Highlights
-
•
Production of highly specific IgY monoclonal antibodies against SARS CoV-2 holds much effective clinical and scientific advantages.
-
•
This method holds higher specificity, consistency and reproducibility.
-
•
Highly scalable manufacturing and lower cross-reactive due to the absence of invariable regions.
Keywords: SARS CoV-2, Spike protein (S), IgY, Phage display, Passive immunotherapy
Abstract
The present state of diagnostic and therapeutic developmental race for vaccines against the SARS CoV-2 (nCOVID-19) focuses on prevention and control of this global pandemic which also represents a critical challenge to the global health community. Although development of novel vaccines can prevent the SARS CoV-2 infections, it is still impeded by several other factors and therefore novel approaches towards treatment and management of this disease is the urgent need. Passive immunotherapy plays a vital role as a possible alternative to meet this challenge and among various antibody sources, chicken egg yolk antibodies (IgY) can be used as an alternative to mammalian antibodies which have been previously studied against SARS CoV outbreak in China. In this review, we discuss the strategies for the use of chicken egg yolk (IgY) antibodies in the development of rapid diagnosis and immunotherapy against SARS CoV-2. Also, IgY antibodies have previously been used against various respiratory bacterial and viral infections in humans and animals. Compared to mammalian antibodies (IgG), chicken egg yolk antibodies (IgY) have greater binding affinity to specific antigens, ease of extraction and lower production costs, hence possessing remarkable pathogen-neutralizing activity of pathogens in respiratory and lungs. We provide an overall importance for the use of monoclonal chicken egg yolk antibodies (IgY) using phage display method describing their potential passive immunotherapeutic application for the treatment and prevention of SARS CoV-2 infection which is simple, fast and safe way of approach for treating patients effectively.
1. Introduction
The novel severe acute respiratory syndrome SARS CoV-2 (nCOVID-19) is a novel coronavirus infection that has emerged as global pandemic threat in todays world. This SARS CoV-2 infection was caused by SARS-associated coronaviruses [1], [2], [3] where the spike protein ‘S’ were found to be the major domain responsible for mediating the membrane fusion during the infection [4], [5]. Also, the SARS CoV-2 S protein belongs to large membrane glycoprotein and the angiotensin converting enzyme 2 (ACE-2) present in the human cell membranes found to possess the functional receptor for this virus [6]. The conformational changes induced by this complex spike protein (S) with the ACE-2 receptor leads to membrane fusion and furthermore, due to the higher antigenicity nature of S protein, it efficiently elicits the neutralizing antibodies within the system by protecting them from infection [7], [8]. Hence, this spike protein domain (S) will be a good candidate for developing antiviral drugs, vaccines and that generating high specific antibodies to recognize this domain would be much more valuable. Some of the reported studies with SARS CoV shows that immunization of mice with recombinant spike protein (S) can protect them from infection effectively [7], [8]. Therefore, generating monoclonal antibodies that are specifically to recognize the spike protein (S) is the need of the hour. However, we have to overcome the difficulties in generating highly specific monoclonal antibodies from the traditional way of antibody generation [9], phage display method of monoclonal antibody generation would be safe and effective procedure because of the in vitro process of antibody generation by cloning the specific repertoires and subsequent screening and isolation of monoclonal antibodies from the antibody libraries [10], [11]. Although there are many ways of recombinant antibody generation, the single-chain variable fragments (scFv) of the whole antibody will be an effective region that can be generated in phage display system [12], [13], [14].
Antibody production in chickens is the simplest, easier and efficient way of generating monoclonal antibodies using phage display library selection for scFv gene constructs with higher affinity against targetted pathogens [15], [16]. Also, it has been reported that monospecific scFv antibodies generated using phage display technology has high neutralizing effect against SARS CoV infection that have been generated from non-immunized individuals and convalescent SARS infected individuals [17], [18]. This review focuses to show that the monoclonal IgY scFv antibodies raised against SARS CoV-2 spike protein (S) isolated from chickens using phage display technology would be a potential model for large scale production of high-affinity antibodies effectively.
2. SARS CoV-2 structure and its etiology
The SARS-CoV-2 (nCOVID-19) are enveloped viruses with round or pleiomorphic structure of approximately 80 to 120 nm in diameter containing positive single-stranded RNA genome of 30 kb size [19], [20]. The RNA genome is complexed with basic nucleocapsid protein (N) to form a helical viral protein and these are spike proteins (S) which are the Type-I glycoprotein that forms the peplomers on the virion surface giving it a crown-like structure (Fig. 1 ). The membrane protein (M) which spans three time the viral membrane has a short N-terminal ectodomain and a cytoplasmic tail. The small membrane protein (E) is found to be highly hydrophobic in nature [21] and this spans twice the viral membrane has both N and C terminals on the interior part of the virion [22]. There are still many other minor proteins present in the viral structure yet undetected and the genomes of all the coronaviruses were found to have similar stuctural characteristics [23]. For all coronaviruses, the structural proteins are encoded in order of S-E-M-N within the one third of the genome and each group of coronaviruses encodes a group of unique small proteins while these are non-essential proteins and have been found to serve as accessory proteins to interact or interfere with the host innate immune responses which has not been demonstrated for any of these proteins [24], [25]. Untranslated regions of coronaviruses (UTRs) on both 5′ and 3′ ends of the genome were believed to interact with the host and control the RNA replication process and the viral transcription has been reviewed in recent studies [26].
Fig. 1.
Morphological representation of SARS CoV-2 coronavirus and its Spike protein (S) with structural binding domain regions.
3. Spike protein (S) of SARS-CoV-2
Transmembrane spike (S) protein of the coronavirus mediates host cell entry. S protein is a glycoprotein that forms homotrimers [27]. Each monomer is about 180 kDa. Because it is surface-exposed on part of the virus and easily accessible for neutralizing antibodies, it becomes a potential drug target and interest of several structural studies. SARS-CoV-2 S protein is capable of triggering protease-independent and receptor-dependent syncytium formation; this enhances virus spreading through cell-cell fusion and the rapid progress of the diseases [28], [29]. The functional domains of S protein are distributed among the two subunits, the receptor-binding domain (RBD) is in the S1 subunit [Fig. 1]. SARS CoV and SARS CoV-2 interact directly with human angiotensin-converting enzyme 2 (hACE2) through domain B (SB). The cryoelectron microscopy structures of the SARS-CoV-2 S ectodomain trimer reveal that it adopts multiple SB conformations as like SARS-CoV [30], [31], [32]. The S proteins of SARS CoV-2 share the amino acid similarity of about 76% and 97% with SARS CoV, and RaTG13 (bat coronavirus- BatCoV RaTG13) respectively. Interestingly, the RBD has only 74% and 90.1% similarity with SARS CoV, and RaTG13 respectively [33]. This partially explains the difference in the binding affinity of these viruses towards their respective host targets and the efficient transmission. In most of the coronavirus, these two subunits are non-covalently bound in the prefusion conformation [34], [35], [36]. Additionally, the S protein structure is covered with N-linked glycans, this perhaps helps to maintain its integrity and acts as a shield to escape the host immune system [36], [37]. The neutralizing antibodies have to slot in between the glycans to attach with the spike protein.
