Abstract
Purpose
Dioscin, a natural glycoside derived from many plants, has been proved to exert anti-cancer activity. Several studies have found that it reverses TGF-β1-induced epithelial–mesenchymal transition (EMT). Whether dioscin can reverse EMT by pathways other than TGF-β is still unknown.
Methods
We used network-based pharmacological methods to systematically explore the potential mechanisms by which dioscin acts on lung cancer. Cell Counting Kit-8 assay, scratch healing, Transwell assay, Matrigel invasion assay, immunofluorescence assay, and Western blotting were employed to confirm the prediction of key targets and the effects of dioscin on EMT.
Results
Here, using network-based pharmacological methods, we found 42 possible lung cancer-related targets of dioscin, which were assigned to 98 KEGG pathways. Among the 20 with the lowest p-values, the PI3K-AKT signaling pathway is involved and significantly related to EMT. AKT1 and mTOR, with high degrees (reflecting higher connectivity) in the compound-target analysis, participate in the PI3K-AKT signaling pathway. Molecular docking indicated the occurrence of dioscin-AKT1 and dioscin-mTOR binding. Functional experiments demonstrated that dioscin suppressed the proliferation, migration, invasion, and EMT of human lung adenocarcinoma cells in a dose-dependent manner, without TGF-β stimulation. Furthermore, we determined that dioscin downregulated p-AKT, p-mTOR and p-GSK3β in human lung adenocarcinoma cells without affecting their total protein levels. The PI3K inhibitor LY294002 augmented these changes.
Conclusion
Dioscin suppressed proliferation, invasion and EMT of lung adenocarcinoma cells via the inactivation of AKT/mTOR/GSK3β signaling, probably by binding to AKT and mTOR, and inhibiting their phosphorylation.
Keywords: dioscin, lung adenocarcinoma, epithelial–mesenchymal transition, network-based pharmacology
Introduction
As the most common cause of cancer-related mortality worldwide,1 the prognosis of patients with lung cancer remains poor. Lymphatic and hematogenous metastases are the major causes of high mortality in patients with lung cancer. Thus, blocking the metastatic process is critically important for cancer treatment. Epithelial–mesenchymal transition (EMT) is thought to be involved in lung cancer metastasis.2
EMT, the process by which polar epithelial cells transform into mesenchymal cells under particular physiological and pathological conditions, participates in the invasion and metastasis of tumor cells in tumor tissues.3 Studies have shown that there are many signaling pathways involved in EMT,4–6 including transforming growth factor beta (TGF-β), SMAD-dependent/independent, Wnt/β-catenin, Matrix, PI3K/AKT/mTOR, and AKT/GSK3β/β-catenin signaling pathways.
Dioscin, with the molecular formula C45H72O16 and a molecular weight of 869.05 g/mol, is widely found in plants, such as Dioscoreae, Liliaceae, Dianthus, and Rosaceae. Among these plants, Dioscorea zingiberensis has the highest content of dioscin and is the main plant material used in China to produce dioscin, which has extremely high medicinal value. Numerous studies have reported that dioscin exerts a strong anti-tumor activity.7,8 Recently, several studies have revealed that dioscin reverses EMT.9,10 Whether dioscin could reverse EMT by pathways other than TGF-β remains unclear. To identify other pathways by which dioscin can reverse EMT, here we employed network-based pharmacological methods to explore the possible targets of dioscin and used experimental approaches to verify some pathways of the predicted targets.
Materials and Methods
Collection of Lung Cancer-Related Targets of Dioscin
CTD (http://ctdbase.org/),11 Similarity Ensemble Approach (http://sea16.docking.org/)12 and SwissTargetPrediction (http://swisstargetprediction.ch/)13 were employed to collect the dioscin-related targets. For SwissTargetPrediction, the targets with a probability value≥0.5 were selected. The lung cancer-related targets were extracted from GeneCards (https://www.genecards.org/)14 with “lung cancer” as a search term, and the 500 with the highest scores were retained. The targets matching those obtained from the above described approach were identified as potential lung cancer-related targets of dioscin. The gene symbols for all candidates were verified by the UniProt (https://www.uniprot.org/).15
Construction of Compound-Target Networks
The interactions between the above-mentioned potential targets of dioscin were analyzed using the STRING database (https://string-db.org/),16 and interactions with a combined score higher than 0.4 were screened. The compound-target network was generated based on the PPI data (protein–protein interaction) and was visualized using Cytoscape-v3.7.1 software. The network characteristics were analyzed by the applied plug-in Network Analyzer. The degree of freedom was used as a topological index, which is often used to describe the importance of the network node. The larger the value, the more critical the node is in the network.
Enrichment Analysis of Dioscin Lung Cancer-Related Target Pathway
Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were completed utilizing the DAVID program (https://david.ncifcrf.gov/).17
Molecular Docking Verification
The chemical structure of dioscin was obtained from the PubChem (https://pubchem.ncbi.nlm.nih.gov/),18 saved in its SDF format, and converted to the mol2 format by Discovery Studio 3.0. The PDB IDs of the candidates AKT1 and mTOR were derived from the UniProt database, and the corresponding protein three-dimensional structure was obtained from the RCSB PDB (http://www.rcsb.org/)19 database and saved in PDB format. Molecular docking was performed using Autodock Tools-1.5.6, and the docking score was used to assess the binding affinity of the target to the dioscin molecule. The two-dimensional plan of the docking results was presented by Discovery Studio 2019 Client.
Cell Lines and Reagents
Lung adenocarcinoma cell lines A549 and H1299 were obtained from the American Type Culture Collection (Manassas, VA, USA). All media were supplemented with 10% fetal bovine serum (FBS) (ExCell Bio Inc., Shanghai, China) and 100 U/mL penicillin-streptomycin mixture (Gibco, Grand Island, NY, USA). The cells were maintained at 37°C in a humidified atmosphere of 95% air and 5% CO2.
Dioscin with purity of 98% (Lot No. MUST-16090203) was obtained from Chengdu Must Biotechnology Co., Ltd. (Chengdu, China), which was dissolved in dimethyl sulfoxide (DMSO) at a concentration of 10 µM and stored at −20°C. For use, stock solutions were freshly diluted with medium (DMSO<0.1%). Antibodies against ZO-1, vimentin, N-cadherin, E-cadherin, ZEB1, GAPDH, AKT, p-AKT, mTOR, p-mTOR, GSKβ, p-GSK3β, SNAIL, SLUG, claudin-1, and peroxidase-conjugated and 488-conjugated secondary antibodies were obtained from Cell Signaling Technology, Inc. (Beverly, MA, USA). CCK8 was obtained from Shanghai Beibo Biological Co. (Shanghai, China). The Immobilon western chemiluminescent horseradish peroxidase (HRP) substrate was obtained from Millipore Co. (Billerica, MA, USA).
