Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is the most prevalent pathogen causing osteomyelitis. The tendency of MRSA to evade standard antibiotic treatment by hiding inside bone cells and biofilms is a major cause of frequent osteomyelitis recurrence. In this study, we developed a lipid-polymer hybrid nanoparticle loading the antibiotic linezolid (LIN-LPN), and focused on evaluating if this new nanoantibiotic can achieve significant in vitro activities against these intracellular and biofilm-embedded MRSA. The optimal LIN-LPN formulation demonstrated both high linezolid payload (12.0% by weight of nanoparticles) and controlled release characteristics (gradually released the entrapped antibiotic in 120 h). Although it achieved lower activities against bacteria including USA300-0114, CDC-587, RP-62A in planktonic form, it was substantially superior against the intracellular MRSA reservoir inside osteoblast cells. The differences of intracellular activities between LIN-LPN and linezolid were 87.0-fold, 12.3-fold, and 12.6-fold in CFU/ml (p<0.05 or <0.01) at 2 μg/ml, 4 μg/ml, and 8 μg/ml linezolid concentrations, respectively. LIN-LPN also suppressed the MRSA biofilm growth to 35-60% of the values achieved with free linezolid (p<0.05). These enhanced intracellular and anti-biofilm activities of LIN-LPN were likely contributed by the extensive accumulation of LIN-LPN inside the MRSA-infected osteoblasts and biofilms as revealed in the confocal microscope images. The study thus validates the feasibility of exploiting the good nanoparticle-host cell and nanoparticle-biofilm interactions for improving the antibiotic drug activities against the poorly accessible bacteria, and supports LIN-LPN as a new alternative therapy for preventing the recurrence of MRSA-mediated bone infections.
Keywords: Osteomyelitis, Nanoparticles, Antibiotic, Drug-resistant bacteria
1. INTRODUCTION
Osteomyelitis is one of the most challenging and insidious infectious diseases to treat [1–3]. The majority of osteomyelitis cases are associated with bacterial infections, with Staphylococcus aureus (S. aureus) being the most prevalent pathogen [4]. Current drug therapy of osteomyelitis typically includes 4-6 weeks of systemic antibiotics following by months of oral antibiotic use [5–6]. Despite lengthy, costly antibiotic treatment; the recurrence rate of chronic osteomyelitis remains as high as 30% at 12 months [2]. There is clearly a need for a more effective, efficient form of antibacterial therapy for this challenging medical condition.
In addition to inconsistent bone penetration by many antibiotics [1,2,7], the effectiveness of conventional antibiotic therapy is further limited by the growth behaviors of some antibiotic-resistant bacteria, e.g. methicillin-resistant S. aureus (MRSA), that cause osteomyelitis [8–9]. These bacteria can reduce exposure to the antibiotic treatment and evade host defenses by hiding intracellularly and in their biofilms [10]. For instance, S. aureus can be internalized into human osteoblasts via receptor-mediated pathway using the cytoskeletal elements [11,12]. Meanwhile, thick biofilms can easily grow on the surface of prosthetic and bone tissue, thereby serve as diffusion barriers against antibiotic penetration and promote innate antibiotic resistance [1,13].
Recently, alternative antimicrobial therapies such as antibiotics delivered by nanoparticles, aka nanoantibiotics, were explored to improve osteomyelitis treatment [14–16]. The nanoantibiotics can either be immobilized in matrix/cement for local use or injected parenterally in free form [17–22]. While the former can release drug by gradual elution to lower the risk of bone infections [18,19], they require expertise for surgical implantation. Besides, the eluted drug molecules may still face difficulties to diffuse to the intracellular compartment and biofilm matrix. In comparison, free nanoantibiotics with selected biomaterials like phospholipids may inherently adsorb onto the biofilm or bacteria surface due to their resemblance to biological surface structure [14,20]. For treatment of those persistent MRSA in the infected bones, these nanoparticle-cell and nanoparticle-biofilm interactions are potentially valuable.
Linezolid is an antibiotic often used against MRSA-bone infections that respond poorly to vancomycin [23], but was shown to achieve inconsistent drug levels in bone tissue and biofilms [19, 23]. Up to date, only a few linezolid nanoformulations have been reported and they were not tailored for bone MRSA infections [24,25]. In this study, we aimed to develop and optimize a new lipid-polymer hybrid nanoparticle (LPN) system suitable for delivering linezolid, and evaluate if this nanotherapy could interact well with osteoblasts and biofilms and be more effective against the MRSA residing there. We hypothesized that (see Fig. 1A for detail): (i) with the hybrid design of LPN, the PLGA core of LPN can efficiently entrap the polar, well-diffusible linezolid molecules while the phospholipid/lipid coating will prevent their uncontrolled release, and (ii) with a phospholipid surface, LPN is expected to have good interactions with the bone cells and biofilms. As a result, this linezolid-loaded LPN (LIN-LPN) could achieve enhanced therapeutic activities against the intracellular or biofilm-embedded MRSA.
Figure 1.

(A) Design of a linezolid-loaded lipid polymer hybrid nanoparticles (LIN-LPN) and its expected effects. Orange lipid: phospholipid; Blue lipid: PEGylated phospholipid; Red: cholesterol as stabilizer; Orange yellow core: PLGA core; D: linezolid. (i) Release of linezolid to treat planktonic bacteria; (ii) Internalized by osteoblasts to treat intracellular bacteria; (iii) Accumulated in biofilms to treat biofilm-embedded bacteria. (B) Size distribution of LIN-LPNs as measured by dynamic light scattering. (C) Morphology of LIN-LPNs as revealed by scanning electron microscope.
2. MATERIAL AND METHODS
2.1. Materials
Linezolid was purchased from LKT Laboratories (St. Paul, MN); DSPE-mPEG (1,2-Distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000]), cholesterol and DPPE-lissamine rhodamine B (1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) from Avanti Polar Lipids (Alabaster, AL); ), lecithin (from soybean), gentamicin from Sigma-Aldrich (St. Louis, MO); PLGA (poly(D,L-lactic-co-glycolic acid), 50:50 monomer ratio, ester cap, MW 15kDa) from Polyscitech (West Lafayette, IN). Tryptic soy broth (TSB) and agar were supplied by Becton-Dickinson (Cockeysville, MD). Wheat germ agglutinin-Alexa Fluor 633 conjugate for cell plasma membrane labeling and DAPI (4’,6-diamidino-2-phenylindole) as nuclear dye, Fluoromount aqueous mounting medium, HEPES (N-(2-hydroxyethyl)piperazine-N’-2-ethanesulfonic acid) and most solvents used were bought from Thermo-Fisher Scientific (Pittsburg, PA, USA). Triton X-100 was purchased from Bio-Rad (Hercules, CA).
2.2. Preparation of linezolid-loaded LPN (LIN-LPN)
Linezolid-loaded LPNs (LIN-LPNs) were prepared based on nanoprecipitation technique [26]. Briefly, lipid components including lecithin, DSPE-mPEG(2000), and cholesterol were dissolved in chloroform in the molar ratio of 7:3:3.5. The mixture was blow-dried by nitrogen gas and stored in vacuum overnight. After that, the dried film was rehydrated with 5% (v/v) ethanol solution. The mixture was sonicated for 1 min and then stirred under 65°C water bath until completely dissolved. To form the polymeric phase, PLGA was dissolved in acetonitrile as 15 mg/ml solution. For drug loading, linezolid was dissolved in PLGA/acetonitrile solution. The two phases were combined by slowly adding PLGA solution into the lipid solution under stirring. The typical weight ratio of PLGA/lipids used was 4:1. The mixture was vortexed for 1 min then gently stirred under vacuum for 2 h at room temperature. Unencapsulated drug and any remaining organic solvent were washed with deionized water 3 times by centrifugal filter tube using Amicon® Ultra-4 (100k molecular weight cut-off or MWCO, Sigma-Aldrich, St. Louis, MO) at 4000g for 20 min. LPN was rehydrated using 5% sucrose or culture medium for experiments. For comparison, lipid-free, PLGA only nanoparticles were also prepared by excluding the lipid components.
2.3. Particle size, zeta potential and morphology analysis
The size distribution, polydispersity index (PDI) and zeta potential values of nanoparticle suspensions were determined by dynamic light scattering (Zetasizer® 3000HS, Malvern, UK). All samples (25 μl) were diluted with 1 ml of deionized water and measurements were performed at 25 °C, with medium viscosity set at 0.89 mPa.s and refractive index at 1.330. Particle size values by intensity, PDI and zeta potential values were recorded in triplicate with samples prepared separately. PDI values were calculated by the manufacturer’s software which analyzed a cumulants fit of the correlation function as defined in the ISO standard document 13321:1996 E and 22412 [27]. The values reported were normalized between 0 and 1 (the higher the value, the broader the size distribution). To characterize the structure, electron microscope images were taken by JEOL F200 S/TEM (Peabody, MA).
2.4. Determination of drug loading and in vitro drug release profile
High performance liquid chromatograpy (HPLC) analyses were performed using Agilent 1100 system (Santa Clara, CA) and the separation was carried out on a reverse-phase Phenomenex Gemini C18 column (5 μm, 150 × 4.6 mm, Torrance, CA). The mobile phase was pumped at the rate of 1.0 mL/min with a mixture of acetonitrile and 2.67 mM acetic acid aqueous solution in 25:75 volume ratio. The run time was 10 min and injection volume was 10 μl. The UV absorbance of eluent was monitored over the spectrum range 190-300 nm and processed at 253 nm.
