Abstract
Hepatocellular carcinoma (HCC) is one of the most lethal malignant diseases worldwide. Despite advances in the diagnosis and treatment of HCC, its overall prognosis remains poor. Recent studies have shown that long noncoding RNAs (lncRNAs) play crucial roles in various pathophysiological processes, including liver cancer. In the current study, we report that lncRNA SLC2A1‐AS1 is frequently downregulated in HCC samples, as shown by quantitative real‐time polymerase chain reaction analysis. SLC2A1‐AS1 deletion is significantly associated with recurrence‐free survival in HCC. By performing glucose uptake, lactate production and ATP detection assays, we found that SLC2A1‐AS1‐mediated glucose transporter 1 (GLUT1) downregulation significantly suppressed glycolysis of HCC. In vitro Cell Counting Kit‐8, colony formation, transwell assays as well as in vivo tumorigenesis and metastasis assays showed that SLC2A1‐AS1 overexpression significantly suppressed proliferation and metastasis in HCC through the transcriptional inhibition of GLUT1. Results from fluorescence in situ hybridization, ChIP and luciferase reporter assays demonstrated that SLC2A1‐AS1 exerts its regulatory role on GLUT1 by competitively binding to transketolase and signal transducer and activator of transcription 3 (STAT3) and inhibits the transactivation of Forkhead box M1 (FOXM1) via STAT3, thus resulting in inactivation of the FOXM1/GLUT1 axis in HCC cells. Our findings will be helpful for understanding the function and mechanism of lncRNA in HCC. These data also highlight the crucial role of SLC2A1‐AS1 in HCC aerobic glycolysis and progression and pave the way for further research regarding the potential of SLC2A1‐AS1 as a valuable predictive biomarker for HCC recurrence.
Keywords: FOXM1, GLUT1, glycolysis, hepatocellular carcinoma, long noncoding RNA, STAT3
Long noncoding RNAs play crucial roles in various pathophysiological processes including liver cancer. In this study, we report that antisense lncRNA SLC2A1‐AS1 is downregulated in HCC. Overexpression of SLC2A1‐AS1 inhibits aerobic glycolysis and progression of HCC. Mechanistically, SLC2A1‐AS1 interacts with transketolase and signal transducer and activator of transcription 3 and inhibits Forkhead box M1/glucose transporter 1 axis activation in HCC cells.
Abbreviations
- FISH
fluorescence in situ hybridization
- FOXM1
Forkhead box M1
- GLUT1
glucose transporter 1
- HCC
hepatocellular carcinoma
- HE
haematoxylin and eosin
- IHC
immunohistochemistry
- lncRNAs
long noncoding RNAs
- qRT‐PCR
quantitative real‐time polymerase chain reaction
- STAT3
transketolase and signal transducer and activator of transcription 3
1. Introduction
Hepatocellular carcinoma (HCC) is the second leading cause of cancer‐related death worldwide (Armengol et al., 2018). Despite advances in diagnosis and treatments, the overall prognosis for HCC remains extremely low due to the high rate of recurrence and distant metastasis (Xia et al., 2013). Therefore, clarifying the underlying molecular mechanism of HCC progression is urgently needed to find effective therapeutic targets and recurrence predictors.
Metabolic reprogramming is a vital property of cancer cells. Unlike normal cells, cancer cells prefer to use the glycolysis pathway for metabolizing glucose upon enhanced glucose uptake and lactate production. This phenomenon, referred to as the Warburg effect, confers cancer cells with great advantages regarding tumour growth, apoptosis resistance, metastasis and immune escape (Gatenby et al., 2006; Kroemer and Pouyssegur, 2008). These extensive connections between cancer metabolism and aggressive cancer features suggest that targeting metabolic pathways may be a promising effective method for treating cancer patients. Recent research demonstrated that cancer metabolism reprogramming can be regulated by both coding and noncoding genes via targeting glycolysis‐related transporters, enzymes and signalling pathways (Cairns et al., 2011; Lin et al., 2018; Yang et al., 2014). A previous study and our recent publication demonstrated that a key glycolytic transporter, glucose transporter 1 (GLUT1), is specifically overexpressed in HCC and promotes HCC cell glycolysis and progression (Amann et al., 2009; Shang et al., 2017). We further revealed that GLUT1 is transactivated by the transcription factor Forkhead box M1 (FOXM1) in different cancer types (Shang et al., 2017; Wang et al., 2016). However, knowledge of how noncoding genes regulate GLUT1 expression is lacking.
Recently, long noncoding RNAs (lncRNAs), which are defined as a class of RNAs composed of more than 200 nucleotides and with no protein‐coding ability, have been identified to participate in numerous physiological and pathological processes (Sahu et al., 2015). lncRNAs have been reported to be dysregulated in various types of cancers and play multiple roles in cancer cell proliferation, metastasis and metabolism. Increasing evidence indicates that a subtype of lncRNAs—antisense lncRNAs—play critical roles in cancer progression via regulating the expression of genes on the opposite strand with protein‐coding ability (Li et al., 2016; Sun et al., 2016). However, the role and specific regulatory mechanism of critical oncogene‐related antisense lncRNAs in HCC are largely unknown.
In this study, we reported the expression and function of a previously identified but unannotated antisense lncRNA, SLC2A1‐AS1, in HCC. We found that SLC2A1‐AS1 was frequently downregulated in HCC tissues. SLC2A1‐AS1 inhibited HCC cell aerobic glycolysis and progression through inactivating the key glycolytic transporter GLUT1, which is coded by the opposite strand coding gene SLC2A1. In detail, competitive binding of SLC2A1‐AS1 to the transcriptional factor STAT3 inactivates the FOXM1/GLUT1 axis in HCC cells. To the best of our knowledge, our study is the first to investigate the role and mechanism of lncRNA SLC2A1‐AS1 in HCC.
2. Materials and methods
2.1. Cell lines and culture conditions
The human nontumour liver cell line HL‐02 and human HCC cell lines MHCC97‐H, Huh7, HepG2 and Hep3B (obtained from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Grand Island, NY, USA) supplemented with 10% FBS and 100 U·mL−1 penicillin/streptomycin (HyClone, Logan, UT, USA) at 37° C in a humidified 5% CO2 atmosphere.
2.2. Clinical samples
Thirty‐three paired HCC samples and surrounding nontumour tissues were obtained during surgical resection at Xijing Hospital, Fourth Military Medical University, from 2011 to 2015. None of the patients received radiotherapy or chemotherapy before resection. All samples were immediately frozen in liquid nitrogen after resection and further analysed by quantitative real‐time polymerase chain reaction (qRT‐PCR). The study was approved by the Ethics Committee of Xijing Hospital, informed written consent to participate in this study was obtained from each patient, and the study methodologies conformed to the standards set by the Declaration of Helsinki.
2.3. RNA extraction and qRT‐PCR
Total RNA was extracted from cells or tissues using TRIzol reagent (Invitrogen Inc., San Diego, CA, USA) and converted to cDNA with a PrimeScript RT Reagent Kit (TaKaRa, Tokyo, Japan). qRT‐PCR was performed with SYBR Green Premix Ex Taq (TaKaRa) and detected using an iQ‐5 Real‐Time PCR Detection System (Bio‐Rad, Hercules, CA, USA). All results were normalized to β‐actin. The primers for qRT‐PCR are listed in Table S1.
2.4. Western blot analysis
Whole cells and tissues were lysed in RIPA Lysis Buffer (Beyotime, Shanghai, China) with protease inhibitors. The extracted proteins were loaded onto SDS/PAGE gels for separation and then transferred to nitrocellulose membranes. The signal was visualized by a ChemiDoc™ XRS + System (Bio‐Rad). Primary antibody information is listed in Table S2.
2.5. Plasmids, siRNA and cell transfection
Full‐length SLC2A1‐AS1 was constructed and inserted in the PEX‐3 vector (Genecreate, Wuhan, China). siRNAs and negative control siRNAs were synthesized by GenePharm (Sunnyvale, CA, USA). All plasmids and siRNAs were transfected using Lipofectamine 2000 (Invitrogen Inc.).
