Highlights
-
•
Porcine epidemic diarrhea virus (PEDV) causes acute diarrhea, dehydration and high mortality in neonatal pig.
-
•
S INDEL and non-S INDEL strains were detected during the diarrheal disease outbreak in US swine in 2013–2014.
-
•
The non-S INDEL PEDV was highly virulent, whereas the S INDEL PEDV caused milder disease.
-
•
The main PEDV transmission route is fecal–oral, but airborne transmission via the fecal–nasal route may also occur.
-
•
PEDV-specific IgA effector and memory B cells in orally primed sows play a critical role in sow lactogenic immunity.
Keywords: Porcine epidemic diarrhea virus, PEDV, Pathogenesis, Prevention, Coronavirus, Pigs
Abstract
Porcine epidemic diarrhea virus (PEDV), a member of the genus Alphacoronavirus in the family Coronaviridae, causes acute diarrhea and/or vomiting, dehydration and high mortality in neonatal piglets. Two different genogroups of PEDV, S INDEL [PEDV variant containing multiple deletions and insertions in the S1 subunit of the spike (S) protein, G1b] and non-S INDEL (G2b) strains were detected during the diarrheal disease outbreak in US swine in 2013–2014. Similar viruses are also circulating globally. Continuous improvement and update of biosecurity and vaccine strains and protocols are still needed to control and prevent PEDV infections worldwide. Although the non-S INDEL PEDV was highly virulent and the S INDEL PEDV caused milder disease, the latter has the capacity to cause illness in a high number of piglets on farms with low biosecurity and herd immunity. The main PEDV transmission route is fecal–oral, but airborne transmission via the fecal–nasal route may play a role in pig-to-pig and farm-to-farm spread. PEDV infection of neonatal pigs causes fecal virus shedding (alongside frequent detection of PEDV RNA in the nasal cavity), acute viremia, severe atrophic enteritis (mainly jejunum and ileum), and increased pro-inflammatory and innate immune responses. PEDV-specific IgA effector and memory B cells in orally primed sows play a critical role in sow lactogenic immunity and passive protection of piglets. This review focuses on the etiology, transmission, pathogenesis, and prevention and control of PEDV infection.
1. Introduction
Porcine epidemic diarrhea virus (PEDV), a member of the genus Alphacoronavirus in the family Coronaviridae of the order Nidovirales, causes acute diarrhea, vomiting, dehydration and high mortality in neonatal piglets. The disease was reported in the European and Asian pig industries over the last 30 years, with the virus first appearing in England (Wood, 1977) and Belgium (Pensaert and de Bouck, 1978) in the late 1970s. PEDV was first reported in the US in 2013 (Stevenson et al., 2013). Within months, the virus had spread rapidly nationwide (Cima, 2013), causing significant economic losses (Jung and Saif, 2015). The disease, porcine epidemic diarrhea (PED), in the US was caused by two different genogroups (G) of PEDV, S INDEL [PEDV variant containing multiple deletions and insertions in the S1 subunit of the spike (S) protein, G1b] and non-S INDEL (G2b) strains (Lee, 2015; Vlasova et al., 2014).
The family Coronaviridae is composed of four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (Woo et al., 2012). The alphacoronavirus transmissible gastroenteritis virus (TGEV), which is clinically similar to PEDV, was first described in the US in 1946 (Saif et al., 2012). Since the emergence of porcine respiratory coronavirus (PRCV) in 1984, a natural mutant of TGEV with deletions in the spike protein, TGEV infections have decreased (Saif et al., 2012). A new deltacoronavirus, porcine deltacoronavirus (PDCoV), which is clinically similar to PEDV and TGEV, was detected in diarrheic US pigs in 2014 (Wang et al., 2014a). In China in 2016–2017, another new alphacoronavirus (bat HKU2-like), genetically distinct but clinically similar to the others, was identified in diarrheic pigs and was designated swine acute diarrhea syndrome coronavirus (SADS-CoV) (Gong et al., 2017; Pan et al., 2017; Zhou et al., 2018). Clinical signs caused by these four porcine enteric coronaviruses are indistinguishable. Therefore, differential diagnosis is critical to control viral epidemic diarrheas in pigs. This review focuses on updating the current understanding of the etiology, transmission, pathogenesis, and prevention and control of PEDV infection.
2. Etiology
2.1. PEDV structure and genomic organization
PEDV is enveloped, pleomorphic and 95–190 nm in diameter, including the projections, which are approximately 18 nm in length (Pensaert and de Bouck, 1978). Details of the viral structure and genome, epidemiology, evolution, and replication of PEDV are described by Thiel and colleagues in another review in this current special issue. PEDV has a single-stranded positive-sense RNA genome of approximately 28 kb in size (excluding the poly A-tail) that encodes four structural proteins, namely, S, envelope (E), membrane (M) and nucleocapsid (N) proteins, sixteen nonstructural proteins (nsp1-nsp16) and an accessory protein ORF3 (Kocherhans et al., 2001). Many nonstructural and structural proteins inhibited type I and III interferon responses (IFNs) in vitro (Hou and Wang, 2019; Koonpaew et al., 2019; Zhang et al., 2018b; Zhang and Yoo, 2016). The S protein is critical for interactions with the specific host cell receptor to mediate viral binding and entry and the formation of syncytia, and for inducing neutralizing antibodies (Lin et al., 2016). The S protein is divided into S1 [amino acids (aa) 1–726 based on PEDV CV777] and S2 (aa 727–1386) subunits. The N-terminal S1 subunit contains the receptor binding domain and the C-terminal S2 subunit is responsible for membrane fusion (Li et al., 2016). The accessory protein ORF3 is an ion channel protein and is dispensable for virus replication in vitro (Wang et al., 2012).
2.2. Emergence of non-S INDEL (G2b) and S INDEL (G1b) PEDV strains in the US
The US G2b non-S INDEL and G1b S INDEL strains were first detected in April–May 2013 and January 2014, respectively (Huang et al., 2013; Stevenson et al., 2013; Wang et al., 2014b). The US G2b strains were genetically closest to PEDV strains that emerged in China in 2010 (Chen et al., 2014; Huang et al., 2013; Vlasova et al., 2014; Wang et al., 2014b). The G2b non-S INDEL PEDV strains that emerged in China in 2010 caused large PED outbreaks (Lee, 2015; Pensaert and Martelli, 2016) and was highly virulent (Lin et al., 2016; Stevenson et al., 2013; Wang et al., 2013). In China, the S INDEL PEDV strains were detected in 2011 (Li et al., 2012), probably resulting from recombination events between the G1a CV777-lineage classical and the G2 strains of PEDV (Lee, 2015).
2.3. Serological cross-reactivity and cross-neutralization between non-S INDEL (G2b) and classical PEDV (G1a) or S INDEL (G1b) PEDV strains in vitro
Researchers evaluated cross-reactivity or cross-neutralization between G2b non-S INDEL and G1b S INDEL PEDV using convalescent sera from pigs infected with each strain in immunofluorescent assays (IFA) or cell culture IF (CCIF) assays and virus neutralization (VN) tests in vitro (Chen et al., 2016b; Lin et al., 2015b). The IFA or CCIF PEDV antibody titers of the two convalescent sera against both PEDV strains were similar by the assays using S INDEL PEDV antigen, but the titers differed slightly (higher antibody titers against non-S INDEL than for S INDEL strain) when using non-S INDEL PEDV antigen (Chen et al., 2016b), confirming the serological cross-reactivity of G2b non-S INDEL and G1b S INDEL PEDV. Despite the nucleotide and amino acid differences in their S gene (Sato et al., 2018), there was cross-neutralization of G2b non-S INDEL and G1b S INDEL PEDV in vitro (Chen et al., 2016b; Lin et al., 2015b). There was also a varying but moderate degree of cross-reactivity and cross-neutralization between the two strains and the G1a classical CV777 PEDV strain (Lin et al., 2015b). However, there were 4- to 16-fold differences in the CCIF and VN antibody titers of pig antisera between either of the two strains and G1a CV777 (Lin et al., 2015b).
2.4. Cross-protection between non-S INDEL (G2b) and S INDEL (G1b) PEDV in vivo
One study reported that four sows exposed via feedback to a G1b S INDEL PEDV at approximately seven months pre-farrowing and then re-exposed to a G2b non-S INDEL PEDV at day 109 of gestation provided long-term (7 months), passive immune protection of piglets against challenge with the same G2b non-S INDEL PEDV (Goede et al., 2015). There was 0% mortality rate of piglets (with 57 % reduced incidence of diarrhea), compared with mean 33 % mortality and 100 % morbidity rates of piglets born to non-immunized sows. However, another study revealed that more than 80 % of the piglets inoculated orally with a G1b S INDEL PEDV (Iowa106 strain) at 3−4 days of age had diarrhea after challenging with a G2b non-S INDEL PEDV at 24 days of age, whereas none of the piglets inoculated previously and then challenged with the same non-S INDEL PEDV showed diarrhea (Annamalai et al., 2017; Lin et al., 2015a).
2.5. Serological properties and cross-reactivity of PEDV with other swine or animal coronaviruses
Antigenic cross-reactivity or cross-neutralization between PEDV and feline infectious peritonitis virus (FIPV) was detected by enzyme linked immunosorbent assay (ELISA), immunoblotting and immune-precipitation (Zhou et al., 1988), or VN tests (Zhao et al., 2019). Despite no serological cross-neutralization detected between PEDV and TGEV (Hofmann and Wyler, 1989; Pensaert and de Bouck, 1978), investigators found some antigenic cross-activity between PEDV and TGEV (Miller, but not Purdue strain) based on at least two conserved epitopes on the N-terminal region of their N proteins, as well as via their M proteins or whole virus particles (Gimenez-Lirola et al., 2017; Lin et al., 2015b; Xie et al., 2019). Truncation of the N-terminal region of the N protein or avoidance of M- or purified whole virus-based serologic tests may eliminate PEDV and TGEV shared epitope(s) or reduce the possible cross-reactivity for serologic assays (Gimenez-Lirola et al., 2017; Xie et al., 2019). Polyclonal hyperimmune antisera against PDCoV did not cross-react with PEDV (Jung et al., 2015c). Among N, M and E proteins and whole PEDV particles, only the M protein of PEDV showed cross-reactivity in 1/12 convalescent sera from PDCoV-infected pigs (Gimenez-Lirola et al., 2017). However, one study reported antigenic cross-reactivity between US PDCoV and PEDV strains, possibly due to at least one shared epitope in the N-terminal region of their N proteins (Ma et al., 2016). For SADS-CoV, monoclonal antibody against the N protein did not cross-react with PEDV, TGEV or PDCoV (Pan et al., 2017).
3. Transmission
3.1. Direct or indirect contact transmission
The fecal–oral route is the main means of direct transmission of PEDV via the feces and/or vomitus of infected pigs (Jung and Saif, 2015). Indirect contact transmission of PEDV is also frequent within and between farms, particularly, with a low biosecurity, via other contaminated fomites including (Jung and Saif, 2015; Kim et al., 2017b): transport trailers (Lowe et al., 2014), farm workers’ hands, boots and clothes (Kim et al., 2017b), feed (Bowman et al., 2015a; Dee et al., 2014; Schumacher et al., 2017), feed ingredients and additives, such as spray-dried porcine plasma (Pasick et al., 2014; Perri et al., 2018), and feed totes used for transporting bulk feed or feed ingredients (Anon., 2015; Scott et al., 2016). PEDV remained infectious on tote material for 35 days at room temperature (Scott et al., 2016). PEDV cross-contamination also occurred during feed manufacturing (Schumacher et al., 2018).
3.2. Aerosol (indirect contact) transmission
The fecal–nasal route is another route of pig-to-pig, or farm-to-farm (up to 10 miles away) airborne transmission of PEDV via aerosolized PEDV particles that are infectious in nursing pigs (Alonso et al., 2014, 2015; Beam et al., 2015; Gallien et al., 2018a; Li et al., 2018). Airborne PEDV transmission occurs within farrowing herds where newborn piglets, highly susceptible to the virus, are raised (Alonso et al., 2015; Niederwerder et al., 2016). The nasal cavity of naïve pigs housed at a distance from clinical pigs was frequently positive for PEDV RNA (Niederwerder et al., 2016). Aerosolized PEDV does not infect only the intestine of pigs (Alonso et al., 2015), but it also infects the epithelium lining the nasal cavity (Li et al., 2018). Li et al. (2018) reported that dendritic cells in the lamina propria of the nasal mucosa or lymphoid tissue carry and transfer PEDV to CD3+ T cells. PEDV-loaded T cells may reach the intestine via unknown specialized endothelial venules during blood circulation. The virus loaded onto T cells had the capacity to infect intestinal epithelial cells via cell-to-cell contact and transfer infection (Li et al., 2018). Compared with neonatal piglets, however, higher doses of aerosolized PEDV may be required to infect weaned and older pigs (Niederwerder et al., 2016). In the study, none of aerosol contact, 28-day-old pigs (0/5 pigs) was infected, although their nasal cavities (5/5 pigs) were positive for PEDV RNA.
3.3. Variation in direct contact or aerosol transmission rates of PEDV among farms
The severity of PEDV infection and the disease, and the transmissibility of PEDV, depends on the overall immunity and health status of the pig population and the levels of biosecurity on farms (Pensaert and Martelli, 2016). Nevertheless, transmissibility of PEDV via direct or aerosol contact also varies dependent on the PEDV genogroup. Direct contact or aerosol transmission rates were significantly higher in pigs infected with non-S INDEL PEDV compared with S INDEL-infected pigs (Gallien et al., 2018a). Gallien et al. (2018) revealed that despite the presence of aerosolized S INDEL PEDV, none of aerosol contact pigs (0/10 pigs) was infected, whereas 10/10 aerosol contact pigs were infected by non-S INDEL PEDV (Gallien et al., 2018a).