In order to measure drug loading, blank nanoparticles and LIN-LPN formulations with different initial drug feeds were passed through spin desalting column for 5 min under 1500 × g (Zeba, 40K MWCO, Thermo Scientific, Waltham, MA). Nanoparticles were then collected and dissolved in methanol and water (1:4 v/v, with 0.05 N hydrochloride acid and 0.05% Triton-X100) before drug quantification with HPLC. Drug encapsulation efficiency (EE%) was determined as: [Amount of encapsulated drug × 100%]/Initial drug feed.
To study the drug release characteristics, 3 ml of the following groups: free drug, blank LPN or LIN-LPN were transferred into dialysis tubes with MWCO at 10 kDa. Dialysis was performed against 1L phosphate buffered saline (pH 7.4) at 37 °C, and the buffer was changed every 24 h to maintain sink condition. At each selected time point, 100 μl of drug or LPN sample was collected from the dialysis tube and then centrifuged through the desalting column to remove any unencapsulated free drug. Linezolid was quantified by HPLC and the % drug released was calculated based on the proportion of linezolid released into the release buffer.
2.5. Dispersion stability of LPNs
Dispersion stability of LPNs was evaluated to learn if the nanoparticles may prematurely aggregate in a serum-rich environment like culture medium or blood plasma. The nanoparticles were diluted to 1 mg/ml with PBS supplemented with or without 10% fetal bovine serum and incubated at 37 °C and at room temperature for up to 2 days. For each measurement, 25 μl sample was collected, and the LPN size distribution were measured as above-mentioned in triplicate.
2.6. Osteoblast cell culture and bacterial culture
We obtained MC3T3-E1 osteoblast cell line from American Type Culture Collection (ATCC, Manassas, USA). This cell line was derived from mouse calvariae and has been studied for bone matrix synthesis, mineralization, changes in morphology and metabolism during differentiation [28]. The cells were maintained in alpha minimum essential medium supplemented with 10% FBS and penicillin-streptomycin 100 U/ml-100 μg/ml at 37 °C in a 5% CO2 incubator, and subcultivated at 1:6 ratio once the cells reached confluence. Three strains of osteomyelitis-causing bacteria including USA-300-0114 (the predominant community acquired MRSA strains in the US [29]), CDC-587 (a methicillin-sensitive S. aureus) and RP-62A (S. epidermidis) were purchased from ATCC. Each liquid suspension of bacteria used for experiments was obtained by incubating a single colony overnight at 37 °C in 10 ml of tryptic soy broth.
2.7. Minimum inhibitory concentration (MIC)
MIC assays were determined by the broth dilution method in 96-well plates. LIN-LPN and free linezolid were assayed by dissolved as 500 μg/ml stock solution in TSB, and serially diluted for the assay. Each well of a sterile 96-well polystyrene plate was inoculated with 5×105 CFU/ml bacteria per well from the overnight liquid bacterial cultures. Positive controls included TSB plus seeded bacteria without drug added. Plates were incubated at 37 °C for 24 h. The determination of growth was completed by measuring the absorbance at 600 nm (OD600) using a microplate reader. Each group was repeated nine times. MIC values were calculated using Lambert et al’s method [30]. The concentration values were converted to their logarithm equivalents, and the data were fit to a Gompertz model and analyzed with Graphpad prism software.
2.8. MRSA internalization into osteoblasts
After MC3T3-E1 osteoblasts reached confluence growing in 12-well plates, the cells were washed three times with Dulbecco’s phosphate-buffered saline (DPBS) and fresh drug-free growth medium was added to each well. The cells were then inoculated with 5×108 MRSA bacteria per well for 1 h at 37 °C. After bacterial internalization, the supernatants were removed and the osteoblasts were washed three times. Growth medium containing 25 μg/ml gentamicin, an antibiotic that cannot efficiently permeate across cell membrane, was added to each well to eliminate any remaining extracellular bacteria. To quantify intracellular MRSA, after 3 h incubation at 37°C, the infected osteoblasts were washed with DPBS and lysed with 0.1% Triton X-100. Suspensions of serially diluted lysate were plated on tryptic soy agar overnight at 37 °C. The supernatants were also similarly plated to confirm the eradication of extracellular bacteria.
2.9. Antimicrobial assay of intracellular MRSA
After MRSA internalization, osteoblasts were washed twice with DPBS and grown in fresh antibiotic-free growth medium. The cells were then treated with serially diluted LIN-LPNs, blank LPNs or linezolid free drug solution. Following incubation at 37 °C for 6 h, treated cell cultures were washed and then lysed by incubation with 0.1% Triton X-100 for 5 min to release the internalized bacteria. The internalized bacteria were quantified by plating the serially diluted lysates on tryptic soy agar overnight at 37 °C as in 2.7.
2.10. Visualization of LPN internalization into osteoblasts
To examine if LIN-LPN could be internalized by osteoblasts, LPNs were labeled with 0.25% DPPE-lissamine rhodamine B (by weight of their lipid content). We performed size measurements (as in 2.3), MTT assay (using MC3T3-E1 osteoblasts as in 2.11) and MIC assay (using USA300-0114 as in 2.7) and no significant differences were observed comparing unlabeled and labeled LIN-LPNs (data not shown). We thus assumed that the labeling did not have an impact on the nanoparticles’ biological effects. Osteoblasts were seeded and grown on sterilized coverslips in a 35 mm poly-lysine coated cell culture dish. After osteoblasts reached confluence, 50 μl of 10 mg/ml of fluorescence labeled nanoparticles were introduced to each well. At the end of 6 h incubation at 37 °C, the osteoblast nuclei and cell membrane were stained with DAPI and wheat germ agglutinin (Alexa Fluor 633 Conjugate), respectively. The cells were then fixed with 4% formaldehyde for 10 min and mounted on slides with Fluoromount. Nanoparticle internalization was visualized with a Leica DM IRE2 confocal microscope with a TCS SL system.
2.11. Cytotoxicity assay
The intrinsic toxicity of LPNs against osteoblasts was evaluated using MTT (3-(4, 5 dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide) assay. Briefly, the cells were seeded at density of 8,000 per well in a 96-well plate, and incubated at 37 °C for 16 h. After aspiration of the supernatant, the cells were incubated with LIN-LPNs, blank LPNs or free linezolid drug solution of different drug concentrations for 16 h. Drug-free medium and 0.5% Triton-X served as the negative and positive controls, respectively. The cells were then washed and incubated in MTT containing medium for 4 h. Formazan formed was dissolved in dimethyl sulfoxide. Absorbance readings at 570 nm were taken by Spectramax M2 microplate reader (Molecular Devices) using 630 nm as reference.
2.12. Biofilm microplate Assay
Overnight cultures of S. aureus grown in TSB medium were added to TSB+1% sucrose at a concentration of 5 × 106 CFU/ml (or normalized to OD600 = 0.02). The bacterial suspension was inoculated into a 96-well tissue culture plate. After overnight incubation, LIN-LPN or free linezolid solution were added at different concentrations for 12 h. The culture plate was washed with water three times to remove any planktonic bacteria and non-embedded antibiotics. Fresh TBS was added into the culture plate for another overnight incubation at 37 °C. Crystal violet staining of biofilm was then performed as follows. [31] Non-adherent cells were washed off by water 3 times, and adherent biofilm was stained with 0.1% crystal violet solution at room temperature for 15 min. After washing with water 3 times, the stained biofilm was dried overnight and mixed with 30% acetic acid (v/v) for 10 min. The absorbance was measured at 550 nm with SpectraMax M2 microplate reader.
2.13. Confocal laser-scanning microscopy (CLSM) of LPNs in biofilm
Biofilm formation and eradication was visualized by CLSM. Wheat germ agglutinin-fluorescent Alexa Fluor 633, which binds to polysaccharide intercellular adhesin, was used to help indicate the biofilm structure. The overnight bacterial suspension was inoculated in optically clear bottom 96-well plate. DPPE-lissamine rhodamine B labeled LPNs at different concentrations were added into the wells. After 12 h incubation, each well was washed with PBS three times and stained with 10 μg/ml wheat germ agglutinin-Alexa Fluor 633 for 15 min. To counterstain the bacterial cells in biofilms, LIVE/DEAD BacLight Bacterial Viability kit was used. The cell was fixed with 4% formaldehyde before scanned with Operetta CLS High-Content Analysis System (PerkinElmer, Walham, MA).
2.14. Measurement of linezolid levels in animals’ bones
All animal works were approved by Institutional Animal Care and Use Committee and animals were cared for in accordance with institutional guidelines. Sprague-Dawley rats (250±25 g, male) purchased were given full access to food and water ad libtitum and were acclimatized for 7 days before use. Animals were randomized into three groups: vehicle control (dextrose for injection), free linezolid and LIN-LPN (the latter two at 20 mg/kg linezolid dose level). Rats were injected via tail-vein, and after 24 h their tibia were extracted, rinsed, pad-dried and immersed in chloroform to dissolve any nanoparticles to release the linezolid. The linezolid levels were measured by HPLC as in 2.4.
2.15. Statistical analysis
Results are presented as mean±S.D. from a minimum of three independent experiments. Statistical analysis was performed with GraphPad Prism 7 (La Jolla, CA). Unpaired student’s t-test or one-way ANOVA test were performed to determine the difference of means among groups. A value of p<0.05 was considered statistically significant.
