Abstract
Stereolithography (SL) is emerging as an attractive alternative to soft lithography for fabricating microfluidic devices due to its low cost and high design efficiency. Low molecular weight poly(ethylene glycol)diacrylate (MW = 258) (PEG-DA-258) has been used for SL 3D-printing of biocompatible microdevices at submillimeter resolution. However, 3D-printing resins that simultaneously feature high transparency, high biocompatibility, and high resolution are still lacking. It is found that photosensitizer isopropyl thioxanthone can, in a concentration-dependent manner, increase the absorbance of the resin (containing PEG-DA-258 and photoinitator Irgacure-819) by over an order of magnitude. This increase in absorbance allows for SL printing of microdevices at sub pixel resolution with commercially available desktop printers and without compromising transparency or biocompatibility. The assembly-free, rapid (<15 h) 3D-printing of a variety of complex 3D microfluidic devices such as a 3D-fluid router, a passive chaotic micro-mixer, an active micro-mixer with pneumatic microvalves, and high-aspect ratio (37:1) microchannels of single pixel width is demonstrated. These manufacturing capabilities are unavailable in conventional microfluidic rapid prototyping techniques. The low absorption of small hydrophobic molecules and microfluidic labeling of cultured mammalian cells in 3D-printed PEG-DA-258 microdevices is demonstrated, indicating the potential of PEG-DA-based fabrication of cell-based assays, drug discovery, and organ-on-chip platforms.
Keywords: 3D printing, microfluidic, poly(ethylene glycol)diacrylate, rapid prototyping, stereolithography
1. Introduction
Stereolithography (SL) is emerging as an attractive alternative to soft lithography due to its low-cost improvements in design efficiency and rapid prototyping. With SL, designs can be digitally inspected, adjusted, annotated, and cloud-shared with collaborators, which result in improvements toward design turn-around time, cost, and performance. By comparison with other manufacturing fields, micro fluidics has been slow to adopt SL than traditional soft lithography. Microfluidic chips are still designed largely from scratch; the materials are often manually mixed and poured into a mold to form 2D-layer replicas, and the mold replicas are manually aligned and bonded to form the final device. The 2D layers are usually constructed by micromolding thermoset or thermoplastic polymers chosen for their high transparency and biocompatibility, such as poly(dimethyl siloxane) (PDMS),[1] poly(methyl methacrylate) (PMMA),[2] cyclic olefin copolymer (COC),[3] and polystyrene (PS).[4] The molding procedures for these polymers have been optimized for microfluidics applications for decades.
Micromolding approaches have three major limitations that have hindered the growth of microfluidics. First, they are not ideal for R&D prototyping because making the photomasks and the molds lengthens the cycle time. Second, launching the production of microfluidic devices by micromolding is expensive, which can impede progress even when the molds’ costs can be recovered later by selling large numbers of assembled device copies. Third, 3D structures are difficult/costly to fabricate by micromolding because channels and chambers must be fabricated by bonding (and usually aligning) at least two layers. In summary, the production of microfluidic devices by micromolding, while being optimized for mass manufacturing, cannot be optimized at the same time for design variety. These limitations are difficult for researchers to assimilate because micromolding has been the prevalent mode of microfluidics manufacturing for over two decades. The vast majority of microfluidic devices designed for biomedical applications are still fabricated in PDMS using soft lithography.[5] PDMS has been used to build microvalves[6] and micropumps,[7] passive[8] and active[9] micromixers, pneumatic microvortexers,[10] and devices for screening cells,[11] droplets,[12] and small organisms,[13] among others. PDMS is an optically clear, water-impermeable, gas-permeable, and biocompatible elastomer that can be used to make microdevices from molds fabricated by photolithography.[14] However, PDMS molding is labor-intensive (thus is slow and expensive), requires personnel with specialized training, and has severe constraints on 3D design complexity. In addition, connectors for fluid feeds and pressure sources must be manually integrated with the microdevice, even if specialized multi-connectors are adopted.[15,16]
SL is a form of additive manufacturing (popularly known as 3D-Printing) invented in the 1980s[17] that does not have any of the three previous limitations of micromolding technologies. SL allows for the assembly-free production of quasi-arbitrary 3D shapes in a single polymeric material (at once) from a liquid photoresin precursor by means of a focused laser[18–20] or a digital light-processing (DLP) projector.[21,22] Two-photon micro-SL has been used to produce micron-resolution structures of a variety of materials, but the instruments are prohibitively expensive.[23–25] More recently, desktop SL systems based on DLP projectors are emerging as an attractive alternative to soft lithography for fabricating microfluidic devices due to their high resolution and low cost (the patent has expired) and ability to pattern cavities.[26,27] The molar absorptivity of the resin determines how far the light penetrates into the resin and is thus a critical determinant of the Z-resolution of the apparatus; the XY-resolution is a function of the projected size of the projector’s pixels and of the diffusive broadening of the reactants.
Therefore, 1) SL is inherently rapid, because it bypasses the step(s) involved in fabricating/replicating from a mold; 2) SL is very economical, as it circumvents costly mold-making and connects efficient digital design with efficient additive manufacturing; and 3) SL is not limited to 2D-layered geometries, rather, it generates quasi-arbitrary 3D architectures. As a result, SL is inherently suited for design variety and design customization. The economics of 3D-printing are well suited for microfluidics because, as opposed to molding approaches, the cost per device does not scale up with its 3D complexity (complexity is free) and is insensitive to the size of the production batch, i.e., 3D-printing is ideal for project customization (variety is free).[28] Also, since SL equipment is largely automated, training personnel for SL equipment is much faster and inexpensive than for molding processes.
Poly(ethylene glycol)diacrylate (PEG-DA) is a photopolymerizable material that is very commonly used to prepare biocompatible hydrogels by SL for cell encapsulation and tissue engineering.[29] Several groups including ours have demonstrated microfluidic devices made of 3D-printed low molecular weight (MW = 258) PEG-DA (PEG-DA-258).[30,31] The addition of biomolecules such as antibodies, enzymes, or peptides to the PEG-DA resin can endow the prints with biosensing functionalities[32] and cell-adhesive properties.[33] We have shown that these 3D-printed PEG-DA-258 microfluidic devices can be made both transparent and cytocompatible[30]; however, the resolution was limited by the absorption of the resin. Here we present and characterize a high-absorption PEG-DA-258-based resin formulation and use it to fabricate transparent 3D microfluidic devices compatible with cell culture, single-pixel, 27 µm wide microchannels, as well as to produce sub-pixel (≈5–10 µm wide) features, which altogether show that it is possible to 3D-print PEG-DA-258 biomicrofluidic devices at high precision.
2. Results and Discussion
2.1. Resin Formulation
SL-printing of transparent microfluidic devices requires 1) a transparent monomer that polymerizes into water-impermeable plastic structures, 2) a colorless photoinitiator system that is soluble in the monomer, and 3) high photochemical efficiency and low penetration (high absorbance) at the wavelength of the projector’s light source. Previously, we have argued that a monochromatic (385 nm) UV-LED projector is more adequate than a visible-light projector for high-resolution printing with a transparent resin because the resin appears highly absorptive to the UV source (but transparent to the human eye).[30] (Note that the 385 nm UV-LED is the lowest wavelength UV light source that is available with commercial DLP systems.) Using 385 nm UV-LED DLP SL, we printed transparent microfluidic devices and cell-culture compatible Petri dishes with low molecular weight (MW = 258) PEG-DA (PEG-DA-258; Figure 1a) as our monomer, photoinitiator Irgacure-819 (IRG; Figure 1b), and photosensitizer isopropyl thioxanthone (ITX; Figure 1c). IRG is a light yellow solid powder that absorbs strongly at 385 nm. A relatively low concentration of IRG (0.4% w/w) is sufficient for polymerizing PEG-DA-258 (albeit with low absorption, see below), thereby ensuring that the final prints remain transparent with low pigmentation. Unless otherwise stated, the PEG-DA-258 resin used for SL-printing the devices described in this paper contained 0.4% (w/w) IRG as the photoinitiator. The absorbance graph for IRG at 0.4% w/w is shown in Figure 1d (red curve). A qualitative transparency comparison of SL-printed 500 µm thick PEG-DA-258 blocks using a range of increasing % ITX concentrations (0.4% IRG + 0% ITX, 0.4% IRG + 0.2% ITX, 0.4% IRG + 0.4% ITX, and 0.6% IRG + 0.6% ITX) shows little increasing pigmentation (Figure S2, Supplementary Information).
Figure 1.