4. Disease mechanisms and pathogenesis of PEDV
4.1. Cellular receptor for PEDV
Porcine small intestinal villous enterocytes express aminopeptidase N (APN), a 150-kDa glycosylated transmembrane protein, which was tentatively identified as the cellular receptor for PEDV (Li et al., 2007; Liu et al., 2015a; Nam and Lee, 2010). However, porcine APN may not be the major cell surface receptor for PEDV (Ji et al., 2018; Li et al., 2017; Shirato et al., 2016). Concomitantly, APN knockout pigs were susceptible to infection with PEDV, but not with TGEV that uses APN as the cellular receptor (Whitworth et al., 2019; Zhang et al., 2019). Cell membrane cholesterol or two cell surface molecules, such as sialic acids and occludin expressed on the apical surface of secretary (goblet) or absorptive enterocytes, respectively, were involved partially in binding and entry of PEDV into enterocytes (Deng et al., 2016; Jeon and Lee, 2017; Li et al., 2016; Luo et al., 2017). The strains of PEDV, such as cell culture-adapted vs. wild-type, may be a factor involved in the cellular entry via sialic acids (Deng et al., 2016; Li et al., 2016). Intracytoplasmic localization of PEDV antigen in goblet cells of infected pigs and porcine intestinal enteroids in vitro (Jung and Saif, 2017; Li et al., 2019) raises questions on whether or how goblet cells are utilized for PEDV replication, or if they only transfer PEDV to absorptive enterocytes.
4.2. Tissue tropism of PEDV in the gastrointestinal tract
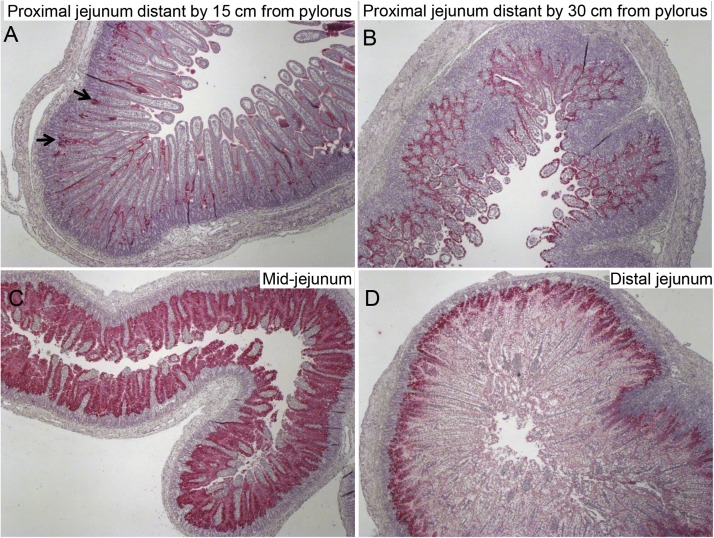
We demonstrated the tissue tropism of PEDV to certain intestinal locations (Jung et al., 2018). During acute PEDV infection [post-inoculation hour (PIH) 12–24] in nursing pigs, PEDV initially infected mainly the mid-jejunum and ileum and to a lesser extent, the proximal and distal jejunum and duodenum (Jung et al., 2018) (Fig. 1 ). The pylorus was not the site of acute PEDV infection (Jung et al., 2018), whereas the villous enterocytes of the large intestine were frequently infected; however, infected colonic enterocytes did not undergo necrosis (Jung et al., 2015a, 2014). Unlike colonic enterocytes, PEDV-infected small intestinal villous enterocytes undergo acute necrosis and exfoliation from the lamina propria, leading to marked villous atrophy or fusion in the small intestine (Jung et al., 2018, 2014). Like TGEV (Kim et al., 2000) and PDCoV (Jung et al., 2016), PEDV may not induce apoptotic death of intestinal villous enterocytes in vivo (Jung and Saif, 2015). However, PEDV-infected Vero cells in vitro underwent apoptosis (Kim and Lee, 2014).
Fig. 1.
Immunohistochemical detection and distribution of PEDV antigen-positive cells in the proximal jejunum, mid-jejunum, or distal jejunum of a gnotobiotic 9-day-old pig inoculated with the original US non-S INDEL PEDV strain PC21A. The pig exhibited vomiting (but no diarrhea) at post-inoculation hour (PIH) 16 and was euthanized. (A) Proximal jejunum distant by 15 cm from the pylorus, showing low numbers of PEDV antigen-positive cells (red color) in the villus-crypt interface (arrows). (B) Proximal jejunum distant by 30 cm from the pylorus, showing high numbers of PEDV antigen-positive cells (red color) in most of the villous epithelium. (C) Mid-jejunum [mid-location of the small intestine (duodenum to ileum)], showing extremely high numbers of PEDV antigen-positive cells (dark red color) in most of the epithelium of mildly atrophied villi. (D) Distal jejunum, showing moderate numbers of PEDV antigen-positive cells (red color) in the villus-crypt interface. Original magnification, all ×40. Fast Red, Gill’s hematoxylin counterstain.
4.3. Cellular tropism of PEDV to the intestinal villus-crypt interface during acute infection
During acute PEDV infection (PIH 12–24) of nursing pigs, early localization of PEDV antigen was evident in the villous-crypt interface of the small intestine, rather than the villous tips (Jung et al., 2018) (Fig. 1). Immature enterocytes were the major site of initial PEDV infection, although the exact reason remains obscure (Jung et al., 2018). The PEDV antigen-positive regions subsequently expanded to the upper and then the entire villous epithelium of the jejunum to ileum (<24 h after oral inoculation) (Jung et al., 2018). The villous-crypt interface is close to blood vessels in the submucosa. Although PEDV-related viremia might be a result of diffusion of replicated PEDV from the acutely infected intestine to blood (Jung et al., 2018), further studies are needed to investigate whether PEDV or PEDV-loaded CD3 + T cells circulating in blood reach and infect the villous-crypt interface or other villous regions, and other types of cells of extra-intestinal origin (Jung et al., 2018; Li et al., 2018).
4.4. Interactions of PEDV with other cells besides villous epithelial cells
PEDV antigens were also frequently detected in the intestinal crypt cells or antigen presenting cells, such as macrophages, in the lamina propria or Peyer's patches (Debouck et al., 1981; Jung et al., 2014; Lin et al., 2015a; Madson et al., 2016; Stevenson et al., 2013; Sueyoshi et al., 1995). PEDV remains infectious in dendritic cells for <24 h in vitro (Gao et al., 2015), but dendritic cells may not be a site of PEDV replication (Wang et al., 2017). Instead, PEDV appeared to employ dendritic cells to cross the epithelial barrier of the nasal cavity (Li et al., 2018). Lung tissues of oronasally infected pigs were negative for PEDV antigen (Debouck et al., 1981; Jung et al., 2014; Stevenson et al., 2013; Sueyoshi et al., 1995). However, the upper respiratory tract may be the site of PEDV infection, because the epithelium lining the nasal cavity was positive for PEDV antigens (Li et al., 2018). Although acute viremia was frequently noted in infected pigs during the acute or incubation stage of infection (Jung et al., 2018, 2014; Madson et al., 2016), PEDV antigens were not detected in other major organs, such as liver and kidneys (Debouck et al., 1981; Jung et al., 2014; Stevenson et al., 2013; Sueyoshi et al., 1995). The reproductive organs of experimentally infected boars, such as Cowpers’s glands, were also negative for PEDV RNA, but their semen was transiently positive for PEDV RNA (Gallien et al., 2018b, 2019). Nevertheless, whether the PEDV RNA-positive semen contained infectious viral particles is unclear.
4.5. Intestinal replication of PEDV during disease progression
During the incubation period (<PIH 16) in nursing pigs, a few to 50 % of the villous epithelial cells positive for PEDV antigen were seen in the villous-crypt interface and upper villous epithelium of the small intestine (Debouck et al., 1981;Jung et al., 2018). During this period, infected enterocytes underwent necrosis, but complete villous atrophy was not observed (Jung et al., 2018). When clinical signs were first seen (PIH 16–24), moderate to large numbers of infected villous epithelial cells were observed throughout the small intestine (Debouck et al., 1981). During this period, up to 100 % of the epithelial cells were positive for PEDV antigen in the mid-jejunum to ileum where moderate to severe villous atrophy was observed (Jung et al., 2018). During the middle [post-inoculation day (PID) 1–3] to late (PID 4−5) stages of PEDV infection of nursing pigs (5–9-day-old), regardless of the locations (except for the duodenum with milder villous atrophy), all small intestinal segments exhibited similar, extensive villous atrophy (mean 1−2 intestinal villous height to crypt depth ratios, compared with 5–9 of uninfected pigs) (Chen et al., 2016a; Jung et al., 2015a). During this period, large numbers of PEDV-infected epithelial cells could still be observed (Debouck et al., 1981).
4.6. Pathophysiology
Diarrhea induced by PEDV is a consequence of malabsorption and maldigestion due to massive loss and functional disorders of infected enterocytes, followed by decreased brush border membrane-bound digestive enzymes (Coussement et al., 1982; Jung et al., 2006). Dehydration is exacerbated by vomiting and decreased appetite (Jung and Saif, 2015). Serotonin is involved in induction of vomiting, and increased pro-inflammatory cytokine responses in part led to decreased appetite (Jung et al., 2018). PEDV-infected piglets also showed hyperkalemia and acidosis due to the loss of bicarbonate (Jung and Saif, 2015). The cardiac contractility is impaired by acidosis and dehydration. In the small intestine of clinically infected pigs, the tight or adherens junctions of the villous enterocytes were destroyed (Jung et al., 2015b). Expression of the junction proteins, such as zonula occludin-1, was also decreased or altered, followed by decreased transepithelial resistance (Curry et al., 2017; Jung et al., 2015b; Luo et al., 2017; Zong et al., 2019). During the clinical period, there were remarkably reduced numbers of goblet cells in the small intestine (Jung and Saif, 2017), followed by decreased amounts of mucins (Curry et al., 2017; Jung and Saif, 2017). The damaged gut integrity may lead to uptake of luminal food and bacteria causing allergic reactions and co-infections. The impaired gut integrity or digestive function usually recovered by the second week after infection in survived pigs (Curry et al., 2017).
4.7. Onset of diarrhea, fecal virus shedding and peak viral titers
In infected gnotobiotic or conventional nursing piglets (1−10-day-old), a few (<5%) to all of the inoculated pigs began to show diarrhea at PID 1, which mostly coincided with or was detected several hours later than the first detection of viral RNA in serum or feces (Chen et al., 2016a; Gerber et al., 2016; Jung et al., 2015a, 2018; Liu et al., 2015c; Madson et al., 2016; Thomas et al., 2015a). Fecal viral RNA titers shed peaked by PID 1, or occasionally, 1–3 days later, and the titers decreased remarkably during the middle to late stages of infection and then titers remained low (with at least one more recurrent peak, possibly due to PED re-infection of regenerating enterocytes) during the recovery stage of infection (Chen et al., 2016a; Jung et al., 2015a; Lin et al., 2015a; Madson et al., 2016). A one-three-day delay in onset of diarrhea was observed in nursing pigs inoculated with S INDEL PEDV compared with the non-S INDEL counterparts (Gallien et al., 2018a; Lin et al., 2015a), indicating a longer incubation time for S INDEL PEDV. In infected conventional weaned to feeder pigs (21−56-day-old), a few (<5%) to all of the inoculated pigs began to show diarrhea at PID 1–4, which coincided with or was detected 1−2 days later than the first detection of viral RNA in feces (Gallien et al., 2018a; Gerber et al., 2016; Jung et al., 2015a; Lohse et al., 2017; Madson et al., 2014; Niederwerder et al., 2016; Thomas et al., 2015a) (Table 1 ). Fecal viral RNA titers shed peaked by 1–3 days after diarrhea was first detected (Gallien et al., 2018a; Jung et al., 2015a; Lohse et al., 2017; Madson et al., 2014; Niederwerder et al., 2016; Thomas et al., 2015a). Detailed data from boars or sows are summarized in Table 1.
Table 1.
Summary of clinical signs and fecal viral RNA shedding in conventional weaned and feeder pigs, and boars and sows experimentally infected with different genogroups of porcine epidemic diarrhea virus (PEDV) strains via oral inoculation, or direct or indirect aerosol contact.