3. RESULTS
3.1. Effects of composition on LPN physicochemical properties.
LPN as a hybrid nanosystem with lipids coating a polymeric core (PLGA) (as illustrated in Fig. 1A) was successfully prepared. Table 1A shows that the higher the PLGA molecular weight grade, the smaller the resulting LPN (from 192.1±4.5 nm using 5000-10000Da PLGA down to 80±12.4 nm using 45000-55000Da). Considering that at 45000-55000 Da grade the polydispersity index (PDI) began to noticeably increase, we chose 35000-45000Da PLGA for the remaining studies. Table 1B shows that low lipid:polymer ratio generally resulted in smaller LPN size; however, the PDI increased considerably when the ratio was <0.2 likely due to reduced stability. We therefore eventually selected 0.2-0.25 as the optimal range of lipid:polymer ratio. Table 1C shows that within this range, lower soy lecithin (PC): phosphoethanolamine (PE) ratio (or higher degree of PEGylation) led to smaller LPN size. When the PC:PE molar ratio was around 7:3 and lipid:polymer ratio at 20%, the LPNs formed were ~110 nm with low PDI (0.13). Using this as our optimal LPN composition, we then fed different amounts of linezolid into LPN to form LIN-LPN (from 0 to 20% by total weight). Table 1D shows that the size, PDI and zeta potential values all remained similar, indicating that linezolid loading did not affect the particle characteristics and stability. Fig. 1B shows the size distribution of a representative sample of LIN-LPN as measured using dynamic light scattering. The morphology of LIN-LPN fed with 15% linezolid was revealed in the SEM image (Fig. 1C). The nanoparticles were generally spherical, and the size was in the range of 50-150 nm, confirming the size distribution result.
Table 1A.
Effects of various formulation parameters on the characteristics of LIN-LPN Effect of molecular weight of PLGA polymer used
| PLGA MW (Da) | Size (nm) ± SD | PDI ± SD |
|---|---|---|
| 5000-10000 | 192.1 ± 4.5 | 0.145 ± 0.037 |
| 10000-15000 | 150.0 ± 2.9 | 0.127 ± 0.025 |
| 35000-45000 | 113.6 ± 4.3 | 0.125 ± 0.010 |
| 45000-55000 | 80.0 ± 12.4 | 0.184 ± 0.051 |
MW – molecular weight; PDI – Polydispersity Index; PLGA - poly(D,L-lactic-co-glycolic acid)
Table 1B.
Effects of various formulation parameters on the characteristics of LIN-LPN Effect of Lipid/Polymer ratio
| Lipid/PLGA (w/w) | Size (nm)±SD | PDI±SD |
|---|---|---|
| 1:9 (or 0.1) | 95±5.6 | 0.187±0.064 |
| 1:4 (or 0.2) | 113.6±4.3 | 0.125±0.01 |
| 1:2 (or 0.33) | 133.2±11.4 | 0.22±0.031 |
| 1:1 (or 0.5) | 162.6±15.5 | 0.21±0.077 |
| 1:0.5 (or 0.67) | 150.3±21.4 | 0.27±0.031 |
Table 1C.
Effects of various formulation parameters on the characteristics of LIN-LPN Effect of lipid composition and PEGylation (i.e. amount of DSPE-PEG)
| Soy Lecithin | DSPE-PEG | Cholesterol | PC:PE (molar) | Lipid/Polymer (weight ratio) | Size (nm)±SD | PDI±SD | Zeta potential (mV)±SD |
|---|---|---|---|---|---|---|---|
| 0.46 | 0.74 | 0.06 | 7:3 | 0.2 | 111.77±1.37 | 0.13±0.02 | −43.97±0.58 |
| 0.57 | 0.63 | 0.08 | 10:3 | 0.2 | 110±3.19 | 0.18±0.06 | −43.6±1.16 |
| 0.65 | 0.55 | 0.09 | 13:3 | 0.2 | 169.2±3.27 | 0.26±0.01 | −39.17±0.9 |
| 0.73 | 0.47 | 0.10 | 17:3 | 0.2 | 141.5±4.25 | 0.16±0.02 | −41.7±0.43 |
| 0.58 | 0.92 | 0.08 | 7:3 | 0.25 | 115.77±2.11 | 0.19±0.07 | −41.97±0.21 |
| 0.71 | 0.79 | 0.10 | 10:3 | 0.25 | 136.17±3.67 | 0.19±0.02 | −44.73±0.45 |
| 0.81 | 0.69 | 0.11 | 13:3 | 0.25 | 136.57±4.96 | 0.2±0.04 | −44.67±0.87 |
| 0.91 | 0.59 | 0.13 | 17:3 | 0.25 | 143.07±2.57 | 0.15±0.01 | −43.9±1.02 |
In each of the above samples, PLGA used was 6 mg. PC – Phosphatidylcholine (i.e. soy lecithin); PE – Phosphoethanolamine. DSPE-PEG – 1,2-Distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000]
Table 1D.
Effects of various formulation parameters on the characteristics of LIN-LPN Effect of drug feeding (amount of drug added during LPN preparation by % wt)
| Drug Fed (%wt of LPN) | Size (nm)±SD | PDI±SD | Zeta-potential (mV)±SD |
|---|---|---|---|
| 0 | 104.8±0.8 | 0.13±0.01 | −43.6±3.4 |
| 5 | 98.1±1.1 | 0.13±0.01 | −44.9±1.9 |
| 10 | 107.3±0.1 | 0.14±0.02 | −43.9±4.7 |
| 15 | 109.3±1.4 | 0.14±0.01 | −43.9±2.2 |
| 20 | 115.7±0.4 | 0.16±0.05 | −43.0±1.6 |
3.2. Drug loading and release properties of LPN
Figs. 2A and B show the drug loading and drug release properties of LIN-LPN, respectively. As shown in Fig. 2A, the drug payload (% of linezolid actually encapsulated, by weight of LPN) generally increased as the initial drug feeding (i.e. the amount of linezolid added to LPN preparation) increased up to 15%. The payloads were 3.4±0.2%, 7.1±0.2%, 12.0±0.4% and %EE were 68.6±4.6%, 71.4±2.2%, 79.9±2.33% when the drug feeding levels were 5%, 10% and 15%, respectively (calculated based on payload/drug fed x 100%). Higher drug feeding level was tried (20%), but it was counterproductive. The payload actually declined to 10.3% and %EE to 51.4±1.33% probably due to drug precipitation. Overall, the highest drug payload that could be achieved was 12.0% by weight of LPN at 15% initial drug feeding. For comparison, lipid-free, PLGA only nanoparticles were studied as well. The linezolid payloads were 3.1%, 5.7% and 9.4% at drug feeding levels were at 5%, 10% and 15%, respectively (not shown in Figure).
Figure 2.
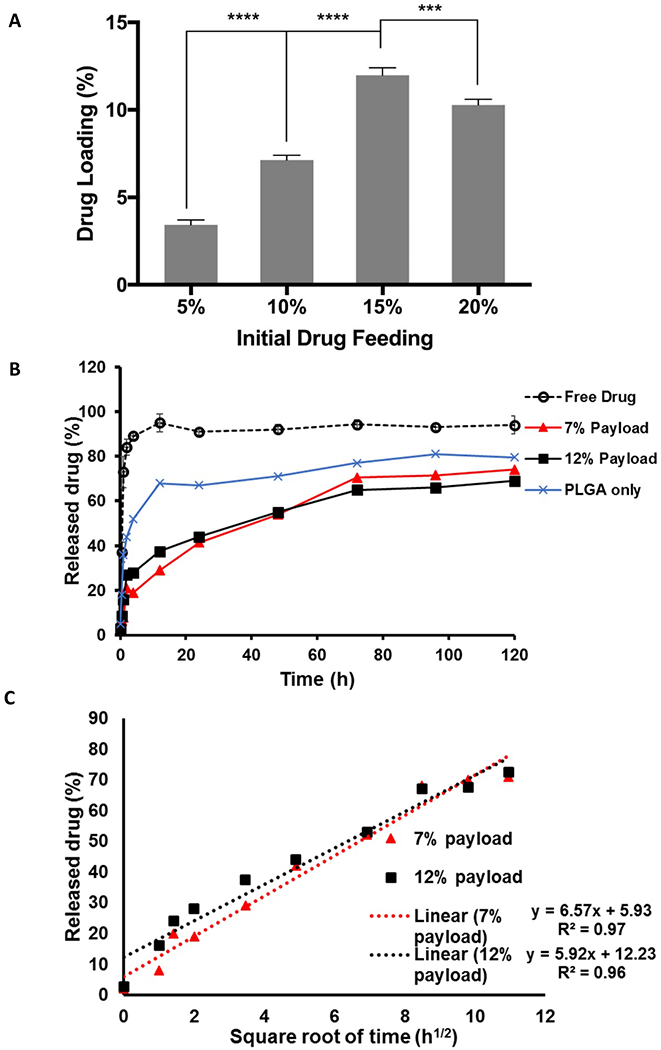
Drug loading and drug release properties of LIN-LPN. (A) Actual % of drug loading (by weight of LPN) in preparation fed with different amounts of linezolid (i.e. initial drug feeding, by weight of LPN). *** P<0.05; **** P<0.01, Mean+SD (N=3) shown; (B) Drug release profiles of LIN-LPN. LIN-LPNs with 7% or 12% drug payload (equivalent to 10% or 15% drug feeding) were studied. We also included PLGA-only nanoparticles with 9.4% payload for comparison. (C) Plot of % drug released versus square root of time to evaluate if the drug release follows Higuchi’s model. Mean±SD (N=3) shown.