S protein is the main target for neutralizing antibodies and vaccine design which plays a vital role in studying the viral entry, determination of virulence, understanding the range of hosts, a pseudotype system with S protein of SARS-CoV-2 [38], [39]. Though there is considerable homology between S protein of SARS CoV and SARS CoV-2, polyclonal rabbit anti-SARS S1 antibodies T62 did not completely neutralize the SARS-CoV-2 and there is less affinity of binding with SARS-CoV-2 S protein [39]. Previous studies postulated that major immune-epitopes for anti-SARS S1 antibody T62 may lie in the region of RBD. Moreover, the experiment with convalescent sera from SARS and COVID-19 patients has shown only moderate cross-neutralization and the sera from SARS-CoV infected individual targets SB site of the SARS-CoV-2 S protein [40]. Few studies had reported the lack of cross-reactivity among these SB-directed antibodies [41], [42]. Most of SARS-CoV neutralizing antibodies target the SB domain and several of them recognize the receptor binding domain regions and prevent receptor engagement [43], [44], [45]. This suggests that those previously recovered from SARS-CoV infection may not be fully protected against SARS-CoV-2 infection, and vice versa. The more proper understanding of the S protein and its accessible antigenic regions will help to develop more effective antibodies or drugs to fight against SARS-CoV-2 effectively.
4. Insights of IgY antibodies
Antibodies are also called as immunoglobulins, which are associated with the humoral immune response [37]. Producing antibodies from mammals involves stress to animals, such as immunization, blood collection and sometimes sacrifice [46]. An alternative for the mammalian antibodies is the IgY antibody which is being purified from eggs of immunized birds (mostly hens and ducks). Looking closely, into the functions, IgY antibodies are similar to that of mammalian IgG’s [47] and are transported from the blood serum to the yolk plasma [48]. Chicken egg yolk antibodies (IgY) from hyperimmunized eggs against tetanus toxin had the ability to neutralize the lethality in in vivo experiments [49]. These neutralizing antibodies from chickens were later termed as IgY in 1969 [50]. Chicken egg yolk antibodies (IgY) were considered to be the largest source of major nutrients and the chicken eggs also contains various other biological substances in it that forms the basic unit of life [51], [52].
In recent years, the use of 3R’s (Reducing, Refining and Reusing) has been well accepted by industries producing antibodies and IgY clearly defines the use of 3R’s, which has allowed the production of IgY antibodies for research, diagnostic and therapeutic purposes. IgY antibodies are highly effective in neutralizing the bacterial and viral diseases of respiratory system, causing no side effects [52]. The response of immunoglobulins in chickens to the highly conserved mammalian proteins is robust showing higher affinity, thereby potentially targeting broad spectrum of epitopes on protein immunogens [49].
Molecular weight of IgY is around 180 kDa and is slightly greater than IgG (150 kDa) as the IgY heavy chain has an additional constant domain and higher amounts of carbohydrates [53]. Hinge region is absent in the heavy chain of IgY. Sequence analysis of IgY and IgG showed that CH3 and CH4 domains of IgY are closely related to CH2 and CH3 domains of IgG [54]. Irrespective of IgY being a protein molecule, it shows resistant to heat at temperatures ranging from 30 to 70 °C and is active at pH 3–11 [55]. The isoelectric point (pI) of IgY is between 5.4 and 7.6 and are stable for a period of 10 years at 4 °C, at room temperature for 6 months and for 1 month at 37 °C, without loss of any activity. Since antibodies from chickens are harvested from egg yolks, chickens show higher advantages to conventional mammalian species such as rabbits, goats and sheep. Chickens are able to produce 100–150 mg of antibodies per yolk, of which 5–10% is the specific antibody [56], [46]. A chicken can lay up to 300–325 eggs every year which yields 20–30 g of IgY production.
Antibody fragments are originally the proteolytic cleavage molecular fragments of the full length antibodies and their properties makes them amenable for developing recombinant antibody libraries and such selection of scFv antibodies recognises the unique specific target of the antigens [47] and it is clear that the antibody fragments developed using phage display technology is a powerful tool in the molecular study targets. Attribution to the chicken egg olk IgY monoclonal antibody production using phage display model will be more likely to obtain a robust immune response against various highly conserved mammalian protein molecules from the chicken hosts [57] and due to the presence of an additional constant region (CH2) in the heavy chain they lack the flexible hinge region [58]. The scFv libraries of recombinant chicken egg yolk monoclonal antibody fragments can be generated easily by cloning a single pair of oligonucleotides of either heavy and light chain gene receptors effectively [57] (Fig. 2 ).
Fig. 2.
Structural comparison of mammalian antibody (IgG) and chicken egg yolk antibody (IgY) and their recombinant scFv fragment structures.
5. Use of IgY antibodies in passive immunization therapy
The administration of preformed antibodies or immunoglobulins to treat various infectious diseases is known as passive immunization therapy [58], [59], [60] and transferring of such pathogen specific derived antibodies to non-immune subjects by oral, intravenous and systemic route of passive immunization. Owing to such immediate, short-term development of immunity, continuous and mass production of highly specific antibodies can be produced by hyperimmunization of chickens with target antigens will be the need of the hour [61] for large scale production of antibodies ensuring higher purity and constant supply of antibodies [62]. Intranasal route of passive immunotherapy proves to be highly promising and effective in treating many viral infections because it binds directly to the virus, acts rapidly to neutralize the virus and one such has also been reported with the use of IgY antibodies against SARS CoV outbreak in china [63], [64], [65].
6. IgY monoclonal antibody production using phage display method
In recent years the use of monoclonal chicken IgY antibodies derived from phage display technology has been widely studied and promising results of producing neutralizing antibodies has been observed by different research groups. It was in 1985 when George smith first used this technology, which used an expression vector, having a capability of presenting the foreign amino acid sequences, which could bind to the antibody [66]. This technology was considered as an alternative to the conventional production of monoclonal antibodies using hybridoma technology. The major advantage of using PDT is that the generation of scFv/ Fab fragments specific to a particular antigen can be performed within few weeks [67]. The immunoglobulin repertoire of chickens is ideally suited for phage display, as it is generated from a single set of VH and VL germ line sequence [68]. The phage display system is considered as a fast and valuable technique for monoclonal antibody production against broad viral infections [69]. In the conventional monoclonal antibody production through hybridoma technology, the lymphocytes are isolated, immortalized by hybridization with myeloma cells which paves a way for in vitro cloning in eukaryotic cell culture system. Whereas in the phage display technology, the prokaryotic system is used for cloning the cells.
