Abstract
The recent adaptation of the CRISPR/Cas9 system for targeted genome engineering has led to its widespread applications in many fields worldwide. In order to better understand the design rules of CRISPR/Cas9 systems, several groups have carried out large library-based screens leading to some insight into sequence preferences among highly active target sites. To facilitate CRISPR/Cas9 design these studies have spawned a plethora of gRNA design tools with algorithms based solely on direct or indirect sequence features. Here we demonstrate that the predictive power of these tools is poor, suggesting that sequence features alone cannot accurately inform the cutting efficiency of a particular CRISPR/Cas9 gRNA design. Furthermore we demonstrate that DNA target site accessibility influences the activity of CRISPR/Cas9. With further optimisation we hypothesise that it will be possible to increase the predictive power of gRNA design tools by including both sequence and target site accessibility metrics.
Introduction
The development of designer nucleases has revolutionised the field of biotechnology by allowing site specific DNA modifications with single nucleotide resolution (Kim et al., 1996; Urnov et al., 2005; Miller et al., 2011; Mussolino et al., 2011). In particular, the clustered regulatory interspaced short palindromic repeats (CRISPR), CRISPR associated (Cas) system has been widely adopted as the designer nuclease of choice. The CRISPR/Cas system is a prokaryotic immune system present in bacteria and archaea that confers resistance to foreign DNA via a type of acquired immunity (Barrangou et al., 2007; Marraffini & Sontheimer, 2008). The critical target specific component of the CRISPR/Cas9 system is the guide RNA (gRNA), a short single stranded RNA sequence with an 80nt constant region and a short 20nt target specific sequence with binds to a DNA target via Watson-Crick base pairing. By altering the sequence of the gRNA, it is possible to specifically target almost any 20bp sequence in the human genome (Jinek et al., 2012; Cong et al., 2013; Mali et al., 2013). However, early studies found that different gRNAs had different levels of DNA targeting with some gRNAs capable of targeting 21% of alleles in a population of cells while others achieved targeting rates of 77% (Cradick et al., 2013; Lee et al., 2017). Several studies have attempted to elucidate rules for rational design of highly active gRNAs (Doench et al., 2014; Chari et al., 2015; Moreno-Mateos et al., 2015; Wong et al., 2015; Xu et al., 2015). However, most design tools have not been experimentally validated for single gene targeting. Here we demonstrate that existing CRISPR/Cas9 design tools do not accurately predict the activity of single gene targeting gRNA sequences. Furthermore we report that target site accessibility may influence Cas9 activity and suggest that future gRNA design algorithms need to incorporate locus specific metrics.
gRNA Design tools for predicting activity
The CRISPR/Cas9 system adopted from Streptococcus pyogenes (Spy) has been widely used in many genome editing applications. By altering the short 20nt sequence it is possible for SpyCas9 to target virtually any locus in the human genome. However, different gRNAs have varying degrees of activity. While some gRNAs are capable of disrupting almost every target allele in a population of cells, others display no detectable activity. This discrepancy can lead to a significant amount of work in the search for a highly active gRNA design. This has led to the recent development of several CRISPR/Cas9 design tools. The studies underlying these design tools typically use high-throughput methods which screen >1,000 gRNAs based on experimental data. The first high throughput study analysed the activity of 1,841 gRNAs in an effort to establish a common set of design rules which could be used for the rational design of highly active gRNAs (Doench et al., 2014), resulting in the first publicly available design tool sgRNA Designer. This was later updated with a new algorithm derived from machine learning based predictive modelling (Doench et al., 2016). There are now several other design tools available (Ren et al., 2014; Chari et al., 2015; Moreno-Mateos et al., 2015; Wong et al., 2015; Xu et al., 2015). However, many of these studies do not directly measure gRNA activity. For example, some studies determine gRNA efficiency by the loss of a surface marker thereby counting only those cells that have undergone biallelic frameshift mutations (Doench et al., 2014). These studies have also been conducted in a wide range of different organism and tissues, including human cells, mouse cells, C. elegans, D. melanogaster, and D. rerio. Although these algorithms display some significance within their own data sets (R2= 0.51 +/− 0.12) when they are cross validated with other data sets the predictive power diminishes significantly (R2= 0.23 +/− 0.14) (Haeussler et al., 2016). To assess the ability of these publicly available design tools to predict the efficiency of gRNAs for single gene targeting experiments we tested 198 gRNAs individually and compared their predicted score with the observed level of activity (Fig. 1A). The best performing algorithm had an R2 of 0.3 demonstrating poor predictive capability (Fig. 1B).
Figure 1. Performance of gRNA prediction algorithms.
