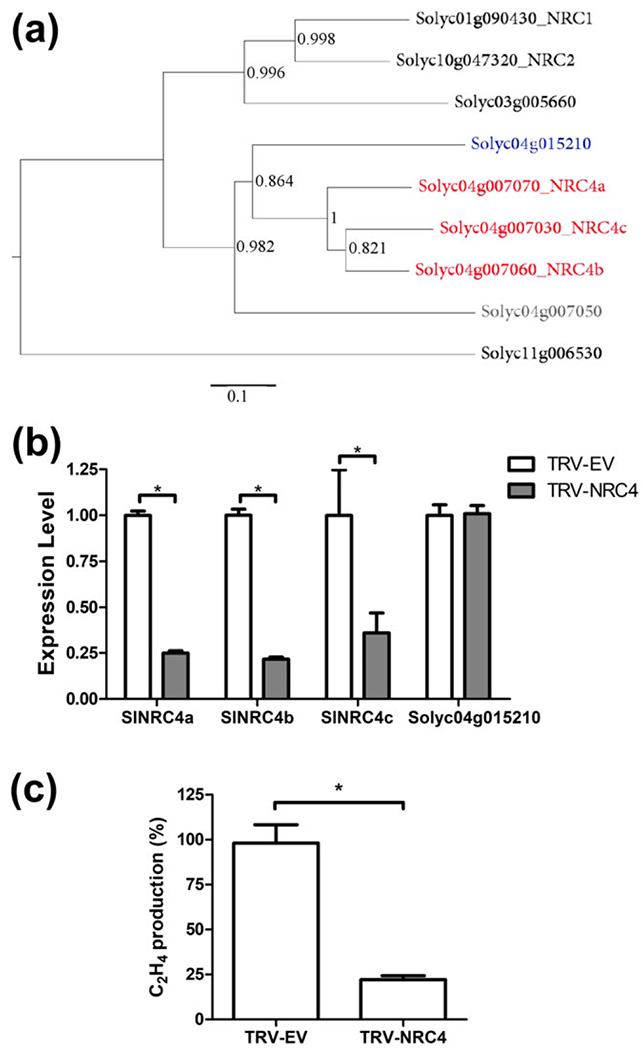
Silencing SINRC4 family members in tomato abolishes LeEIX2-mediated defence responses. (a) Phylogenetic analysis of tomato CC-NBS-LRRs genes. The genomic DNA sequences of SINRC4a’s closest homologs were used to generate maximum likelihood phylogeny (Dereeper et al., 2008). Labels show Sol Genomics Network accession numbers and gene annotation for each gene; SINRC4 genes targeted by virus-induced gene silencing are indicated in red, an SINRC4 homolog that is not predicted to be targeted is indicated in blue, and an SINRC4 homolog that is not predicted to be expressed (Andolfo et al., 2014) is indicated in grey. Silencing SINRC4 family members: Wild-type M82 tomato plants were pre-infected with tobacco rattle virus (TRV) to silence SINRC4a-c (TRV-SlNRC4) or TRV empty vector (TRV-EV) as a control. Six-week-old plants were used for assessing gene expression and physiological assays. (b) RT-PCR confirming the silencing of SINRC4a in tomato using virus induced gene silencing. Solyc04g015210, the nontargeted homolog of SINRC4, served as a control for the silencing specificity. Genes were normalized to the control gene SlCyclophilin (Solyc01g111170). Error bars represent the average ± SEM values of three independent experiments, n = 9 each. Asterisks indicate significant differences with control (two-way analysis of variance, *P < 0.0001). (c) Effect of SINRC4 silencing on EIX-mediated ethylene induction. Tomato leaf disks were floated on a 250 mM of sorbitol solution with 1 μg ml−1 of EIX. Ethylene biosynthesis was measured after 4 hr. Error bars represent the average ± SEM of three different experiments, and asterisks indicate significant differences with control treatment (N = 18, t test, P < 0.0001). (Marchler-Bauer et al., 2017)
We measured ethylene production upon EIX elicitation in control and SlNRC4a,b,c-silenced plants. Silencing of SINRC4a,b,c transcripts significantly compromised the EIX-mediated response. SINRC4a,b,c-silenced plants showed close to an 80% reduction in ethylene production compared with the control plants (Figure 5c). This result suggests an important role for SlNRC4 clade in EIX-mediated defence responses.
3.6 |. SlNRC4a’s coiled-coil domain associates with LeEIX2 and is sufficient to enhance EIX-mediated defence responses
Several studies have shown that overexpression of the N-terminal CC and TIR domains from various NLRs in solanaceous plants is sufficient to trigger an effector-independent HR, indicating the ability of this domain to directly participate in signalling (Bernoux et al., 2011; Cesari et al., 2016; Maekawa et al., 2011; Wang et al., 2015). On the basis of National Center for Biotechnology Information (NCBI)-conserved domain prediction (Marchler-Bauer et al., 2017), we generated a truncated version of the SlNRC4a protein, composed of its N-terminal 125 amino acids, the CC domain (SlNRC4a-CCd, Figure 6a,b). Similarly to the crystal-solved structure of the S. tuberosum CNL protein—StRx, the CC domain of SlNRC4a possesses four predicted α-helices and a conserved EDxxD motif located in the third α-helix (Figure 6b; Hao, Collier, Moffett, & Chai, 2013). The EDxxD motif is reported to be involved in NB-LRR binding (Rairdan et al., 2008). Overexpression of SlNRC4a-CCd by itself was not sufficient to trigger an elicitor-independent HR.
FIGURE 6.