2.6. Glucose uptake, lactate production and ATP detection assays
Glucose, lactate and ATP assay kits (Nanjing Jiancheng) were used to measure the glucose uptake, lactate production and intracellular ATP levels of HCC cells according to the manufacturer’s instructions. All the results were normalized to the protein quantification values.
2.7. Cell proliferation and colony formation assays
A Cell Counting Kit‐8 (CCK‐8) assay was used for cell proliferation analysis. In brief, five replicate cell samples were seeded into 96‐well plates at a density of 2.0 × 103 cells per well. At the indicated time points, 90 μL of culture medium containing 10% serum and 10 μL of CCK‐8 reagent were added to each well. Following incubation for 2 h at 37 °C, the absorbance was measured at 450 nm. For the colony formation assay, cells were seeded at a density of 300 cells/well in a 6‐well plate and cultured in 3 mL of DMEM supplemented with 10% FBS for 2 weeks. Then, the colonies were fixed in 95% EtOH and stained with a 4 g·L−1 crystal violet solution for counting.
2.8. Cell migration assays
Cell migration ability was analysed with cell culture chambers. Briefly, 3 × 104 transfected HCC cells were serum‐starved and seeded onto the membrane (24‐well insert) in the upper chamber with serum‐free medium. After incubation for 24 h, the cells were fixed with methanol for 5 min and stained with a 0.1% crystal violet solution for 30 min. The invaded and migrated cells were counted at 200× magnification from five random fields of each filter.
2.9. Animal studies
The protocols for the animal studies were approved by the Animal Experiment Administration Committee of the Fourth Military Medical University. Four‐week‐old male BALB/C nude mice were used for the animal studies. For tumourigenesis assays, transfected MHCC97‐H cells (1 × 106) were injected subcutaneously into the flanks of nude mice, with five mice per group. Tumours were harvested and weighed at 4 weeks after injection. For the metastasis assays, transfected MHCC97‐H‐luc cells (2 × 106) were injected through the tail vein of the mice. Thirty days later, the mice were anaesthetized and injected intraperitoneally with d‐luciferin (Caliper), and the bioluminescence was detected 15 min later. Then, the mice were euthanized, and the lungs were excised, fixed and paraffin‐embedded for haematoxylin and eosin (H&E) and immunohistochemistry (IHC) staining.
2.10. RNA fluorescence in situ hybridization (FISH)
Fluorescence‐labelled probes for SLC2A1‐AS1, 18S rRNA and U6 RNA were synthesized by Genecreate Company. The FISH assays were performed using a Ribo™ Fluorescent In Situ Hybridization Kit (RiboBio, Guangzhou, China) according to the manufacturer’s instructions, and images were acquired using an Olympus Fluoview laser scanning confocal microscope.
2.11. Luciferase reporter assay
Luciferase reporter assays were performed using a luciferase assay kit (Promega, Madison, WI, USA). The wild‐type and mutant GLUT1 promoters were cloned into a pGL3 basic vector and cotransfected with the Renilla luciferase reporter and SLC2A1‐AS1 overexpression plasmids. The relative firefly luciferase activity was measured by a Dual Luciferase Assay System (Promega) at 48 h after transfection.
2.12. Chromatin immunoprecipitation (ChIP) assay
ChIP assays were conducted in Huh7 and MHCC97‐H cells as the manufacturers instructed (Millipore Corp., Billerica, MA, USA). A STAT3 antibody (Cell Signal Technology, Danvers, MA, USA) was used to perform the immunoprecipitation experiment. PCR primer sequences for the DNA fragments targeting the FOXM1 and GLUT1 promoters are listed in Table S3.
2.13. RNA immunoprecipitation (RIP) assay
RNA immunoprecipitation assays were performed using an EZ‐Magna RIP™ RNA‐Binding Protein Immunoprecipitation Kit (Millipore Corp.). Briefly, Huh7 cells were lysed and incubated with a STAT3 antibody (Cell Signal Technology) or normal mouse IgG (Millipore Corp.). Then, qRT‐PCR assays were conducted using the RNA fractions.
2.14. Statistical analysis
Statistical analyses were conducted using graphpad prism 5 software (GraphPad Software Inc., La Jolla, CA, USA). Representative data are shown as the mean ± SD. Two independent groups were analysed by two‐tailed Student’s t‐tests. The correlation between SLC2A1‐AS1 and GLUT1 expression was analysed using Spearman's rank correlation. Survival curves were calculated using the Kaplan–Meier method and statistically compared using the log‐rank test. All technical repeats were performed at least three times.
3. Results
3.1. SLC2A1‐AS1 is downregulated in HCC and inversely correlated with GLUT1 expression and HCC recurrence
SLC2A1‐AS1 is located at chromosomal band 1p34.2 (https://www.ncbi.nlm.nih.gov/; Fig. S1A), which has limited protein‐coding potential (https://lncipedia.org/; Fig. S1B). To investigate the expression levels of SLC2A1‐AS1 in HCC, qRT‐PCR analysis was performed on a total of 33 HCC patient RNA samples that were extracted from both tumour samples and their matched adjacent nontumour counterparts. As shown in Fig. 1A,B, the expression of SLC2A1‐AS1 was significantly downregulated in 63.6% (21/33) of the HCC samples compared with that in the paired nontumour samples. In vitro, we also found that SLC2A1‐AS1 expression was lower in HCC cell lines than in normal liver cell line (Fig. 1C). Moreover, a Kaplan–Meier analysis indicated that patients with low SLC2A1‐AS1 expression exhibited higher recurrence rates (Fig. 1D).
Fig. 1.
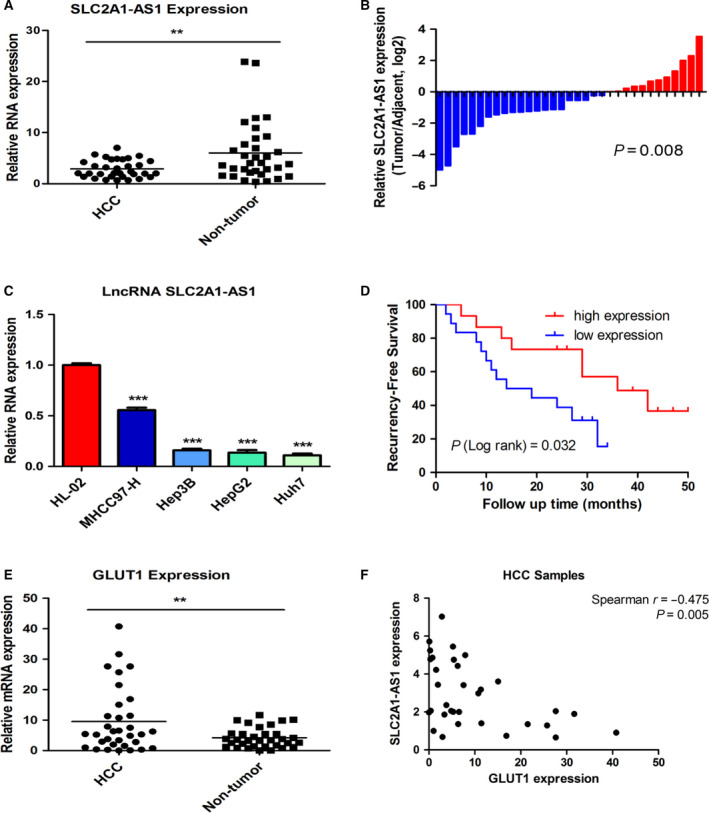
SLC2A1‐AS1 is downregulated in HCC and correlated with tumour recurrence in HCC patients. (A, B) qRT‐PCR analysis of SLC2A1‐AS1 expression levels in 33 HCC tissues and paired adjacent nontumour tissues. (C) Relative expression of SLC2A1‐AS1 in normal liver cell lines and HCC cell lines according to qRT‐PCR analysis. (D) Kaplan–Meier curve for recurrence‐free survival based on SLC2A1‐AS1 expression level. (E, F) qRT‐PCR analysis of GLUT1 mRNA expression in 33 HCC tissues and their paired adjacent nontumour tissues (E). The correlation of GLUT1 mRNA expression and SLC2A1‐AS1 expression in HCC samples (F). Expression of SLC2A1‐AS1 in HCC samples was analysed with Wilcoxon signed‐rank test; comparisons of the relative SLC2A1‐AS1 levels between the groups were analysed with ANOVA followed by post hoc correction; Kaplan–Meier survival analysis was analysed with log‐rank test; the correlation between GLUT1 and SLC2A1‐AS1 expression was analysed with Spearman's rank correlation. **P < 0.01, ***P < 0.001.
