Abstract
The fucosylated carbohydrate moieties on intestinal epithelial cells (IECs) are involved in the creation of an environmental niche for commensal and pathogenic bacteria. Core fucosylation catalyzed by fucosyltransferase 8 (Fut8) is the major fucosylation pattern on the N-glycans of the surface glycoproteins on IECs, however, the role of IECs core fucosylation during infection remains unclear. This study was conducted to investigate the interaction between IECs core fucosylation and gut microbiota, and the effects of this interaction on protecting Salmonella enterica subsp. enterica serovar Typhi (S. Typhi) infection. Firstly, the Fut8+/+ and Fut8+/– mice were infected with S. Typhi. The level of IECs core fucosylation and protein expression of intestinal mucosa were then detected by LCA blot and Western blot, respectively. The gut microbiota of Fut8+/+ and Fut8+/– mice before and after S. Typhi infection was assessed by 16S rRNA sequencing. Our results showed that core fucosylation was ubiquitous expressed on the intestinal mucosa of mice and had significant effects on their gut microbiota. Fut8+/– mice was more susceptive to S. Typhi infection than Fut8+/+ mice. Interestingly, infection of S. Typhi upregulated the core fucosylation level of IECs and increased the abundances of beneficial microorganisms such as Lactobacillus and Akkermansia spp. Further in vitro and in vivo studies demonstrated that Wnt/β-catenin signaling pathway mediated the elevation of IECs core fucosylation level upon infection of S. Typhi. Taken together, our data in this study revealed that the IECs core fucosylation plays an important role in protecting against S. Typhi infection via up-regulating the biological antagonism of intestinal microbiota.
Keywords: core fucosylation, gut microbiota, S. Typhi infection, Wnt signaling pathway, Lactobacillus
Introduction
Intestinal mucosa creates a basic and physical barrier between the gut microbes and the host tissues. The fucosylation of intestinal epithelial cells (IECs) is closely related to the immune function of intestinal mucosa, and can prevent against pathogen infection, and the development of colitis and colon cancer (Goto et al., 2016; Yu et al., 2019). The core fucosylation catalyzed by fucosyltransferase 8 (Fut8) is the major fucosylation pattern on the N-glycans of the surface glycoproteins on IECs. Fut8 transfers the guanosine diphosphate-fucose (GDP-Fucose) to the sixth carbon atom on the N-acetylglucosamine (GlcNAc) of the N-glycan, forming a α-1, 6-glycosidic bond which is called as core fucosylation (Miyoshi et al., 1999; Calderon et al., 2016). Although many studies support that Fut8 gene knockout has an great impact on the function of glycoproteins which is exerting an enormous function on immune responses, such as cell recognition and information transferation, no study has investigated the role of IECs core fucosylation during infection (Liang et al., 2018).
Intestinal commensal bacteria play an important role in immune system. IECs send signals which are constituted by glycosylation, protein decoration and other signals to mucosal immune cells to make an interaction with commensal bacteria and IECs to establishing intestinal immunological homeostasis (Goto and Kiyono, 2012). The glycosylation of IECs contributes to the resistance to intestinal pathogens, and has a significant impact on shaping commensal bacteria (Hooper et al., 1999). Studies have shown that commensal bacteria are able to utilize fucoses, especially Bacteroides spp., as components of the bacterial outer membrane, or using them as a nutrient (Leis et al., 1997; Coyne et al., 2005). However, the interaction between IECs core fucosylation and gut microbiota, and whether this interaction affects immune responses during infection remain unclear.
The infection of pathogenic Salmonella spp. is a major public health concern all around the world. It’s chronic colonization in human intestine is also a risk factor for the development of colorectal cancer (CRC) and inflammatory bowel disease (IBD) (Gradel et al., 2009; Kato et al., 2013). Salmonella enterica serotype typhi mainly cause typhoid fever, a systemic infection, at least 16 million new cases and 600,000 deaths each year (Maskalyk, 2003). Changes of gut microbiota and signaling pathways of IECs were both found involved in Salmonella spp. infection (Liu et al., 2010; Wang et al., 2018). Among them, the Wnt signaling pathway was found to be involved in Salmonella infection, enhanced accumulation of nuclear β-catenin and in turn activated the transcription of its target genes (Lawhon et al., 2011; Kogut and Arsenault, 2015). Wnt signaling pathway was also found to be correlated with the hyper-expression of core fucosylation in breast cancer through activation of the Fut8 gene transcription (Yang et al., 2017). It was also known to play an important role in the development and differentiation of immune cells, and regulate pathogen-induced inflammation and mucosal tolerance by antigen presenting cells (APCs) (Staal et al., 2008; Maloy and Powrie, 2011; Rescigno, 2011; Swafford et al., 2018). We therefore asked that whether Salmonella infection induce alteration of IECs core fucosylation through activating Wnt signaling pathway, and whether this alteration can affect gut homeostasis.
In this study, we provide the first confirmation that the deficiency of core fucosylation in mice resulted in severe infection when challenged by Salmonella enterica subsp. enterica serovar Typhi (S. Typhi), and this was correlated with changing of gut microbiota and activated Wnt signaling pathway in IECs. The S. Typhi-induced activation of Wnt signaling pathway contributed to the rapid elevation of IECs core fucosylation, which in turn promoted the dominant growth of fucose-utilizing bacteria such as Lactobacillus and Akkermansia, to resist the invasion of S. Typhi. Our study thus suggests that Wnt signaling pathway is involved in the regulation of IECs core fucosylation, which contributes to the protection against S. Typhi infection via up-regulating the biological antagonism of intestinal microbiota.
Materials and Methods
Mice
Fut8+/– mice were generated according to the methods from the studies of Wang et al. (2005) and Li et al. (2006). Wild type Fut8+/+ and heterozygous Fut8+/– mice with the Institute of Cancer Research (ICR) background were maintained in a room illuminated for 12 hours (h) (08:00–20:00) and kept at 24 ± 1°C with free access to food and water in the specific pathogen-free laboratory animal facility of Dalian Medical University, Dalian, China. Animal experiments of S. Typhi-infected mouse model were performed by using specific pathogen-free ICR mice that were 6–7 weeks old. All Fut8+/+ and Fut8+/– mice were obtained by crossing Fut8+/– female and male mice, and they were nurtured by Fut8+/– maternal mice to exclude the influence of maternal milk glycans on gut microbiota of the neonates. Due to the low body weight, low birth rate and high death rate of Fut8–/– mice, we adopted the Fut8+/– mice in this study to establish to the infection models as the growth of these mice are comparable with Fut8+/+ mice.
Mice were infected with the indicated S. Typhi strain by oral gavage at a concentration of 1 × 108 colony-forming units (CFU)/ml, 0.1 ml once a day, for 7 days, and were weighted at 9:00 am everyday. Mice were sacrificed on the morning of day 8 for intestinal tissue, intestinal mucosal and intestinal content collection. Tissue sections were prepared for hematoxylin and eosin (H&E) and immunohistochemical staining. All the animal experiments were conducted according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publication No. 8023).
Antibodies
Anti-Frizzled 7 (ab64636), anti-β-catenin (ab32572), and FITC-labeled goat anti-rabbit IgG (ab6717) were obtained from Abcam; anti-GAPDH (BP009232MO) were obtained from Cusabio; biotinylated lens culinaris agglutinin (LCA) (Z1010) and fluorescein labeled LCA (FL1041) were purchased from Vector; horseradish peroxidase (HRP)-labeled goat anti-mouse IgG (A0218), HRP-labeled goat anti-rabbit IgG (A0208) and HRP-labeled Streptavidin (A0303) were purchased from Beyotime. Andy Fluor 647 streptavidin secondary antibody (SA2K1469A) was purchased from GeneCopoeia.
Fecal DNA Extraction, PCR, and 16S rRNA Amplicon Data Processing
Microbial genome DNA was extracted from fecal samples of mice using E.Z.N.A.® Stool DNA kit (Omega Bio-tek, Inc.) according to the manufacturer’s instructions. A Nanodrop 2000 spectrophotometer was used to evaluate the purity and concentration of isolated DNA. Universal primers 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 518R (5′- ATTACCGCGGCTGCTGG -3′) were used to amplify the V3 hypervariable region of the 16S rRNA from metagenomic DNA in mice feces. The 50-μl PCR mixture contained the following components: 5 μl DNA template, 25 μl PCR 2 × Easy Taq Super mix (HotStarTaq Plus DNA polymerase, dNTPs, MgCl2 and reaction buffer), 1 μl of each primer and 18 μl deionized water. The PCR program consisted of an initial step at 95°C for 5 min; 30 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 60 s; and a final extension at 72°C for 8 min. PCR amplicons were sequenced and the data were analyzed by Illumina MiSeq (Novogene Bioinformatics Technology Co., Ltd., Beijing, China) (Li et al., 2017). The sequencing data were deposited in NCBI SRA under the accession number PRJNA625618.
