Abstract
The newly identified SARS-CoV-2 has now been reported from around 185 countries with more than a million confirmed human cases including more than 120,000 deaths. The genomes of SARS-COV-2 strains isolated from different parts of the world are now available and the unique features of constituent genes and proteins need to be explored to understand the biology of the virus. Spike glycoprotein is one of the major targets to be explored because of its role during the entry of coronaviruses into host cells. We analyzed 320 whole-genome sequences and 320 spike protein sequences of SARS-CoV-2 using multiple sequence alignment. In this study, 483 unique variations have been identified among the genomes of SARS-CoV-2 including 25 nonsynonymous mutations and one deletion in the spike (S) protein. Among the 26 variations detected in S, 12 variations were located at the N-terminal domain (NTD) and 6 variations at the receptor-binding domain (RBD) which might alter the interaction of S protein with the host receptor angiotensin-converting enzyme 2 (ACE2). Besides, 22 amino acid insertions were identified in the spike protein of SARS-CoV-2 in comparison with that of SARS-CoV. Phylogenetic analyses of spike protein revealed that Bat coronavirus have a close evolutionary relationship with circulating SARS-CoV-2. The genetic variation analysis data presented in this study can help a better understanding of SARS-CoV-2 pathogenesis. Based on results reported herein, potential inhibitors against S protein can be designed by considering these variations and their impact on protein structure.
Keywords: SARS-CoV-2, Spike protein, Sequence analysis, COVID-19, Genomic variants
Graphical abstract
Highlights
-
•
There are 483 unique variations among SARS-CoV-2 genomes.
-
•
In SARS-CoV-2 S glycoprotein, there are 40 variations
-
•
14 synonymous and 25 non-synonymous mutations were observed in S glycoprotein.
-
•
12 non-synonymous mutations are located at the N-terminal domain of S glycoprotein.
-
•
6 non-synonymous mutations exist in at the receptor-binding domain (RBD).
1. Introduction
Coronavirus disease (COVID-19) is a pandemic manifesting respiratory illness and first reported in Wuhan, Hubei province of China in December 2019. The death toll rose to more than 68,000 among 1,250,000 confirmed cases around the Globe (until April 4, 2020) (WHO (World Health Organization), 2020). The virus causing COVID-19 is named as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Based on the phylogenetic studies, the SARS-CoV-2 is categorized as a member of the genus Betacoronavirus, the same lineage that includes SARS coronavirus (SARS-CoV) (Gorbalenya et al., 2020) that caused SARS (Severe Acute Respiratory Syndrome) in China during 2002 (Ksiazek et al., 2003). Recent studies showed that SARS-CoV-2 has a close relationship with bat SARS-like CoVs (Li et al., 2005; Zhou et al., 2020), though the intermediate hosts for zoonotic transmission of SARS-CoV-2 from bats to humans remain undiscovered.
SARS-CoV-2 has been identified as an enveloped, single-stranded positive-sense RNA virus with a genome size of approximately 29.9 kb encoding 27 proteins from 14 ORFs including 15 non-structural, 8 accessory, and 4 major structural proteins. Two-thirds of the viral RNA harbors the first ORF (ORF1ab) dedicated for translating polyprotein 1a (pp1a) and polyprotein 1ab (pp1ab), which later undergo proteolytic cleavage to form 15 non-structural proteins. Spike glycoprotein (S), membrane (M), envelope (E) and nucleocapsid (N) are the four major structural proteins of SARS-CoV-2 (Wu et al., 2020a; Wu et al., 2020b). Interestingly, S glycoprotein is characterized as the critical determinant for viral entry into host cells which consists of two functional subunits namely S1 and S2. The S1 subunit recognizes and binds to the host receptor through the receptor-binding domain (RBD) whereas S2 is responsible for fusion with the host cell membrane (Wrapp et al., 2020; Walls et al., 2020; Chen et al., 2020). MERS-CoV uses dipeptidyl peptidase-4 (DPP4) as an entry receptor (Raj et al., 2013) whereas SARS-CoV and SARS-CoV-2 utilize ACE-2 (angiotensin-converting enzyme 2) (Li et al., 2003), abundantly available in lung alveolar epithelial cells and enterocytes, suggesting S glycoprotein as a potential drug target to halt the entry of SARS-CoV-2 (Letko et al., 2020).