Recent investigations demonstrated that antisense transcripts may function as inhibitors or enhancers of corresponding gene expression. Therefore, we further assessed the mRNA expression of the coding gene SLC2A1 (GLUT1), which is on the opposite strand of SLC2A1‐AS1. As expected, the mRNA expression of the oncogene SLC2A1 was dramatically upregulated in HCC samples (Figs 1E and S2A) as well as in HCC cell lines (Fig. S2B). The Kaplan–Meier analysis indicated that patients with high GLUT1 mRNA expression exhibited higher recurrence rates (Fig. S2C). To evaluate whether SLC2A1‐AS1 expression and GLUT1 expression were independently predictive of recurrence‐free survival in HCC patients, we performed multivariate Cox analysis. As shown in Table S4, SLC2A1‐AS1 expression was an independent prognostic factor for recurrence‐free survival in patients with HCC.
Moreover, we observed a significant negative correlation between SLC2A1‐AS1 expression and GLUT1 mRNA expression (Fig. 1F, r = −0.475, P = 0.005), which implies a potential regulation relationship between SLC2A1‐AS1 and GLUT1.
Accordingly, these results suggested that SLC2A1‐AS1 loss in HCC was correlated with HCC recurrence and inversely associated with GLUT1 expression.
3.2. SLC2A1‐AS1 inhibits HCC glycolysis by negatively regulating GLUT1 expression
To investigate whether SLC2A1‐AS1 regulates GLUT1 expression, we induced SLC2A1‐AS1 expression in HCC cell lines. We transfected the HCC cells with either 5 μg (Fig. 2A) or 0.5 μg (Fig. S3A) SLC2A1‐AS1 overexpression plasmids for 24 h and then assessed the relative expression of SLC2A1‐AS1 and GLUT1 mRNA. We found that SLC2A1‐AS1 overexpression in varying degrees consistently reduced GLUT1 mRNA expression in four HCC cell lines (Figs 2A and S3A). Western blot results further confirmed the reduction in GLUT1 at the protein level after SLC2A1‐AS1 transfection (Figs 2B and S3B). As a crucial glycolysis‐related transport protein, GLUT1 has been confirmed to be upregulated and participate in the activation of HCC aerobic glycolysis. Therefore, we investigated whether SLC2A1‐AS1 could regulate HCC glycolysis by inhibiting GLUT1 expression. We transfected Huh7 and MHCC97‐H cells with SLC2A1‐AS1 and GLUT1 plasmids alone or in combination and then detected GLUT1 protein expression (Fig. 2C). Moreover, we evaluated glycolytic activity by measuring glucose uptake, lactate production and intracellular ATP content. Notably, SLC2A1‐AS1 overexpression prominently suppressed glucose uptake and lactate production and decreased intracellular ATP content in the two HCC cell lines (Fig. 2D). Intriguingly, GLUT1 upregulation reversed the effect of SLC2A1‐AS1 overexpression in HCC glycolysis (Fig. 2D). These findings confirmed that SLC2A1‐AS1 inhibited HCC glycolysis by negatively regulating GLUT1 expression.
Fig. 2.
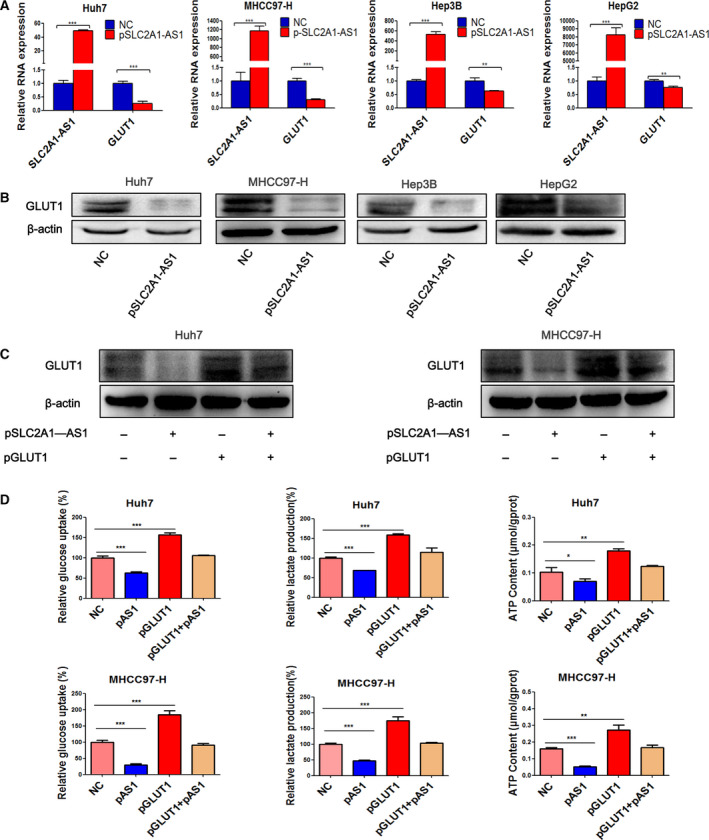
SLC2A1‐AS1 regulates GLUT1 expression and HCC cell aerobic glycolysis. (A, B) Overexpression of SLC2A1‐AS1 decreased the expression of GLUT1 at both the mRNA (A) and protein (B) levels in HCC cell lines. Data are expressed as mean ± SD of three independent experiments conducted in triplicate; they were analysed with Student’s t‐test. (C, D) Overexpression of SLC2A1‐AS1 inhibits HCC cell glucose uptake, lactate production and intracellular ATP content, while overexpression of GLUT1 reversed these effects. Data are shown as means ± SD, and ANOVA followed by post hoc correction is used to calculate significance. pAS1, pSLC2A1‐AS1. *P < 0.05, **P < 0.01, ***P < 0.001.
3.3. SLC2A1‐AS1 suppresses HCC cell proliferation and metastasis in vitro and in vivo
Metabolic reprogramming in cancer cells has been confirmed to contribute to tumour progression through a variety of mechanisms. Hence, we further explored whether SLC2A1‐AS1 affects tumour progression accompanying metabolic changes. To determine the role of SLC2A1‐AS1 in HCC cell proliferation in vitro, CCK‐8 and clone formation assays were performed in HCC cell lines after transfection with vector control plasmids or SLC2A1‐AS1 overexpression plasmids. Meanwhile, the GLUT1 overexpression plasmid was also transfected in the same cells as positive control. As shown in Fig. 3A‐D, the proliferation ability was clearly reduced in MHCC97‐H, Huh7, Hep3B and HepG2 cells when SLC2A1‐AS1 was overexpressed. In contrast, overexpression of GLUT1 accelerated proliferation in these cells. Transwell assays revealed that SLC2A1‐AS1 suppressed the migration of HCC cells while GLUT1 induction promoted HCC migration. (Fig. 3C,E).
Fig. 3.