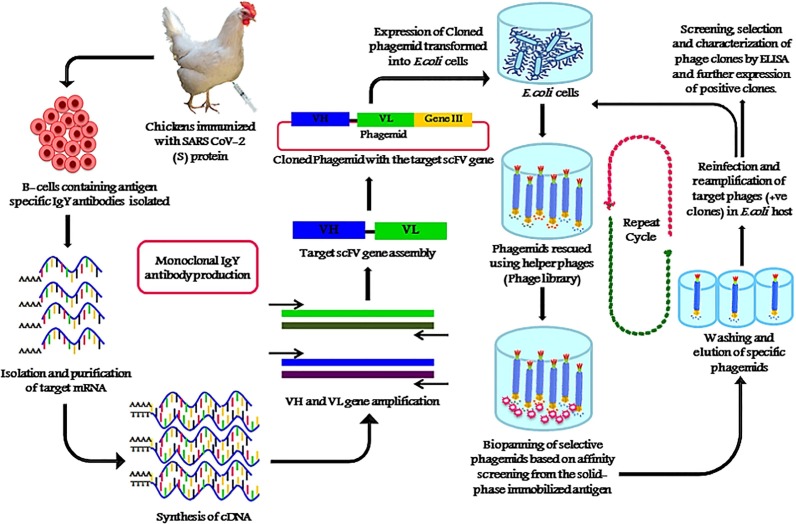
In brief, for phage display technology, first the lymphocytes are being isolated from spleen of the host animal, followed by encoding the DNA of the binding region of immunoglobulins in new expression system. The RNA is prepared from lymphocytes and reverse transcripted into cDNA. The variable regions of immunoglobulin antibody cDNA are amplified by PCR. The DNA is then ligated into suitable vectors, which are then used for transformation of E. coli. The next step, the latter are infected with helper phages. The result is that the bacteria expresses and releases phage particles containing the vector DNA, showing the engineered antibody protein on their surfaces [70] (Fig. 3 ).
Fig. 3.
An overall schematic outline of generation, isolation and screening of anti-SARS CoV-2 monoclonal scFv IgY antibodies against the spike protein (S) of SARS CoV-2 antigen using phage display technology.
The scFv antibody (IgY) library from immune spleen cells of chickens against virulent infectious bursal disease virus (vvIBDV) strain CS89 for detection and differentiation of vvIBDV from other IBDV strains [71]. IgY antibodies against the SARS coronavirus strain (BJ01) has been proven to be effective in neutralizing the virus at an ELISA titre of 1:640 against the particular virus [72]. Development of monoclonal IgY scFv antibodies against the spike (S) protein of SARS-CoV, proved to neutralize the virus under in vitro conditions which confirmed that the epitope (S protein) is a suitable candidate for development of vaccine and for rapid detection of viral infections [65]. It has also been stated that using the biotin-tagged phage conjugated with streptavidin coated quantum dot has the ability to detect as few as 10 cells/ml [65], [73]. IgY antibodies purified from ostrich eggs can aid in development of antibody vaccines for corona virus, SARS- CoV and MERS-CoV [74]. For selection of phage display systems, moreover E. coli filamentous bacteriophages, such as the M13 phages are commonly used [75]. Most commonly, for construction of library, the following steps are widely preferred,
-
i.
Preparation of primary library or amplification of the existing one
-
ii.
Exposing phage particles to target (cell surface proteins/vascular endothelium)
-
iii.
Removing non-specific binders (washing/perfusion)
-
iv.
Recovery of the target bound by elution or direct bacterial infection
-
v.
Amplification of recovered phages.
-
vi.
Repeating step (i) until highest binding population is reached, bio-panning is repeated.
7. Mode of action of monoclonal IgY antibodies
Currently, there are various ongoing clinical trials which involves different drugs leading to the discovery of potential vaccine or the potential drug to treat SARS CoV-2 pandemic which would then lead to the rapid development of vaccine and its application as a powerful means in preventing the global pandemic situation. Inspite of these, vaccine developments are little slower compared to the human transmission of SARS CoV-2 infection there is still an urgent need and necessity for vaccine development [76]. Also, it is clear that the epidemic situation became pandemic and it is appearing to be transformed likely to a flu-like seasonal infection from human to human transmission which is still rapidly co-existing with human hosts for long time.
The above schematic representation explains the mode of action of the chicken monoclonal scFv IgY antibodies developed against SARS CoV-2 spike protein as an immunotherapeutic tool. Currently, the most commonly used recombinant antibody fragments are the single-chain variable (scFv) fragments which is of approximately 25 kDa in size. These scFv antibody fragments are composed of VL and VH domains of light and heavy chains of the full length antibody molecule and are linked to specific linkers. Both these domains can be generated against the target protein to develop scFv antibody molecules to recognize the specific antigens. Also, the smaller size of the scFv antibody molecules holds greater advantage in higher yielding of specific antibodies which can be successfully attained from expressing in the host vectors like E. coli and the antibody affinity can be enhanced to target the antigen by multiple cycles of screening process and by using random mutagenesis of the CDR regions effectively. Further, these repeated phage selection screening cycles improves the selection of positive antibody scFv fragments and the coupling of various selection strategies during in vitro screening of phages containing positive clones of antibodies will be a powerful method to tailor the scFv monoclonal IgY antibodies fragment targeting the specific antigen of interest in this perspectives. Hence, recombinant antibody production of monoclonal antibodies involved with variable fragment genes (VH/VL) obtained by PCR amplification from the B-cell repetoires are assembled here forming a full length single chain variable antibody fragment (scFv) [Fig. 3].
These scFv regions are cloned and expressed into vectors (E. coli) for generating antigen-specific IgY antibodies expressing the antibody library and it is constructed [77], [78], [79] which ensures that the antibody library constructs are not diversified during amplification [80], [81]. It has also been reported thar recent studies with production of IgY monoclonal antibodies designed based on ezymatic methods for assembling scFv IgY antibodies [82]. This method of chicken IgY monoclonal antibody production against SARS CoV-2 spike protein (S) would be more standardized, reproducible and suitable for large scale production of antigen specific antibodies (anti-SARS CoV-2 IgY antibodies) which inhibits the antigen (SARS CoV-2) to the binding receptor (ACE2) present on the human cell membrane and thereby leading to prevention of viral entry or replication in the host cell effectively (Fig. 4 ). Additionally this technology offers lesser methodological variabilities when compared to that of bacterial display technology [83] and also ensures that the expressed scFv antibody targets are structurally and functionally correct with higher capability of target antigen binding for effective neutralization [71]. Currently, immune and naïve derived antibody libraries from avian sources using phage display method can be easily established and the distinctive immunological benefits of chicken antibodies enables development of highly specific unique antibodies creating an efficient immunotherapeutic and diagnostic tool that allows the capture of distinct conformational structural changes of immunoglobulin molecules pertaining to their protein structures.
Fig. 4.
Diagrammatic representation of mode of action of anti-SARS CoV-2 scFv IgY monoclonal antibodies which inhibits the attachment check points of the virus to the ACE2 receptors present on the cell membrane and prevents the entry into the cell.