Cell Proliferation Assay
Cells in the logarithmic growth phase were collected as cell suspensions, and 100 µL of the cell suspension was added to a 96-well plate (2000 cells/well). The experimental wells were filled with different concentrations of dioscin (0–8 μM, DMSO<0.1%) medium, and the control wells were filled with complete medium. After 24–48 h, 10µL of CCK8 solution was added per well, and incubated for 2–4 h. The optical density (OD) at 450 nm was measured using a microplate reader, and cell viability (cell viability = experimental well/control well × 100%) was calculated.
Scratch Healing Assay
Lung adenocarcinoma A549 or H1299 cells were collected and made into cells suspensions, which were seeded on a six-well plate (at about 6×105 cells/well) to ensure that the cells could spread over the entire well overnight. The monolayer was scratched across the center of each well utilizing the tip of a 200 µL pipette. The cells were then cultured in a serum-free medium with concentrations of dioscin at 0, 1, and 2 µM for 24 h. Each scratch wound was visualized by microscopy and five random fields (×100) were chosen to assess the cell migration ability.
Transwell Migration Assay
After 12 h of serum-free starvation, cells in the logarithmic growth phase were digested, collected, and resuspended in serum-free medium, and adjusted to a cell density of 1×106 cells/mL. Cells in 200 µL of serum-free medium containing different concentrations of Dioscin were seeded on the upper chamber, and 600 µL of medium supplemented with 15% fetal calf serum (FCS) was placed in the lower chamber. After 24 h of incubation, the chamber was fixed with 4% paraformaldehyde and subsequently dyed with 0.1% crystal violet. The upper chamber was carefully wiped with cotton swabs. The numbers of cells that had migrated to the lower surface were enumerated in five randomly selected visual fields (×200).
In vitro Invasion Assay
The effect of dioscin on the invasion of NSCLC cells was assessed using in vitro invasion assays in a Transwell chamber (Corning Inc., Corning, NY, USA). Matrigel was applied to the membrane (polycarbonate membrane, 8 µm pore size, 6.5 mm diameter) in the upper compartment. Then, 200 µL of pre-warmed serum-free medium was added to the Transwell chamber and allowed to stand at 37°C for 30 min to gel the Matrigel. Other procedures resembled those of the Transwell migration assay.
Western Blotting
Lung adenocarcinoma cells were collected 24 h after dioscin stimulation, lysed on ice for 30 min with radio-immunoprecipitation assay (RIPA) buffer (Beyotime, Jiangsu, China), collected in a 1.5 mL centrifuge tube, and centrifuged at 4°C for 10 min. The supernatant was retained and the protein concentration was quantified utilizing a BCA protein quantitative kit (Keygen, Changchun, China). After being supplemented with 5× loading buffer, the protein was heated in a metal bath for 10min and was stored at −80°C until use. The samples (20 μg per lane) were separated using 10% SDS-PAGE gel electrophoresis and transferred (300 mA, 100 min) onto a polyvinylidene fluoride (PVDF) membrane, which was blocked in 5% skim milk at room temperature for 2 h. Subsequently, the primary antibodies were added and incubated at 4°C overnight. The membranes were then incubated with secondary antibodies for 1 h. A fully automated chemiluminescence image analysis system was used to image the immunoreactive protein bands. The following antibodies were used in this study: rabbit monoclonal anti-N-cadherin (13116T), anti-E-cadherin (3195T), anti-vimentin (5741T), anti-ZEB1 (3396T), anti-ZO-1 (8193T), anti-Slug (9585T), anti-Snail (3879T), anti-claudin-1 (13255T), anti-β-catenin (8480T), anti-MMP9 (13667T), anti-GAPDH (2118T), anti-AKT (4691T), anti-phospho-AKT (4060T), anti-GSK3β (12456T), anti-phospho-GSK3β (5558T), anti-mTOR (2983T), and anti-phospho-mTOR (5536T), all of which were from Cell Signaling Technology (Boston, MA, USA). HRP Goat Anti-Rabbit IgG (AS014) was from Abclonal (Wuhan, China).
Immunofluorescence Assay
A 12-well circular scaffold was placed in a 12-well plate and the slides were rinsed with a medium. The cells were seeded on the 12-well plate at the density of 3×104 cells/well. At 24 h after dioscin stimulation, the cells were fixed with 4% paraformaldehyde for 20 min and then blocked with a solution containing 0.3% TritonX-100 10% BSA. The blocking solution was discarded, and the primary antibody (in blocking solution, 1:100) was added and incubated overnight at 4°C in a wet box. The fluorescent secondary antibody (prepared in blocking solution, 1:1000) was added and the slides were incubated at room temperature for 1 h in the dark. Finally, the cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) at room temperature for 10 min in the dark and the images were collected under a laser confocal microscope. The following antibodies were used in this study: rabbit monoclonal anti-E-cadherin (3195T; Cell Signaling Technology), rabbit monoclonal anti-vimentin (5741T; Cell Signaling Technology) and Alexa Fluor 488 conjugated anti-rabbit IgG (4412S; Cell Signaling Technology).
Statistical Analysis
SPSS13.0 statistical software (IBM Corp., Armonk, NY, USA) and GraphPad Prism (version 5.0, GraphPad Inc., La Jolla, CA, USA) were used for statistical analysis. One-way analysis of variance (ANOVA) was used to determine the significance of differences among the groups. A p-value of 0.05 was considered statistically significant.
Results
Network Pharmacology-Based Target Prediction for Dioscin
To uncover the mechanism behind dioscin’s anti-tumor effects, especially EMT, we employed CTD, Similarity Ensemble Approach (SEA), Swiss Target Prediction, GeneCards, and STRING to identify the lung cancer-related targets of dioscin based on text-mining, ligand topology, and protein structure similarity. Forty-two lung cancer-related targets of Dioscin were identified (Table 1). The compound-target network was constructed and was shown in Figure 1A. Specifically, TP53, CASP3, AKT1, BCL2L1, MAPK3, VEGFA, MAPK8, CCND1, IL6, MAPK1, CASP9, CASP8, SRC, TNF, and mTOR represent the crucial targets of dioscin based on degree value (reflecting their level of connectivity within the network).