| Pig | Age (days or years) at inoculation or exposure | PEDV strain and inoculum dose | Infection or exposure route | Onset of diarrhea (PID)* | Duration of diarrhea (PID) | Onset of fecal viral RNA shedding (PID) | Duration of fecal viral RNA shedding | Peak viral RNA titers in feces or rectal swab fluids | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Weaned | 21 days | US non-S INDEL, USA/Iowa/18984/2013, 1 mL of 1 × 103 PFU/mL* | Oral | PID 2 in 8/32 pigs (25 %) | PID 2−7 | PID 1 | Up to PID 24 | 4 × 104 PFU/mL at PID 4 | Madson et al., 2014 |
| Weaned | 21 days | US non-S INDEL, USA/IN19338/2013, 10 mL of 5.6, 56, or 560 TCID50/mL* | Oral | PID 2 in 1/6 pigs (16.7 %) | PID 2−7 | PID 2 | Up to PID 21 | Approximately 5−7.5 log10 RNA copies/mL at PID 5−6 | Thomas et al., 2015a |
| Weaned | 28 days | US non-S INDEL, USA/2014/Iowa, jejunal homogenate from infected pigs, 5 mL of 108 RNA copies/mL | Oral | PID 1 (2/2; 100 %) | NR¶ (but with sporadic vomiting at PID 1−31 and lethargy at PID 2) | PID 1 | Up to PID 21 | 11.5 log10 RNA copies/mL at PID 2−3 | Gallien et al., 2018a |
| Direct contact | PID 2 (6/8; 75 %) | PID 1−2 | Up to PID 42 | 11.1−11.4 log10 RNA copies/mL at PID 2−5 | |||||
| Indirect aerosol contact | PID 2 (8/10; 80 %) | NR (but with sporadic vomiting at PID 11−46) | PID 2 | Up to PID 49 | 10.5−12.2 log10 RNA copies/mL at PID 2−6 | ||||
| European S INDEL, FR/001/2014, jejunal homogenate from infected pigs, 5 mL of 108 RNA copies/mL | Oral | PID 4 | PID 4−25 (with sporadic vomiting and lethargy) | PID 2 | Up to PID 21 | 9.7−10.2 log10 RNA copies/mL at PID 4−6 | |||
| Direct contact | PID 4 | PID 4−14 | Up to PID 49 | 6.6−9.3 log10 RNA copies/mL at PID 6−21 | |||||
| Indirect aerosol contact | No clinical signs in 10 pigs (0/10) | No clinical signs | No fecal shedding | No fecal shedding | No fecal shedding | ||||
| Weaned to feeder | 28 days | US non-S INDEL, USA/KS/2013, 5 mL of 1.42 × 105 TCID50/mL | Oral | PID 3 | PID 3−8 | PID 2 | Up to PID 28 (1/12 pigs) | Titers not determined, but lowest Ct values in fecal and nasal swabs at PID 5−6 | Niederwerder et al., 2016 |
| Direct contact | PID 3 | PID 3−8 | PID 3 (with nasal shedding at PID 2) | Up to PID 21 (2/5 pigs) | |||||
| Indirect aerosol contact | No clinical signs in 5 pigs (0/5) | No clinical signs | No fecal shedding | No fecal shedding | No fecal shedding | ||||
| Weaned to feeder | 35 days (5 weeks) | US non-S INDEL, jejunal homogenate from a pig in Iowa | Oral | PID 4 | NR | PID 1−2 | Up to PID 18 | Titers not determined, but lowest Ct values at PID 2−6 | Lohse et al., 2017 |
| European, classical S INDEL, cell culture-grown Br1/87, 2 mL of 1 × 106.8 TCID50/mL | Oral | PID 4 | NR | PID 2−3 | Up to PID 18 | Titers not determined, but lowest Ct values at PID 3−8 | |||
| Feeder vs. nursing | 56 days (8 weeks) vs. 10 days | US non-S INDEL, USA/Colorado/2013, 10 mL of 9 × 104 PFU/mL | Oral in feeder pigs | PID 2 in 3 of 40 pigs (7.5 %) | NR | PID 1 in 9 of 48 pigs (18.7 %) | At least up to PID 14 in 9/20 pigs (45.0 %) | NR | Gerber et al., 2016 |
| Oral in nursing pigs | PID 1 in 2 of 43 pigs (4.6 %) | NR | PID 1 in 8 of 43 pigs (18.6 %) | At least up to PID 14 in 20/33 pigs (60.6%) | NR | ||||
| Boars | 1.5−2.5 years | US non-S INDEL, USA/2014/Iowa, jejunal homogenate from infected pigs, 5 mL | Oral | PID 2 | PID 2−5 and PID 25−28 (with vomiting at PID 2 and 11) | PID 2 | Up to PID 21−28 | 6.8 × 108 to 5.7 × 109 RNA copies/mL at PID 3−4 | Gallien et al., 2018b |
| Sows | Multiparous sows | European S INDEL, German cell culture-grown strain, 6 mL of 3.16 × 105 TCID50/mL | Oral | No diarrhea | No diarrhea (but with slight depression in 2/2 sows for 4 days) | PID 4−5 | Up to PID 10−14 | NR | Leidenberger et al., 2017 |
| European S INDEL, jejunal homogenate from a German pig | Oral | PID 1 (with vomiting) | NR | PID 4 | Up to PID 12−13 | NR | |||
| Gilts | Pregnant gilts§ | US non-S INDEL, PC22A, 1 × 105 PFU/pig | Oral | PID 2−4 | PID 2−6 | PID 2−4 | At least up to PID 14 | Approximately 8.5−10.0 log10 RNA copies/mL at PID 4−6 | Langel et al., 2019a |
Note no mortality of PEDV-infected nursing, weaned, or feeder pigs reported in these studies.
TCID50, 50 % tissue culture infectious dose; PFU, plaque forming unit; PID, post-inoculation day.
NR, not reported.
Pregnant gilts were inoculated with PEDV at the early (day 19–22), middle (day 57–59), or late (day 96–97) gestation periods.
4.8. Duration of diarrhea and fecal virus shedding
Duration of diarrhea in infected nursing pigs varied from 2 to 8 days, with a higher variability for the US S INDEL strain (2−7 days) compared with the US non-S INDEL (7–8 days) (Lin et al., 2015a). Prolonged fecal viral RNA shedding (at least PID 14) was evident in 20/33 (61 %) US non-S INDEL PEDV-infected nursing (10-day-old) pigs (Gerber et al., 2016) (Table 1). Diarrhea in weaned to feeder pigs (21−56-day-old) inoculated orally with US non-S INDEL PEDV was observed for approximately 6 days (Madson et al., 2014; Niederwerder et al., 2016; Thomas et al., 2015a). Prolonged fecal viral RNA shedding was evident in non-S INDEL or S INDEL PEDV-infected weaned or feeder pigs for 15–26 days (Gallien et al., 2018a; Lohse et al., 2017; Madson et al., 2014; Niederwerder et al., 2016; Thomas et al., 2015a). Duration of fecal shedding was similar between non-S INDEL- (15–26 days) (Gallien et al., 2018a; Lohse et al., 2017; Madson et al., 2014; Niederwerder et al., 2016; Thomas et al., 2015a) and S INDEL-infected weaned or feeder pigs (15–18 days) (Gallien et al., 2018a; Lohse et al., 2017). Longer fecal viral RNA shedding was observed in weaned pigs infected via direct contact (42−46 days) or aerosol contact (48 days) compared with the orally infected counterparts (18-21 days) (Crawford et al., 2015; Gallien et al., 2018a). However, infectious PEDV was shed in feces only for the initial 2 weeks tested using sentinel naïve pigs (Crawford et al., 2015).
4.9. Higher susceptibility of neonatal pigs to non-S INDEL PEDV infection compared with older pigs
The minimum infectious dose [0.56 median tissue culture infectious dose (TCID50)] of non-S INDEL PEDV required to infect 5-day-old nursing pigs was lower than for 3-week-old weaned pigs (56 TCID50) (Thomas et al., 2015a). Similarly, 3 plaque-forming unit (PFU)/pig of the non-S INDEL PC22A strain caused diarrhea in 100 % of 4-day-old Cesarean-derived, colostrum-deprived (CDCD) piglets (Liu et al., 2015c). Compared with nursing pigs that began to show diarrhea, villous atrophy, and fecal virus shedding at PID 1, a longer incubation time of non-S INDEL PEDV was required for weaned pigs to show fecal virus shedding (1 more day), or diarrhea and lesions (2 more days) (Jung et al., 2015a). One factor contributing to the higher susceptibility of nursing pigs to non-S INDEL PEDV infection is immaturity of gut innate immunity, as evident by a functional defect in natural killer cells in the ileum and blood (Annamalai et al., 2015). The lack of diversity and decreased Firmicutes to Bacteroidetes ratio in the gut microbiota of neonatal piglets contributed to the immature gut innate immunity (Zhao et al., 2015). Compared with grower pigs (gastric pH 2−3), neonatal pigs have a higher gastric pH (4−6) that allows PEDV to survive in the stomach (Mavronmichalis, 2016).
4.10. Lower vulnerability of older pigs to PED compared with neonatal pigs
Compared with nursing pigs, several factors that influence a shorter recovery of weaned pigs from PED include: 1) the faster regeneration of enterocytes in weaned pigs compared with nursing pigs (Moon et al., 1973); and 2) the anatomically and physiologically fully developed large intestine of weaned pigs where water reabsorption is greater (Jung et al., 2015a). There was a lack of LGR5 (leucine-rich repeat-containing G protein-coupled receptor 5, marker for crypt stem cells)-positive crypt base columnar cells (LGR5+ cells) and low proliferation of crypt cells in the small intestine of naïve nursing piglets (9-day-old), causing the slower turnover of enterocytes (Jung et al., 2015a). Unlike nursing pigs, naïve weaned pigs (3-week-old) exhibited high proliferation of intestinal crypt cells and large numbers of LGR5+ cells in the crypts, leading to the higher turnover rate of enterocytes (Jung et al., 2015a). After PEDV infection, large numbers of LGR5+ cells and high proliferation of crypt cells were maintained, possibly relating to the rapid recovery from PED in weaned pigs.
4.11. Viremia
Detection of viremia where viral RNA in serum ranged from 4.5 to 8.6 log10 genomic equivalents (GE)/mL was identified in gnotobiotic neonatal (5/5; 100 %), or conventional 9-day-old nursing (16/16; 100 %) and 26-day-old weaned pigs (11/20; 55 %) infected with a US non-S INDEL PEDV strain at PID 1−5 (Jung et al., 2015a, 2014). PEDV RNA was detected in all sera from 20 nursing pigs infected with either one of three different US non-S INDEL strains or a US S INDEL strain at PID 3 (Chen et al., 2016a). The classical PEDV Br1/87 also induced viremia in 5/13 inoculated weaned pigs (38.5 %) at PID 2−7 (Lohse et al., 2017). In experimentally infected nursing pigs, the frequency of viremia and serum viral RNA titers peaked at PID 1, then decreased progressively, and ceased at PID 14–21 (Chen et al., 2016a; Jung et al., 2015a; Opriessnig et al., 2014). A significantly higher frequency of viremia in weaned pigs infected via direct or aerosol contact, or orally with non-S INDEL PEDV was identified, compared with the S INDEL counterparts (Gallien et al., 2018a). The role of viremia in the pathogenesis of PEDV is still obscure, although PEDV identified in the serum could be infectious (Li et al., 2018; Pasick et al., 2014; Perri et al., 2018).
4.12. Intestinal microbiota and PEDV infection
During the clinical period, alteration in the abundance or proportion of gut microbiota (dysbiosis) occurred in nursing pigs and their dams (Huang et al., 2018; Koh et al., 2015; Liu et al., 2015b; Song et al., 2017; Tan et al., 2019). The transfer of a healthy gut microbiota from infected sows to their offspring was impaired (Song et al., 2017). The dysbiosis was characterized by reduced numbers of beneficial bacteria (Verrucomicrobia) and increased numbers of harmful bacteria (Fusobacterium and Proteobacteria) (Liu et al., 2015b; Tan et al., 2019), followed by decreased cellular transport and catabolism (Huang et al., 2018). A study showed that feeding Bacillus subtilis to 2-week-old pigs infected with PEDV reduced intestinal pathology (but not clinical disease) (Canning et al., 2017). Similarly, live or cell-free supernatants of some Lactobacillus plantarum strains had antiviral effects against PEDV infection in Vero cells (Sirichokchatchawan et al., 2017). However, this feed additive method (1/77 herds surveyed; 1.2 %) was rarely used to mitigate PED on US pig farms during the 2013–2017 epidemic (Niederwerder and Hesse, 2018).
4.13. S INDEL PEDV has lower infectivity compared with non-S INDEL PEDV
Early studies verified high enteropathogenicity of the G1a CV777 in nursing pigs (Coussement et al., 1982; Pensaert and Martelli, 2016). The S1 of the S protein and the rest of the genome of the S INDEL PEDV (G1b) was genetically closest to that of G1a and the highly virulent G2 PEDV, respectively (Lee, 2015; Pensaert and Martelli, 2016; Vlasova et al., 2014), the strain was initially thought to be virulent. However, during the initial outbreaks by S INDEL PEDV in the US and Europe, the low to zero mortality rates of pigs in the field were distinct from those of US non-S INDEL infections with up to 100 % mortality rates in nursing pigs (Boniotti et al., 2016; Goede et al., 2015; Stevenson et al., 2013; Wang et al., 2014b). Nevertheless, difference in virulence of the two types of strains in the field was not often observed (Grasland et al., 2015; Mesquita et al., 2015; Stadler et al., 2015; Steinrigl et al., 2015). A study using a wild-type strain of S INDEL PEDV reproduced as severe PED in infected sows and their piglets as that observed in the field (Leidenberger et al., 2017). Other studies showed generally milder pathogenicity of S INDEL PEDV in 3−7-day-old conventional pigs, compared with non-S INDEL PEDV (Chen et al., 2016a; Lin et al., 2015a; Yamamoto et al., 2015). However, the virulence of S INDEL PEDV was increased in the PEDV challenged suckling piglets when the mothers became sick, corresponding to poor immune, nutritional or health conditions (Lin et al., 2015a). The mortality rates of piglets increased from 0% to 75 %, and the peak fecal PEDV RNA titers shed were also as high as those of non-S INDEL strain.
4.14. Clinical disease, fecal shedding, or lesions of pigs inoculated orally with attenuated PEDV
The virulence of PEDV was attenuated by: 1) high cell-culture passage (93rd–160th) in the cell line of non-swine origin, Vero cells (with trypsin in the cell culture medium) (Chen et al., 2015; Kweon et al., 1999; Lin et al., 2017a; Sato et al., 2011; Song et al., 2003); 2) modified cell culture conditions or additives for PEDV growth [glycochenodeoxycholic acid (GCDCA) instead of trypsin] (Kim et al., 2017a); and 3) modified viral genes by reverse genetics (Beall et al., 2016; Deng et al., 2019; Hou et al., 2019a, 2017; Hou et al., 2019b; Teeravechyan et al., 2016). In addition, PEDV variants of mild virulence, such as the S1 N-terminal domain (NTD)-Del variant, also occurred naturally (Su et al., 2018; Suzuki et al., 2016; Zhang et al., 2018a). PEDV strains fully adapted to Vero cells and serially passaged at least 70 times (with trypsin or GCDCA in the cell culture medium) tended to replicate less efficiently in the intestine of pigs (Chen et al., 2015; Kim et al., 2017a; Lin et al., 2017a; Sato et al., 2011; Wu et al., 2019) (Table 2 ) and elicited low serum PEDV-specific antibody titers (Lin et al., 2017a). Twelve-week-old pigs orally inoculated with 8.2 log10 TCID50 of G1a PEDV 83P-5 (100th passage in Vero cells) exhibited only intermittent fecal viral RNA shedding in 3 of 4 pigs (Sato et al., 2011). None of the 10-day-old pigs orally inoculated with 6 log10 TCID50 of G2b PEDV YN (144th passage in Vero cells) showed fecal viral RNA shedding; however, PEDV antigen was detected in the small intestine (Chen et al., 2015). Compared with G2b PEDV YN (144th) and PC22A (140–160th passage in Vero cells) grown with trypsin, G2b PEDV 8aa (70th–105th passage in Vero cells) grown with GCDCA and attenuated at lower cell passage numbers, caused no or little fecal viral RNA shedding in 1-day-old pigs (6 log10 TCID50/pig) (Kim et al., 2017a). Four-day-old CDCD pigs orally inoculated with 2 log10 PFU of PC22A (140th–160th) showed low to moderate fecal viral RNA shedding titers, with mild villous atrophy (Lin et al., 2017a) (Table 2). Six-day-old conventional pigs orally inoculated with 6 log10 TCID50 of G2b PEDV CT (120th passage in Vero cells) showed mild diarrhea and moderate fecal viral RNA shedding titers, with mild villous atrophy (Wu et al., 2019). On the other hand, pregnant sows orally inoculated with G1a PEDV DR13 (100th passage in Vero cells) showed no clinical signs but high VN antibody levels in the colostrum (Song et al., 2007). Attenuated PEDV strains (highly passaged in Vero cells) were less infectious, replicative, and immunogenic in the intestine of pigs (with mild or no clinical disease or lesions) compared with the corresponding virulent PEDV strains.