8. Clinical perspectives and potential use of monoclonal IgY antibodies
Development of chicken monoclonal IgY antibodies using phage display technology for its promising applications in diagnosis and therapeutic use widely to treat various pathogenic infections especially viral diseases like the current pandemic SARS CoV-2 infection. Still, there is need for numerous areas of research required for the development and use of high-affinity monoclonal IgY antibodies. Immunization of these developed antibodies can be performed effectively to reduce the immunogenecity of the developed antigen-specific monoclonal IgY antibodies against SARS CoV-2 as a parameter that should be considered in using these antibodies in use as immunodiagnostic and immunotherapeutic mode of treatments and humanization of monoclonal IgY antibodies has been reported to be successful tool as immunotherapeutic administration [84], [85]. Additionally, various studies has been reported that the use of chimeric scFv chicken IgY antibodies can be extended further to other mammalian species in veterinary applications and the more recent use of chicken IgY monoclonal antibodies seems to have more potential in immunotherapy [86], [87]. Hence, molecular characterization also allows researchers to determine and correlate target specific parameters of scFv IgY genes in relation to its properties like thermal stability, aggregation capacity, solubility, binding efficacy and its expression stability [88], [89]. The residual changes of scFv IgY antibodies facilitates the bioconjugation chemistries of chicken scFv IgY immunoglobulin when it reacts with certain chemicals like chelating agents and prodrugs used in certain immunotherapeutic treatments [90]. Therefore, these approaches and reported studies reinforces that the need of future research in contribution to better understanding and knowledge of chicken IgY monoclonal antibodies against specific antigens like SARS CoV-2 explained here can be developed further from the molecular level to the large scale production of antibodies with their effective applications thereof effectively.
It has been proven that antibody receptors that are specifically engineered to a target antigen with combined specific sites were used in human therapy and reported to be effective with these single-chain antigen binding molecules [91]. Also, chicken immunoglobulin fragments like chFcR/L receptor molecules were identified to be the first member of this avian antibody family which plays a vital role in mammalian immune system that makes it a significant and important tool in immunoregulatory family and additionally exploring these specific mechanisms would be certainly contributing a new perspective on antibody small molecule receptor interactions [92]. Further analysis provides evidences that some of the major shifts in the mode of Fc, scFv receptor binding regions during immunoglobulin evolutions undergoes various structural changes which may lead to develop novel functions during the evolution of immunoglobulins in mammals [93].
The development of chicken IgY monoclonal antibodies using phage display method against various target pathogens have been reported to be effective in enriching the target specific scFv IgY antibodies from the phage displayed antibody libraries [93]. Also, it is noted that the feasibility and effectiveness in generating highly specific scFv IgY monoclonal antibodies using phage display method in chickens is significantly potential and the ability in generating mass production of these highly specific monoclonal scFv IgY antibodies will facilitate the investigations of their ability to quantitative and qualitative response to the target pathogen contributing to be a valuable tool in immunotherapeuti applications [94]. Further to this, these IgY antibodies were even developed against snake venoms which has the potential to replace equine antibodies because they do not cause any adverse effects upon therapeutic administration [95]. Recently, it has also been reported that anti-ZIKV-E IgY antibodies has been a potential target for development of vaccine and immunodiagnostic assays which were derived from the antibody libraries and these scFv antibody fragments effectively played a vital role in inhibition and detection assays [96], [97].
9. Conclusion and future perspectives
To face the current global pandemic situation of SARS CoV-2, it is the need of the hour to have better diagnostic and therapeutic modes for overcoming this health problem. Hence, production of highly specific monoclonal antibodies using chicken egg yolk antibodies (IgY) by phage display method against SARS CoV-2 will be an effective platform which holds much effective clinical and scientific advantages [98], [15], [99]. The use of monoclonal IgY antibodies promises the need for effective and continuous research to improve further in various aspects and development of monoclonal IgY antibodies using phage display system will combine the benefits of both mammalian monoclonal antibodies (IgG) and avain antibodies (IgY). Also chicken monoclonal IgY antibodies proved to be more specific when compared to polyclonal antibodies in recognizing a single unique epitope. Thus, the identified epitope of SARS CoV-2 spike protein (S) will be an effective potential candidate for antibody based vaccine development as immunotherapeutic agent in preventing and treating the infection and thereby playing a vital role in both scientific and clinical applications. It is foreseen that chicken scFv IgY antibodies developed against SARS CoV-2 spike protein (S) using phage display technology can be a potential model for effective mass production of high-affinity monoclonal IgY antibodies to treat this disease with standardized preparation for its effective use in long term.
Author contributions
All authors contributed equally and have read and approved the complete manuscript.
Funding and conflict of interest
There are no funding support and no conflict of interest to report.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.intimp.2020.106654.
Contributor Information
Rajeswari Somasundaram, Email: rajeswaris69@yahoo.com.
Ankit Choraria, Email: ankitchoraria91@gmail.com.
