Abstract
Group A rotavirus (RVA) and bovine coronavirus (BCoV) are the two main viral enteropathogens associated with neonatal calf diarrhea. The aim of the present survey was to investigate the epidemiology and the role of RVA and BCoV in the presentation of dairy and beef calf diarrhea in Lerma Valley of Salta province, within the Northwest region of Argentina. Stool samples of calves with or without diarrhea younger than 2 months of age were collected from 19 dairy farms and 20 beef farms between the years 2014 and 2016. Stool samples were screened for RVA and BCoV detection by ELISA. Heminested multiplex RT-PCR was used for RVA typing and RT-PCR to confirm BCoV. Positive samples were submitted to sequencing analysis. Bovine RVA and BCoV were circulating in 63% (12/19) and 10.52% (2/19) of the dairy farms, respectively, where 9.5% (46/484) of the calves were positives to RVA and 0.4% (2/484) to BCoV. In beef herds, RVA was detected in 40% (8/20) of the farms and in 6.75% (21/311) of the calves, without positives cases of BCoV. Molecular analysis showed that in dairy farms, G6P[11] and G10P[11] were the prevalent RVA strains, while in beef farms, G10P[11] was the prevalent. The main finding was the detection for the first time of a G15P[11] causing diarrhea in beef calves of Argentina that represents a new alert to be consider for future vaccine updates. Analysis of detected BCoV showed that it is related to the other circulating strains of Argentina.
Keywords: Argentina, Coronavirus, Neonatal calf diarrhea, Rotavirus
Introduction
Group A rotavirus (RVA) and bovine coronavirus (BCoV) are the two main viral enteropathogens associated with neonatal calf diarrhea (Afshari Safavi et al. 2012). Rotavirus is a non-enveloped virus, with a triple-layered capsid with eleven segments of double-stranded RNA. In bovine hosts, only G6, G8, and G10 RVAs associated with P[5], P[11], and P[1] were found to be epidemiologically relevant (Badaracco et al. 2013a). Bovine CoV is an enveloped virus that possesses a non-segmented single-stranded RNA genome of positive polarity. The ribonucleoprotein is formed by the RNA genome with N nucleoprotein in helical symmetry with an envelope constituted with several membrane proteins (M, E, HE, S) (Bok et al. 2015). For molecular characterization, the hypervariable region of the subunits 1 (N-terminal half) of the S protein is commonly used (Brandão et al. 2006).
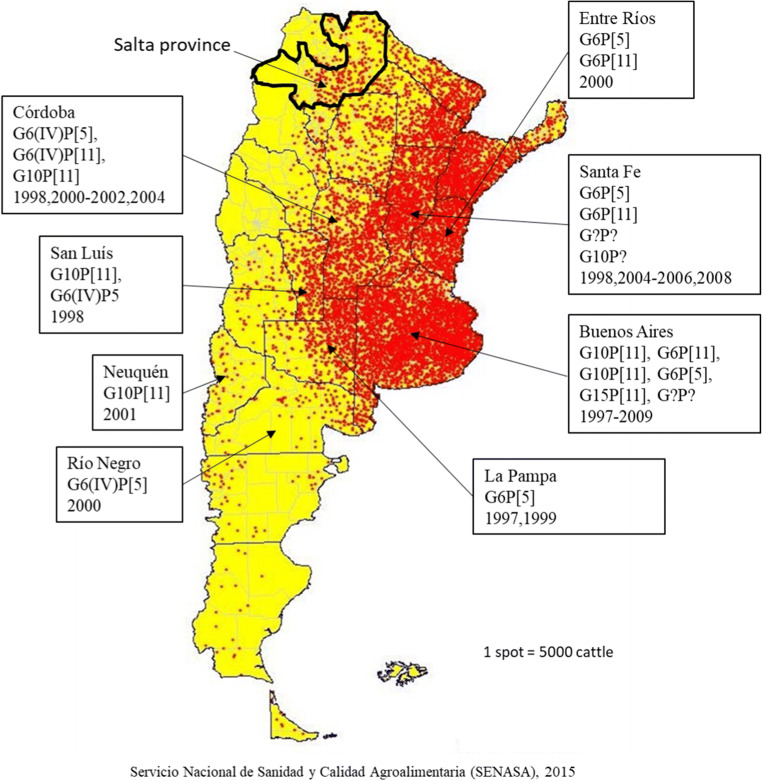
At present, the surveys of bovine RVA and BCoV in Argentina come from herds located in the Pampa, Mesopotamic, Patagonic, and Cuyo regions (Fig. 1b), where the most widely distributed genotypes of bovine RVA were G6 and G10 associated to P[5] and P[11], where the combinations G6P[11] and G10P[11] were found in dairy herds, while in beef farms, G6P[5] was the prevalent strain (Badaracco et al. 2013a, b). Phylogenetics studies of BCoV from Argentinean strains showed that they are distantly related to Mebus reference strain included in the available commercial vaccines, but all strains conform a unique serogroup (Bok et al. 2015).