Bacterial Strains and Growth Condition
The S. Typhi strain used in this study is Salmonella enterica subsp. enterica serovar Typhi CMCC (B) 50071 CICC 10871 strain. The S. Typhi strain was provided by professor Yongliang Yang at Dalian University of Technology, China. The culture was prepared by inoculating 10 ml of Luria–Bertani broth with 10 μl of a stationary-phase culture followed by overnight incubation (∼18 h) at 37°C.
Detection of Intestinal sIgA by ELISA
The content of the mouse small intestine was rinsed with PBS (5 ml) and centrifuged at 10,000 rpm for 10 min, and the supernatant was collected for sIgA detection by ELISA kit (Shanghai Langton Biological Technology Co., Ltd., China) according to the manufacturer’s instructions. The absorbance was measured at 450 nm using a computer-interfaced microplate reader (Bio-Rad, Houston, TX, United States).
Western Blot and Lectin Blot Analysis
Cells were washed with phosphate buffer solution (PBS) and then lysed with ice-cold buffer (50 mM Tris-HCl, 150 mM of NaCl, 1% Triton X-100, 2 mM EDTA, supplemented with 0.1 mM phenylmethylsulfonyl fluoride). The intestinal mucosa of mice was lysed with ice-cold buffer (50 mM Tris-HCl, 150 mM of NaCl, 1% Triton X-100, 2 mM EDTA, supplemented with 0.1 mM phenylmethylsulfonyl fluoride). Total protein concentrations were determined by bicinchoninic acid protein assay (BCA Protein Assay Kit, Pierce, Rockford, IL, United States). 10 μl of proteins was subjected to SDS-PAGE. After SDS-PAGE, the proteins were transferred to polyvinylidene difluoride (PVDF) membranes immunoblot or lectin blot. Following incubation with the appropriate primary antibodies first antibody anti-Frizzled 7, anti-β-catenin, anti-GAPDH or the biotinylated LCA, which preferentially recognizes the core fucose, followed by incubation with HRP-conjugated secondary antibody or HRP-conjugated streptavidin for 1h at room temperature. After washing, the membranes were visualized by chemiluminescence using an ECL kit (Pierce, Rockford, IL, United States).
Immunofluorescence
Formalin-fixed ovarian tissue specimens were paraffin-embedded. For immunohistochemical analysis, the specimens were deparaffinized twice in xylene and hydrated through a graded series of ethanol to PBS. The endogenous peroxidase activity was blocked with 3% H2O2 for 10 min and then in 5% BSA in PBS for 30 min to reduce non-specific background. The specimens were incubated with biotinylated LCA (1:600) or anti-β-catenin (1: 200) for 10 to 12 h at 4°C. Then the specimens were incubated with Andy Fluor 647 streptavidin and FITC-labeled goat anti-rabbit IgG secondary antibody for 1 h at 37°C, and incubated with DAPI (C0060, Solarbio) for 10 min at room temperature. Other specimens were directly incubated with fluorescein labeled LCA for 10 to 12 h at 4°C. Finally, the slides were examined under a fluorescence microscope (Leica, United Kingdom) at a 200× magnification using the appreciate filter. The specimens were analyzed by hematoxylin-eosin (H&E) staining.
Histopathological Evaluation
The specimens were strained with hematoxylin-eosin (H&E) and pathology was quantified as previously described, evaluating submucosal edema, PMN infiltration, goblet cells and epithelial damage yielding a total score of 0–13 points (Hapfelmeier et al., 2008).
Cells and Culture Conditions
Caco-2 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), and maintained at 37°C with 5% CO2 as previously described (Liu et al., 2011).
Cells were inoculated in 6-well plates for 24 h and then replaced. The cells were divided into control group (group 0) (106 cells) and experimental group (group 10, 100, and 200 ng) (106 cells) according to the completely random method. The group 10 ng was given 103 ng/mL Dkk-1 (HY-P7155A, MCE) 10 μl, the group 100 ng was given 104 ng/mL Dkk-1 10 μl, the group 200 ng was given 2 × 104 ng/mL Dkk-1 10 μl, and the same volume PBS was added into group 0. Four groups of cells were taken after 48 h of intervention. After rinsing with PBS and centrifugation at 1,200 rpm/min for 3 min, cell lysis was added. The total protein was extracted by lysis on ice for 30 min.
S. Typhi Invasion Assay
To determine the effect of S. Typhi infection to Caco-2 cells, Caco-2 cells were incubated with the S. Typhi strain (∼20 CFU/cell) for 30 min, washed, and incubated in fresh DMEM with 10% FBS for 30, 60, or 120 min (Wang et al., 2018). Control group indicates without bacterial treatment. After rinsing with PBS and centrifugation at 1,200 rpm/min for 3 min, cell lysis was added. The total protein was extracted by lysis on ice for 30 min.
Caco-2 cells were classified into Control group (106 cells without any treatment), Dkk-1 group (106 cells were treated by 2 × 104 ng/mL, 10 μl Dkk-1 for 48 h), S. Typhi group (106 cell were incubated with S. Typhi strain for 30 min, and incubated with medium for 60 min), Sal + Dkk-1 group (106 cell were incubated with S. Typhi strain for 60 min, and incubated with medium for 60 min then treated by 2 × 104 ng/mL, 10 μl Dkk-1 for 48 h).
Chromatin Immunoprecipitation-qPCR (ChIP-qPCR) Assay
Chromatin immunoprecipitation (ChIP) was performed by using the ChIP Assay Kit (Beyotime Biotechnology) and anti-β-catenin antibody, as previously described (Bernt et al., 2011). Caco-2 cells were classified into Control group (13.7 × 106 cells without any treatment) and S. Typhi group (13.7 × 106 cells were incubated with S. Typhi strain for 30 min, and incubated with medium for 60 min), crosslinked with 1% formalin, then lysed in SDS buffer and sonication was used to fragment the DNA. ChIP for TCF was performed using an anti-β-catenin antibody. Eluted DNA fragments were analyzed by qPCR. To amplify the human Fut8 promoter region (−1416 bp to −1409 bp) containing the TCF-binding site, the following primer sets were used: Fut8 ChIP forward, 5′-CACCCCTTCTTGCTCTTGGC-3′ and Fut8 ChIP reverse, 5′-GACTGTCAGCCATGGAAGCAT-3′.
Statistical Analysis
Each experiment was performed at least three times and the mean was used to calculate significance. Data was expressed as mean values ± standard error mean (SEM). Data analysis was performed with the statistical software package Graph Pad Prism 5. Student’s t-test was used for statistical analysis between two groups, and the quantitative multiple group comparisons was performed using one-way ANOVA followed by Tukey’s test (compare all pairs of columns). Differences were considered to be statistically significant at ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Results
The Ubiquitous Core Fucosylation of IECs Affects Intestinal Microbiota in Mouse
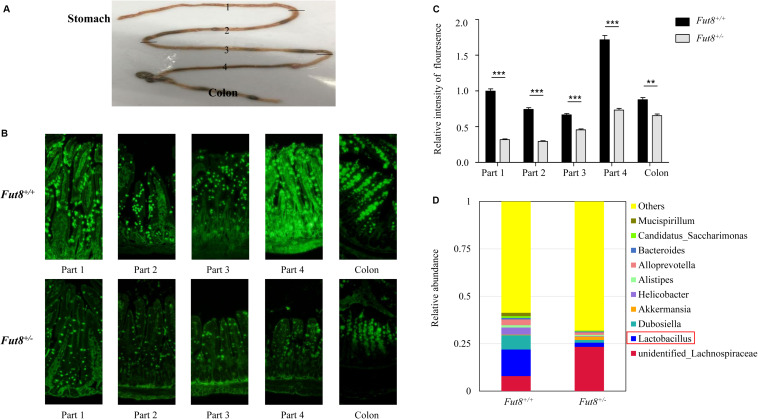
To assess the level of core fucosylation throughout mouse intestine, we first divided the small intestine into four parts (part 1, 2, 3, and 4) equally, from the proximal (duodenum) to the distal (ileum) ends (Figure 1A), and investigated all the 4 parts and the colon by an immunofluorescence assay. Our result showed that core fucosylation was ubiquitous in mouse intestine, especially in colon and the part 1 and part 4 of small intestine (Figures 1B,C). Compared to Fut8+/+ mice, a significant decrease of the core fucosylation was detected in the intestine of Fut8+/– mice.