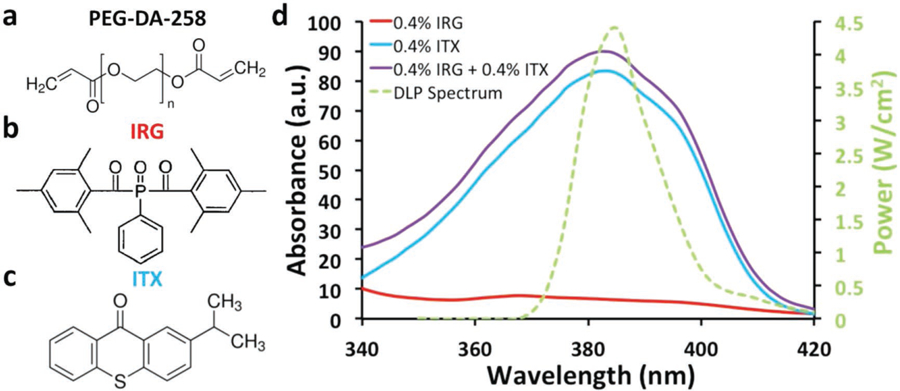
Resin composition and absorbance spectrum. a) Molecular structure of the monomer used in our resin: poly(ethylene glycol)diacrylate (PEG-DA-258). b) Molecular structure of the photoinitiator used in our resin: phenylbis(2,4,6-trimethylbenzoyl)phosphine oxide, known as Irgacure-819 (IRG). c) Molecular structure of the photosensitizer used in our resin: isopropylthioxanthone (ITX). d) Absorbance spectra of ITX at 0.4% (blue), IRG 0.4% (red), and IRG 0.4% + ITX 0.4% (purple) plotted along with the power spectrum of the 385 nm UV-LED DLP projector (green). The percentages indicate % of weight in isopropyl alcohol.
As quantitatively discussed in our earlier paper, the absorption of the photoinitiator system (at 385 nm) is a critical determinant of the Z-resolution of microfluidic channels that can be SL-printed.[30] This relationship is explained by a derivation from the Beer–Lambert law,[34] where absorbance (A), which is defined as the logarithmic ratio between the radiation intensity entering the sample (Io) and the radiation intensity emerging from it (I), is equal to the product of the path length through the sample (l), the concentration of the sample (c), and the molar absorptivity (ε):
| (1) |
From Equation (1), it is straightforward to show (see Supplementary Materials) that the cure depth, or the depth to which the resin is photopolymerized (zr), is linearly related to the logarithm of the exposure time (tr) according to the following equation[34]:
| (2) |
Increasing the concentration of the photoinitiator, IRG, can increase the absorption of the resin and therefore produce prints with improved Z-resolution; however, an increased concentration will also make the prints colored (yellow). A complementary strategy is to use a photosensitizing additive that strongly absorbs at 385 nm and may, in some instances, transfer the energy to another reactive molecule. UV absorbing dyes like Sudan I and 2-nitrophenyl phenyl sulfide (NPS) have been used to increase absorption and Z-resolution, but these dyes render the prints colored (deep orange in the case of Sudan I,[35] yellow in the case of NPS[36]). The cytocompatibility of these dyes has not been studied.
Our goal was to enhance the resolution of SL-printed microdevices without compromising the transparency and cytocompatibility of the prints. Therefore, we first searched for an additive that absorbs strongly at 385 nm but does not impart a strong color to the resin. Note that a highly absorptive photochemical species can potentially increase the Z-resolution at concentrations low enough to not significantly color the SL-printed devices. We surveyed the absorption spectra of commercially available photosensitive free-radical initiating chemicals as well as UV-absorbers and matched them with the UV-LED spectrum of our 385 nm DLP light source. We also took into account the chemical nature and solubility profiles of the chemicals—for example, a highly hydrophobic chemical or one that is sparingly miscible with alcohols or water was discarded. Among the few available candidate molecules, we decided to further examine ITX (Figure 1c), which is a photosensitizer typically used in Type II photopolymerization systems[37,38] and has a secondary absorption peak at 384 nm (Figure 1d). ITX dissolves in PEG-DA-258 when heated at 70 °C for 10 min. The absorbance plot for 0.4% (w/w) ITX is shown in Figure 1d (blue curve). Since the molar absorptivity of ITX is 7.9 times higher than that of IRG, at equal concentrations (0.4%), the final absorbance is dominated by ITX. By increasing the concentration of ITX, we can further enhance the total absorbance of the resin. At 385 nm, the addition of ITX to IRG (0.4% IRG + 0.4% ITX; purple curve) is exactly 13.97 times higher in absorbance than without the addition of ITX (0.4% IRG; red curve) (Figure 1d). Photosensitizers such as ITX absorb light, become energized to an excited triplet state, and transfer this energy to nearby hydrogen-donor molecules (also known as co-initiators) like amines, thiols, or alcohols, which then form free-radicals.[39] However, our resin formulation lacks efficient co-initiators. Therefore, we believe that ITX, due to its high molar absorption coefficient, contributes primarily as a strong absorber at 385 nm and therefore helps in increasing the Z-resolution of printing with minimal pigmentation. For a blend of photosensitive compounds, the absorbance (A; Equation (1)) is linearly related to the sum of the products of molar absorptivity (ε) and concentration (c) of the individual photosensitive components in the resin.[40] Since ITX is a light yellow powder, the liquid resin becomes yellow-tinged and can add color to the prints at higher concentrations. However, as we show later, at ITX concentrations less than 0.4% w/w, the SL-printed structures are transparent and remain effectively colorless (the transmittance of 4 mm thick 3D-printed parts at visible spectrum wavelengths higher than 425 nm is greater than 80%; see Figure S1, Supplementary Information). A qualitative comparison of 500 µm thick PEG-DA-258 blocks with a range of increasing % ITX concentrations (0.4% IRG + 0% ITX, 0.4% IRG + 0.2% ITX, 0.4% IRG + 0.4% ITX, and 0.6% IRG + 0.6% ITX) shows slight increases in yellow pigmentation, but still maintains transparency (Figure S2, Supplementary Information).
2.2. Resolution Characterization
We first examined the highest XY-resolution at which we could reliably SL-print PEG-DA-258 patterns using a commercial 3D-printer (Ilios HD). For any given resin, the XY-resolution primarily depends on the nominal pixel resolution (52 µm for this printer) and optical precision of our 3D-printer. The printer’s UV-LED DLP is based on a Digital Mirror Display (DMD) chip that consists of square-shaped deflectable-micromirrors arranged in a rectangular array (see Figure S3a,b, Supplementary Information). Each micromirror projects one pixel of the DLP; we tested whether sub-structures in each micromirror (such as the central attachment point of each mirror and the gaps between mirrors) could also be projected and printed. We printed ≈200 µm high patterns of 1–10 pixel wide lines (horizontal, vertical, or diagonal) in four 50 µm Z-layers; the lines were separated by gaps equal to the linewidths (Figure S4, Supplementary Information; Figure 2a,b). In Figure 2c–f, sub-pixel features created by the topology of the micromirrors in the DMD can be clearly resolved. The scanning electron microscope (SEM) pictures in Figure 2e,f show the ≈10 µm-diameter circular dimples at the center of each pixel and ≈5–10 µm wide lines between the pixels. The resulting features show that the projection optics have the resolution to project sub-pixel features and that the resin polymerization kinetics are much faster than the diffusion rate of the reactants,[41] suggesting that this resin would produce better prints with a higher resolution printer.
Figure 2.
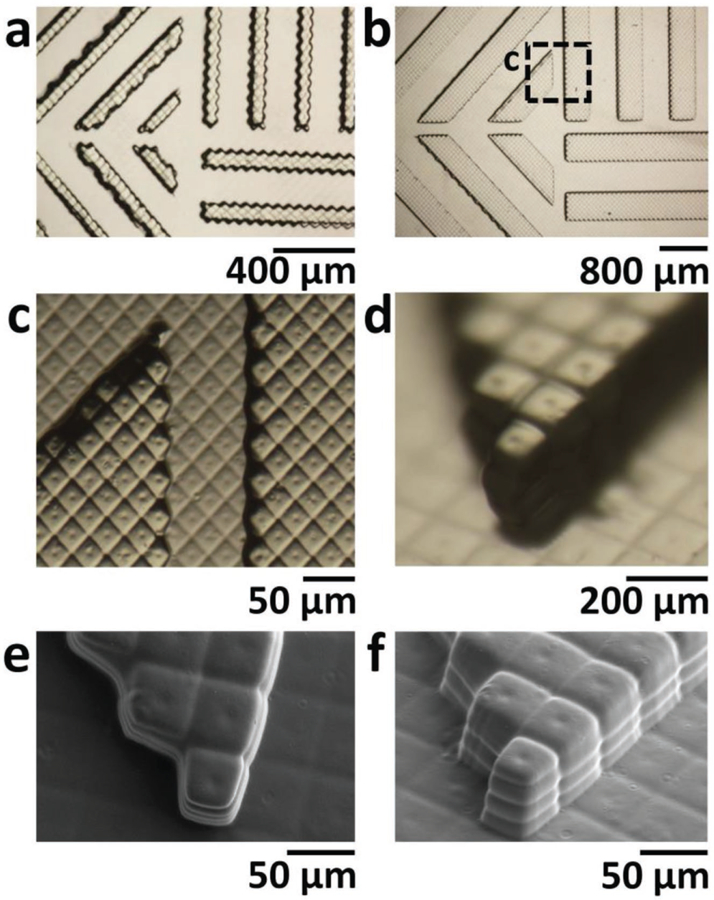
X–Y resolution. a,b) Bright field micrographs of a) 2 pixel wide and b) 8 pixel wide, 200 µm tall lines that were 3D-printed horizontally, vertically, and diagonally in 50 µm thick Z-layers with 0.4% IRG and 0.4% ITX PEG-DA-258 resin. c) Magnification of the area boxed in (b). (d) Oblique-view bright-field micrograph of SL-printed 200 µm tall pixels at the corner of an 8 pixel wide diagonal line. Note how sub-pixel features in (c) and (d) are well defined. e) Top-view SEM image of pixels at the corner of an 8 pixel wide diagonal line. The image reveals the shape of sub-pixel features created by the topology of the DMD micromirrors: ≈10 µm diameter circular dimples at the center of each pixel and ≈5–10 µm wide lines between the pixels. f) Oblique-view SEM image of pixels at the end of an 8 pixel wide diagonal line where individual 50 µm thick layers can be seen as well as the sub-pixel features mentioned above.