Table 2.
Summary of in vivo results from conventional or caesarean-derived, colostrum-deprived (CDCD) pigs inoculated orally with attenuated porcine epidemic diarrhea virus (PEDV) strains.
| Strain (cell passage number) | Genogroup | Dose per pig | Age of pigs inoculated | Animal number | Mortality (%) | Clinical signs | Fecal viral RNA shedding | Gross or histologic lesions | PEDV antigen in the intestine | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| 83P-5 (100th) | 1a (classical S INDEL) | 8.2 log10 TCID50* | 12-week | 4 | NR¶ | No diarrhea at PID 1−14 | Intermittent detection of viral RNA in the feces of 3/5 pigs at PID 3−9*, as tested at PID 1−14 | NR | NR | Sato et al., 2011 |
| YN144 (144th) | 2b (non-S INDEL) | 6 log10 TCID50 | 10-day | 4 | NR | No diarrhea at PID 1−5 | No detection [possibly due to no necrosis of virus-infected (antigen-positive) enterocytes, as claimed by the authors] at PID 1 and 5 | No gross lesions at PID 5 | Low to moderate numbers of positive enterocytes lining the villous epithelium at PID 5 | Chen et al., 2015 |
| PEDV 8aa (70th) (with GCDCA)§ | 2b (non-S INDEL) | 6 log10 TCID50 | 1-day | 15 | 0 at PID 1−14 | Loose stool at PID 1−2, as monitored at PID 1−14 | No or little viral RNA shedding at PID 1−14 | NR | NR | Kim et al., 2017a |
| PEDV 8aa (105th) (with GCDCA)§ | 2b (non-S INDEL) | 6 log10 TCID50 | 1-day | 16 | 0 at PID 1−14 | No diarrhea at PID 1−14 | No or little viral RNA shedding at PID 1−14 | NR | NR | |
| PC22A (120th) | 2b (non-S INDEL) | 2 log10 PFU* | 4-day CDCD | 6 | 0 at PID 1−10 or 15 | Mild diarrhea in 3/4 pigs (75 %) at PID 1−10 or 15 | High (with mean peak viral RNA shedding titer of 10.19 log10 GE/mL)*, as tested at PID 1−10 or 15 | Moderate villous atrophy at PID 9 (mean VH:CD ratio of 3.6 vs. 6.8 in the control)* | Low numbers of positive enterocytes lining the villous epithelium at PID 9 | Lin et al., 2017a |
| PC22A (140th) | 2b (non-S INDEL) | 2 log10 PFU | 4-day CDCD | 6 | 0 at PID 1−10 or 15 | Loose stool in 2/4 pigs (50 %) at PID 1−10 or 15 | Moderate (with mean peak viral RNA shedding titer of 7.65 log10 GE/mL), as tested at PID 1−10 or 15 | Mild villous atrophy at PID 9 (mean VH:CD ratio of 5.0 vs. 6.8 in the control) | No detection of PEDV antigen at PID 9 | |
| PC22A (160th) | 2b (non-S INDEL) | 2 log10 PFU | 4-day CDCD | 7 | 0 at PID 1−10 or 15 | Loose stool in 2/5 pigs (40 %) at PID 1−10 or 15 | Low (with mean peak viral RNA shedding titer of 5.00 log10 GE/mL), as tested at PID 1−10 or 15 | Mild villous atrophy at PID 9 (mean VH:CD ratio of 5.2 vs. 6.8 in the control) | No detection of PEDV antigen at PID 9 | |
| CT (120th) | 2b (non-S INDEL) | 6 log10 TCID50 | 6-day | 5−6 | 0 at PID 1−10 | Loose stool in some pigs at PID 4−5, at monitored at PID 1−10 | Moderate (with mean peak viral RNA shedding titer of approximate 6−7 log10 GE/mL at PID 4−5), as tested at PID 1−10 | Mild villous atrophy in a pig at PID 3 | No or little detection of PEDV antigen in a pig at PID 3 | Wu et al., 2019 |
TCID50, 50 % tissue culture infectious dose; PFU, plaque forming unit; PID, post-inoculation day; GE, genome equivalents; VH:CD ratio, villous height to crypt depth ratio.
NR, not reported.
Among PEDV strains, only PEDV 8aa was grown in Vero cells treated with glycochenodeoxycholic acid (GCDCA) instead of trypsin.
4.15. Innate or pro-inflammatory cytokine responses of pigs to PEDV infection
Despite in vitro observations where PEDV inhibits type I and type III IFN responses (Deng et al., 2019; Hou et al., 2019a; Koonpaew et al., 2019; Zhang and Yoo, 2016), there were increased systemic innate (IFNα and IL-22) and pro-inflammatory cytokine (TNFα, IL-6, IL-8, IL-12, and IL-17) responses in acutely infected neonatal pigs (Annamalai et al., 2015; Jung et al., 2018). This coincided with increased numbers of PEDV antigen-positive cells (Gao et al., 2015; Madson et al., 2016) or inflammatory cells (lymphocytes or neutrophils) in the gut-associated lymphoid tissue (GALT) (Coussement et al., 1982; de Arriba et al., 2002b; Debouck et al., 1981; Jung and Saif, 2015), and the onset of necrosis and exfoliation of infected enterocytes (Jung et al., 2018). Necrosis of enterocytes is in part involved in the innate or pro-inflammatory cytokine responses in infected pigs, as also observed in vitro (Koonpaew et al., 2019; Lin et al., 2017b). The increased systemic innate and pro-inflammatory cytokine responses that occur during acute PEDV infection are beneficial for differentiation of naïve T cells into cytotoxic or helper T cells, leading to removal of infected cells or production of VN antibodies, respectively (Siegrist, 2013). However, highly increased, prolonged pro-inflammatory cytokines that can cause decreased appetite, etc. are also detrimental (Jung et al., 2018; Langhans, 2000). Increased systemic IFNα and IFNγ cytokine responses were also observed in PEDV-infected gilts at PID 13–17 and PID 6–8, respectively (Langel et al., 2019b).
4.16. Humoral immune responses of pigs to PEDV infection
Clinically infected pigs responded to a primary PEDV infection and seroconverted at PID 7–14 (mean PID 14) (PID 14–21 for serum PEDV-specific IgA antibodies), with peak serum PEDV-specific IgG or IgA antibody titers at PID 21 (Annamalai et al., 2017; Chen et al., 2016b; de Arriba et al., 2002a,c; Gallien et al., 2018a; Gerber et al., 2016; Niederwerder et al., 2016; Opriessnig et al., 2014; Thomas et al., 2015a). There were lower serum IgG or VN antibodies in S INDEL-infected pigs compared with the non-S INDEL counterpart (Annamalai et al., 2017; Chen et al., 2016b). Serum VN antibodies were detected 7 days earlier than serum PEDV-specific IgG antibodies (Chen et al., 2016b; Thomas et al., 2015a). There were low titers of PEDV-specific secretory IgA (sIgA) in the feces of PEDV-infected pigs (10-day or 8-week-old) by PID 14 (Gerber et al., 2016). Serum PEDV-specific IgA and IgG antibodies in naturally infected sows remained high for 6 months, whereas fecal PEDV-specific sIgA antibodies disappeared 1−2 months after infection (Ouyang et al., 2015). PEDV-specific IgA or VN antibody titers of sows infected at 3 months pre-farrowing peaked in the colostrum at post-farrowing day (PFD) 1, decreased rapidly in milk by PFD 3, and then declined only gradually during PFD 4–19 (Song et al., 2016). Memory B cell responses were first identified by in vitro secondary stimulation in mononuclear cells from PEDV CV777-infected nursing pigs at PID 21 (de Arriba et al., 2002a). The number of PEDV-specific IgA antibody-positive pigs also rose from 30 % at the day of re-infection to 70 % 3 days later (Gerber et al., 2016). Similarly, the number of PEDV-specific IgA antibody secreting cells of sows infected at 3−4 months pre-farrowing and then after re-exposure to PEDV at PFD 3–5, increased in the milk at PFD 8–14, compared with PFD 0 (colostrum) or PFD 3–5, and then peaked at PFD 15–22 (Langel et al., 2019b). Previously infected sows showed increased VN and PEDV-specific IgG and IgA antibodies in the colostrum and milk after vaccination with an inactivated PEDV at 5 and 2 weeks pre-farrowing, whereas naïve sows showed no or little VN or PEDV-specific IgG and IgA antibody responses in the colostrum and milk after the same vaccination (Gillespie et al., 2018). Memory B cells, following PEDV infection or oral vaccination, may play a critical role in induction of rapid systemic and mucosal humoral immune responses to PEDV upon re-infection or booster vaccination.
5. Enhanced biosecurity as a preventive and control strategy
5.1. Herd and farm management
Enhanced biosecurity, such as decontamination and disinfection of transport trailers and other potential contaminated fomites, supply of secure feed or feed additives (Gordon et al., 2019; Niederwerder and Hesse, 2018), and maintenance of strong hygiene measures on swine farms and farm workers (Kim et al., 2017b), is critical to prevent direct or indirect contact transmission of PEDV. Transport trailers (19/72 herds surveyed; 26.4 %) or feed (21/72 herds surveyed; 29.2 %) were thought to be the main contaminated fomites (55.6 % in total) in US swine farms during the 2013–2017 epidemic (Niederwerder and Hesse, 2018). Spillover of PEDV from PEDV-infected farms to others or wildlife should also be controlled through high quarantine protocols that should include no movement of pigs until the diarrhea is terminated and the disease is controlled (Niederwerder and Hesse, 2018). Indeed, 28 of 287 fecal samples (9.8 %) of wild boars in South Korea (2010–2011) were positive for PEDV RNA (Lee et al., 2016), and 8 of 7997 serum samples (0.1 %) of US feral pigs during the 2013–2014 epidemic were PEDV-seropositive (Bevins et al., 2018). However, the role of cross-species PEDV spillover in the maintenance and transmission of PEDV needs to be further studied.
5.2. Disinfectants for PEDV inactivation, and sanitation of contaminated trailers
Use of 0.5 % Virkon S and 2.06 % Clorox that reduced the quantity of PEDV RNA were the best disinfectants for PEDV inactivation in feces (Bowman et al., 2015b). Super-oxidized water (pH 6.0) also effectively inactivated PEDV at room temperature after 10−90 min contact (Chen et al., 2017). Washing trailers with a high pressure power washer, followed by disinfection and drying, was effective for their sanitation and decontamination (Baker et al., 2018; Thomas et al., 2015b). Accelerated hydrogen peroxide or peroxygen-based disinfectants effectively inactivated PEDV on contaminated metal (trailer) surfaces at 20 °C after 30 min contact (Holtkamp et al., 2017) and at −10 °C after 40−60 min contact (Baker et al., 2017), or at -10 to 4 °C after 10−30 min contact (Baker et al., 2018), respectively. PEDV RNA was still detectable after disinfection, but no infectious virus was detected.
6. Immunoprophylaxis as a preventive and control strategy
6.1. An historic overview of the gut-mammary-sIgA axis
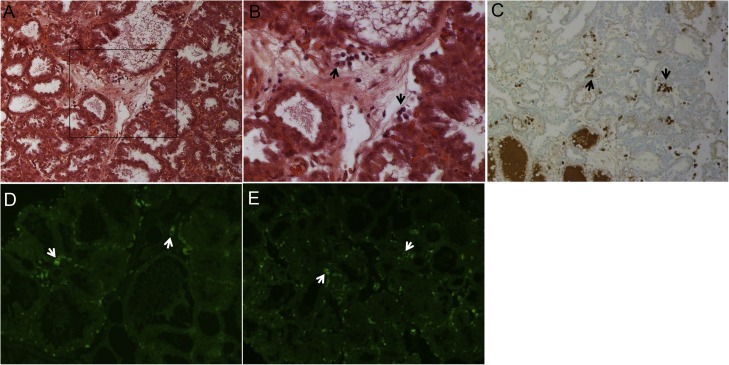
The levels of PEDV-specific sIgA antibody in the colostrum/milk of dams are key to determine the extent of passive immunity of their piglets against PEDV infection (Langel et al., 2019a,b; Langel et al., 2016, 2020; Saif et al., 2012). The sIgA antibody is resistant to proteolytic enzymes and is highly efficient at neutralizing PEDV in the intestinal mucosa (Langel et al., 2016). In the mammary glands of female swine, however, there is an absence of lymphoid follicles for antigen processing and IgA or IgG antibody production (Fig. 2 A and B). The PEDV-specific sIgA antibodies in the colostrum and milk are produced by effector B cells or plasmablasts that migrate from the GALT of the PEDV-infected intestine to the mammary gland (Langel et al., 2016, 2020; Saif et al., 2012). This is defined as the gut-mammary-sIgA axis, that was first described by Saif and Bohl in studies of immunity to TGEV [reviewed in (Saif et al., 2012)]. The details were reviewed previously (Langel et al., 2016, 2020; Saif et al., 2012). The endothelium in the mammary glands contains specialized endothelial venules that attract and bind to the specific integrins on the surface of the IgA-plasmablasts, such as α4β7 and chemokine receptor 9 (CCR9) (Bourges et al., 2008; Jiang et al., 2016; Salmon, 2000). Mucosal addressin cellular adhesion molecule 1 (MAdCAM-1) and chemokine ligand 25 expressed in the specialized venules (Fig. 2D and E) interact with α4β7 and CCR9, respectively. Extravasated sIgA antibody-secreting cells reside in the alveoli-supporting fibrous stroma of the mammary glands (Fig. 2A–C). The number of PEDV-specific sIgA antibody-secreting cells may positively correlate with the amount and density of these endothelial surface molecules in the mammary glands (Salmon, 2000), although the specific role of each surface molecule in the gut-mammary-sIgA axis needs to be better characterized.