Fig. 1.
Salta province and distribution of bovine stocks in Argentina. Black arrows pointing different provinces of Argentina with RVA circulation
The objective of this work was to study the relevance of RVA and BCoV and their association to diarrheas in beef and dairy calves from herds of Lerma Valley, province of Salta, in the northwest of Argentina.
Materials and methods
Lerma Valley is located in Salta province from Argentina, covering an area of 17,000 Km2, between 1,100 and 1,450 m above sea level (Martinez 2015) (Fig. 1). There are a total of 51 dairy farms (32 commercial and the rest considered small subsistence economies), with approximately 6500 Holstein cows in milking each (Suarez and Martinez 2015). Beef herds represent 11% of the total bovine stock of the province where farmers breed Bradford, Brangus, and autochthonous cattle under extensive production systems, where they do not use vaccines to prevent neonatal calf diarrhea.
To conduct the survey, the size of the sample was estimated using the proMESA.mdb software with 95% confidence interval and 20% standard error considering a 14% diarrhea rate over a total of 3615 dairy calves while in beef herds over a total of 1055 calves with a 25% diarrhea rate. The estimated sample size was 508 and 227 samples from dairy and beef herds, respectively (Geurden et al. 2008). Between the years 2014 and 2016, calves with or without diarrhea younger than 2 months of age from each farm were randomly selected and sampled (19 dairy farms and 20 beef farms). Samples were taken directly from the rectum. One aliquot of each sample was stored in vials in a 10% (W/V) suspension in phosphate-buffered saline pH 7.4 (PBS) and preserved at – 70 and – 20 °C until BCoV and RVA detection, respectively.
The RVA antigen detection was carried out using a polyclonal double sandwich ELISA (Garaicoechea et al. 2006), and BCoV antigen detection was conducted by a monoclonal capture ELISA kindly supplied by Dr. LJ Saif, Food Animal Health Research Program, The Ohio State University, USA (Bok et al. 2015). Rotaviruses G-, P- and E-type were determined by heminested multiplex RT-PCR as previously described (Gouvea et al. 1994; Badaracco et al. 2012; Louge Uriarte et al. 2014). Coronavirus was determined by RT-PCR of S1 and S2 glycoprotein gene following standard protocols described elsewhere (Bok et al. 2015).
Bovine RVA (INDIANA, G6(IV)P[5]; B223, G10P[11]; NCDV-Cody I801, G8P[1]) and BCoV (Mebus) were included as positive controls for typing analysis. The Argentinean field strains RVA/Cow-wt/ARG/B61/1997/G6P[11] (Garaicoechea et al. 2006) and BCoV Arg95 (Bok et al. 2015) were also included as local positive controls. The nucleotide sequencing of VP7 and VP8 encoding genes of selected samples was performed in an ABI-Prism 377 DNA sequencer (Perkin-Elmer, Applied Biosystems) at the sequencing service of the Biotechnology Institute (INTA, Argentina), with the same primers used in the RVA and BCoV RT-PCR assays.
The accession numbers for the RVA G6 sequences are MN177508-MN177509, for the G10 sequences are MN177510-MN177512, for the G15 sequences are MN177505- MN177507, for the P[11] sequences are MN177513-MN177517, for the E12 sequences are MN199234, and for BCoV S2 gene is MN199235.
