Abstract
Background
Neonatal diet has a large influence on child health and might modulate changes in fecal microbiota and metabolites.
Objectives
The aim is to investigate fecal microbiota and metabolites at different ages in infants who were breastfed (BF), received dairy-based milk formula (MF), or received soy-based formula (SF).
Methods
Fecal samples were collected at 3 (n = 16, 12, and 14, respectively), 6 (n = 20, 19, and 15, respectively), 9 (n = 12, 11, and 12, respectively), and 12 mo (n = 14, 14, and 15, respectively) for BF, MF, and SF infants. Infants that breastfed until 9 mo and switched to formula were considered as no longer breastfeeding at 12 mo. Microbiota data were obtained using 16S ribosomal RNA sequencing. Untargeted metabolomics was conducted using a Q-Exactive Hybrid Quadrupole-Orbitrap mass spectrometer. The data were analyzed using R (version 3.6.0) within the RStudio (version 1.1.463) platform.
Results
At 3, 6, and 9 mo of age BF infants had the lowest α-diversity, SF infants had the highest diversity, and MF was intermediate. Bifidobacterium was 2.6- to 5-fold lower in SF relative to BF infants through 1 y of life. An unidentified genus from Ruminococcaceae higher in the SF (2%) than in the MF (0.4%) and BF (0.08%) infants at 3 mo of age was observed. In BF infants higher levels of butyric acid, d-sphingosine, kynurenic acid, indole-3-lactic acid, indole-3-acetic acid, and betaine were observed than in MF and SF infants. At 3 mo Ruminococcaceae was positively correlated to azelaic, gentisic, isocitric, sebacic, and syringic acids. At 6 mo Oscillospira was negatively correlated with 3-hydroxybutyric-acid, hydroxy-hydrocinnamic acid, and betaine whereas Bifidobacterium was negatively associated with 5-hydroxytryptamine. At 12 mo of age, Lachnospiraceae was negatively associated with hydroxyphenyllactic acid.
Conclusions
Infant diet has a large impact on the fecal microbiome and metabolome in the first year of life.
This study was registered at clinicaltrials.gov as NCT00616395.
Keywords: breastfeeding, formula diets, microbiota, metabolites, immune system
Introduction
Exclusive breastfeeding is recommended by the WHO for the first 6 mo of life. There is no other period in life where nutrition comes from a sole source (breast milk or infant formula), and the health implications of early-life nutrition have been well described (1–4). For example, breastfeeding is associated with lower risks of infection during infancy, as well as a long-term decreased risk of atopy and obesity (1–4). Although the macro- and micronutrient composition of infant formula meets the requirements of a rapidly growing infant, there remain many bioactive components within human milk that have not been incorporated into the formula. Several of these components, such as proteins and oligosaccharides, remain partially undigested as they enter into the large intestine and thus influence the developing lower-gut microbiome (5, 6).
There is a growing body of literature describing the ability of the gut microbiome to modulate health (7). The infant's gut microbiome is predominately seeded after birth (6). Breastfed (BF) infants have lower α-diversity and altered β-diversity measures compared with their formula-fed (FF) counterparts during the first year of life (8–10). Thus, the nutrition provided at this early age can significantly affect bacterial colonization and development. Whereas increased bacterial diversity is considered to be beneficial for adults (11), lower diversity displayed by BF infants is thought to be beneficial to the developing gut and immune system (12). In addition, breastfeeding is associated with increased abundances of specific bacteria, primarily Bifidobacteria and Bacteroidetes (6). Gut microbiota have been found to produce metabolites; specifically, SCFAs benefit to the colonic environment by improving barrier function, increase Treg cells to maintain homeostasis, and also protect from colonizing pathogenic bacteria (13, 14). These findings highlight that understanding the gut metabolite milieu is just as important as cataloging microbiome shifts, in terms of understanding microbe–host communication and physiology. It is increasingly appreciated that neonatal feeding and gut microbiota can influence the metabolites within urine, feces, and the sera (15, 16). Thus, an infant's microbiota and metabolite profile may be another link in the effect of early nutrition on healthy growth and development.
In this study, how early-life diet alters the lower-gut microbiome and host metabolism was examined. Toward this aim, we examined fecal samples from a clinical cohort that examined the effect of early infant diet on growth and body composition. Aside from examining BF compared with FF infants, this study contains 2 types of FF infants: those fed with a cow milk–based (MF) or a soy-based (SF) formula. To the best of our knowledge, an investigation into the fecal microbiome and metabolome of SF infants has not been carried out previously. The infant fecal microbiome and metabolome at 3, 6, 9, and 12 mo of age were determined.
Methods
Samples originated from a cross-sectional subset of a Central Arkansas cohort study, referred to as the Beginnings Study (NCT00616395). The study was approved by the institutional review board at the University of Arkansas for Medical Sciences and the study design has been reported previously (17). Inclusion criteria were that infants were born from healthy pregnancies and with no medical diagnosis (e.g., diabetes or pre-eclampsia) or had no medications that can affect fetal or infant growth (e.g., selective serotonin reuptake inhibitors or thyroid replacement). All women were nonsmokers, with no alcohol consumption and no usage of soy products or estrogenic compounds during pregnancy and lactation (if breastfeeding). An additional exclusion criterion was change of formula between 2 and 12 mo of age. Participants 1–2 mo of age were recruited between 2002 and 2010. Before enrollment, parents chose the diet to feed their infant with advice from their pediatricians. Infants were either BF or provided a milk (MF) (Similac Advance or Enfamil Lipil) or soy (SF) (Similac SoyIsomil or Enfamil Prosobee) infant formula for the first 12 mo of life. Breastfeeding was encouraged until 12 mo of age. When this was not possible, infants were switched to MF and considered as no longer breastfeeding (NLB). In this study, a majority of the infants were switched to MF after 9 mo, thus only 12-mo NLB samples were included in the analysis. The study visits presented in these analyses were conducted at ages 3 (n = 16, 12, and 14, respectively), 6 (n = 20, 19, and 15, respectively), 9 (n = 12, 11, and 12, respectively), and 12 mo (n = 14, 14, and 15, respectively) for BF, MF, and SF infants. NLB at 12 mo had 10 samples (Figure 1). Complementary food intake could start from 4 mo. Anthropometric measures were obtained at each study visit using standard methods that had been previously published (18). Briefly, weight was measured with the infant wearing only a diaper to the nearest 0.01 kg using a tared scale (model 727; SECA Corp.). Length was measured to the nearest 0.1 cm on a length board (Easy Glide Bearing Infantometer; Perspective Enterprises) (18). Sterile tubes were provided to families and fecal samples were collected either during the study visit or at home. If fecal samples were collected at home, the sample was frozen (−20°C) and delivered back to the research facility on ice. Samples were kept at −80°C until further analyses. Self-reported maternal prepregnancy weight and measured height at the 3-mo visit were used to compute maternal BMI (in kg/m2). Infant race, gestational age, birth weight, and length at birth were also self-reported at the 3-mo visit.
FIGURE 1.
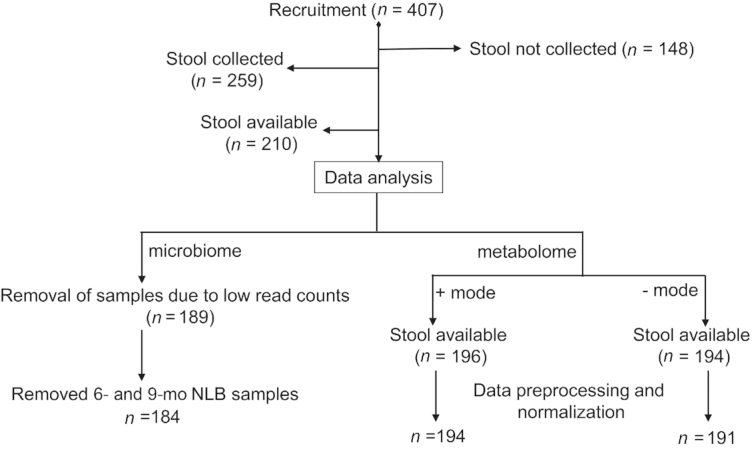
Flow diagram of the study participants showing enrollment, the number of stool samples collected, and the numbers of available samples for microbiota and metabolome data acquisition. The samples with low read counts and also the 6- and 9-mo NLB group for the microbiome data were excluded from analysis. After preprocessing and normalization of the metabolomics data, 2 samples in positive mode and 3 samples in negative mode were dropped. NLB, no longer breastfeeding.
16S ribosomal RNA amplicon sequencing
Fecal samples were homogenized and the PowerSoil® DNA Isolation Kit (MOBIO Laboratories, Inc.) was used for DNA extraction. The variable region 4 of bacterial 16S ribosomal RNA (rRNA) was amplified using 515F/806R forward and reverse primers (18). A ∼30% PhiX DNA was added to pooled amplicons and paired-end sequences (2 × 250 bp) using Illumina MiSeq. Sequence reads were analyzed using QIIME 1.9.1 as described previously (19). Standard QIIME-based scripts were used for quality assessment of sequence reads, operational taxonomic unit (OTU) picking, and sequence alignment. A similarity threshold of 97% was used to cluster amplicon sequencing followed by taxonomic annotation of OTUs using the Greengenes 16S rRNA database at a confidence threshold of 80%.
