Abstract
The novel coronavirus disease 2019 (COVID-19) emerged in Wuhan, China, and has spread rapidly to nearly every part of the world. Unfortunately, no drug or vaccine has been accepted for the treatment of this pandemic. Researchers have established the efficacy of some existing antiviral drugs to control COVID-19 in vitro. Some of them are remdesivir (GS-5734), chloroquine, hydroxychloroquine, theaflavin. Topological indices are mathematical interpretations of a molecule generated by an algorithm implemented to a given molecular representation. Topological indices are used to model different physicochemical properties and biological activities of chemical compounds. In this work, some degree-based and neighborhood degree sum-based topological indices are investigated for the aforesaid antiviral drugs using polynomial approach. The results obtained can aid in the design of new medicine for the treatment of COVID-19.
MSC (2010): Primary: 05C35, Secondary: 05C07, 05C40
Keywords: Chloroquine, COVID-19, hydroxychloroquine, M-polynomial, molecular graph, remdesivir (GS-5734), theaflavin, topological index
1. Introduction
Historically, epidemics of multiple infectious diseases with millions dying have been recorded in the past few centuries. The most terrific was pandemics due to the plague, flu, cholera, etc. Currently, the COVID-19 pandemic is disrupting human health and the economy around the world. It is originated in a Wuhan1 seafood market but has rapidly spread in and beyond China. As of 3 April 2020, there were 1,116,643 confirmed cases, including 59,158 deaths worldwide (as per world meter information). The novel corona virus (2019-nCoV) is a betacoronavirus and shares genetic sequence and viral structure with severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV). No specific medication for the new disease is currently available. It is, therefore, urgent to identify appropriate antiviral agents to combat the pathogen. An effective experiment to drug discovery is to test whether existing antiviral drugs are efficient in the treatment of related viral diseases. Researchers tested some existing antiviral agents2–6 and got positive results to inhibit the infection and transmission of the 2019-nCoV in vitro. Some of these antiviral compounds are remdesivir (GS5734), chloroquine, hydroxychloroquine and theaflavin. Remdesivir is a nucleotide analog drug having broad spectrum activity developed to prevent Ebola virus infection.7 It is also highly efficient to prevent 2019-nCoV in vitro.2 The clinical trial is currently underway in several hospitals and tests on efficacy are awaited. Chloroquine is a broadspectrum antiviral drug8,9 effective for treating malaria and autoimmune disease. Many randomized controlled trials were performed to assess the impact of chloroquine in the treatment of COVID-19. Therapeutic results in terms of fever control, enhanced CT imaging and delayed disease progression have been reported. Hydroxychloroquine has antiviral activity very similar to that of chloroquine. Both exhibit immune modulating activity, which can improve their antiviral effect in vivo. As per Forbes report in 30 March 2020, FDA approves chloroquine and hydroxychloroquine for emergency coronavirus treatment. Hydroxychloroquine reduce the acute evolution of COVID-19 by suppressing the cytokine storm by inhibiting T cell activation. Theaflavin, a polyphenol chemical in black tea, is found liable for the medical benefit of black tea. Theaflavin has shown a wide range of antiviral activity against many viruses, including influenza A, B and hepatitis C viruses.10,11 In the work of Lung et al.4, it is found that theaflavin may be used as a lead compound for the production of an inhibitor of 2019-nCoV. In pharmaceutical drug design, information related to physicohemical properties, biological activities of molecular graph of compounds are necessary. These properties can be predicted without using any weight lab by a well-known tool of chemical graph theory known as topological index. By molecular graph12,13 we mean a simple connected graph where nodes and edges between them are considered as atoms and chemical bonds between them, respectively. Topological indices are mathematical measure of molecular graph that remain invariant under graph isomorphism. It was originated in 194714 and then its journey started. A great variety of topological indices has been developed and a lot of work has been performed on computing indices of various molecular graphs and networks. Nilanjan De15 computed the reformulated first Zagreb index of some chemical graphs as an application of generalized hierarchical product of graphs. Gao et al.16 obtained some degree-based indices of networks derived from Honey comb networks. In the