According to recent reports, neutralizing antibodies are generated in response to the entry and fusion of surface-exposed S protein (mainly RBD domain) which is predicted to be an important target for vaccine candidates (Chen et al., 2020; Hoffmann et al., 2020; Tian et al., 2020). However, SARS-CoV-2 has emerged with remarkable properties like glutamine-rich 42 aa long exclusive molecular signature (DSQQTVGQQDGSEDNQTTTIQTIVEVQPQLEMELTPVVQTIE) in position 983–1024 of polyprotein 1ab (pp1ab) (Cárdenas-Conejo et al., 2020), diversified receptor-binding domain (RBD), unique furin cleavage site (PRRAR↓SV) at S1/S2 boundary in S glycoprotein which could play roles in viral pathogenesis, diagnosis, and treatment (Coutard et al., 2020). To date, few genomic variations of SARS-CoV-2 are reported (Ceraolo and Giorgi, 2020; Lu et al., 2020). There are growing evidences that spike protein, a 1273 amino acid long glycoprotein having multiple domains, possibly plays a major role in SARS-CoV-2 pathogenesis. Viral entry to the host cell is initiated by the receptor-binding domain (RBD) of S1 head. Upon receptor-binding, proteolytic cleavage occurs at S1/S2 cleavage site and two heptad repeats (HR) of S2 stalk form a six-helix bundle structure triggering the release of the fusion peptide. As it comes into close proximity to the transmembrane anchor (TM), the TM domain facilitates membrane destabilization required for fusion between virus-host membranes (Bosch et al., 2003; Liu et al., 2004). Insights into the sequence variations of S glycoprotein among available genomes are key to understanding the biology of SARS-CoV-2 infection, developing antiviral treatments and vaccines. In this study, we have analyzed 320 genomic sequences of SARS-CoV-2 to identify mutations between the available genomes followed by the amino acid variations in the glycoprotein S to foresee their impact on the viral entry to host cell from the structural biology viewpoint.
2. Methods and materials
2.1. Dataset
All available sequences (320 whole genome and surface glycoprotein amino acid sequences of SARS-CoV-2) related to the COVID-19 pandemic were retrieved from NCBI Virus Variation Resource repository (https://www.ncbi.nlm.nih.gov/labs/virus/) (Hatcher et al., 2017). Among the protein sequences, 11 were discarded due to incomplete sequence coverage. In addition, all 40 S glycoprotein sequences from different coronavirus families were retrieved for phylogenetic analysis. The NCBI reference sequence of SARS-CoV-2 S glycoprotein, accession number YP_009724390 was used as the canonical sequence for the analyses of spike protein variants.
2.2. Phylogenetic analysis
Variant analyses of SARS-CoV-2 genomes were performed in the Genome Detective Coronavirus Typing Tool Version 1.13 which is specially designed for this virus (https://www.genomedetective.com/app/typingtool/cov/) (Cleemput et al., 2020). For multiple sequence alignment (MSA), Genome Detective Coronavirus Typing Tool uses a reference dataset of 431 whole genome sequences (WGS) where 386 WGS were from known nine coronavirus species. The dataset was then aligned with MUSCLE (Edgar, 2004). Entropy (H(x)) plot of nucleotide variations in SARS-CoV-2 genome was constructed using BioEdit (Hall et al., 1999). MEGA X (version 10.1.7) was used to construct the MSAs and the phylogenetic tree using pairwise alignment and neighbor-joining methods in ClustalW (Kumar et al., 2018; Saitou and Nei, 1987). Tree structure was validated by running the analysis on 1000 bootstraps (Efron et al., 1996) replications dataset and the evolutionary distances were calculated using the Poisson correction method (Zuckerkandl and Pauling, 1965).
2.3. Homology modeling of S glycoprotein
Variant sequences of SARS-CoV-2 were modeled in Swiss-Model (Andrew et al., 2018) using the Cryo-EM spike protein structure of SARS-CoV-2 (PDB ID 6VSB; (Wrapp et al., 2020)) as a template. The overall quality of models was assessed in RAMPAGE server (Prisant et al., 2003) by generating Ramachandran plots (Supplementary Table 1). PyMol and BIOVIA Discovery Studio were used for structure visualization and superpose (DeLano et al., 2002).