Michael Antonysamy, Email: amichela2000@gmail.com.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Peiris J.S.M., Lai S.T., Poon L.L.M., Guan Y., Yam L.Y.C., Lim W., Nicholls J. Coronavirus as a possible cause of severe acute respiratory syndrome. The Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drosten Christian, Günther Stephan, Preiser Wolfgang, Van Der Werf Sylvie, Brodt Hans-Reinhard, Becker Stephan, Rabenau Holger. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. New England J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 3.Ksiazek Thomas G., Erdman Dean, Goldsmith Cynthia S., Zaki Sherif R., Peret Teresa, Emery Shannon, Tong Suxiang. A novel coronavirus associated with severe acute respiratory syndrome. New England J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 4.Peigang Wang, Jian Chen, Aihua Zheng, Yuchun Nie, Xuanling Shi, Wei Wang, Guangwen Wang, et al., Expression cloning of functional receptor used by SARS coronavirus, Biochem. Biophys. Res. Commun. 315(2) (2004) 439–444. https://doi.org/10.1016/j.bbrc.2004.01.076. [DOI] [PMC free article] [PubMed]
- 5.Tripet Brian, Howard Megan W., Jobling Michael, Holmes Randall K., Holmes Kathryn V., Hodges Robert S. Structural characterization of the SARS-coronavirus spike S fusion protein core. J. Biol. Chem. 2004;279(20):20836–20849. doi: 10.1074/jbc.M400759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Wenhui, Moore Michael J., Vasilieva Natalya, Sui Jianhua, Wong Swee Kee, Berne Michael A., Somasundaran Mohan. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Zhi-yong, Kong Wing-pui, Huang Yue, Roberts Anjeanette, Murphy Brian R., Subbarao Kanta, Nabel Gary J. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428(6982):561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bisht Himani, Roberts Anjeanette, Vogel Leatrice, Bukreyev Alexander, Collins Peter L., Murphy Brian R., Subbarao Kanta, Moss Bernard. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. 2004;101(17):6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groves D.J., Morris B.A. Veterinary sources of nonrodent monoclonal antibodies: interspecific and intraspecific hybridomas. Hybridoma. 2000;19(3):201–214. doi: 10.1089/02724570050109602. [DOI] [PubMed] [Google Scholar]
- 10.Carlos F. Barbas, Angray S. Kang, Richard A. Lerner, Stephen J. Benkovic, Assembly of combinatorial antibody libraries on phage surfaces: the gene III site, Proc. Natl. Acad. Sci. 88(18) (1991) 7978–7982. https://doi.org/10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed]
- 11.Winter Greg, Griffiths Andrew D., Hawkins Robert E., Hoogenboom Hennie R. Making antibodies by phage display technology. Annu. Rev. Immunol. 1994;12(1):433–455. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- 12.Chi X. Sherry, Landt Yvonne, Crimmins Dan L., Dieckgraefe Brian K., Ladenson Jack H. Isolation and characterization of rabbit single chain antibodies to human Reg Iα protein. J. Immunol. Meth. 2002;266(1–2):197–207. doi: 10.1016/S0022-1759(02)00117-5. [DOI] [PubMed] [Google Scholar]
- 13.Wang Shi-Hua, Zhang Ji-Bin, Zhang Zhi-Ping, Zhou Ya-Feng, Yang Rui-Fu, Chen Jia, Guo Yong-Chao, You Fan, Zhang Xian-En. Construction of single chain variable fragment (ScFv) and BiscFv-alkaline phosphatase fusion protein for detection of Bacillus Anthracis. Anal. Chem. 2006;78(4):997–1004. doi: 10.1021/ac0512352. [DOI] [PubMed] [Google Scholar]
- 14.Pavoni Emiliano, Flego Michela, Dupuis Maria Luisa, Barca Stefano, Petronzelli Fiorella, Anastasi Anna Maria, D'Alessio Valeria. Selection, affinity maturation, and characterization of a human scFv antibody against CEA protein. BMC Cancer. 2006;6(1):41. doi: 10.1186/1471-2407-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kyung Je Park, Dong Woon Park, Chun Hee Kim, Beom Ku Han, Tae Sub Park, Jae Yong Han, Hyun Soon Lillehoj, Jin-Kyoo Kim, Development and characterization of a recombinant chicken single-chain Fv antibody detecting Eimeria acervulina sporozoite antigen, Biotechnol. Lett. 27(5) (2005) 289–295. https://doi.org/10.1007/s10529-005-0682-8. [DOI] [PubMed]
- 16.Finlay William J.J., Shaw Iain, Reilly Joanna P., Kane Marian. Generation of high-affinity chicken single-chain Fv antibody fragments for measurement of the Pseudonitzschia pungens toxin domoic acid. Appl. Environ. Microbiol. 2006;72(5):3343–3349. doi: 10.1128/AEM.72.5.3343-3349.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sui Jianhua, Li Wenhui, Murakami Akikazu, Tamin Azaibi, Matthews Leslie J., Wong Swee Kee, Moore Michael J. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. 2004;101(8):2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang Xiaoping, Yang Bao-an, Hu Yuyang, Zhao Hui, Xiong Wei, Yang Yinhui, Si Bingyin, Zhu Qingyu. Human neutralizing Fab molecules against severe acute respiratory syndrome coronavirus generated by phage display. Clin. Vaccine Immunol. 2006;13(8):953–957. doi: 10.1128/CVI.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lomniczi B. Biological properties of avian coronavirus RNA. J. General Virol. 1977;36(3):531–533. doi: 10.1099/0022-1317-36-3-531. [DOI] [PubMed] [Google Scholar]
- 20.Bond Clifford W., Leibowitz Julian L., Robb James A. Pathogenic murine coronaviruses II. Characterization of virus-specific proteins of murine coronaviruses JHMV and A59V. Virology. 1979;94(2):371–384. doi: 10.1016/0042-6822(79)90468-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maeda Junko, Repass John F., Maeda Akihiko, Makino Shinji. Membrane topology of coronavirus E protein. Virology. 2001;281(2):163–169. doi: 10.1006/viro.2001.0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.David A. Brian, Brenda G. Hogue, Thomas E. Kienzle, The coronavirus hemagglutinin esterase glycoprotein, in: The Coronaviridae, Springer, Boston, MA, 1995, pp. 165–179. https://doi.org/10.1007/978-1-4899-1531-3_8.
- 23.An Sungwhan, Chen Chun-Jen, Yu Xin, Leibowitz Julian L., Makino Shinji. Induction of apoptosis in murine coronavirus-infected cultured cells and demonstration of E protein as an apoptosis inducer. J. Virol. 1999;73(9):7853–7859. doi: 10.1128/JVI.73.9.7853-7859.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arden Katherine E., Nissen Michael D., Sloots Theo P., Mackay Ian M. New human coronavirus, HCoV-NL63, associated with severe lower respiratory tract disease in Australia. J. Med. Virol. 2005;75(3):455–462. doi: 10.1002/jmv.20288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D.A. Brian, R.S. Baric, Coronavirus genome structure and replication, in: Coronavirus replication and reverse genetics, Springer, Berlin, Heidelberg, 2005, pp. 1–30. https://doi.org/10.1007/3-540-26765-4_1.