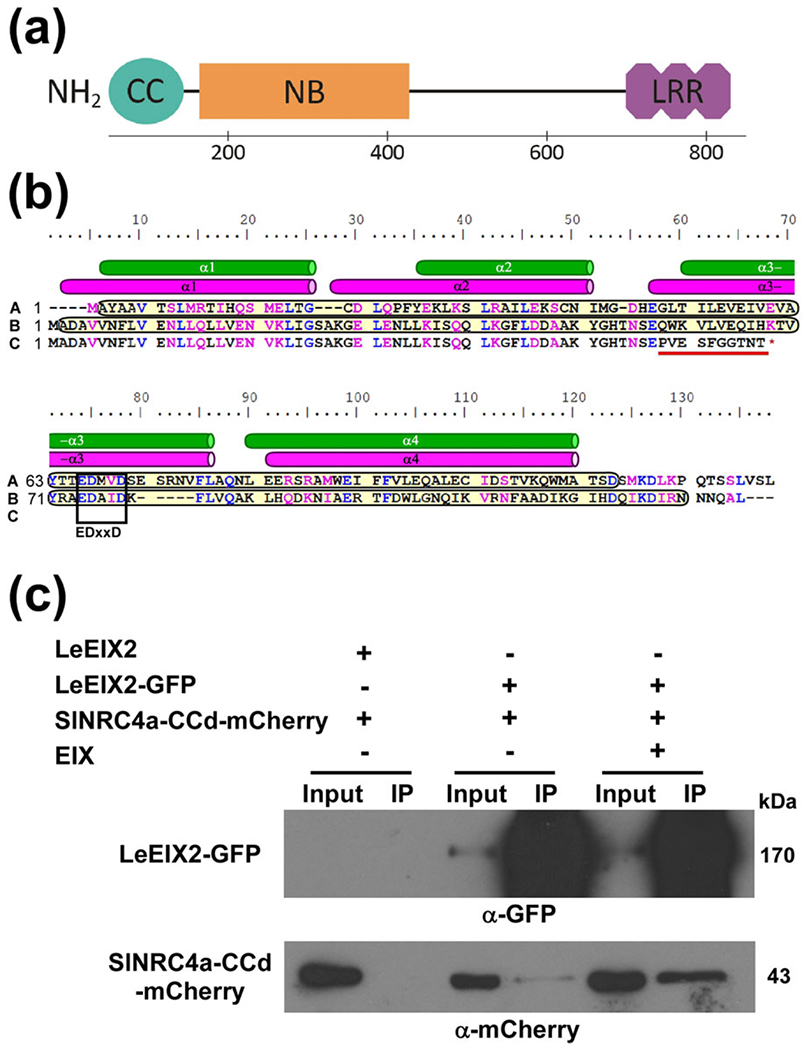
SlNRC4a’s coiled-coil domain associates with LeEIX2 in planta. (a) Schematic representation of SlNRC4a’s domain architecture. The Sol Genomics Network (Fernandez-Pozo et al., 2015) was used to plot the SlNRC4a protein sequence. Domains were identified using NCBI-conserved domains: coiled-coil 1–125 aa, nucleotide binding 159–437 aa, and leucine-rich repeat 696–838 aa (Marchler-Bauer et al., 2017). (b) SlNRC4a’s coiled-coil domain secondary structure. Sequence alignment of StRx 1–130 aa (A), SlNRC4a 1–130 aa (B), and slnrc4a-2 1–67 aa (C). Similar and identical aa are marked in magenta and blue, respectively. NCBI prediction of coiled-coil domains is marked in yellow (Marchler-Bauer et al., 2017). StRx-resolved and SlNRC4a-predicted α-helices shown as green and magenta cylinders, respectively (Hao et al., 2013). Conserved EDxxD motif is boxed. Mutated aa are underlined in red. (c) Co-IP of SlNRC4a’s coiled-coil domain with LeEIX2 in Nicotiana benthamiana. Plants were transiently cotransformed with LeEIX2-GFP or LeEIX2 and SlNRC4a-CCd-mCherry. Leaves were harvested and treated with either EIX or water (mock) at the petiole for 15 min. Triton X-100 soluble membrane protein fractions were immunopurified using GFP affinity beads. Input and immunopurified (IP) proteins were subjected to sodium dodecyl sulphate-polyacrylamide gel electrophoresis and immunoblot with anti-GFP and anti-mCherry
Next, we examined the ability of SlNRC4a-CCd to associate with LeEIX2 and positively regulate EIX-elicited responses. We performed Co-IP experiments in N. benthamiana as described above using mCherry-tagged SlNRC4a-CCd. As in the case of the previous Co-IP, LeEIX2-GFP is highly concentrated in IPs, indicating a high efficiency pull down. SlNRC4a-CCd was successfully pulled down when using LeEIX2-GFP, whereas no co-immunoprecipitated fraction was detected in the controls (Figures 6c and S3d). The association between the two proteins was significantly strengthened upon EIX perception, suggesting an EIX-dependent interaction (Figure 6c).
Although SlNRC4a-CCd is not able to trigger an elicitor-independent HR, it is sufficient to interact with LeEIX2. Therefore, we examined the effect of SlNRC4a-CCd overexpression on EIX-elicited signalling. We measured ROS burst and ethylene production caused by EIX elicitation. SlNRC4a-CCd enhances the response by a 73% increase in total ROS burst, with more than a 50% increase at peak levels and a more than 25% rise in ethylene induction (Figure 7a,b). Hence, SlNRC4a-CCd is able to enhance EIX-mediated defence responses to a level comparable with full-length SlNRC4a.
FIGURE 7.
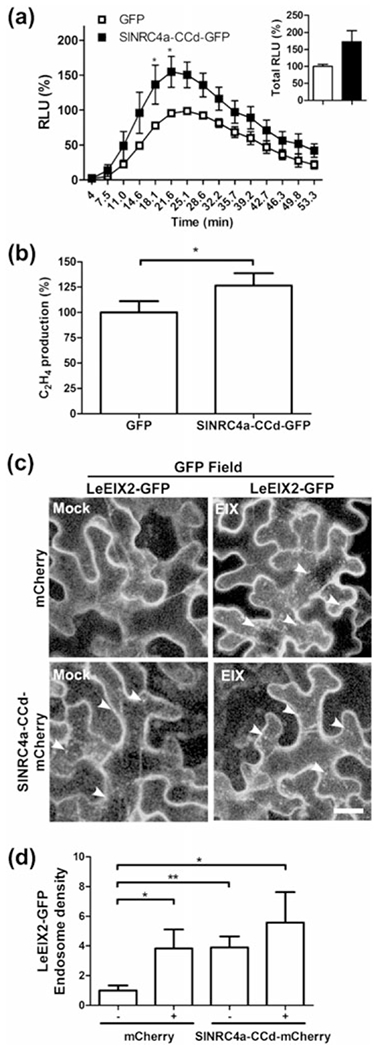
Effect of overexpression of SlNRC4a’s coiled-coil domain on LeEIX2-mediated defence responses in Nicotiana tabacum. (a) Leaf disks of transiently transformed N. tabacum leaves with SlNRC4a-CCd-GFP or control (free GFP) as indicated (24 hr after transformation) were floated on water for ~24 hr and then replaced with luminescence solution containing 1 μg ml−1 of EIX. Luminescence (RLU) was immediately measured to track the ROS burst. Average ± SEM values of three independent experiments, n = 12 each. Asterisks indicate significant differences with control treatment (two-way analysis of variance, *P < 0.05). (b) Leaf disks of transiently transformed N. tabacum leaves with SlNRC4a-CC-GFP domain or control (free GFP) as indicated (40 hr after transformation) were floated on a 250 mM of sorbitol solution with 1 μg ml−1 of EIX. Ethylene biosynthesis was measured after 4 hr. Ethylene induction was defined as the ΔEthylene (Ethylene+EIX – Ethylene−EIX). Control induction average value was defined as 100%. Error bars represent the average ± SEM of three independent experiments, with asterisks denoting significant differences with control treatment (N = 21, t test, P < 0.0001). Effect of overexpression of SlNRC4a-CCd on LeEIX2 endosomes in N. benthamiana: Leaves transiently expressing LeEIX2-GFP and free mCherry (Control) or SlNRC4a-CCd-mCherry, 40 hr after transformation, leaves were treated with EIX (1 μg g−1 tissue) or water (mock) at the petiole and visualized after 15 min. (c) Representative Z-projections were acquired using the Zeiss LSM 780 confocal microscope (20× objective, arrowheads indicate endosomes. Scale bar 30 μm). (d) The number of LeEIX2 endosomes was quantified using 3D object counter (Fiji-ImageJ). Error bars represent the average ± SEM of three independent replicates, four images each. Asterisks indicate significant differences from the control (N = 12, t test, *P < 0.05, **P < 0.001)
EIX elicitation induces LeEIX2 endocytosis (Sharfman et al., 2011), and full-length SlNRC4a increases the number and density of LeEIX2-tagged endosomes. Thus, we examined whether SlNRC4a-CCd is able to affect LeEIX2 endosomes. When overexpressing both SlNRC4a-CCd and LeEIX2-GFP in N. benthamiana, we observed an increase in the basal level of both the number and density of GFP-tagged endosomes as well as an increase in the density of LeEIX2 endosomes after EIX application. This result agrees with the increase in EIX-elicited defence responses caused by SlNRC4a-CCd (Figures 7c,d and S5). EIX treatment did not alter overall protein levels of any of the co-expressed proteins (Figure S5b,c).