We then explored different resin formulations (with different concentrations of the photoinitiator and the photosensitizer) to study the impact of different exposure parameters on roof thickness. For each resin, we SL-printed 2 mm wide, 5 mm long, 500 µm high walls with 500 µm wide gaps. Between the walls, roof structures were exposed with increasing exposure time per roof (Figure 3a,b). Here the measured roof thickness is equal to the cure depth zr in Equation (2); therefore, the roof thickness zr is log-linear with respect to the exposure time tr. Hence, the characteristic penetration depth (hd), equal to 1/(2.303εc), can be found as the slope of the plot between zr and ln(tr). Thus, the slope of the plot determines the hd of the resin formulation. Both the hd and zr decreased over five times as we increased the concentrations of IRG and ITX from 0.4% IRG + 0.1% ITX to 0.8% IRG + 0.8% ITX for the exposure range of 100 ms < tr < 300 ms. Lower hd (lower slopes in Figure 3c) are more advantageous because they allow for more precise control over the roof thickness as the exposure is varied. With higher concentrations of IRG and ITX, the hd decreases, resulting in lesser curing of the void underneath the roof, thereby producing unobstructed channels with thinner roofs. The minimum roof height that we were able to build was ≈15 µm (Figure 3c, blue line). These data indicate that a high-absorption resin (i.e., a resin that features a low hd) enables the 3D-printing of shallow unobstructed channels. We also printed these structures without the addition of ITX to demonstrate decreasing resolution when lacking the photosensitizer (Figure S5, Supplementary Information). The wall structures were printed with a 1 mm gap and 2 mm tall height in order to show gradual occlusion of the channel, as the roof thickness was much higher for similar times. When printed without ITX (0.4% IRG) and exposed at 200 ms, the roof thickness was ≈741 µm, but with ITX (0.4% IRG + 0.2% ITX), the roof thickness was over seven times thinner (≈103 µm). This control using 0.4% IRG resin shows increasingly thicker roofs when compared with the addition of ITX (0.4% IRG + 0.4% ITX), suggesting that the addition of ITX leads to more precise control over Z resolution.
Figure 3.
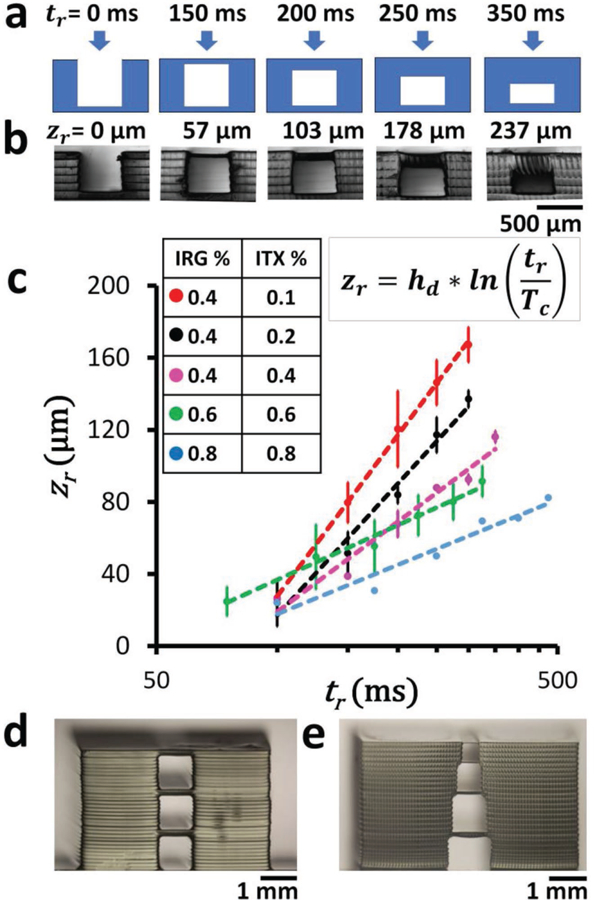
Z resolution. a) Cross-sectional schematic of the photopolymerization of flat roof layers over a 500 µm wide gap supported by 200 µm wide walls. Different times of exposure (tr) will result in roofs of different thicknesses, according to Equation (2). b) Phase-contrast micrographs of the side views of 3D-printed structures printed with 0.4% IRG and 0.2% ITX PEG-DA-258 resin. The measured thickness of the polymerized roof structures zr is displayed above each image corresponding to the different times of exposure tr shown in (a). c) Log-linear plot of the thickness of 3D-printed roofs (zr) with respect to exposure times (tr) for different resin compositions ([0.4% IRG + 0.1% ITX], [0.4% IRG + 0.2% ITX], [0.4% IRG + 0.4% ITX], [0.6% IRG + 0.6% ITX], [0.8% IRG + 0.8% ITX]). Error bars denote standard deviations. d) 3D-printed stacked 1 mm square cross-sectional channels, separated by 100 µm roofs. The roofs are printed with a single 100 µm thick Z-layer using PEG-DA-258 resin with 0.4% IRG and 0.4% ITX. e) 3D-printed stacked channels with progressively reduced square cross sections (1, 0.8, 0.6, and 0.4 mm) separated by 50 µm roofs. The roofs are printed with a single 50 µm thick Z-layer using PEG-DA-258 resin with 0.4% IRG and 0.4% ITX.
In complex 3D microfluidic architectures with channels in different layers, irradiation of the resin for building the upper channel walls can potentially photopolymerize the underlying channels that are still filled with resin. Therefore, we tested the 3D-printability of directly stacked 1 mm wide square channels with ≈150 µm thick roofs (Figure 3d). We investigated whether a pyramidal stack of channels with progressively reduced widths will decrease the width of the underlying channels because of occlusion along the channel walls. We printed a stack of square channels starting at 1 mm wide with widths decreasing by 20%, each overlaid with a ≈50 µm thick roof, and the channel widths remained within 3.4% of the designed widths (Figure 3e). Our ability to print stacked channels would enable assembly-free fabrication of 3D microfluidic devices.
2.3. 3D Microfluidic Devices
Having shown that we could stack channels on top of each other (Figure 3d,e), we designed a microfluidic “router” that demonstrates the monolithic SL fabrication of stacked, overlapping microchannels. The router exchanges the order of three microchannels by crisscrossing the channels in three different fluid layers and merges the three channels (filled with blue, yellow, and red dyes) into one (Figure 4a). All the channels were 500 µm in height. The three different fluid layers were separated by 250 µm of PEG-DA-258 along the Z-axis. All the channels were 500 µm wide before merging at the outlet, and the outlet channel was 1.5 mm wide. The integrated fluid ports were designed as 1/16 in. barb connectors. High-magnification micrographs of the laminar flow at the outlet (Figure 4b) and of the overlapping of the fluids without mixing on the inlet side (Figure 4c) are shown.
Figure 4.