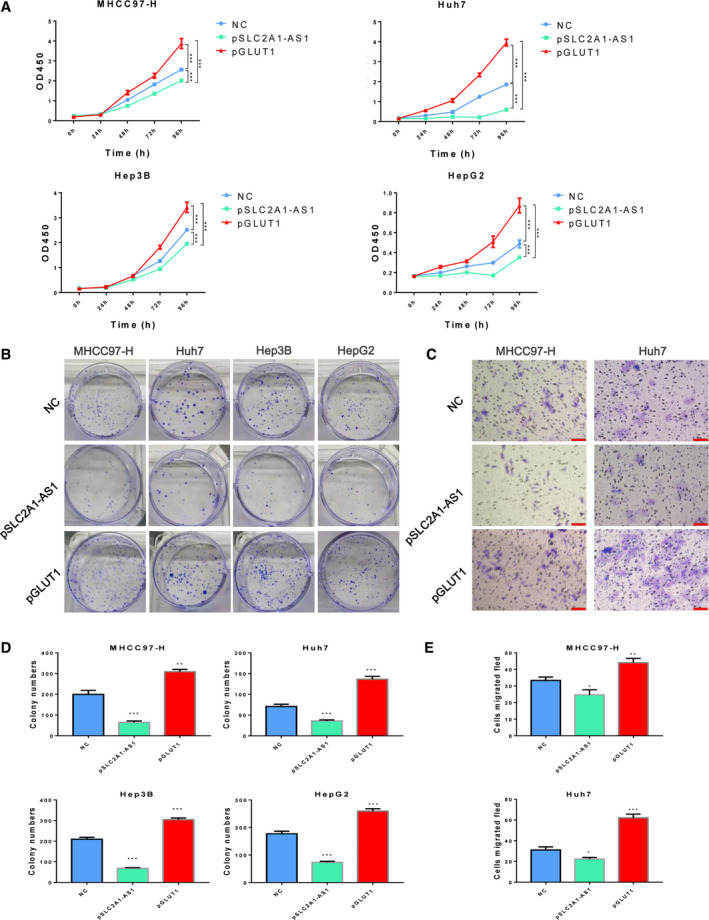
SLC2A1‐AS1 inhibits HCC cell proliferation and metastasis in vitro. (A) HCC cell proliferation was assessed using CCK‐8 assays after SLC2A1‐AS1 overexpression. (B) Overexpression of SLC2A1‐AS1 significantly reduced colony formation in HCC cells. (C) Expression of SLC2A1‐AS1 significantly reduced HCC cell migration. Scale bar = 100 μm. (D, E) Quantification of colonies (D) and migrated cells (E) with or without SLC2A1‐AS1 overexpression. Data are shown as means ± SD, and ANOVA followed by post hoc correction is used to calculate significance. *P < 0.05, **P < 0.01, ***P < 0.001.
To further validate the impact of SLC2A1‐AS1 on tumour proliferation in vivo, MHCC97‐H cells overexpressing SLC2A1‐AS1 were injected into the flanks of immunodeficient BALB/C nude mice to generate a subcutaneous xenotransplantation model. We observed that mice in the SLC2A1‐AS1 overexpression group developed smaller tumours than mice in the control group (Fig. 4A,B). The overexpression of SLC2A1‐AS1 in cell line‐derived tumour mass was confirmed by qRT‐PCR analysis (Fig. 4C). Subsequently, western blot and immunohistochemical staining showed that GLUT1 expression was significantly downregulated in the SLC2A1‐AS1 overexpression group (Fig. 4D,E). Additionally, staining for the proliferation marker Ki67 was much weaker in the SLC2A1‐AS1 overexpression group than in the NC group (Fig. 4E).
Fig. 4.
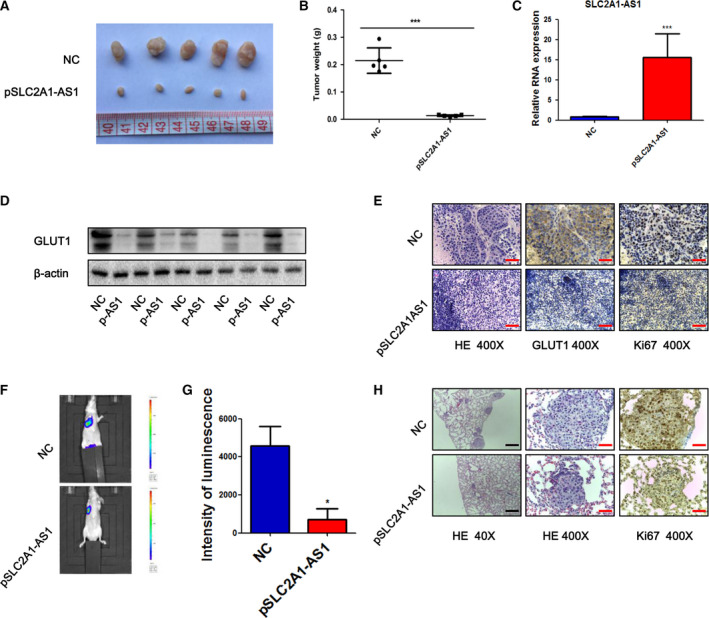
SLC2A1‐AS1 inhibits tumour growth and metastasis in vivo. (A) Representative photograph of mice from each group injected with MHCC97‐H‐control or MHCC97‐H‐SLC2A1‐AS1 cells. (B) Tumour weights were measured at day 28 in the control and SLC2A1‐AS1 overexpression groups. (C) Representative SLC2A1‐AS1 expression was analysed by qRT‐PCR. (D) Expression of GLUT1 was analysed by western blotting. (E) Representative images of H&E staining and immunohistochemical staining for Ki‐67 and GLUT1. Scale bar = 50 μm. (F, G) Overexpression of SLC2A1‐AS1 significantly reduced the lung metastasis luminescence intensity. (H) Representative H&E and IHC staining for Ki‐67 in lung metastatic nodules are shown. Scale bar (black) = 500 μm, Scale bar (red) = 50 μm. pAS1, pSLC2A1‐AS1. Student’s t‐test was performed for statistical comparisons. *P < 0.05, ***P < 0.001.
We also studied the effects of SLC2A1‐AS1 expression on tumour metastasis in vivo. Luciferase‐labelled MHCC97‐H cells transfected with SLC2A1‐AS1 or control plasmid were injected into nude mice via their tail vein. The bioluminescent signal was detected after the completion of 30 days postinjection. Representative bioluminescent images showed intense fluorescence in the NC group but very weak fluorescence in the SLC2A1‐AS1 group (Fig. 4F,G). Histological analysis further confirmed that the number of metastatic lung nodules and cell proliferation ability were decreased in the SLC2A1‐AS1‐overexpressing group (Fig. 4H).
Altogether, these results confirmed that SLC2A1‐AS1 inhibits HCC cell proliferation and metastasis in vivo as well as in vitro.
3.4. Overexpression of SLC2A1‐AS1 inhibits GLUT1 transcriptional activation
To study the mechanism of GLUT1 expression regulation by SLC2A1‐AS1, we first detected the subcellular distribution of SLC2A1‐AS1 in HCC cells. The RNA FISH results revealed that SLC2A1‐AS1 was localized mainly in the cytoplasm, but only very weak expression was shown (Fig. 5A). Intriguingly, a large amount of SLC2A1‐AS1 accumulated in the cell nucleus when HCC cells were transfected with the SLC2A1‐AS1 overexpression plasmid (Fig. 5A,B). This phenomenon was further confirmed by qRT‐PCR analysis of the nuclear/cytoplasmic RNA fractions (Fig. 5C). These results suggest that SLC2A1‐AS1 could traffic through the nuclear envelope and may exert its function in the nucleus when overexpressed. Therefore, we next investigated whether SLC2A1‐AS1 can regulate GLUT1 transcription. In this case, 1000 base pairs of the GLUT1 upstream promoter were cloned into the pGL3‐basic vector and used to perform a Dual‐Luciferase Reporter Assay. As shown in Fig. 5D, SLC2A1‐AS1 overexpression significantly decreased the promoter activity of GLUT1 in both Huh7 and MHCC97‐H cells, suggesting an inhibitory role of SLC2A1‐AS1 on GLUT1 transcription.
Fig. 5.