Table 1.
Predicted Lung Cancer-Related Targets of Dioscin
| No. | Gene Symbol | No. | Gene Symbol | No. | Gene Symbol |
|---|---|---|---|---|---|
| 1 | CASP3 | 15 | IL6 | 29 | IL1B |
| 2 | CASP9 | 16 | KRT18 | 30 | KDR |
| 3 | MAPK1 | 17 | AKT1 | 31 | KEAP1 |
| 4 | MAPK3 | 18 | BCL2L1 | 32 | MAPK8 |
| 5 | PARP1 | 19 | BIRC5 | 33 | mTOR |
| 6 | TNF | 20 | CASP8 | 34 | NFE2L2 |
| 7 | ABCB1 | 21 | CCNB1 | 35 | NFKBIA |
| 8 | BAK1 | 22 | CCND1 | 36 | PTK2 |
| 9 | BAX | 23 | CDK1 | 37 | RELA |
| 10 | BCL2 | 24 | CDKN1A | 38 | SRC |
| 11 | CTNNB1 | 25 | CHEK2 | 39 | TNFRSF10B |
| 12 | CYP2E1 | 26 | FAS | 40 | TNFRSF1A |
| 13 | ESR1 | 27 | FASLG | 41 | TP53 |
| 14 | ESR2 | 28 | HMOX1 | 42 | VEGFA |
Figure 1.
Analysis of dioscin compound-target network. (A) The overlapped targets of lung cancer and dioscin-related targets were analyzed by STRING to generate the compound-target network. The diameter of the nodes was determined by the degree of freedom. The larger the diameter, the more critical the node is in the network. (B) GO functional annotation of potential targets of dioscin. Biological processes (BP), Cellular components (CC) and Molecular functions (MF) were ranked according to -logP values. (C) KEGG enrichment analysis for potential targets of dioscin. The top 20 with lower P-value was shown.
We further used GO and KEGG pathway enrichment analyses to assess the characteristics of these protein targets. GO analysis showed that most of the targets were present in the cytosol and nucleus with protein homodimerization or heterodimerization activity. These targets were particularly associated with in the biological processes, such as extrinsic apoptotic signaling pathway via death domain receptors, intrinsic apoptotic signaling pathway in response to DNA damage, negative regulation of apoptotic process, and peptidyl-threonine phosphorylation (Figure 1B). In addition, the 42 targets were shown to contribute to 98 pathways using KEGG analysis. The 20 targets with the lowest p-values were shown in Figure 1C. Among them, the PI3K-Akt signaling pathway has been confirmed to be related to EMT and plays a crucial role in tumorigenesis and metastasis.20 AKT and mTOR are crucial participants in the PI3K-Akt signaling pathway in EMT.6,21 Notably, AKT and mTOR had a higher degree (reflecting greater network connectivity) in the compound-target analysis, as shown in Figure 1A. We therefore hypothesized the inhibition of EMT by dioscin occurred via targeting AKT and mTOR.
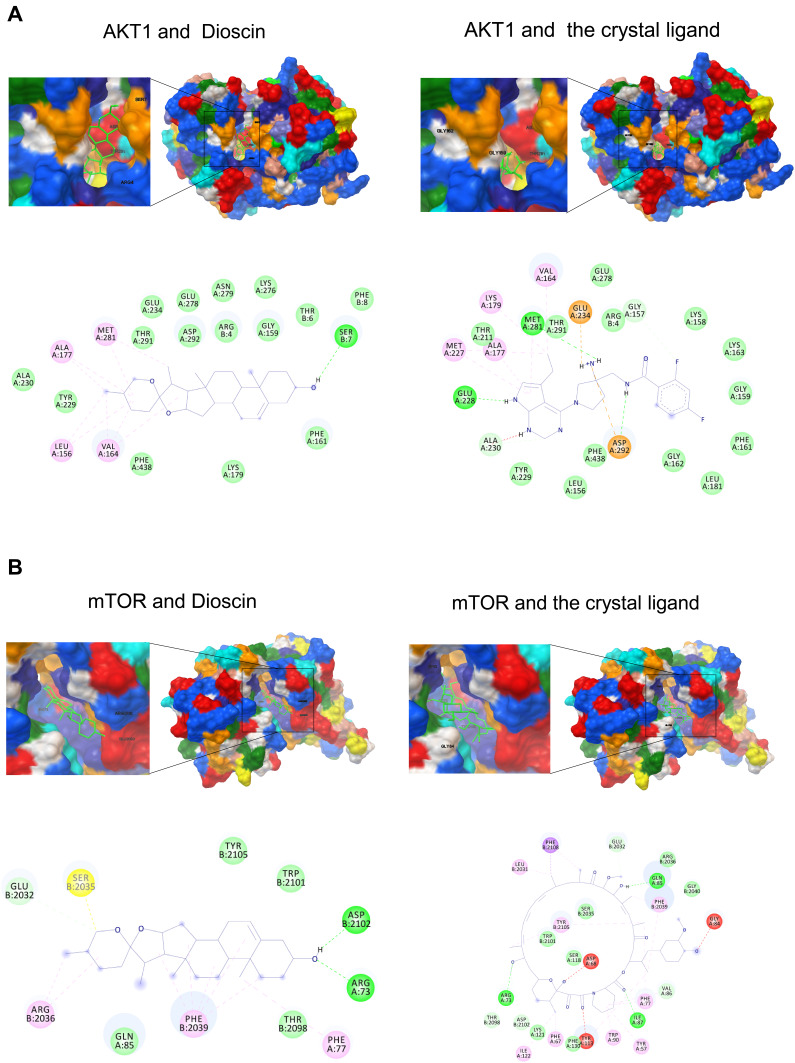
To address this hypothesis, we analyzed the interactions of AKT–dioscin and mTOR–dioscin using molecular docking. Our results showed that the binding energy of AKT–dioscin was −8.91 kcal/mol, compared with −10.89 for AKT and the original crystal structure ligand. The binding energy of mTOR-Dioscin was −9.62 kcal/mol, compared with −18.57 for mTOR and the crystal ligand. The AKT–dioscin and mTOR–dioscin docking models were shown in Figure 2A and B. These findings indicated that dioscin could possibly bind to AKT and mTOR.