3. Results
3.1. Genomic variations of SARS-CoV-2
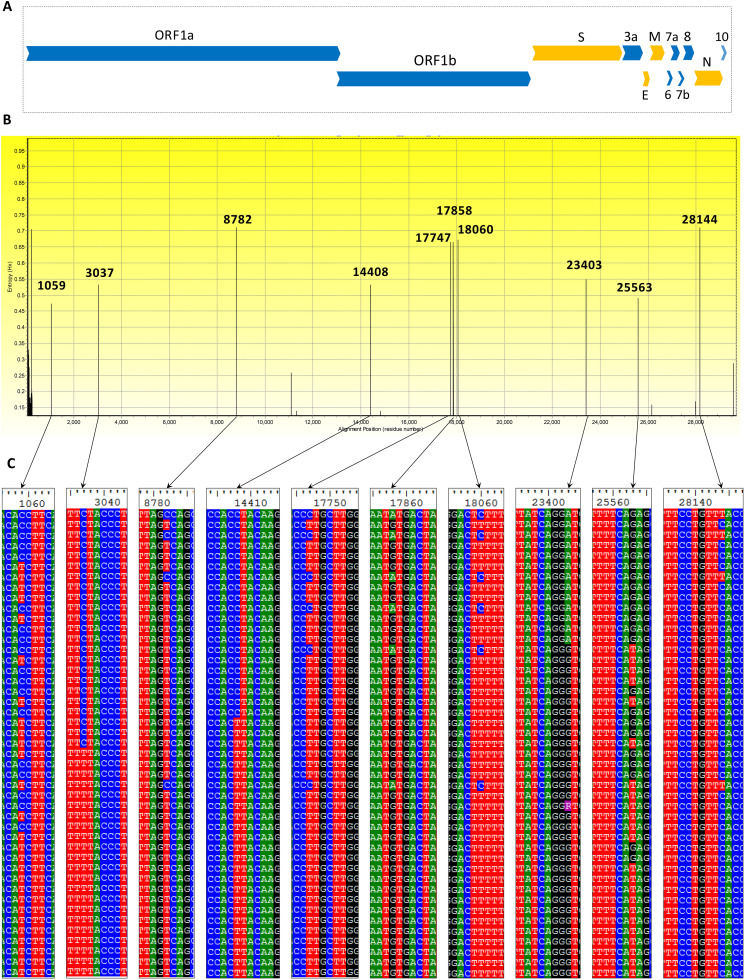
Multiple sequence alignment of the available 320 genomes of SARS-CoV-2 were performed and 483 variations were found throughout the 29,903 bp long SARS-CoV-2 genome with in total 115 variations in UTR region, 130 synonymous variations that cause no amino acid alteration, 228 non-synonymous variations causing change in amino acid residue, 16 INDELs, and 2 variations in non-coding region (Supplementary Table 2). Among the 483 variations, 40 variations (14 synonymous, 25 non-synonymous mutations and one deletion) were observed in the region of ORF S that encodes S glycoprotein which is responsible for viral fusion and entry into the host cell (Li, 2015). Notably, most of the SARS-CoV-2 genome sequences were deposited from the USA (250) and China (50) (Supplementary Fig. 1). Positional variability of the SARS-CoV-2 genomes was calculated from the MSA of 320 SARS-CoV-2 whole genomes as a measure of Entropy value (H(x)) (Manaresi et al., 2017). Excluding 5′ and 3′ UTR, ten hotspots of hypervariable positions were identified, of which seven were located at ORF1ab (1059C > T, 3037C > T, 8782C > T, 14408C > T, 17747C > T, 17858A > G, 18060C > T), and one at ORF S (23403A > G), ORF3a (25563G > T), and ORF8 (28,144 T > C), respectively. The variability at position 8782 and 28,144 were found to be the highest among the other hotspots (Fig. 1 ).
Fig. 1.