Untargeted metabolomics analyses
Fecal samples were subjected to untargeted metabolomics analyses. Optima grade reagents and all equipment and instrumentation were obtained from Thermo Fisher Scientific unless otherwise noted. Fecal samples (100 mg) were dried overnight under a low-flow nitrogen stream to remove water (20), resuspended in 500 µL 50% aqueous methanol plus 100 μL recovery standard D4-glycocholic acid solution (2.1 mM), and homogenized using a Precellys 24 homogenizer (Bertin Corp) at 5300 rpm for two 30-s cycles. Homogenates were then extracted in 1 mL ice-cold acetonitrile. Experimental pools, used for quality control (QC) samples, were prepared by pooling equal volumes of each sample extract (10 µL). Samples and QC extracts were evaporated to dryness under a nitrogen stream and reconstituted in 300 µL 5% aqueous methanol containing an internal standard [Lorazepam (10 nM), (Sigma Aldrich)]. Chromatography was performed on a Dionex Ultimate 3000 UHPLC, using an XSelect CSH C18 reversed phase column (2.1 × 100 mm, 2.5 µm) kept at 49°C as previously described (21). Solvent A was 0.1% formic acid in water and solvent B was 0.1% formic acid in acetonitrile. Metabolites were eluted using the following gradient at a flow of 0.4 mL/min: 0–2 min, 0%–1% B; 2–6.5 min, 1%–20% B; 6.5–11.5 min, 20%–95% B; 11.5–13.5 min, 95%–99% B; 13.5–16.5 min, 99%–1% B; 16.5–20 min, 1% B; 20–21 min, 1%–0% B; 21–22 min, 0% B. Injection volumes were set to 5 μL. Samples from all the groups were assayed in mixed and random order.
Detection was carried out on a Q-Exactive Hybrid Quadrupole-Orbitrap mass spectrometer with data acquisition executed using Xcalibur 4.0 software as previously described (21). All samples were analyzed by positive and negative electrospray ionization (ESI +/−) Full-MS scan mode. Nitrogen as sheath, auxiliary, and sweep gas was set at 50, 13, and 3 units, respectively. Other conditions were as follows: resolution, 70,000 full width half maximum (FWHM); automatic gain control (AGC) target, 3 × 106 ions; maximum injection time, 200 ms; scan range, 50–750 m/z; capillary temperature, 320°C; source temperature, 425°C; and spray voltages, 2.7 and 3.6 kV for negative and positive modes, respectively. ESI +/− data-dependent MS2 spectra were generated for QC pool samples using the following conditions: resolution, 17,500 FWHM; AGC target, 1 × 105 ions; maximum injection time, 50 ms; loop count, 5; isolation window, 4.0 m/z; and normalized collision energy, 30.
The acquired data set, composed of full MS and data-dependent MS2 raw files, was processed by Compound Discoverer 3.0 using an untargeted metabolomics workflow including retention time alignment, unknown compound detection, compound-grouping across all samples, gap filling, and metabolite identification using online and “in-house” data-dependent MS2-fragmentation spectral databases (ddMS2). The software parameters for alignment were as follows: adaptive curve model, 5 ppm mass tolerance, and 0.2-min maximum shift for alignment. The software parameters for detecting unknown compounds were as follows: mass tolerance for detection 10 ppm, intensity tolerance 30%, S/N threshold of 3, and minimum peak height 1 × 106, and the parameters for compound groups were mass tolerance 10 ppm and retention time tolerance 0.15 min. Gap filling was performed across all samples using the Real Peak detection method, a mass tolerance of 10 ppm, and S/N threshold of 1.5. Metabolites were identified by using MassList (accurate mass ± 5 ppm, retention time ± 15 s to a known standard), mzCloud (online ddMS2 database), and mzVault (in-house ddMS2 database). Supplemental Table 1 lists the metabolite standards used to generate the MassList and mzVault databases. Metabolite identification confidence levels are as follows: level 1: accurate mass, retention time, and MS2 spectra matching to mzVault or mzCloud; level 2: accurate mass, retention time matching to known standard, no ddMS2 information.
Statistical analysis
All data preprocessing and analyses were conducted in R (version 3.6.0; R Development Core Team) within the RStudio (version 1.1.463) platform.
Microbiome
Total sampling depth was 3,192,176 sequencing reads. Mean sample depth per sample was 19,216 sequencing reads and sequences were not rarefied before analysis (22). Data were initially screened to remove samples with sequence reads < 1000 counts (n = 21). Low abundant OTUs were then removed if <20% of samples did not have ≥10 sequencing reads. OTUs in duplicated samples (n = 16) were analyzed for CV values and were removed if mean CV > 40%. For duplicated samples with mean CV < 40%, OTUs were averaged and rounded to the nearest whole number for subsequent data analysis. A total of 184 samples remained after preprocessing of the 16S rRNA sequencing data, with an average sampling depth of 11,414 reads. All data analysis was performed at the genus level. The phyloseq package was used to merge, filter, and calculate α-diversity indexes and Bray–Curtis distance matrices (23). α-Diversity (within-sample diversity) was estimated for taxa richness (total number of taxa, Chao1 index), evenness (Shannon Index), and dominance (Simpson Index). These α-diversity estimates were analyzed with either a 2-factor or 1-factor ANOVA, depending on whether the main effect of age was included (i.e., 2-factor if all samples were analyzed together and 1-factor if assessing diet within age). β-Diversity (i.e., between-sample differences) was estimated using Bray–Curtis dissimilarities. Permutational multivariate ANOVA (PERMANOVA) was used to determine dietary and age differences of β-diversity in all samples, followed by pairwise assessment of diet within each age using PERMANOVA. These analyses were all adjusted for child sex and birth weight (kg) and conducted using the R vegan package (24). Only 22 of 98 infants had fecal samples at >2 ages; therefore, repeated measures were not conducted in these analyses. Post hoc analysis of α-diversity indexes was assessed using contrasts on estimated marginal means using the emmeans package (25). β-Diversity was visualized using principal coordinate analysis. Dietary differences in genera within age groups were assessed by identifying intersecting results by 2 statistical approaches: 1) Kruskal–Wallis and Dunn's tests were assessed on percentage relative abundance data (full results in Supplemental Table 2), and 2) Wald's test for pairwise comparisons (full results in Supplemental Table 3) on sequence counts as implemented in the DESeq2 package (22, 26). P values for all tests were adjusted for multiple comparisons using Benjamini and Hochberg's false discovery rate (FDR) correction unless noted. Statistical significance was determined at Padj ≤ 0.05 (27).
Metabolomics
Data from the negative and positive modes were processed separately. Raw peak areas were first adjusted by dry fecal weights of samples. CVs of QC samples were then assessed and metabolites with CVs > 40% were removed. The overall and group variance of raw data were calculated and compared against variance estimates using several normalization procedures (e.g., vector sum, single internal standard, compensating multiple internal standards). Compensating multiple internal standards had lower overall and within-group variance than did the raw data and still maintained the metabolite distribution within samples; therefore, this normalization procedure was applied to the data using the crmn package (28). Sample outliers were visually assessed using principal component analysis (PCA) and removed if the sample PCA score was >3 SD along components 1 and/or 2 (n = 7). Univariate metabolite outliers were determined using an iterative assessment of Grubb's test for outliers at α < 0.01 (n = 23 and 29 in negative and positive modes, respectively). All missing samples were imputed using the K-Nearest Neighbor algorithm (29). Totals of 96 and 117 metabolites were used for statistical testing in the negative and positive modes, respectively. The effect of diet within each age group was then analyzed with the Kruskal–Wallis test followed by post hoc analysis by Dunn's Multiple Comparison test (full results in Supplemental Tables 4, 5). P values derived from Kruskal–Wallis and Dunn's Multiple Comparison tests were adjusted for FDR correction.
Functional gene prediction of gut microbiota of infants fed with either breast-milk or formula diets
The functional gene prediction of the gut microbiota of the BF, MF, and SF infants was carried out using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) (30). A closed-reference OTU was generated in QIIME and taxonomy assignment was made with the Greengenes database with a 97% similarity. To obtain the functional prediction of the metagenome, the OTU data were normalized with the PICRUSt at level 3 pathways. Statistical analysis was conducted using the STAMP version 2.1.3 (Statistical Analysis of Metagenomic Profiles) software package (31) with the nonparametric Welch test to investigate and illustrate alterations in microbial functions between groups.
Correlation analysis of 16S rRNA sequencing and metabolomics data
16S rRNA data were adjusted for correlation-based analyses (e.g., PCA and Pearson's correlations) using a centered log ratio (CLR) transformation (32). Zeros were imputed using the geometric Bayesian multiplicative methods using the zComposition package (33). Metabolomics and adjusted 16S rRNA data were separately assessed by PCA for dimension reduction and visualization. Components from each PCA were correlated and plotted. Correlations between metabolites altered by diet and CLR-transformed genera were assessed using Pearson's correlations, and correlations with FDR-adjusted P values < 0.05 were visualized using heat maps.
Results
The cohort was 19.1% African American, 88.0% Caucasian, and 7.1% reporting >1 race. Girls made up 50.2% of the infants. Table 1 shows gestational age, birth weight, and length, as well as maternal BMI.
TABLE 1.