Fig. 2B presents the drug release profiles of LPN with 7% (or 10% drug feeding) and 12% (or 15% drug feeding) linezolid payloads. Both profiles were in a biphasic manner. About 30-40% of the drug was released relatively fast in the first 12 h, following by gradual release of the remaining payload until 120 h. More than 70% of the payload was released by the end of the experiment. Free drug solution was also evaluated as a control. Around 90% of free linezolid compound was quickly released within 4 h. This indicates that the observed controlled drug release profiles were mainly due to the inherent properties of LPN instead of limited diffusion across the dialysis membrane used in the experimental setup. Fig. 2C plots the % drug released against square root of time. High linearity for both LIN-LPNs was achieved, suggesting that the drug release followed Higuchi’s diffusion-based model [32]. In brief, LPN was able to achieve controlled release primarily by diffusion-limited kinetics for at least 120 h or 5 days.
Fig. 2B also presents linezolid release from PLGA only nanoaparticles. Much quicker initial drug release was observed. About 70% of the payload was released in the first 12 h.
3.3. Dispersion stability of LPN
Fig. 3 presents the size change of LPN incubated in medium at pH7.2 at 37 °C with or without supplement with 10% fetal bovine serum and PBS. In all cases, both drug-free and linezolid-loaded LPNs showed modest size increases in the first 2 h. Comparing the serum positive with negative groups, the size increases in the former were larger. This suggests that the modest size increases were partly contributed by interactions with the serum proteins. The particle size remained consistent at around 140 nm since then (PDI remained <0.3 throughout the whole study). The LPNs were generally stable when dispersed in an environment that simulated blood plasma.
Figure 3.
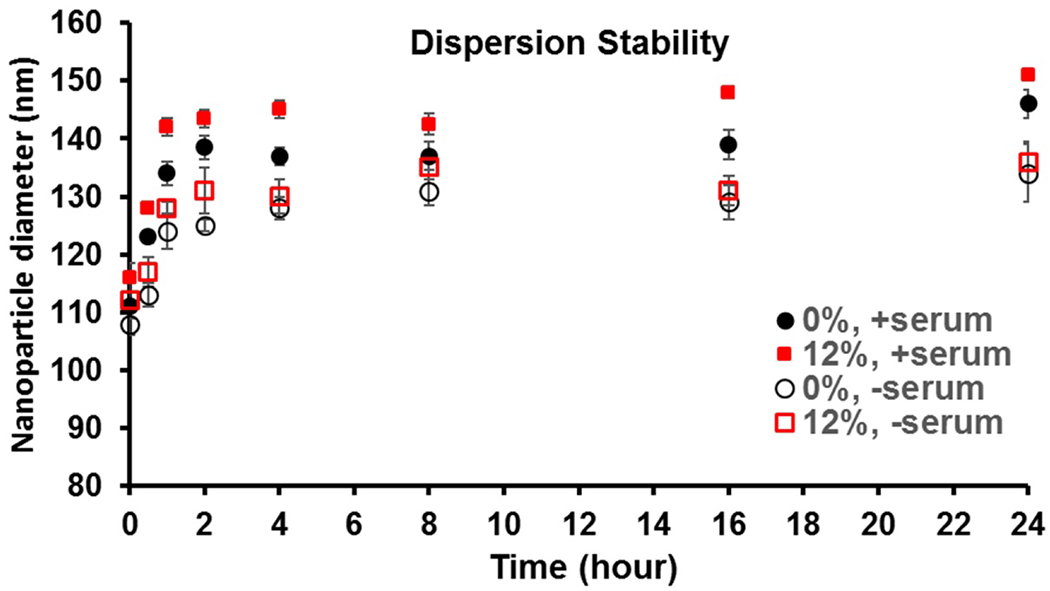
Monitoring of nanoparticle size of LIN-LPN in serum-supplemented medium at 37 °C. Nanoparticles without drug (0%) or with linezolid (12% payload) were studied.
3.4. Activities on planktonic bacteria
Three bacteria strains, including two strains of S.aureus and one strain of Staphylococcus epidermidis (MRSA: USA300-0114 and CDC-587, S. epidermidis RP-62A), were treated with free linezolid or LIN-LPN for 24 h and MIC measured. Table 2 presents the MIC assays result. In all cases the free drug was more effective. The MIC50 and MIC90 values of free linezolid were approximately 40-50% of the values of LIN-LPN at corresponding drug concentrations. We also evaluated the activity of drug-free, blank LPN. No significant antibacterial bacterial activity was observed (not shown), indicating that the observed antibacterial effects mostly came from the antibiotic linezolid.
Table 2.
Minimum inhibitory concentration values after 12 h treatment with free linezolid or LIN-LPN
| Bacterial strain | MIC50** (μg/ml) | MIC90** (μg/ml) | ||
|---|---|---|---|---|
| Free linezolid | LIN-LPN | Free linezolid | LIN-LPN | |
| USA300-0114 (MRSA) | 0.42±0.06 | 0.91±0.08 | 0.95±0.10 | 1.67±0.19 |
| CDC-587 (MSSA) | 0.64±0.04 | 1.38±0.11 | 1.41±0.15 | 1.89±0.22 |
| RP-62A (S. epidermidis) | 0.25±0.05 | 0.53±0.07 | 0.44±0.08 | 0.91±0.10 |
MIC50 and MIC90 values were defined as the lowest concentration of linezolid (as free or encapsulated form) at which 90 and 50% of the isolates were inhibited, respectively. MRSA – methicillin-resistant S. aureus; MSSA – methicillin-sensitive S. aureus. All values expressed as mean±SD (n=3).
3.5. LPN inhibition of MRSA intracellular infection and cytotoxicity effects to osteoblasts
Fig. 4 shows the interactions of LIN-LPN with MRSA-infected osteoblasts. Fig. 4A presents the antibacterial effects of LIN-LPN against the intracellular MRSA (USA-300-0114 chosen for this purpose for its high intracellular virulence [33]). The intracellularly grown MRSA were released, cultured and the colonies formed quantified as CFU/ml in Fig. 4A. At dosing levels of 2 μg/ml or higher, LIN-LPN was substantially more effective in reducing the intracellular MRSA counts than the free linezolid control. It should be noted that the bacteria colony counts are expressed in log10 units instead of linear scale at the y-axis. The differences comparing LIN-LPN with free linezolid were: at 2 μg/ml linezolid, antilog (2.52 vs 4.46); at 4 μg/ml, antilog (1.34 vs 2.43); at 8 μg/ml, antilog (0 vs 1.10), translating into 87.0-fold, 12.3-fold, and 12.6-fold differences in CFU/ml (p<0.05). Higher antibiotic concentrations resulted in complete clearance of intracellular MRSA so the results were not shown. Blank LPN + free linezolid co-administration group was also included for comparison. Its inhibitory effect on the intracellular MRSA was similar to free linezolid only (Fig. 4A).
Figure 4.