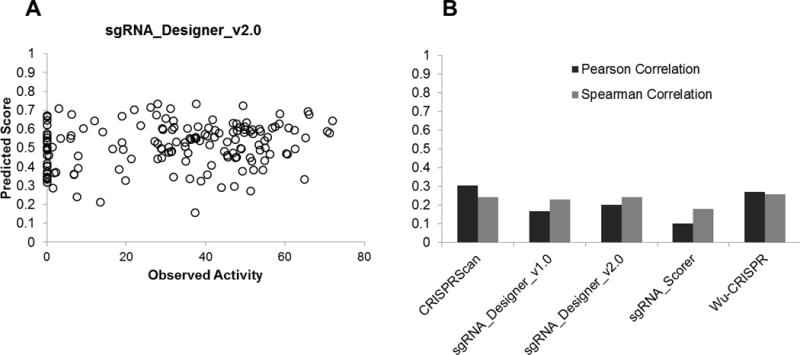
(a) Plot of gRNA activity and predicted scores obtained from the sgRNA Designer v2.0 design tool. (b) Correlation of gRNA activity and predicted scores from 5 different gRNA design tools.
Locus Effects on CRISPR/Cas9 Activity
One thing these design algorithms have in common is heavy weighting of gRNA sequence features e.g. what nucleotide is favoured at each position of highly active gRNAs. Other algorithms also include metrics for di- and tri-nucleotides. Some also include other metrics such as duplex binding (ΔG) and the secondary structure or folding properties of the gRNA (Wong et al., 2015), both of which are gRNA sequence dependent. We have begun to investigate the effect of non-sequence dependent features in order to better understand what influences CRISPR/Cas9 activity and to enable more accurate prediction of gRNAs prior to beginning any experimental work. In particular we hypothesise that DNA accessibility and chromatin state plays a role in the ability of CRISPR/Cas9 to generate a DNA double strand break at the target locus. Indeed, recent reports have shown that DNA packaged as nucleosomes is protected from CRISPR/Cas9 cleavage in vitro (Hinz et al., 2015; Horlbeck et al., 2016; Isaac et al., 2016). However, nucleosomes can be repositioned or restructured by ATP-dependent chromatin remodelers (Clapier & Cairns, 2009; Narlikar et al., 2013) exposing the DNA target site to CRISPR/Cas9 in the process (Isaac et al., 2016). However, the dynamic nature of nucleosomes makes it difficult to translate in vitro findings into a cellular context.
In order to directly study the effect on DNA accessibility in mammalian cells we designed a series of gRNAs that target repetitive elements thus removing any bias which may occur from using gRNAs that target different sequences. These gRNAs have between 4 and 20,000 target sites in the human genome. If chromatin state and DNA accessibility has no effect of CRISPR/Cas9 then the activity observed at each locus should be the same. However, if the activity differs significantly across different loci it would strongly suggest that DNA accessibility influences CRISPR/Cas9 activity. To query individual loci, we isolated individual target sites using barcoded primers unique to each locus. This limits the sites that can be analysed by deep sequencing to repeats that are shorter than 500bp or that have sufficient sequence divergence. For one of these gRNAs, CTS_30, we successfully isolated 52 distinct genomic loci for deep sequencing. The level of CRISPR activity varied significantly across all 52 sites (Fig. 2A, Supplementary Table S1) suggesting that CRISPR/Cas9 activity is influenced by locus specific features independent of the target sequence. Previous work has shown that microhomologies were associated with 39.6% of all mutations induced by CRISPR/Cas9 gRNAs designed to target different sequences (Bae et al., 2014). Interestingly, here we show that the nature of the mutations induced by a single gRNA (CTS30) at multiple different loci were identical across all the active loci suggesting that this sequence microhomology effect on the mutation spectrum following NHEJ repair is not just heavily influenced by the target sequence but is solely dependent on the target sequence(Fig. 2B).
Figure 2. Multi-target gRNA activity.
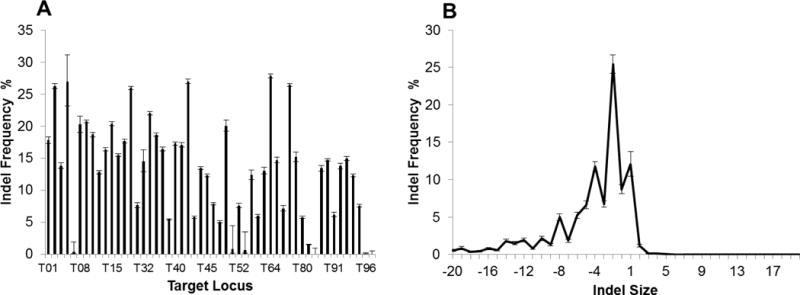
(a) The activity of gRNA CTS_30 at 52 distinct genomic loci. Error bars derived from 95% Confidence Intervals. (b) The frequency of different mutation sizes observed at 52 CTS_30 target sites. The error bars reflect the variation in indel spectra across all 52 active sites. N=52, error bars ± standard deviation.