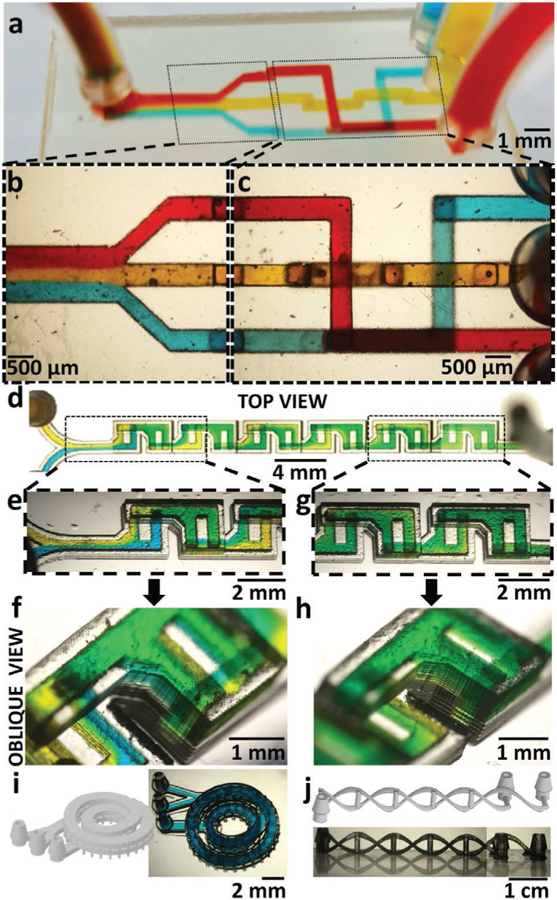
3D microfluidics. a) Oblique view of a 3D-printed 3-way microfluidic “3D-router” with red, yellow, and blue dye flowing through 500 µm square channels. b) Magnified top-view micrograph of the 3-way microfluidic router channels at the outlet area, showing the laminar flow of the three dyes as they merge in the outlet channel. c) Magnified top-view micrograph of the 3D-router showing the channels at the inlet area. The image shows channels crossing each other (blue and yellow, red and yellow) or overlaying each other (red and blue), without the dyes mixing. d) Top-view micrograph of a 3D-printed F-mixer (500 µm wide, 500 µm tall channels) with blue and yellow dyes flowing at 6 mL h−1. e) Top-view micrograph of the laterally inverted, stacked F-units next to the inlets. The fluid in the upper (second) F-unit appears green. In the subsequent lower F-unit, the fluid splits back into separate blue and yellow streams, confirming the vertical lamination of flow in the previous F-unit. f) Oblique-view micrograph of the F-units next to the outlet. The fluid in the last F-unit appears almost homogeneously green, confirming the effectiveness of this mixer design at mixing fluids. g) Top-view micrograph of the first F-unit showing the vertical lamination of blue and yellow dyes visible through the translucent side-walls of the device. h) Oblique-view micrograph of the last F-unit showing almost complete mixing of the blue and yellow dyes. i) (Left) 3D-CAD of a two-turn coil microchannel nested inside another two-turn coil microchannel. (Right) Oblique-view micrograph of the 3D-printed device filled with blue dye. The channels are 1 mm wide and 300 µm tall. j) (Top) 3D-CAD of two microchannels intertwined like a double-helix DNA, with the “nucleotide” side channels acting as points of fluidic contact between the two main channels. (Bottom) Side-view micrograph of the 3D-printed double-helix DNA microfluidic device. The main channels are 400 µm × 750 µm in cross section and the side channels are 500 µm in diameter.
3D fabrication can be especially useful for enhancing mixing, since the laminar flow regime predominates in microchannels, with mixing limited to diffusive processes.[42] Hence, we SL-printed a highly efficient passive mixer known as the chaotic F-mixer (Figure 4d)[43]; the alternating, laterally inverted, stacked upper and lower F-shaped units combine chaotic advection and splitting/recombination methods of micromixing.[43] Previous F-mixers, C-mixers, and L-mixers have been produced by molding[8,43] and by laser cutting,[44] which requires precise assembly, alignment, and bonding of at least two parts, plus inlet fabrication. Our SL-printed F-mixer was printed as a single part without alignment or bonding and included integrated barb fluidic connectors. The F-mixer had 12 F-units (500 µm × 500 µm channels) arranged in series and was operated at a flow rate of 6 mL h−1 with different colored dyes (blue and yellow in Figure 4d–h). The transparency of the device printed with our PEG-DA-258-based resin and the absence of extra bulk material allowed us to characterize the mixing of the blue and yellow dyes within the F-mixer with straightforward methods compared with traditional (molded) devices. When we observed the mixing of the dyes (at 6 mL h−1) from a top view, the fluid in the upper F-units appeared mixed (i.e., the unit looked green); however, the fluid in the subsequent lower F-unit appeared to split back into blue and yellow (Figure 4e,f). When the F-units are observed obliquely through one of the translucent lateral walls, blue and yellow dyes flows were seen to overlay on top of each other through the side wall of the device (Figure 4f). The splitting/ recombination of the dyes could be observed till the last pair of F-units, where the dyes appeared mixed in both the top and lateral views (Figure 4g,h). This device demonstrates the potential of SL for assembly-free fabrication and microscopic observation of 3D microfluidic devices that feature complex flow patterns.
We have also printed two microfluidic devices which demonstrate the level of complexity enabled by SL that cannot be afforded by micromolding. First, we have 3D-printed a two-turn coil microchannel nested inside another two-turn coil microchannel (Figure 4i, left). The channels are 1 mm wide and 300 µm tall and have been filled with blue dye to demonstrate channel clearance (Figure 4i, right). We envision this device could be used for particle separation using Dean flow. We have also 3D-printed two microchannels intertwined like a DNA-like double helix, with “nucleotide” side channels (500 µm in diameter) acting as points of fluidic contact between the two main channels (400 µm × 750 µm in cross section) (Figure 4j). We believe that the quasi-absence of 3D design restrictions in SL (compared to micromolding), combined with advances in new materials and resins, will open the field to an unprecedented range of microfluidic device functionalities.
2.4. Single-Pixel High-Aspect-Ratio Microdevices
A high-aspect-ratio channel can be used to protect the UV penetration with the resin itself, as long as space is available on top of the channel. For experiments, we used an Asiga Pico2 HD UV printer, which features 27 µm XY resolution. As shown in Figure 5a, we designed and printed a microfluidic device consisting of two channels which are orthogonally connected by a series of parallel microchannels that are 500 µm long, 1 mm tall, and 27 µm wide (1 pixel wide) and are separated by 54 µm (2 pixel wide) walls (Figure 5b). Note that the microchannels, filled with blue dye, have a 37:1 aspect ratio. It is interesting to note that the fabrication of high-aspect-ratio microstructures, which are difficult to achieve by photolithography, is very easily achieved with SL. Furthermore, with our high-absorbance resin, it is straightforward to construct a roof over very deep channels because the UV light becomes absorbed within the very top portion of the channel volume. Although errors in CAD slicing by the printer’s STL file conversion software result in occasional omission/addition of pixels (black arrow in Figure 5a), the printing error had no noticeable effect on the overall functionality of the device. Importantly, as more researchers apply 3D-printing to microfluidics, we envision that in a near future, there will be a bigger incentive for software developers to produce improved slicing algorithms.
Figure 5.
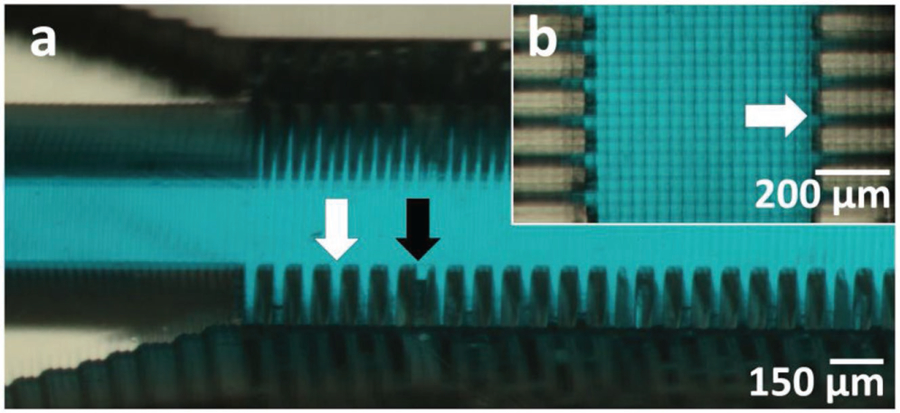
High-resolution, high aspect ratio channels. a) Oblique-view micrograph of 1 pixel (27 µm) wide, 1 mm tall channels separated by 2 pixel (54 µm) wide walls. The channels (white arrow) are printed using the Asiga Pico2 HD Printer and subsequently filled with blue dye to demonstrate channel clearance. Errors in CAD slicing (black arrow) by the printer’s STL file conversion software resulted in omission/addition of pixels, which led to inaccuracies in the printing of some wall structures but had no detectable effect on the overall functionality of the device. b) Top-view micrograph of 1 pixel wide channels (white arrow) filled with blue dye.