Figure 2.
Modeling of dioscin with the potential targets. (A) Dioscin-AKT docking model with predicted binding energy of −8.91 kcal/mol. The Docking model of AKT and the crystal ligand has predicted binding energy of −10.89 kcal/mol. (B) Dioscin-mTOR docking model with predicted binding energy of −9.62 kcal/mol. The Docking model of AKT and the crystal ligand has predicted binding energy of −18.57 kcal/mol.
Dioscin Suppressed Proliferation, Migration, and Invasion of Human Lung Adenocarcinoma Cells
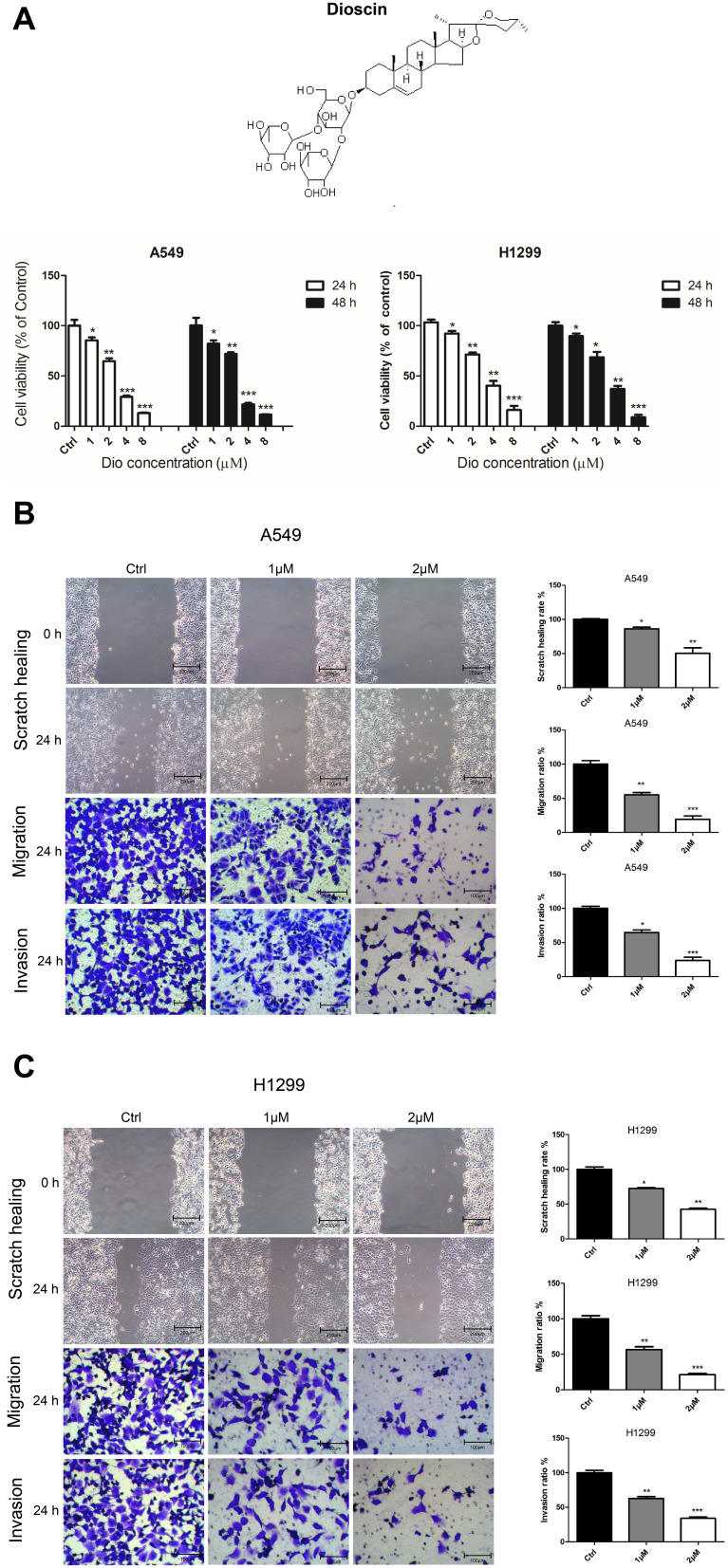
The cytotoxicity of dioscin (1, 2, 4, and 8µM) in human lung adenocarcinoma cells, A549 and H1299, was determined using a Cell Counting Kit-8 assay (CCK8) after 24 and 48 h. The results showed that, compared to the control, the inhibitory effects of 1 and 2 µM dioscin on A549 and H1299 cells were lower than 50% at a cell density >90% (Figure 3A). Therefore, 2 and 1 µM dioscin were selected as suitable doses in subsequent experiments to exclude interference from dioscin’s suppression of the proliferation of lung adenocarcinoma cells.
Figure 3.
Dioscin inhibited the proliferation, migration, and invasion of A549 and H1299 cells. (A) The cell viability of A549 and H1299 cells treated with dioscin (1, 2, 4, and 8 µM) for 24 h and 48 h was determined using a Cell Counting Kit 8 assay. The width of scratches was measured in cells treated or not with dioscin, at 0 and 24 h in A549 (B) and H1299 (C) cells. Additionally, using a Transwell migration assay and Matrigel invasion assay, the number of migrated or invaded cells treated with dioscin per chamber was counted and compared with that in the control group from three independent experiments (*p < 0.05; **p< 0.01; ***p< 0.001).
To determine whether dioscin affected the metastatic ability of A549 and H1299 cells, we performed scratch healing and Transwell assay experiments. The scratch healing assay showed that the scratches in dioscin‐treated A549 and H1299 cells were markedly larger than those in the control cells at 24 h (Figure 3B and C). Meanwhile, Transwell migration assays revealed that dioscin significantly suppressed the migration rates of A549 and H1299 cells, which could be observed by comparison with their corresponding control cells (Figure 3B and C). In addition, Matrigel invasion assays proved that dioscin can suppress the invasion abilities of A549 and H1299 cells by comparison with their corresponding control cells (Figure 3B and C). These observations confirmed that dioscin could effectively inhibit lung adenocarcinoma cell migration and invasion.