FIGURE 1.
The ubiquitous core fucosylation of IECs affects intestinal microbiota in mouse. (A) Mouse intestine was divided into the small intestine and colon parts. And the small intestine was further divided into four parts (parts 1, 2, 3, and 4) equally, from the proximal (duodenum) to the distal (ileum) ends. (B) The core fucosylation expression and localization of wild type Fut8+/+ and heterozygous Fut8+/– mice in intestinal mucosa was detected by fluorescein labeled LCA. (C) The relative intensity of fluorescence was detected by ImageJ. The core fucosylation level was found higher in part 1, part 4 of small intestine and in the colonic tissues compared to other parts. And there was a significant decrease of the core fucosylation expression of Fut8+/– mice compared to Fut8+/+ (WT) mice. Data are shown as mean values ± SEM (n = 3; **p < 0.01; ***p < 0.001). (D) The relative abundances of fecal bacterial genera of Fut8+/– and Fut8+/– mice.
To examine the effects of core fucosylation on gut microbiota, we compared the microbial structure of Fut8+/+ and Fut8+/– mice by 16S rRNA sequencing. The major bacterial genera in gut of Fut8+/+ and Fut8+/– mice are shown in Figure 1D. Several genera, including the unidentified Lachnospiraceae (7.92%), Lactobacillus (13.99%), Dubosiella (7.56%), and Akkermansia (0.30%) were found dominant in gut of Fut8+/+ mice. In contrast, the Fut8+/– mice exhibited an altered gut microbial structure. Compared with Fut8+/+ mice, a significant increase in the proportion of the unidentified Lachnospiraceae (23.37%) was detected in gut of Fut8+/– mice, which also harbors remarkably reduced abundance of Lactobacillus spp. (2.17%) (Figure 1D). Our data suggested that the core fucosylation of IECs in mice have a strong effect on their gut microbiota.
Fut8+/– Mice Is Susceptive to S. Typhi Infection
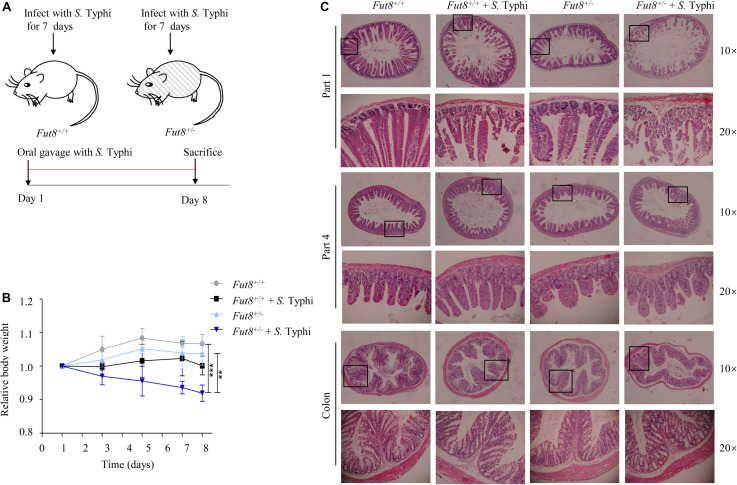
To investigate the role of core fucosylation during Salmonella infection, we infected mice with the S. Typhi strain CMCC (B) 50071 CICC 10871 by oral gavage for 7 days (Figure 2A). S. Typhi infected Fut8+/– mice gained a significant weight loss compared to uninfected Fut8+/– mice (p < 0.01) and Fut8+/+ mice (p < 0.001) (Figure 2B). To assess the susceptibility of Fut8+/+ and Fut8+/– mice, we stained the small intestine and colon sections of mice of different experimental groups, such as Fut8+/+, Fut8+/+ + S. Typhi, Fut8+/– and Fut8+/– + S. Typhi groups. The inflammation which was expressed in the disruption and swelling of intestinal villus, cell infiltration and thickening of muscular layer were observed in the intestinal tissues of the S. Typhi-infected Fut8+/– mice, which was found more severe compared to the S. Typhi-infected Fut8+/+ mice (Figure 2C and Supplementary Figure S2), suggested that low core fucosylation in Fut8+/– mice increased the susceptibility to S. Typhi infection.
FIGURE 2.
Fut8+/– mice is susceptive to S. Typhi infection. (A) Fut8+/– and Fut8+/– mice were infected with S. Typhi every day by oral gavage for 7 days at a concentration of 1 × 108 CFU/ml. (B) Following oral infection with S. Typhi, mice were weighed daily and the percent change in weight from days 1 to 8 is shown. Data were analyzed with a one-way ANOVA using Bonferroni correction (Fut8+/+, n = 3; Fut8+/+ + S. Typhi, n = 3; Fut8+/–, n = 3; Fut8+/– + S. Typhi, n = 3; **p < 0.01; ***p < 0.001). (C) Sections from the mouse intestines were stained with H&E. The infection of the S. Typhi-infected Fut8+/– mice was found more severe compared to the S. Typhi-infected Fut8+/+ mice. And the infection was expressed in the disruption and swelling of intestinal villus, cell infiltration and thickening of muscular layer.
S. Typhi Infection Upregulated the Core Fucosylation Level of IECs in Mice
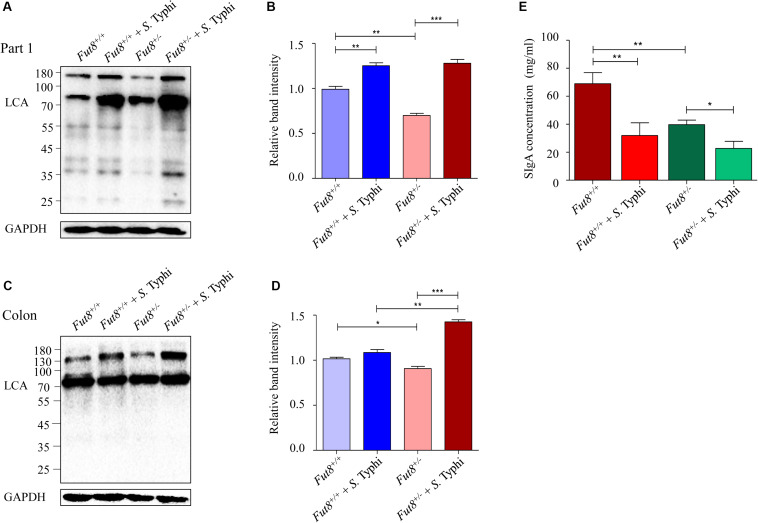
To further investigate the impact of S. Typhi infection on IECs core fucosylation, we next examined the levels of core fucosylation in intestinal mucosa of the infected mice. The core fucosylation of Fut8+/– mice intestinal mucosa was found significantly lower than that of the Fut8+/+ mice, especially in the part 1 of small intestine (p = 0.002, Figures 3A,B) and colon (p = 0.0234, Figures 3C,D). After infection, the core fucosylation of the part 1 of small intestine and colon of Fut8+/+ mice showed an up-regulated pattern, significantly up-regulated in (part 1 of small intestine, p = 0.0047, Figures 3A,B), but without statistic significance in colon (p = 0.1357, Figures 3C,D). While the core fucosylation of Fut8+/– mice was significantly up-regulated in the part 1 of small intestine (p = 0.0003) and colon (p = 0.0001). Overall, our data showed that S. Typhi infection significantly up-regulated the IECs core fucosylation in mice. In addition, we also analyzed the concentration of sIgA in intestinal content. The concentration of sIgA was obviously down-regulated after S. Typhi infection in both Fut8+/+ (p = 0.098) and Fut8+/– mice (p = 0.0172) (Figure 3E), while it was essentially lower in Fut8+/– mice (p = 0.0035).
FIGURE 3.