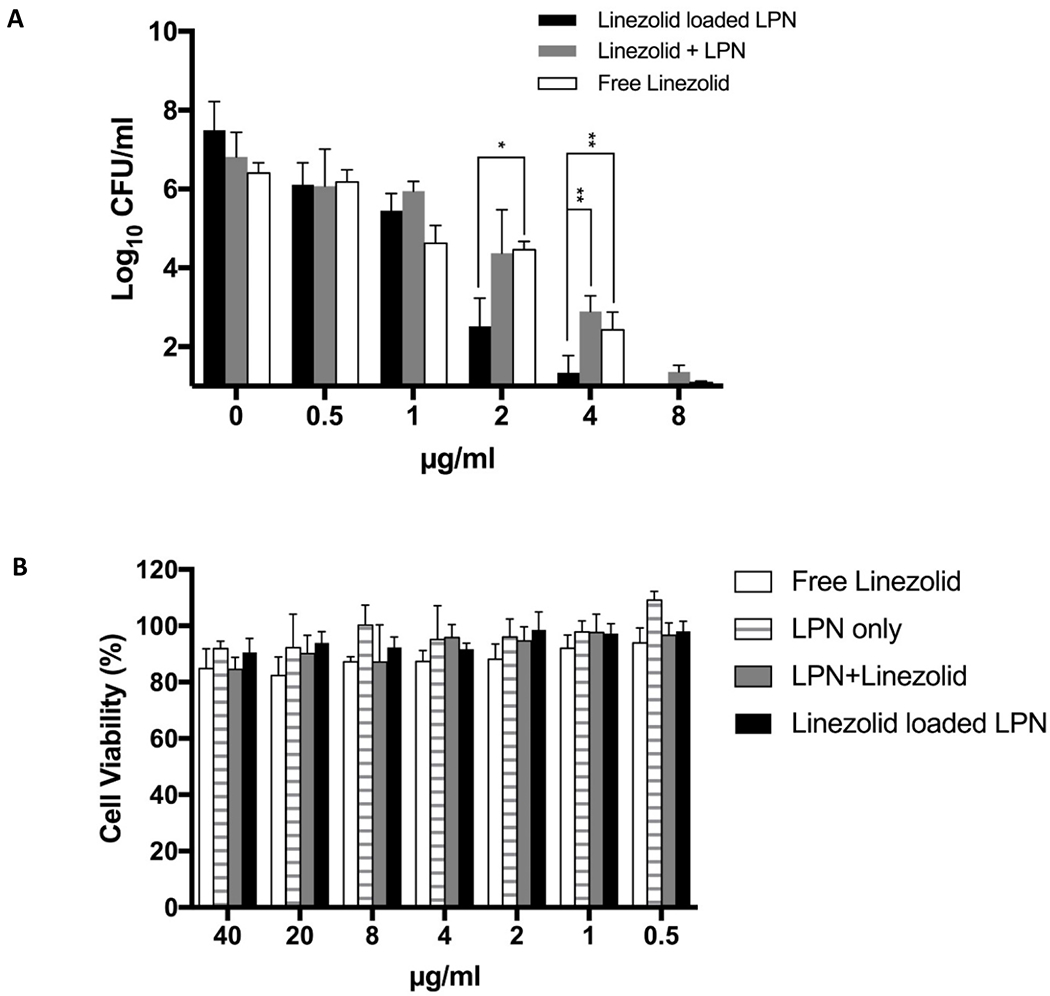
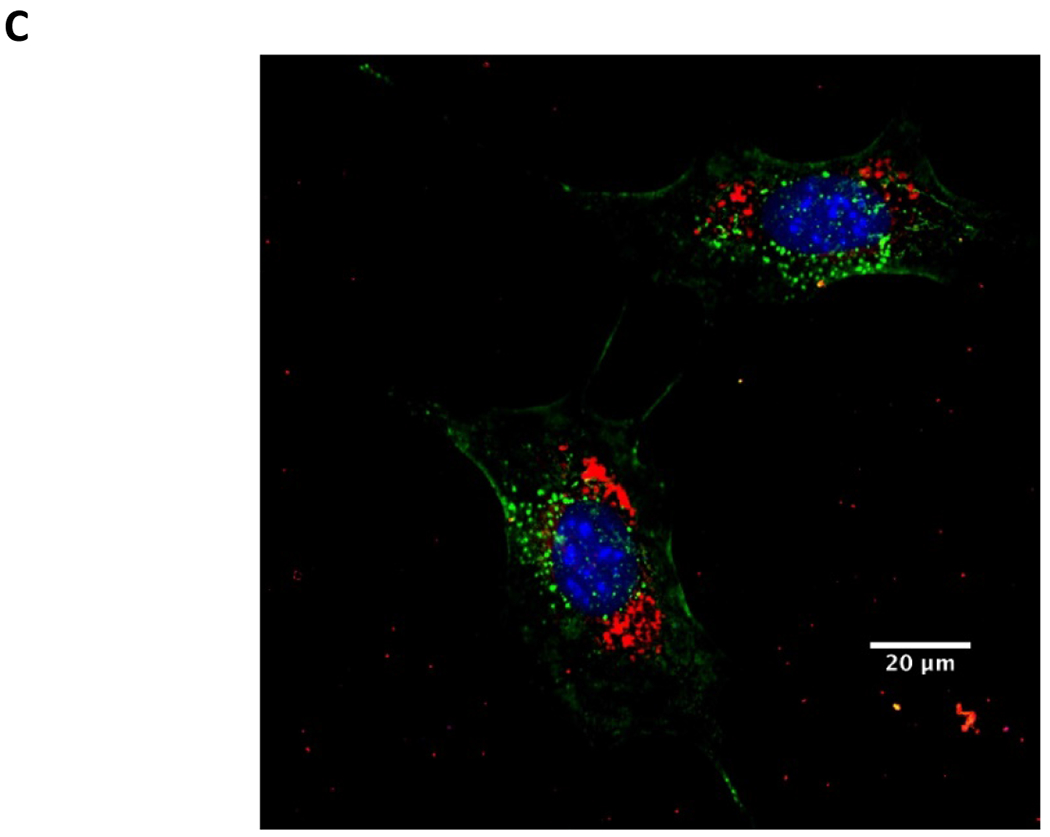
Treatment of intracellular MRSA with LIN-LPN. (A) Comparison of activities to eradicate intracellular MRSA (USA300–0114) grown inside osteoblasts (MC3T3-E1). Colony counts are shown in log10 scale at y-axis. Linezolid concentrations in μg/ml as x-axis. Results expressed as mean±SD (N=3). * P<0.05, ** P<0.01 (B) MTT assay evaluating the viability rates of osteoblasts after treatment with LIN-LPN. Results expressed in mean±SD (N=3). (C) Confocal image of MC3T3-E1 cells treated with LIN-LPN. Red: LIN-LPN, green: cell membrane, blue: cell nuclei.
To learn if the observed higher efficacy of LIN-LPN was a misleading result caused by the death of the host cells instead of the MRSA, we performed MTT assays to evaluate the viability of the infected MC3T3 osteoblasts. Fig. 4B presents the cell viability (in %) of osteoblasts after they were treated with blank LPN only, LIN-LPN, free linezolid only and blank LPN/free linezolid combination. All groups showed minimum cytotoxicity against osteoblasts, with no significant differences among these groups even the drug concentration was as high as 40 μg/ml (far exceeding the concentration range used in the study as Fig. 4A shown).
Fig. 4C presents the confocal microscopy images of MC3T3 osteoblasts after 6 h exposure to fluorescently-labeled LIN-LPN. The LPN were shown in red, nuclei in blue and cell membranes in green. The image shows that LPNs were extensively internalized by the osteoblasts and accumulated at the cytosolic region, suggesting that the enhanced intracellular antibacterial effects could be contributed by cell internalization of LIN-LPN.
3.6. Anti-biofilm activities of LPN
Fig. 5A presents the result of biofilm retention assay. In this assay, higher anti-biofilm activity resulted in less biofilm left, i.e. reduced biofilm retention, after treatment. Our result showed that LIN-LPN was consistently more effective than free linezolid for eradicating the MRSA biofilm. Comparing the two groups, the drug in nanoantibiotic form (i.e. LIN-LPN) suppressed the biofilm to 35%-60% of the value achieved using the free drug (linezolid) at the same drug concentration (all p<0.05). To rule out if the observed stronger anti-biofilm effect of LIN-LPN was caused by the inherent toxicity of the nanoparticles themselves, free linezolid + blank LPN group was included as a control. We found that the efficacy of this control was statistically similar to the free linezolid group. The blank LPN was considered not therapeutically active.
Figure 5.
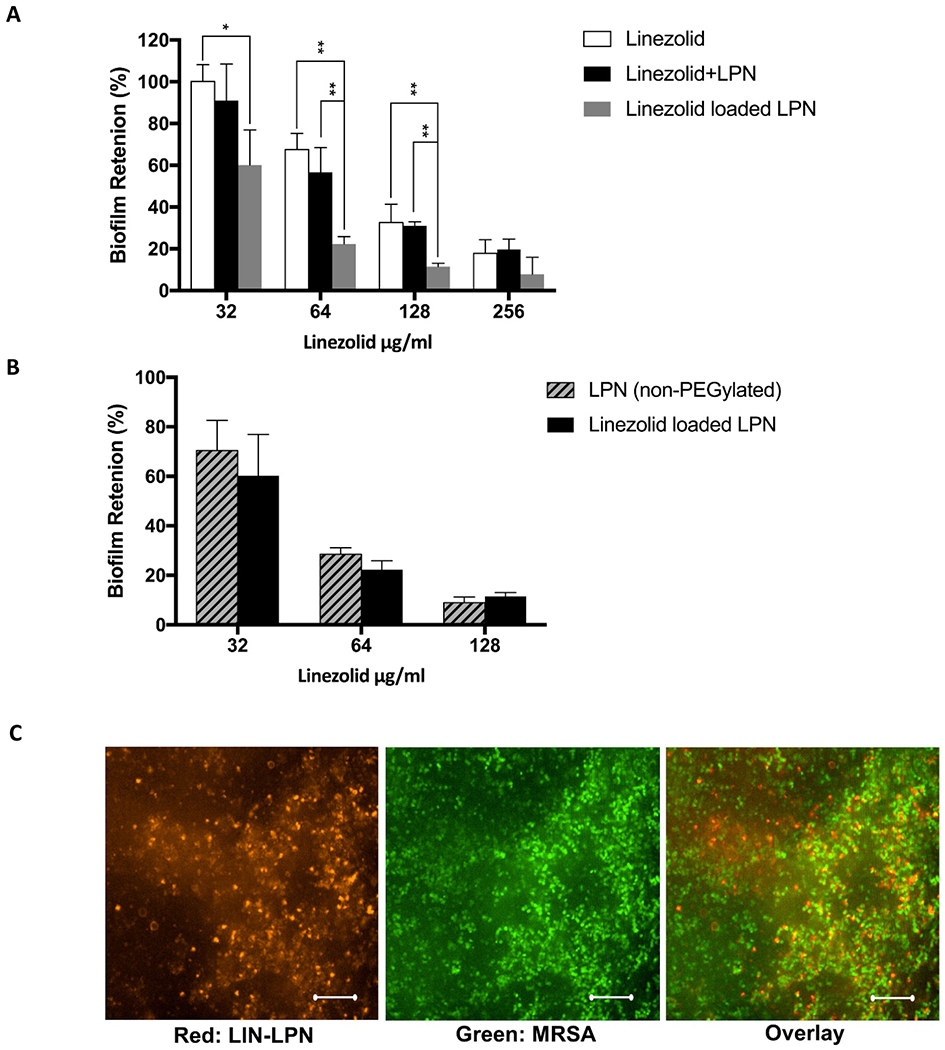
Treatment of MRSA biofilms with LIN-LPN. (A) % of biofilm retained after treatment with LIN-LPN or free linezolid. Results expressed as mean±SD (N=3). * P<0.05, ** P<0.01. (B) Comparing anti-biofilm effects between normal LIN-LPN and LIN-LPN without PEGylated coating (non-PEGylated LPN). (C) Confocal image of biofilm treated with LIN-LPN after multiple washing with fresh medium. Red: clusters of labeled LIN-LPN, Green: bacteria stained. Scale bar = 20 μm.