work of Anjum and Safdar,17 different structural properties of nanostructures are investigated in terms of several topological indices. Some multiplicative degree-based indices for Bismuth Tri-Iodide chain and sheet are investigated by Shao et al.18 Topological indices of Graphene and chemical compound widely used in the manufacture of anticancer drugs are derived in studies of Mondal et al.19 and Zheng et al.20 To overcome the laborious approach of computing of a certain type of indices of a specific category of graphs, many algebraic polynomials are found in the literature. For instance, the Hosoya polynomial plays the key role in the field of distance-based topological indices.21 Its differentiation at 1 yields Wiener and hyper-Wiener index.22 The Tratch–Stankevich–Zefirov index can be obtained similarly.23 Some more polynomials in the area of chemical graph theory are the Clar covering polynomial,24 PI Polynomial,25 Schultz polynomial,26 theta polynomial,27 Tutte polynomial,28 etc. In case of degree-based topological indices,29 the M-polynomial has a significant role to compute the indices. It was introduced by Deutsch and Klavzar in 201530 where its role to compute degree-based indices was shown to be parallel to the role of the Hosoya polynomial for distance-based indices. After that, numerous works on M-polynomial have been done. Munir et al. computed M-polynomial and degree-based topological indices for nanostar dendrimers,31 single-walled titania nanotubes,32 armchair and zigzag polyhex nanotubes,33 and family of circulant graphs.34 Present authors derived M-polynomial of para-line graphs of some graceful structures and hence recovered some degree-based indices.35 Topological indices of V-phylenic nanotubes and nanotori are investigated by Chel Kwun et al.36 using M-polynomial approach. Gao et al.37 put their attention to compute topological indices of crystallographic structure of cuprous oxide and titanium difluoride using M-polynomial. General expressions of M-polynomial for triangular, hourglass and jagged-rectangle benzenoid systems are evaluated in Chel Kwun et al.38 which produce several indices based on degree. Similar work is performed for cactus chains, benzene ring embedded in P-type surface network, zigzag and rhombic benzenoid and 3-layered probabilistic neural network.39–43 Let denote the degree of vertex u of a graph G. The M-polynomial of G is defined as,
| (1) |
Neighborhood M-polynomial44 play the similar role for neighborhood degree sum-based indices.45–48 Some topological indices based on neighborhood degree sum for bismuth tri-iodide chain and sheet are derived using neighborhood M-polynomial method.49 Let denote the degree sum of all vertices of G that are adjacent to u. We call as neighborhood degree sum of u in G. The neighborhood M-polynomial of G is defined as,
| (2) |
Degree-based (D) and neighborhood degree sum-based (ND) topological indices defined on edge set E(G) of a graph G can be expressed as
where the formulation of and are given in Table 1.
Table 1.
Description of some topological indices.
| D | ND | . | |
|---|---|---|---|
| First Zagreb index (M1)50 | Third version of Zagreb index (59 | ||
| Second Zagreb index (M2)50 | Neighborhood second Zagreb index ()46 | ||
| Forgotten topological index (F)51 | Neighborhood forgotten topological index ()46 | ||
| Second modified Zagreb index ()52 | Neighborhood second modified Zagreb index ()49 | ||
| General Randić index ()53 | Neighborhood general Randić index ()49 | ||
| Redefined third Zagreb index (ReZG3)54 | Third NDe index47 (ND3) | ||
| Symmetric division deg index (SDD)55 | Fifth NDe index47 (ND5) | ||
| Harmonic index (H)56 | Neighborhood Harmonic index (NH)49 | ||
| Inverse sum indeg index (I)57 | Neighborhood inverse sum index (NI)49 | ||
| Augmented Zagreb index (A)58 | Sanskruti index (S)60 |
The first and second Zagreb indices were determined to be effective for the measurement of the total π-electron energy of molecules.61 The Randić index has been related to a huge amount of structural properties of atoms. SDD is used to assess the total surface area of polychlorobiphenyls.62 The augmented Zagreb index provides the best estimation of the heat of formation of alkanes.58 Neighborhood degree sum-based indices can predict different physicochemical properties with powerful accuracy.46,47,49,59,60 The goal of this work is to evaluate some topological indices of the antiviral compounds remdesivir, chloroquine, hydroxychloroquine and theaflavin. First, we obtain M-polynomial and NM-polynomial of the structures and then recover some degree-based and neighborhood degree sum-based indices. Further, surface representation of M and NM polynomials are depicted. In addition, some results are compared graphically.