3.7 |. CRIPSR-Cas9 editing of SlNRC4a generates a truncated allele that enhances EIX-mediated defence responses
In addition to silencing SINRC4 clade genes by VIGS, we applied CRISPR-Cas9 methodology to generate stable SINRC4a edited tomato lines. Two independent lines (slnrc4a-2 and slnrc4a-5) were demonstrated to be bi-allelic homozygous mutants in SINRC4a. Sequence analysis revealed a single insertion after the first 171 nucleotides of SINRC4a. A cytosine insertion occurred in slnrc4a-2, and a thymine insertion occurred in slnrc4a-5. In both lines, the nucleotide insertion led to a frame shift producing 10 mutated amino acids (aa) followed by an early stop codon, resulting in a 67 aa truncated protein (Figures 6b and S6a). The protein product resulting from the gene editing consists of the two first predicted α-helices, thus lacking the EDxxD motif (Figures 6b and S6a). This resulting nonsense mutation did not affect SINRC4a transcript level, or the mRNA level of its closest homolog, SINRC4b (Figure S6b).
As the CRISPR-Cas9 methodology manipulates the gene at DNA level (whereas VIGS silencing occurs at the RNA level), we expected stable and complete loss of function of the SINRC4a gene. Surprisingly, the 67 aa mutated and truncated SlNRC4a protein, generated by editing (having only part of the CC domain), produced the opposite effect and significantly enhanced EIX-elicited defence responses. The sInrc4a-2 edited gene led to a 219% increase in total ROS burst, with more than 250% increase at peak levels and more than 260% rise in ethylene induction upon EIX treatment (Figure 8a,b). Similar results were obtained for slnrc4a-5 (Figure S7). Hence, the mutated and truncated slnrc4a is able to enhance EIX-elicited defence responses in a similar manner but at a higher amplitude than overexpression of the full-length SlNRC4a protein.
FIGURE 8.
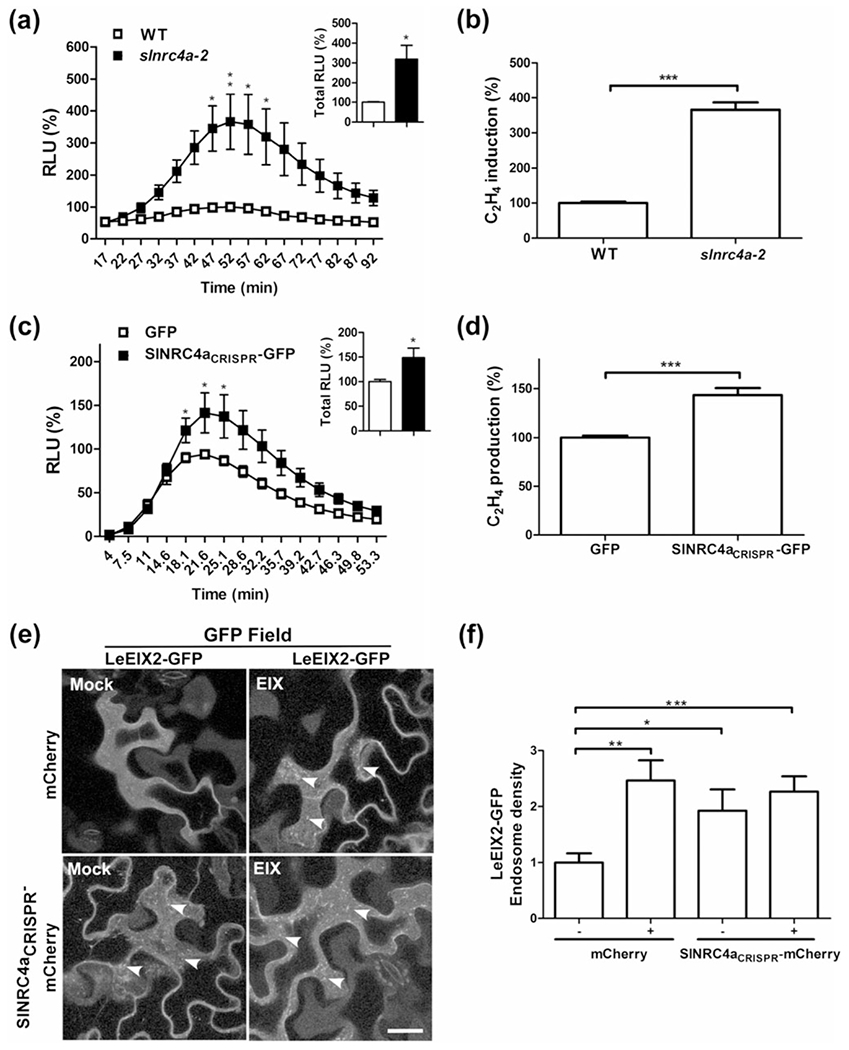
Effect of SlNRC4a CRISPR-Cas9 tomato editing on defence responses. (a) Leaf disks of slnrc4a-2 CRISPR-Cas9 edited or WT plants (M82 control) were taken from leaves 5-7. Leaf disks were floated on water for ~4 hr and then replaced with luminescence solution containing 1 μg ml−1 of EIX. Luminescence (RLU) was immediately measured to track the ROS burst. Average ± SEM values of three independent experiments, n = 12 each. Asterisks indicate significant differences with control treatment (two-way analysis of variance, **P < 0.001, *P < 0.05). (b) Leaf disks of slnrc4a-2 CRISPR-Cas9 edited or WT tomato plants (M82 control) were taken from leaves 5-7. Leaf disks were floated on a 250 mM of sorbitol solution with 1 μg mL−1 of EIX. Ethylene biosynthesis was measured after 5 hr. Ethylene induction was defined as the ΔEthylene (Ethylene+EIX – Ethylene−EIX). Control induction average value was defined as 100%. Error bars represent the average ± SEM of three independent experiments, significant differences to control treatment (N = 33, t test, ***P < 0.0001). Effect of overexpression of SlNRC4a CRISPR edited fragment on LeEIX2 mediated defence responses in Nicotiana tabacum: (c) Leaf disks of transiently transformed N. tabacum leaves with SlNRC4aCRISPR-GFP or control (free GFP) as indicated (24 hr after transformation) were floated on water for ~24 hr and then replaced with luminescence solution containing 1 μg ml−1 of EIX. Luminescence (RLU) was immediately measured. Average ± SEM values of five independent experiments, n = 12 each. Asterisks indicate significant differences with control treatment (two-way analysis of variance, *P < 