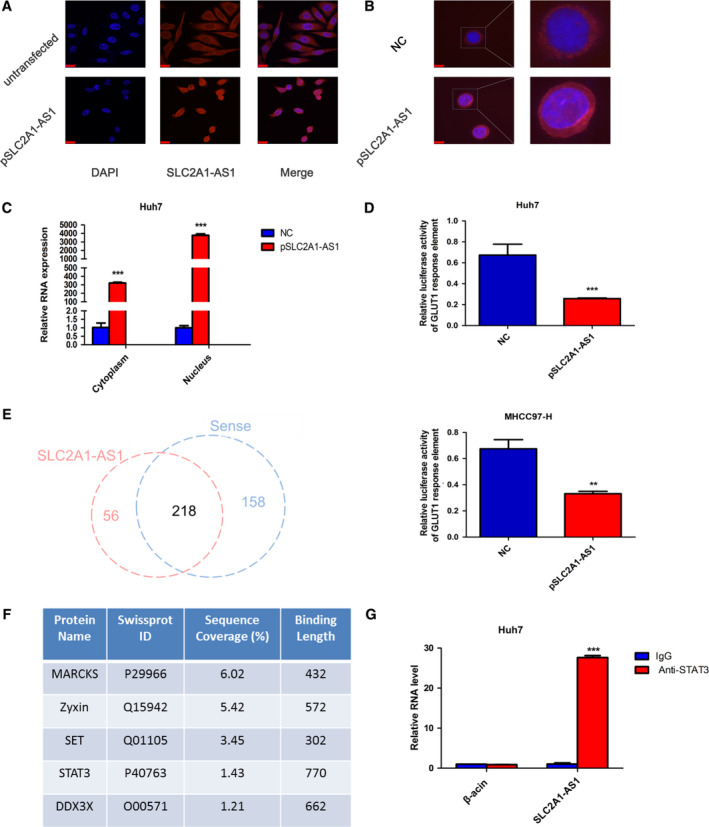
SLC2A1‐AS1 regulates GLUT1 transcription and interacts with STAT3. (A) Subcellular distribution of SLC2A1‐AS1 in Huh7 cells before and after SLC2A1‐AS1 plasmid transfection by RNA FISH. Scale bar = 1900 μm. (B) Subcellular distribution of SLC2A1‐AS1 in Huh7‐control and Huh7‐SLC2A1‐AS1 cells by RNA FISH. Scale bar = 800 μm. (C) Cytoplasmic and nuclear fractions of Huh7 cells transfected with control or SLC2A1‐AS1 plasmid were subjected to qRT‐PCR assays. (D) GLUT1 promoter activity in Huh7 and MHCC97‐H cells was assessed by luciferase reporter assays after SLC2A1‐AS1 transfection. (E) Venn diagram showing SLC2A1‐AS1‐specific binding proteins from a biotin‐labelled RNA pulldown assay combined with mass spectrometric analysis. (F) SLC2A1‐AS1‐bound transcriptional factors identified by mass spectrometric analysis. (G) Cell lysates from Huh7 cells were immunoprecipitated with STAT3 or IgG antibody, and qRT‐PCR assays were subsequently performed to measure relative SLC2A1‐AS1 expression. DAPI, 4’, 6‐diamidino‐2‐phenylindole. Student’s t‐test was performed for statistical comparisons. **P < 0.01, ***P < 0.001.
To further explore the specific mechanism of how SLC2A1‐AS1 inhibits GLUT1 transcription, we first analysed the SLC2A1‐AS1 sequence and found a potential ‘head‐to‐head’ pairing pattern composed of SLC2A1‐AS1 and GLUT1 with 529 nucleotides with full complementarity (Fig. S1C). The ‘head‐to‐head’ or ‘head‐to‐tail’ complementary pairing pattern has been shown to play a crucial role in the regulation of antisense lncRNAs and their opposite strand coding genes (Chen et al., 2018; Li et al., 2012). Hence, we constructed two plasmids containing the complementary pairing sequence of SLC2A1‐AS1 (pSLC2A1‐AS1‐part1) and the noncomplementary sequence of SLC2A1‐AS1 (pSLC2A1‐AS1‐part2) separately for cell transfection. Unexpectedly, the qRT‐PCR results revealed that overexpression of pSLC2A1‐AS1‐part2, but not only pSLC2A1‐AS1‐part1, also reduced GLUT1 mRNA expression (Fig. S1D). Moreover, the reduction in GLUT1 in the pSLC2A1‐AS1‐part1‐transfected group was no more than that in the pSLC2A1‐AS1‐part2 overexpression group. These results indicate that ‘head‐to‐head’ pairing is not the principal regulatory mechanism of SLC2A1‐AS1 and GLUT1.
3.5. SLC2A1‐AS1 interacts with the transcriptional factor STAT3
To further explore the major regulatory mechanism by which SLC2A1‐AS1 affects GLUT1 transcriptional activity, we focused on the transcriptional factors that interact with SLC2A1‐AS1. We performed a biotin‐labelled RNA pulldown assay and then subjected the precipitants to mass spectrometric analysis to identify proteins that interact with SLC2A1‐AS1 in HCC cells. We found that 56 proteins interact with SLC2A1‐AS1 uniquely (Fig. 5E and Table S5). Among these proteins, we screened five potential transcriptional factors (Fig. 5F). We focused particularly on STAT3, which has been demonstrated to be a key regulator of glucose metabolism in HCC (Li et al., 2017; Liu and Yu, 2018; Wang et al., 2012). To further validate the interactions between SLC2A1‐AS1 and STAT3, we conducted an RNA immunoprecipitation assay and subsequent qRT‐PCR analysis. The results from these assays confirmed the enrichment of SLC2A1‐AS1 in the STAT3 complex when compared with IgG (Fig. 5G).
3.6. SLC2A1‐AS1/STAT3 interaction inhibits the activity of the FOXM1/GLUT1 axis
The underlying regulatory mechanism of the SLC2A1‐AS1/STAT3 interaction in GLUT1 transcription was further examined. First, we explored whether the SLC2A1‐AS1/STAT3 interaction directly inhibits the transcriptional activation of GLUT1 via STAT3. Although STAT3 has been confirmed to participate in aerobic glycolysis in HCC, there is no evidence demonstrating that GLUT1 is a direct target of STAT3. Therefore, we first analysed the sequence of the GLUT1 promoter to identify potential STAT3‐binding sites. The bioinformatics analysis results revealed three potential STAT3‐binding sites present in the GLUT1 promoter (Fig. S4A). However, the ChIP assay results showed that GLUT1 chromatin did not specifically immunoprecipitate with antibodies against STAT3 when compared to IgG (Fig. S4B).
Next, we explored whether the SLC2A1‐AS1/STAT3 interaction inhibits GLUT1 transcription via an indirect mechanism. The transcription factor FOXM1 has been demonstrated as a direct target of STAT3 (Mencalha et al., 2012). Previously, we found that FOXM1 regulates the transcriptional activation of GLUT1 in HCC and ovarian cancer (Shang et al., 2017; Wang et al., 2016). Therefore, we hypothesized that FOXM1 mediates the regulation of GLUT1 via the SLC2A1‐AS1/STAT3 interaction. To verify our hypothesis, we first assessed whether the SLC2A1‐AS1/STAT3 interaction altered FOXM1 expression. As shown in Fig. 6A, HCC cells transfected with SLC2A1‐AS1 showed a significant decrease in FOXM1 expression at both the mRNA and protein levels. However, the expression of STAT3 did not change at different time points after transfection. Moreover, the trend of altered GLUT1 expression at different time points after SLC2A1‐AS1 transfection was consistent with the FOXM1 expression levels. Next, we cloned the sequence containing the FOXM1‐binding site in the GLUT1 promoter (ACAAATAA) or the corresponding mutant sequence (AtAcgatc) into the pGL3‐basic vector for transfection and performed luciferase reporter assays. We found that FOXM1 overexpression significantly increased wild‐type GLUT1 reporter activity, and this effect was reversed when SLC2A1‐AS1 was overexpressed (Fig. 6B). Notably, these effects were abolished with the introduction of the mutant reporter construct (Fig. 6B). These results suggest that FOXM1 mediates the regulation of GLUT1 transcription via the SLC2A1‐AS1/STAT3 interaction.