Clinical cohort characteristics at 3, 6, 9, and 12 mo of age from either BF, MF, NLB, or SF infants1
| 3 Mo | 6 Mo | 9 Mo | 12 Mo | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameters | BF | MF | SF | BF | MF | SF | BF | MF | SF | BF | MF | NLB | SF |
| Infants, n | 16 | 12 | 14 | 20 | 19 | 15 | 12 | 11 | 12 | 14 | 14 | 10 | 15 |
| Self-reported ethnicity, % | |||||||||||||
| White | 100 | 83.3 | 64.3 | 100 | 84.2 | 80 | 100 | 100 | 75 | 92.9 | 78.6 | 100 | 86.7 |
| African-American | 16.7 | 21.4 | 15.8 | 20 | 25 | 21.4 | 13.3 | ||||||
| >1 race | 7.1 | 7.1 | |||||||||||
| Not reported | 7.1 | ||||||||||||
| Sex, % female | 62.5 | 16.7 | 50 | 70 | 57.9 | 46.7 | 66.7 | 45.5 | 33.3 | 64.3 | 42.9 | 70 | 26.7 |
| Maternal BMI, kg/m2 | 24.9 ± 4.5 | 26.7 ± 4.3 | 27.0 ± 4.2 | 25.0 ± 4.7 | 26.5 ± 5.8 | 29.2 ± 8.2 | 24.1 ± 5.0 | 28.9 ± 6.2 | 23.3 ± 4.9 | 24.6 ± 3.2 | 27.9 ± 6.0 | 24.2 ± 2.7 | 26.6 ± 6.7 |
| Birth length, cm | 51.0 ± 2.0 | 50.6 ± 2.5 | 51.4 ± 2.1 | 50.7 ± 1.5 | 50.9 ± 2.3 | 50.6 ± 1.8 | 50.2 ± 2.5 | 51.3 ± 1.9 | 51.0 ± 2.8 | 51.2 ± 2.8 | 51.0 ± 1.9 | 50.8 ± 1.2 | 51.0 ± 2.6 |
| Birth weight, kg | 3.5 ± 0.3 | 3.3 ± 0.3 | 3.4 ± 0.4 | 3.4 ± 0.4 | 3.4 ± 0.3 | 3.4 ± 0.4 | 3.4 ± 0.3 | 3.4 ± 0.3 | 3.4 ± 0.4 | 3.5 ± 0.3 | 3.4 ± 0.3 | 3.4 ± 0.4 | 3.4 ± 0.4 |
| Gestational age, wk | 39.6 ± 0.6 | 39.2 ± 0.8 | 39.2 ± 0.8 | 39.5 ± 0.6 | 39.2 ± 1.0 | 39.0 ± 0.8 | 39.5 ± 0.6 | 39.6 ± 0.5 | 39.3 ± 0.8 | 39.5 ± 0.5 | 39.2 ± 0.9 | 39.6 ± 1.0 | 38.9 ± 0.8 |
n = 184. Values are percentages or means ± SDs unless otherwise indicated. BF, breastfed; MF, cow milk–based formula; NLB, no longer breastfeeding; SF, soy-based formula.
BF infants have lower α-diversity through the first 9 mo of life than FF infants
To determine if infant diet, age, sex, or birth weight influence microbiota richness and diversity, a 2-factor ANCOVA was performed on all samples with covariate adjustments for child sex, and birth weight (Supplemental Table 6) at genus level. After covariate adjustment, a significant effect of diet was noted for all α-diversity indexes (Supplemental Table 6). Furthermore, a significant effect of age was seen with Observed (P < 0.01) measures. Child sex was also statistically significant in the Shannon (P < 0.05) and Simpson (P < 0.05) indexes.
Investigation of dietary differences within age showed differences in α-diversity measures between the diet groups at 3, 6, and 9 mo of age (Table 2). At 3 mo of age the BF group showed significantly lower Shannon and Simpson diversity measures than the MF and SF groups (Table 2). At 6 mo of age, the BF group had lower Observed and Chao1 measures than did the 2 formula groups. Interestingly, the Shannon and Simpson indexes were significantly different between BF and SF at 6 mo of age. At 9 mo of age only the Shannon index was significantly lower in the BF than in the SF group. At 3, 6, and 9 mo, the SF infants showed higher diversity than MF infants (Table 2). No differences were observed between diet groups at 12 mo of age (Table 2).
TABLE 2.
Estimated marginal means (95% CIs) of fecal α-diversity indexes at 3, 6, 9, and 12 mo of age from either BF, MF, NLB, or SF infants1
| Age | Index | BF | n | MF | n | NLB | n | SF | n | Diet | Birth weight | Sex |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | Observed | 22.0 (19.9, 24.1) | 16 | 24.7 (22.1, 27.3) | 12 | 23.7 (21.6, 25.9) | 14 | 0.306 | 0.084 | 0.513 | ||
| 3 | Chao1 | 24.1 (22.1, 26.1) | 24.5 (22.0, 27.1) | 25.1 (23.0, 27.2) | 0.738 | 0.208 | 0.054 | |||||
| 3 | Shannon | 1.3 (1.2, 1.5)a | 1.8 (1.6, 2.0)b | 2.0 (1.9, 2.2)c | <0.01 | 0.241 | 0.052 | |||||
| 3 | Simpson | 0.6 (0.6, 0.7)a | 0.8 (0.7, 0.8)b | 0.8 (0.7, 0.9)c | <0.01 | 0.241 | 0.018 | |||||
| 6 | Observed | 21.2 (19.6, 22.9)a | 20 | 23.8 (22.2, 25.5)b | 19 | 24.6 (22.8, 26.4)c | 15 | 0.026 | 0.626 | 0.112 | ||
| 6 | Chao1 | 22.2 (20.4, 24.0)a | 25.1 (23.3, 26.8)b | 25.1 (23.2, 27.1)c | 0.048 | 0.992 | 0.241 | |||||
| 6 | Shannon | 1.5 (1.3, 1.7)a | 1.7 (1.6, 1.9)a | 2.0 (1.8, 2.2)b | <0.01 | 0.958 | 0.571 | |||||
| 6 | Simpson | 0.7 (0.6, 0.7) | 0.7 (0.7, 0.8) | 0.8 (0.7, 0.8) | 0.066 | 0.94 | 0.778 | |||||
| 9 | Observed | 22 (18.9, 25.0) | 12 | 23.2 (19.8, 26.5) | 11 | 25 (21.9, 28.1) | 12 | 0.174 | 0.699 | 0.139 | ||
| 9 | Chao1 | 22.6 (19.8, 25.4) | 23.8 (20.8, 26.9) | 25.4 (22.6, 28.2) | 0.198 | 0.619 | 0.238 | |||||
| 9 | Shannon | 1.5 (1.3, 1.7)a | 1.8 (1.5, 2.0)a | 1.9 (1.7, 2.2)b | 0.032 | 0.707 | 0.534 | |||||
| 9 | Simpson | 0.6 (0.6, 0.7) | 0.7 (0.7, 0.8) | 0.7 (0.7, 0.8) | 0.069 | 0.715 | 0.726 | |||||
| 12 | Observed | 24.9 (23.5, 26.2) | 14 | 25.3 (24.0, 26.7) | 14 | 26.0 (24.4, 27.5) | 10 | 25.9 (24.6, 27.3) | 15 | 0.765 | 0.026 | 0.783 |
| 12 | Chao1 | 25.3 (24.0, 26.7) | 25.8 (24.5, 27.2) | 26.2 (24.6, 27.8) | 26.2 (24.9, 27.5) | 0.894 | 0.025 | 0.759 | ||||
| 12 | Shannon | 1.7 (1.5, 1.9) | 1.7 (1.5, 1.9) | 1.8 (1.6, 2.1) | 1.9 (1.7, 2.1) | 0.470 | 0.144 | 0.085 | ||||
| 12 | Simpson | 0.7 (0.6, 0.8) | 0.7 (0.6, 0.7) | 0.7 (0.6, 0.8) | 0.7 (0.7, 0.8) | 0.615 | 0.373 | 0.059 |
Differences in dietary intake assessed by ANCOVA, blocked for gender and birth weight as covariate. Estimated marginal means in the same row without a common letter differ. P value provided for main effect (diet), covariate (birth weight), and blocking variable (sex). Post hoc pairwise assessment performed by contrast analysis. Sample sizes range from 10 to 20 per diet group at 3, 6, 9, and 12 mo of age. BF, breastfed; MF, cow milk–based formula; NLB, no longer breastfeeding; SF, soy-based formula.