Dioscin Reversed EMT in Human Lung Adenocarcinoma Cells
To further determine how dioscin inhibited cell migration and invasion, we investigated whether it could reverse EMT. Using Western blotting and immunofluorescence assays, we detected the protein levels of EMT-associated biomarkers after dioscin treatment alone. Our data showed that the protein levels of N-cadherin, vimentin, MMP-9, β-catenin, zinc finger E-box binding homeobox 1 (ZEB1), SLUG (snail family transcriptional repressor 2), and SNAIL (snail family transcriptional repressor 1) were significantly reduced by dioscin stimulation in a dose-dependent manner, whereas the levels of E-cadherin, claudin-1, and ZO-1 (zona occludens 1) were markedly increased (Figure 4A). Besides, the immunofluorescent staining results showed that dioscin treatment clearly increased the intensive green fluorescence expression of the epithelial marker E-cadherin and significantly reduced the mesenchymal marker vimentin compared with those in control cells (Figure 4B). These findings indicated that EMT could be reversed by dioscin.
Figure 4.
Effect of dioscin on the expression of EMT markers in H1299 and A549 cells. (A) Cells were treated with dioscin (1 and 2 µM) for 24 h, and the EMT-associated biomarkers were detected using Western blotting. Typical graphs and a histogram (mean ± SD) are shown, n = 3. *p < 0.05; **p< 0.01; ***p< 0.001 compared with the control group. (B) After treatment with dioscin (1 and 2 µM) for 24 h, immunofluorescence staining was performed to determined Vimentin and E-cadherin expression in H1299 and A549 cells using confocal microscopy. Blue fluorescence indicates DAPI-labeled nuclei, and Green fluorescence indicates Vimentin or E-cadherin positive expression.
The Inhibitory Effect of Dioscin on EMT in Human Lung Adenocarcinoma Cells via the AKT/GSK3β/mTOR Signaling Pathways
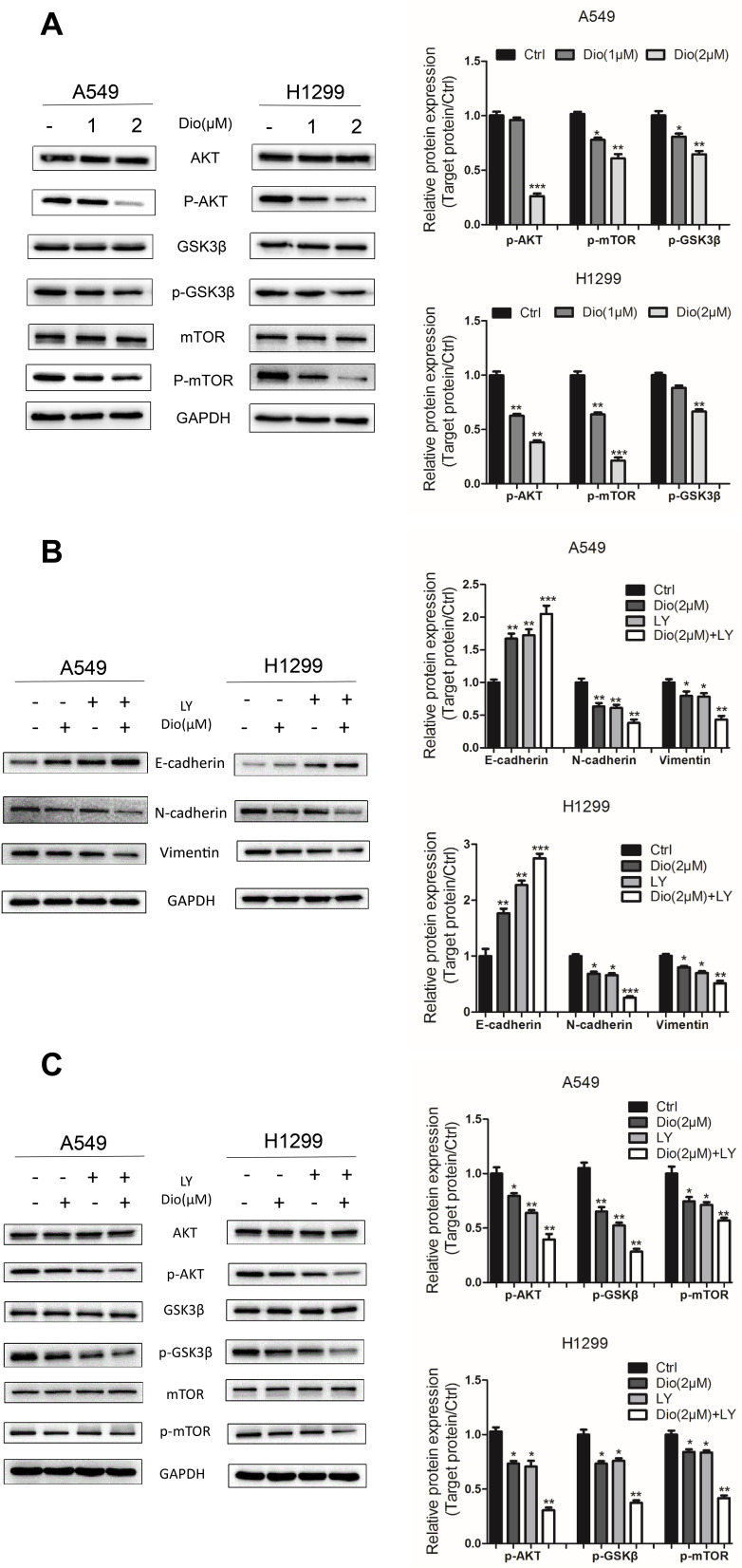
To test whether the binding of AKT and mTOR to dioscin affects their function, we detected the protein expression of AKT, phosphorylated AKT (p-AKT), mTOR, and phosphorylated mTOR (p-mTOR) in the selected cells after dioscin treatment. Our data showed dioscin markedly downregulated p-AKT and p-mTOR in both H1299 and A549 cells in a dose-dependent manner, without altering their total protein level (Figure 5A). p-GSK3β is a downstream protein of p-AKT in EMT.5 Consistent with this, dioscin reduced p-GSK3β without affecting GSK3β total protein (Figure 5A).
Figure 5.