2. Methodology
Our main findings include topological indices of some antiviral drug structures with the help of algebraic polynomials. The chemical structures of remdesivir, chloroquine, hydroxychloroquine and theaflavin are collected from pubchem.ncbi.nlm.nih.gov. We consider hydrogen suppressed molecular graphs of compounds since the vertices representing hydrogen atom make no contribution to graph isomorphism. We utilize combinatorial computation, edge partition method, graph theoretical tools, analytical techniques and degree counting method to derive our results. At first patterns of edge partitions of hydrogen deleted molecular graph of the aforesaid compounds are constructed based on degree and neighborhood degree sum of end vertices. Using those partitions, some closed forms of M-polynomial and NM-polynomial are derived. The surface plotting of the polynomials are made by using Maple 2015. Degree-based and neighborhood degree sum-based indices are computed from M-polynomial and NM-polynomial using some mathematical operators and Table 2 as defined in previous section. Numerical results are compared graphically using MATLAB 2017.
Table 2.
Derivation of topological indices from M-polynomial and NM-polynomial.
| Topological index | Derivation from M(G) | Topological index | Derivation from NM(G). |
|---|---|---|---|
| M1 | |||
| M2 | |||
| F | |||
| ReZG3 | ND3 | ||
| SDD | ND5 | ||
| H | NH | ||
| I | NI | ||
| A | S |
Here,
3. Results and discussion
In this section, we give our main computational results. We compute the M-polynomial and NM-polynomial of molecular graph of remdesivir (GS-5734) in the following theorem.
Theorem 1.
Let G be the molecular graph of remdesivir (GS-5734). Then, we have,
Proof 1.
Let G be the molecular graph of remdesivir (Figure 1). It has 44 number of edges. Let be the set of all edges with degree of end vertices i, j, i.e., . Let be the number of edges in . From Figure 1, it is clear that . From the Equation (1), the M-polynomial of G is obtained as follows.
Putting the values of “s, we obtain
Let be the set of all edges with neighborhood degree sum of end vertices i, j, i.e., . Let be the number of edges in . From Figure 1, we have . From the Equation (2), the NM-polynomial of G is obtained bellow.
After putting the values of ‘s, we obtain the required result (Figure 2). □
Figure 1.
Chemical structure of remdesivir (GS-5734).
Figure 2.
Plotting of (a) M-polynomial and (b) NM-polynomial of remdesivir (GS-5734).
Now using the M and NM polynomials, we calculate some degree-based and neighborhood degree some-based topological indices of the molecular graph of remdesivir in the following theorem.
Theorem 2.
Let G be the molecular graph of remdesivir. Then, we have,
,
,
,
,
,
,
,
,
,
.
Proof 2.