BF infants have distinct microbiota through the first 9 mo of life compared with FF infants
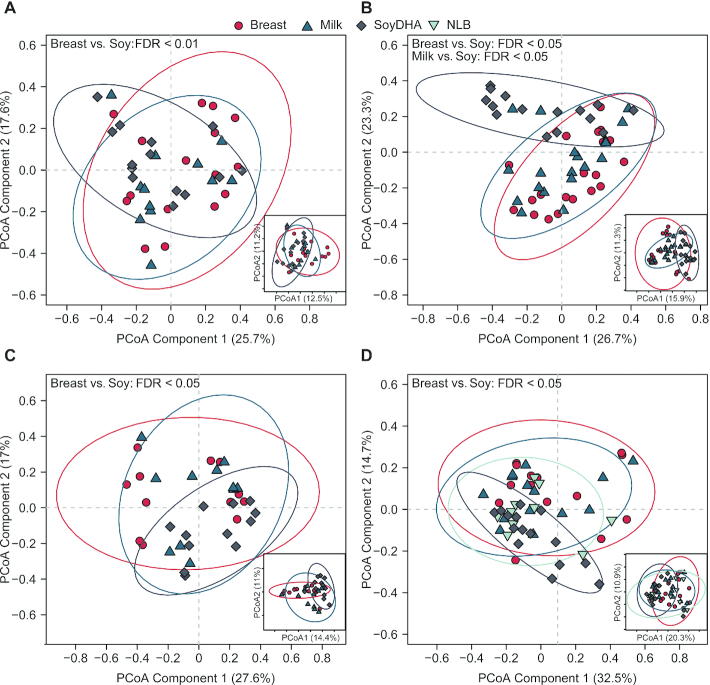
When comparing the effect of diet and age, PERMANOVA results showed diet (P < 0.05) and age (P < 0.05) effects with no interaction, and child sex was a significant confounder (P = 0.05) (Supplemental Table 7). Next, the diet effect on community structure at genus level within time was determined. Figure 2 displays the first 2 components from PCA ordinations of genus data at 3, 6, 9, and 12 mo of age. Based on Bray–Curtis dissimilarities, in comparison with BF infants, SF infants had an altered microbial profile at all ages (Figure 2A–D). The distinguishing of SF infants was most apparent at 6 mo of age, where the microbial profile of SF infants was also altered when compared with MF infants (Figure 2B). There were no statistical differences between BF and MF infants at any ages. In addition, BF and MF were both different from SF at 3, 6, and 9 mo of age, whereas only BF was different from SF at 12 mo of age at OTU level (Figure 2).
FIGURE 2.
Visualization of β-diversity (between-sample diversity) by PCoA in infants consuming differential diets at 3 (A), 6 (B), 9 (C), and 12 (D) mo of age. Data generated from 16S ribosomal RNA amplicon sequencing on fecal samples. β-Diversity estimated by Bray–Curtis dissimilarities at both genus and OTU levels. The first 2 components of PCoA scores (i.e., individual samples) are displayed for genera (large plots) and OTUs (inserts). Confidence regions of group clusters are presented as 95% confidence ellipses based on Hotelling's T2 statistic. Red circles indicate breastfed infants, dark blue triangles indicate cow milk formula-fed infants, grey diamonds indicate soy-formula-fed infants, and light blue upside-down triangles indicate infants that were no longer breastfeeding. Statistical difference in diet within an age group was assessed by pairwise comparison of diets by permutational multivariate ANOVA. P values were adjusted by FDR to control for the family-wise error rate. Sample numbers were n = 10–20 per group and age. FDR, false discovery rate; OTU, operational taxonomic unit; PCoA, principal coordinate analysis.
Microbiota relative abundance is significantly altered in BF in comparison with FF infants
The significant differences in relative abundances of genera due to infant diet were determined with both Kruskal–Wallis with post hoc Dunn's test and Wald test from Deseq2 (Supplemental Tables 2, 3). Bacteroides and Bifidobacterium were the most abundant genera observed in all 3 diet groups at all ages (Supplemental Tables 2, 3). Table 3 presents the statistically significant differences obtained from both analyses. Diet-associated differences were consistent across multiple ages. Bifidobacterium was significantly different by diet at 3, 6, 9, and 12 mo of age. BF infants had a higher relative abundance of Bifidobacterium than did the SF infants. At 3 and 6 mo of age BF infants had lower abundance of unidentified genera from the Clostridiaceae family than did SF infants. An unidentified genus belonging to the Peptostreptococcaceae family was significantly lower in the BF than in the SF infants at 3 and 9 mo of age. Another unidentified genus belonging to the Clostridiales order was of lower abundance in the BF and MF infants than in the SF infants at 6 and 9 mo of age. SMB53 was significantly lower in the BF and MF infants at 3, 6, and 9 mo of age than in the SF infants. In addition, at 3 mo of age, in comparison with BF and MF, an unidentified genus in the Ruminococcaceae family was significantly enriched in the SF infants. At 6 mo of age, relative to the BF or MF infants, an unidentified genus in the Lachnospiraceae family, unidentified genera from the order Clostridiales, Coprococcus, and Roseburia were all higher in the SF infants. At 9 mo of age, an unidentified genus in the Veillonellaceae family was higher in MF infants than in the BF or SF infants. In addition, Clostridium was significantly higher in BF infants than in the MF or SF infants. Finally, at 12 mo of age in the BF, Veillonella was significantly higher whereas an unidentified genus in the Lachnospiraceae family was lower than in SF infants.
TABLE 3.
Median [IQR] percentage relative abundances of sequencing counts representing fecal bacterial genera at 3, 6, 9, and 12 mo of age from either BF, MF, NLB, or SF infants1
| Age | Genus | BF | NLB | MF | SF | FDR |
|---|---|---|---|---|---|---|
| 3 | Bifidobacterium | 13.791 [10–33]a | 24.195 [14–35]a | 7.919 [2–12]b | 0.021 | |
| 3 | f__Clostridiaceae | 0.026 [0–0]a | 0.279 [0–1]b | 0.672 [0–3]b | 0.002 | |
| 3 | f__Peptostreptococcaceae | 0.053 [0–0]a | 0.219 [0–1]a | 2.225 [1–4]b | <0.001 | |
| 3 | f__Ruminococcaceae | 0.087 [0–1]a | 0.448 [0–1]b | 2.081 [1–4]c | <0.001 | |
| 3 | SMB53 | 0.015 [0–0]a | 0.057 [0–0]a | 0.373 [0–1]b | <0.001 | |
| 6 | Bifidobacterium | 23.625 [15–39]a | 27.52 [12–34]a | 3.361 [0–7]b | <0.001 | |
| 6 | Coprococcus | 0.021 [0–0]a | 0.293 [0–1]b | 0.481 [0–2]b | <0.001 | |
| 6 | Enterococcus | 0.044 [0–0]a | 0.04 [0–0]a | 0 [0–0]b | 0.017 | |
| 6 | f__Clostridiaceae | 0.024 [0–0]a | 0.158 [0–1]ab | 0.571 [0–5]b | 0.007 | |
| 6 | f__Lachnospiraceae | 0.115 [0–1]a | 0.901 [0–2]b | 4.17 [2–7]c | <0.001 | |
| 6 | o__Clostridiales | 0 [0–0]a | 0 [0–0]a | 0.226 [0–0]b | <0.001 | |
| 6 | Roseburia | 0 [0–0]a | 0 [0–0]a | 0.088 [0–2]b | 0.013 | |
| 6 | SMB53 | 0.007 [0–0]a | 0.071 [0–0]a | 0.616 [0–1]b | <0.001 | |
| 9 | Akkermansia | 0 [0–0]a | 0.017 [0–0]b | 0.107 [0–2]b | 0.038 | |
| 9 | Bifidobacterium | 17.951 [11–37]a | 11.847 [10–30]a | 6.065 [3–17]b | 0.038 | |
| 9 | Clostridium | 1.096 [0–4]a | 0.047 [0–0]b | 0.032 [0–0]b | 0.026 | |
| 9 | f__Peptostreptococcaceae | 0.064 [0–1]a | 0.282 [0–1]a | 2.447 [1–7]b | 0.005 | |
| 9 | f__Veillonellaceae | 0.027 [0–2]a | 7.207 [1–18]b | 0 [0–0]a | 0.01 | |
| 9 | o__Clostridiales | 0 [0–0]a | 0 [0–0]a | 0.228 [0–0]b | <0.001 | |
| 9 | SMB53 | 0 [0–0]a | 0.076 [0–0]a | 1.227 [1–2]b | <0.001 | |
| 12 | Bifidobacterium | 14.202 [13–19]a | 8.631 [4–12]ab | 15.18 [6–22]a | 4.604 [2–6]b | 0.008 |
| 12 | f__Lachnospiraceae | 1.267 [1–2]a | 1.473 [1–3]ab | 2.894 [2–5]bc | 3.836 [2–6]c | 0.012 |
| 12 | Veillonella | 0.836 [0–4]a | 0.415 [0–1]ab | 0.36 [0–1]ab | 0.25 [0–0]b | 0.047 |
Featured genera were found to be significantly altered by diet by both Kruskal–Wallis and DESeq2 Wald assessments. Zeros indicate abundances of genera lower than the quartiles. See Supplemental Tables 4 and 5 for full assessments of each statistical output. Only the FDRs for the Kruskal–Wallis tests are provided. Groups with different letters differ according to Dunn's test. Sample sizes range from 10 to 20 per diet group at 3, 6, 9, and 12 mo of age. BF, breastfed; FDR, false discovery rate; MF, cow milk–based formula; NLB, no longer breastfeeding; SF, soy-based formula.