S. Typhi infection up-regulated the core fucosylation level of IECs in mice. (A) The core fucosylation was up-regulated in intestinal mucosa in the part 1 of small intestine after infected with S. Typhi. The core fucosylation levels of intestine tissues detected by means of LCA blotting. (B) Expression levels of the core fucosylation in the part 1 of small intestine is assessed by relative band intensity of LCA. Data are shown as mean values ± SEM (Fut8+/+, n = 3; Fut8+/+ + S. Typhi, n = 3; Fut8+/–, n = 3; Fut8+/– + S. Typhi, n = 3; ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001). (C) The core fucosylation was up-regulated in intestinal mucosa of colon after S. Typhi infection. (D) Expression levels of the core fucosylation in colon was assessed by relative band intensity of LCA. Data are shown as mean values ± SEM (Fut8+/+, n = 3; Fut8+/+ + S. Typhi, n = 3; Fut8+/–, n = 3; Fut8+/– + S. Typhi, n = 3; ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001). (E) The concentration of sIgA was reduced after S. Typhi infection in both Fut8+/+ and Fut8+/– mice, which was measured by sIgA ELISA kit. The concentration of sIgA was found essentially lower in Fut8+/– mice. Data are shown as mean values ± SEM (Fut8+/+, n = 3; Fut8+/+ + S. Typhi, n = 3; Fut8+/–, n = 3; Fut8+/– + S. Typhi, n = 3; ns, not significant; *p < 0.05, **p < 0.01).
S. Typhi Infection Induced Changes of Gut Microbiota in Fut8+/+ and Fut8+/– Mice
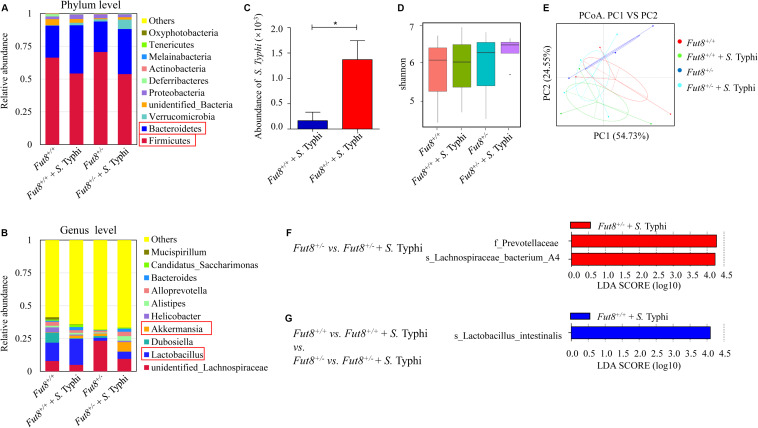
To assess the interaction between IECs core fucosylation and gut microbiota, we investigated the gut microbiota of Fut8+/+ and Fut8+/– mice before and after infection. The relative abundance of the major bacterial phyla in mice belonged to the four experimental groups (Fut8+/+, Fut8+/+ + S. Typhi, Fut8+/–, and Fut8+/– + S. Typhi) were shown in Figure 4A. The major bacterial phyla in gut of mice are Firmicutes and Bacteroides, they occupied more than 60% of the total bacteria in both Fut8+/+ and Fut8+/– mice, with the abundance of Firmicutes obviously higher than that of Bacteroides. Interestingly, post infection, the abundance of Firmicutes was found decreased in Fut8+/+ (66.37% vs. 54.28% before and post infection) and Fut8+/– mice (70.68% vs. 53.78%), in contrast to the elevation of Bacteroides abundance in Fut8+/+ (24.37% vs. 36.75%) and Fut8+/– mice (23.23% vs. 34.38%) (Supplementary Figures S3A,B). Notably, at genus level (Figure 4B), an obvious increase of Lactobacillus was detected in both Fut8+/+ (13.99% vs. 19.76%) and Fut8+/– mice (2.17% vs. 5.46%) post S. Typhi infection, which was accompanied with an increase of Akkermansia in these mice (0. 3% vs. 1.98% in Fut8+/+ mice; 1.74% vs. 7.25% in Fut8+/– mice) (Supplementary Figures S3C,D). Also, we detected more colonization of S. Typhi in Fut8+/– mice post infection when compared with Fut8+/+ + S. Typhi (p = 0.0417, Figure 4C). Overall, although the alpha diversity indicated by Shannon index did not reveal a significant difference among the four groups (Figure 4D), a principal-coordinate analysis (PCoA) revealed a significant clustering pattern among them (Figure 4E), after infection the microbiota patterns of Fut8+/+ and Fut8+/– mice shift toward the same direction. All the information about alpha and beta diversity analyses were shown in Supplementary Figures S4, S5. The bacterial groups that showed significant differences between each group were then analyzed by the LEfSe (linear discriminant analysis effect size) method (Figures 4F,G). The family of Prevotellaceae and species of Lachnospiraceae family A4 were found significantly more abundant in Fut8+/– + S. Typhi group compared with Fut8+/– group (Figure 4F). In addition, comparisons between all the four groups revealed that Lactobacillus intestinalis was significantly more abundant in Fut8+/+ + S. Typhi group (Figure 4G), which suggested an important role of Lactobacillus in gut of mice during S. Typhi infection.
FIGURE 4.
S. Typhi infection induced changes of gut microbiota in Fut8+/+ and Fut8+/– mice. (A) The relative abundances of the major fecal bacterial phyla in mice. (B) The relative abundances of fecal bacterial genera, Lactobacillus and Akkermansia, were found increased post infection. (C) S. Typhi was found more abundant in Fut8+/– + S. Typhi compared with Fut8+/+ + S. Typhi. Data are shown as mean values ± SEM (Fut8+/+ + S. Typhi, n = 3; Fut8+/– + S. Typhi, n = 3; *p < 0.05). (D) The alpha diversity indicated by Shannon index in each group. (E) The beta diversity of fecal microbiota analyzed by principal-coordinate analysis (PCoA). (F,G) The bacterial groups that showed significant differences between each group analyzed by the LEfSe method (linear discriminant analysis effect size).
Wnt Signaling Pathway Along With Core Fucosylation Is Activated After S. Typhi Infection
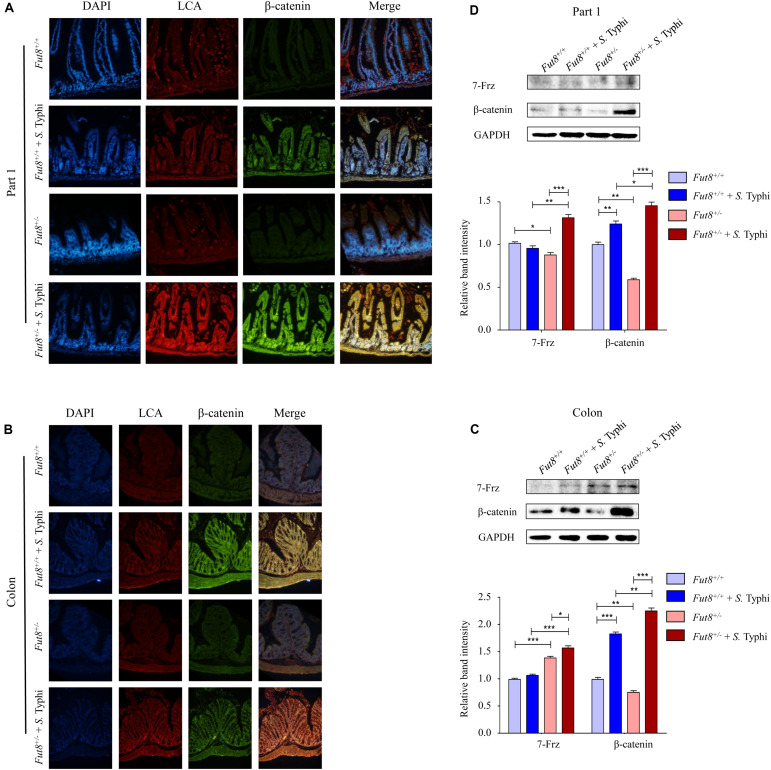
Next, we assessed the expression of core fucosylation and Wnt signaling pathway of Fut8+/+ and Fut8+/– mice before and after infection in the part 1 and part 4 of small intestine and colon by immunofluorescence and western blot. Frizzled 7 (7-Frz) and β-catenin are the representative protein molecules of Wnt signaling pathway. The immunofluorescence indicated the expression and localization in mucosa of core fucosylation and β-catenin of four groups in part 1 (Figure 5A) and part 4 (Supplementary Figure S1) of small intestine and colon (Figure 5B). Our data showed that core fucosylation and β-catenin were both up-regulated post infection of S. Typhi no matter in Fut8+/+ mice or in Fut8+/– mice (Figures 5A,B). 7-Frz was found significantly up-regulated in part 1 of small intestine (Figure 5C, p = 0.0012) and colon (Figure 5D, p = 0.0189) in Fut8+/– mice after S. Typhi infection. In addition, β-catenin was also up-regulated in part 1 of small intestine of Fut8+/+ (p = 0.0063) and Fut8+/– mice (p < 0.0001, Figure 5C), and colon of Fut8+/+ (p < 0.0001) and Fut8+/– mice (p < 0.0001) (Figure 5D) after S. Typhi infection. Overall, our data showed that Wnt signaling pathway and core fucosylation were activated simultaneously after S. Typhi infection.