Nucleotide sequence variation among 320 SARS-CoV-2 whole genomes. A. Positional organization of major structural protein-encoding genes in orange color (S = Spike protein, E = Envelope protein, M = Membrane protein, N = Nucleocapsid protein) and accessory protein ORFS in blue colors. B. Variability within 320 SARS-CoV-2 genomic sequences represented by entropy (H(x)) value across genomic location. Two highest frequency of alterations were found at position 8785 of ORF1a and 28,144 of ORF8. C. The respective alignment view of each highly variable regions. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. Phylogenetic analysis of S glycoprotein
The phylogenetic analysis of a total of 66 sequences (26 unique SARS-CoV-2 and 40 different coronavirus S glycoprotein sequences) was performed. The evolutionary distances showed that all the SARS-CoV-2 spike proteins cluster in the same node of the phylogenetic tree confirming the sequences were similar to Refseq YP_009724390 (Fig. 2 ). Bat coronaviruses have a close evolutionary relationship as different strains were found in the nearest outgroups and clades (Bat coronavirus BM48–31, Bat hp-beta coronavirus, Bat coronavirus HKU9) conferring that coronavirus has a vast geographical spread and bat is the most prevalent host (Fig. 2). In other clades, the clusters were speculated through different hosts which may describe the evolutionary changes of surface glycoprotein due to cross-species transmission. Viral hosts reported from different spots at different times are indicative of possible recombination.
Fig. 2.
Sequence phylogeny of SARS-CoV-2 spike glycoprotein variants and other coronavirus spike proteins based on amino acid sequences, retrieved from NCBI database using neighbor-joining methods in ClustalW and tree structure was validated by running the analysis on 1000 bootstraps. The branch length is indicated in the scale bar. The accession number YP_009724390 represents identical sequences out of SARS-CoV-2 spike proteins.
3.3. SARS-CoV-2 spike protein variation analysis
The S glycoprotein sequences of SARS-CoV-2 were retrieved from the NCBI Virus Variation Resource repository and aligned using ClustalW. The relative positions of SARS-CoV-2 spike protein domains were measured by aligning with the SARS-CoV spike protein (Fig. 3 ) (Yuan et al., 2017; Gui et al., 2017). From the sequence identity matrix, 26 unique variants among unique SARS-CoV-2 spike glycoprotein sequences were identified to have 25 substitutions and a deletion (Fig. 4A and Supplementary Table 3). 215 sequences were found identical with SARS-CoV-2 S protein reference sequence (YP_009724390) while 64 sequences were identical with the same variation of D614G (Supplementary Table 4, 5). Among all the variations, twelve (Y28N, T29I, H49Y, L54F, N74K, E96D, D111N, Y145Del, F157L, G181V, S221W, and S247R) were located at the N-terminal domain (NTD). Another 6 variations (A348T, R408I, G476S, V483A, H519Q, A520S) were found at the receptor-binding domain (RBD) while only two variations (A930V, and D936Y) were found at the heptad repeat 1 (HR1) domain. Single variations were found in signal peptide (L5F) domain, sub-domain-2 (D614G), sub-domain-3 (A1078V), heptad repeat 2 domain (D1168H), and cytoplasmic tail domain (D1259H) each. Notably, the substitution of Cystine by Phenylalanine was observed at 19 amino acids upstream of the fusion peptide domain (Fig. 4A). The mutation of Aspartic acid to Glycine at position 614 was observed 71 times with entropy value over 0.5 among the available 320 SARS-CoV-2 spike protein sequences (Supplementary Fig. 2).
Fig. 3.
Overall architecture of the SARS-CoV-2 S glycoprotein. A.Schematic diagram of the SARS-CoV S glycoprotein showing domain organization (Reconstructed from Y. Yuan et al., 2017 and M. Gui et al., 2017). B. Schematic domain organization diagram of the SARS-CoV-2 S glycoprotein constructed by aligning with SARS-CoV S protein domain. C. Homology model of SARS-CoV-2 S protein reference sequence YP_009724390 with PDB:6VSB. S protein trimer with two protomers surface shadowed (left). Ribbon diagram of SARS-CoV-2 S glycoprotein monomer from B. Here, NTD: N-terminal domain; RBD: receptor-binding domain; SD: subdomain; CR: connecting region; HR: heptad repeat; CH: central helix; BH: β-hairpin; FP: fusion peptide; TM: transmembrane domain; CT: cytoplasmic tail.
Fig. 4.