First, we compute the degree-based indices. Let Then, we have,
Using Table 2, we have
For neighborhood degree sum-based indices, we consider . Then, applying the above operations and Table 2, we can easily obtain the neighborhood degree sum-based indices. This completes the proof. □
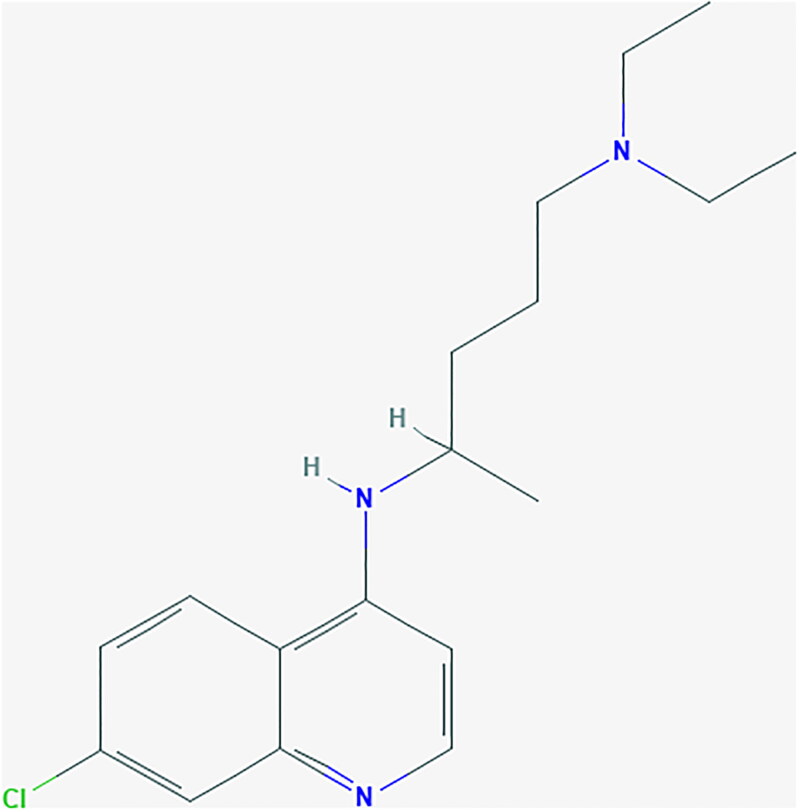
We evaluate the M-polynomial and NM-polynomial of the molecular graph of chloroquine in the following theorem.
Theorem 3.
Let G be the molecular graph of chloroquine. Then, we have,
Proof 3
. Let G be the molecular graph of chloroquine (Figure 3). It has 23 number of edges. From Figure 3, its edge partition based on degree of end vertices is as follows: Using the edge partition of G, the M-polynomial can be evaluated easily like previous. The edge partition based on neighborhood degree sum of end vertices is as follows: Using the edge partition of G, the NM-polynomial can be obtained easily. □
Figure 3.
Chemical structure of chloroquine.
Now using the M and NM polynomials, we calculate some degree-based and neighborhood degree some-based indices of chloroquine in the following theorem like the Proof 2.
Theorem 4.
Let G be the molecular graph of chloroquine. Then, we have (Figure 4),
,
,
,
,
,
,
,
,
,
.
M-polynomial and NM-polynomial of the molecular graph of hydroxychloroquine are evaluated below (Figures 5 and 6).
Figure 4.
Plotting of (a) M-polynomial and (b) NM-polynomial of chloroquine.
Figure 5.
Chemical structure of hydroxychloroquine.
Figure 6.
Plotting of (a) M-polynomial and (b) NM-polynomial of hydroxychloroquine.
Theorem 5.
Let G be the molecular graph of hydroxychloroquine. Then, we have,
Proof 4.
Let G be the molecular graph of hydroxychloroquine (Figure 5). It has 24 number of edges. From Figure 5, its edge partition based on degree of end vertices is as follows: Using the edge partition of G, the M-polynomial can be evaluated easily like previous. The edge partition based on neighborhood degree sum of end vertices is as follows: Using the edge partition of G, the NM-polynomial can be obtained easily. □
Now using the M and NM polynomials, we evaluate the degree-based and neighborhood degree sum-based indices of hydroxychloroquine in the following theorem like the same method as Proof 2.
Theorem 6.
Let G be the molecular graph of hydroxychloroquine. Then, we have,
,
,
,
,
,
,
,
,
,
.
M-polynomial and NM-polynomial of the molecular graph of theaflavin are computed as follows.
Theorem 7.
Let G be the molecular graph of theaflavin. Then, we have,
Proof 5.
Let G be the molecular graph of theaflavin (Figure 7). It has 46 number of edges. Its edge partition based on degree of end vertices is as follows: Using the edge partition of G, the M-polynomial can be evaluated easily like previous. The edge partition based on neighborhood degree sum of end vertices is as follows: Using the edge partition of G, the NM-polynomial can be obtained easily. □
Figure 7.