FIGURE 5.
Wnt signaling pathway and core fucosylation were activated in mice after S. Typhi infection. (A) The core fucosylation and β-catenin expression and localization in mucosa of part 1 of mouse intestine. (B) The core fucosylation and β-catenin expression and localization in mucosa of mouse colon. (C) Expression levels of 7-Frz and β-catenin in the part 1 of small intestine assessed by Western blotting in intestinal tissues. Data are shown as mean values ± SEM (Fut8+/+, n = 3; Fut8+/+ + S. Typhi, n = 3; Fut8+/–, n = 3; Fut8+/– + S. Typhi, n = 3; ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001). 7-Frz and β-catenin were found up-regulated in intestinal tissues of the part 1 of small intestine after infected with S. Typhi. (D) Expression levels of 7-Frz and β-catenin in colon. Data are shown as mean values ± SEM (Fut8+/+, n = 3; Fut8+/+ + S. Typhi, n = 3; Fut8+/–, n = 3; Fut8+/– + S. Typhi, n = 3; ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001). 7-Frz and β-catenin were up-regulated in colon after infected with S. Typhi.
Wnt Signaling Pathway Mediates the Up-Regulation of Caco-2 Cell Core Fucosylation Stimulated by S. Typhi in vitro
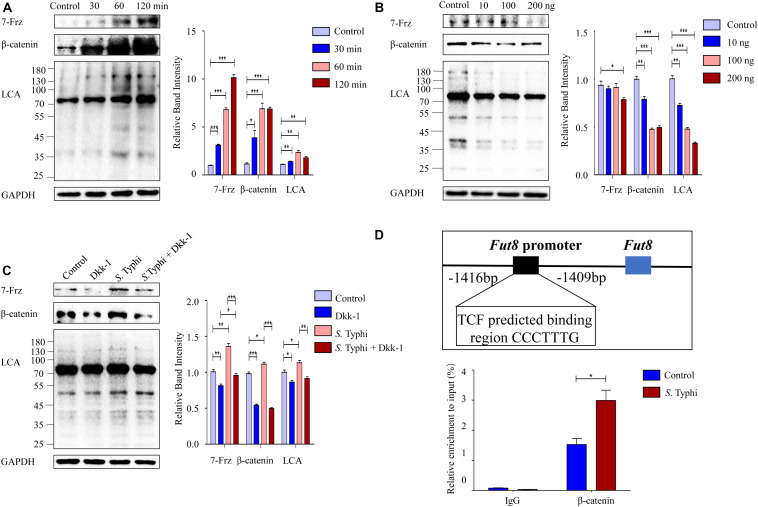
To determine whether core fucosylation is regulated through Wnt signaling pathway, we infected Caco-2 cells with S. Typhi and found that the expression levels of 7-Frz and β-catenin were significantly increased post S. Typhi infection with a time-dependent pattern (Figure 6A). Consistently, we also found up-regulation of the core fucosylation levels of Caco-2, suggesting that the up-regulation of the core fucosylation may correlate with Wnt signaling pathway. To further determine this correlation, we investigated the effect of Wnt signaling antagonist, Dickkopf-1 (Dkk-1), which functions by binding to the LRP5/LRP6 helper receptor and inhibiting canonical Wnt signaling pathway (Nakamura et al., 2003). The expression of 7-Frz and β-catenin were significantly decreased in Caco-2 after the application of Dkk-1 with a concentration-dependent pattern (Figure 6B). Consistently, we also found a similar down-regulation of the core fucosylation levels in Caco-2. Next, we analyzed the effect of S. Typhi infection after the application of Dkk-1. As shown in Figure 6C, Caco-2 cells were divided into four group (group Control, Dkk, S. Typhi, and S. Typhi + Dkk-1), we found that S. Typhi no longer increase the expression of 7-Frz, β-catenin and the expression of the core fucosylation with the application of Dkk-1. Therefore, we sought to clarify whether β-catenin directly regulates Fut8 expression or not. There are the promoter regions of Fut8 gene upstream of the transcription initiation site. To further validate Fut8 is one of the downstream targets of the Wnt/β-catenin signaling pathway, we predicted a putative β-catenin-T cell factor (TCF)-binding site (5′-CCCTTTG-3′) on Fut8 promoter regions, and infected Caco-2 cells with S. Typhi. In the CHIP experiments, the β-catenin specifically bound to the Fut8 promoter region (−1416 bp to −1409 bp) (p = 0.021) (Figure 6D). Collectively, these results indicated that Wnt signaling pathway mediated the up-regulation of core fucosylation stimulated by S. Typhi.
FIGURE 6.
Wnt signaling pathway mediates the core fucosylation of Caco-2 by S. Typhi infection in vitro. (A) The expression of 7-Frz, β-catenin and core fucosylation of Caco-2 cells were up-regulated post infection. Caco-2 cells were incubated with S. Typhi for 30 min, washed, and incubated in fresh DMEM with 10% fetal bovine serum for 30, 60, or 120 min. Control group indicates without bacterial treatment. Data are shown as mean values ± SEM (Control, n = 3; 30 min, n = 3; 60 min, n = 3; 120 min, n = 3; ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001). (B) The expression of 7-Frz and β-catenin of Caco-2 cells was up-regulated after application of Dkk-1. The cells were divided into control group (group Control) and experimental group (group 10, 100, and 200 ng) according to the completely random method. The dose of Dkk-1 for experimental group: 10, 100, and 200 ng. Data are shown as mean values ± SEM (Control, n = 3; 10 ng, n = 3; 100 ng, n = 3; 200 ng, n = 3; ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001). (C) Caco-2 cells were classified into Control group (cells without any treatment), Dkk group (cells were treated by 2 × 104 ng/mL, 10 μl Dkk-1 for 48 h), S. Typhi group (cell were incubated with S. Typhi strain for 30 min, and incubated with medium for 60 min), S. Typhi + Dkk group (cell were incubated with S. Typhi strain for 60 min, and incubated with medium for 60 min then treated by 2 × 104 ng/mL, 10 μl Dkk-1 for 48 h). Data are shown as mean values ± SEM (Control, n = 3; Dkk-1, n = 3; S. Typhi, n = 3; S. Typhi + Dkk-1, n = 3; ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001). (D) The interaction between β-catenin/TCF complexes and Fut8 promoter was analyzed by ChIP analysis. TCF can directly bound to the Fut8 promoter, and S. Typhi infection increased the inputs. Caco-2 cells were incubated with S. Typhi for 30 min, washed, and incubated in fresh DMEM with 10% fetal bovine serum for 60 min. Control group indicates without bacterial treatment. Normalized inputs of chromatin DNA from Caco-2 cells were pulled down with anti-β-catenin or negative IgG antibodies. Data are shown as mean values ± SEM (Control, n = 3; S. Typhi, n = 3; *p < 0.05).