Sequences were aligned using default parameters of BioEdit 7.1.3.0 (Hall 1999). The best-fit model of sequence evolution was estimated (Kalyaanamoorthy et al. 2017) for an inferred maximum likelihood tree using MEGA software versión 7.0.21 (Kumar et al. 2016) and IQ-TREE multicore version 1.6.7 using 1.000 bootstrap replicates as statistical support (Hoang et al. 2017). Trees were edited and drawn with FigTree 1.4.3 (http://figtree.googlecode.com/).
The comparisons between neonatal calf diarrhea and virus infection rates found in dairy vs. beef herds were evaluated by Chi-square method. Risk factors’ significance were evaluated by Fisher’s Exact Test using the GraphPad Prism version® for Windows 5.00 statistical software; p value < 0.05 was considered for significance.
Results
The total samples collected at the end of the study covered 97% (794/819) of the estimated sample size (dairy, 484 calves, 19 farms; beef, 311 calves, 20 farms). The amount of calves sampled represented 17% of the total estimated stock of calves in the Lerma Valley at that moment and 59% (19 out of 32) of the commercial dairy farms of the Valley. Regarding the beef farms, we were able to study 20 herds including the largest farm in the region and other different herds considered small subsistence economies. Bovine RVA rate was 9.5 (46/484) and 63% (12/19) of the calves and dairy farms, respectively, while BCoV rates in calves and dairy farms were 0.4 (2/484) and 10.5% (2/19), respectively. In beef herds, RVA rate was 40% (8/20) and in beef calves was 6.7% (21/311). There was no detection BCoV in beef farms. Of the dairy and beef calves shedding RVA, 58.7 (27/46) and 38.1% (8/21) were diarrheic, respectively, and they showed higher risk of suffering diarrhea than not infected calves OR 2.8 (p = 0.001, two-sided) and OR 2.09 (p = 0.04, two-sided), respectively.