Fig. 6.
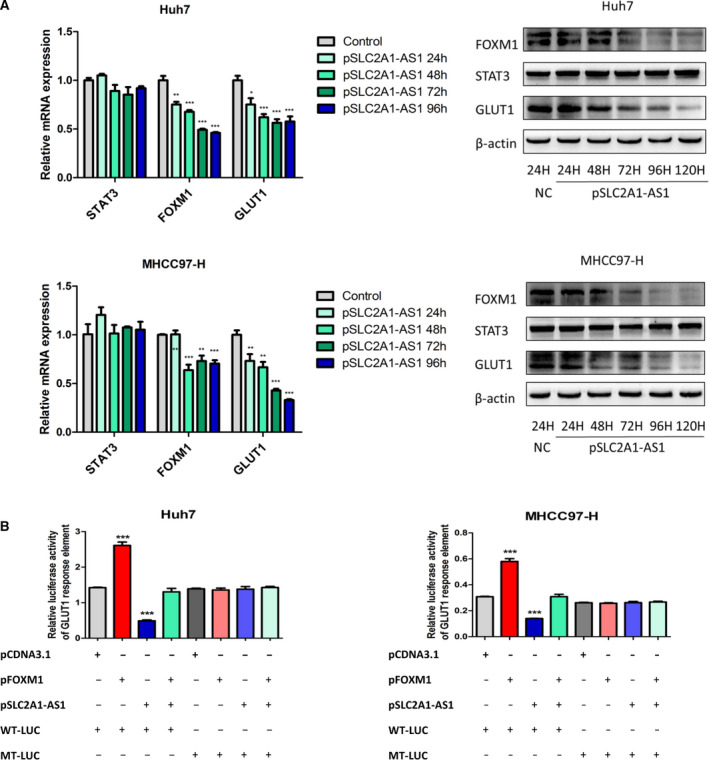
FOXM1 mediates the transcriptional regulation of GLUT1 via SLC2A1‐AS1/STAT3 interaction. (A) mRNA and protein expression levels of STAT3, FOXM1 and GLUT1 were assessed 24–96 h or 24–120 h after SLC2A1‐AS1 plasmid transfection. (B) Reporter plasmids containing wild‐type (WT‐LUC) or mutant (MT‐LUC) FOXM1‐binding sites in the GLUT1 promoter were transfected in combination with control plasmid, FOXM1 overexpression plasmid, SLC2A1‐AS1 overexpression plasmid or combined FOXM1 and SLC2A1‐AS1 plasmids into Huh7 and MHCC97‐H cells. Relative luciferase activity was detected using a luciferase assay kit. Data were analysed with ANOVA followed by post hoc correction. *P < 0.05, **P < 0.01, ***P < 0.001.
To further validate FOXM1 as a direct target of STAT3 in HCC, we transfected HCC cells with siRNA targeting STAT3 for further experiments. The qRT‐PCR and western blot assay results indicated that STAT3 knockdown significantly reduced both FOXM1 and GLUT1 expression (Fig. 7A,B). Moreover, downregulation of STAT3 significantly decreased the activity of the FOXM1 promoter (Fig. 7D). In addition, the ChIP assay results suggested that the region from −480 to −490 of the FOXM1 promoter is a crucial response element of STAT3 in HCC cells (Fig. 7C,E).
Fig. 7.
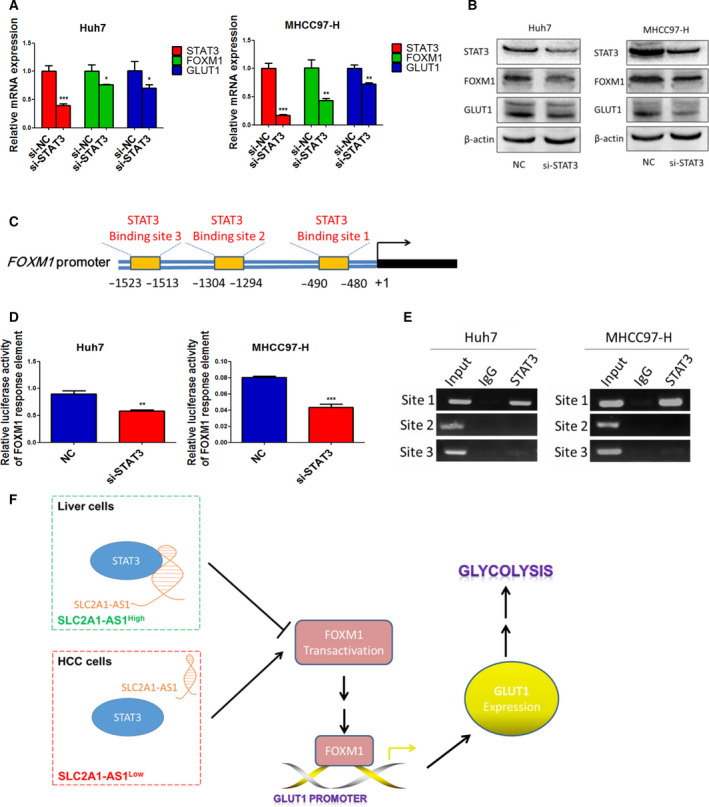
SLC2A1‐AS1/STAT3 interaction inhibits FOXM1 transcriptional activity and FOXM1/GLUT1 axis activation. (A, B) STAT3 knockdown significantly reduced the expression of FOXM1 and GLUT1 at the transcription (A) and translation (B) levels. (C) Putative STAT3‐binding sites in the FOXM1 promoter. (D) STAT3 knockdown significantly reduced the promoter activity of FOXM1. (E) ChIP analysis of the FOXM1 promoter using antibodies against STAT3 or IgG in Huh7 and MHCC97‐H cells. (F) A schematic model of the role and mechanism of SLC2A1‐AS1 in HCC glycolysis. Student’s t‐test was performed for statistical comparisons. *P < 0.05, **P < 0.01, ***P < 0.001.
Next, to confirm whether FOXM1 is required for the SLC2a1‐AS1‐mediated GLUT1 downregulation, we conducted cell transfection with control plasmids and SLC2A1‐AS1 overexpression plasmids, or cotransfected with SLC2A1‐AS1 and FOXM1 overexpression plasmids in HCC cells. As shown in Fig. S5, overexpression of FOXM1 abrogated the SLC2a1‐AS1‐mediated GLUT1 downregulation without altering SLC2A1‐AS1 expression.
In summary, the above results indicate that the SLC2A1‐AS1/STAT3 interaction inhibits the transcriptional activation of FOXM1 via STAT3 and then reduces the activity of the FOXM1/GLUT1 axis.
4. Discussion
It has been well established that lncRNAs can affect coding gene expression through several mechanisms, including pretranscriptional, transcriptional and post‐transcriptional processing. Various studies have uncovered the crucial roles of lncRNAs in regulating carcinogenesis and progression. However, the roles of lncRNAs in tumour metabolism remain obscure. Specifically, the involvement of lncRNAs in HCC glycolysis and progression is not widely studied. In this study, we identified a previously unstudied lncRNA, SLC2A1‐AS1 (Ensembl ID: ENSG00000227533), that is frequently downregulated in HCC cell lines and tissues and associated with recurrence‐free survival. This molecule acts as an antisense RNA of GLUT1. We further validated that SLC2A1‐AS1 inhibits HCC cell glycolysis via negatively regulating GLUT1 expression. In addition, we observed that SLC2A1‐AS1 overexpression markedly decreased the proliferation and metastasis of HCC cells. Moreover, we found SLC2A1‐AS1 accumulation in the cell nucleus when it was overexpressed. We also found an interaction between SLC2A1‐AS1 and STAT3, the latter of which is a transcription factor that plays crucial roles in tumour glycolysis. Furthermore, we determined that the SLC2A1‐AS1/STAT3 interaction weakens the activation of the FOXM1/GLUT1 axis.