0.05). (d) Leaf disks of transiently transformed N. tabacum leaves with SlNRC4aCRISPR-GFP or control (free GFP) as indicated (40 hr after transformation) were floated on a 250 mM of sorbitol solution with 1 μg ml−1 of EIX. Ethylene biosynthesis was measured after 4 hr. Ethylene induction was defined as the ΔEthylene (Ethylene+EIX – Ethylene−EIX). Control induction average value was defined as 100%. Error bars represent the average ± SEM of three independent experiments, with asterisks denoting significant differences with control treatment (N = 18, t test, ***P < 0.0001). Effect of overexpression of SlNRC4a CRISPR-edited fragment on LeEIX2 endosomes in N. benthamiana: Leaves transiently expressing LeEIX2-GFP and free mCherry (Control) or SlNRC4aCRISPR-mCherry as indicated, 40 hr after transformation, leaves were treated with EIX (1 μg g−1 tissue) or water (mock) at the petiole and visualized after 15 min. (e) Representative Z-projections were acquired using Zeiss LSM 780 confocal microscope (40× objective, arrowheads indicate endosomes. Scale bar 30 μm). (f) Number of LeEIX2 endosomes was quantified using 3D object counter (Fiji-ImageJ). Error bars represent the average ± SEM of three independent replicates, four images each. Asterisks indicate significant differences from the control (N = 12, t test, *P < 0.05, **P < 0.001, ***P < 0.0001)
In order to verify that the LeEIX2 phenotype observed in the SINRC4a genome-edited lines is due to an alteration in SlNRC4a and not the result of the insertion of the CRISPR-Cas9 system, we generated the same 67 aa peptide (SlNRC4aCRISPR) via transient expression. When we transiently expressed SlNRC4aCRISPR in N. tabacum (which contains the endogenous NtNRC4a), we observed a 49% increase in total ROS burst, with more than a 50% increase at peak levels and more than a 40% rise in ethylene induction upon EIX treatment (Figure 8c,d). These results indicate that the enhancement of LeEIX2 signalling observed in the genome-edited plants is indeed a result of SlNRC4a gene editing. Tomato WT and slnrc4a-2 plants show a slower development of ROS burst, peaking 30 min after the peak observed when transiently expressing in tobacco (Figure 8a,c). These time signature distinctions likely originate from organism-based responsiveness properties. In order to visualize a possible effect of the edited fragment on LeEIX2 endosomes, we created SlNRC4aCRISPR tagged with mCherry. When SlNRC4aCRISPR was co-expressed with LeEIX2-GFP in N. benthamiana, we observed an increase in both the number and density of GFP-tagged endosomes independent of EIX application (Figures 8e,f and S8). This result agrees with the increase in EIX-elicited defence responses caused by SlNRC4aCRISPR. EIX elicitation did not affect protein levels of any of the co-expressed proteins (Figure S8b,c).
3.8 |. SlNRC4a gene editing results in increased fungal resistance
The enhanced immune responses identified upon mutation of SlNRC4a highlight the possibility of editing NRCs to boost diverse immune responses. In order to examine this hypothesis, we employed the necrotrophic fungi B. cinerea (De Meyer & Hofte, 1997). The slnrc4a-2-edited gene resulted in a restriction in the lesion observed in response to B. cinerea 24 hr after fungal inoculation, demonstrating increased resistance to B. cinerea (Figure 9a,b). Similar results were obtained for slnrc4a-5 (Figure S9). This result indicates that editing the SlNRC4a gene not only enhances EIX-elicited defence responses but also confers enhanced resistance to biotic attack.
FIGURE 9.