Fig. 5B compares the effects of regular LPN and LPN without PEGylated phospholipids (i.e. DSPE-PEG) on biofilms. No statistically significant differences between the two groups were observed.
Fig. 5C presents the confocal microscope image of MRSA biofilms after exposure to labeled LIN-LPN. USA-300-0114 bacteria were in green and LIN-LPN in red. The image indicates extensive retention of the nanoparticles in the biofilms even after multiple buffer washing. We also repeated this experiment using LPN without PEGylation, similar nanoparticle retention was observed (image not shown).
3.7. Improved linezolid delivery to bone using LIN-LPN
We compared the bone levels of linezolid as delivered by LIN-LPN versus as free linezolid solution (Fig. 6). It was shown that LIN-LPN could increase the bone linezolid level to over 4-fold of the free linezolid group (p<0.05), No signs of acute toxicity was observed in all animals tested.
Figure 6.
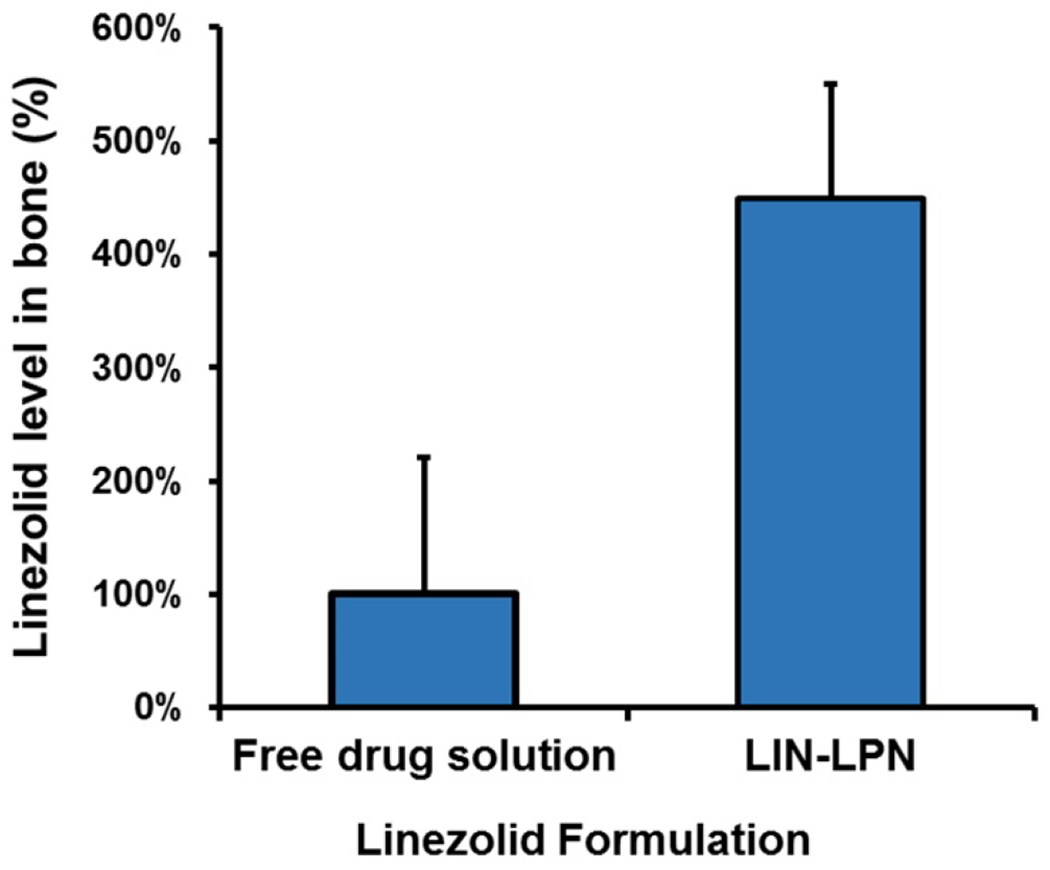
Comparison of in vivo linezolid levels in the bones of rats treated with different formulations. Rats were injected with free linezolid or linezolid-loaded LPN (both dosed at 20 mg/kg linezolid). After 24 h the tibia were extracted, rinsed, pad-dried and immersed in chloroform to dissolve any nanoparticles in the bones to release linezolid for measurement. The results were normalized (divided by the bone weight) and presented as mean±SD (N=3 per group).
4. DISCUSSION
In this study, our general research question is: considering the potential to directly interact with the bone cells and biofilms, will a nanoantibiotic in its non-matrix immobilized form be more effective than free antibiotic molecules against the intracellular or biofilm-embedded MRSA? We thus developed LIN-LPN to test and implement this strategy. Overall, our findings indicate significantly enhanced antibacterial activities against these less accessible, more antibiotic-resistant pathogens that often cause osteomyelitis chronicity and recurrence [12, 34].
Our first challenge in developing a parenteral linezolid nanoformulation is to ensure that it can efficiently entrap the well-diffusible linezolid molecules (considering its low molecular weight at 337 Da and fair water-solubility at 3 mg/ml [35]) and prevent their burst release, while keeping its size small enough for systemic circulation and extravasation from the blood vessels. As a multi-component system with high versatility, researchers have pointed that the various parameters such as the type and proportion of the ingredients in a lipid-polymer hybrid nanosystem need to be well optimized to achieve the desirable physicochemical and therapeutic properties [36]. Table 1A to 1D summarizes the related data. Contrary to the results of Budhian et al [37] which showed modest increase in their nanoparticle size using PLGA of higher molecular weight, we observed the opposite trend (Table 1A). This may be one of the differences between a lipid-coated hybrid nanocarrier and a PLGA only nanoparticle. We next evaluated the impact of polymer-to-lipid ratio (Table 1B). We observed that clinically useful size (around 100 nm) was obtained when the ratio is at 1:9 to 1:4. This is consistent with the findings by Farokhzad’s group, which showed the optimal lipid/polymer ratio for their hybrid nanocarrier was 15% (w/w), or equivalent to 1:5.7 polymer-to-lipid ratio [38,39]. With the ratio defined, the effects of PEGylation were investigated (Table 1C), and it was shown that higher degree of PEGylation tended to reduce the LPN size in the manner similar to the findings by Zheng et al [40]. The higher surface density of PEG groups likely avoided aggregation of the droplets of nanoparticle ingredients during preparation by steric stabilization.
By manipulating the above-mentioned several aspects of the nanoparticle composition, and after showing that the amount of linezolid loaded did not have substantial impact (Table 1D), we successfully obtained LPNs with size around 110 nm and PDI value around 0.2. This is considerably smaller than previous linezolid nanoformulations, e.g. Parisi et al’s polymeric linezolid nanoparticle was 259.6 nm in diameter [41]. The LPN size was in fact close to the two most clinically used parenteral nanoformulations, Doxil (85 nm) and Abraxane (130 nm) [42], which both have reliable track records of staying in circulation and extravasating well.
We hypothesized that the polymer-lipid hybrid nature of LPN could contribute to both significant entrapment (by PLGA core) and controlled drug release (by lipid coating) of the polar linezolid molecules. The data indicated that high linezolid payload up to 12% by weight of LPN could be achieved. The payload values were reduced but remained high in the PLGA-only, lipid-free nanoparticles (e.g. from 12.0% to 9.4%) at same drug feeding levels. This suggests that the linezolid was primarily entrapped in the PLGA core and the lipid coating likely reduces the drug loss into the solvent during the LPN formation process. Meanwhile, controlled drug release profile (quicker release of ~25% payload in first 4 h, followed by ~70% release of payload in 120 h) were observed. The faster drug release from the PLGA-only nanoparticles also confirms the role of lipid coating as a diffusion barrier against uncontrolled release of the linezolid molecules. Overall, the findings support our hypothesis and show that even with reasonably small particle size, the hybrid design of LPN remained effective for holding up the well-diffusible linezolid molecules. Moreover, LIN-LPNs also maintained their size and stayed well dispersed in serum-supplemented medium at 37 °C (Fig. 3), ruling out the concern of nanoparticle instability and aggregation after injection.