Breastfeeding alters the metabolite profile relative to formula feeding in infants through 1 y of life
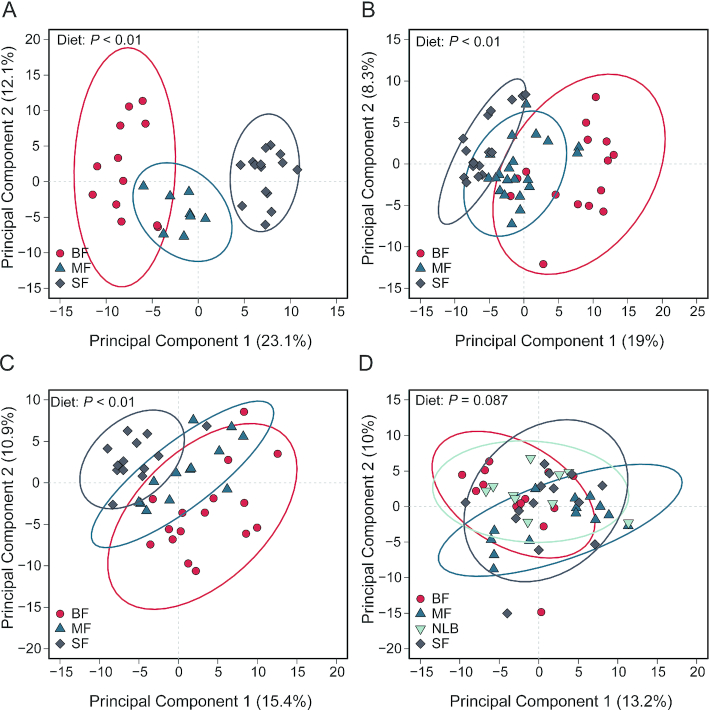
Effects of dietary changes on the fecal metabolome within each time point were investigated using PCA. Figure 3 displays the first 2 components within each age group. There was a significant effect of diet at 3, 6, and 9 mo within both principal component (PC) 1 and PC2 (Supplemental Table 8). The SF group is different than the BF and the MF infants at 3, 6, and 9 mo of age (P < 0.001). In order to determine differences due to diet within each age group, a Kruskal–Wallis test followed by post hoc (Dunn test) analysis with the metabolites detected in negative or positive mode was performed. A large number of metabolites across all ages were found to be significantly affected by infant diet (Supplemental Tables 4, 5). Table 4 provides a select number of these metabolites categorized by functional category determined by descriptions found in the Human Metabolome Database (34). Butyric acid is significantly higher in BF than in MF and SF infants. Tryptophan metabolism also appears to be affected by infant diet with significantly increased kyneurenic acid in the BF compared with the MF and SF infants. Fecal dopamine and sphingosine were increased in the BF relative to the MF and SF infants. Not surprisingly, relative to BF or MF infants higher abundance of plant-based metabolites (jasmonic acid and genistein) were observed in SF infants. For the complete list of metabolites altered by early infant diet, please see Supplemental Tables 4 and 5, which display the negative and positive mode metabolite results, respectively.
FIGURE 3.
Visualization of variance associated with infant diet in fecal metabolomics data from infants consuming differential diets at 3 (A), 6 (B), 9 (C), and 12 (D) mo of age. Fecal metabolomics data were generated from LC-MS. Results from positive and negative modes were combined, log transformed, and scaled to unit variance before assessment by PCA. PCA scores (i.e., individual samples) for PCA components 1 and 2 are displayed. Confidence regions of group clusters are presented as 95% confidence ellipses based on Hotelling's T2 statistic. Red circles indicate BF infants, dark blue triangles indicate MF infants, grey diamonds indicate SF infants, and light blue upside-down triangles indicate NLB infants. Scores from PCA component 1 were assessed for differences in diet group by 1-factor ANOVA. Sample numbers were n = 10–20 per group and age. BF, breastfed; MF, cow milk formula fed; NLB, no longer breastfeeding; PCA, principal component analysis; SF, soy-based formula fed.
TABLE 4.
Fecal metabolites and their associated functional metabolic categories at 3, 6, 9, and 12 mo of age from either BF, MF, or SF infants1
| Metabolite | Functional category | Diet effect | Month |
|---|---|---|---|
| Butyric acid | SCFA | BF > MF; BF > SF | 3, 6, 9; 3, 6 |
| Propionic acid | SCFA | BF < FF | 3, 6 |
| Isovaleric acid | SCFA | BF > FF | 3, 6 |
| Isobutyric acid | SCFA | BF < FF | 3, 6 |
| Dopamine | Neurotransmitter | BF > MF; BF > SF | 3, 6, 9; 3, 6 |
| d-sphingosine | Involved in immune cell trafficking | BF > FF | 3, 6, 9 |
| Kyneuric acid | Tryptophan metabolism | BF > FF | 3, 6, 9 |
| Indole-3-lactic acid | Tryptophan metabolism | BF > SF | 3, 6 |
| Indole-3-acetic acid | Tryptophan metabolism | BF > SF | 3 |
| Betaine | Helps reduce Th17 cells, IL-6 production | BF > FF | 3, 6, 9 |
| Genistein | Soy-derived isoflavone | BF, MF < SF | 3, 6, 9 |
| Jasmonic acid | Plant stress hormone | BF, MF < SF | 3, 6, 9 |
| 3,5-Dihydroxybenzoic acid | Phenyl alanine metabolism in bacteria | BF > FF | 9, 12 |
| 1-Methyluric acid | Microbial metabolite of methylxanthines | BF > FF | 3, 6, 9 |
| Azelaic acid | Plant product | BF, MF < SF | 3, 6, 9, 12 |
See Supplemental Tables 6 (metabolites detected in positive ion mode) and 7 (metabolites detected in negative ion mode) for a complete list of the metabolites affected by diet. Statistical differences were assessed by Kruskal–Wallis test followed by correction for multiple comparisons using Benjamini and Hochberg's false discovery rate adjustment. The functional categories of the metabolites were obtained from the Human Metabolome Database. BF, breastfed; MF, cow milk–based formula; SF, soy-based formula.
The predictive metagenome analysis shows significant differences between BF and FF infants
Several microbial metabolic pathways were predicted to be differentially abundant in BF and FF infants across all the ages (Supplemental Figures 1–7). Relative to BF, porphyrin and chlorophyll metabolism of microbiota was higher in SF across all the ages, suggesting gut bacterial activity due to the soy diet. Glutathione metabolism was higher in BF microbiota than in FF infant microbiota at all ages. PICRUSt analysis provided predictive information of microbial metabolism but did not necessarily integrate the metabolite data, thus correlation analysis was conducted.
Fecal metabolites show significant association with microbiota in infants fed either breast-milk or formula diets
In order to examine global differences in fecal metabolites and their associations with the microbiota, PCA on metabolite data was performed across all age groups and correlated with CLR-transformed microbiome PCA data. A strong correlation was observed between the taxa in PC1 and the metabolites within PC1 (coeff. = −0.582; P < 0.001) (Figure 4). Some of the metabolites within PC1 that separated on the positive end included isovaleric acid, valeric acid, indole-3-acetic acid, indole-3-propionic acid, and dopamine, whereas kynurenic acid and betaine appeared to separate out on the negative end (Supplemental Figure 8). Some of the bacteria that separated within PC1 on the positive side included Bifidobacterium, Clostridium, Bacteroides, Streptococcus, Ruminococcus, and an unspecified genus in the Enterobacteriaceae family, whereas on the negative end SMB53 and unspecified genera in the Lachnospiraceae and Ruminococcaceae families and also genera in the Clostridiales order were observed (Supplemental Figure 9).
FIGURE 4.
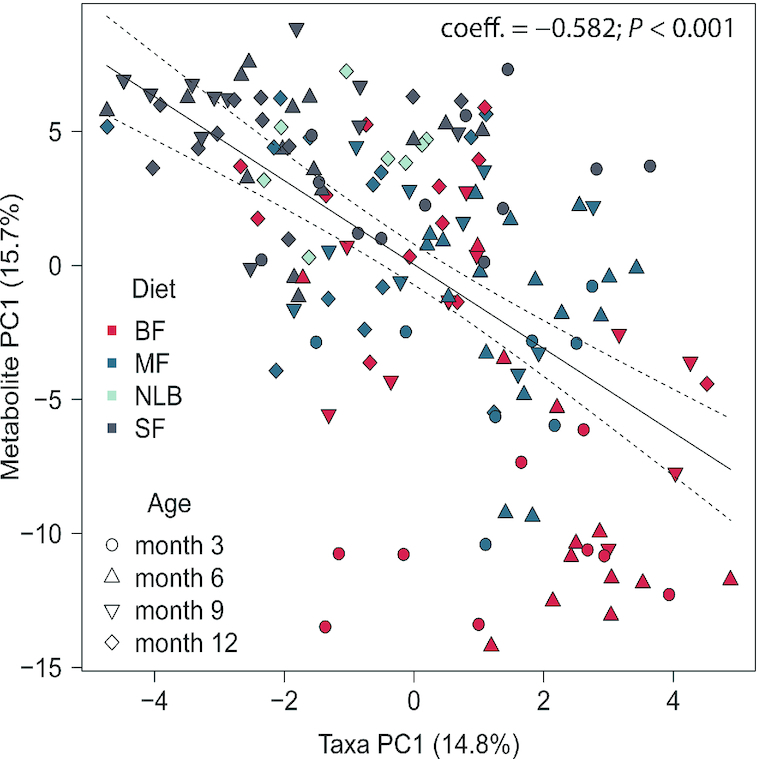
Correlation between fecal microbiota and metabolome in infants consuming differential diets at 3, 6, 9, and 12 mo of age. Genus-level microbial sequencing data and metabolomics data were assessed separately by PCA. Before PCA, sequencing data were transformed by center-log ratio to ensure the data were linearly related, whereas metabolomics data were log transformed and scaled to unit variance. PCA scores (individual samples) were plotted along components 1 and 2 and variation associated with diet was determined along the first components in both PCAs. The percentage explained variance for each component is provided in parentheses. The shapes of the PCA scores represent age (3 mo: circle; 6 mo: triangle; 9 mo: upside down triangle; 12 mo: diamond) and colors represent diet (red: BF; blue: MF; light blue: NLB; grey: SF). The linear relation between the first components for the microbial and metabolomics PCA is shown. The solid line is the best-fit line whereas the dotted lines represent 95% CIs. Pearson's r coefficient and P value are provided in the figure. Sample numbers were n = 10–20 per group and age. BF, breastfed; MF, cow milk formula fed; NLB, no longer breastfeeding; PC, principal component; PCA, principal component analysis; SF, soy-based formula fed.