In dairy farms, 50% (6/12) of samples were classified as G6P[11] and G10P[11]. A mixed infection of G6 + G10P[11] was detected only in one case. Partial typing (G?P[11]) was encountered once. In beef farms, G10P[11] was the prevalent strain (38% 3/8), while G6P[11] was 12.5% (1/8), mixed infections were 12.5% (1/8), and co-infections was 25% (2/8). Typing results of the VP7 and VP4 encoding genes of RVA strains were confirmed by sequence analysis, and 13 sequences were obtained (Table 1). Circulation of a G15P[11] strain was detected in one beef herd, where 62% (20/32) of the calves were diarrheic of which 30% (6/20) were positive to RVA and three samples (SVLG4, SVLG5, and SVLG12) were confirmed as G15P[11] by sequence analysis.
Table 1.
RVA detection rate and odds ratio associated to neonatal calf diarrhea and RVA G and P strain typing characterization in dairy and beef farms of Lerma Valley, Salta Province, Argentina (from 2014 to 2016)
| Productive system | RVA positives calves with diarrhea | Risk of diarrhea | RVA Genotypes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Odds ratio | CI 95% | p value | G6P[11] | G10P[11] | G?P[11] | *Mixed infection | **Co-infection | Untyped samples (negative) | Total | Sequences analyzed by farm | ||
| Dairy | 27/46 | 2.8 | 1.5, 5.2 | 0.001 | 6 | 1 | 1 | 1* | 0 | 3 | 12 | 7 |
| Beef | 8/21 | 2.1 | 1.1, 3.8 | 0.04 | 1 | 3 | 0 | 1** | 2*** | 1 | 8 | 6 |
| Total | 35/67 | – | – | – | 7 | 4 | 1 | 2 | 2 | 4 | 20 | 13 |
CI 95%, confidence interval 95%
Mixed infection, two calves in the same herd excreting different strains: * G10P[11] and G6P[11]; ** G15P[11] and G6P[11]
Co-infection, one calf excreting two strains: *** G10 P[11] + G6 P[11]
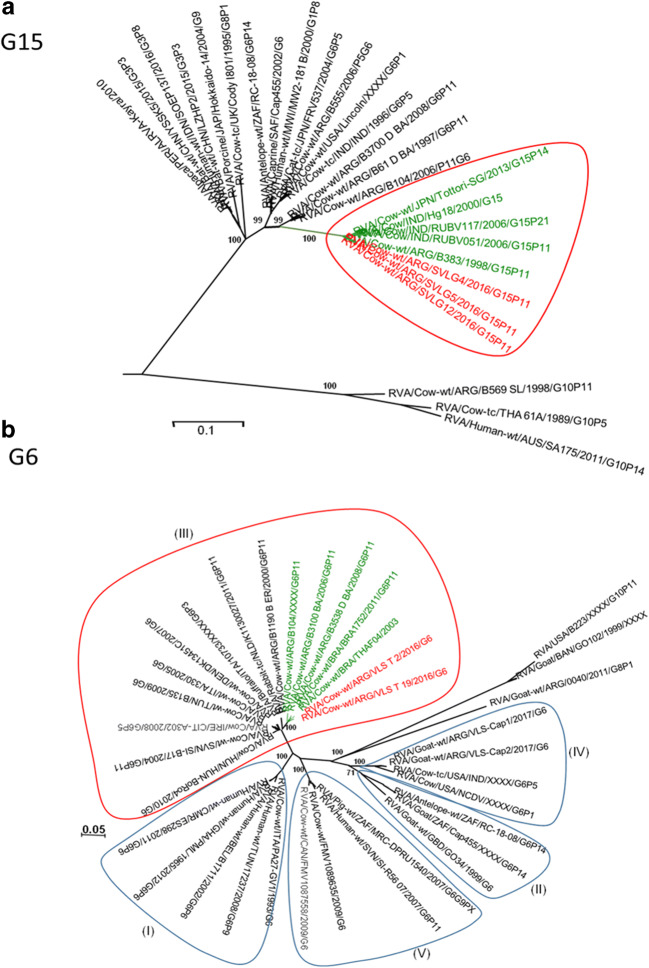
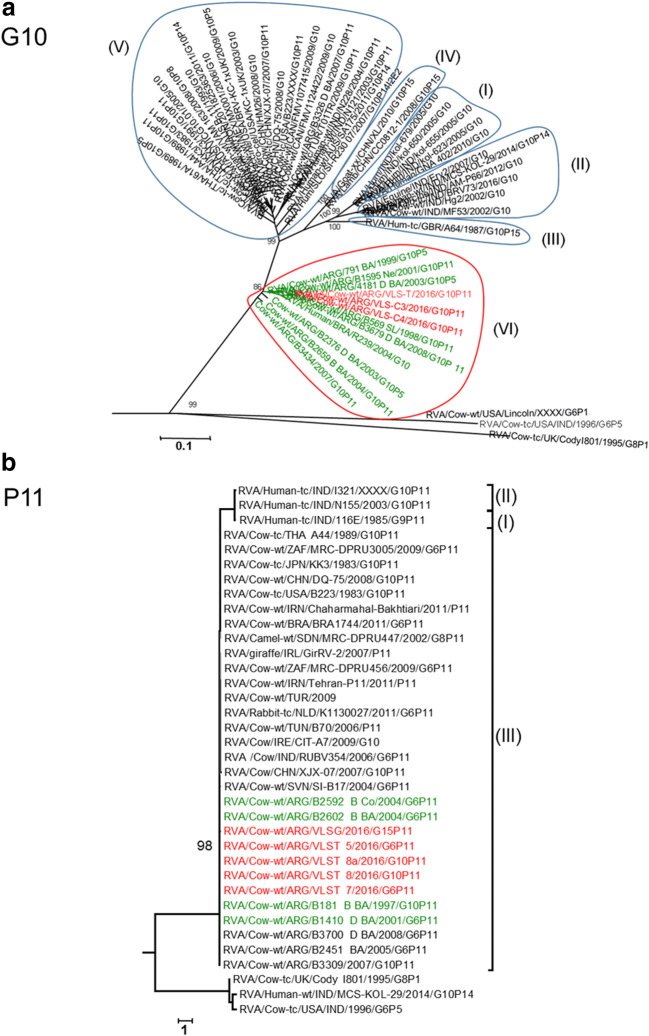
The phylogenetic analysis of the G15 strains showed that they clustered together in a monophyletic group (bootstrap of 100%). These viruses were different from the previously G15 strains detected in India (85% of similarity) and Japan (87% of similarity) (Fig. 2a). The phylogenetic analysis showed that VLST_2 and VLST_19 strains belonged to G6 genotype and clustered (bootstrap of 99%) within lineage III (Hun-4 like) (Fig. 2b), and G10 genotype strains showed that one strain from a dairy farm (VLS–T) and two strains from beef farms (VLS-C3 and VLS-C4) grouped within the IV lineage, together with strains from Argentina and Brazil (bootstrap of 86%) (Fig. 3a).