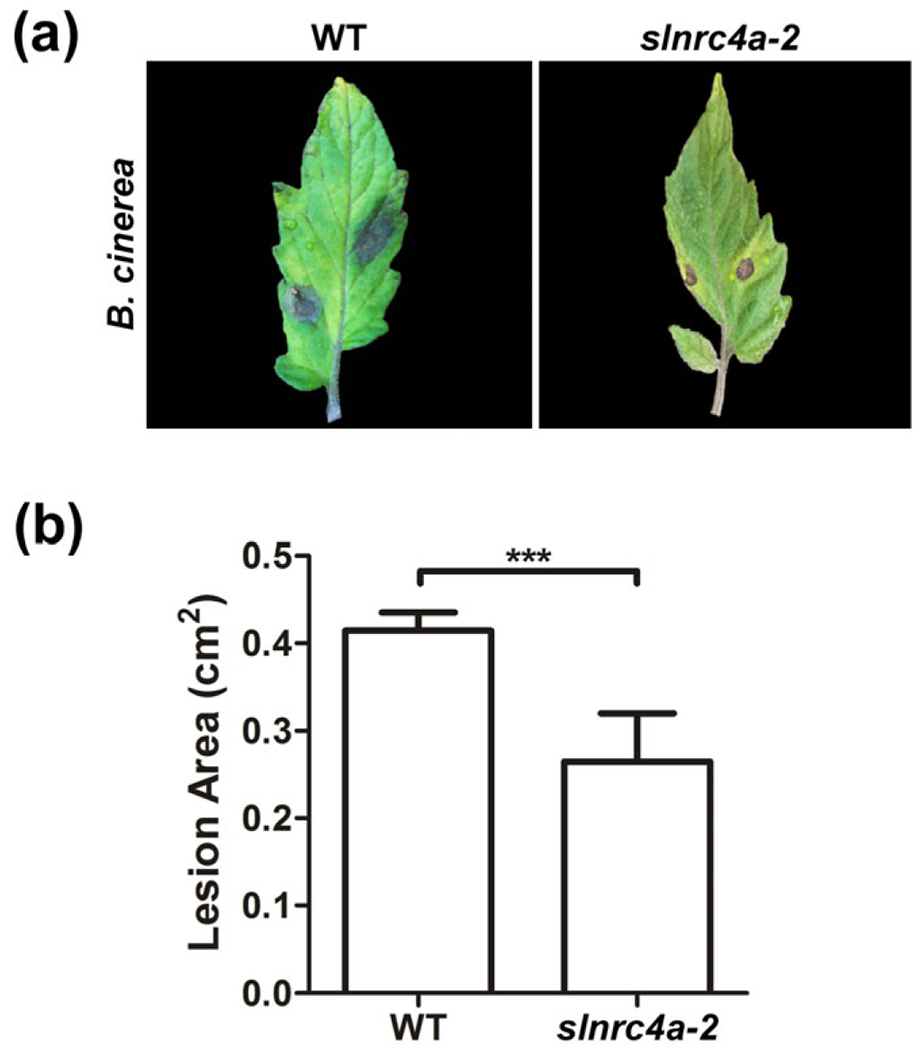
Effect of SlNRC4a CRISPR-Cas9 tomato editing on pathogen resistance. slnrc4a-2 CRISPR-Cas9 edited or WT plants (M82 control) were inoculated with Botrytis cinerea, and leaves were monitored for symptomatic development of disease. (a) Infected leaves were photographed 48 hr postinoculation. (b) Lesion area was quantified using Fiji-ImageJ. Error bars represent the average ± SEM. Asterisks indicate significant differences from the control (N = 16, t test, ***P < 0.0001)
4 |. DISCUSSION
Plants rely on a bilayered defence mechanism against pathogens. Cell surface localized receptors possessing transmembrane domains (PRRs) act to recognize conserved microbial features. Thus, pathogens must be able to suppress this first layer of defence. Pathogens have developed effector molecules to suppress the PTI immune signalling and enable plant colonization (Thomma et al., 2011; Toruno, Stergiopoulos, & Coaker, 2016). Plants also possess intracellular NLR immune receptors that activate ETI (Bonardi et al., 2012; Cui et al., 2015). Using a proteomic approach, we have identified and characterized the tomato SlNRC4a NLR as a component of LeEIX2-mediated responses.
In this work, we demonstrate the ability of SlNRC4a to associate with LeEIX2 and positively regulate EIX-elicited defence responses. SlNRC4a was also able to interact with another PRR, the well-characterized flagellin receptor AtFLS2 (Gomez-Gomez & Boller, 2000; Schlecht et al., 1999; Zipfel et al., 2004), and enhanced flagellin-mediated defence response. Moreover, we show that SlNRC4a is able to directly associate with PRRs and enhance PTI signalling in the absence of effectors. As such, SlNRC4a can be defined as a noncanonical positive regulator of immunity, capable of affecting both RLK and RLP signalling. PTI and ETI can elicit similar defence responses, such as defence gene induction, ROS burst, and in some cases the HR (Thomma et al., 2011; Thomma & Bignell, 2016). However, early signalling overlap between PTI and ETI remains largely unknown. Previously, Mantelin and colleagues (2011) demonstrated that the extracellular RLK—SlSERK1—is required for full responses of the NLR Mi-1.2. Mi-1.2 is a CNL that confers resistance to root knot nematodes as well as three genera of phloem-feeding insects (Casteel, Walling, & Paine, 2006; Milligan et al., 1998; Nombela, Williamson, & Muniz, 2003). SlSERK1 was able to associate with Mi-1.2 in planta and was required for Mi-1.2-mediated aphid resistance (Mantelin et al., 2011). Another example is SlNRC1, which is required for the extracellular RLP—Cf4-mediated HR (Gabriels et al., 2007). Recent work has demonstrated the contribution of additional NRC clade members (NbNRC2 and NbNRC3) in Cf4-triggered HR (Wu, Belhaj, Bozkurt, Birk, & Kamoun, 2016). These results, coupled with our own, highlight the overlap between extracellular perception and NLR intracellular immune signalling.
An emerging concept proposes that NLRs can function together, where “sensor” NLR proteins require “helper” NLRs to initiate immune signalling (Cesari, Bernoux, Moncuquet, Kroj, & Dodds, 2014). Recently, exciting work by Wu et al. (2017) demonstrated that NbNRC family members are “helper” NLRs, required for the function of multiple “sensor” CNLs with various degrees of redundancy and specificity, creating a complex signalling network (Wu et al., 2017). NbNRC4s were required for the function of the Rpi-blb2, Mi-1.2, and R1 sensor CNLs (Wu et al., 2017). Here, we demonstrate that SlNRC4a enhances plant immune responses upon MAMP perception by the RLP LeEIX2 and RLK AtFLS2. We propose that SlNRC4a, like NbNRC4, acts as a conserved “helper” NLR to facilitate immune responses from diverse PRRs that act as “sensor” (Caplan Padmanabhan, & Dinesh-Kumar, 2008; Wu et al., 2017).
The N-terminal TIR or CC domains of multiple NLRs have beer shown to be both necessary and sufficient for triggering an HR (Zhang, Dodds, & Bernoux, 2017). These data suggest that many NLRs signal through their TIR/CC domain to activate immune signalling. In some cases, the CC domain was shown to trigger effector-independent cell death (Cesari et al., 2016; Collier, Hamel, & Moffett, 2011; Maekawa et al., 2011; Wang et al., 2015). Some resistance genes also encode fragments of typical NLRs, including those consisting only of the TIR/CC domain which still mediate resistance (Nandety et al., 2013 Nishimura et al., 2017; Wang, Devoto, Turner, & Xiao, 2007; Xiao et al., 2001). The SlNRC4a CC domain alone (SlNRC4a-CCd) is unable to trigger EIX-independent HR, but is able to bind LeEIX2 and enhance EIX-elicited defence responses as efficiently as the full length SlNRC4a. Intriguingly, a SlNRC4a CRISPR-Cas9-edited plant, encoding a 67 aa truncated variant of SlNRC4a, displayed significant increases in defence responses upon EIX elicitation. Congruently, overexpression of the same 67 aa protein in N. tabacum also enhanced the response to EIX elicitation. This demonstrates that the enhancement of EIX-elicited defence responses is indeed a result of the edited SlNRC4a gene. The enhancement of EIX-elicited defence responses caused by both SlNRC4a-CCd and SlNRC4aCRISPR fragment indicates that a SlNRC4a partial or complete coiled-coil domain is sufficient to alter the LeEIX2-PTI signalling pathway in an EIX-dependent manner. Interestingly, although we employed CRISPR technology with the intention of creating a loss of function allele, the result was the generation of a mutant expressing a truncated peptide with enhanced signalling activities. As the SlNRC4aCRISPR fragment is able to enhance EIX-elicited defence responses but still requires the PRR “sensor” to do so, our data imply that in the EIX-mediated signalling pathway, the first two α-helices of SlNRC4a are sufficient to perform the “helper” activity.