Discussion
Core fucosylation is a common type of post-translational protein modification, and it is involved in a variety of physiological and pathological processes, such as Crohn’s disease and colorectal cancer (Muinelo-Romay et al., 2008; Franke et al., 2010; McGovern et al., 2010). Understanding the mechanisms of regulation of Fut8 gene expression and core fucosylation, one of the major glycosylation events in IECs, is therefore of great important. Our results proved that core fucosylation is ubiquitous in IECs, and core fucosylation is closely related to abundance of Lactobacillus spp. It has been reported that the expression of epithelial fucosylation is higher in the ileum (part 4 of intestine), which harbors more abundant gut microbes than in the duodenum (part 1) (Barker et al., 2007; Goto et al., 2014). Our results also proved that the core fucosylation of part 4 is significantly higher than other parts of intestine. Our recent study also revealed that Lactobacillus and Bifidobacterium specifically organisms synthesize α-L-fucosidase that cleave core fucose residue, and once inside the bacteria, the fucose was catabolized. Therefore, the core fucosylated N-glycans can facilitate the growth of Lactobacillus in gut of infants (Li et al., 2019).
Given that the commensal bacteria of the gastrointestinal tract play an important role in protecting the host from pathogenic bacteria infection to maintain intestinal homeostasis (Mueller and Macpherson, 2006; Backhed et al., 2012). The imbalance of microbiota caused by pathogen infection is considered to be closely related to inflammation, obesity, fucosylation, sialylation and other metabolic factors (Boulange et al., 2016; Zhou et al., 2020). It has been reported that Salmonella infection enhanced the levels of proinflammatory cytokines IL-1β, IL-8, and LITNF. And the imbalance of microbiota can increase susceptibility to pathogenic infection (Brown et al., 2013; Moreira et al., 2017). For instance, antibiotic treatment alters the microbial community and predisposes the host to Salmonella infection (Sekirov et al., 2008). Our study showed that the deficiency of core fucosylation affected intestinal microbiota in mouse, and Fut8+/– mice was more susceptive to S. Typhi infection when compared with Fut8+/+ mice, this suggested an interaction between IECs core fucosylation and gut microbiota. Fut2-mediated epithelial fucosylation (α1,2 fucosylation) is initiated by direct interaction between commensals and IECs (Goto et al., 2014). Commensal bacteria, for example the Bacteroides spp., can use focuses as components of the bacterial outer membrane, or use them as a nutrient (Leis et al., 1997; Coyne et al., 2005). Moreover, some glycans are able to resist pathogens and shape commensal bacterial structure (Hooper et al., 1999). As we found that the deficient of core fucosylation, Lactobacillus is decreased in Fut8+/– mice, therefore, the epithelial core fucose reacts as a mediator between the host and commensal microbiota. Based on these data, it can be concluded that core fucosylation can promote the colonization of commensal bacteria like Bacteroides fragilis, Lactobacillus, and Akkermansia. When assessing the microbiota community structure of Fut8+/+ and Fut8+/– mice, we also found that the abundance of Lactobacillus and Akkermansia was increased in mice after S. Typhi infection. It has been reported that lactic acid and hydrogen peroxide, the main metabolites of Lactobacillus, act in a cooperative way to resist enteric and vaginosis-associated pathogens (Servin, 2004; Fayol-Messaoudi et al., 2007). Akkermansia not only has the capacity to degrade mucins, but also to stimulate mucin synthesis to clear environmental antigens, acting as innate host defense and protection against to infections (McGuckin et al., 2011; Shin et al., 2014). Furthermore, the mucin degradation activity of Akkermansia leads to the production of short-chain fatty acids (SCFAs), which can form an acidic environment to inhibit the growth of S. Typhi (Derrien et al., 2004; Shin et al., 2014). It can be concluded that the increase of Lactobacillus and Akkermansia is antagonistic to S. Typhi infection. Furthermore, there are some studies showed that bacterial flagellin of S. Typhi can induce Th17 cells which promotes inflammation, and Lactobacillus have the potential to induce Treg cells which promotes immune tolerance (Hansson and Johansson, 2010; Round and Mazmanian, 2010; Round et al., 2011; Goto and Kiyono, 2012). Moreover, intestinal APCs such as dendritic cells (DCs) and macrophages (MPs) play a pivotal role in suppressing microbiota-induced inflammation and mediating mucosal tolerance (Staal et al., 2008; Maloy and Powrie, 2011; Rescigno, 2011; Swafford et al., 2018). It has also been reported that cytokines interleukin-22 and lymphotoxin producing type 3 innate lymphoid cells (ILC3) regulate epithelial fucosylation on IECs to inhibit S. Typhimurium infection (Goto et al., 2014). Secretory IgA (sIgA) functioned by limiting pathogen access to the mucosal surface and protecting immunity toward Salmonella infection (Endt et al., 2010). In addition, the damaged muscular layer was observed in the intestinal tissues of the S. Typhi-infected Fut8+/– mice, and the concentration of sIgA is obviously reduced after S. Typhi infection by a damage of mucosal immune function (Tokuhara et al., 2010). Our data thus suggest that S. Typhi induced up-regulation of IECs core fucosylation, which promoted the abundance of core-fucose utilizing bacteria such as Lactobacillus and Akkermansia, to resist the invasion of S. Typhi, and this interaction contributed to the improvement of the intestinal barrier function and immune responses subsequently.
It has been reported that Salmonella infection causes the activation of canonical Wnt signaling pathway which plays an important role in embryonic development and tumorigenesis, such as intestinal renewal, inflammation and colorectal cancer (Liu et al., 2010; Wang et al., 2018). Wnt signaling pathway was also significantly activated after S. enterica serovar Enteritidis infection, characterized by GSK-3β and β-catenin phosphorylated (Kogut and Arsenault, 2015). The most relevant limitation of this study is the use of pharmacological drugs as unique tool to demonstrate the mechanism underlying the fucosylation modification on IECs, although we used Dickkopf-1 (Dkk-1), a wnt signaling antagonist, which functions by binding to the LRP5/LRP6 helper receptor and inhibiting canonical Wnt signaling pathway (Nakamura et al., 2003). Our study showed that the Wnt signaling pathway is activated post S. Typhi infection, and the application of Wnt signaling pathway antagonist Dkk-1suppressed IECs core fucosylation. Our study also demonstrated that β-catenin can directly bound to the Fut8 promoter regions of Fut8 gene. These data indicated that the S. Typhi-induced activation of Wnt signaling pathway leads to the accumulation of β-catenin and enhances Fut8 transcription, contributes to the rapid elevation of IECs core fucosylation.
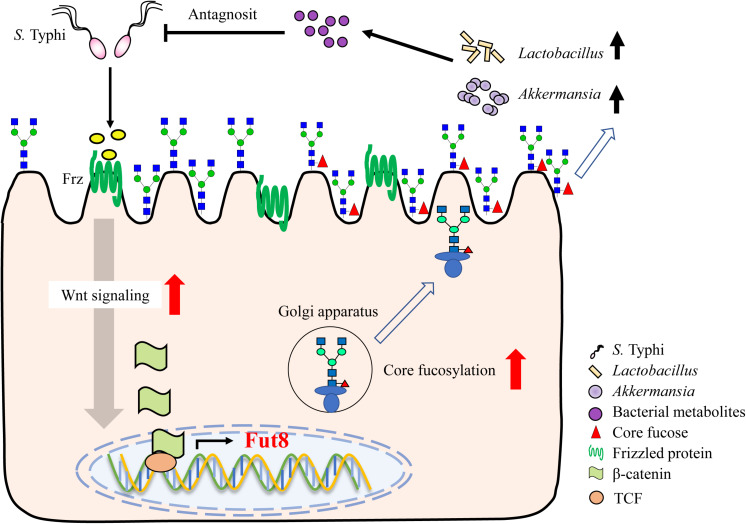
An immense structural diversity of IECs glycans contribute to the intestinal microbiota formation. Thus, their importance for both host and intestinal bacteria in long-term commensal interactions is beginning to be appreciated. A key finding of our study is the core fucosylation on IECs is closely associated with the intestinal microbiota formation and invasion of S. Typhi. During S. Typhi infection, the Wnt signaling pathway, represented by the expression of Frizzled protein and β-catenin, is rapidly up-regulated, resulted in elevation of epithelial core fucosylation. The abundant core fucose on the surface of IECs facilitates the growth of fucose-utilizing commensal bacteria such as Lactobacillus and Akkermansia, who producing lactic acid and other metabolites to resist the invasion of S. Typhi (Figure 7).
FIGURE 7.
Core fucosylation of IECs protects against S. Typhi infection via up-regulating the biological antagonism of intestinal microbiota.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
All animal work was approved by the Ethics Committee (approval No. AEE17013) at the Dalian Medical University, Dalian, China.