Fig. 2.
a Phylogenetic tree of the G15 genotype constructed with the NJ method (bootstrap of 1000). Nucleotide sequences are obtained in this study and GenBank available sequences. Red semicircle shows where Salta bovine strains grouped. Red letters are bovine strains from Salta. b Phylogenetic tree of the G6 genotype is constructed with the NJ method (bootstrap of 1000). Nucleotide sequences from strains are obtained in this survey and GenBank available sequences. Semicircles show different G6 lineages. Red semicircle shows where Salta bovine strains grouped. Red letters are bovine RVA strains from Salta
Fig. 3.
a Phylogenetic tree of the G10 genotype constructed with the NJ method (bootstrap of 1000). Nucleotide sequences are obtained in this study and GenBank available sequences. Brackets on the right show different G10 lineages. Semicircles show different G10 lineages. Red semicircle shows where Salta bovine strains grouped. Red letters are bovine strains from Salta. b Phylogenetic tree of the P[11] genotype is constructed with the NJ method and a bootstrap of 1000, showing amino acid sequences obtained in this study and Gen Bank available sequences. Brackets in the right show different P[11] lineages. Blue square shows where Salta bovine strains grouped. Red letters are bovine RVA strains from Salta
The phylogenetic analysis of the VP4 genes shows that all strains from dairy (VLST_5; VLST_7; VLST_8a; VLST_8) and beef (VLSG- G15P[11]) farm from Salta belonged to P[11] genotype. All strains were grouped in a single branch within the lineage III (bootstrap 98%), whit other strains detected in Argentina (Fig. 3b).
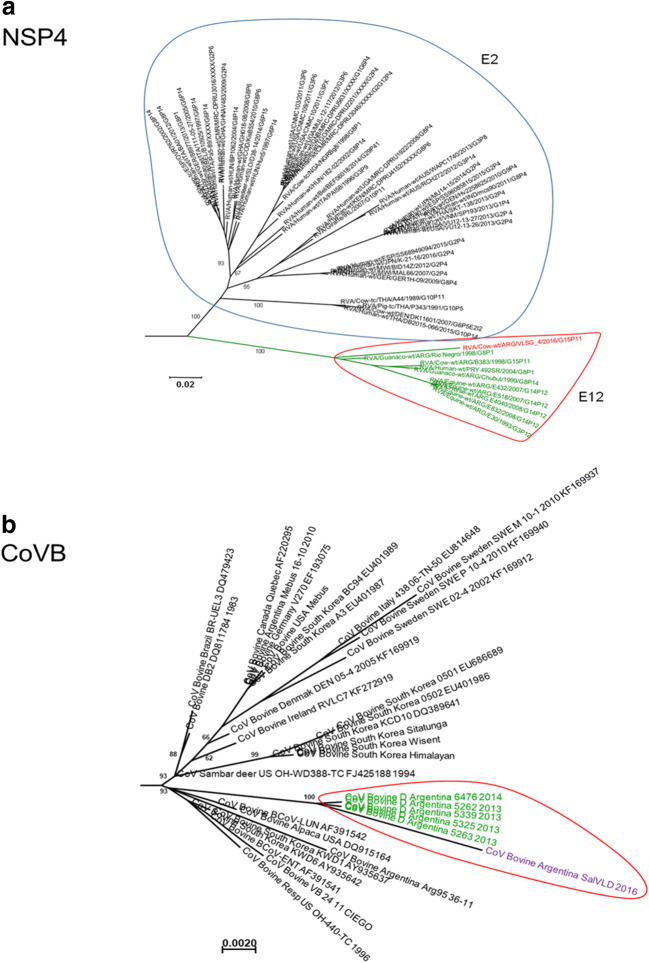
The NSP4 gene of SVLG4 (G15P[11]) strain was classified as E12 genotype and grouped together (bootstrap of 100%) with other RVA strains detected in cattle, guanacos, and horses from Argentina (Fig. 4a).