Although both CRISPRed plants and overexpression of the SlNRC4aCRISPR 67 aa protein enhanced EIX-mediated defence responses, the amplitude of responses in CRISPRed plants was significantly higher. This could stem from differences in the ability of tomato and tobacco to elicit defence responses or difference in expression of the two truncated proteins. In addition, when overexpressing SlNRC4aCRISPR in the background of the endogenous NtNRC4 tobacco ortholog, regions of the full-length NtNRC4, absent in the CRISPRed plants, could attenuate signalling (van Ooijen, van den Burg, Cornelissen, & Takken, 2007). Additional investigations into the function of SlNRC4a will address this issue. The enhanced defence responses identified upon EIX elicitation and the increased resistance to B. cinerea in the SlNRC4a CRISPRed plants give rise to the possibility of editing NRCs in the Solanaceae to examine the possibility of creating agricultural improved varieties with wide spectrum pathogen resistance. Deeper analysis is required to determine the range of SlNRC4a-mediated biotic resistance and decipher the signalling mechanism allowing amplification of diverse immune responses.
Endocytosis is required for LeEIX2 signalling: Inhibition of endosome formation reduces EIX-mediated responses while arresting endosomal trafficking increases the responses (Sharfman et al., 2011). When we studied the effect of overexpressing SlNRC4a on LeEIX2 endosomes prior to EIX elicitation, we observed a marked increase in the number and density of LeEIX2-positive endosomes. After EIX elicitation, both SlNRC4a overexpressing and control samples showed a similar increase relative to the control prior to EIX elicitation. A similar pattern was detected using SlNRC4a-CCd and SlNRC4a−CRISPR. SlNRC4a modifies LeEIX2 subcellular localization, leading to an early onset of LeEIX2 endosome induction. These data suggest that the increase in LeEIX2 endosome number primes the cell, enabling enhanced endosomal signalling and cellular responses upon EIX elicitation.
SlNRC4a emerges from our work as a positive regulator of plant defence in PTI, whereas so far N. benthamiana NRC4 orthologs have been shown to participate in ETI (Wu et al., 2017). We have identified a conserved solanaceous NLR that is involved in PTI signal propagation, highlighting the overlap in downstream signalling networks upon activation of diverse immune extracellular receptors. The separation between PTI/ETI is not always clear and can be considered as more of a continuum requiring specific shared signalling mechanisms (Thomma et al., 2011). Our findings indicate that SlNRC4a may be such a conserved signalling regulator required for diverse immune receptors. Future work will address this broad functionality and shed light on mechanisms common to different immune receptor types.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Mily Ron (UC Davis) for kindly providing plasmids, Prof. Yigal Elad and Dalia Rav-David for kindly providing B. cinerea isolate Bc-16, and Dr. James M. Elmore (Iowa State University) for mass spectrometry data analyses. This work was partly supported by Research Grant Award IS-4842-15 R from BARD, the United States-Israel Binational Agriculture Research and Development Fund, the United States-Israel Binational Science Foundation (2013227), and Chief Scientist of the Israel Ministry of Agriculture and Rural Development (Grant 13-37-0001). No conflict of interest declared.
Funding information
Chief Scientist of the Israel Ministry of Agriculture and Rural Development, Grant/Award Number: 13-37-0001; United States-Israel Binational Science Foundation, Grant/Award Number: 2013227; United States-Israel Binational Agriculture Research and Development Fund, Grant/Award Number: IS-4842-15 R
Footnotes
Current Address
Donald Danforth Plant Science Center, Saint Louis, Missouri, USA
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- Andolfo G, Jupe F, Witek K, Etherington GJ, Ercolano MR, & Jones JD (2014). Defining the full tomato NB-LRR resistance gene repertoire using genomic and cDNA RenSeq. BMC Plant Biology, 14, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni A, Bailey BA, Mattoo AK, & Anderson JD (1994). Induction of ethylene biosynthesis in Nicotiana tabacum by a Trichoderma viride xylanase is correlated to the accumulation of 1-aminocyclopropane-1-carboxylic acid (ACC) synthase and ACC oxidase transcripts. Plant Physiology, 106, 1049–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey BA, Dean JF, & Anderson JD (1990). An ethylene biosynthesis-inducing endoxylanase elicits electrolyte leakage and necrosis in Nicotiana tabacum cv Xanthi leaves. Plant Physiology, 94, 1849–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar M, & Avni A (2009). EHD2 inhibits ligand-induced endocytosis and signaling of the leucine-rich repeat receptor-like protein LeEix2. The Plant Journal, 59, 600–611. [DOI] [PubMed] [Google Scholar]
- Bar M, Sharfman M, Schuster S, & Avni A (2009). The coiled-coil domain of EHD2 mediates inhibition of LeEix2 endocytosis and signaling. PLoS one, 4, e7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M, Ve T, Williams S, Warren C, Hatters D, Valkov E, … Dodds PN (2011). Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host & Microbe, 9, 200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm H, Albert I, Fan L, Reinhard A, & Nurnberger T (2014). Immune receptor complexes at the plant cell surface. Current Opinion in Plant Biology, 20, 47–54. [DOI] [PubMed] [Google Scholar]
- Bonardi V, Cherkis K, Nishimura MT, & Dangl JL (2012). A new eye on NLR proteins: Focused on clarity or diffused by complexity? Current Opinion in Immunology, 24, 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V, & Dangl JL (2012). How complex are intracellular immune receptor signaling complexes? Frontiers in Plant Science, 3, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan J, Padmanabhan M, & Dinesh-Kumar SP (2008). Plant NB-LRR immune receptors: From recognition to transcriptional reprogramming. Cell Host & Microbe, 3, 126–135. [DOI] [PubMed] [Google Scholar]
- Casteel CL, Walling LL, & Paine TD (2006). Behavior and biology of the tomato psyllid, Bactericerca cockerelli, in response to the Mi-1.2 gene. Entomologia Experimentalis et Applicata, 121, 67–72. [Google Scholar]