Author Contributions
WL, ML, and SH designed the research. SH, QF, YB, HF, and JZ performed the experiments. SH analyzed the experimental data and wrote the manuscript. ML, WL, TF, and JG corrected the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by grants from the National Natural Science Foundation of China (31870797, 31570797, and 31900920), the Nature Science Foundation of Liaoning Province, China (2019-MS-085), and the Nutrition and Care of Maternal & Child Research Fund Project of Guangzhou Biostime Institute of Nutrition & Care (2019BINCMCF02, 2018BINCMCF39, and 2016BINCMCF1102). This work was also supported by Liaoning Provincial Program for Top Discipline of Basic Medical Sciences.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01097/full#supplementary-material
Wnt signaling pathway along with core fucosylation is activated after S. typhi infection in part 4 of small intestine. (A) The core fucosylation and β-catenin expression and localization in mucosa of part 4 of intestine.
The pathology score of small intestine and colon. (A) The pathology score of part 1 of small intestine. (B) The pathology score of part 4 of small intestine. (C) The pathology score of colon. Data are shown as mean values ± SEM (Fut8+/+, n = 3; Fut8+/+ + S. Typhi, n = 3; Fut8+/–, n = 3; Fut8+/– + S. Typhi, n = 3; ns, not significant; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
(A) Relative abundance of Firmicutes. (B) Relative abundance of Bacteroidetes. (C) Relative abundance of Lactobacillus. (D) Relative abundance of Akkermansia. Data are shown as mean values ± SEM (Fut8+/+, n = 3; Fut8+/+ + S. Typhi, n = 3; Fut8+/–, n = 3; Fut8+/– + S. Typhi, n = 3; ns, not significant).
The alpha diversity of fecal microbiota. (A) The alpha diversity indicated by ACE index in each group. (B) The alpha diversity indicated by chao 1 index in each group. (C) The alpha diversity indicated by goods coverage index in each group. (D) The alpha diversity indicated by observed species index in each group. (E) The alpha diversity indicated by PD whole tree index in each group. (F) The alpha diversity indicated by simpson index in each group.
The beta diversity of fecal microbiota. (A) The beta diversity indicated by weighted unifrac index in each group. (B) The beta diversity of fecal microbiota analyzed by NMDS. (C) The beta diversity of fecal microbiota analyzed by PCA. (D) The beta diversity of fecal microbiota analyzed by heatmap.
References
- Backhed F., Fraser C. M., Ringel Y., Sanders M. E., Sartor R. B., Sherman P. M., et al. (2012). Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe 12 611–622. 10.1016/j.chom.2012.10.012 [DOI] [PubMed] [Google Scholar]
- Barker N., van Es J. H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., et al. (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449 1003–1007. 10.1038/nature06196 [DOI] [PubMed] [Google Scholar]
- Bernt K. M., Zhu N., Sinha A. U., Vempati S., Faber J., Krivtsov A. V., et al. (2011). MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 20 66–78. 10.1016/j.ccr.2011.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulange C. L., Neves A. L., Chilloux J., Nicholson J. K., Dumas M. E. (2016). Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 8:42. 10.1186/s13073-016-0303-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. E., Libby S. J., Moreland S. M., McCoy M. W., Brabb T., Stepanek A., et al. (2013). Salmonella enterica causes more severe inflammatory disease in C57/BL6 Nramp1G169 mice than Sv129S6 mice. Vet. Pathol. 50 867–876. 10.1177/0300985813478213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon A. D., Liu Y., Li X., Wang X., Chen X., Li L., et al. (2016). Substrate specificity of FUT8 and chemoenzymatic synthesis of core-fucosylated asymmetric N-glycans. Org. Biomol. Chem. 14 4027–4031. 10.1039/c6ob00586a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne M. J., Reinap B., Lee M. M., Comstock L. E. (2005). Human symbionts use a host-like pathway for surface fucosylation. Science 307 1778–1781. 10.1126/science.1106469 [DOI] [PubMed] [Google Scholar]
- Derrien M., Vaughan E. E., Plugge C. M., de Vos W. M. (2004). Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54(Pt 5), 1469–1476. 10.1099/ijs.0.02873-0 [DOI] [PubMed] [Google Scholar]
- Endt K., Stecher B., Chaffron S., Slack E., Tchitchek N., Benecke A., et al. (2010). The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea. PLoS Pathog. 6:e1001097. 10.1371/journal.ppat.1001097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayol-Messaoudi D., Coconnier-Polter M. H., Moal V. L., Atassi F., Berger C. N., Servin A. L. (2007). The Lactobacillus plantarum strain ACA-DC287 isolated from a Greek cheese demonstrates antagonistic activity in vitro and in vivo against Salmonella enterica serovar Typhimurium. J. Appl. Microbiol. 103 657–665. 10.1111/j.1365-2672.2007.03293.x [DOI] [PubMed] [Google Scholar]
- Franke A., McGovern D. P., Barrett J. C., Wang K., Radford-Smith G. L., Ahmad T., et al. (2010). Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 42 1118–1125. 10.1038/ng.717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y., Kiyono H. (2012). Epithelial barrier: an interface for the cross-communication between gut flora and immune system. Immunol. Rev. 245 147–163. 10.1111/j.1600-065X.2011.01078.x [DOI] [PubMed] [Google Scholar]
- Goto Y., Obata T., Kunisawa J., Sato S., Ivanov I. I., Lamichhane A., et al. (2014). Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science 345:1254009. 10.1126/science.1254009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y., Uematsu S., Kiyono H. (2016). Epithelial glycosylation in gut homeostasis and inflammation. Nat. Immunol. 17 1244–1251. 10.1038/ni.3587 [DOI] [PubMed] [Google Scholar]
- Gradel K. O., Nielsen H. L., Schonheyder H. C., Ejlertsen T., Kristensen B., Nielsen H. (2009). Increased short- and long-term risk of inflammatory bowel disease after Salmonella or campylobacter gastroenteritis. Gastroenterology 137 495–501. 10.1053/j.gastro.2009.04.001 [DOI] [PubMed] [Google Scholar]
- Hansson G. C., Johansson M. E. (2010). The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Gut Microbes 1 51–54. 10.4161/gmic.1.1.10470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapfelmeier S., Muller A. J., Stecher B., Kaiser P., Barthel M., Endt K., et al. (2008). Microbe sampling by mucosal dendritic cells is a discrete, MyD88-independent step in DeltainvG S. Typhimurium colitis. J. Exp. Med. 205 437–450. 10.1084/jem.20070633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper L. V., Xu J., Falk P. G., Midtvedt T., Gordon J. I. (1999). A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl. Acad. Sci. U.S.A. 96 9833–9838. 10.1073/pnas.96.17.9833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato I., Boleij A., Kortman G. A., Roelofs R., Djuric Z., Severson R. K., et al. (2013). Partial associations of dietary iron, smoking and intestinal bacteria with colorectal cancer risk. Nutr. Cancer 65 169–177. 10.1080/01635581.2013.748922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogut M. H., Arsenault R. J. (2015). A role for the non-canonical Wnt-beta-catenin and TGF-beta signaling pathways in the induction of tolerance during the establishment of a Salmonella enterica serovar enteritidis persistent cecal infection in chickens. Front. Vet. Sci. 2:33. 10.3389/fvets.2015.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawhon S. D., Khare S., Rossetti C. A., Everts R. E., Galindo C. L., Luciano S. A., et al. (2011). Role of SPI-1 secreted effectors in acute bovine response to Salmonella enterica serovar Typhimurium: a systems biology analysis approach. PLoS One 6:e26869. 10.1371/journal.pone.0026869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leis O., Madrid J. F., Ballesta J., Hernandez F. (1997). N- and O-linked oligosaccharides in the secretory granules of rat Paneth cells: an ultrastructural cytochemical study. J. Histochem. Cytochem. 45 285–293. 10.1177/002215549704500213 [DOI] [PubMed] [Google Scholar]