Fig. 4.
a Phylogenetic tree of the E12 genotype constructed with the NJ method and a bootstrap of 1000, showing nucleotide sequences obtained in this study and Gen Bank available sequences. Red semicircle shows where Salta bovine strains grouped. Red letters are bovine RVA strains from Salta. b Phylogenetic tree of the S2 protein of BCoV is constructed with the NJ method and a bootstrap of 1000, showing nucleotide sequences obtained in this study and Gen Bank available sequences. Red semicircle shows where Salta bovine strains grouped. Violet letters are bovine RVA strains from Salta
Bovine CoV was confirmed by the sequencing of a segment of the S2 gene of SalVLD strain from a dairy farm and grouped together with other BCoV strains circulating in Argentina, with high similarity to Arg95 strain (96%) (Fig. 4b).
Discussion
The RVA detection rate in dairy farms observed in this study (63%) was similar to the prevalence reported in other countries and in other regions of Argentina (Barbosa et al. 1998; Bartels et al. 2010; Badaracco et al. 2012; Miño et al. 2013). Furthermore, RVA detection rate in calves of dairy farms (9.5%) was similar to rates reported in Brazil and in other regions of Argentina (Caruzo Ramos et al. 2010; Ammar et al. 2014). In beef farms, RVA detection rate in calves (6.7%) was similar to that reported in Japan (Abe et al. 2009) but lower than rates reported in France and Brazil (Bendali et al. 1999; Alfieri et al. 2006). Higher rates in dairy and beef farms were obtained in other regions of Argentina (Badaracco et al. 2012), but those rates were estimated only from diarrheic calves.
In our survey, all the strains detected in dairy and beef herds were P[11](III) and clustered together with other strains from Argentina (Badaracco et al. 2013a, b). Interestingly, in contrast with the findings reported in other regions of the country, we did not detect P[5] strains circulating in Salta province. However, there was a broad variation of the VP7 gene and lineages, where G6 lineage III and G10 lineage VI strains were found in dairy farms, with G6P[11] being the most frequent association, while in beef farms, G10 lineage VI, G6 lineage III, and G15 genotypes were detected, with G10P[11] strains being the most frequent combination found. There was a close genetic relationship among the strains isolated in this survey and other strains from Argentina, maybe for the fact that dairy production systems in Salta province are similar to those found in the rest of the country, and the cattle stock reposition comes from the regions where previous studies were conducted. The result of the RVA epidemiology found in beef herds was quite different to the results of the Pampa region. The beef farms from Salta show the same profile than the dairy herds (from Salta and the rest of the country). One possible reason for this finding is the fact that males stay with the females leading to calf births throughout the year, allowing the presence of susceptible calves all year around resulting in a most diverse RVA genotype, supporting our previous hypothesis (Badaracco et al. 2012; 2013a, b).
The detection of RVA G15P[11] as the established strain in a beef farm is an important finding. This is the first time that a G15P[11] RVA is associated with an outbreak of diarrhea in beef herds from Argentina, demonstrating that this strain is able to infect, spread, and cause diarrhea among our calves, as reported only in India (Ghosh et al. 2008). This uncommon genotype was previously reported once in Argentina in 2006, from a dairy calf without other epidemiological significance (Garaicoechea et al. 2006).
Bovine CoV infection rate in dairy herds was low (0.48%) but similar to other regions of Argentina (Bok et al. 2015). In beef herds, negative results may be related to the production system where a less overcrowding, better colostral immunoglobulins levels and environmental conditions, can have a negative impact in the biological cycle of BCoV. Phylogenetic analysis of the S2 BCoV gene of the SalVLD strain showed a high similarity (96%) with other BCoV found in different regions of Argentina and also grouped separately from the Mebus strain as has been previously observed (Bok et al. 2015).
Data reported here emphasize the importance of conducting surveys in unexplored regions of the country, since these viruses affect the productivity of bovine production systems and human health. Also, it is an important highlights that epidemiology of rotavirus and coronavirus could be affected by the type of the region and productive system.