- 26.Bredenbeek Peter J., Pachuk Catherine J., Noten Ans F.H., Charité Jeroen, Luytjes Willem, Weiss Susan R., Spaan Willy J.M. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucl. Acids Res. 1990;18(7):1825–1832. doi: 10.1093/nar/18.7.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tortorici M. Alejandra, Veesler David. Structural insights into coronavirus entry. Adv. Virus Res. 2019;105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Xiaobo, Yu Yuan, Xu Jiqian, Shu Huaqing, Liu Hong, Wu Yongran, Zhang Lu. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respirat Med. 2020 doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li-Li Ren, Ye-Ming Wang, Zhi-Qiang Wu, Zi-Chun Xiang, Li Guo, Teng Xu, Yong-Zhong Jiang, et al., Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study, Chinese Med. J. (2020) https://doi.org/10.1097/CM9.0000000000000722. [DOI] [PMC free article] [PubMed]
- 30.Kirchdoerfer Robert N., Wang Nianshuang, Pallesen Jesper, Wrapp Daniel, Turner Hannah L., Cottrell Christopher A., Corbett Kizzmekia S., Graham Barney S., McLellan Jason S., Ward Andrew B. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018;8(1):1–11. doi: 10.1038/s41598-018-34171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song Wenfei, Gui Miao, Wang Xinquan, Xiang Ye. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathogens. 2018;14(8) doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ge Xing-Yi, Li Jia-Lu, Yang Xing-Lou, Chmura Aleksei A., Zhu Guangjian, Epstein Jonathan H., Mazet Jonna K. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Peng, Yang Xing-Lou, Wang Xian-Guang, Ben Hu., Zhang Lei, Zhang Wei, Si Hao-Rui. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Millet Jean Kaoru, Whittaker Gary R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park Jung-Eun, Li Kun, Barlan Arlene, Fehr Anthony R., Perlman Stanley, McCray Paul B., Gallagher Tom. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc. Natl. Acad. Sci. 2016;113(43):12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walls Alexandra C., Alejandra Tortorici M., Snijder Joost, Xiong Xiaoli, Bosch Berend-Jan, Rey Felix A., Veesler David. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. 2017;114(42):11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walls Alexandra C., Xiong Xiaoli, Young-Jun Park M., Tortorici Alejandra, Snijder Joost, Quispe Joel, Cameroni Elisabetta. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176(5):1026–1039. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong Xiaoli, Alejandra Tortorici M., Snijder Joost, Yoshioka Craig, Walls Alexandra C., Li Wentao, McGuire Andrew T., Rey Félix A., Bosch Berend-Jan, Veesler David. Glycan shield and fusion activation of a deltacoronavirus spike glycoprotein fine-tuned for enteric infections. J. Virol. 2018;92(4):e01628–e1717. doi: 10.1128/JVI.01628-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walls Alexandra C., Young-Jun Park M., Tortorici Alejandra, Wall Abigail, McGuire Andrew T., Veesler David. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ou Xiuyuan, Liu Yan, Lei Xiaobo, Li Pei, Mi Dan, Ren Lili, Guo Li, Guo Ruixuan, Chen Ting, Hu Jiaxin, Xiang Zichun, Mu Zhixia, Chen Xing, Chen Jieyong, Hu Keping, Jin Qi, Wang Jianwei, Qian Zhaohui. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11(1) doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Hong, Wang Guangwen, Li Jian, Nie Yuchun, Shi Xuanling, Lian Gewei, Wang Wei. Identification of an antigenic determinant on the S2 domain of the severe acute respiratory syndrome coronavirus spike glycoprotein capable of inducing neutralizing antibodies. J. Virol. 2004;78(13):6938–6945. doi: 10.1128/JVI.78.13.6938-6945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tian Xiaolong, Li Cheng, Huang Ailing, Xia Shuai, Lu Sicong, Shi Zhengli, Lu Lu, Jiang Shibo, Yang Zhenlin, Wu Yanling, Ying Tianlei. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microb. Infect. 2020;9(1):382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wrapp Daniel, Wang Nianshuang, Corbett Kizzmekia S., Goldsmith Jory A., Hsieh Ching-Lin, Abiona Olubukola, Graham Barney S., McLellan Jason S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rockx Barry, Corti Davide, Donaldson Eric, Sheahan Timothy, Stadler Konrad, Lanzavecchia Antonio, Baric Ralph. Structural basis for potent cross-neutralizing human monoclonal antibody protection against lethal human and zoonotic severe acute respiratory syndrome coronavirus challenge. J. Virol. 2008;82(7):3220–3235. doi: 10.1128/JVI.02377-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rockx Barry, Donaldson Eric, Frieman Matthew, Sheahan Timothy, Corti Davide, Lanzavecchia Antonio, Baric Ralph S. Escape from human monoclonal antibody neutralization affects in vitro and in vivo fitness of severe acute respiratory syndrome coronavirus. J. Infect. Dis. 2010;201(6):946–955. doi: 10.1086/651022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee Warren, Syed Atif Ali, Tan Soo Choon, Leow Chiuan Herng. Insights into the chicken IgY with emphasis on the generation and applications of chicken recombinant monoclonal antibodies. J. Immunol. Meth. 2017;447:71–85. doi: 10.1016/j.jim.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 47.Pereira E.P.V., van Tilburg M.F., Florean E.O.P.T., Guedes M.I.F. Egg yolk antibodies (IgY) and their applications in human and veterinary health: a review. Int. Immunopharmacol. 2019;73:293–303. doi: 10.1016/j.intimp.2019.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aymn Talat Abbas, Sherif Aly El-Kafrawy, Sayed Sartaj Sohrab, Esam Ibraheem Ahmed Azhar, IgY antibodies for the immunoprophylaxis and therapy of respiratory infections, Human Vacc. Immunotherap. 15(1) (2019) 264–275. https://doi.org/10.1080/21645515.2018.1514224. [DOI] [PMC free article] [PubMed]
- 49.Lesnierowski Grzegorz, Stangierski Jerzy. What's new in chicken egg research and technology for human health promotion?-a review. Trends Food Sci. Technol. 2018;71:46–51. doi: 10.1016/j.tifs.2017.10.022. [DOI] [Google Scholar]
- 50.Klemperer Felix. About natural immunity and its use for immunization therapy. Arch. Exp. Pathol. Pharmacol. 1893;31(4–5):356–382. doi: 10.1007/BF01832882. [DOI] [Google Scholar]
- 51.Leslie Gerrie A., Clem L.W. Phylogeny of immunoglobulin structure and function: III. Immunoglobulins of the chicken. J. Exp. Med. 1969;130(6):1337–1352. doi: 10.1084/jem.130.6.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.S. Rajeswari, Ankit Choraria, Michael Antonysamy, Xiao-Ying Zhang, Applications of chicken egg yolk antibodies (Igy) in healthcare-a review, Biomed. J. Sci. Tech. Res. 2(1) (2018) 2161–2163. https://doi.org/10.26717/BJSTR.2018.02.000649.
- 53.William J.J. Finlay, Laird Bloom, Joanne Grant, Edward Franklin, Deirdre Ní Shúilleabháin, Orla Cunningham, Phage display: a powerful technology for the generation of high-specificity affinity reagents from alternative immune sources, in: Protein Chromatography, Humana Press, New York, NY, 2017, pp. 85–99. https://doi.org/10.1007/978-1-4939-6412-3_6. [DOI] [PMC free article] [PubMed]
- 54.A. Alvarez, Y. Montero, P. Parrilla, C. Malave, N. Zerpa, Poultry IgY Alternatives to Antivenom Production, Springer, Dordrecht, 2013. https://doi.org/10.1007/978-94-007-6647-1_1-1.