2.5. Active Microfluidic Devices
Many high-throughput microfluidic assays require automation (microvalves and/or micropumps), but present designs of 3D-printed microfluidic valves occupy large amounts of real estate[45] and/or are printed with non-transparent resins.[31] Our resin formulation has allowed us to 3D-print a (500 µm diameter) open-at-rest “pinch” microvalve[46] which consists of a membrane that is deflected by the pneumatic application of a small positive pressure (≈2 psi) via a “control channel” overlaid orthogonally atop the flow channel (Figure 6a–c). The valve devices were printed using a 25 µm Z-layer thickness. The membrane consists of a single 25 µm-thick Z-layer between the control and the flow channels, and when ≈2 psi of pressure is applied, the membrane contacts the bowl-shaped seat of the valve, closing the valve (Figure 6c). Because PEG-DA-258 is less flexible than PDMS (the Young’s Modulus of PEG-DA-258 is about 130 MPa[47] and that of PDMS is less than 1 MPa[48]), the membrane’s restoring force allows for reopening the valve without negative pressure.[46] Using our microvalve design, we have been able to fabricate a rotary mixer (similar to the design by Quake)[49,50] controlled by three individually actuated microvalves for pumping and three additional microvalves for fluid routing (Figure 6d), labeled V1–V6 (Figure 6e). The flow and control channels are 300 µm wide.[46] We have modified the control channel design by replacing the bulky outlets (which are plugged during operation to resemble dead-ended PDMS valves) with space-saving 300 µm diameter vents that remain open at all times. The vents allow for clearing of uncured resin from the channels after printing, effectively halving the required number of pneumatic inlets. Remarkably, the air leak through the small vents effectively constitutes a high-resistance flow path and has an insignificant effect on the pressure required for closing the valves, as the same closing pressure of 7 psi was used both with vents and with blocked outlets for comparison. The individually controlled microvalves allow for rapid mixing (under 1.5 min) of yellow (inlet S1) and blue (inlet S2) dyes by sequentially opening and closing V3, V4, and V5, thus mixing the fluid in a counterclockwise flow pattern (Video 1, Supplementary Information). Laminar flow is demonstrated by applying equal pressure to the control valves and flowing the blue and yellow dyes at 3 psi (Figure 6f). For V1–V5, the control channel pressure was 3 psi, while for V6, the control channel was 7 psi to prevent leaking to the mixed-fluid outlet (M1). Valve 6 is closed during mixing and is opened to flow mixed green dye to the M1 by individual control valve actuation (Figure 6g). With the fabrication of a rotary mixer, we demonstrate our ability to 3D-print functional microfluidic components with active elements. The pump presently requires three pneumatic control lines, but in principle, a pump design based on three membranes of dissimilar sizes would only require a single control line.[7] Other active mixing schemes based on different configurations of pneumatic elements are also possible.[9]
Figure 6.
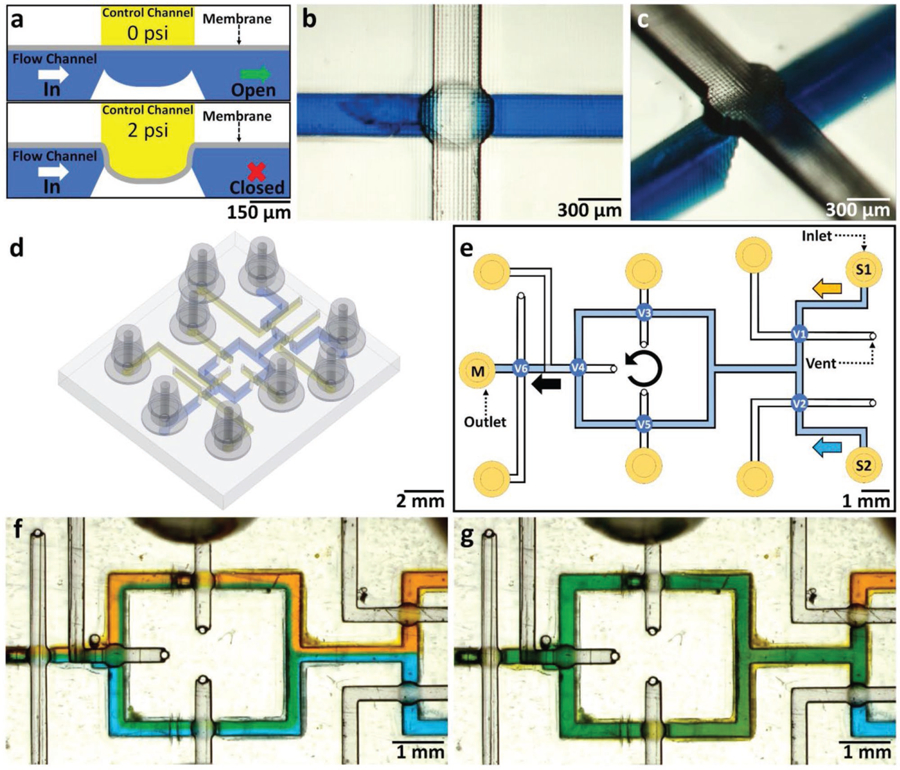
Active microfluidic devices—microvalves and rotary mixer. a) Cross-sectional schematic of a single 3D-printed 500 µm diameter circular “pinch” valve. The valve consists of two orthogonal channels (the flow channel and a control channel) separated by a thin (20 µm thick) membrane. The valve is closed by applying a pneumatic pressure of 2 psi to the membrane via the control channel, pressing the membrane on the valve seat, thus closing the flow channel below. b) CAD design of the circular pinch valve, showing the flow channel (blue) and control channel (yellow). c) Oblique-view micrograph of a single 3D-printed circular pinch valve at the intersection of a 300 µm wide flow channel (filled with blue dye) and a 300 µm wide control channel (transparent). d) Oblique view of CAD design of rotary mixer. e) Top-view CAD design of rotary mixer channel layout with six individually actuated valves (V1–V6). Yellow (S1) and blue (S2) dyes are flown with V6 closed at 7 psi. V3–V5 are sequentially opened and closed at 3 psi to mix fluids in a counterclockwise motion. V6 is opened to flow mixed fluid to the outlet (M1). f) Magnified top-view micrograph of the rotary mixer device, showing the laminar flow of the two dyes as they are initially flown with V6 closed at 7 psi. g) Magnified top-view micrograph of the rotary mixer device after 1.5 min of mixing, showing green dye flowing to the outlet (M1).
Video 1 (Supplementary Information):
The rotary mixer is controlled by six individually actuated microvalves (Figure 6d), labeled V1–V6 (Figure 6e). The individually controlled microvalves allow for rapid mixing (under 1.5 min) of yellow and blue dyes by sequentially opening and closing V3, V4, and V5, mixing the fluid in a counterclockwise flow pattern. The control channel pressure was 3 psi for V1–V5, while for V6 it was 7 psi, to prevent leaking to mixed-fluid outlet (M1). Valve 6 is closed during mixing and is opened to flow mixed green dye to the M1 by individual control valve actuation (Figure 6g).
2.6. Absorption of Small Hydrophobic Molecules
The absorption of small non-polar molecules (drugs, hormones) into the bulk polymer of a biomicrofluidic device used for cell-based assays or high-throughput drug screening studies can limit the utility and biological relevance of these platforms.[51] The loss of these molecules into the bulk polymer not only reduces their effective concentration in the delivery channels, but can also contaminate a fluidically separated compartment.[52] Several groups have demonstrated the sequestration of small hydrophobic molecules (e.g., Nile Red, Rhodamine G, estrogen, diazepam) into the walls of microfluidic devices made with PDMS.[51–54] PEG-DA-258 prints, however, contain hydrophilic ethylene oxide chains that should help reduce the absorption of hydrophobic molecules.[55] To confirm that our 3D-printed formulation of PEG-DA-258 absorbs small hydrophobic molecules less than PDMS does, we filled wells made of conventionally molded PDMS (Sylgard-184) or 3D-printed PEG-DA-258 with a hydrophobic fluorescent dye (Nile Red, 1 mm) for 90 min, rinsed the wells, and then measured the retained fluorescence (Figure 7). The total area under the PEG-DA-258 peak was 8.25 times smaller than that of PDMS. In order to determine the diffusivity of Nile Red (D) into the polymer, we fitted the fluorescent intensity data (R2 > 0.99) to the 1D analytical solution of Fick’s second law of diffusion,[52] assuming semi-infinite boundary conditions (concentration Cx → 0 at large distances from the constant source boundary, where Cx = C0):
| (3) |
Using Equation (3), we obtain the diffusivity of Nile Red in PEG-DA-258 (6.54 × 10−9 cm2 s−1) to be ≈640 times lower than in PDMS (4.2 × 10−6 cm2 s−1).
Figure 7.