Fig. 2.
Histologic (A and B) and immunohistochemical (C) localization of mammary IgA-secreting cells, and expression of mucosal addressin cellular adhesion molecule 1 (MAdCAM-1) (D) and chemokine ligand 25 (CCL25) (E) by immunofluorescent (IF) staining in the mammary gland tissue of PEDV naïve gilts. (A) Hematoxylin and eosin–stained mammary gland tissue of a gilt at 4 days post-farrowing, showing fully expanded alveoli, with mostly empty lumens. (B) Higher magnification of Panel A (inset), showing clusters of plasma cells characterized by the typical peripheral nuclei in the abundant cytoplasm in the interlobular stroma. Original magnification, ×200 (A) and ×400 (B). (C) Immunohistochemistry (IHC)-stained mammary gland tissue of a gilt at 5 days post-farrowing, showing a moderate number of IgA-positive cells (dark brown) in the intralobular or interlobular stroma (arrows). Original magnification, ×200. IHC was done using a polyclonal goat antibody to pig IgA conjugated with peroxidase (Bio-RAD) and DAB substrate. (D) IF-stained mammary gland tissue of a gilt at 4 days pre-farrowing, showing moderate expression of MAdCAM-1 (green) on the endothelial cells lining the intralobular and interlobular vessels (arrows). Original magnification, ×200. (E) IF-stained mammary gland tissue of a gilt at 4 days post-farrowing, showing moderate expression of CCL25 (green) on the endothelial cells lining the intralobular and interlobular vessels (arrows). Original magnification, ×200. IF staining was done using monoclonal antibodies against human MAdCAM-1 and CCL25 (Novus Biologicals).
6.2. Whole-herd feedback
Immunization of pregnant sows is important in the control of epidemic PED and to reduce the number of deaths of suckling piglets (Jung and Saif, 2015; Langel et al., 2020). The most common practice used to initiate herd immunity in US pig farms during the 2013–2017 epidemic when no PEDV vaccines were available, was use of whole-herd feedback using a load-close-expose protocol (59/77 herds surveyed; 76.6 %), in which a controlled exposure for gilt acclimation (23/77 herds surveyed; 29.9 %) was included (Niederwerder and Hesse, 2018), whereas 19/77 herds surveyed (24.7 %) used vaccination. There is no standard feedback protocol for any type of pig production systems to have the most consistent and highest efficacy (Niederwerder and Hesse, 2018). Immunization of pregnant sows, or gilts expected to be bred (gilt acclimation), is undertaken by exposure to virulent autogenous virus, such as minced intestines from infected neonatal piglets negative for other infectious agents (Jung and Saif, 2015; Niederwerder and Hesse, 2018). Feedback stimulated lactogenic immunity via the gut-mammary-sIgA axis in sows (Saif et al., 2012), as well as prolonged, passive immune protection of piglets against PEDV infection (Goede et al., 2015). In our study, strong protective immunity (100 % survival rate of the piglets challenged with PC22A) was induced in gilts inoculated orally once with virulent non-S INDEL PEDV strain PC22A (105 PFU/gilt) during mid-gestation (day 57–59) (Langel et al., 2019a). Gilts exposed to the virus earlier (day 19–22) or later (day 96–97) in the gestation period showed less protective immunity, as evident by 87.2 % or 55.9 % survival rates of their piglets, respectively. Sows given feedback or orally inoculated with virulent PEDV during mid-gestation also discontinued fecal PEDV shedding prior to farrowing (Langel et al., 2019a; Leidenberger et al., 2017). Whether prolonged PEDV shedding post-exposure and during farrowing and lactation may cause infection of piglets, remains unclear. However, the most efficient and safest timing for feedback should be mid-gestation.
6.3. Vaccination as safer alternative to feedback
Because of the frequent inefficiency or lack of safety, feedback should be replaced by oral immunization with high, consistent doses of live attenuated PEDV that is minimally replicative, but immunogenic in the intestine of pigs (Song et al., 2007). Pregnant sows orally inoculated with live attenuated PEDV (DR13 strain) vaccine twice at 2 and 4 weeks pre-farrowing exhibited higher PEDV-specific IgA and VN antibody levels in the colostrum compared with the counterpart sows administered the same vaccine IM (Song et al., 2007). Although the IM route has also been used as an administration option using live PEDV vaccine (Kweon et al., 1999), to what extent PEDV-specific IgA memory B cells are induced in sows initially primed via an IM route with live PEDV vaccine, has not been confirmed. The efficacy of PEDV IM boost vaccination is dependent on induction of IgA memory B cells in initially field exposed or orally primed sows (Gillespie et al., 2018), similar to the success of TGEV oral prime/parenteral boost vaccine strategies (Saif et al., 2012). Live attenuated strains of PEDV have been isolated from the feces of pigs, suggesting that recombinant or mutated strains of PEDV might have emerged (Gerdts and Zakhartchouk, 2017; Guo et al., 2016).
6.4. Maintenance and booster of herd immunity
For maintaining or boosting herd immunity initiated through feedback or oral live vaccines in farrowing herds, IM administration with either live PEDV (Sato et al., 2018) or inactivated, adjuvanted whole-virus PEDV vaccine (Gillespie et al., 2018) once or twice at 4−5 or 2 weeks pre-farrowing was effective in boosting lactogenic immunity in sows positive for PEDV-specific IgA memory B cells in GALT. Many vaccine candidates or products have been tested and developed in the US and worldwide. The details were reviewed previously (Crawford et al., 2016; Gerdts and Zakhartchouk, 2017). IM live vaccine booster plus feedback was more effective in reducing the number of deaths of suckling piglets, compared with either IM live vaccine alone or feedback alone (Sato et al., 2018). Prime and boost IM vaccination with inactivated PEDV provided incomplete protection of piglets against PEDV infection (Gerdts and Zakhartchouk, 2017; Niederwerder and Hesse, 2018), because of poor lactogenic immunity induced in their vaccinated dams (Gillespie et al., 2018). Despite the safety issue, therefore, feedback alone, or prime and boost vaccination via oral and/or IM routes with live PEDV have been frequently used for maternal immunization against PEDV infection.
7. Conclusions
PEDV remains a threat to the swine industry in the US and worldwide because no effective vaccines are currently available. Regardless of the genogroup, PEDV can be deadly in pig populations with no immunity or low immunity or health status. The extent of PEDV transmission and PED on farms has a reverse relationship with the levels of biosecurity. High biosecurity to control PED should be practiced prior to seasonal or expected epidemics. Concurrently, pregnant sows are immunized by either feedback or vaccination. However, the current feedback or vaccination protocols are frequently inefficient or unsafe, due to: 1) no standardized protocol for feedback; 2) poor capacities of current IM vaccines (live or killed) to induce lactogenic immunity; 3) antigenic differences of vaccine vs. epidemic strains; and 4) possibility of continuous re-infection by PEDV used for feedback within herds. In addition, current live vaccines, which were generated by conventional cell culture-adaptation methods, may revert to virulent PEDV or recombine with field PEDV strains to generate new strains after they are applied in the field. Therefore, better vaccination protocols and vaccine strains (attenuated oral vaccines) are required for prevention and control of PED. PEDV-specific IgA memory B cells in orally primed sows play a critical role in boosting lactogenic immunity of sows and passive protection of piglets when piglets are exposed to PEDV during the suckling period. Feedback can be replaced by oral immunization with live attenuated PEDV vaccines that are minimally replicative, but immunogenic in the intestine of pregnant sows, thereby inducing PEDV-specific IgA memory B cells in the GALT or other mucosal and systemic lymphoid tissues and lactogenic immunity. Lastly, ideal live attenuated vaccines should not revert to virulent PEDV or recombine with field PEDV strains to generate new virulent PEDV strains, which remains an obstacle in vaccine development (Hou and Wang, 2019).
Declaration of Competing Interest
None of the authors of this paper has a financial or personal relationship with other people or organizations that could inappropriately influence or bias the content of the paper.
Acknowledgements
We sincerely thank all the authors cited in our review for their efforts and contributions to understand PEDV. We apologize to authors whose contributions to PEDV research may have been inadvertently and unintentionally omitted, because of the page limits. Salaries and research support were provided by state and federal funds appropriated to the Ohio Agricultural Research and Development Center, College of Food, Agricultural and Environmental Sciences, The Ohio State University, Wooster, Ohio, USA. Part of this work was supported by a grant from the National Institute of Child Health and Human Development, National Institutes of Health, Grant HD095881-01 (L.J. Saif and A. Vlasova, co-PIs).
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.virusres.2020.198045.
Contributor Information
Kwonil Jung, Email: jung.221@osu.edu.
Linda J. Saif, Email: saif.2@osu.edu.
Qiuhong Wang, Email: wang.655@osu.edu.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Alonso C., Goede D.P., Morrison R.B., Davies P.R., Rovira A., Marthaler D.G., Torremorell M. Evidence of infectivity of airborne porcine epidemic diarrhea virus and detection of airborne viral RNA at long distances from infected herds. Vet. Res. 2014;45(1):73. doi: 10.1186/s13567-014-0073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso C., Raynor P.C., Davies P.R., Torremorell M. Concentration, size distribution, and infectivity of airborne particles carrying swine viruses. PLoS One. 2015;10(8) doi: 10.1371/journal.pone.0135675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annamalai T., Saif L.J., Lu Z., Jung K. Age-dependent variation in innate immune responses to porcine epidemic diarrhea virus infection in suckling versus weaned pigs. Vet. Immunol. Immunopathol. 2015;168(3–4):193–202. doi: 10.1016/j.vetimm.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annamalai T., Lin C.M., Gao X., Liu X., Lu Z., Saif L.J., Wang Q. Cross protective immune responses in nursing piglets infected with a US spike-insertion deletion porcine epidemic diarrhea virus strain and challenged with an original US PEDV strain. Vet. Res. 2017;48(1):61. doi: 10.1186/s13567-017-0469-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anon Feed tote bags implicated in pig disease spread. J. Am. Vet. Med. Assoc. 2015;247(10):1083–1084. [PubMed] [Google Scholar]
- Baker K.L., Thomas P.R., Karriker L.A., Ramirez A., Zhang J., Wang C., Holtkamp D.J. Evaluation of an accelerated hydrogen peroxide disinfectant to inactivate porcine epidemic diarrhea virus in swine feces on aluminum surfaces under freezing conditions. BMC Vet. Res. 2017;13(1):372. doi: 10.1186/s12917-017-1300-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K.L., Mowrer C.L., Zhang J., Chen Q., Ramirez A., Wang C., Karriker L.A., Holtkamp D.J. Evaluation of a peroxygen-based disinfectant for inactivation of porcine epidemic diarrhea virus at low temperatures on metal surfaces. Vet. Microbiol. 2018;214:99–107. doi: 10.1016/j.vetmic.2017.12.019. [DOI] [PubMed] [Google Scholar]
- Beall A., Yount B., Lin C.M., Hou Y., Wang Q., Saif L., Baric R. Characterization of a pathogenic full-length cDNA clone and transmission model for porcine epidemic diarrhea virus strain PC22A. mBio. 2016;7(1):e01451–1415. doi: 10.1128/mBio.01451-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beam A., Goede D., Fox A., McCool M.J., Wall G., Haley C., Morrison R. A porcine epidemic diarrhea virus outbreak in one geographic region of the United States: descriptive epidemiology and investigation of the possibility of airborne virus spread. PLoS One. 2015;10(12) doi: 10.1371/journal.pone.0144818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevins S.N., Lutman M., Pedersen K., Barrett N., Gidlewski T., Deliberto T.J., Franklin A.B. Spillover of swine coronaviruses, United States. Emerging Infect. Dis. 2018;24(7):1390–1392. doi: 10.3201/eid2407.172077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniotti M.B., Papetti A., Lavazza A., Alborali G., Sozzi E., Chiapponi C., Faccini S., Bonilauri P., Cordioli P., Marthaler D. Porcine epidemic diarrhea virus and discovery of a recombinant swine enteric coronavirus, Italy. Emerg. Infect. diseases. 2016;22(1):83–87. doi: 10.3201/eid2201.150544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourges D., Meurens F., Berri M., Chevaleyre C., Zanello G., Levast B., Melo S., Gerdts V., Salmon H. New insights into the dual recruitment of IgA(+) B cells in the developing mammary gland. Mol. Immunol. 2008;45(12):3354–3362. doi: 10.1016/j.molimm.2008.04.017. [DOI] [PubMed] [Google Scholar]
- Bowman A.S., Krogwold R.A., Price T., Davis M., Moeller S.J. Investigating the introduction of porcine epidemic diarrhea virus into an Ohio swine operation. BMC Vet. Res. 2015;11:38. doi: 10.1186/s12917-015-0348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman A.S., Nolting J.M., Nelson S.W., Bliss N., Stull J.W., Wang Q., Premanandan C. Effects of disinfection on the molecular detection of porcine epidemic diarrhea virus. Vet. Microbiol. 2015;179(3–4):213–218. doi: 10.1016/j.vetmic.2015.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canning P., Ruston C., Madson D., Bates J., Skoland K., Davenport J., Gaul S., Wang C., Chen Q., Zhang J., Karriker L. Effect of direct-fed microbial Bacillus subtilis C-3102 on enteric health in nursery pigs after challenge with porcine epidemic diarrhea virus. J. Swine Health Prod. 2017;25(3):129–137. [Google Scholar]