Effects of dioscin on the AKT/GSK3β/mTOR pathway in A549 and H1299 cells. (A) Cells were treated with dioscin (1 and 2 µM) for 24 h. The levels of AKT/GSK3/mTOR related pathway proteins, including phosphorylated (p)-AKT, AKT, p-GSK3, GSK3, p-mTOR, and mTOR, were examined using Western blotting analysis. (B, C) Cells were pre-treated with 10 µM LY294002 (a PI3K/AKT inhibitor, LY) for 2 h and then treated with dioscin (1 or 2 µM) for 24 h. Western blotting was then employed to determine the protein levels of Vimentin, E-cadherin, and N-cadherin (B) and p-AKT, AKT, p-GSK3, GSK3, p-mTOR, and mTOR (C). GAPDH was used as a loading control. Typical graphs and a histogram (mean±SD) are shown, n=3. *p < 0.05; **p< 0.01; ***p< 0.001 compared with the control group.
We further explored the effect of dioscin on the AKT/GSK3β/mTOR signaling pathways using the specific pharmacological inhibitor LY294002 (LY, a PI3K inhibitor). A549 and H1299 cells were pre-incubated with 10 µM LY29400222 for 2h and then co-incubated with Dioscin (1 and 2 µM) for 24 h. The dioscin-mediated upregulation of E-cadherin and downregulation of N-cadherin and vimentin were augmented by LY treatment in A549 and H1299 cells (Figure 5B). Similarly, dioscin-mediated downregulation of p-AKT, p-GSK3β, and p-mTOR was augmented by LY treatment (Figure 5C).
Collectively, these findings revealed that dioscin suppresses EMT of lung adenocarcinoma cells by inactivating of the AKT/GSK3β/mTOR signaling pathway.
Discussion
Most patients with lung cancer are diagnosed with non-small-cell lung cancer (NSCLC), of which lung adenocarcinoma represents the major subtype. Metastasis is the major cause of the high mortality in lung cancer patients. In terms of the factors affecting metastasis, it is closely related to tumor size, growth rate, and invasiveness. In recent years, increasing evidence has shown that EMT is involved in the metastatic process.23 Moreover, many studies have underlined the contribution of EMT to lung cancer.24,25 EMT is the method by which epithelial cells acquire migration ability, and has been revealed as an important pathway by which more than 90% of malignant epithelial cells undergo carcinogenesis in adult humans. Therefore, the development of a new anti-metastasis drug for EMT-related pathways is of great significance to treat patients with metastatic lung adenocarcinoma.
During EMT, tumor cells downregulate epithelial markers (E-cadherin) and tight junction proteins (ZO-1 and claudin-1), as well as upregulating mesenchymal markers (N-cadherin and vimentin) and transcription factors (SNAIL, SLUG, and ZEB1). E-cadherin is a typical epithelial marker of EMT and is thought to be a switch for EMT. When tumor cells showed a downregulation of E-cadherin expression, β-catenin from the membrane was seen to relocate to the nucleus, causing a high expression of interstitial cell markers such as N-cadherin and vimentin, which promote tumor cell metastasis.26 Studies have shown that many signaling pathways are involved in EMT, including transforming growth factor beta (TGF-β), Wnt/β-catenin, PI3K/AKT/mTOR, and AKT/GSK3β/β-catenin signaling pathways.27–31 AKT, known as protein kinase B (PKB), is a downstream signaling molecule of PI3K (Phosphatidylinositol-3 kinase). GSK3β mediates the phosphorylation of Snail, and increases its cytoplasmic retention and degradation. AKT promotes the phosphorylation of GSK3β to ubiquitinate and degrade, thereby stabilizing Snail and enhancing the inhibition of E-cadherin expression.32 AKT directly phosphorylates mTOR on Ser(2448).33 mTOR can integrate multiple signal stimuli to affect transcription and protein synthesis, which have important links with cell proliferation, apoptosis, differentiation and EMT.6 Tumor cells also increase the secretion of matrix metalloproteinases (MMPs), which leads to the degradation of extracellular matrix components, the remodeling of the cytoskeleton, and the invasion and metastasis of tumor cells from the original site.34
Dioscin is an extracted component of Chinese yam, which was also called “Dioscorea nipponica” in ancient times. Dioscin has a wide range of effects, including lipid-lowering,35 antihypertensive,36 anti-inflammatory,37 desensitizing,38 liver protective,39 antiviral,40 anti-tumor,7,8 and immunoregulatory activities.41 Recently, several studies have revealed that dioscin reverses TGF-β1 induced epithelial–mesenchymal transition (EMT).9,10 In the current investigation, we observed dioscin’s remarkable effects on upregulating E-cadherin and tight junction proteins (ZO-1 and claudin-1), downregulating N-cadherin and vimentin and its transcription factors (SNAIL, SLUG, and ZEB1), as well as β-catenin without TGF-β stimulation, which suggests that dioscin could reverse EMT by other pathways than the participation of TGF-β and its receptors.
Network-based pharmacology, derived from systems biology, which can address the connectivity and interdependence of multiple drug targets, is commonly used to decipher mechanisms of drug activity and drug-target relationships. It shares a holistic philosophy similar to that of Traditional Chinese Medicine (TCM).42,43 Using network-based pharmacological approaches, we found that the lung cancer-related targets of Dioscin were involved in pathways including the TNF signaling pathway, PIK3-AKT signaling pathway, NOD-like receptor signaling pathway, and HIF-1 signaling pathway. As AKT and mTOR had higher degrees in the compound-target network, we focused on the AKT-mTOR signaling pathway. Subsequently, our experiments revealed that dioscin downregulated the phosphorylation levels of AKT, mTOR, and GSK3β in a dose-dependent manner, which suggested that dioscin could inhibit their phosphorylation. Furthermore, we used LY294002, the PI3K inhibitor, which has been shown to block PI3 kinase-dependent Akt phosphorylation and kinase activity.44 We found that the effects of dioscin on the AKT/GSK3β/mTOR signaling pathway were similar to those of LY294002 (Figure 5), which could support the inhibition of AKT phosphorylation by dioscin. Interestingly, we found that LY294002 enhanced the effect of dioscin on p-AKT, p-mTOR, and p-GSK3β. This also suggested that the manner of inhibition of AKT phosphorylation by dioscin differed from that of LY294002. The molecular docking analysis in the current study showed that dioscin could bind to AKT and mTOR. However, more experiments are needed to further validate the binding of dioscin-AKT and dioscin-mTOR, and to explore whether AKT or mTOR is the direct target of dioscin in the future.