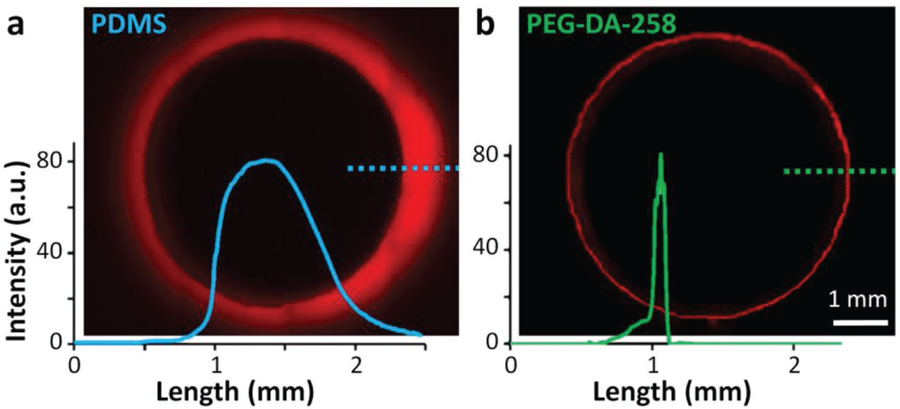
Absorption of small molecules. Residual fluorescence of Nile Red (1 mm) in the walls of a) thermally cured 5 mm diameter Sylgard-184 PDMS well and b) 3D-printed 5 mm diameter PEG-DA-258 well, after the wells were incubated with the dye for 90 min. The associated graphs show the fluorescence intensity profile along the dashed lines.
2.7. Microfluidic Labeling of Cells
We next set out to demonstrate that 3D-printed PEG-DA-258 microfluidic devices can be used for tissue culture–based biomedical applications where flow is present (e.g., organs-on-chip, cell-based biosensors, drug testing, etc.; Figure 8). We designed and SL-printed a 3-inlet microfluidic device, with a 1 mm (W) × 1 mm (H) × 2 cm (L) cell culture channel (Figure 8a,b). We have previously shown that by extracting the toxic leachates and uncured monomers, we were able to render 3D-printed PEG-DA-258 Petri dishes compatible for CHO-K1 and primary mammalian hippocampal neuron culture.[30] However, since the leaching of toxic monomers and photoinitiators are the primary cause of cytotoxicity, confined microchannels with a high surface-to-volume ratio can accumulate a higher concentration of the leachates and present a more challenging test of viability for cells. Therefore, as described in the Section 4, enclosed microchannels required extended flow-based post-processing before cell culture. CHO-K1 cells cultured inside the post-processed 3D-printed microfluidic channel had viability and proliferation rates similar to 3D-printed or commercial Petri dishes. The optimal plating density of the cells for the channels to be uniformly filled to confluence in ≈48 h was found to be ≈20% (Figure 8c). Higher plating density led to compromised cell viability and non-uniform coverage of the channel surface with cells. Note that the channel walls were directly built on glass slides so that the cells are attached on a glass surface, which enables excellent phase-contrast microscopy (Figure 8d) and fluorescence microscopy (Figure 8e). The clarity of the pictures is remarkable given that the roof of the channel contains visibly pixelated structures; however, the focal plane of the pixels is ≈1 mm above that of the cells in these images so they are too out-of-focus to be discernible.
Figure 8.
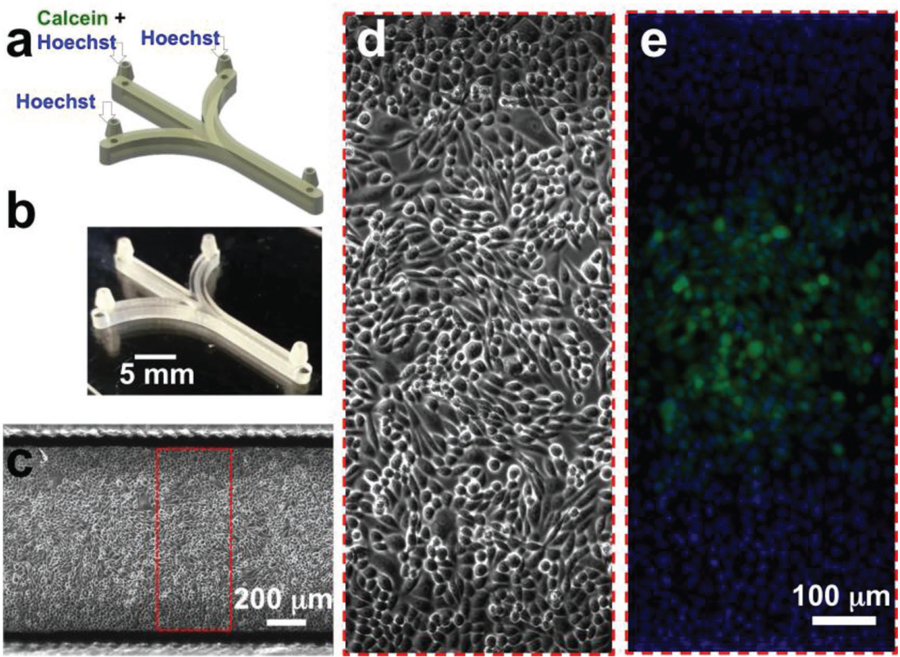
Cell labeling in 3D-printed microchannels. a) CAD design of the microfluidic device for a simple cell labeling experiment. The device integrates three inlet and one outlet barb connectors. For focal labeling of the cells, Calcein Green AM (5 µm) and Hoechst 33342 (1 µm) were flowed through the middle inlet, while only Hoechst 33342 (1 µm) was introduced through the side inlets. b) Oblique-view photograph of the 3D-printed microfluidic device. c) Stitched phase-contrast micrograph of a confluent layer of Chinese hamster ovary (CHO-K1) cells growing in the device. d) Magnified phase-contrast micrograph of CHO-K1 cells in the channel. e) Magnified fluorescence micrograph of CHO-K1 cells in the channel focally labeled with Calcein Green AM. All the cells in the channel are also labeled with the nuclear dye, Hoechst 33342.
To show that 3D-printed PEG-DA microfluidic channels could be used for exposing selected regions of a cell culture monolayer to a biomolecule, we used a standard acetoxymethyl (AM) ester of a cell-permeable dye, Calcein Green AM, to label live cells in the central region of the 1 mm wide channel (Figure 8e). Hoechst 33342 was flowed through all the channels to label the nuclei of cells. While we have illustrated here the applicability of first-generation 3D-printed PEG-DA microfluidic channels to demonstrate the viability of mammalian cell cultures in microchannels, more sophisticated designs incorporating small roof openings or porous printed layers[56] to enhance gas permeability, and/or 3D-printable PEG-DA micropumps[31,46] to automate perfusion protocols, could in principle be implemented for more sensitive cells such as primary neurons and stem cells.
3. Conclusion
We have developed a new SL-printable PEG-DA-based resin based on the addition of ITX, a UV photosensitizer, with which we have demonstrated the fabrication of transparent biomicrofluidic devices down to the single-pixel resolution afforded by present commercial desktop SL printers. The observation of sub-pixel features provides evidence that the resin should allow for printing at higher resolutions as rapidly improving printers become available, and further modifications to the resin chemistry could enhance resolution even further. We also fabricated both passive and active functional microfluidic devices that leverage the 3D-design capabilities provided by the 3D-printing technology. We can potentially organize arrays of microvalves into multiplexers[57] to generate combinatorial mixtures of drugs[58] or high-throughput immunoassays.[59] We envision that this cell-culture-compatible and low small-molecule absorbing resin will enable users to SL-print microfluidic systems with complex functionalities and 3D architectures such as organ-on-a-chip,[60] tumor-on-a-chip,[61] patch clamp chips,[62] biosensors,[63] droplet/digital microfluidics,[64–67] and diagnostics.[59]
4. Experimental Section
Resin Components:
SL resin in the present study consists of the monomer, PEG-DA-258 (Sigma Aldrich), the photoinitiator, phenylbis(2,4,6-trimethylbenzoyl)phosphine oxide, also known as Irgacure-819 (IRG) (Esstech Inc.), and a photosensitizer, ITX (Esstech Inc.). Photoinitiators and photosensitizers were dissolved in PEG-DA-258 at varying concentrations (0.4–0.8% w/w and 0.1–0.8% w/w, respectively). Selection of an appropriate photoinitiator and photosensitizer depends on the solubility in the monomer PEG-DA-258, absorbance spectrum, and the color imparted on the prints. A PEG-DA-258 resin with 0.4% IRG and 0.4% ITX was used for building all the devices unless specified otherwise, as it provided the best balance between resolution and transparency of prints. After briefly vortexing the components of the resin, the resin was placed in an oven at 70 °C for 10 min to facilitate dissolving the components. All steps were done in a yellow-light, UV-filtered dark room to avoid photopolymerization with ambient light.