- Li J., Zhao F., Wang Y., Chen J., Tao J., Tian G., et al. (2017). Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 5:14. 10.1186/s40168-016-0222-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Bai Y., Zhou J., Huang W., Yan J., Tao J., et al. (2019). Core fucosylation of maternal milk N-glycan evokes B cell activation by selectively promoting the l-fucose metabolism of gut Bifidobacterium spp. and Lactobacillus spp. mBio 10:e00128-19. 10.1128/mBio.00128-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Nakagawa T., Koyama N., Wang X., Jin J., Mizuno-Horikawa Y., et al. (2006). Down-regulation of trypsinogen expression is associated with growth retardation in alpha1,6-fucosyltransferase-deficient mice: attenuation of proteinase-activated receptor 2 activity. Glycobiology 16 1007–1019. 10.1093/glycob/cwl023 [DOI] [PubMed] [Google Scholar]
- Liang W., Mao S., Sun S., Li M., Li Z., Yu R., et al. (2018). Core fucosylation of the T cell receptor is required for T cell activation. Front. Immunol. 9:78. 10.3389/fimmu.2018.00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Lu R., Wu S., Sun J. (2010). Salmonella regulation of intestinal stem cells through the Wnt/beta-catenin pathway. FEBS Lett. 584 911–916. 10.1016/j.febslet.2010.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wu S., Xia Y., Li X. E., Xia Y., Zhou Z. D., et al. (2011). Wingless homolog Wnt11 suppresses bacterial invasion and inflammation in intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 301 G992–G1003. 10.1152/ajpgi.00080.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy K. J., Powrie F. (2011). Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 474 298–306. 10.1038/nature10208 [DOI] [PubMed] [Google Scholar]
- Maskalyk J. (2003). Typhoid fever. CMAJ 169:132. [PMC free article] [PubMed] [Google Scholar]
- McGovern D. P., Jones M. R., Taylor K. D., Marciante K., Yan X., Dubinsky M., et al. (2010). Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn’s disease. Hum. Mol. Genet. 19 3468–3476. 10.1093/hmg/ddq248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuckin M. A., Linden S. K., Sutton P., Florin T. H. (2011). Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 9 265–278. 10.1038/nrmicro2538 [DOI] [PubMed] [Google Scholar]
- Miyoshi E., Noda K., Yamaguchi Y., Inoue S., Ikeda Y., Wang W., et al. (1999). The alpha1-6-fucosyltransferase gene and its biological significance. Biochim. Biophys. Acta 1473 9–20. 10.1016/s0304-4165(99)00166-x [DOI] [PubMed] [Google Scholar]
- Moreira A. C., Neves J. V., Silva T., Oliveira P., Gomes M. S., Rodrigues P. N. (2017). Hepcidin-(In)dependent mechanisms of iron metabolism regulation during infection by Listeria and Salmonella. Infect. Immun. 85:e00353-17. 10.1128/iai.00353-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller C., Macpherson A. J. (2006). Layers of mutualism with commensal bacteria protect us from intestinal inflammation. Gut 55 276–284. 10.1136/gut.2004.054098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muinelo-Romay L., Vazquez-Martin C., Villar-Portela S., Cuevas E., Gil-Martin E., Fernandez-Briera A. (2008). Expression and enzyme activity of alpha(1,6)fucosyltransferase in human colorectal cancer. Int. J. Cancer 123 641–646. 10.1002/ijc.23521 [DOI] [PubMed] [Google Scholar]
- Nakamura T., Sano M., Songyang Z., Schneider M. D. (2003). A Wnt- and beta -catenin-dependent pathway for mammalian cardiac myogenesis. Proc. Natl. Acad. Sci. U.S.A. 100 5834–5839. 10.1073/pnas.0935626100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rescigno M. (2011). Dendritic cells in oral tolerance in the gut. Cell. Microbiol. 13 1312–1318. 10.1111/j.1462-5822.2011.01626.x [DOI] [PubMed] [Google Scholar]
- Round J. L., Lee S. M., Li J., Tran G., Jabri B., Chatila T. A., et al. (2011). The toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332 974–977. 10.1126/science.1206095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round J. L., Mazmanian S. K. (2010). Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. U.S.A. 107 12204–12209. 10.1073/pnas.0909122107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I., Tam N. M., Jogova M., Robertson M. L., Li Y., Lupp C., et al. (2008). Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 76 4726–4736. 10.1128/IAI.00319-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servin A. L. (2004). Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 28 405–440. 10.1016/j.femsre.2004.01.003 [DOI] [PubMed] [Google Scholar]
- Shin N. R., Lee J. C., Lee H. Y., Kim M. S., Whon T. W., Lee M. S., et al. (2014). An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 63 727–735. 10.1136/gutjnl-2012-303839 [DOI] [PubMed] [Google Scholar]
- Staal F. J., Luis T. C., Tiemessen M. M. (2008). WNT signalling in the immune system: WNT is spreading its wings. Nat. Rev. Immunol. 8 581–593. 10.1038/nri2360 [DOI] [PubMed] [Google Scholar]
- Swafford D., Shanmugam A., Ranganathan P., Hussein M. S., Koni P. A., Prasad P. D., et al. (2018). Canonical Wnt signaling in CD11c(+) APCs regulates microbiota-induced inflammation and immune cell homeostasis in the colon. J. Immunol. 200 3259–3268. 10.4049/jimmunol.1701086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuhara D., Yuki Y., Nochi T., Kodama T., Mejima M., Kurokawa S., et al. (2010). Secretory IgA-mediated protection against V. cholerae and heat-labile enterotoxin-producing enterotoxigenic Escherichia coli by rice-based vaccine. Proc. Natl. Acad. Sci. U.S.A. 107 8794–8799. 10.1073/pnas.0914121107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Lu R., Fu X., Dan Z., Zhang Y. G., Chang X., et al. (2018). Novel regulatory roles of Wnt1 in infection-associated colorectal cancer. Neoplasia 20 499–509. 10.1016/j.neo.2018.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Inoue S., Gu J., Miyoshi E., Noda K., Li W., et al. (2005). Dysregulation of TGF-beta1 receptor activation leads to abnormal lung development and emphysema-like phenotype in core fucose-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 102 15791–15796. 10.1073/pnas.0507375102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. F., Yu M., Jin H. D., Yao J. Q., Lu Z. L., Yabasin I. B., et al. (2017). Fentanyl promotes breast cancer cell stemness and epithelial-mesenchymal transition by upregulating alpha1, 6-fucosylation via Wnt/beta-catenin signaling pathway. Front. Physiol. 8:510. 10.3389/fphys.2017.00510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Cui X., Wang H., Liu J., Qin H., Liu S., et al. (2019). FUT8 drives the proliferation and invasion of trophoblastic cells via IGF-1/IGF-1R signaling pathway. Placenta 75 45–53. 10.1016/j.placenta.2018.11.005 [DOI] [PubMed] [Google Scholar]
- Zhou J., Wang Y., Fan Q., Liu Y., Liu H., Yan J., et al. (2020). High levels of fucosylation and sialylation of milk N-glycans from mothers with gestational diabetes mellitus alter the offspring gut microbiome and immune balance in mice. FASEB J. 34 3715–3731. 10.1096/fj.201901674R [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Wnt signaling pathway along with core fucosylation is activated after S. typhi infection in part 4 of small intestine. (A) The core fucosylation and β-catenin expression and localization in mucosa of part 4 of intestine.
The pathology score of small intestine and colon. (A) The pathology score of part 1 of small intestine. (B) The pathology score of part 4 of small intestine. (C) The pathology score of colon. Data are shown as mean values ± SEM (Fut8+/+, n = 3; Fut8+/+ + S. Typhi, n = 3; Fut8+/–, n = 3; Fut8+/– + S. Typhi, n = 3; ns, not significant; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
(A) Relative abundance of Firmicutes. (B) Relative abundance of Bacteroidetes. (C) Relative abundance of Lactobacillus. (D) Relative abundance of Akkermansia. Data are shown as mean values ± SEM (Fut8+/+, n = 3; Fut8+/+ + S. Typhi, n = 3; Fut8+/–, n = 3; Fut8+/– + S. Typhi, n = 3; ns, not significant).
The alpha diversity of fecal microbiota. (A) The alpha diversity indicated by ACE index in each group. (B) The alpha diversity indicated by chao 1 index in each group. (C) The alpha diversity indicated by goods coverage index in each group. (D) The alpha diversity indicated by observed species index in each group. (E) The alpha diversity indicated by PD whole tree index in each group. (F) The alpha diversity indicated by simpson index in each group.
The beta diversity of fecal microbiota. (A) The beta diversity indicated by weighted unifrac index in each group. (B) The beta diversity of fecal microbiota analyzed by NMDS. (C) The beta diversity of fecal microbiota analyzed by PCA. (D) The beta diversity of fecal microbiota analyzed by heatmap.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.