Antisense lncRNA SLC2A1‐AS1 is located on the opposite strand of SLC2A1 (GLUT1) on chromosome 1. Antisense transcripts have been demonstrated to regulate the expression of corresponding sense‐strand mRNAs by affecting their transcription or mRNA stability (Nolasco et al., 2012; Pan et al., 2018). In the current study, we found that SLC2A1‐AS1 inhibits the mRNA and protein expression of GLUT1, which indicates that SLC2A1‐AS1 exerts its functional roles via regulating GLUT1 expression. The role of GLUT1 has been well established in cancer cells, especially in cancer metabolism. Cancer cells prefer to use the glycolysis pathway to meet their increased bioenergetics and biosynthetic demands, and they exhibit increased glucose uptake and lactate production. The glucose transporter (GLUT) family plays a vital role in glucose transport across the plasma membrane, which is the initial step of glycolysis. In different cancer cells, GLUT1, a member of the GLUT family, is frequently aberrantly expressed, which has potential effects on the glycolysis process in cancer (Szablewski, 2013). GLUT1 is abnormally overexpressed in HCC and promotes HCC cell glycolysis and progression (Amann et al., 2009). Our results show that the decreased glycolysis ability in HCC cells caused by SLC2A1‐AS1 overexpression can be reversed by GLUT1 overexpression. This observation indicates that SLC2A1‐AS1 decreased HCC glycolysis through reducing GLUT1 expression.
As a vital connection between the tumour microenvironment and tumour progression, the process of tumour metabolic reprogramming has been regarded as cancer’s Achilles’ heel because this process contributes to tumour progression by promoting cell growth, invasion, metastasis, tumour angiogenesis and immunosurveillance (Kroemer and Pouyssegur, 2008). The primary benefit of glycolysis in tumour cells is that increased glucose levels provide a carbon source for anabolic reactions. Moreover, lactate, the principal end product of aerobic glycolysis, provides a favourable acidic microenvironment for tumour invasion and metastasis (Koukourakis et al., 2006). Similarly, we found that SLC2A1‐AS1 inhibits the production of lactate as well as glucose uptake in HCC cells. The results from in vitro and in vivo assays further demonstrate the inhibitory role of SLC2A1‐AS1 in HCC proliferation and metastasis. These results indicate that SLC2A1‐AS1 contributes to HCC glycolysis and then manipulates cell growth and migration abilities.
Although antisense lncRNAs have been shown to act at nearly every level of gene regulation, including pretranscription, transcription and post‐transcription (Villegas and Zaphiropoulos, 2015), the different subcellular localization of lncRNAs determines the specific regulatory molecular mechanism (Guttman and Rinn, 2012). lncRNAs accumulating in the cytoplasm commonly act as sponges for microRNAs and proteins and participate in post‐transcription regulation (Kim et al., 2016; Liang et al., 2015). However, in the nucleus, lncRNAs are involved in gene transcription (Zong et al., 2011). SLC2A1‐AS1 expression was relatively low in HCC cells and was located predominantly in the cytoplasm. However, we observed SLC2A1‐AS1 accumulation in the cell nucleus when it was overexpressed. Hence, we speculated that SLC2A1‐AS1 affected GLUT1 transcription. This hypothesis was validated by a luciferase reporter assay.
Recently, sense–antisense pairing was identified as an important mechanism of the regulation between antisense lncRNAs and their corresponding coding genes (Chen et al., 2018; Hung and Chang, 2010; Li et al., 2012; Morris et al., 2008). We also found a ‘head‐to‐head’ pairing pattern between SLC2A1‐AS1 and GLUT1. Nevertheless, we found that overexpression of both the pairing part and the nonpairing part of SLC2A1‐AS1 reduced the mRNA expression of GLUT1. This indicates that ‘head‐to‐head’ pairing is not the principal regulatory mechanism of SLC2A1‐AS1 and GLUT1.
Using a biotin‐labelled RNA pulldown assay and RNA immunoprecipitation assay, we revealed an interaction between SLC2A1‐AS1 and the STAT3 protein. STAT3 is an important transcriptional factor that has been identified to regulate cancer initiation and progression (Ihle, 1996; Wang et al., 2011). It has also been reported to play a role in cancer glycolysis (Li et al., 2017; Li et al., 2015; Wang et al., 2013). We found that although SLC2A1‐AS1 did not directly regulate the transcriptional activity of GLUT1, it inhibited the transcription of FOXM1, which we previously determined to be a crucial transcriptional activator of GLUT1 (Shang et al., 2017; Wang et al., 2016). These results therefore indicated a possible molecular link among SLC2A1‐AS1, STAT3 and the FOXM1/GLUT1 axis, but the precise binding site between SLC2A1‐AS1 and STAT3 requires further study.
In summary, we report antisense lncRNA SLC2A1‐AS1 as a tumour suppressor gene. SLC2A1‐AS1 interacts with STAT3 and inhibits FOXM1/GLUT1 axis activation in HCC cells. We also characterized the suppressive effects that SLC2A1‐AS1 exerts in glycolysis through inactivating GLUT1, which in turn inhibits HCC progression. Therefore, our study reveals the function and mechanism of SLC2A1‐AS1 in HCC and highlights its prognostic and therapeutic significance for HCC patients.
5. Conclusion
SLC2A1‐AS1 inhibits glycolysis and progression in HCC via the STAT3/FOXM1/GLUT1 axis. SLC2A1‐AS1 can be considered a valuable predictive biomarker for HCC recurrence.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
RS, MW and BD performed most experiments and analysed the data. RS wrote the manuscript. JD and JW performed prognosis and data analysis. ZL and SQ participated in the in vitro study. XY, JL participated in the in vivo study. CX collected HCC tissue samples and clinical data. YL, DW and LW designed the overall study, supervised the experiments and revised the paper.
Supporting information
Fig. S1 . Characterization of the antisense lncRNA SLC2A1‐AS1.
Fig. S2. GLUT1 is upregulated in HCC and associated with tumour recurrence in HCC patients.
Fig. S3. SLC2A1‐AS1 inhibits GLUT1 expression.
Fig. S4 . STAT3 cannot bind to the GLUT1 promoter in HCC cells.
Fig. S5 . FOXM1 is required for SLC2A1‐AS1‐mediated GLUT1 downregulation.
Table S1 . Primers used in qRT‐PCR assays.
Table S2 . List of primary Antibodies.
Table S3 . PCR primer sequences of DNA fragments targeted FOXM1 and GLUT1 promoters.
Table S4 . Univariate and multivariate Cox regression analyses of recurrence‐free survival in 33 patients with HCC.
Table S5 . Summary of proteins interacting with SLC2A1‐AS1 uniquely by mass spectrometric analysis.
Acknowledgement
This work was supported by National Natural Science Foundation of China (Grant No. 81872129 and 81672341).
Runze Shang, Miao Wang and Bin Dai contributed equally to this work
Contributor Information
Lin Wang, Email: fierywang@163.com.
Desheng Wang, Email: wangdesh@163.com.
Yu Li, Email: liyu@nwpu.edu.cn.