- Cesari S, Bernoux M, Moncuquet P, Kroj T, & Dodds PN (2014). A novel conserved mechanism for plant NLR protein pairs: The “integrated decoy” hypothesis. Frontiers in Plant Science, 5, 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari S, Moore J, Chen C, Webb D, Periyannan S, Mago R, … Dodds PN (2016). Cytosolic activation of cell death and stem rust resistance by cereal MLA-family CC-NLR proteins. Proceedings of the National Academy of Sciences of the United States of America, 113, 10204–10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Bauer Z, Regenass M, Boller T, & Felix G (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell, 18, 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou KC, & Shen HB (2010). Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS one, 5, e11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier SM, Hamel LP, & Moffett P (2011). Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Molecular Plant-Microbe Interactions, 24, 918–931. [DOI] [PubMed] [Google Scholar]
- Couto D, & Zipfel C (2016). Regulation of pattern recognition receptor signalling in plants. Nature Reviews. Immunology, 16, 537–552. [DOI] [PubMed] [Google Scholar]
- Cui H, Tsuda K, & Parker JE (2015). Effector-triggered immunity: From pathogen perception to robust defense. Annual Review of Plant Biology, 66, 487–511. [DOI] [PubMed] [Google Scholar]
- De Meyer G, & Hofte M (1997). Salicylic acid produced by the rhizobacterium Pseudomonas aeruginosa 7NSK2 induces resistance to leaf infection by Botrytis cinerea on bean. Phytopathology, 87, 588–593. [DOI] [PubMed] [Google Scholar]
- van der Hoorn RA, Wulff BB, Rivas S, Durrant MC, van der Ploeg A, de Wit PJ, & Jones JD (2005). Structure-function analysis of cf-9, a receptor-like protein with extracytoplasmic leucine-rich repeats. Plant Cell, 17, 1000–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, … Gascuel O (2008). Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Research, 36, W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, & Rathjen JP (2010). Plant immunity: Towards an integrated view of plant-pathogen interactions. Nature Reviews. Genetics, 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Elad Y, & Yunis H (1993). Effect of microclimate and nutrients on development of cucumber gray mold (Botrytis cinerea) . Phytoparasitica, 21, 257–268. [Google Scholar]
- van Engelen FA, Molthoff JW, Conner AJ, Nap J-P, Pereira A, & Stiekema WJ (1995). pBINPLUS: An improved plant transformation vector based on pBIN19. Transgenic Research, 4, 288–290. [DOI] [PubMed] [Google Scholar]
- Eshed Y, & Zamir D (1995). An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics, 141, 1147–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Pozo N, Menda N, Edwards JD, Saha S, Tecle IY, Strickler SR, … Mueller LA (2015). The Sol Genomics Network (SGN)—From genotype to phenotype to breeding. Nucleic Acids Research, 43, D1036–D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriels SH, Vossen JH, Ekengren SK, van Ooijen G, Abd-El-Haliem AM, van den Berg GC, … Joosten MH (2007). An NB-LRR protein required for HR signalling mediated by both extra- and intracellular resistance proteins. The Plant Journal, 50, 14–28. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, & Boller T (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis . Mol Cell, 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Gust AA, & Felix G (2014). Receptor like proteins associate with SOBIR1-type of adaptors to form bimolecular receptor kinases. Current Opinion in Plant Biology, 21, 104–111. [DOI] [PubMed] [Google Scholar]
- Hanania U, & Avni A (1997). High-affinity binding site for ethylene-inducing xylanase elicitor on Nicotiana tabacum membranes. Plant Journal, 12, 113–120. [Google Scholar]
- Hao W, Collier SM, Moffett P, & Chai J (2013). Structural basis for the interaction between the potato virus X resistance protein (Rx) and its cofactor Ran GTPase-activating protein 2 (RanGAP2). The Journal of Biological Chemistry, 288, 35868–35876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry E, Yadeta KA, & Coaker G (2013). Recognition of bacterial plant pathogens: Local, systemic and transgenerational immunity. The New Phytologist, 199, 908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawchuk LM, Hachey J, Lynch DR, Kulcsar F, van Rooijen G, Waterer DR, … Prufer D (2001). Tomato Ve disease resistance genes encode cell surface-like receptors. Proceedings of the National Academy of Sciences of the United States of America, 98, 6511–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot M, Karniel U, Gelbart D, Fogel D, Evenor D, Kutsher Y, … Levin I (2015). A novel route controlling begomovirus resistance by the messenger RNA surveillance factor pelota. PLoS Genetics, 11, e1005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibman-Markus M, Schuster S, & Avni A (2017). LeEIX2 interactors’ analysis and EIX-mediated responses measurement In Shan L, & He P (Eds.), Plant pattern recognition receptors: Methods and protocols (pp. 167–172). New York, NY: Springer New York. [DOI] [PubMed] [Google Scholar]
- Liebrand TW, van den Berg GC, Zhang Z, Smit P, Cordewener JH, America AH, … Joosten MH (2013). Receptor-like kinase SOBIR1/EVR interacts with receptor-like proteins in plant immunity against fungal infection. Proceedings of the National Academy of Sciences of the United States of America, 110, 10010–10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Lin ZJ, & Coaker G (2011). A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host & Microbe, 9, 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, & Dinesh-Kumar SP (2002). Virus-induced gene silencing in tomato. The Plant Journal, 31, 777–786. [DOI] [PubMed] [Google Scholar]
- Macho AP, & Zipfel C (2014). Plant PRRs and00 the activation of innate immune signaling. Molecular Cell, 54, 263–272. [DOI] [PubMed] [Google Scholar]
- Maekawa T, Cheng W, Spiridon LN, Toller A, Lukasik E, Saijo Y, … Schulze-Lefert P (2011). Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host & Microbe, 9, 187–199. [DOI] [PubMed] [Google Scholar]
- Mantelin S, Peng HC, Li B, Atamian HS, Takken FL, & Kaloshian I (2011). The receptor-like kinase SlSERK1 is required for Mi-1-mediated resistance to potato aphids in tomato. The Plant Journal, 67, 459–471. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S Bryant SH (2017). CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Research, 45, D200–D203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marone D, Russo MA, Laido G, De Leonardis AM, & Mastrangelo AM (2013). Plant nucleotide binding site-leucine-rich repeat (NBS-LRR) genes: Active guardians in host defense responses. International Journal of Molecular Sciences, 14, 7302–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S, Niedermeyer J, Fry J, Barnason A, Horsch R, & Fraley R (1986). Leaf disc transformation of cultivated tomato (L. esculentum) using Agrobacterium tumefaciens. Plant Cell Reports, 5, 81–84. [DOI] [PubMed] [Google Scholar]