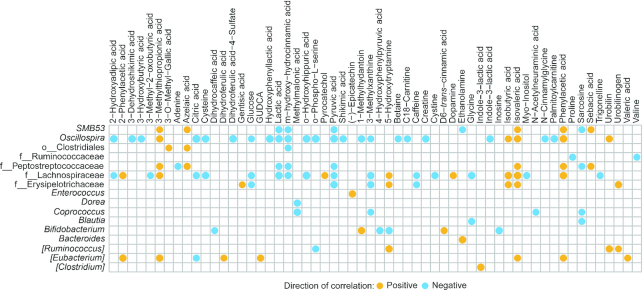
Next we performed an examination within each age group by correlating the metabolites found to be significant from the aforementioned Kruskal–Wallis and Dunn's tests to the CLR-transformed taxa. At 3 mo, genera from the family Clostridaceae were positively correlated to dihydroferulic acid. In addition, genera from the family Ruminococcaceae were positively correlated to azelaic acid, gentisic acid, isocitric acid, sebacic acid, and syringic acid (Figure 5). Figure 6 displays the significant correlations at 6 mo of age at a cutoff of P < 0.01. Supplemental Figure 10 shows the full list of significant correlations. At 6 mo of age, there were multiple correlations between metabolites and bacteria that are also affected by diet. At 6 mo Oscillospira was negatively correlated with 3-hydroxybutyric-acid, hydroxy-hydrocinnamic acid (lower in SF than in MF and BF), and betaine (higher in BF than in MF and SF). Interestingly, at 6 mo of age Oscillospira was enriched owing to the formula diets (Supplemental Table 3). All 3 diet groups were different from each other and the Oscillospira abundance was highest in the SF and lowest in the BF infants (Supplemental Tables 2, 3). Bifidobacterium, which was found to be significantly less abundant in the SF than in the MF and BF infants, but not different between the MF and BF infants (Supplemental Table 2), was negatively associated with 5-hydroxytryptamine and inosine at 6 mo. These metabolites were higher in the SF than in the MF or BF infants (Supplemental Tables 4, 5). An unspecified genus in the Clostridiales order that was enriched in the SF group was positively associated with azelaic acid (higher in SF than in MF and BF at all ages).
FIGURE 5.
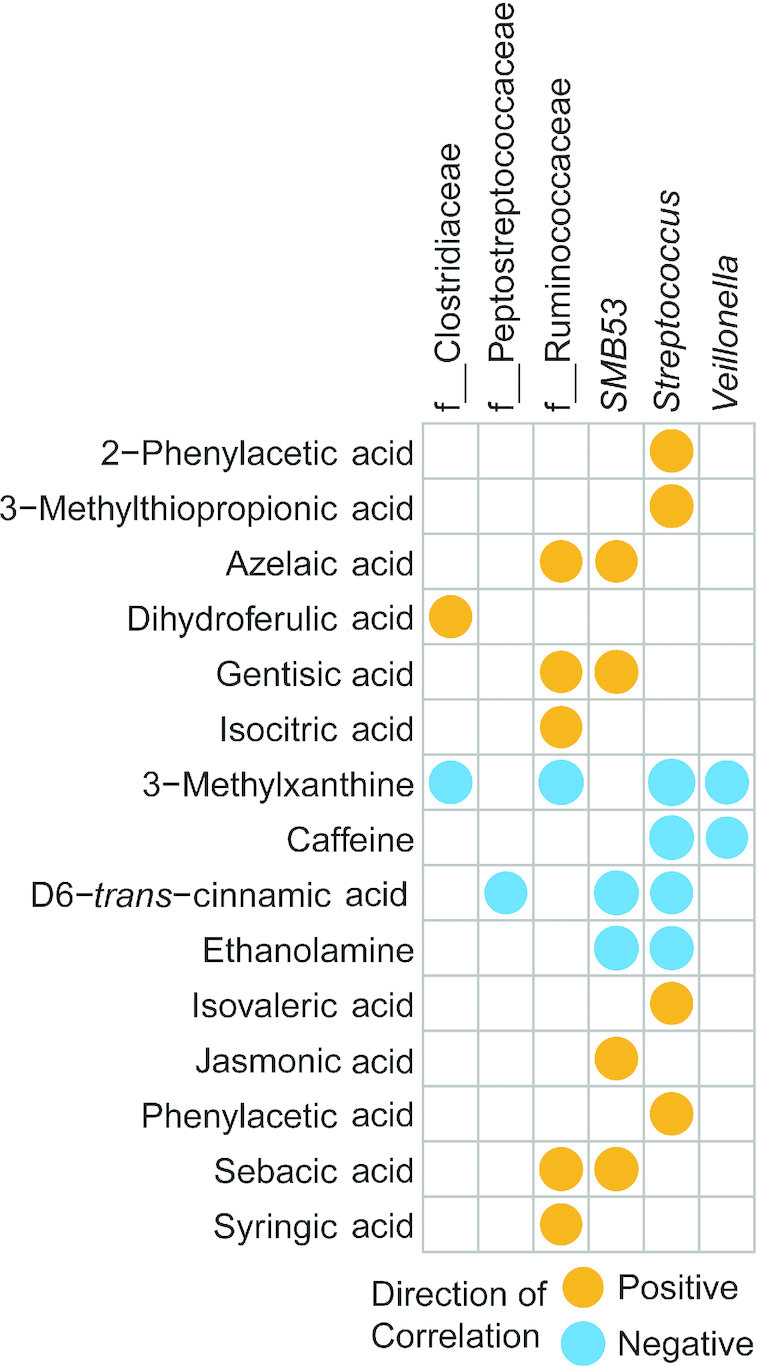
Correlation heatmap of genera and metabolites from infants fed either breast milk, milk-based formula, or soy-based formula at 3 mo of age (n = 161). Circles represent significant Pearson's correlations (FDR-adjusted P values < 0.05). Orange circles indicate positive relations and blue circles indicate negative relations. Supplemental Table 9 gives Pearson's r coefficients, raw P values, and FDR-corrected P values of significant correlations. FDR, false discovery rate.
FIGURE 6.
Correlation heatmap of genera and metabolites from infants fed either breast milk, milk-based formula, or soy-based formula at 6 mo of age (n = 160). Circles represent significant Pearson's correlations (FDR-adjusted P values < 0.05). Orange circles indicate positive relations and blue circles indicate negative relations. Supplemental Table 9 gives Pearson's r coefficients, raw P values, and FDR-corrected P values of significant correlations. FDR, false discovery rate; GUDCA, glycoursodeoxycholic acid.
At 9 mo of age Ruminococcus and genera in the Lachnospiraceae family were negatively associated with hexadecanoic acid (data not shown). Interestingly, only Lachnospiraceae was significantly higher in SF than in BF or MF infants (Table 3). At 12 mo of age, genera in the Lachnospiraceae family were negatively associated with hydroxyphenyllactic acid (Figure 7). See Supplemental Table 9 for all the quantitative data on correlations.
FIGURE 7.
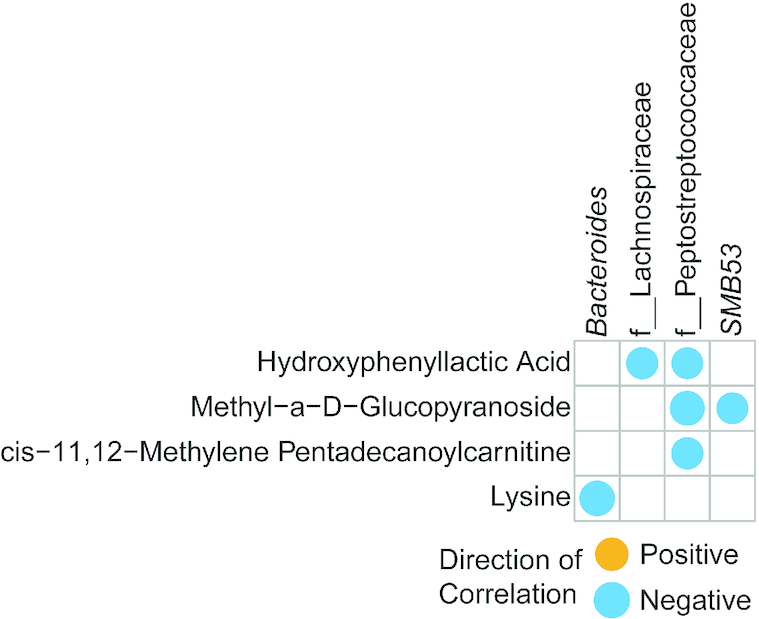
Correlation heatmap of genera and metabolites from infants fed either breast milk, milk-based formula, or soy-based formula at 12 mo of age (n = 72). Circles represent significant Pearson's correlations (FDR-adjusted P values < 0.05). Orange circles indicate positive relations and blue circles indicate negative relations. Supplemental Table 9 gives Pearson's r coefficients, raw P values, and FDR-corrected P values of significant correlations. FDR, false discovery rate.