- Chen Q., Li G., Stasko J., Thomas J.T., Stensland W.R., Pillatzki A.E., Gauger P.C., Schwartz K.J., Madson D., Yoon K.J., Stevenson G.W., Burrough E.R., Harmon K.M., Main R.G., Zhang J. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 2014;52(1):234–243. doi: 10.1128/JCM.02820-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Zhu Y., Wu M., Ku X., Ye S., Li Z., Guo X., He Q. Comparative genomic analysis of classical and variant virulent parental/attenuated strains of porcine epidemic diarrhea virus. Viruses. 2015;7(10):5525–5538. doi: 10.3390/v7102891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Gauger P.C., Stafne M.R., Thomas J.T., Madson D.M., Huang H., Zheng Y., Li G., Zhang J. Pathogenesis comparison between the United States porcine epidemic diarrhoea virus prototype and S-INDEL-variant strains in conventional neonatal piglets. J. Gen. Virol. 2016;97(5):1107–1121. doi: 10.1099/jgv.0.000419. [DOI] [PubMed] [Google Scholar]
- Chen Q., Thomas J.T., Gimenez-Lirola L.G., Hardham J.M., Gao Q., Gerber P.F., Opriessnig T., Zheng Y., Li G., Gauger P.C., Madson D.M., Magstadt D.R., Zhang J. Evaluation of serological cross-reactivity and cross-neutralization between the United States porcine epidemic diarrhea virus prototype and S-INDEL-variant strains. BMC Vet. Res. 2016;12:70. doi: 10.1186/s12917-016-0697-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Zhang C., Liu Y., Liu G. Super-oxidized water inactivates major viruses circulating in swine farms. J. Virol. Methods. 2017;242:27–29. doi: 10.1016/j.jviromet.2017.01.002. [DOI] [PubMed] [Google Scholar]
- Cima G. Fighting a deadly pig disease. J. Am. Vet. Med. Assoc. 2013;243(4):467–470. [PubMed] [Google Scholar]
- Coussement W., Ducatelle R., Debouck P., Hoorens J. Pathology of experimental CV777 coronavirus enteritis in piglets. I. Histological and histochemical study. Vet. Pathol. 1982;19(1):46–56. doi: 10.1177/030098588201900108. [DOI] [PubMed] [Google Scholar]
- Crawford K., Lager K., Miller L., Opriessnig T., Gerber P., Hesse R. Evaluation of porcine epidemic diarrhea virus transmission and the immune response in growing pigs. Vet. Res. 2015;46:49. doi: 10.1186/s13567-015-0180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K., Lager K.M., Kulshreshtha V., Miller L.C., Faaberg K.S. Status of vaccines for porcine epidemic diarrhea virus in the United States and Canada. Virus Res. 2016;226:108–116. doi: 10.1016/j.virusres.2016.08.005. [DOI] [PubMed] [Google Scholar]
- Curry S.M., Schwartz K.J., Yoon K.J., Gabler N.K., Burrough E.R. Effects of porcine epidemic diarrhea virus infection on nursery pig intestinal function and barrier integrity. Vet. Microbiol. 2017;211:58–66. doi: 10.1016/j.vetmic.2017.09.021. [DOI] [PubMed] [Google Scholar]
- de Arriba M.L., Carvajal A., Pozo J., Rubio P. Isotype-specific antibody-secreting cells in systemic and mucosal associated lymphoid tissues and antibody responses in serum of conventional pigs inoculated with PEDV. Vet. Immunol. Immunopathol. 2002;84(1–2):1–16. doi: 10.1016/S0165-2427(01)00386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Arriba M.L., Carvajal A., Pozo J., Rubio P. Lymphoproliferative responses and protection in conventional piglets inoculated orally with virulent or attenuated porcine epidemic diarrhoea virus. J. Virol. Methods. 2002;105(1):37–47. doi: 10.1016/S0166-0934(02)00063-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Arriba M.L., Carvajal A., Pozo J., Rubio P. Mucosal and systemic isotype-specific antibody responses and protection in conventional pigs exposed to virulent or attenuated porcine epidemic diarrhoea virus. Vet. Immunol. Immunopathol. 2002;85(1–2):85–97. doi: 10.1016/s0165-2427(01)00417-2. [DOI] [PubMed] [Google Scholar]
- Debouck P., Pensaert M., Coussement W. The pathogenesis of an enteric infection in pigs, experimentally induced by the coronavirus-like agent, Cv-777. Vet. Microbiol. 1981;6(2):157–165. [Google Scholar]
- Dee S., Clement T., Schelkopf A., Nerem J., Knudsen D., Christopher-Hennings J., Nelson E. An evaluation of contaminated complete feed as a vehicle for porcine epidemic diarrhea virus infection of naive pigs following consumption via natural feeding behavior: proof of concept. BMC Vet. Res. 2014;10(1):176. doi: 10.1186/s12917-014-0176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng F., Ye G., Liu Q., Navid M.T., Zhong X., Li Y., Wan C., Xiao S., He Q., Fu Z.F., Peng G. Identification and comparison of receptor binding characteristics of the spike protein of two porcine epidemic diarrhea virus strains. Viruses. 2016;8(3):55. doi: 10.3390/v8030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., van Geelen A., Buckley A.C., O’Brien A., Pillatzki A., Lager K.M., Faaberg K.S., Baker S.C. Coronavirus endoribonuclease activity in porcine epidemic diarrhea virus suppresses type I and type III interferon responses. J. Virol. 2019;93(8) doi: 10.1128/JVI.02000-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallien S., Andraud M., Moro A., Lediguerher G., Morin N., Gauger P.C., Bigault L., Paboeuf F., Berri M., Rose N., Grasland B. Better horizontal transmission of a US non-InDel strain compared with a French InDel strain of porcine epidemic diarrhoea virus. Transbound. Emerg. Dis. 2018;65(6):1720–1732. doi: 10.1111/tbed.12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallien S., Moro A., Lediguerher G., Catinot V., Paboeuf F., Bigault L., Berri M., Gauger P.C., Pozzi N., Authie E., Rose N., Grasland B. Evidence of porcine epidemic diarrhea virus (PEDV) shedding in semen from infected specific pathogen-free boars. Vet. Res. 2018;49(1):7. doi: 10.1186/s13567-018-0505-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallien S., Moro A., Lediguerher G., Catinot V., Paboeuf F., Bigault L., Gauger P.C., Pozzi N., Berri M., Authie E., Rose N., Grasland B. Limited shedding of an S-InDel strain of porcine epidemic diarrhea virus (PEDV) in semen and questions regarding the infectivity of the detected virus. Vet. Microbiol. 2019;228:20–25. doi: 10.1016/j.vetmic.2018.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Zhao S., Qin T., Yin Y., Yang Q. Effects of porcine epidemic diarrhea virus on porcine monocyte-derived dendritic cells and intestinal dendritic cells. Vet. Microbiol. 2015;179(3–4):131–141. doi: 10.1016/j.vetmic.2015.05.016. [DOI] [PubMed] [Google Scholar]
- Gerber P.F., Xiao C.T., Lager K., Crawford K., Kulshreshtha V., Cao D., Meng X.J., Opriessnig T. Increased frequency of porcine epidemic diarrhea virus shedding and lesions in suckling pigs compared to nursery pigs and protective immunity in nursery pigs after homologous re-challenge. Vet. Res. 2016;47(1):118. doi: 10.1186/s13567-016-0402-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdts V., Zakhartchouk A. Vaccines for porcine epidemic diarrhea virus and other swine coronaviruses. Vet. Microbiol. 2017;206:45–51. doi: 10.1016/j.vetmic.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie T., Song Q., Inskeep M., Stone S., Murtaugh M.P. Effect of booster vaccination with inactivated porcine epidemic diarrhea virus on neutralizing antibody response in mammary secretions. Viral Immunol. 2018;31(1):62–68. doi: 10.1089/vim.2017.0023. [DOI] [PubMed] [Google Scholar]
- Gimenez-Lirola L.G., Zhang J., Carrillo-Avila J.A., Chen Q., Magtoto R., Poonsuk K., Baum D.H., Pineyro P., Zimmerman J. Reactivity of porcine epidemic diarrhea virus structural proteins to antibodies against porcine enteric coronaviruses: diagnostic implications. J. Clin. Microbiol. 2017;55(5):1426–1436. doi: 10.1128/JCM.02507-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goede D., Murtaugh M.P., Nerem J., Yeske P., Rossow K., Morrison R. Previous infection of sows with a “mild” strain of porcine epidemic diarrhea virus confers protection against infection with a “severe” strain. Vet. Microbiol. 2015;176(1–2):161–164. doi: 10.1016/j.vetmic.2014.12.019. [DOI] [PubMed] [Google Scholar]
- Gong L., Li J., Zhou Q., Xu Z., Chen L., Zhang Y., Xue C., Wen Z., Cao Y. A new Bat-HKU2-like coronavirus in swine, China, 2017. Emerging Infect. Dis. 2017;23(9) doi: 10.3201/eid2309.170915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon R.K., Kotowski I.K., Coulson K.F., Link D., MacKenzie A., Bowling-Heyward J. The role of non-animal origin feed ingredients in transmission of viral pathogens of swine: a review of scientific literature. Front. Vet. Sci. 2019;6:273. doi: 10.3389/fvets.2019.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasland B., Bigault L., Bernard C., Quenault H., Toulouse O., Fablet C., Rose N., Touzain F., Blanchard Y. Complete genome sequence of a porcine epidemic diarrhea s gene indel strain isolated in france in december 2014. Genome Announc. 2015;3(3) doi: 10.1128/genomeA.00535-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Hu H., Chen F., Li Z., Ye S., Cheng S., Zhang M., He Q. iTRAQ-based comparative proteomic analysis of Vero cells infected with virulent and CV777 vaccine strain-like strains of porcine epidemic diarrhea virus. J. Proteomics. 2016;130:65–75. doi: 10.1016/j.jprot.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M., Wyler R. Quantitation, biological and physicochemical properties of cell culture-adapted porcine epidemic diarrhea coronavirus (PEDV) Vet. Microbiol. 1989;20(2):131–142. doi: 10.1016/0378-1135(89)90036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtkamp D.J., Myers J., Thomas P.R., Karriker L.A., Ramirez A., Zhang J., Wang C. Efficacy of an accelerated hydrogen peroxide disinfectant to inactivate porcine epidemic diarrhea virus in swine feces on metal surfaces. Can. J. Vet. Res. 2017;81(2):100–107. [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Wang Q. Emerging highly virulent porcine epidemic diarrhea virus: molecular mechanisms of attenuation and rational design of live attenuated vaccines. Int. J. Mol. Sci. 2019;20(21) doi: 10.3390/ijms20215478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Lin C.M., Yokoyama M., Yount B.L., Marthaler D., Douglas A.L., Ghimire S., Qin Y., Baric R.S., Saif L.J., Wang Q. Deletion of a 197-amino-acid region in the N-terminal domain of spike protein attenuates porcine epidemic diarrhea virus in piglets. J. Virol. 2017;91(14) doi: 10.1128/JVI.00227-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Ke H., Kim J., Yoo D., Su Y., Boley P., Chepngeno J., Vlasova A.N., Saif L.J., Wang Q. Engineering a live attenuated porcine epidemic diarrhea virus vaccine candidate via inactivation of the viral 2’-O-methyltransferase and the Endocytosis Signal of the spike protein. J. Virol. 2019;93(15) doi: 10.1128/JVI.00406-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Meulia T., Gao X., Saif L.J., Wang Q. Deletion of both the tyrosine-based endocytosis signal and the endoplasmic reticulum retrieval signal in the cytoplasmic tail of spike protein attenuates porcine epidemic diarrhea virus in pigs. J. Virol. 2019;93(2) doi: 10.1128/JVI.01758-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.W., Dickerman A.W., Pineyro P., Li L., Fang L., Kiehne R., Opriessnig T., Meng X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio. 2013;4(5):e00737–00713. doi: 10.1128/mBio.00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M.Z., Wang S.Y., Wang H., Cui D.A., Yang Y.J., Liu X.W., Kong X.J., Li J.Y. Differences in the intestinal microbiota between uninfected piglets and piglets infected with porcine epidemic diarrhea virus. PLoS One. 2018;13(2) doi: 10.1371/journal.pone.0192992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J.H., Lee C. Cellular cholesterol is required for porcine nidovirus infection. Arch. Virol. 2017;162(12):3753–3767. doi: 10.1007/s00705-017-3545-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji C.M., Wang B., Zhou J., Huang Y.W. Aminopeptidase-N-independent entry of porcine epidemic diarrhea virus into Vero or porcine small intestine epithelial cells. Virology. 2018;517:16–23. doi: 10.1016/j.virol.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Hu J., Thirumalai D., Zhang X. Immunoglobulin transporting receptors are potential targets for the immunity enhancement and generation of mammary gland bioreactor. Front. Immunol. 2016;7:214. doi: 10.3389/fimmu.2016.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Saif L.J. Porcine epidemic diarrhea virus infection: etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet. J. 2015;204(2):134–143. doi: 10.1016/j.tvjl.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Saif L.J. Goblet cell depletion in small intestinal villous and crypt epithelium of conventional nursing and weaned pigs infected with porcine epidemic diarrhea virus. Res. Vet. Sci. 2017;110:12–15. doi: 10.1016/j.rvsc.2016.10.009. [DOI] [PubMed] [Google Scholar]