Variability within 320 SARS-CoV-2 S protein sequences. A. Schematic representation of mutations across the spike protein domain organization. Blue, red, and black color represents charge of the amino acid residue as positive, negative, and neutral respectively. B—N, Superposed structures of SARS-CoV-2 spike protein variants with the Cryo-EM structure of SARS-CoV-2 Spike Protein (PDB: 6VSB). Template residues are indicated by green color and variants' residues are indicated as red color. Here, B: Y28N, C: T29I, D: H49Y, E: L54F, F: D111N, G: S221W, H: A348T, I: R408I, J: H519Q, K: A520S, L: D614G, M: F797C, N: A930V, O: D936Y, and P: A1078V. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Alterations of amino acid residual charge from positive to neutral (H49Y, R408I, H519Q), negative to neutral (D111N, D614G, D936Y), negative to positive (D1168H, D1259H), and neutral to positive (N74K, S247R) were seen in variants QHW06059, QHS34546, QIS61422, QIS61338, QIK50427, QIS30615, QIS60978, QIS60582, QIO04367, and QHR84449 respectively due to the substitution of amino acids that differs in charge. The remaining 15 variants were mutated with the amino acids that are similar in charge (Fig. 4 A). The SARS-CoV-2 spike protein variants were superposed with the cryo-electron microscopic structure of SARS-CoV-2 spike protein (Wrapp et al., 2020). L5F, N74K, E96D, F157L, G181V, S247R, G476S, V483A, D1168H, and D1259H variants were excluded from superposition due to absence of respective residues in the 3D structure of the template (PDB: 6VSB). The superposition showed that most of the residual change was causing incorporation of bulky amino acid residues (T29I, H49Y, L54F, S221W, A348T, H519Q, A520S, A930V, D936Y, and A1078V) in place of smaller sized residues except for Y28N, D111N, R408I, D614G, and F797C (Fig. 4 B—P).
The sequence comparison of spike glycoprotein between SARS-CoV-2 variants and SARS-CoV (Uniprot ID: P59594) revealed nearly 77.46% similarity and identified the presence of an additional 22 amino acids in SARS-CoV-2 spike protein variants resulting from a total of 5 insertions (Supplementary Fig. 3). Among these, the major insertion consisting of 7 amino acids (GTNGTKR) at position 72–78 followed by 4 amino acids (NKSW) at position 149–152 and 6 amino acids (SYLTPG) at 247–252 occurred in the N-terminal domain. Insertion of Glycine at 482 was found in receptor binding domain, preceding another insertion of 4 amino acids (NSPR) at position 679–682, just upstream of S1/S2 cleavage site that leads to form a furin-like cleavage site (PRRARS) in the S protein variants of SARS-CoV-2 (Supplementary Fig. 3). The S2 subunit of spike protein, especially the heptad repeat region 2, fusion peptide domain, transmembrane domain, and cytoplasmic tail were found to be highly conserved in the SARS-CoV and the SARS-CoV-2 variants while the S1 subunit was more diverse, specifically the N-terminal domain (NTD) and receptor-binding domain (RBD).
The spatial distribution of S protein sequences having different variations over time reveals that most of the variants (17 out of 240 S glycoprotein sequences) were reported from the US followed by 3 out of 2 sequences (including Y145 deletion) and 2 out of 50 sequences from India and China, respectively (Fig. 5 ). Only one variant was found out of only one available sequence in the repository from Sweden, Australia, South Korea and Peru. Interestingly, all the sequences are unique among countries from where they were reported except D614G, which was found both in the US and Peru (Fig. 5). Moreover, we have also analyzed sequences from Brazil, Italy, Nepal, Pakistan, Spain, Taiwan, and Vietnam but no variation in the S glycoprotein sequences were found when compared to the Refseq YP_009724390.
Fig. 5.
The spatial distribution of variations found in S glycoprotein over time. The surface plot illustrates the frequency distribution of each variation over time. The geographic location of the sample is presented with flags and if the frequency of each variation (if more than one from a single country) is shown below the respective flag.