Chemical structure of theaflavin (ZINC3978446).
The M-polynomial and the NM-polynomial are the wealth of information about degree-based and neighborhood degree sum-based indices, respectively. We expect that a deeper analysis of the properties of the M-polynomial and the NM-polynomial would open up novel general insights in the study of topological indices. To visualize the polynomials, their surface plotting is made by Maple 2015. The expressions of M and NM-polynomials for remdesivir, chloroquine, hydroxychloroquine and theaflavin are depicted in Figures 2, 4, 6 and 8, respectively. For different surfaces, different colors are assigned. First, we construct a horizontal grid by considering the parameters x, y and then, the surface is built on that grid. This plot reveals that the polynomials show different behaviors corresponding to different parameters. We can control topological indices and hence different properties and activities by regulating M and NM-polynomials through those parameters.
Figure 8.
Plotting of (a) M-polynomial and (b) NM-polynomial of theaflavin (ZINC3978446).
Now applying the M and NM polynomials, we can easily obtain the degree-based and neighborhood degree sum-based indices of the molecular graph of theaflavin in the following theorem like the Proof 2.
Theorem 8.
Let G be the molecular graph of theaflavin. Then, we have,
,
,
,
,
,
,
,
,
,
.
4. Conclusion
In this article, we have studied some topological properties of some chemical structures used to inhibit the outbreak and transmission of COVID-19 in terms of some degree-based and some neighborhood degree sum-based indices. It includes remdesivir (GS-5734), chloroquine, hydroxychloroquine and theaflavin. First, we evaluate M-polynomial and NM-polynomial of those structures with graphical representations. Then, we compute some topological indices from the expression of polynomials. We also made a graphical comparison of and for aforesaid structures in Figure 9. As considered topological indices are able to predict different properties and activities such as boiling point, entropy, enthalpy, acentric factor, critical pressure, etc., our findings can be helpful in designing new drug and vaccine for the treatment of COVID-19.
Figure 9.
Plotting of and for different molecular structures under consideration. In vertical axis, logarithmic values of indices are considered to show the comparison clearly.
Funding Statement
This work was supported by Department of Science and Technology (DST), Government of India (INSPIRE Fellowship).
Disclosure statement
There are no conflicts of interest.
References
- 1.Huang Chaolin, Wang Yeming, Li Xingwang, Ren Lili, Zhao Jianping, Hu Yi, Zhang Li, Fan Guohui, Xu Jiuyang, Gu Xiaoying, et al. , “Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China,” The Lancet 395, no. 10223 (2020): 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Manli, Cao Ruiyuan, Zhang Leike, Yang Xinglou, Liu Jia, Xu Mingyue, Shi Zhengli, Hu Zhihong, Zhong Wu, Xiao Gengfu, et al. “Remdesivir and Chloroquine Effectively Inhibit the Recently Emerged Novel Coronavirus (2019-nCoV) in Vitro,” Cell Research 30, no. 3 (2020): 269–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou Dan, Dai Sheng-Ming, and Tongg Qiang, “COVID-19: A Recommendation to Examine the Effect of Hydroxychloroquine in Preventing Infection and Progression,” Journal of Antimicrobial Chemotherapy dkaa114: (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lung