Absorbance Measurements:
Equation (1) was used to calculate the molar absorptivity (ε) from the absorbance (A), the light’s path length through the sample (l), and the concentration of the sample (c). The absorbance spectrum of the resins was measured using a NanoDrop 2000c Spectrophotometer (Thermo Scientific).
3D-Printers:
The minimum feature size (resolution) that was achievable by traditional SL was dependent on the pixel size of the light source, on the absorption spectrum of the photoresin, and on the exposure time required to achieve photopolymerization (which causes diffusive broadening of all the reactants).[40] Two different 3D-printers were used for this work. For some prints, an Ilios HD 3D-printer with an HT stepper motor having a nominal Z-layer resolution of 12.5 µm and controlled by an Arduino board was used. The printer was fitted with a 385 nm Wintech Digital PRO4500 UV-LED DLP projector with a printing area of 65.6 mm × 41.0 mm and a resolution of 51.25 µm × 51.25 µm (1280 pixels × 800 pixels). An Asiga Pico2 HD DLP SL-printer that has a nominal Z-layer resolution of 10 µm was also used. The printer uses a 385 nm projector with a printing area of 52 mm × 29 mm and a nominal pixel resolution of 27 µm × 27 µm (1925 pixels × 1074 pixels). The XY pixel resolution was determined by the projection size of the DMD micromirrors in each respective printer. In both of these printers, prints were built in a “bat configuration”, where the final print was printed upside down as the light source was below the vat,[68] which contained the resin. Thus, object height was not limited by vat depth, as the print was made layer by layer and was begun from the bottom-most layer.
3D-Printing Process:
All objects were designed with Autodesk Inventor and exported in STL format. For the Ilios HD printer, Creative Workshop software was used to slice the objects and convert them into an image sequence, dependent on layer thickness. The whole 3D printing process was controlled by custom-made software written in MATLAB (Mathworks) to control the Arduino board and the DLP projectors. The custom software allows us to precisely and independently control parameters such as LED intensity, exposure time, speed of approach and separation, intermediate pausing, and layer thickness for each individual slice. The printing process has been described in more detail in earlier work.[30] The 3D object was built layer-by-layer by using selective light exposure to photopolymerize a precursor resin collected in a vat, the housing for the photocurable resin. To produce transparent prints, glass slides (75 mm × 50 mm × 1 mm) cleaned sequentially with acetone, isopropyl alcohol, and deionized water were used as substrates for printing. To ensure the attachment of the PEG-DA-258 prints to the glass substrate, the cleaned glass slides were derivatized with 3-(trimethoxysilyl)propyl methacrylate (TMSPMA) (Sigma-Aldrich).[30] To print a device, the photocurable resin was poured into the vat. With the vat covered, blocking ambient light, a silanized glass slide was “glued” to the build plate by coating one side with a thin film of PEG-DA-258 resin and briefly exposing with UV light using a broadband UV lamp (B-100 A, UVP). After the print was completed, the glass slide can be detached from the build plate using a thin razor blade.
Post-Processing:
In SL, a microchannel was built by photopolymerizing first the layers that build the channel walls and then the layers that constitute the roof, leaving trapped uncured resin in the channels. This uncured resin was removed before the print was brought to ambient light; exposing the print to ambient light at this point would otherwise cause photopolymerization of the uncured resin inside the channel. Immediately after printing, a pressure source (e.g., a vacuum line) was used to remove the uncured resin, and a series of washes with water to remove unreacted monomers.
Microfluidic Setup and Operation:
For microfluidic operation, a Fusion 720 syringe injection pump (Chemyx Inc.) was used. All the microfluidic devices with integrated barb-type 1/16 in. fluidic connectors were printed, which can be directly connected to silicone tubing (1/16 in. ID; Cole-Parmer). Plastic syringes (5 or 10 mL) (BD Biosciences) filled with fluids were connected to the 3D-printed devices and run using the syringe-pump at the desired flow rates.
Microscopy:
Bright-field images of the 3D-printed parts and devices were captured using a Nikon SMZ1500 stereoscope fitted with a Canon EOS Rebel 5D Mark II DSLR camera. Previously, a similar formulation of PEG-DA-258 was shown to have very low levels of autofluorescence, allowing for fluorescence microscopy observations.[30] Phase-contrast and fluorescence images of cells, as well as high-resolution bright-field images of 3D-printed parts, were taken with a Nikon TE3000 epifluorescence microscope. SEM images were captured by mounting samples on a multi-stub holder that was then placed in an FEI XL830 Dual Beam FIB/SEM. The field-emission electron source allows observation of 3D prints down to 4 nm resolution.
Cell Culture in Microfluidic Channels:
Prior to cell culture, the 3D-printed microfluidic channels required post-processing to remove toxic leachates. First, DI water was passed through the channels at ≈15 mL h−1 for 36 h, while keeping the entire device immersed under water. The devices were then exposed to UV for ≈12 h under a UV gel box (High-Performance Trans-illuminator TFL-40, UVP), which also sterilized the devices. The channels were filled with cell culture media (dulbecco’s modified eagle medium (DMEM) + 10% fetal bovine serum (FBS)) and kept in the humidified incubator at 37 °C overnight (≈12 h).
Chinese hamster ovary (CHO-K1) cell lines were cultured in DMEM media (Invitrogen) supplemented with 10% FBS (Hyclone) and grown in a 5% CO2 atmosphere at 37 °C and 100% humidity. The density of cells injected into the microfluidic channel was calculated to cover ≈20% of the channel surface area. Media was loaded into Luer-lock connectors attached to the fluid inlets. The entire device was kept in a covered and humidified Petri dish to prevent evaporation. The media was replaced every 24 h. After ≈48 h, the cells reached confluence inside the channels.
Cell Labeling:
Prior to labeling with cell-permeable fluorescent dyes, the channels were washed once with phosphate-buffered saline to remove any serum and then filled with pre-warmed serum-free Fluorobrite DMEM (Invitrogen). A 5 mL syringe (BD Falcon) filled with 5 µm Calcein Green AM (Invitrogen) and 1 µm Hoechst 33342 (Invitrogen) in serum-free Fluorobrite DMEM was connected to the central channel inlet. Syringes filled with 1 µm Hoechst 33342 (Invitrogen) in serum-free Fluorobrite DMEM were connected to the flanking channel inlets. The syringes were run for 1 h at 1 mL h−1 using a syringe pump (Fusion 720; Chemyx). The microfluidic device was placed in a microscope stage incubator (Pathology Devices Inc.) while the cells were being labeled with the dyes.
Supplementary Material
Acknowledgements
A.P.K. and N.B. contributed equally to this work. A.F. oversaw and directed the research. A.P.K. and N.B. developed the resin, designed, and 3D-printed devices for fluid routing, mixing, and cell culture. Y.-S.L. designed and 3D-printed the rotary mixer. Y.T.K. fabricated the single-pixel channels. K.C. obtained the SEM images. A.P.K., N.B., and A.F. wrote the manuscript, with contributions from all authors. All authors have given approval to the final version of the manuscript. This research was partially supported by the National Institutes of Health (Grant number: 5R01NS064387) and the BioNano Health-Guard Research Center funded by the Ministry of Science, ICT (MSIT) of Korea as Global Frontier Project (Grant number: H-GUARD_2014M3A6B2060302).
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Yuan-Sheng Lee, Department of Mechanical Engineering University of Washington, Seattle, WA 98195, USA.
Kurt Castro, Department of Bioengineering, University of Washington Seattle, WA 98195, USA.
Yong Tae Kim, Department of Bioengineering, University of Washington Seattle, WA 98195, USA; National NanoFab Center (NNFC), 291 Daehak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Albert Folch, Department of Bioengineering, University of Washington Seattle, WA 98195, USA.