- 55.Thirumalai Diraviyam, Senthil Visaga Ambi, Ricardo S. Vieira-Pires, Zhang Xiaoying, Saravanan Sekaran, Umamaheswari Krishnan, Chicken egg yolk antibody (IgY) as diagnostics and therapeutics in parasitic infections–a review, Int. J. Biol. Macromol. (2019) https://doi.org/10.1016/j.ijbiomac.2019.06.118. [DOI] [PubMed]
- 56.Shofiqur Rahman, Sa Van Nguyen, Faustino C. Icatlo Jr, Kouji Umeda, Yoshikatsu Kodama, Oral passive IgY-based immunotherapeutics: a novel solution for prevention and treatment of alimentary tract diseases, Human Vacc. Immunotherap. 9(5) (2013) 1039–1048. https://doi.org/10.4161/hv.23383. [DOI] [PMC free article] [PubMed]
- 57.Bossy-Wetzel Ella, Schwarzenbacher Robert, Lipton Stuart A. Molecular pathways to neurodegeneration. Nat. Med. 2004;10(7):S2–S9. doi: 10.1038/nm1067. [DOI] [PubMed] [Google Scholar]
- 58.Harley Carol, Vieira-Pires Ricardo S. Antibody fragment technology and avian IgY antibodies: a powerful combination. Drug Target Rev. 2016;3(1):62–66. [Google Scholar]
- 59.Parvari Ruti, Avivi Aaron, Frida Lentner E., Ziv S., Tel-Or Y. Burstein, Schechter I. Chicken immunoglobulin gamma-heavy chains: limited VH gene repertoire, combinatorial diversification by D gene segments and evolution of the heavy chain locus. EMBO J. 1988;7(3):739–744. doi: 10.1002/j.1460-2075.1988.tb02870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kovacs-Nolan Jennifer, Mine Yoshinori. Egg yolk antibodies for passive immunity. Annu. Rev. Food Sci. Technol. 2012;3:163–182. doi: 10.1146/annurev-food-022811-101137. [DOI] [PubMed] [Google Scholar]
- 61.Müller Sandra, Schubert Andreas, Zajac Julia, Dyck Terry, Oelkrug Christopher. IgY antibodies in human nutrition for disease prevention. Nutr. J. 2015;14(1):109. doi: 10.1186/s12937-015-0067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Casadevall Arturo, Scharff Matthew D. Return to the past: the case for antibody-based therapies in infectious diseases. Clin. Infect. Dis. 1995;21(1):150–161. doi: 10.1093/clinids/21.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chalghoumi Raja, Marcq Christopher, Thewis André, Portetelle Daniel, Beckers Yves. Effects of feed supplementation with specific hen egg yolk antibody (immunoglobin Y) on Salmonella species cecal colonization and growth performances of challenged broiler chickens. Poultry Sci. 2009;88(10):2081–2092. doi: 10.3382/ps.2009-00173. [DOI] [PubMed] [Google Scholar]
- 64.Gadde U., Rathinam T., Lillehoj Hyun S. Passive immunization with hyperimmune egg-yolk IgY as prophylaxis and therapy for poultry diseases–a review. Animal Health Res. Rev. 2015;16(2):163–176. doi: 10.1017/S1466252315000195. [DOI] [PubMed] [Google Scholar]
- 65.Fu Chao-Yang, Huang He, Wang Xiao-Mei, Liu Yong-Gang, Wang Zhi-Guo, Cui Shang-Jin, Gao Hong-Lei, Li Zan, Li Jing-Peng, Kong Xian-Gang. Preparation and evaluation of anti-SARS coronavirus IgY from yolks of immunized SPF chickens. J. Virol. Meth. 2006;133(1):112–115. doi: 10.1016/j.jviromet.2005.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perlman Stanley, Netland Jason. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arap Marco Antonio. Phage display technology: applications and innovations. Genet. Mol. Biol. 2005;28(1):1–9. doi: 10.1590/S1415-47572005000100001. [DOI] [Google Scholar]
- 68.Carmen Sara, Jermutus Lutz. Concepts in antibody phage display. Briefings Funct. Genom. 2002;1(2):189–203. doi: 10.1093/bfgp/1.2.189. [DOI] [PubMed] [Google Scholar]
- 69.William J.J. Finlay, Laird Bloom, Orla Cunningham, Phage display: a powerful technology for the generation of high specificity affinity reagents from alternative immune sources, in: Protein Chromatography, Humana Press, 2011, pp. 87–101. https://doi.org/10.1007/978-1-60761-913-0_6. [DOI] [PMC free article] [PubMed]
- 70.Cortay Jean-Claude, Gerlier Denis, Iseni Frédéric. Selection of single-chain antibodies that specifically interact with vesicular stomatitis virus (VSV) nucleocapsid and inhibit viral RNA synthesis. J. Virol. Meth. 2006;131(1):16–20. doi: 10.1016/j.jviromet.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 71.Schmitz U., Versmold A., Kaufmann P., Frank H.-G. Phage display: a molecular tool for the generation of antibodies—a Review. Placenta. 2000;21:S106–S112. doi: 10.1053/plac.1999.0511. [DOI] [PubMed] [Google Scholar]
- 72.Sapats Sandra I., Trinidad L., Gould G., Heine H.G., Van den Berg T.P., Eterradossi N., Jackwood D., Parede L., Toquin D., Ignjatovic J. Chicken recombinant antibodies specific for very virulent infectious bursal disease virus. Arch. Virol. 2006;151(8):1551–1566. doi: 10.1007/s00705-006-0729-8. [DOI] [PubMed] [Google Scholar]
- 73.Yu-Ching Lee, Sy-Jye C. Leu, Han-Chang Hung, Hsueh-Hsia Wu, I-Jen Huang, Wen-Shyang Hsieh, Wen-Ta Chiu, Ming-Song Hsieh, Tsui-Fen Cheng, Yi-Yuan Yang, A dominant antigenic epitope on SARS-CoV spike protein identified by an avian single-chain variable fragment (scFv)-expressing phage, Veterinary Immunol. Immunopathol. 117(1-2) (2007) 75–85. https://doi.org/10.1016/j.vetimm.2007.02.001. [DOI] [PMC free article] [PubMed]
- 74.Yu-Ching Lee, Sy-Jye C. Leu, Chaur-Jong Hu, Neng-Yao Shih, I-Jen Huang, Hsueh-Hsia Wu, Wen-Shyang Hsieh, Bor-Luen Chiang, Wen-Ta Chiu, Yi-Yuan Yang, Chicken single-chain variable fragments against the SARS-CoV spike protein, J. Virol. Meth. 146(1-2) (2007) 104–111. https://doi.org/10.1016/j.jviromet.2007.06.010. [DOI] [PMC free article] [PubMed]
- 75.Tsukamoto Yasuhiro, Nakano Yuna, Adachi Kazuhide. Protection against infectious bronchitis virus, a corona virus infection, using ostrich antibodies. Health. 2018;10(10):1294–1308. doi: 10.4236/health.2018.1010100. [DOI] [Google Scholar]
- 76.Bazan Justyna, Całkosiński Ireneusz, Gamian Andrzej. Phage display—a powerful technique for immunotherapy: 1. Introduction and potential of therapeutic applications. Human Vacc. Immunotherap. 2012;8(12):1817–1828. doi: 10.4161/hv.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Neher Richard A., Dyrdak Robert, Druelle Valentin, Hodcroft Emma B., Albert Jan. Impact of seasonal forcing on a potential SARS-CoV-2 pandemic. medRxiv. 2020 doi: 10.1101/2020.02.13.20022806. [DOI] [PubMed] [Google Scholar]
- 78.Finlay W.J.J., DeVore N.C., Dobrovolskaia E.N., Gam A., Goodyear C.S., Slater J.E. Exploiting the avian immunoglobulin system to simplify the generation of recombinant antibodies to allergenic proteins. Clin. Exp. Allergy. 2005;35(8):1040–1048. doi: 10.1111/j.1365-2222.2005.02307. [DOI] [PubMed] [Google Scholar]