Jrhau, Lin Yu‐Shih, Yang Yao‐Hsu, Chou Yu‐Lun, Shu Li‐Hsin, Cheng Yu‐Ching, Liu Hung Te, and Wu Ching‐Yuan, “The Potential Chemical Structure of anti-SARS-CoV-2 RNA-Dependent RNA Polymerase,” Journal of Medical Virology (2020): 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morse Jared S., Lalonde Tyler, Xu Shiqing, and Liu Wenshe Ray, “Learning from the past: Possible Urgent Prevention and Treatment Options for Severe Acute Respiratory Infections Caused by 2019-nCoV,” Chembiochem: A European Journal of Chemical Biology 21, no. 5 (2020): 730–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Xintian, Chen Ping, Wang Jingfang, Feng Jiannan, Zhou Hui, Li Xuan, Zhong Wu, and Hao Pei, “Evolution of the Novel Coronavirus from the Ongoing Wuhan Outbreak and Modeling of Its Spike Protein for Risk of Human Transmission,” Science China. Life Sciences 63, no. 3 (2020): 457–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warren Travis K., Jordan Robert, Lo Michael K., Ray Adrian S., Mackman Richard L., Soloveva Veronica, Siegel Dustin, Perron Michel, Bannister Roy, Hui Hon C., et al. “Therapeutic Efficacy of the Small Molecule GS-5734 against Ebola Virus in Rhesus Monkeys,” Nature 531, no. 7594 (2016): 381–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savarino Andrea, Di Trani Livia, Donatelli Isabella, Cauda Roberto, and Cassone Antonio, “New Insights into the Antiviral Effects of Chloroquine,” The Lancet. Infectious Diseases 6, no. 2 (2006): 67–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan Zhen, Zou Yiwu, Sun Yang, Li Xiao, Xu Kai-Feng, Wei Yuquan, Jin Ningyi, and Jiang Chengyu, “Anti-Malaria Drug Chloroquine is Highly Effective in Treating Avian Influenza a H5N1 Virus Infection in an Animal Model,” Cell Research 23, no. 2 (2013): 300–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Zi-Feng, Bai Li-Ping, Huang Wen-Bo, Li Xu-Zhao, Zhao Sui-Shan, Zhong Nan-Shan, and Jiang Zhi-Hong, “Comparison of in Vitro Antiviral Activity of Tea Polyphenols against Influenza A and B Viruses and Structure–Activity Relationship Analysis,” Fitoterapia 93 (2014): 47–53. [DOI] [PubMed] [Google Scholar]
- 11.Chowdhury Pritom, Sahuc Marie-Emmanuelle, Rouillé Yves, Rivière Céline, Bonneau Natacha, Vandeputte Alexandre, Brodin Priscille, Goswami Manoranjan, Bandyopadhyay Tanoy, Dubuisson Jean, et al. “Theaflavins, Polyphenols of Black Tea, Inhibit Entry of Hepatitis C Virus in Cell Culture,” PLoS One 13, no. 11 (2018): e0198226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trinajstić Nenad. Chemical Graph Theory, 2nd ed (Boca Raton: CRC Press, 1983). [Google Scholar]
- 13.Gutman Ivan and Polansky Oskar E., Mathematical Concepts in Organic Chemistry (Berlin: Springer, 1986). [Google Scholar]
- 14.Wiener Harold. “Structural Determination of Paraffin Boiling Points,” Journal of the American Chemical Society 69, no. 1 (1947): 17–20. [DOI] [PubMed] [Google Scholar]
- 15.De Nilanjan. “ Computing Reformulated First Zagreb Index of Some Chemical Graphs as an Application of Generalized Hierarchical Product of Graphs,” Open Journal of Mathematical Sciences 2, no. 1 (2018): 338–50. [Google Scholar]
- 16.Gao Wei, Asif Muhammad, and Nazeer Waqas, “The Study of Honey Comb Derived Network via Topological Indices,” Open Journal of Mathematical Analysis 2, no. 2 (2018): 10–26. [Google Scholar]
- 17.Anjum Muhammad Shahzad and Safdar Muhammad Usman, “K Banhatti and K hyper-Banhatti Indices of Nanotubes,” Engineering and Applied Science Letters 2, no. 1 (2019): 19–37. [Google Scholar]