Discussion
This study investigated the role of neonatal diet on fecal microbiota and metabolite profile during the first year of life. The novel aspect of our study includes 1) fecal metabolite assessment from infant groups fed 3 neonatal diets through the first year of life and 2) association of metabolites to microbiota profiles to determine the host–microbiota crosstalk with different neonatal diets. Significant differences in bacterial diversity and abundance due to infant diets were observed at multiple ages, similar to previous publications (8, 35). Specifically, BF infants showed the lowest α-diversity measurements in comparison with both groups of FF infants. Previously, a study by Baumann-Dudenhoeffer et al. (36) investigated microbiota differences between breast milk, cow milk formula, and soy formula from 0 through 8 mo of age and found similar results in which the SF group had the highest α-diversity. In the same line, the current study showed that the SF group has higher bacterial diversity and a greater number of altered metabolic pathways than have the BF and MF groups, suggesting that the components of neonates’ diet uniquely promote microbiota growth and development. At 3, 6, and 9 mo of age, a large proportion of the total genera consisted of Bacteroides and Bifidobacterium. Interestingly, higher abundance of Bifidobacterium at 5–16 wk of age showed a positive association to bacille Calmette-Guérin, tetanus toxoid, and polio vaccine-specific response at 2 y of age (37). At 12 mo of age the proportion of Bacteroides increased and Bifidobacterium decreased in this study. Studies from different cohorts across the world have reported that some BF infants tend to have higher Bacteroides than Bifidobacterium (8, 38–43). Interestingly, Bacteroides spp. utilize human milk oligosaccharides (44). These shifts in microbiome correspond to what has previously been described in the literature, in which large changes are in part due to the human milk oligosaccharides used by Bacteroides and also the introduction of solid food (44–47).
Our group has published studies on the effect of infant formulas, both soy and dairy based, on the microbiota and immune system within a piglet model (48). The soy group was found to have higher relative abundances of Bilophila, Ruminococcus, and Clostridium. However, how SF affects microbiota development in infants has not been thoroughly investigated. In the current study, an unidentified genus from Ruminococcaceae was enriched in the SF group compared with the MF and BF groups at 3 mo of age. In addition, unidentified genera in the Clostridiales order were higher in the SF group than in the MF or BF groups at 6 and 9 mo of age, supporting previously observed data from the animal model. Thus, both the neonatal piglet model and the data from this study suggest that the components of soy formula may differentially affect gut microbiota and metabolism.
Infant nutrition strongly influences the gut environment during infancy, which in turn modulates the developing immune system. Indeed, human milk oligosaccharides have been shown to improve epithelial barrier function and alter microbiota profile and SCFA production in vitro (49). In our study, the levels of butyric acid present in feces of BF infants were higher than in FF infants. Butyric acid has been shown to increase Treg cell production in the colon and periphery (50–52) and reduced levels of butyrate increase salmonella infection in the gut (14). In addition, d-sphingosine, the precursor of sphingosine 1-phosphate (which regulates immune cell trafficking) (53) and related microbial metabolic pathways such as the glycosphingolipid biosynthesis series, was significantly higher in BF infants than in both FF groups, suggesting an advantage to BF infants in fighting infections during the infancy period. Furthermore, in the current study betaine was significantly higher in BF than in both groups of FF infants. This is an important observation because betaine has been shown to inhibit dendritic cell inflammatory cytokine production (IL-6) and Th17 differentiation (54), suggesting a role for T cell function in BF relative to FF infants. In our study, Bifidobacterium was significantly higher in BF than in SF infants. A high prevalence of Bifidobacterium has been shown to be negatively correlated with the acquisition of antimicrobial-resistant genes (55). The microbiota of infants has been shown to influence sensitization to atopy. Fujimura et al., study grouped infants by risk of atopy (3). Infants grouped into the highest risk of atopy had lower abundances of Bifidobacterium, Akkermansia, and Faecalibacterium. When adult-population of T-cells were cultured with fecal water containing the fecal material from this high-risk group, the cells displayed higher concentrations of IL-4 (implicated in inflammation) expression and lower differentiation into the T regulatory (CD25+ FOXP3+) phenotype (3), suggesting again benefits of Bifidobacterium in BF infants. Serum and urine metabolites after breastfeeding or formula feeding in rhesus macaques have been previously investigated (15). In this model, microbiota and metabolites were altered as seen using PCA plots, and separation by feeding method was observed at 2 wk. FF monkeys had higher urine and serum galactose and amino acid levels (15). In support of this notion, our results highlighted that breastfeeding induced greater numbers of microbial genes related to d-glutamine and d-glutamate metabolism; cysteine and methionine metabolism; valine, leucine, and isoleucine biosynthesis; tyrosine metabolism; and tryptophan metabolism. These results highlight that gut microbiota and metabolites produced by bacteria can play a role in the developing immune system.
The kynurenine pathway for tryptophan metabolism has been implicated in developing tolerance within the immune system (56). Along with kynurenic acid, changes in tryptophan-related metabolites were observed that were due to the infant diet. Kynurenic acid, indole-3-lactic acid, and indole-3-acetic acid were increased in BF compared with FF infants. In vitro models have shown that both kynurenic acid and synthetic analogs influence the expression of inflammatory and noninflammatory cytokines, albeit differently depending on the type of infection investigated (56). When monocyte cells were infected with heat-inactivated Staphylococcus aureus, there was attenuated TNF-α (inflammatory) secretion and increased tumor necrosis factor-stimulated gene-6 (TSG-6) mRNA (anti-inflammatory) expression. A similar phenotype was seen in Chlamydia pneumonia–infected cells with kynurenic acid, but not the synthetic analog (57). Thus, differences in these metabolites may have functional consequences for the developing immune system and, further, allergy outcomes (56). Within this study, multiple metabolites were altered by neonatal diet, suggesting metabolite-mediated communication with the microbiota and the immune system possibly affecting cellular mechanisms in the gut and other organs.
Limitations
It is important to note that different species of Bifidobacterium and Bacteroides colonize the guts of BF and FF infants. Within this study, the relative abundance of Bifidobacterium between the BF and MF infants was not different and no significant effect of diet was observed on Bacteroides abundance. However, the species-level differences were not captured due to the limitations of 16S rRNA sequencing. The other limitations include lack of data on mode of delivery, weight gain, and perinatal consumption of probiotics/antibiotics. These factors can influence the microbiota composition. These data are a secondary analysis of a clinical cohort, and the same subjects were not available at all the time points to enable longitudinal assessments of microbiota and metabolites. Furthermore, the absence of breast-milk samples limited our ability to determine mothers’ secretor or nonsecretor status, as well as the possible association of secretor status with microbiota composition. Nonetheless, this work further demonstrates the large influence infant diet has on the developing microbiota and metabolome through the first year of life.
Future mechanistic studies are needed to determine the role of components of breast milk, microbiota, and specific gut-derived metabolites on regulating the developing infant's immune and other physiological systems.
Supplementary Material
Acknowledgments
We thank the ACNC Clinical Core team for subject recruitment and sample collection.
The authors’ responsibilities were as follows—LY: designed the research; LRB, BDP, and AE: conducted the data analysis; LB and LY: wrote the manuscript; AKB and KSM: conducted technical aspects of the research; SVC and KS: acquired the microbiota data; LP, SHA, and KEM: generated the metabolomics data; TMB and AA: were principal investigators of the Beginnings cohort; LY: had primary responsibility for the final content and for editing the manuscript; and all authors: read and approved the final manuscript. The authors report no conflicts of interest.
Notes
Supported by USDA-Agricultural Research Service Project 6026-51000-010-05S (to LY), NIH grant P20GM121293 (to LY, SVC, and BDP), and National Institute of Allergy and Infectious Diseases grant 1R21AI146521 (to LY).
Supplemental Figures 1–10 and Supplemental Tables 1–9 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/ajcn/.
All the raw data will be available upon request.
Abbreviations used: BF, breastfed; CLR, centered log ratio; ESI, electrospray ionization; FDR, false discovery rate; FF, formula-fed; FHWM, full width half maximum; MF, milk formula; NLB, no longer breastfeeding; OTU, operational taxonomic unit; PC, principal component; PCA, principal component analysis; PERMANOVA, permutational multivariate ANOVA; PICRUSt, Phylogenetic Investigation of Communities by Reconstruction of Unobserved States; QC, quality control; rRNA, ribosomal RNA; SF, soy-based formula.