With a proper LPN system established, we focused on our main goal of this study, i.e. to study if the non-immobilized nanodelivery system can achieve significant activities against the intracellular and biofilm-residing MRSA. Prior to these studies we first performed a standard MIC assay to establish the baseline anti-bacterial activities of LIN-LPN against three strains of osteomyelitis-causing bacteria, including MRSA (USA-300), methicillin-sensitive S. aureus (CDC-587) and S. epidermidis (RP-62A), in their planktonic form. As expected, in all cases free linezolid was 40-50% more effective than LIN-LPN containing the same drug concentration (Table 2). This is reasonable considering that the treatment time was 12 h (technically difficult to extend the experiment due to fast bacteria growth in the control), and LIN-LPN only released ~30% of its total drug payload within this time period (see Fig. 2B). It should be noted that free linezolid in human body is eliminated in hours (mean half life ~ 5 hr in adults) [43], while nanoparticles tend to accumulate and stay at the diseased peripheral site for days. Our early in vivo biodistribution study of LIN-LPN actually supported this trend (Fig. 6, showing >4-fold higher in vivo linezolid bone level using LIN-LPN). In clinical situation, the planktonic bacteria in the bone will thus likely expose to a high linezolid level delivered by LPNs for a much longer duration than when free drug is used, and be better treated as a result. At this stage, this finding at least shows that the antibacterial activity of the linezolid in LIN-LPN was well preserved with no sign of degradation.
In comparison to the planktonic bacteria, an opposite trend was clearly observed when LIN-LPNs were used to treat MRSA residing intracellularly or in their biofilms. As shown in Fig. 4A, when the linezolid concentration was in the range of 2 to 8 μg/ml, the activity of LIN-LPN against intracellular MRSA was substantially higher than free linezolid, ranging from 12.3-fold to as high as 87.0-fold. For information, in a pharmacokinetics study of linezolid bone penetration [44], linezolid 600 mg 12 hourly was given orally to ten patients over 48 h before operation and intravenously 1 h before induction of anesthesia. Their bone linezolid levels were shown to be highly variable (estimated range: 3.6 to 22.5 μg/ml). It should be noted that the bone linezolid levels of four of these ten patients fell within the 2 to 8 μg/ml range, where LPN was shown to perform significantly superior. In other words, considering the high variability of bone penetration by linezolid, a sizeable portion of osteomyelitis patients will likely benefit from the higher efficacy of LIN-LPN for eradication of the intracellular MRSA.
It may be questioned that the observed higher suppression of intracellular MRSA count by LPN could be a misleading result caused by cytotoxic death of the host osteoblasts. The viability assay result (Fig. 4B) rules out this possibility as LIN-LPN and free linezolid led to similar impact on the host cell viability. We also included the free linezolid/blank LPN combination as a control. Its activities were similar to the linezolid only group, suggesting that the antibiotic needs to be physically loaded into the nanoparticle to gain the enhanced intracellular activities. The confocal microscope image (Fig. 4C) has confirmed extensive internalization and accumulation of LPNs in the osteoblasts, supporting nanoparticle-mediated drug trafficking into the cells as the key reason of the observed intracellular antibacterial activities.
Even though linezolid is effective against planktonic bacteria, its activity against biofilm isolates has been questionable [45]. In this study, we hypothesized that considering the track record of nanoparticle-biofilm interactions, LPNs could increase the activity of linezolid for biofilm eradication. This hypothesis is supported by our finding in Fig. 5A, which shows significantly improved reduction in biofilms in the drug concentration range of 32-128 μg/ml. As above-mentioned, even in individual patients with high bone penetration by free linezolid [41], the bone drug level barely exceeded 20 μg/ml. Considering that at 32 μg/ml concentration, free linezolid practically achieved no activities against the MRSA biofilms, it is expected that once biofilms of MRSA establish on the bone or prosthetic surfaces, the chance of treatment failure by standard linezolid therapy is high. In comparison, LIN-LPN at 32 μg/ml already shows moderate activity after one treatment. Considering that our in vivo data (Fig. 6) showed that LIN-LPN actually could substantially increase the bone linezolid level, the LIN-LPN therapy reported here should at least have a higher potential to tackle the MRSA biofilms that make osteomyelitis so frequently resistant to antibiotic therapy.
The higher anti-biofilm activity of LIN-LPN over free linezolid was likely caused by accumulation of the nanoparticles in the biofilm matrix, as shown in our CLSM image (Fig. 5C). This brought us to ponder about what led to the LPN accumulation in the biofilms. One of the possible explanations is the that PEGylated shell of LPN may achieve increased biofilm affinity so the LIN-LPNs could get well entangled in the biofilm matrix [46]. We thus also prepared LIN-LPN without PEGylated lipid and compared with the standard PEGylated LIN-LPN. Our data (Fig. 5B) revealed no significant differences in their anti-biofilm activities. Hence, further investigation is still needed in future to identify the manipulable parameters that are suitable for optimization of the LPN-biofilm interactions. At a minimum, the current LIN-LPN version is already significantly more effective than free linezolid to eradicate intracellular and biofilm-embedded MRSA. The LIN-LPN nanotherapy thus serves as a promising alternative to achieve osteomyelitis treatment with a lower risk of recurrence.
5. CONCLUSIONS
A new nanocarrier known as LIN-LPN has been developed for delivery of linezolid. This nanoantibiotic was designed for treatment of MRSA that frequently cause persistent, recurrent bone infections. The high propensity of bacteria like MRSA to evade treatment and host defense by harboring inside bone cells and biofilms has been a key factor contributing to the high recurrence rate of osteomyelitis. In this study we kept our focus on evaluating the therapeutic activities of LIN-LPN for treatment of these hidden bacteria. Our findings showed significantly enhanced efficacy of LIN-LPN over free linezolid for treating intracellular and biofilm-embedded MRSA. The advantage was particularly substantial as seen in the infected osteoblasts. In general, the study validated the feasibility of exploiting the good nanoparticle-cell and nanoparticle-biofilm interactions to enhance the antibiotic drug activities against those tough-to-treat, poorly accessible bacteria. Specifically, this study has validated LIN-LPN as a new alternative therapy that can be more effective for lowering the risk of the recurrence of MRSA-mediated bone infections. Our future studies will focus on further enhancing the bone penetrating capability of LPN, and evaluate the in vivo efficacy and biodistribution of this promising form of nanoantibiotic.
ACKNOWLEDGEMENTS
The study was supported by National Institute of Health/ National Institute of Allergy and Infectious Diseases R01 grant (1R01AI132852).
ABBREVIATIONS
- CLSM
Confocal laser-scanning microscopy
- DSPE-mPEG
1,2-Distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000]
- %EE
encapsulation efficiency
- HEPES
(N-(2-hydroxyethyl)piperazine-N’-2-ethanesulfonic acid)
- HPLC
high performance liquid chromatograpy
- LIN-LPN
linezolid-loaded lipid polymer hybrid nanoparticle
- LPN
lipid-polymer hybrid nanoparticle
- MIC
Minimum inhibitory concentration
- MRSA
methicillin resistant Staphylococcus aureus
- MTT
(3-(4, 5 dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide)
- MWCO
molecular weight cut-off
- PDI
polydispersity index
- PLGA
poly(D,L-lactic-co-glycolic acid)
- SCV
small colony variants
- TSB
tryptic soy broth
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Lew DP, Waldvogel Osteomyelitis FA, The Lancet 364(9431) (2004) 369–379. [DOI] [PubMed] [Google Scholar]
- [2].Hatzenbuehler J, Pulling TJ, Diagnosis and management of osteomyelitis, American family physician 84(9) (2011) 1027. [PubMed] [Google Scholar]
- [3].Spellberg B, Lipsky BA, Systemic antibiotic therapy for chronic osteomyelitis in adults, Clin Infect Dis 54(3) 2012. 393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kremers HM, Nwojo ME, Ransom JE, Wood-Wentz CM, Melton LJ III, Huddleston PM III, Trends in the epidemiology of osteomyelitis: a population-based study, 1969 to 2009, The Journal of bone and joint surgery American 97(10) (2015) 837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Drancourt M, Stein A, Argenson JN, Zannier A, Curvale G, Raoult D, Oral rifampin plus ofloxacin for treatment of Staphylococcus-infected orthopedic implants, Antimicrobial agents and chemotherapy 37(6) (1993) 1214–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lew DP, Waldvogel FA, Use of quinolones in osteomyelitis and infected orthopaedic prosthesis, Drugs 58(2) (1999) 85–91. [DOI] [PubMed] [Google Scholar]