- 18.Shao Zehui, Virk Abaid ur Rehman, Javed Muhammad Samar, Rehman M. A., and Farahani Mohammad Reza, “Degree Based Graph Invariants for the Molecular Graph of Bismuth Tri-Iodide,” Engineering and Applied Science Letters 2, no. 1 (2019): 1–11. [Google Scholar]
- 19.Mondal Sourav, De Nilanjan, and Pal Anita, “Topological Properties of Graphene Using Some Novel Neighborhood Degree-Based Topological Indices,” International Journal of Mathematics for Industry 11, no. 01 (2019): 1950006. [Google Scholar]
- 20.Zheng Lina, Wang Yiqiao, and Gao Wei, “Topological Indices of Hyaluronic Acid-Paclitaxel Conjugates’ Molecular Structure in Cancer Treatment,” Open Chemistry 17, no. 1 (2019): 81–7. [Google Scholar]
- 21.Hosoya Haruo, “On Some Counting Polynomials in Chemistry,” Discrete Applied Mathematics. 19, no. 1–3 (1988): 239–57. [Google Scholar]
- 22.Gutman Ivan, “Some Properties of the Wiener Polynomials,” Graph Theory Notes N.Y 125 (1993): 13–8. [Google Scholar]
- 23.Bruckler Franka Miriam, Doslić Tomislav, Graovac Ante, and Gutman Ivan., “On a Class of Distance-Based Molecular Structure Descriptors,” Chemical Physics Letters 503, no. 4–6 (2011): 336–8. [Google Scholar]
- 24.Zhang Heping, and Zhang Fuzi, “The Clar Covering Polynomial of Hexagonal Systems I,” Discrete Applied Mathematics. 69, no. 1–2 (1996): 147–67. [Google Scholar]
- 25.Alamian Vahid, Bahrami Amir, and Edalatzadeh Behrooz, “PI Polynomial of V-Phenylenic Nanotubes and Nanotori,” International Journal of Molecular Sciences 9, no. 3 (2008): 229–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassani F., Iranmanesh Ali, and Mirzaie Samaneh, “Schultz and Modified Schultz Polynomials of C100 Fullerene,” MATCH Communications in Mathematical and in Computer Chemistry 69 (2013): 87–92. [Google Scholar]
- 27.Mohammad Reza Farahani. “Computing Theta Polynomial, and Theta Index of V-Phenylenic Planar, Nanotubes and Nanotoris,” Journal of Theoretical Chemistry 1, no. 1 (2013): 01–9. [Google Scholar]
- 28.Doslić Tomislav. “Planar Polycyclic Graphs and Their Tutte Polynomials,” Journal of Mathematical Chemistry 51 (2013): 1599–607. [Google Scholar]
- 29.Gutman Ivan. “Degree-Based Topological Indices,” Croatica Chemica Acta 86, no. 4 (2013): 351–61. [Google Scholar]
- 30.Deutsch Emeric and Klavzar Sandi, “M-Polynomial, and Degree-Based Topological Indices,” Iranian Journal of Mathematical Chemistry 6, no. 2 (2015): 93–102. [Google Scholar]
- 31.Munir Mobeen, Nazeer Waqas, Rafique Shazia, and Kang Shin, “M-Polynomial and Related Topological Indices of Nanostar Dendrimers,” Symmetry 8, no. 9 (2016): 97–108. [Google Scholar]
- 32.Munir Mobeen, Nazeer Waqas, Nizami Abdul, Rafique Shazia, and Kang Shin, “M-Polynomials and Topological Indices of Titania Nanotubes,” Symmetry 8, no. 11 (2016): 117–25. [Google Scholar]
- 33.Munir Mobeen, Nazeer Waqas, Rafique Shazia, and Kang Shin, “M-Polynomial and Degree-Based Topological Indices of Polyhex Nanotubes,” Symmetry 8, no. 12 (2016): 149–56. [Google Scholar]
- 34.Munir Mobeen, Nazeer Waqas, Shahzadi Zakia, and Kang Shin, “Some Invariants of Circulant Graphs,” Symmetry 8, no. 11 (2016): 134–41. [Google Scholar]
- 35.Mondal Sourav, De Nilanjan, and Pal Anita, “The M-Polynomial of Line Graph of Subdivision Graphs,” Communications Faculty of Sciences University of Ankara Series A1-Mathematics and Statistics 68, no. 2 (2019): 2104–16. [Google Scholar]