- Jung K., Ahn K., Chae C. Decreased activity of brush border membrane-bound digestive enzymes in small intestines from pigs experimentally infected with porcine epidemic diarrhea virus. Res. Vet. Sci. 2006;81(3):310–315. doi: 10.1016/j.rvsc.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Jung K., Wang Q., Scheuer K.A., Lu Z., Zhang Y., Saif L.J. Pathology of US porcine epidemic diarrhea virus strain PC21A in gnotobiotic pigs. Emerg. Infect. Dis. 2014;20(4):662–665. doi: 10.3201/eid2004.131685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Annamalai T., Lu Z., Saif L.J. Comparative pathogenesis of US porcine epidemic diarrhea virus (PEDV) strain PC21A in conventional 9-day-old nursing piglets vs. 26-day-old weaned pigs. Vet. Microbiol. 2015;178(1–2):31–40. doi: 10.1016/j.vetmic.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Eyerly B., Annamalai T., Lu Z., Saif L.J. Structural alteration of tight and adherens junctions in villous and crypt epithelium of the small and large intestine of conventional nursing piglets infected with porcine epidemic diarrhea virus. Vet. Microbiol. 2015;177(3–4):373–378. doi: 10.1016/j.vetmic.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Eyerly B., Lu Z., Chepngeno J., Saif L.J. Pathogenecity of 2 porcine deltacoronavirus strains in gnotobiotic pigs. Emerg. Infect. Dis. 2015;21(4):650–654. doi: 10.3201/eid2104.141859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Saif L.J. Porcine deltacoronavirus induces apoptosis in swine testicular and LLC porcine kidney cell lines in vitro but not in infected intestinal enterocytes in vivo. Vet. Microbiol. 2016;182:57–63. doi: 10.1016/j.vetmic.2015.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Miyazaki A., Saif L.J. Immunohistochemical detection of the vomiting-inducing monoamine neurotransmitter serotonin and enterochromaffin cells in the intestines of conventional or gnotobiotic (Gn) pigs infected with porcine epidemic diarrhea virus (PEDV) and serum cytokine responses of Gn pigs to acute PEDV infection. Res. Vet. Sci. 2018;119:99–108. doi: 10.1016/j.rvsc.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Lee C. Porcine epidemic diarrhea virus induces caspase-independent apoptosis through activation of mitochondrial apoptosis-inducing factor. Virology. 2014;460–461:180–193. doi: 10.1016/j.virol.2014.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B., Kim O., Tai J.H., Chae C. Transmissible gastroenteritis virus induces apoptosis in swine testicular cell lines but not in intestinal enterocytes. J. Comp. Pathol. 2000;123(1):64–66. doi: 10.1053/jcpa.2000.0386. [DOI] [PubMed] [Google Scholar]
- Kim Y., Oh C., Shivanna V., Hesse R.A., Chang K.O. Trypsin-independent porcine epidemic diarrhea virus US strain with altered virus entry mechanism. BMC Vet. Res. 2017;13(1):356. doi: 10.1186/s12917-017-1283-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Yang M., Goyal S.M., Cheeran M.C., Torremorell M. Evaluation of biosecurity measures to prevent indirect transmission of porcine epidemic diarrhea virus. BMC Vet. Res. 2017;13(1):89. doi: 10.1186/s12917-017-1017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocherhans R., Bridgen A., Ackermann M., Tobler K. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes. 2001;23(2):137–144. doi: 10.1023/A:1011831902219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh H.W., Kim M.S., Lee J.S., Kim H., Park S.J. Changes in the swine gut microbiota in response to porcine epidemic diarrhea infection. Microbes Environ. 2015;30(3):284–287. doi: 10.1264/jsme2.ME15046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonpaew S., Teeravechyan S., Frantz P.N., Chailangkarn T., Jongkaewwattana A. PEDV and PDCoV pathogenesis: the interplay between host innate immune responses and porcine enteric coronaviruses. Front. Vet. Sci. 2019;6:34. doi: 10.3389/fvets.2019.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweon C.H., Kwon B.J., Lee J.G., Kwon G.O., Kang Y.B. Derivation of attenuated porcine epidemic diarrhea virus (PEDV) as vaccine candidate. Vaccine. 1999;17(20–21):2546–2553. doi: 10.1016/S0264-410X(99)00059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langel S.N., Paim F.C., Lager K.M., Vlasova A.N., Saif L.J. Lactogenic immunity and vaccines for porcine epidemic diarrhea virus (PEDV): historical and current concepts. Virus Res. 2016;226:93–107. doi: 10.1016/j.virusres.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langel S.N., Paim F.C., Alhamo M.A., Buckley A., Van Geelen A., Lager K.M., Vlasova A.N., Saif L.J. Stage of gestation at porcine epidemic diarrhea virus infection of pregnant swine impacts maternal immunity and lactogenic immune protection of neonatal suckling piglets. Front. Immunol. 2019;10:727. doi: 10.3389/fimmu.2019.00727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langel S.N., Paim F.C., Alhamo M.A., Lager K.M., Vlasova A.N., Saif L.J. Oral vitamin A supplementation of porcine epidemic diarrhea virus infected gilts enhances IgA and lactogenic immune protection of nursing piglets. Vet. Res. 2019;50(1):101. doi: 10.1186/s13567-019-0719-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langel S.N., Wang Q., Vlasova A.N., Saif L.J. Host factors affecting generation of immunity against porcine epidemic diarrhea virus in pregnant and lactating swine and passive protection of neonates. Pathogens. 2020;9:130. doi: 10.3390/pathogens9020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhans W. Anorexia of infection: current prospects. Nutrition. 2000;16(10):996–1005. doi: 10.1016/s0899-9007(00)00421-4. [DOI] [PubMed] [Google Scholar]
- Lee C. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol. J. 2015;12:193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.U., Kwon T., Je S.H., Yoo S.J., Seo S.W., Sunwoo S.Y., Lyoo Y.S. Wild boars harboring porcine epidemic diarrhea virus (PEDV) may play an important role as a PEDV reservoir. Vet. Microbiol. 2016;192:90–94. doi: 10.1016/j.vetmic.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidenberger S., Schroder C., Zani L., Auste A., Pinette M., Ambagala A., Nikolin V., de Smit H., Beer M., Blome S. Virulence of current German PEDV strains in suckling pigs and investigation of protective effects of maternally derived antibodies. Sci. Rep. 2017;7(1):10825. doi: 10.1038/s41598-017-11160-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.X., Ge J.W., Li Y.J. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virology. 2007;365(1):166–172. doi: 10.1016/j.virol.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li H., Liu Y., Pan Y., Deng F., Song Y., Tang X., He Q. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg. Infect. Dis. 2012;18(8):1350–1353. doi: 10.3201/eid1808.120002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., van Kuppeveld F.J.M., He Q., Rottier P.J.M., Bosch B.J. Cellular entry of the porcine epidemic diarrhea virus. Virus Res. 2016;226:117–127. doi: 10.1016/j.virusres.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Luo R., He Q., van Kuppeveld F.J.M., Rottier P.J.M., Bosch B.J. Aminopeptidase N is not required for porcine epidemic diarrhea virus cell entry. Virus Res. 2017;235:6–13. doi: 10.1016/j.virusres.2017.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wu Q., Huang L., Yuan C., Wang J., Yang Q. An alternative pathway of enteric PEDV dissemination from nasal cavity to intestinal mucosa in swine. Nat. Commun. 2018;9(1):3811. doi: 10.1038/s41467-018-06056-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Fu F., Guo S., Wang H., He X., Xue M., Yin L., Feng L., Liu P. Porcine intestinal enteroids: a new model for studying enteric coronavirus porcine epidemic diarrhea virus infection and the host innate response. J. Virol. 2019;93(5) doi: 10.1128/JVI.01682-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.M., Annamalai T., Liu X., Gao X., Lu Z., El-Tholoth M., Hu H., Saif L.J., Wang Q. Experimental infection of a US spike-insertion deletion porcine epidemic diarrhea virus in conventional nursing piglets and cross-protection to the original US PEDV infection. Vet. Res. 2015;46:134. doi: 10.1186/s13567-015-0278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.M., Gao X., Oka T., Vlasova A.N., Esseili M.A., Wang Q., Saif L.J. Antigenic relationships among porcine epidemic diarrhea virus and transmissible gastroenteritis virus strains. J. Virol. 2015;89(6):3332–3342. doi: 10.1128/JVI.03196-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.M., Saif L.J., Marthaler D., Wang Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016;226:20–39. doi: 10.1016/j.virusres.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.M., Hou Y., Marthaler D.G., Gao X., Liu X., Zheng L., Saif L.J., Wang Q. Attenuation of an original US porcine epidemic diarrhea virus strain PC22A via serial cell culture passage. Vet. Microbiol. 2017;201:62–71. doi: 10.1016/j.vetmic.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Li B., Chen L., Ma Z., He K., Fan H. Differential protein analysis of IPEC-J2 cells infected with porcine epidemic diarrhea virus pandemic and classical strains elucidates the pathogenesis of infection. J. Proteome Res. 2017;16(6):2113–2120. doi: 10.1021/acs.jproteome.6b00957. [DOI] [PubMed] [Google Scholar]
- Liu C., Tang J., Ma Y., Liang X., Yang Y., Peng G., Qi Q., Jiang S., Li J., Du L., Li F. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J. Virol. 2015;89(11):6121–6125. doi: 10.1128/JVI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Zhao L., Zhai Z., Zhao W., Ding J., Dai R., Sun T., Meng H. Porcine epidemic diarrhea virus infection induced the unbalance of gut microbiota in piglets. Curr. Microbiol. 2015;71(6):643–649. doi: 10.1007/s00284-015-0895-6. [DOI] [PubMed] [Google Scholar]
- Liu X., Lin C.M., Annamalai T., Gao X., Lu Z., Esseili M.A., Jung K., El-Tholoth M., Saif L.J., Wang Q. Determination of the infectious titer and virulence of an original US porcine epidemic diarrhea virus PC22A strain. Vet. Res. 2015;46:109. doi: 10.1186/s13567-015-0249-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse L., Krog J.S., Strandbygaard B., Rasmussen T.B., Kjaer J., Belsham G.J., Botner A. Experimental infection of young pigs with an early european strain of porcine epidemic diarrhoea virus and a recent US strain. Transbound. Emerg. Dis. 2017;64(5):1380–1386. doi: 10.1111/tbed.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J., Gauger P., Harmon K., Zhang J., Connor J., Yeske P., Loula T., Levis I., Dufresne L., Main R. Role of transportation in spread of porcine epidemic diarrhea virus infection, United States. Emerging Infect. Dis. 2014;20(5):872–874. doi: 10.3201/eid2005.131628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Guo L., Zhang J., Xu Y., Gu W., Feng L., Wang Y. Tight junction protein occludin is a porcine epidemic diarrhea virus entry factor. J. Virol. 2017;91(10) doi: 10.1128/JVI.00202-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhang Y., Liang X., Oglesbee M., Krakowka S., Niehaus A., Wang G., Jia A., Song H., Li J. Two-way antigenic cross-reactivity between porcine epidemic diarrhea virus and porcine deltacoronavirus. Vet. Microbiol. 2016;186:90–96. doi: 10.1016/j.vetmic.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson D.M., Magstadt D.R., Arruda P.H., Hoang H., Sun D., Bower L.P., Bhandari M., Burrough E.R., Gauger P.C., Pillatzki A.E., Stevenson G.W., Wilberts B.L., Brodie J., Harmon K.M., Wang C., Main R.G., Zhang J., Yoon K.J. Pathogenesis of porcine epidemic diarrhea virus isolate (US/Iowa/18984/2013) in 3-week-old weaned pigs. Vet. Microbiol. 2014;174(1–2):60–68. doi: 10.1016/j.vetmic.2014.09.002. [DOI] [PubMed] [Google Scholar]
- Madson D.M., Arruda P.H., Magstadt D.R., Burrough E.R., Hoang H., Sun D., Bower L.P., Bhandari M., Gauger P.C., Stevenson G.W., Wilberts B.L., Wang C., Zhang J., Yoon K.J. Characterization of porcine epidemic diarrhea virus isolate US/Iowa/18984/2013 infection in 1-Day-Old cesarean-derived colostrum-deprived piglets. Vet. Pathol. 2016;53(1):44–52. doi: 10.1177/0300985815591080. [DOI] [PubMed] [Google Scholar]
- Mavronmichalis I. WATTAgNet; 2016. How Piglet Gastric pH Development Affects Gut Health. May 26. [Google Scholar]
- Mesquita J.R., Hakze-van der Honing R., Almeida A., Lourenco M., van der Poel W.H., Nascimento M.S. Outbreak of porcine epidemic diarrhea virus in Portugal, 2015. Transbound. Emerg. Dis. 2015;62(6):586–588. doi: 10.1111/tbed.12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon H.W., Norman J.O., Lambert G. Age dependent resistance to transmissible gastroenteritis of swine (TGE). I. Clinical signs and some mucosal dimensions in small intestine. Can. J. Comp. Med. 1973;37(2):157–166. [PMC free article] [PubMed] [Google Scholar]
- Nam E., Lee C. Contribution of the porcine aminopeptidase N (CD13) receptor density to porcine epidemic diarrhea virus infection. Vet. Microbiol. 2010;144(1–2):41–50. doi: 10.1016/j.vetmic.2009.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederwerder M.C., Hesse R.A. Swine enteric coronavirus disease: a review of 4 years with porcine epidemic diarrhoea virus and porcine deltacoronavirus in the United States and Canada. Transbound. Emerg. Dis. 2018;65(3):660–675. doi: 10.1111/tbed.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederwerder M.C., Nietfeld J.C., Bai J., Peddireddi L., Breazeale B., Anderson J., Kerrigan M.A., An B., Oberst R.D., Crawford K., Lager K.M., Madson D.M., Rowland R.R., Anderson G.A., Hesse R.A. Tissue localization, shedding, virus carriage, antibody response, and aerosol transmission of Porcine epidemic diarrhea virus following inoculation of 4-week-old feeder pigs. J. Vet. Diagn. Invest. 2016;28(6):671–678. doi: 10.1177/1040638716663251. [DOI] [PubMed] [Google Scholar]