Conclusion
Our results suggest that dioscin restrains the migration and invasion, and converts EMT through regulating the AKT/GSK3β/mTOR signaling pathway, likely by binding to AKT, and mTOR and inhibiting their phosphorylation. Thus dioscin has a potential therapeutic value and may provide a theoretical basis for further clinical treatment of lung adenocarcinoma.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 81873154) and the 2017-annual project of construction of high-level university (Grant No. A1-AFD018171Z11003). We thank Professor Guoan Chen from South University of Science and Technology for his revision of the manuscript.
Abbreviations
EMT, epithelial–mesenchymal transition; DMSO, dimethyl sulfoxide; OD, optical density; MMP, matrix metalloproteinases; PVDF, polyvinylidene fluoride; DAPI, 4’,6-diamidino-2-phenylindole; ANOVA, one-way analysis of variance; PPI, protein-protein interaction; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; CCK8, Cell Counting Kit-8 assay; Dio, dioscin; ZEB1, E-box binding homeobox 1; SLUG, snail family transcriptional repressor 2; SNAIL, snail family transcriptional repressor 1; ZO-1, zona occludens 1; LY, LY294002; SEA, Similarity Ensemble Approach; NSCLC, non-small-cell lung cancer; p-AKT, phosphorylated AKT; p-mTOR, phosphorylated mTOR; p-GSK3β, phosphorylated GSK3β; TGF-β, transforming growth factor beta; FCS, fetal calf serum.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 2.Sung WJ, Kim H, Park KK. The biological role of epithelial-mesenchymal transition in lung cancer (Review). Oncol Rep. 2016;36(3):1199–1206. doi: 10.3892/or.2016.4964 [DOI] [PubMed] [Google Scholar]
- 3.Thompson EW, Newgreen DF, Tarin D. Carcinoma invasion and metastasis: a role for epithelial-mesenchymal transition? Cancer Res. 2005;65(14):5991–5995. doi: 10.1158/0008-5472.CAN-05-0616 [DOI] [PubMed] [Google Scholar]
- 4.Feng YL, Chen DQ, Vaziri ND, Guo Y, Zhao YY. Small molecule inhibitors of epithelial-mesenchymal transition for the treatment of cancer and fibrosis. Med Res Rev. 2019;40(1):54–78. [DOI] [PubMed] [Google Scholar]
- 5.Lin Y, Yang Z, Xu A, et al. PIK3R1 negatively regulates the epithelial-mesenchymal transition and stem-like phenotype of renal cancer cells through the AKT/GSK3β/CTNNB1 signaling pathway. Sci Rep. 2015;5(1):8997. doi: 10.1038/srep08997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karimi Roshan M, Soltani A, Soleimani A, Rezaie Kahkhaie K, Afshari AR, Soukhtanloo M. Role of AKT and mTOR signaling pathways in the induction of epithelial-mesenchymal transition (EMT) process. Biochimie. 2019;165:229–234. doi: 10.1016/j.biochi.2019.08.003 [DOI] [PubMed] [Google Scholar]
- 7.Chen H, Xu L, Yin L, et al. iTRAQ-based proteomic analysis of dioscin on human HCT-116 colon cancer cells. Proteomics. 2014;14(1):51–73. doi: 10.1002/pmic.201300101 [DOI] [PubMed] [Google Scholar]
- 8.Zhang G, Zeng X, Zhang R, et al. Dioscin suppresses hepatocellular carcinoma tumor growth by inducing apoptosis and regulation of TP53, BAX, BCL2 and cleaved CASP3. Phytomedicine. 2016;23(12):1329–1336. doi: 10.1016/j.phymed.2016.07.003 [DOI] [PubMed] [Google Scholar]
- 9.Chen B, Zhou S, Zhan Y, et al. Dioscin inhibits the invasion and migration of hepatocellular carcinoma hepG2 cells by reversing TGF-β1-induced epithelial-mesenchymal transition. Molecules. 2019;24(12):222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim WC, Kim H, Kim YJ, et al. Dioscin suppresses TGF-β1-induced epithelial-mesenchymal transition and suppresses A549 lung cancer migration and invasion. Bioorg Med Chem Lett. 2017;27(15):3342–3348. doi: 10.1016/j.bmcl.2017.06.014 [DOI] [PubMed] [Google Scholar]
- 11.Davis AP, Grondin CJ, Johnson RJ, et al. The comparative toxicogenomics database: update 2019. Nucleic Acids Res. 2019;47(D1):D948–d954. doi: 10.1093/nar/gky868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007;25(2):197–206. doi: 10.1038/nbt1284 [DOI] [PubMed] [Google Scholar]
- 13.Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357–w364. doi: 10.1093/nar/gkz382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Safran M, Dalah I, Alexander J, et al. GeneCards Version 3: the human gene integrator. Database (Oxford). 2010;2010:baq020. doi: 10.1093/database/baq020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.UniProt C. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1):D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–d613. doi: 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim S, Chen J, Cheng T, et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47(D1):D1102–d1109. doi: 10.1093/nar/gky1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burley SK, Berman HM, Bhikadiya C, et al. RCSB protein data bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019;47(D1):D464–d474. doi: 10.1093/nar/gky1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ginnebaugh KR, Ahmad A, Sarkar FH. The therapeutic potential of targeting the epithelial-mesenchymal transition in cancer. Exp Opin Ther Targets. 2014;18(7):731–745. doi: 10.1517/14728222.2014.909807 [DOI] [PubMed] [Google Scholar]
- 21.Fernandez-Rozadilla C, Alvarez-Barona M, Schamschula E, et al. Early colorectal cancers provide new evidence for a lynch syndrome-to-CMMRD phenotypic continuum. Cancers (Basel). 2019;11(8):1081. doi: 10.3390/cancers11081081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shao L, Li H, Chen J, et al. Irisin suppresses the migration, proliferation, and invasion of lung cancer cells via inhibition of epithelial-to-mesenchymal transition. Biochem Biophys Res Commun. 2017;485(3):598–605. doi: 10.1016/j.bbrc.2016.12.084 [DOI] [PubMed] [Google Scholar]