4. Discussion
COVID-19 is one of the most contagious pandemics the world has ever had with 1,250,000 confirmed cases to date (April 4, 2020) and the cases have increased as high as 5 times in less than a month (WHO (World Health Organization), 2020). Phylogenetic analysis showed that SARS-CoV-2 is a unique coronavirus presumably related to Bat coronaviruss (BM48–31, Hp-betacoronavirus). During this study, we investigated the available genomes of SARS-CoV-2 and found variations in 483 positions resulting in 130 synonymous and 228 non-synonymous variants. Out of them, 25 non-synonymous variants were observed in the spike protein of SARS-CoV-2. Viral spike protein is thought to have a crucial role in drug and vaccine development as reported previously in managing the viruses like SARS-CoV and MERS-CoV (Tian et al., 2020; Kapadia et al., 2005; Elshabrawy et al., 2012; Du et al., 2013). Likewise, a number of studies targeting SARS-CoV-2 spike protein have been undertaken for the therapeutic measures (Xia et al., 2020), but the unique structural and functional details of SARS-CoV-2 spike protein are still under scrutiny. We also found a variant (R408I) at receptor-binding domain (RBD) that mutated from positively charged Arginine residue to neutral and smaller sized Isoleucine residue (Fig. 4 I). This change might alter the interaction of viral RBD with the host receptor because the R408 residue of SARS-CoV-2 is known to interact with the ACE2 receptor for viral entry (Ortega et al., 2020). Similarly, alterations of RBD (G476S, V483A, H519Q, and A520S) also could affect the interaction of SARS-CoV-2 spike protein with other molecules which requires further investigations. QIA98583 and QIS30615 variants were found to have an alteration of Alanine to Valine (A930V), and Aspartic acid to Tyrosine (D936Y) respectively in the alpha helix of the HR1 domain. Previous reports have indicated that HR1 domain plays a significant role in viral fusion and entry by forming helical bundles with HR2, and mutations including alanine substitution by valine (A1168V) in HR1 region are predominantly responsible for conferring resistance to mouse hepatitis coronaviruses against HR2 derived peptide entry inhibitors (Bosch et al., 2008). This study hypothesizes the mutation (A930V) found in that of SARS-CoV-2 might also have a role in the emergence of drug-resistance virus strains. Also, the mutation (D1168H) found in the heptad repeat 2 (HR2) of SARS-CoV-2 could play a vital role in viral pathogenesis. Moreover, we found that 20 variants including one deletion out of 26 were located within S1 especially within NTD and RBD region of glycoprotein S (Fig. 4A), the region is responsible for the preliminary interaction with the host cell receptor ACE2. This indicates that the NTD and RBD are very prone to mutations. However, the NTD and RBD portions harbor potential epitopes that might serve as potential peptide vaccine candidates against SARS-CoV-2 as reported in different studies (Bhattacharya et al., 2020; Rasheed et al., 2020; Rahman et al., 2020). The reason behind choosing the sequences from S protein domain NTD and RBD as antigenic determinants is they are situated in the outer surface of the virus that could be more accessible for the immune system (Fig. 3C). So the variations reported herein within the outer domains of S glycoprotein could help to design effective epitope-based vaccines or antivirals.
The SARS-CoV-2 S protein contains additional furin protease cleavage site, PRRARS, in S1/S2 domain which is conserved among all 320 sequences as revealed during this study (Supplementary Fig. 3). This unique signature is thought to make SARS-CoV-2 more virulent than SARS-CoV and regarded as novel features of the viral pathogenesis (Walls et al., 2020). According to previous reports, the more the host cell proteases able to process the coronavirus S protein, the more acceleration in viral tropism observed (Walls et al., 2020; Klenk and Garten, 1994; Steinhauer, 1999; Millet and Whittaker, 2015). Apart from that, this could also promote viruses to escape antiviral therapies targeting transmembrane protease TMPRSS2 (ClinicalTrials.gov, NCT04321096) which is a well-reported protease to cleave at S1/S2 of S glycoprotein (Fehr and Perlman, 2015). Comparative analyses between SARS-CoV and SARS-CoV-2 spike glycoprotein showed 77% similarity between them where the most diverse region was the N-terminal domain and receptor-binding domain. The insertion of 17 additional amino acid residues in the N-terminal domain of SARS-CoV-2 and its high sequence diversity suggests that it may have a role in binding with other cell receptors in humans. This is because the N-terminal domain could function as the receptor-binding domain of various coronaviruses (Li, 2015; Yuan et al., 2017). A similar phenomenon has been observed in mouse hepatitis coronavirus (MHV) and porcine transmissible gastroenteritis coronavirus (TGEV) where the NTD is reported to attach with the host entry receptor (Williams et al., 1991; Delmas et al., 1992).
The spatial distribution of variations in amino acid sequences of S glycoprotein of SARS-CoV-2 showed that 17 variations out of 26 were found among the sequences deposited from the US. But there were 7 unique variations found among the sequences from 5 countries (from 4 continents namely Australia, Europe, Latin America, and Asia) out of 6 sequences available during the study period (Fig. 5). Although the number of sequences analyzed herein is too small to speculate the exact trend of S glycoprotein evolution, these variants might play a vital role in adaptation to a new geospatial environment.