References
- [1].Whitesides GM, Xia Y, Angew. Chem., Int. Ed 1998, 28, 153. [Google Scholar]
- [2].Chen Y, Zhang L, Chen G, Electrophoresis 2008, 29, 1801. [DOI] [PubMed] [Google Scholar]
- [3].Jena RK, Yue CY, Lam YC, Microsyst. Technol, 2012, 18, 159–166. [Google Scholar]
- [4].Young EWK, Berthier E, Guckenberger DJ, Sackmann E, Lamers C, Meyvantsson I, Huttenlocher A, Beebe DJ, Anal. Chem 2011, 83, 1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Folch A, Introduction to BioMEMS, CRC Press, Boca Raton, FL: 2012. [Google Scholar]
- [6].Grover WH, Skelley AM, Liu CN, Lagally ET, Mathies RA, Sens. Actuators, B 2003, 89, 315. [Google Scholar]
- [7].Lai H, Folch A, Lab Chip 2011, 11, 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liu RH, Stremler MA, Sharp KV, Olsen MG, Santiago JG, Adrian RJ, Aref H, Beebe DJ, J. Microelectromech. Syst 2000, 9, 190. [Google Scholar]
- [9].Hsu CH, Folch A, Appl. Phys. Lett 2006, 89, 114103. [Google Scholar]
- [10].Yang SY, Lin JL, Bin Lee G, J. Micromech. Microeng 2009, 19, 035020. [Google Scholar]
- [11].Yu L, Ng SR, Xu Y, Dong H, Wang YJ, Li CM, Lab Chip 2013, 13, 3163. [DOI] [PubMed] [Google Scholar]
- [12].Kintses B, van Vliet LD, Devenish SRA, Hollfelder F, Curr. Opin. Chem. Biol 2010, 14, 548. [DOI] [PubMed] [Google Scholar]
- [13].Rohde CB, Zeng F, Gonzalez-Rubio R, Angel M, Yanik MF, Proc. Natl. Acad. Sci. U. S. A 2007, 104, 13891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhou Z, Xie S, Chen D, Fundamentals of Digital Manufacturing Science, Springer-Verlag London, London, UK: 2012. [Google Scholar]
- [15].Scott A, Au AK, Vinckenbosch E, Folch A, Lab Chip 2013, 13, 2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cooksey GA, Plant AL, Atencia J, Lab Chip 2009, 9, 1298. [DOI] [PubMed] [Google Scholar]
- [17].Hull C, Mod. Cast 1988, 78, 38. [Google Scholar]
- [18].Zhang X, Jiang XN, Sun C, Sens. Actuators, A 1999, 77, 149. [Google Scholar]
- [19].Bertsch A, Bernhard P, Vogt C, Renaud P, Rapid Prototyping J 2000, 6, 259. [Google Scholar]
- [20].Kaehr B, Ertaş N, Nielson R, Allen R, Hill RT, Plenert M, Shear JB, Anal. Chem 2006, 78, 3198. [DOI] [PubMed] [Google Scholar]
- [21].Sun C, Fang N, Wu DM, Zhang X, Sens. Actuators, A 2005, 121, 113. [Google Scholar]
- [22].Lu Y, Mapili G, Suhali G, Chen S, Roy K, J. Biomed. Mater. Res., Part A 2006, 77A, 396. [DOI] [PubMed] [Google Scholar]
- [23].Nielson R, Kaehr B, Shear JB, Small 2009, 5, 120. [DOI] [PubMed] [Google Scholar]
- [24].Gittard SD, Ovsianikov A, Chichkov BN, Doraiswamy A, Narayan RJ, Expert Opin Drug Deliv 2010, 7, 513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Coenjarts CA, Ober CK, Chem. Mater 2004, 16, 5556. [Google Scholar]
- [26].Bhattacharjee N, Urrios A, Kang S, Folch A, Lab Chip 2016, 16, 1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Waheed S, Cabot JM, Macdonald NP, Lewis T, Guijt RM, Paull B, Breadmore MC, Lab Chip 2016, 16, 1993. [DOI] [PubMed] [Google Scholar]
- [28].Hod Lipson MK, Fabricated: The New World of 3D Printing, 1st ed., John Wiley & Sons, Inc., Hoboken, NJ: 2013, p. 1. [Google Scholar]
- [29].Zhu J, Biomaterials 2010, 31, 4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Urrios A, Parra-Cabrera C, Bhattacharjee N, Gonzalez-Suarez AM, Rigat-Brugarolas LG, Nallapatti U, Samitier J, DeForest CA, Posas F, Garcia-Cordero JL, Folch A, Lab Chip 2016, 16, 2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gong H, Woolley AT, Nordin GP, Lab Chip 2016, 16, 2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mandon CA, Blum LJ, Marquette CA, Anal. Chem 2016, 88, 10767. [DOI] [PubMed] [Google Scholar]
- [33].Chan V, Zorlutuna P, Jeong JH, Kong H, Bashir R, Lab Chip 2010, 10, 2062. [DOI] [PubMed] [Google Scholar]
- [34].Gong H, Beauchamp M, Perry S, Woolley AT, Nordin GP, RSC Adv 2015, 5, 106621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lee MP, Cooper GJT, Hinkley T, Gibson GM, Padgett MJ, Cronin L, Sci. Rep 2015, 5, 9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gong H, Bickham BP, Woolley AT, Nordin GP, Lab Chip 2017, 17, 2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Fouassier JP, Allonas X, Lalevee J, Dietlin C, Photochemistry and Photophysics of Polymer Materials, John Wiley & Sons, Inc., Hoboken, NJ: 2010, p. 351. [Google Scholar]
- [38].Gruber HF, Prog. Polym. Sci 1992, 17, 953. [Google Scholar]
- [39].Allen NS, Catalina F, Moghaddam B, Given PN, Giun WA, Eur. Polym. J 1986, 22, 691. [Google Scholar]
- [40].Fang N, Sun C, Zhang X, Appl. Phys. A 2004, 79, 1839. [Google Scholar]
- [41].Goodner MD, Bowman CN, Chem. Eng. Sci 2002, 57, 887. [Google Scholar]
- [42].Squires TM, Quake SR, Rev. Mod. Phys 2005, 77, 977. [Google Scholar]
- [43].Kim DS, Lee SH, Kwon TH, Ahn CH, Lab Chip 2005, 5, 739. [DOI] [PubMed] [Google Scholar]
- [44].Neils C, Tyree Z, Finlayson B, Folch A, Lab Chip 2004, 4, 342. [DOI] [PubMed] [Google Scholar]
- [45].Au AK, Bhattacharjee N, Horowitz LF, Chang TC, Folch A, Lab Chip 2015, 15, 1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lee Y-S, Bhattacharjee N, Folch A, Lab Chip 2018, 8, 1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Rogers CI, Oxborrow JB, Anderson RR, Tsai LF, Nordin GP, Woolley AT, Sens. Actuators, B 2014, 191, 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Armani D, Liu C, Aluru N, in IEEE Int. MEMS 99 Conf. Twelfth IEEE Int. Conf. Micro Electro Mechanical Systems (Cat. No.99CH36291), Technical Digest, 1999, pp. 222–227. [Google Scholar]
- [49].Chou HP, Unger MA, Quake SR, Biomed. Microdevices 2001, 3, 323. [Google Scholar]
- [50].Hong JW, Studer V, Hang G, Anderson WF, Quake SR, Nat. Biotechnol 2004, 22, 435. [DOI] [PubMed] [Google Scholar]
- [51].Regehr KJ, Domenech M, Koepsel JT, Carver KC, Ellison-Zelski SJ, Murphy WL, Schuler LA, Alarid ET, Beebe DJ, Lab Chip 2009, 9, 2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Shirure VS, George SC, Lab Chip 2017, 17, 681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Toepke MW, Beebe DJ, Lab Chip 2006, 6, 1484. [DOI] [PubMed] [Google Scholar]
- [54].Wang JD, Douville NJ, Takayama S, Elsayed M, Ann. Biomed.. Eng 2012, 40, 1862. [DOI] [PubMed] [Google Scholar]
- [55].Rogers CI, Pagaduan JV, Nordin GP, Woolley AT, Anal. Chem 2011, 83, 6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kim YT, Castro K, Bhattacharjee N, Folch A, Micromachines 2018, 9, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Thorsen T, Maerkl SJ, Quake SR, Science 2002, 298, 580. [DOI] [PubMed] [Google Scholar]
- [58].Cooksey GA, Sip CG, Folch A, Lab Chip 2009, 9, 417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Garcia-Cordero JL, Maerkl SJ, Lab Chip 2014, 14, 2642. [DOI] [PubMed] [Google Scholar]
- [60].Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE, Science 2010, 328, 1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Chang TC, Mikheev AM, Huynh W, Monnat RJ, Rostomily RC, Folch A, Lab Chip 2014, 14, 4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Scott A, Weir K, Easton C, Huynh W, Moody WJ, Folch A, Lab Chip 2013, 13, 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lafleur JP, Jönsson A, Senkbeil S, Kutter JP, Biosens. Bioelectron 2016, 76, 213. [DOI] [PubMed] [Google Scholar]
- [64].Ding Y, Choo J, deMello AJ, Microfluid. Nanofluid 2017, 21, 10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Dressler OJ, Casadevall i Solvas X, deMello AJ, Annu. Rev. Anal. Chem 2017, 10, 1. [DOI] [PubMed] [Google Scholar]
- [66].Fair RB, Khlystov A, Tailor TD, Ivanov V, Evans RD, Srinivasan V, Pamula VK, Pollack MG, Griffin PB, Zhou J, IEEE Des. Test Comput 2007, 24, 10. [Google Scholar]
- [67].Choi K, Ng AHC, Fobel R, Wheeler AR, Annu. Rev. Anal. Chem 2012, 5, 413. [DOI] [PubMed] [Google Scholar]
- [68].Waldbaur A, Rapp H, Länge K, Rapp BE, Anal. Methods 2011, 3, 2681. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