- 23.Williams ED, Gao D, Redfern A, Thompson EW. Controversies around epithelial-mesenchymal plasticity in cancer metastasis. Nat. Rev. Cancer. 2019;19(12):716–732. doi: 10.1038/s41568-019-0213-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Matteis S, Canale M, Verlicchi A, et al. Advances in molecular mechanisms and immunotherapy involving the immune cell-promoted epithelial-to-mesenchymal transition in lung cancer. J Oncol. 2019;2019:7475364. doi: 10.1155/2019/7475364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tulchinsky E, Demidov O, Kriajevska M, Barlev NA, Imyanitov E. EMT: a mechanism for escape from EGFR-targeted therapy in lung cancer. Biochim Biophys Acta Rev Cancer. 2019;1871(1):29–39. doi: 10.1016/j.bbcan.2018.10.003 [DOI] [PubMed] [Google Scholar]
- 26.Scanlon CS, Van Tubergen EA, Inglehart RC, D’Silva NJ. Biomarkers of epithelial-mesenchymal transition in squamous cell carcinoma. J Dent Res. 2013;92(2):114–121. doi: 10.1177/0022034512467352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu YS, Chung I, Wong WF, Masamune A, Sim MS, Looi CY. Paracrine IL-6 signaling mediates the effects of pancreatic stellate cells on epithelial-mesenchymal transition via Stat3/Nrf2 pathway in pancreatic cancer cells. Biochim Et Biophys Acta General Sub. 2017;1861(2):296–306. doi: 10.1016/j.bbagen.2016.10.006 [DOI] [PubMed] [Google Scholar]
- 28.He S, Li Z, Yu Y, et al. Exosomal miR-499a-5p promotes cell proliferation, migration and EMT via mTOR signaling pathway in lung adenocarcinoma. Exp Cell Res. 2019;379(2):203–213. doi: 10.1016/j.yexcr.2019.03.035 [DOI] [PubMed] [Google Scholar]
- 29.Luo Y, Ren Z, Du B, et al. Structure identification of viceninII extracted from and the reversal of TGF-β1-Induced epithelial⁻mesenchymal transition in lung adenocarcinoma cells through TGF-β/smad and PI3K/Akt/mTOR signaling pathways. Molecules. 2019;24(1):144. doi: 10.3390/molecules24010144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hseu Y-C, Lin Y-C, Rajendran P, et al. Antrodia salmonea suppresses invasion and metastasis in triple-negative breast cancer cells by reversing EMT through the NF-κB and Wnt/β-catenin signaling pathway. Food Chem Toxicol. 2019;124:219–230. doi: 10.1016/j.fct.2018.12.009 [DOI] [PubMed] [Google Scholar]
- 31.Song N, Zhong J, Hu Q, et al. FGF18 enhances migration and the epithelial-mesenchymal transition in breast cancer by regulating Akt/GSK3β/Β-catenin signaling. Cell Physiol Biochem. 2018;49(3):1019–1032. doi: 10.1159/000493286 [DOI] [PubMed] [Google Scholar]
- 32.Zhou BP, Deng J, Xia W, et al. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004;6(10):931–940. doi: 10.1038/ncb1173 [DOI] [PubMed] [Google Scholar]
- 33.Navé BT, Ouwens M, Withers DJ, Alessi DR, Shepherd PR. Mammalian target of rapamycin is a direct target for protein kinase B: identification of a convergence point for opposing effects of insulin and amino-acid deficiency on protein translation. Biochem J. 1999;344(Pt 2):427–431. doi: 10.1042/bj3440427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoo YA, Kang MH, Lee HJ, et al. Sonic hedgehog pathway promotes metastasis and lymphangiogenesis via activation of Akt, EMT, and MMP-9 pathway in gastric cancer. Cancer Res. 2011;71(22):7061–7070. doi: 10.1158/0008-5472.CAN-11-1338 [DOI] [PubMed] [Google Scholar]
- 35.Kwon CS, Sohn HY, Kim SH, et al. Anti-obesity effect of Dioscorea nipponica Makino with lipase-inhibitory activity in rodents. Biosci Biotechnol Biochem. 2003;67(7):1451–1456. doi: 10.1271/bbb.67.1451 [DOI] [PubMed] [Google Scholar]
- 36.Sautour M, Mitaine-Offer AC, Miyamoto T, Dongmo A, Lacaille-Dubois MA. A new steroidal saponin from Dioscorea cayenensis. Chem Pharm Bull (Tokyo). 2004;52(11):1353–1355. doi: 10.1248/cpb.52.1353 [DOI] [PubMed] [Google Scholar]
- 37.Li K, Wang Y, Gu J, Chen X, Zhong D. Determination of dioscin in rat plasma by liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;817(2):271–275. doi: 10.1016/j.jchromb.2004.12.026 [DOI] [PubMed] [Google Scholar]
- 38.Cho J, Choi H, Lee J, Kim MS, Sohn HY, Lee DG. The antifungal activity and membrane-disruptive action of dioscin extracted from Dioscorea nipponica. Biochim Biophys Acta. 2013;1828(3):1153–1158. doi: 10.1016/j.bbamem.2012.12.010 [DOI] [PubMed] [Google Scholar]
- 39.Zhao X, Cong X, Zheng L, Xu L, Yin L, Peng J. Dioscin, a natural steroid saponin, shows remarkable protective effect against acetaminophen-induced liver damage in vitro and in vivo. Toxicol Lett. 2012;214(1):69–80. doi: 10.1016/j.toxlet.2012.08.005 [DOI] [PubMed] [Google Scholar]
- 40.Aquino R, Conti C, De Simone F, Orsi N, Pizza C, Stein ML. Antiviral activity of constituents of Tamus communis. J Chemother. 1991;3(5):305–309. doi: 10.1080/1120009X.1991.11739110 [DOI] [PubMed] [Google Scholar]
- 41.Li C, Lu Y, Du S, et al. Dioscin exerts protective effects against crystalline silica-induced pulmonary fibrosis in mice. Theranostics. 2017;7(17):4255–4275. doi: 10.7150/thno.20270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee WY, Lee CY, Kim YS, Kim CE. The methodological trends of traditional herbal medicine employing network pharmacology. Biomolecules. 2019;9(8):362. doi: 10.3390/biom9080362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang R, Zhu X, Bai H, Ning K. Network pharmacology databases for traditional chinese medicine: review and assessment. Front Pharmacol. 2019;10:123. doi: 10.3389/fphar.2019.00123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niehrs C. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol. 2012;13(12):767–779. doi: 10.1038/nrm3470 [DOI] [PubMed] [Google Scholar]