The variation analyses in amino acids indicated the structural features of different domains of the SARS-CoV-2 spike proteins. The variations have multiple effects on the structure resulting in change in stability, favoring various interactions, and conformational diversity. Augmented infection kinetics and viral spreading may have an association with the structural changes and composition of residues in the viral spike protein. However, to identify the actual role of involvement of S glycoprotein, a larger dataset regarding genomics and proteomics of SARS-CoV-2 is required as this protein is vital to understand the viral pathogenicity, evolution and development of therapeutics. Further analyses of all the S glycoprotein and SARS-CoV-2 genomes with different epidemiological aspects are warranted to get a better understanding of the pathogenesis of SARS-CoV-2.
Credit author statement
Syed Mohammad Lokman: Visualization; Formal analysis; Data curation; Writing – draft.
Md. Rasheduzzaman: Investigation; Formal analysis; Writing – draft.
Asma Salauddin: Investigation; Formal analysis.
Rocktim Barua: Investigation; Visualization.
Afsana Yeasmin Tanzina: Formal analysis; Writing – draft.
Meheadi Hasan Rumi: Investigation; Visualization.
Md. Imran Hossain: Investigation; Formal analysis.
AMAM Zonaed Siddiki: Supervision; Writing - review & editing.
Adnan Mannan: Conceptualization; Project administration; Writing - review & editing; Supervision.
Md. Mahbub Hasan: Formal analysis; Validation; Visualization; Methodology; Data curation; Resources; Writing - review & editing; Supervision.
Declaration of Competing Interest
The authors would like to declare that there is no known contending financial interests or personal relationships that could affect the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2020.104389.
Appendix A. Supplementary data
Supplementary material
References
- Andrew W., Martino B., Stefan B., Gabriel S., Gerardo T., Rafal G., Heer Florian T., de Beer Tjaart A.P., Christine Rempfer, Lorenza Bordoli, Rosalba Lepore, Torsten Schwede. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya M., Sharma A.R., Patra P., Ghosh P., Sharma G., Patra B.C., Lee S., Chakraborty C. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): Immunoinformatics approach. J. Med. Virol. 2020;92:618–631. doi: 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., van der Zee R., de Haan C.A.M., Rottier P.J.M. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., Rossen J.W.A., Bartelink W., Zuurveen S.J., de Haan C.A.M., Duquerroy S., Boucher C.A.B., Rottier P.J.M. Coronavirus escape from heptad repeat 2 (HR2)-derived peptide entry inhibition as a result of mutations in the HR1 domain of the spike fusion protein. J. Virol. 2008;82:2580–2585. doi: 10.1128/JVI.02287-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas-Conejo Y., Liñan-Rico A., Garcia-Rodriguez D.A., Centeno-Leija S., Serrano-Posada H. An exclusive 42 amino acid signature in pp1ab protein provides insights into the evolutive history of the 2019 novel human-pathogenic coronavirus (SARS-CoV2) J. Med. Virol. 2020;92:688–692. doi: 10.1002/jmv.25758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceraolo C., Giorgi F.M. Genomic variance of the 2019-nCoV coronavirus. J. Med. Virol. 2020;92:522–528. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Guo Y., Pan Y., Zhao Z.J. Structure analysis of the receptor binding of 2019-nCoV. Biochem. Biophys. Res. Commun. 2020;525(1):135–140. doi: 10.1016/j.bbrc.2020.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleemput S., Dumon W., Fonseca V., Karim W.A., Giovanetti M., Alcantara L.C., Deforche K., de Oliveira T. Bioinformatics; 2020. Genome Detective Coronavirus Typing Tool for rapid identification and characterization of novel coronavirus genomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W.L. Pymol: An open-source molecular graphics tool, CCP4 Newsl. Protein Crystallogr. 2002;40:82–92. [Google Scholar]
- Delmas B., Gelfi J., L’Haridon R., Sjöström H., Laude H. Aminopeptidase N is a major receptor for the enteropathogenic coronavirus TGEV. Nature. 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Kou Z., Ma C., Tao X., Wang L., Zhao G., Chen Y., Yu F., Tseng C.T.K., Zhou Y., Jiang S. A truncated receptor-binding domain of MERS-CoV spike protein potently inhibits MERS-CoV infection and induces strong neutralizing antibody responses: implication for developing therapeutics and vaccines. PLoS One. 2013;8:2–10. doi: 10.1371/journal.pone.0081587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. Proceedings. 2004 IEEE Computational Systems Bioinformatics Conference, 2004. CSB 2004. Institute of Electrical and Electronics Engineers; Stanford, CA, USA: 2004. MUSCLE: multiple sequence alignment with improved accuracy and speed; pp. 728–729. [Google Scholar]