Conclusion
The first-generation CRISPR gRNA design tools, while useful for quickly identifying gRNA target sites, do not accurately predict the cutting activity of Cas9 for single-locus gene modification in human cells. Our recent work focusing on the target locus as opposed to simply the target sequence suggests that there are non-sequence based features that influence Cas9 activity. Indeed, recent in vitro studies demonstrating protection of DNA from Cas9 cleavage supports our findings in human cells and suggest that DNA accessibility at the target locus could be a critical factor. Further characterisation of the locus features that influence CRISPR/Cas9 activity will improve the predictive power of new-generation gRNA design algorithms as well as shed important light on the ability of Cas9 to recognise particular off-target loci. A better understanding of the target locus specific effects will facilitate the studies of efficiency and specificity of CRISPR/Cas9 based genome editing across different cell types.
Supplementary Material
Supplementary Table S1: Deep Sequencing data of gRNA CTS_30 cutting activity. P-values are calculated using Fishers exact test.
New Findings.
What is the topic of this review?
In this review we analyse the performance of recently described tools for CRISPR/Cas9 guide RNA (gRNA) design, in particular, design tools that predict CRISPR/Cas9 activity and specificity.
What advances does it highlight?
Recently, many tools designed to predict CRISPR/Cas9 activity have been reported. However, the majority of these tools lack experimental validation. Our analyses indicate that these tools have poor predictive power. Our preliminary results suggest that target site accessibility should be considered in order to develop better gRNA design tools with improved predictive power.
Acknowledgments
Funding
This work was supported in part by National Institutes of Health grant PN2EY018244 to G.B. and Cancer Prevention and Research Institute of Texas (CPRIT) grant RR140081 to G.B.
Footnotes
Competing interests
None declared.
Author contributions
All experiments performed in the laboratory of G.B. The experiments were conceived by C.M.L. and G.B. The work was performed by C.M.L. and T.H.D. Data analysis was performed by C.M.L., T.H.D., and G.B. The manuscript was drafted and revised by C.M.L., T.H.D., and G.B. All authors approve the final version of this manuscript and agree to be accountable for all aspects of the work. All persons designated as authors qualify for authorship, and all those who qualify for authorship are listed.
References
- Bae S, Kweon J, Kim HS, Kim JS. Microhomology-based choice of Cas9 nuclease target sites. Nat Methods. 2014;11:705–706. doi: 10.1038/nmeth.3015. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Chari R, Mali P, Moosburner M, Church GM. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat Meth. 2015 doi: 10.1038/nmeth.3473. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41:9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, Virgin HW, Listgarten J, Root DE. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014 doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M, Schonig K, Eckert H, Eschstruth A, Mianne J, Renaud JB, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, Joly JS, Concordet JP. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz JM, Laughery MF, Wyrick JJ. Nucleosomes Inhibit Cas9 Endonuclease Activity in Vitro. Biochemistry. 2015;54:7063–7066. doi: 10.1021/acs.biochem.5b01108. [DOI] [PubMed] [Google Scholar]
- Horlbeck MA, Witkowsky LB, Guglielmi B, Replogle JM, Gilbert LA, Villalta JE, Torigoe SE, Tjian R, Weissman JS. Nucleosomes impede Cas9 access to DNA in vivo and in vitro. Elife. 2016;5 doi: 10.7554/eLife.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac RS, Jiang F, Doudna JA, Lim WA, Narlikar GJ, Almeida R. Nucleosome breathing and remodeling constrain CRISPR-Cas9 function. Elife. 2016;5 doi: 10.7554/eLife.13450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Zhu H, Davis TH, Deshmukh H, Bao G. Design and Validation of CRISPR/Cas9 Systems for Targeted Gene Modification in Induced Pluripotent Stem Cells. Methods Mol Biol. 2017;1498:3–21. doi: 10.1007/978-1-4939-6472-7_1. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussolino C, Morbitzer R, Lutge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Yang Z, Xu J, Sun J, Mao D, Hu Y, Yang SJ, Qiao HH, Wang X, Hu Q, Deng P, Liu LP, Ji JY, Li JB, Ni JQ. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep. 2014;9:1151–1162. doi: 10.1016/j.celrep.2014.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Wong N, Liu W, Wang X. WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 2015;16:218. doi: 10.1186/s13059-015-0784-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Xiao T, Chen CH, Li W, Meyer CA, Wu Q, Wu D, Cong L, Zhang F, Liu JS, Brown M, Liu XS. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015;25:1147–1157. doi: 10.1101/gr.191452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1: Deep Sequencing data of gRNA CTS_30 cutting activity. P-values are calculated using Fishers exact test.