Acknowledgments
The authors would further like to thank Dr. Ulrich Desselberger for reviewing this manuscript and his excellent suggestions.
Funding information
This study was funded by the Instituto Nacional de Tecnología Agropecuaria, Argentina.
Compliance with ethical standards
Conflict of interest
The authors declared that they have no conflicts of interest.
Statement of animal rights
This article does not contain any studies with animals performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abe M, Ito N, Morikawa S, Takasu M, Murase T, Kawashima T, Kawai Y, Kohara J, Sugiyama M. Molecular epidemiology of rotaviruses among healthy calves in Japan: Isolation of a novel bovine rotavirus bearing new P and G genotypes. Virus Research. 2009;144:250–257. doi: 10.1016/j.virusres.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Afshari Safavi, E.A., Mohammadi, G.R., Rad, M. and Naghibi, A., 2012. A Case-Control Study of Association between Diarrhea in Newborn Calves and Infection with Rotavirus and Coronavirus in Some Industrial Dairy Herds of Mashhad Aarea , Iran in 2008 Archives of Razi Institute, 67, 35–41
- Alfieri AA, Parazzi ME, Takiuchi E, Médici KC, Alfieri AF. Frequency of group A rotavirus in diarrhoeic calves in Brazilian cattle herds, 1998-2002. Tropical Animal Health and Production. 2006;38:521–526. doi: 10.1007/s11250-006-4349-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammar SSM, Mokhtaria K, Tahar BB, Amar AA, Redha BA, Yuva B, Si Mohamed H, Abdellatif N, Laid B. Prevalence of rotavirus (GARV) and coronavirus (BCoV) associated with neonatal diarrhea in calves in western Algeria. Asian Pacific Journal of Tropical Biomedicine. 2014;4:S318–S322. doi: 10.12980/APJTB.4.2014C778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badaracco, A., Garaicoechea, L., Matthijnssens, J., Louge Uriarte, E., Odeón, A., Bilbao, G., Fernandez, F., Parra, G.I. and Parreño, V., 2013a. Phylogenetic analyses of typical bovine rotavirus genotypes G6, G10, P[5] and P[11] circulating in Argentinean beef and dairy herds. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases, 18, 18–30 (Elsevier B.V.) [DOI] [PubMed]
- Badaracco A, Garaicoechea L, Rodríguez D, Uriarte EL, Odeón A, Bilbao G, Galarza R, Abdala A, Fernandez F, Parreño V. Bovine rotavirus strains circulating in beef and dairy herds in Argentina from 2004 to 2010. Veterinary microbiology. 2012;158:394–9. doi: 10.1016/j.vetmic.2011.12.011. [DOI] [PubMed] [Google Scholar]
- Badaracco A, Matthijnssens J, Romero S, Heylen E, Zeller M, Garaicoechea L, Van Ranst M, Parreño V. Discovery and molecular characterization of a group A rotavirus strain detected in an Argentinean vicuña (Vicugna vicugna) Veterinary Microbiology. 2013;161:247–254. doi: 10.1016/j.vetmic.2012.07.035. [DOI] [PubMed] [Google Scholar]
- Barbosa EF, Pereira Figueiredo HC, García AM, Portela Lobato ZI, Lage AP. Rotavirus do grupo A em Bezerros lactantes no estado de minas gerais Ciencia Rural. Santa María. 1998;28:435–439. doi: 10.1590/S0103-84781998000300014. [DOI] [Google Scholar]
- Bartels, C.J.M., Holzhauer, M., Jorritsma, R., Swart, W.A.J.M. and Lam, T.J.G.M., 2010. Prevalence, prediction and risk factors of enteropathogens in normal and non-normal faeces of young Dutch dairy calves Preventive Veterinary Medicine, 93, 162–169 [DOI] [PMC free article] [PubMed]
- Bendali F, Sanaa M, Bichet H, Schelcher F. Risk factors associated with diarrhoea in newborn calves. Vet. Res. 1999;30:509–522. [PubMed] [Google Scholar]