- Efron B., Halloran E., Holmes S. Bootstrap confidence levels for phylogenetic trees. Proc. Natl. Acad. Sci. 1996;93:13429. doi: 10.1073/pnas.93.23.13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshabrawy H.A., Coughlin M.M., Baker S.C., Prabhakar B.S. Human monoclonal antibodies against highly conserved HR1 and HR2 domains of the SARS-CoV spike protein are more broadly neutralizing. PLoS One. 2012;7(11):e50366. doi: 10.1371/journal.pone.0050366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Methods in Molecular Biology:Coronaviruses-Methods and Protocol. Vol. 1282. Humana Press; 2015. Coronaviruses: an overview of their replication and pathogenesis; pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.E., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M., Song W., Zhou H., Xu J., Chen S., Xiang Y., Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.A. Nucleic Acids Symp. Ser. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT; pp. 95–98. [Google Scholar]
- Hatcher E.L., Zhdanov S.A., Bao Y., Blinkova O., Nawrocki E.P., Ostapchuck Y., Schäffer A.A., Brister J.R. Virus variation resource--improved response to emergent viral outbreaks. Nucleic Acids Res. 2017;45(D1):D482–D490. doi: 10.1093/nar/gkw1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapadia S.U., Rose J.K., Lamirande E., Vogel L., Subbarao K., Roberts A. Long-term protection from SARS coronavirus infection conferred by a single immunization with an attenuated VSV-based vaccine. Virology. 2005;340:174–182. doi: 10.1016/j.virol.2005.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenk H.-D., Garten W. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 1994;2:39–43. doi: 10.1016/0966-842x(94)90123-6. [DOI] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020:1–8. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Fang. Receptor Recognition Mechanisms of Coronaviruses: A Decade of Structural Studies. J Virol. 2015;89(4):1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H. Bats are natural reservoirs of SARS-like coronaviruses. Science (80-.) 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R., Xiong H., Farmar J., Debnath A.K., Tien P. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manaresi E., Conti I., Bua G., Bonvicini F., Gallinella G. A parvovirus B19 synthetic genome: sequence features and functional competence. Virology. 2017;508:54–62. doi: 10.1016/j.virol.2017.05.006. [DOI] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega J.T., Serrano M.L., Pujol F.H., Rangel H.R. Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: an in silico analysis. EXCLI J. 2020;19:410–417. doi: 10.17179/excli2020-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prisant M.G., Richardson J.S., Richardson D.C. Structure validation by Calpha geometry: Phi, psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Rahman M.S., Hoque M.N., Islam M.R., Akter S., Rubayet-Ul-Alam A.S.M., Siddique M.A., Saha O., Rahaman M.M., Sultana M., Hossain M.A. Epitope-based chimeric peptide vaccine design against S, M and E proteins of SARS-CoV-2 etiologic agent of global pandemic COVID-19: an in silico approach. BioRxiv. 2020 doi: 10.1101/2020.03.30.015164. 2020.03.30.015164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Müller M.A., Dijkman R., Muth D., Demmers J.A.A., Zaki A., Fouchier R.A.M. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed M.A., Raza S., Zohaib A., Yaqub T., Rabbani M., Riaz M.I., Awais M., Afzal A. 2020. In Silico Identification of Novel B Cell and T Cell Epitopes of Wuhan Coronavirus (2019-nCoV) for Effective Multi Epitope-Based Peptide Vaccine Production, Prepr. [DOI] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Steinhauer D.A. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology. 1999;258:1–20. doi: 10.1006/viro.1999.9716. [DOI] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. 2020;9(1):382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (World Health Organization) 2020. Coronavirus disease (COVID-2019) situation reports. [Google Scholar]
- Williams R.K., Jiang G.-S., Holmes K.V. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl. Acad. Sci. 1991;88:5533–5536. doi: 10.1073/pnas.88.13.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.aax0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., Ying T., Liu S., Shi Z., Jiang S. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020:1–3. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E., Pauling L. Evolving Genes and Proteins. Academic Press; 1965. Evolutionary Divergence and Convergence in Proteins; pp. 97–166. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material