- Opriessnig T., Xiao C.T., Gerber P.F., Zhang J., Halbur P.G. Porcine epidemic diarrhea virus RNA present in commercial spray-dried porcine plasma is not infectious to naive pigs. PLoS One. 2014;9(8) doi: 10.1371/journal.pone.0104766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang K., Shyu D.L., Dhakal S., Hiremath J., Binjawadagi B., Lakshmanappa Y.S., Guo R., Ransburgh R., Bondra K.M., Gauger P., Zhang J., Specht T., Gilbertie A., Minton W., Fang Y., Renukaradhya G.J. Evaluation of humoral immune status in porcine epidemic diarrhea virus (PEDV) infected sows under field conditions. Vet. Res. 2015;46:140. doi: 10.1186/s13567-015-0285-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Tian X., Qin P., Wang B., Zhao P., Yang Y.L., Wang L., Wang D., Song Y., Zhang X., Huang Y.W. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet. Microbiol. 2017;211:15–21. doi: 10.1016/j.vetmic.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasick J., Berhane Y., Ojkic D., Maxie G., Embury-Hyatt C., Swekla K., Handel K., Fairles J., Alexandersen S. Investigation into the role of potentially contaminated feed as a source of the first-detected outbreaks of porcine epidemic diarrhea in Canada. Transbound. Emerg. Dis. 2014;61(5):397–410. doi: 10.1111/tbed.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58(3):243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M.B., Martelli P. Porcine epidemic diarrhea: a retrospect from Europe and matters of debate. Virus Res. 2016;226:1–6. doi: 10.1016/j.virusres.2016.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perri A.M., Poljak Z., Dewey C., Harding J.C.S., O’Sullivan T.L. An epidemiological investigation of the early phase of the porcine epidemic diarrhea (PED) outbreak in Canadian swine herds in 2014: a case-control study. Prev. Vet. Med. 2018;150:101–109. doi: 10.1016/j.prevetmed.2017.12.009. [DOI] [PubMed] [Google Scholar]
- Saif L.J., Pensaert M.P., Sestak K., Yeo S.G., Jung K. Coronaviruses. In: Zimmerman J.J., Karriker L.A., Ramirez A., Schwartz K.J., Stevenson G.W., editors. Diseases of Swine. 10th ed. Wiley-Blackwell, Iowa State University; 2012. pp. 501–524. [Google Scholar]
- Salmon H. Mammary gland immunology and neonate protection in pigs - Homing of lymphocytes into the MG. Adv. Exp. Med. Biol. 2000;480:279–286. doi: 10.1007/0-306-46832-8_32. [DOI] [PubMed] [Google Scholar]
- Sato T., Takeyama N., Katsumata A., Tuchiya K., Kodama T., Kusanagi K. Mutations in the spike gene of porcine epidemic diarrhea virus associated with growth adaptation in vitro and attenuation of virulence in vivo. Virus Genes. 2011;43(1):72–78. doi: 10.1007/s11262-011-0617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Oroku K., Ohshima Y., Furuya Y., Sasakawa C. Efficacy of genogroup 1 based porcine epidemic diarrhea live vaccine against genogroup 2 field strain in Japan. Virol. J. 2018;15(1):28. doi: 10.1186/s12985-018-0940-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher L.L., Huss A.R., Cochrane R.A., Stark C.R., Woodworth J.C., Bai J., Poulsen E.G., Chen Q., Main R.G., Zhang J., Gauger P.C., Ramirez A., Derscheid R.J., Magstadt D.M., Dritz S.S., Jones C.K. Characterizing the rapid spread of porcine epidemic diarrhea virus (PEDV) through an animal food manufacturing facility. PLoS One. 2017;12(11) doi: 10.1371/journal.pone.0187309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher L.L., Cochrane R.A., Huss A.R., Gebhardt J.T., Woodworth J.C., Stark C.R., Jones C.K., Bai J., Main R.G., Chen Q., Zhang J., Gauger P.C., DeRouchey J.M., Goodband R.D., Tokach M.D., Dritz S.S. Feed batch sequencing to decrease the risk of porcine epidemic diarrhea virus (PEDV) cross-contamination during feed manufacturing. J. Anim. Sci. 2018;96(11):4562–4570. doi: 10.1093/jas/sky320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A., McCluskey B., Brown-Reid M., Grear D., Pitcher P., Ramos G., Spencer D., Singrey A. Porcine epidemic diarrhea virus introduction into the United States: root cause investigation. Prev. Vet. Med. 2016;123:192–201. doi: 10.1016/j.prevetmed.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Maejima M., Islam M.T., Miyazaki A., Kawase M., Matsuyama S., Taguchi F. Porcine aminopeptidase N is not a cellular receptor of porcine epidemic diarrhea virus, but promotes its infectivity via aminopeptidase activity. J. Gen. Virol. 2016;97(10):2528–2539. doi: 10.1099/jgv.0.000563. [DOI] [PubMed] [Google Scholar]
- Siegrist C.A. 2. Vaccine immunology. In: Plotkin S.A., Orenstein W.A., Offit P.A., editors. Vaccines. Elesevier; 2013. pp. 14–32. [Google Scholar]
- Sirichokchatchawan W., Temeeyasen G., Nilubol D., Prapasarakul N. 2017. Protective Effects of Cell-Free Supernatant and Live Lactic Acid Bacteria Isolated from Thai Pigs Against a Pandemic Strain of Porcine Epidemic Diarrhea Virus. Probiotics and Antimicrobial Proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D.S., Yang J.S., Oh J.S., Han J.H., Park B.K. Differentiation of a Vero cell adapted porcine epidemic diarrhea virus from Korean field strains by restriction fragment length polymorphism analysis of ORF 3. Vaccine. 2003;21(17–18):1833–1842. doi: 10.1016/S0264-410X(03)00027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D.S., Oh J.S., Kang B.K., Yang J.S., Moon H.J., Yoo H.S., Jang Y.S., Park B.K. Oral efficacy of Vero cell attenuated porcine epidemic diarrhea virus DR13 strain. Res. Vet. Sci. 2007;82(1):134–140. doi: 10.1016/j.rvsc.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q., Stone S., Drebes D., Greiner L.L., Dvorak C.M.T., Murtaugh M.P. Characterization of anti-porcine epidemic diarrhea virus neutralizing activity in mammary secretions. Virus Res. 2016;226:85–92. doi: 10.1016/j.virusres.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Peng Q., Chen Y., Zhou X., Zhang F., Li A., Huang D., Wu Q., Ye Y., He H., Wang L., Tang Y. Altered gut microbiota profiles in sows and neonatal piglets associated with porcine epidemic diarrhea virus infection. Sci. Rep. 2017;7(1):17439. doi: 10.1038/s41598-017-17830-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler J., Zoels S., Fux R., Hanke D., Pohlmann A., Blome S., Weissenbock H., Weissenbacher-Lang C., Ritzmann M., Ladinig A. Emergence of porcine epidemic diarrhea virus in southern Germany. BMC Vet. Res. 2015;11:142. doi: 10.1186/s12917-015-0454-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinrigl A., Fernandez S.R., Stoiber F., Pikalo J., Sattler T., Schmoll F. First detection, clinical presentation and phylogenetic characterization of Porcine epidemic diarrhea virus in Austria. BMC Vet. Res. 2015;11:310. doi: 10.1186/s12917-015-0624-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson G.W., Hoang H., Schwartz K.J., Burrough E.B., Sun D., Madson D., Cooper V.L., Pillatzki A., Gauger P., Schmitt B.J., Koster L.G., Killian M.L., Yoon K.J. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Investig. 2013;25(5):649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- Su Y., Hou Y., Prarat M., Zhang Y., Wang Q. New variants of porcine epidemic diarrhea virus with large deletions in the spike protein, identified in the United States, 2016-2017. Arch. Virol. 2018;163(9):2485–2489. doi: 10.1007/s00705-018-3874-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueyoshi M., Tsuda T., Yamazaki K., Yoshida K., Nakazawa M., Sato K., Minami T., Iwashita K., Watanabe M., Suzuki Y. An immunohistochemical investigation of porcine epidemic diarrhoea. J. Comp. Pathol. 1995;113(1):59–67. doi: 10.1016/S0021-9975(05)80069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Shibahara T., Yamaguchi R., Nakade K., Yamamoto T., Miyazaki A., Ohashi S. Pig epidemic diarrhoea virus S gene variant with a large deletion non-lethal to colostrum-deprived newborn piglets. J. Gen. Virol. 2016;97(8):1823–1828. doi: 10.1099/jgv.0.000513. [DOI] [PubMed] [Google Scholar]
- Tan Z., Dong W., Ding Y., Ding X., Zhang Q., Jiang L. Changes in cecal microbiota community of suckling piglets infected with porcine epidemic diarrhea virus. PLoS One. 2019;14(7) doi: 10.1371/journal.pone.0219868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeravechyan S., Frantz P.N., Wongthida P., Chailangkarn T., Jaru-Ampornpan P., Koonpaew S., Jongkaewwattana A. Deciphering the biology of porcine epidemic diarrhea virus in the era of reverse genetics. Virus Res. 2016;226:152–171. doi: 10.1016/j.virusres.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J.T., Chen Q., Gauger P.C., Gimenez-Lirola L.G., Sinha A., Harmon K.M., Madson D.M., Burrough E.R., Magstadt D.R., Salzbrenner H.M., Welch M.W., Yoon K.J., Zimmerman J.J., Zhang J. Effect of porcine epidemic diarrhea virus infectious doses on infection outcomes in naive conventional neonatal and weaned pigs. PLoS One. 2015;10(10) doi: 10.1371/journal.pone.0139266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P.R., Karriker L.A., Ramirez A., Zhang J.Q., Ellingson J.S., Crawford K.K., Bates J.L., Hammen K.J., Holtkamp D.J. Evaluation of time and temperature sufficient to inactivate porcine epidemic diarrhea virus in swine feces on metal surfaces. J. Swine Health Prod. 2015;23(2):84–90. [Google Scholar]
- Vlasova A.N., Marthaler D., Wang Q., Culhane M.R., Rossow K.D., Rovira A., Collins J., Saif L.J. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013–February 2014. Emerg. Infect. Dis. 2014;20(10) doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Lu W., Chen J., Xie S., Shi H., Hsu H., Yu W., Xu K., Bian C., Fischer W.B., Schwarz W., Feng L., Sun B. PEDV ORF3 encodes an ion channel protein and regulates virus production. FEBS Lett. 2012;586(4):384–391. doi: 10.1016/j.febslet.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhao P., Guo L., Liu Y., Du Y., Ren S., Li J., Zhang Y., Fan Y., Huang B., Liu S., Wu J. Porcine epidemic diarrhea virus variants with high pathogenicity, China. Emerg. Infect. Dis. 2013;19(12):2048–2049. doi: 10.3201/eid1912.121088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg. Infect. Dis. 2014;20(7):1227–1230. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg. Infect. Dis. 2014;20(5):917–919. doi: 10.3201/eid2005.140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Ohnstad M., Nelsen A., Nelson E. Porcine epidemic diarrhea virus does not replicate in porcine monocyte-derived dendritic cells, but activates the transcription of type I interferon and chemokine. Vet. Microbiol. 2017;208:77–81. doi: 10.1016/j.vetmic.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth K.M., Rowland R.R.R., Petrovan V., Sheahan M., Cino-Ozuna A.G., Fang Y., Hesse R., Mileham A., Samuel M.S., Wells K.D., Prather R.S. Resistance to coronavirus infection in amino peptidase N-deficient pigs. Transgenic Res. 2019;28(1):21–32. doi: 10.1007/s11248-018-0100-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood E.N. An apparently new syndrome of porcine epidemic diarrhoea. Vet. Rec. 1977;100(12):243–244. doi: 10.1136/vr.100.12.243. [DOI] [PubMed] [Google Scholar]
- Wu Y., Li W., Zhou Q., Li Q., Xu Z., Shen H., Chen F. Characterization and pathogenicity of Vero cell-attenuated porcine epidemic diarrhea virus CT strain. Virol. J. 2019;16(1):121. doi: 10.1186/s12985-019-1232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., Ao C., Yang Y., Liu Y., Liang R., Zeng Z., Ye G., Xiao S., Fu Z.F., Dong W., Peng G. Two critical N-terminal epitopes of the nucleocapsid protein contribute to the cross-reactivity between porcine epidemic diarrhea virus and porcine transmissible gastroenteritis virus. J. Gen. Virol. 2019;100(2):206–216. doi: 10.1099/jgv.0.001216. [DOI] [PubMed] [Google Scholar]
- Yamamoto R., Soma J., Nakanishi M., Yamaguchi R., Niinuma S. Isolation and experimental inoculation of an S INDEL strain of porcine epidemic diarrhea virus in Japan. Res. Vet. Sci. 2015;103:103–106. doi: 10.1016/j.rvsc.2015.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Yoo D. Immune evasion of porcine enteric coronaviruses and viral modulation of antiviral innate signaling. Virus Res. 2016;226:128–141. doi: 10.1016/j.virusres.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Yim-Im W., Chen Q., Zheng Y., Schumacher L., Huang H., Gauger P., Harmon K., Li G. Identification of porcine epidemic diarrhea virus variant with a large spike gene deletion from a clinical swine sample in the United States. Virus Genes. 2018;54(2):323–327. doi: 10.1007/s11262-018-1542-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Ke H., Blikslager A., Fujita T., Yoo D. Type III interferon restriction by porcine epidemic diarrhea virus and the role of viral protein nsp1 in IRF1 signaling. J. Virol. 2018;92(4) doi: 10.1128/JVI.01677-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wu Z., Yang H. Aminopeptidase N knockout pigs are not resistant to porcine epidemic diarrhea virus infection. Virol. Sin. 2019;34(5):592–595. doi: 10.1007/s12250-019-00127-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Wang Y., Liu S., Huang J., Zhai Z., He C., Ding J., Wang J., Wang H., Fan W., Zhao J., Meng H. The dynamic distribution of porcine microbiota across different ages and gastrointestinal tract segments. PLoS One. 2015;10(2) doi: 10.1371/journal.pone.0117441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Li W., Schuurman N., van Kuppeveld F., Bosch B.J., Egberink H. Serological screening for coronavirus infections in cats. Viruses. 2019;11(8) doi: 10.3390/v11080743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.L., Ederveen J., Egberink H., Pensaert M., Horzinek M.C. Porcine epidemic diarrhea virus (CV 777) and feline infectious peritonitis virus (FIPV) are antigenically related. Arch. Virol. 1988;102(1–2):63–71. doi: 10.1007/BF01315563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Fan H., Lan T., Yang X.L., Shi W.F., Zhang W., Zhu Y., Zhang Y.W., Xie Q.M., Mani S., Zheng X.S., Li B., Li J.M., Guo H., Pei G.Q., An X.P., Chen J.W., Zhou L., Mai K.J., Wu Z.X., Li D., Anderson D.E., Zhang L.B., Li S.Y., Mi Z.Q., He T.T., Cong F., Guo P.J., Huang R., Luo Y., Liu X.L., Chen J., Huang Y., Sun Q., Zhang X.L., Wang Y.Y., Xing S.Z., Chen Y.S., Sun Y., Li J., Daszak P., Wang L.F., Shi Z.L., Tong Y.G., Ma J.Y. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556(7700):255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Q.F., Huang Y.J., Wu L.S., Wu Z.C., Wu S.L., Bao W.B. Effects of porcine epidemic diarrhea virus infection on tight junction protein gene expression and morphology of the intestinal mucosa in pigs. Pol. J. Vet. Sci. 2019;22(2):345–353. doi: 10.24425/pjvs.2019.129226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.