- Bok M, Miño S, Rodriguez D, Badaracco A, Nuñes I, Souza SP, Bilbao G, Louge Uriarte E, Galarza R, Vega C, Odeon A, Saif LJ, Parreño V. Molecular and antigenic characterization of bovine Coronavirus circulating in Argentinean cattle during 1994-2010. Veterinary Microbiology. 2015;181:221–229. doi: 10.1016/j.vetmic.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandão PE, Gregori F, Richtzenhain LJ, Rosales CAR, Villarreal LYB, Jerez JA. Molecular analysis of Brazilian strains of bovine coronavirus (BCoV) reveals a deletion within the hypervariable region of the S1 subunit of the spike glycoprotein also found in human coronavirus OC43. Archives of virology. 2006;151:1735–48. doi: 10.1007/s00705-006-0752-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruzo Ramos, T.A.R., Diederichsen de Brito, W.M., Munford, V. and Rácz, M.L., 2010. Molecular characterization of G and P-types bovine rotavirus strains from Goiás, Brazil: High frequency of mixed P-type infections Memorias do Instituto Oswaldo Cruz, 105, 1040–1043 [DOI] [PubMed]
- Garaicoechea L, Bok K, Jones LR, Combessies G, Odeón A, Fernandez F, Parreño V. Molecular characterization of bovine rotavirus circulating in beef and dairy herds in Argentina during a 10-year period (1994-2003) Veterinary microbiology. 2006;118:1–11. doi: 10.1016/j.vetmic.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Geurden T, Thomas P, Casaert S, Vercruysse J, Claerebout E. Prevalence and molecular characterisation of Cryptosporidium and Giardia in lambs and goat kids in Belgium. Veterinary Parasitology. 2008;155:142–145. doi: 10.1016/j.vetpar.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Ghosh, S., Samajdar, S., Sinha, M., Kobayashi, N., Taniguchi, K. and Naik, T.N., 2008. Molecular characterization of rare bovine group a rotavirus G15P[11] and G15P[21] strains from eastern India: Identification of simian SA11-like VP6 genes in G15P[21] strains Virus Genes, 37, 241–249 [DOI] [PubMed]
- Gouvea V, Santos N, Do Carmo Timenetsky M. VP4 typing of bovine and porcine group A rotaviruses by PCR. Journal of Clinical Microbiology. 1994;32:1333–1337. doi: 10.1128/JCM.32.5.1333-1337.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, T.A., 1999. BioEdit a user friendly biological sequence alignment editor and analysys program for Windows 95/98/NT
- Hoang, D.T., Chernomor, O., von Haeseler, A., Minh, B.Q. and Vinh, L.S., 2017. UFBoot2: Improving the Ultrafast Bootstrap Approximation Improving the Ultrafast Bootstrap Approximation Diep, 35, 518–522 [DOI] [PMC free article] [PubMed]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular biology and evolution. 2016;33:1870–4. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louge Uriarte, E.L., Badaracco, A., Matthijnssens, J., Zeller, M., Heylen, E., Manazza, J., Miño, S., Van Ranst, M., Odeón, A. and Parreño, V., 2014. The first caprine rotavirus detected in Argentina displays genomic features resembling virus strains infecting members of the Bovidae and Camelidae. Veterinary microbiology, doi: 10.1016/j.vetmic.2014.03.013 [DOI] [PubMed]
- Martinez, G.M., 2015. Caracterización del Valle de Lerma In:, V. H. Suarez and G. M. Martinez (eds), Características y problemáticas productivas - sanitarias de la lechería del Valle de Lerma (Salta), (Buenos Aires), 66
- Miño S, Matthijnssens J, Badaracco A, Garaicoechea L, Zeller M, Heylen E, Van Ranst M, Barrandeguy M, Parreño V. Equine G3P[3] rotavirus strain E3198 related to simian RRV and feline/canine-like rotaviruses based on complete genome analyses. Veterinary Microbiology. 2013;161:239–246. doi: 10.1016/j.vetmic.2012.07.033. [DOI] [PubMed] [Google Scholar]
- Suarez, V.H. and Martinez, G.M., 2015. Características generales de los tambos In:, Victor Humberto Suarez and Gabriela Marcela Martinez (eds), Características y problemáticas productivas - sanitarias de la lechería del Valle de Lerma (Salta)., (Buenos Aires), 66