- [7].Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, Andersson DI, Selection of resistant bacteria at very low antibiotic concentrations, PLoS Pathog 7(7) (2011), < 10.1371/journal.ppat.1002158>. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Boucher HW, Talbot GH, Bradley JS, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America, Clinical infectious diseases 48(1)(2009) 1–12. [DOI] [PubMed] [Google Scholar]
- [9].Rice LB, Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE (2008). [DOI] [PubMed] [Google Scholar]
- [10].Ciampolini J, Harding KG, Pathophysiology of chronic bacterial osteomyelitis: why do antibiotics fail so often? Postgrad Med J 76 (2000) 479–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tuchscherr L, Geraci J, Löffler B, Staphylococcus aureus Regulator Sigma B is important to develop chronic infections in hematogenous murine osteomyelitis model, Pathogens 6(3) (2017) e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tuchscherr L, Kreis CA, Hoerr V, Flint L, Hachmeister M, Geraci J, Bremer-Streck S, Kiehntopf M, Medina E, Kribus M, Raschke M, Pletz M, Peters G, Löffler B B, Staphylococcus aureus develops increased resistance to antibiotics by forming dynamic small colony variants during chronic osteomyelitis, J Antimicrob Chemother 71(2) (2016) 438–448. [DOI] [PubMed] [Google Scholar]
- [13].Brady RA, Leid JG, Calhoun JH, Costerton JW, Shirtliff ME, Osteomyelitis and the role of biofilms in chronic infection, FEMS Immunol Med Microbiol 52 (2008) 13–22. [DOI] [PubMed] [Google Scholar]
- [14].Cheow WS, Chang MW, Hadinoto K, The roles of lipid in anti-biofilm efficacy of lipid-polymer hybrid nanoparticles encapsulating antibiotics. Colloids and Surfaces A 389 (2011) 158–165. [Google Scholar]
- [15].Cheow W, Chang M, Hadinoto K, Antibacterial efficacy of inhalable levofloxacin-loaded polymeric nanoparticles against E. coli biofilm cells: the effect of antibiotic release profile, Pharm. Res 27 (2010) 1597–1609. [DOI] [PubMed] [Google Scholar]
- [16].Halwani M, Hebert S, Suntres ZE, Lafrenie RM, Azghani AO, Omri A, Bismuththiol incorporation enhances biological activities of liposomal tobramycin against bacterial biofilm and quorum sensing molecules production by Pseudomonas aeruginosa, Int. J. Pharm 373 (2009) 141–146. [DOI] [PubMed] [Google Scholar]
- [17].Guo P, Xue HY, Wong HL, Therapeutic nanotechnology for bone infection treatment - State of the art, Curr Drug Deliv 15(7) (2018) 941–952. [DOI] [PubMed] [Google Scholar]
- [18].Wentao Z, Lei G, Liu Y, Wang W, Song T, Fan J, Approach to osteomyelitis treatment with antibiotic loaded PMMA, Microb Pathog 102 (2017) 42–44. [DOI] [PubMed] [Google Scholar]
- [19].Tsiolis P, Giamarellos-Bourboulis EJ, Mavrogenis AF, Savvidou O, Lallos SN, Frangia K, Lazarettos I, Nikolaou V, Efstathopoulos NE, Experimental osteomyelitis caused by methicillin-resistant Staphylococcus aureus treated with a polylactide carrier releasing linezolid, Surg Infect (Larchmt) 12(2) (2011) 131–135. [DOI] [PubMed] [Google Scholar]
- [20].Ahmed K, Gribbon P, Jonest MN, The application of confocal microscopy to the study of liposome adsorption onto bacterial biofilms, J. Liposome Res 12 (2002) 285–300. [DOI] [PubMed] [Google Scholar]
- [21].Robinson AM, Creeth JE, Jones MN, The use of immunoliposomes for specific delivery of antimicrobial agents to oral bacteria immobilized on polystyrene, J. Biomat. Sci. Polym. Ed 11 (2000) 1381–1393. [DOI] [PubMed] [Google Scholar]
- [22].Alhajlan M, Alhariri M, Omri A, Efficacy and safety of liposomal clarithromycin and its effect on Pseudomonas aeruginosa virulence factors, Antimicrob. Agents Chemother 57(2013)2694–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cortés-Penfield NW, Kulkarni PA, The history of antibiotic treatment of osteomyelitis, Open Forum Infect Dis 6(5) (2019) ofzl81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Antony EJ, Shibu A, Ramasamy S, Paulraj MS, Enoch IV, Loading of atorvastatin and linezolid in β-cyclodextrin-conjugated cadmium selenide/silica nanoparticles: A spectroscopic study, Mater Sci Eng C Mater Biol Appl 65 (2016) 194–198. [DOI] [PubMed] [Google Scholar]
- [25].Parisi OI, Fiorillo M, Caruso A, Cappello AR, Saturnino C, Puoci F, Panno A, Dolce V, El-Kashef H, Sinicropi MS, Enhanced cellular uptake by “pharmaceutically oriented devices” of new simplified analogs of Linezolid with antimicrobial activity, Int J Pharm 461(1–2) (2014) 163–170. [DOI] [PubMed] [Google Scholar]
- [26].Liu J, Cheng H, Han L, Qiang Z, Zhang X, Gao W, Zhao K, Song Y, Synergistic combination therapy of lung cancer using paclitaxel- and triptolide-coloaded lipid-polymer hybrid nanoparticles, Drug Des Devel Ther 12 (2018) 3199–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Panalytical Malvern. Common Terms Used In Dynamic Light Scattering – AzoM. https://www.azom.com/article.aspx?ArticleID=9924 (retrieved on April 11, 2020) [Google Scholar]
- [28].Liu M, Fan F, Shi P, Tu M, Yu C, Yu C, Du M, Lactoferrin promotes MC3T3-E1 osteoblast cells proliferation via MAPK signaling pathways, Int J Biol Macromol 107 (2018)137–143. [DOI] [PubMed] [Google Scholar]
- [29].Kourbatova EV, Halvosa JS, King MD, Ray SM, White N, Blumberg HM, Emergence of community-associated methicillin-resistant Staphylococcus aureus USA 300 clone as a cause of health care-associated infections among patients with prosthetic joint infections, American journal of infection control 33(7) (2005) 385–391. [DOI] [PubMed] [Google Scholar]
- [30].Lambert RJ, Pearson J. Susceptibility testing: accurate and reproducible minimum inhibitory concentration (MIC) and non-inhibitory concentration (NIC) values. J Appl Microbiol.88 (2000) 784–790. [DOI] [PubMed] [Google Scholar]
- [31].O’Toole GA, Microtiter dish biofilm formation assay, Journal of visualized experiments 47 (2011)2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Paul DR. Elaborations on the Higuchi model for drug delivery. Int J Pharm. 418 (2011) 13–17. [DOI] [PubMed] [Google Scholar]
- [33].Rollin G, Tan X, Tros F, Dupuis M, Nassif X, Charbit A, Coureuil M, Intracellular Survival of Staphylococcus aureus in endothelial cells: a matter of growth or persistence, Front Microbiol 8 (2017) 1354, < 10.3389/fmicb.2017.01354> e2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Krauss JL, Roper PM, Ballard A, Shih CC, Fitzpatrick JAJ, Cassat JE, Ng PY, Pavlos NJ, Veis DJ. Staphylococcus aureus infects osteoclasts and replicates intracellularly. mBio. 10 (2019) e02447–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hashemian SMR, Farhadi T, Ganjparvar M. Linezolid: a review of its properties, function, and use in critical care. Drug Des Devel Ther. 12 (2018) 1759–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mukheijee A, Waters AK, Kalyan P, Achrol AS, Kesari S, Yenugonda VM. Lipid-polymer hybrid nanoparticles as a next-generation drug delivery platform: state of the art, emerging technologies, and perspectives. Int J Nanomedicine. 14 (2019) 1937–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Budhian A, Siegel SJ, Winey KI. Haloperidol-loaded PLGA nanoparticles: systematic study of particle size and drug content. Int J Pharm. 336 (2007) 367–375. [DOI] [PubMed] [Google Scholar]
- [38].Zhang L, Chan JM, Gu FX, Rhee JW, Wang AZ, Radovic-Moreno AF, Alexis F, Langer R, Farokhzad OC. Self-assembled lipid—polymer hybrid nanoparticles: a robust drug delivery platform. ACS Nano. 2 (2008) 1696–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chan JM, Zhang L, Yuet KP, Liao G, Rhee JW, Langer R R, Farokhzad OC . PLGA-lecithin-PEG core-shell nanoparticles for controlled drug delivery. Biomaterials. 30 (2009) 1627–1634. [DOI] [PubMed] [Google Scholar]
- [40].Zheng Y, Yu W, Weecharangsan W, Piao L, Darby M, Mao Y, Koynova R, Yang X, Li H, Xu S, Lee LJ, Sugimoto Y, Brueggemeier RW, Lee RJ. Transferrin-conjugated lipid-coated PLGA nanoparticles for targeted delivery of aromatase inhibitor 7alpha-APTADD to breast cancer cells. Int J Pharm. 390 (2010) 234–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Parisi OI, Fiorillo M, Caruso A, Cappello AR, Saturnino C, Puoci F, Panno A, Dolce V, El-Kashef H, Sinicropi MS, Linezolid penetration into osteo-articular tissues, Int J Pharm 461(1–2) (2014) 163–170. [DOI] [PubMed] [Google Scholar]
- [42].Wu D, Si M, Xue HY, Wong HL, Nanomedicine applications in the treatment of breast cancer: current state of the art, Int J Nanomedicine 12 (2017) 5879–5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zyvox (linezolid) label information, Pfizer (2015), <http://labeling.pfizer.com/ShowLabeling.aspx?id=649#section-11.3>. [Google Scholar]
- [44].Rana B, Butcher I, Grigoris P, Murnaghan C, Seaton RA, Tobin CM, Linezolid penetration into osteo-articular tissues, J Antimicrob Chemother 50(5) (2002) 747–745. [DOI] [PubMed] [Google Scholar]
- [45].Smith K, Perez A, Ramage G, Gemmell CG, Lang S, Comparison of biofilm-associated cell survival following in vitro exposure of meticillin-resistant Staphylococcus aureus biofilms to the antibiotics clindamycin, daptomycin, linezolid, tigecycline and vancomycin, Int J Antimicrob Agents 33(4) (2009) 374–828. [DOI] [PubMed] [Google Scholar]
- [46].Bahamondez-Canas TF, Zhang H, Tewes F, Leal J, Smyth HDC, PEGylation of tobramycin improves mucus penetration and antimicrobial activity against Pseudomonas aeruginosa biofilms in vitro, Mol Pharm 15(4) (2018) 1643–1652. [DOI] [PubMed] [Google Scholar]