References
- 1. Beaudry M, Dufour R, Marcoux S. Relation between infant feeding and infections during the first six months of life. J Pediatr. 1995;126(2):191–7. [DOI] [PubMed] [Google Scholar]
- 2. Hanson LA, Korotkova M, Telemo E. Breast-feeding, infant formulas, and the immune system. Ann Allergy Asthma Immunol. 2003;90(6 Suppl 3):59–63. [DOI] [PubMed] [Google Scholar]
- 3. Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, Panzer AR, LaMere B, Rackaityte E, Lukacs NW et al.. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016;22(10):1187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Melnik BC, John SM, Carrera-Bastos P, Schmitz G. Milk: a postnatal imprinting system stabilizing FoxP3 expression and regulatory T cell differentiation. Clin Transl Allergy. 2016;6:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Andreas NJ, Kampmann B, Mehring Le-Doare K. Human breast milk: a review on its composition and bioactivity. Early Hum Dev. 2015;91(11):629–35. [DOI] [PubMed] [Google Scholar]
- 6. Davis EC, Wang M, Donovan SM. The role of early life nutrition in the establishment of gastrointestinal microbial composition and function. Gut Microbes. 2017;8(2):143–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90(3):859–904. [DOI] [PubMed] [Google Scholar]
- 8. Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Chari RS, Sears MR, Becker AB, Scott JA, Kozyrskyj AL et al.. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ. 2013;185(5):385–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Azad MB, Konya T, Persaud RR, Guttman DS, Chari RS, Field CJ, Sears MR, Mandhane PJ, Turvey SE, Subbarao P et al.. Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: a prospective cohort study. BJOG. 2016;123(6):983–93. [DOI] [PubMed] [Google Scholar]
- 10. Fan W, Huo G, Li X, Yang L, Duan C. Impact of diet in shaping gut microbiota revealed by a comparative study in infants during the six months of life. J Microbiol Biotechnol. 2014;24(2):133–43. [DOI] [PubMed] [Google Scholar]
- 11. Round JL, Palm NW. Causal effects of the microbiota on immune-mediated diseases. Sci Immunol. 2018;3(20):eaao1603. [DOI] [PubMed] [Google Scholar]
- 12. Wang M, Monaco MH, Donovan SM. Impact of early gut microbiota on immune and metabolic development and function. Semin Fetal Neonatal Med. 2016;21(6):380–7. [DOI] [PubMed] [Google Scholar]
- 13. Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. The role of short-chain fatty acids in health and disease. Adv Immunol. 2014;121:91–119. [DOI] [PubMed] [Google Scholar]
- 14. Rivera-Chávez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, Xu G, Velazquez EM, Lebrilla CB, Winter SE et al.. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe. 2016;19(4):443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. O'Sullivan A, He X, McNiven EM, Haggarty NW, Lönnerdal B, Slupsky CM. Early diet impacts infant rhesus gut microbiome, immunity, and metabolism. J Proteome Res. 2013;12(6):2833–45. [DOI] [PubMed] [Google Scholar]
- 16. Bazanella M, Maier TV, Clavel T, Lagkouvardos I, Lucio M, Maldonado-Gòmez MX, Autran C, Walter J, Bode L, Schmitt-Kopplin P et al.. Randomized controlled trial on the impact of early-life intervention with bifidobacteria on the healthy infant fecal microbiota and metabolome. Am J Clin Nutr. 2017;106(5):1274–86. [DOI] [PubMed] [Google Scholar]
- 17. Andres A, Cleves MA, Bellando JB, Pivik RT, Casey PH, Badger TM. Developmental status of 1-year-old infants fed breast milk, cow's milk formula, or soy formula. Pediatrics. 2012;129(6):1134–40. [DOI] [PubMed] [Google Scholar]
- 18. Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79(17):5112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI et al.. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deda O, Gika HG, Wilson ID, Theodoridis GA. An overview of fecal sample preparation for global metabolic profiling. J Pharm Biomed Anal. 2015;113:137–50. [DOI] [PubMed] [Google Scholar]
- 21. Piccolo BD, Mercer KE, Bhattacharyya S, Bowlin AK, Saraf MK, Pack L, Chintapalli SV, Shankar K, Adams SH, Badger TM et al.. Early postnatal diets affect the bioregional small intestine microbiome and ileal metabolome in neonatal pigs. J Nutr. 2017;147(8):1499–509. [DOI] [PubMed] [Google Scholar]
- 22. McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014;10(4):e1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oksanen J, Guillaume Blanchet F, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos P et al.. vegan: community ecology package. [Internet] 2019. Available from: https://CRAN.R-project.org/package=vegan. [Google Scholar]
- 25. Lenth R. emmeans: estimated marginal means aL-SMRpvhCR-pope 2020; . Available from: https://github.com/rvlenth/emmeans [Google Scholar]
- 26. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125(1–2):279–84. [DOI] [PubMed] [Google Scholar]
- 28. Redestig H, Fukushima A, Stenlund H, Moritz T, Arita M, Saito K, Kusano M. Compensation for systematic cross-contribution improves normalization of mass spectrometry based metabolomics data. Anal Chem. 2009;81(19):7974–80. [DOI] [PubMed] [Google Scholar]
- 29. Hastie T, Tibshirani R, Narasimhan B, Chu G. 2019 doi: 10.1073/pnas.082099299. https://www.bioconductor.org/packages/release/bioc/html/impute.html. impute: impute: Imputation for microarray data. R package version 1.58.0. Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R et al.. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31(9):814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30(21):3123–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gloor GB, Reid G. Compositional analysis: a valid approach to analyze microbiome high-throughput sequencing data. Can J Microbiol. 2016;62(8):692–703. [DOI] [PubMed] [Google Scholar]
- 33. Palarea-Albaladejo J, Martín-Fernández JA. zCompositions — R package for multivariate imputation of left-censored data under a compositional approach. Chemom Intell Lab Syst. 2015;143:85–96. [Google Scholar]
- 34. Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S et al.. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007;35(Database issue):D521–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Borewicz K, Suarez-Diez M, Hechler C, Beijers R, de Weerth C, Arts I, Penders J, Thijs C, Nauta A, Lindner C et al.. The effect of prebiotic fortified infant formulas on microbiota composition and dynamics in early life. Sci Rep. 2019;9(1):2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baumann-Dudenhoeffer AM, D'Souza AW, Tarr PI, Warner BB, Dantas G. Infant diet and maternal gestational weight gain predict early metabolic maturation of gut microbiomes. Nat Med. 2018;24(12):1822–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huda MN, Ahmad SM, Alam MJ, Khanam A, Kalanetra KM, Taft DH, Raqib R, Underwood MA, Mills DA, Stephensen CB. Bifidobacterium abundance in early infancy and vaccine response at 2 years of age. Pediatrics. 2019;143(2):e20181489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Avershina E, Storrø O, Øien T, Johnsen R, Pope P, Rudi K. Major faecal microbiota shifts in composition and diversity with age in a geographically restricted cohort of mothers and their children. FEMS Microbiol Ecol. 2014;87(1):280–90. [DOI] [PubMed] [Google Scholar]
- 39. Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy. 2014;44(6):842–50. [DOI] [PubMed] [Google Scholar]
- 40. Roos S, Dicksved J, Tarasco V, Locatelli E, Ricceri F, Grandin U, Savino F. 454 pyrosequencing analysis on faecal samples from a randomized DBPC trial of colicky infants treated with Lactobacillus reuteri DSM 17938. PLoS One. 2013;8(2):e56710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jost T, Lacroix C, Braegger CP, Chassard C. New insights in gut microbiota establishment in healthy breast fed neonates. PLoS One. 2012;7(8):e44595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huda MN, Lewis Z, Kalanetra KM, Rashid M, Ahmad SM, Raqib R, Qadri F, Underwood MA, Mills DA, Stephensen CB. Stool microbiota and vaccine responses of infants. Pediatrics. 2014;134(2):e362–e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lewis ZT, Totten SM, Smilowitz JT, Popovic M, Parker E, Lemay DG, Van Tassell ML, Miller MJ, Jin YS, German JB et al.. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome. 2015;3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB et al.. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10(5):507–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. de Muinck EJ, Trosvik P. Individuality and convergence of the infant gut microbiota during the first year of life. Nat Commun. 2018;9(1):2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bergström A, Skov TH, Bahl MI, Roager HM, Christensen LB, Ejlerskov KT, Mølgaard C, Michaelsen KF, Licht TR. Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl Environ Microbiol. 2014;80(9):2889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Saraf MK, Piccolo BD, Bowlin AK, Mercer KE, LeRoith T, Chintapalli SV, Shankar K, Badger TM, Yeruva L. Formula diet driven microbiota shifts tryptophan metabolism from serotonin to tryptamine in neonatal porcine colon. Microbiome. 2017;5(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Perdijk O, van Baarlen P, Fernandez-Gutierrez MM, van den Brink E, Schuren FHJ, Brugman S, Savelkoul HFJ, Kleerebezem M, van Neerven RJJ. Sialyllactose and galactooligosaccharides promote epithelial barrier functioning and distinctly modulate microbiota composition and short chain fatty acid production in vitro. Front Immunol. 2019;10:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ et al.. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T et al.. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–50. [DOI] [PubMed] [Google Scholar]
- 52. Geuking MB, McCoy KD, Macpherson AJ. Metabolites from intestinal microbes shape Treg. Cell Res. 2013;23(12):1339–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chi H. Sphingosine-1-phosphate and immune regulation: trafficking and beyond. Trends Pharmacol Sci. 2011;32(1):16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yang C, Lai W, Zhou J, Zheng X, Cai Y, Yang W, Xie S, Gao Y, Du C. Betaine ameliorates experimental autoimmune encephalomyelitis by inhibiting dendritic cell–derived IL-6 production and Th17 differentiation. J Immunol. 2018;200(4):1316–24. [DOI] [PubMed] [Google Scholar]
- 55. Taft DH, Liu J, Maldonado-Gomez MX, Akre S, Huda MN, Ahmad SM, Stephensen CB, Mills DA. Bifidobacterial dominance of the gut in early life and acquisition of antimicrobial resistance. mSphere. 2018;3(5):00441–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Van der Leek AP, Yanishevsky Y, Kozyrskyj AL. The kynurenine pathway as a novel link between allergy and the gut microbiome. Front Immunol. 2017;8:1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mándi Y, Endrész V, Mosolygó T, Burián K, Lantos I, Fülöp F, Szatmári I, Lőrinczi B, Balog A, Vécsei L. The opposite effects of kynurenic acid and different kynurenic acid analogs on tumor necrosis factor-α (TNF-α) production and tumor necrosis factor-stimulated gene-6 (TSG-6) expression. Front Immunol. 2019;10:1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.