- McDowell JM, & Simon SA (2006). Recent insights into R gene evolution. Molecular Plant Pathology, 7, 437–448. [DOI] [PubMed] [Google Scholar]
- Milligan SB, Bodeau J, Yaghoobi J, Kaloshian I, Zabel P, & Williamson VM (1998). The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell, 10, 1307–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandety RS, Caplan JL, Cavanaugh K, Perroud B, Wroblewski T, Michelmore RW, & Meyers BC (2013). The role of TIR-NBS and TIR-X proteins in plant basal defense responses. Plant Physiology, 162, 1459–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura MT, Anderson RG, Cherkis KA, Law TF, Liu QL, Machius M Dangl JL (2017). TIR-only protein RBA1 recognizes a pathogen effector to regulate cell death in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 114, E2053–E2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsch JP, & Nitsch C (1969). Haploid plants from pollen grains. Science, 163, 85–87. [DOI] [PubMed] [Google Scholar]
- Nombela G, Williamson VM, & Muniz M (2003). The root-knot nematode resistance gene Mi-1.2 of tomato is responsible for resistance against the whitefly Bemisia tabaci. Molecular Plant-Microbe Interactions, 16, 645–649. [DOI] [PubMed] [Google Scholar]
- van Ooijen G, van den Burg HA, Cornelissen BJ, & Takken FL (2007). Structure and function of resistance proteins in solanaceous plants. Annual Review of Phytopathology, 45, 43–72. [DOI] [PubMed] [Google Scholar]
- Rairdan GJ, Collier SM, Sacco MA, Baldwin TT, Boettrich T, & Moffett P (2008). The coiled-coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell, 20, 739–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M (2004). Tomato LeEix genes: Isolation, characterization and their participation in the signal transduction leading to “program cell death”. Doctor of Phylosophy: Tel Aviv University. [Google Scholar]
- Ron M, & Avni A (2004). The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell, 16, 1604–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T Cardona A (2012). Fiji: An open-source platform for biological-image analysis. Nature Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlecht I, Keske U, Hierholzer J, & Felix R (1999). High resolution computed tomography in a patient with chronic granulomatous disease of the lung. European Journal of Radiology, 32, 208–210. [DOI] [PubMed] [Google Scholar]
- Sharfman M, Bar M, Ehrlich M, Schuster S, Melech-Bonfil S, Ezer R, … Avni A (2011). Endosomal signaling of the tomato leucine-rich repeat receptor-like protein LeEix2. The Plant Journal, 68, 413–423. [DOI] [PubMed] [Google Scholar]
- Steinhauser MC, Steinhauser D, Gibon Y, Bolger M, Arrivault S, Usadel B Stitt M (2011). Identification of enzyme activity quantitative trait loci in a Solanum lycopersicum × Solanum pennellii introgression line population. Plant Physiology, 157, 998–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange RN, & Scott PR (2005). Plant disease: A threat to global food security. Annual Review of Phytopathology, 43, 83–116. [DOI] [PubMed] [Google Scholar]
- Takken FL, Thomas CM, Joosten MH, Golstein C, Westerink N, Hille J Jones JD (1999). A second gene at the tomato Cf-4 locus confers resistance to Cladosporium fulvum through recognition of a novel avirulence determinant. The Plant Journal, 20, 279–288. [DOI] [PubMed] [Google Scholar]
- Thomma BP, & Bignell E (2016). Editorial overview: The fungal infection arena in animal and plant hosts: Dynamics at the interface. Current Opinion in Microbiology, 32, v–vii. [DOI] [PubMed] [Google Scholar]
- Thomma BP, Nurnberger T, & Joosten MH (2011). Of PAMPs and effectors: The blurred PTI-ETI dichotomy. Plant Cell, 23, 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tor M, Lotze MT, & Holton N (2009). Receptor-mediated signalling in plants: Molecular patterns and programmes. Journal of Experimental Botany, 60, 3645–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toruno TY, Stergiopoulos I, & Coaker G (2016). Plant-pathogen effectors: Cellular probes interfering with plant defenses in spatial and temporal manners. Annual Review of Phytopathology, 54, 419–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GF, Ji J, El-Kasmi F, Dangl JL, Johal G, & Balint-Kurti PJ (2015). Molecular and functional analyses of a maize autoactive NB-LRR protein identify precise structural requirements for activity. PLoS Pathogens, 11, e1004674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Devoto A, Turner JG, & Xiao S (2007). Expression of the membrane-associated resistance protein RPW8 enhances basal defense against biotrophic pathogens. Molecular Plant-Microbe Interactions, 20, 966–976. [DOI] [PubMed] [Google Scholar]
- Win J, Chaparro-Garcia A, Belhaj K, Saunders DG, Yoshida K, Dong S Kamoun S (2012). Effector biology of plant-associated organisms: Concepts and perspectives. Cold Spring Harbor Symposia on Quantitative Biology, 77, 235–247. [DOI] [PubMed] [Google Scholar]
- Wu CH, Abd-El-Haliem A, Bozkurt TO, Belhaj K, Terauchi R, Vossen JH, & Kamoun S (2017). NLR network mediates immunity to diverse plant pathogens. Proceedings of the National Academy of Sciences of the United States of America. 201702041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Belhaj K, Bozkurt TO, Birk MS, & Kamoun S (2016). Helper NLR proteins NRC2a/b and NRC3 but not NRC1 are required for Pto-mediated cell death and resistance in Nicotiana benthamiana. The New Phytologist, 209, 1344–1352. [DOI] [PubMed] [Google Scholar]
- Xiao S, Ellwood S, Calis O, Patrick E, Li T, Coleman M, & Turner JG (2001). Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science, 291, 118–120. [DOI] [PubMed] [Google Scholar]
- Xie K, Minkenberg B, & Yang Y (2015). Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proceedings of the National Academy of Sciences of the United States of America, 112, 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachdav G, Kloppmann E, Kajan L, Hecht M, Goldberg T, Hamp T Rost B (2014). PredictProtein—An open resource for online prediction of protein structural and functional features. Nucleic Acids Research, 42, W337–W343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Tehrim S, Zhang F, Tong C, Huang J, Cheng X Liu S (2014). Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana. BMC Genomics, 15, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Dodds PN, & Bernoux M (2017). What do we know about NOD-like receptors in plant immunity? Annual Review of Phytopathology, 55, 205–229. [DOI] [PubMed] [Google Scholar]
- Zipfel C (2014). Plant pattern-recognition receptors. Trends in Immunology, 35, 345–351. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, & Boller T (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature, 428, 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