References
- Amann T, Maegdefrau U, Hartmann A, Agaimy A, Marienhagen J, Weiss TS, Stoeltzing O, Warnecke C, Scholmerich J, Oefner PJ et al (2009) GLUT1 expression is increased in hepatocellular carcinoma and promotes tumorigenesis. Am J Pathol 174, 1544–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengol C, Sarrias MR and Sala M (2018) Hepatocellular carcinoma: present and future. Med Clin 150, 390–397. [DOI] [PubMed] [Google Scholar]
- Cairns RA, Harris IS and Mak TW (2011) Regulation of cancer cell metabolism. Nat Rev Cancer 11, 85–95. [DOI] [PubMed] [Google Scholar]
- Chen Z, Liu Z, Yang Y, Zhu Z, Liang R, Huang B, Wu D, Yang L, Lu H, Jin D et al (2018) Long non‐coding RNA RAB11B‐AS1 prevents osteosarcoma development and progression via its natural antisense transcript RAB11B. Oncotarget 9, 26770–26786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatenby RA, Gawlinski ET, Gmitro AF, Kaylor B and Gillies RJ (2006) Acid‐mediated tumor invasion: a multidisciplinary study. Can Res 66, 5216–5223. [DOI] [PubMed] [Google Scholar]
- Guttman M and Rinn JL (2012) Modular regulatory principles of large non‐coding RNAs. Nature 482, 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T and Chang HY (2010) Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol 7, 582–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle JN (1996) STATs: signal transducers and activators of transcription. Cell 84, 331–334. [DOI] [PubMed] [Google Scholar]
- Kim J, Abdelmohsen K, Yang X, De S, Grammatikakis I, Noh JH and Gorospe M (2016) LncRNA OIP5‐AS1/cyrano sponges RNA‐binding protein HuR. Nucleic Acids Res 44, 2378–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukourakis MI, Giatromanolaki A, Harris AL and Sivridis E (2006) Comparison of metabolic pathways between cancer cells and stromal cells in colorectal carcinomas: a metabolic survival role for tumor‐associated stroma. Can Res 66, 632–637. [DOI] [PubMed] [Google Scholar]
- Kroemer G and Pouyssegur J (2008) Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell 13, 472–482. [DOI] [PubMed] [Google Scholar]
- Li J, Liu T, Zhao L, Chen W, Hou H, Ye Z and Li X (2015) Ginsenoside 20(S)Rg3 inhibits the Warburg effect through STAT3 pathways in ovarian cancer cells. Int J Oncol 46, 775–781. [DOI] [PubMed] [Google Scholar]
- Li M, Jin R, Wang W, Zhang T, Sang J, Li N, Han Q, Zhao W, Li C and Liu Z (2017) STAT3 regulates glycolysis via targeting hexokinase 2 in hepatocellular carcinoma cells. Oncotarget 8, 24777–24784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Su Z, Xu X, Liu G, Song X, Wang R, Sui X, Liu T, Chang X and Huang D (2012) AS1DHRS4, a head‐to‐head natural antisense transcript, silences the DHRS4 gene cluster in cis and trans. Proc Natl Acad Sci USA 109, 14110–14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B, Peng C et al (2016) Upregulation of long noncoding RNA ZEB1‐AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene 35, 1575–1584. [DOI] [PubMed] [Google Scholar]
- Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF et al (2015) The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget 6, 22513–22525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YH, Wu MH, Huang YH, Yeh CT, Cheng ML, Chi HC, Tsai CY, Chung IH, Chen CY and Lin KH (2018) Taurine up‐regulated gene 1 functions as a master regulator to coordinate glycolysis and metastasis in hepatocellular carcinoma. Hepatology 67, 188–203. [DOI] [PubMed] [Google Scholar]
- Liu B and Yu S (2018) Amentoflavone suppresses hepatocellular carcinoma by repressing hexokinase 2 expression through inhibiting JAK2/STAT3 signaling. Biomed Pharmacother 107, 243–253. [DOI] [PubMed] [Google Scholar]
- Mencalha AL, Binato R, Ferreira GM, Du Rocher B and Abdelhay E (2012) Forkhead box M1 (FoxM1) gene is a new STAT3 transcriptional factor target and is essential for proliferation, survival and DNA repair of K562 cell line. PLoS ONE 7, e48160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV, Santoso S, Turner AM, Pastori C and Hawkins PG (2008) Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet 4, e1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolasco S, Bellido J, Goncalves J, Tavares A, Zabala JC and Soares H (2012) The expression of tubulin cofactor A (TBCA) is regulated by a noncoding antisense Tbca RNA during testis maturation. PLoS ONE 7, e42536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W, Zhang N, Liu W, Liu J, Zhou L, Liu Y and Yang M (2018) The long noncoding RNA GAS8‐AS1 suppresses hepatocarcinogenesis by epigenetically activating the tumor suppressor GAS8. J Biol Chem 293, 17154–17165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu A, Singhal U and Chinnaiyan AM (2015) Long noncoding RNAs in cancer: from function to translation. Trends Cancer 1, 93–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang R, Pu M, Li Y and Wang D (2017) FOXM1 regulates glycolysis in hepatocellular carcinoma by transactivating glucose transporter 1 expression. Oncol Rep 37, 2261–2269. [DOI] [PubMed] [Google Scholar]
- Sun J, Wang X, Fu C, Wang X, Zou J, Hua H and Bi Z (2016) Long noncoding RNA FGFR3‐AS1 promotes osteosarcoma growth through regulating its natural antisense transcript FGFR3. Mol Biol Rep 43, 427–436. [DOI] [PubMed] [Google Scholar]
- Szablewski L (2013) Expression of glucose transporters in cancers. Biochem Biophys Acta 1835, 164–169. [DOI] [PubMed] [Google Scholar]
- Villegas VE and Zaphiropoulos PG (2015) Neighboring gene regulation by antisense long non‐coding RNAs. Int J Mol Sci 16, 3251–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Hsu SH, Frankel W, Ghoshal K and Jacob ST (2012) Stat3‐mediated activation of microRNA‐23a suppresses gluconeogenesis in hepatocellular carcinoma by down‐regulating glucose‐6‐phosphatase and peroxisome proliferator‐activated receptor gamma, coactivator 1 alpha. Hepatology 56, 186–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lafdil F, Kong X and Gao B (2011) Signal transducer and activator of transcription 3 in liver diseases: a novel therapeutic target. Int J Biol Sci 7, 536–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TX, Zhang ZQ, Cong Y, Shi XY, Liu YH and Zhao FL (2013) Prosapogenin A induces apoptosis in human cancer cells in vitro via inhibition of the STAT3 signaling pathway and glycolysis. Oncol Lett 6, 1323–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yun Y, Wu B, Wen L, Wen M, Yang H, Zhao L, Liu W, Huang S, Wen N et al (2016) FOXM1 promotes reprogramming of glucose metabolism in epithelial ovarian cancer cells via activation of GLUT1 and HK2 transcription. Oncotarget 7, 47985–47997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L, Huang W, Tian D, Zhu H, Qi X, Chen Z, Zhang Y, Hu H, Fan D, Nie Y et al (2013) Overexpression of forkhead box C1 promotes tumor metastasis and indicates poor prognosis in hepatocellular carcinoma. Hepatology 57, 610–624. [DOI] [PubMed] [Google Scholar]
- Yang F, Zhang H, Mei Y and Wu M (2014) Reciprocal regulation of HIF‐1alpha and lincRNA‐p21 modulates the Warburg effect. Mol Cell 53, 88–100. [DOI] [PubMed] [Google Scholar]
- Zong X, Tripathi V and Prasanth KV (2011) RNA splicing control: Yet another gene regulatory role for long nuclear noncoding RNAs. RNA Biol 8, 968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 . Characterization of the antisense lncRNA SLC2A1‐AS1.
Fig. S2. GLUT1 is upregulated in HCC and associated with tumour recurrence in HCC patients.
Fig. S3. SLC2A1‐AS1 inhibits GLUT1 expression.
Fig. S4 . STAT3 cannot bind to the GLUT1 promoter in HCC cells.
Fig. S5 . FOXM1 is required for SLC2A1‐AS1‐mediated GLUT1 downregulation.
Table S1 . Primers used in qRT‐PCR assays.
Table S2 . List of primary Antibodies.
Table S3 . PCR primer sequences of DNA fragments targeted FOXM1 and GLUT1 promoters.
Table S4 . Univariate and multivariate Cox regression analyses of recurrence‐free survival in 33 patients with HCC.
Table S5 . Summary of proteins interacting with SLC2A1‐AS1 uniquely by mass spectrometric analysis.