- 79.Shin Iwamoto, Norihisa Nishimichi, Yoshiko Tateishi, Yuko Sato, Hiroyuki Horiuchi, Shuichi Furusawa, Tatsuya Sawamura, Haruo Matsuda, Generation and characterization of chicken monoclonal antibodies against human LOX-1, in: MAbs, vol. 1, no. 4, Taylor & Francis, 2009, pp. 357–363. https://doi.org/10.4161/mabs.1.4.8919.x. [DOI] [PMC free article] [PubMed]
- 80.Hof D., Hoeke M.O., Raats J.M.H. Multiple-antigen immunization of chickens facilitates the generation of recombinant antibodies to autoantigens. Clin. Exp. Immunol. 2008;151(2):367–377. doi: 10.1111/j.1365-2249.2007.03569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pansri Potjamas, Jaruseranee Nanthnit, Rangnoi Kuntalee, Kristensen Peter, Yamabhai Montarop. A compact phage display human scFv library for selection of antibodies to a wide variety of antigens. BMC Biotechnol. 2009;9(1):6. doi: 10.1186/1472-6750-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Green Stefan J., Venkatramanan Raghavee, Naqib Ankur. Deconstructing the polymerase chain reaction: understanding and correcting bias associated with primer degeneracies and primer-template mismatches. PloS one. 2015;10(5) doi: 10.1371/journal.pone.0128122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kato Mieko, Hanyu Yoshiro. Construction of an scFv library by enzymatic assembly of VL and VH genes. J. Immunol. Meth. 2013;396(1–2):15–22. doi: 10.1016/j.jim.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 84.Daugherty Patrick S. Protein engineering with bacterial display. Curr. Opin. Struct. Biol. 2007;17(4):474–480. doi: 10.1016/j.sbi.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 85.Nishibori Nahoko, Horiuchi Hiroyuki, Furusawa Shuichi, Matsuda Haruo. Humanization of chicken monoclonal antibody using phage-display system. Mol. Immunol. 2006;43(6):634–642. doi: 10.1016/j.molimm.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 86.Tsurushita Naoya, Park Minha, Pakabunto Kanokwan, Ong Kelly, Avdalovic Anamarija, Fu Helen, Jia Audrey, Vásquez Max, Kumar Shankar. Humanization of a chicken anti-IL-12 monoclonal antibody. J. Immunol. Meth. 2004;295(1–2):9–19. doi: 10.1016/j.jim.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 87.Schusser Benjamin, Yi Henry, Collarini Ellen J., Izquierdo Shelley Mettler, Harriman William D., Etches Robert J., Leighton Philip A. Harnessing gene conversion in chicken B cells to create a human antibody sequence repertoire. PLoS One. 2013;8:11. doi: 10.1371/journal.pone.0080108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Leighton Philip A., Schusser Benjamin, Yi Henry, Glanville Jacob, Harriman William. A diverse repertoire of human immunoglobulin variable genes in a chicken B cell line is generated by both gene conversion and somatic hypermutation. Front. Immunol. 2015;6:126. doi: 10.3389/fimmu.2015.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Leeying Wu, Katarzyna Oficjalska, Matthew Lambert, Brian J. Fennell, Alfredo Darmanin-Sheehan, Deirdre Ní Shúilleabháin, Bénédicte Autin, et al., Fundamental characteristics of the immunoglobulin VH repertoire of chickens in comparison with those of humans, mice, and camelids, J. Immunol. 188(1) (2012) 322–333. https://doi.org/10.4049/jimmunol.1102466. [DOI] [PubMed]
- 90.Paul J. Conroy, Ruby H.P. Law, Sarah Gilgunn, Stephen Hearty, Tom T. Caradoc-Davies, Gordon Lloyd, Richard J. O'Kennedy, James C. Whisstock, Reconciling the structural attributes of avian antibodies, J. Biol. Chem. 289(22) (2014) 15384–15392. https://doi.org/10.1074/jbc.M114.562470. [DOI] [PMC free article] [PubMed]
- 91.Aerin Yoon, Jung Won Shin, Soohyun Kim, Hyori Kim, Junho Chung, Chicken scFvs with an artificial cysteine for site-directed conjugation,“PloS one 11(1) (2016) https://dx.doi.org/10.1371%2Fjournal.pone.0146907. [DOI] [PMC free article] [PubMed]
- 92.Sutton B.J. Immunoglobulin structure and function: the interaction between antibody and antigen. Curr. Opin. Immunol. 1989;2(1):106–113. doi: 10.1016/0952-7915(89)90105-2. [DOI] [PubMed] [Google Scholar]
- 93.Taylor Alexander I., Beavil Rebecca L., Sutton Brian J., Calvert Rosaleen A. A monomeric chicken IgY receptor binds IgY with 2: 1 stoichiometry. J. Biol. Chem. 2009;284(36):24168–24175. doi: 10.1074/jbc.M109.020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Taylor Alexander I., Sutton Brian J., Calvert Rosaleen A. Mutations in an avian IgY-Fc fragment reveal the locations of monocyte Fc receptor binding sites. Develop. Comp. Immunol. 2010;34(2):97–101. doi: 10.1016/j.dci.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen Hong-Xiu, He Fan, Sun Yuan, Luo Yuzi, Qiu Hua-Ji, Zhang Xiao-Ying, Sutton Brian J. Generation and characterization of chicken-sourced single-chain variable fragments (scFvs) against porcine interferon-gamma (pIFN-γ) J. Immunoassay Immunochem. 2015;36(1):27–44. doi: 10.1080/15321819.2014.892511. [DOI] [PubMed] [Google Scholar]
- 96.Choraria Ankit, Somasundaram Rajeswari, Gautam Mrinmoy, Ramanathan Muthiah, Paray Bilal Ahmad, Al-Sadoon Mohammad K., Michael A. Experimental antivenoms from chickens and rabbits and their comparison with commercially available equine antivenom against the venoms of Daboia russelii and Echis carinatus snakes. Toxin Rev. 2020:1–12. doi: 10.1080/15569543.2020.1756858. [DOI] [Google Scholar]
- 97.Pharaoh Fellow Mwale, Chi-Hsin Lee, Liang-Tzung Lin, Sy-Jye Leu, Yun-Ju Huang, Liao-Chun Chiang, Yan-Chiao Mao, Yi-Yuan Yang, Expression, purification, and characterization of anti-Zika virus envelope protein: polyclonal and chicken-derived single chain variable fragment antibodies, Int. J. Mol. Sci. 21(2) (2020) 492. https://doi.org/10.3390/ijms21020492. [DOI] [PMC free article] [PubMed]
- 98.Palaniyappan A., Das D., Kammila S., Suresh M.R., Sunwoo H.H. Diagnostics of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) nucleocapsid antigen using chicken immunoglobulin Y. Poultry Sci. 2012;91(3):636–642. doi: 10.3382/ps.2011-01916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fehrsen J., Van Wyngaardt W., Mashau C., Potgieter A.C., Chaudhary V.K., Gupta A., Jordaan F.A., Du Plessis D.H. Serogroup-reactive and type-specific detection of bluetongue virus antibodies using chicken scFvs in inhibition ELISAs. J. Virol. Meth. 2005;129(1):31–39. doi: 10.1016/j.jviromet.2005.04.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.