- 36.Chel Kwun Young, Munir Mobeen, Nazeer Waqas, Rafique Shazia, and Min Kang Shin, “M-Polynomials and Topological Indices of V-Phenylenic Nanotubes and Nanotori,” Scientific Reports 7, no. 1 (2017): 8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao Wei, Younas Muhammad, Farooq Adeel, Mahboob Abid, and Nazeer Waqas, “M-Polynomials and Degree-Based Topological Indices of the Crystallographic Structure of Molecules,” Biomolecules 8, no. 4 (2018): 107–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chel Kwun Young, Ali Ashaq, Nazeer Waqas, Ahmad Chaudhary Maqbool, and Kang Shin Min, “M-Polynomials and Degree-Based Topological Indices of Triangular, Hourglass, and Jagged-Rectangle Benzenoid Systems,” Journal of Chemistry. 2018 (2018): 1–8. [Google Scholar]
- 39.Basavanagoud Bommanahal, and Barangi Anand P., “M-Polynomial of Some Cactus Chains and Their Topological Indices,” Open Journal of Discrete Applied Mathematics 2, no. 2 (2019): 59–67. [Google Scholar]
- 40.Ali Ashaq, Nazeer Waqas, Munir Mobeen, and Min Kang Shin, “M-Polynomials and Topological Indices of Zigzag and Rhombic Benzenoid Systems,” Open Chemistry 16, no. 1 (2018): 73–8. [Google Scholar]
- 41.Yang Hong, Baig A. Q., Khalid W., Farahani Mohammad Reza, and Zhang Xiujun, “M-Polynomial and Topological Indices of Benzene Ring Embedded in P-Type Surface Network,” Journal of Chemistry 2019 (2019): 1–9. [Google Scholar]
- 42.Javaid Muhammad, Raheem Abdul, Abbas Mujhaid, and Cao Jinde, “M-Polynomial Method for Topological Indices of 3-Layered Probabilistic Neural Networks,” TWMS Journal of Applied and Engineering Mathematics 9, no. 4 (2019): 864–75. [Google Scholar]
- 43.Liu Guoshun, Jia Zhiyang, and Gao Wei “Ontology Similarity Computing Based on Stochastic Primal Dual Coordinate Technique,” Open Journal of Mathematical Sciences 2, no. 1 (2018): 221–7. [Google Scholar]
- 44.Changyin Gan, Kamran Siddiqui Muhammad, Mondal Sourav, and De Nilanjan, “On Topological Indices of Crystallographic Structures,” Preprint. [Google Scholar]
- 45.Mondal Sourav, De Nilanjan, and Pal Anita, “On Neighbourhood Zagreb Index of Product Graphs,” arXiv:1805.05273 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mondal Sourav, De Nilanjan, and Pal Anita, “On Some New Neighbourhood Degree Based Indices,” Acta Chemica Iasi 27, no. 1 (2019): 31–46. [Google Scholar]
- 47.Mondal Sourav, De Nilanjan, and Pal Anita, “QSPR Analysis of Some Novel Neighborhood Degree Based Topological Descriptors,” arXiv:1906.06660 (2019). [Google Scholar]
- 48.Mondal Sourav, De Nilanjan, and Pal Anita, “On Some General Neighborhood Degree Based Indices,” International Journal of Applied Mathematics 32, no. 6 (2019): 1037–49. [Google Scholar]
- 49.Verma Ashish, Mondal Sourav, De Nilanjan, and Pal Anita, “Topological Properties of Bismuth Tri-Iodide Using Neighborhood M-Polynomial,” International Journal of Mathematics Trends and Technology 67, no. 10 (2019): 83–90. [Google Scholar]
- 50.Gutman Ivan and Trinajstić Ninad, “Graph Theory and Molecular Orbitals Total Π-Electron Energy of Alternant Hydrocarbons,” Chemical Physics Letters 17, no. 4 (1972): 535–8. [Google Scholar]
- 51.Furtula Boris and Gutman Ivan, “A Forgotten Topological Index,” Journal of Mathematical Chemistry 53, no. 4 (2015): 1184–90. [Google Scholar]
- 52.Dhanalakshmi K., Jerline J. Amalorpava, and Benedict Michael Raj L., “Modified Zagreb Index of Some Chemical Structure Trees,” International Journal of Mathematics and Its Applications 5, no. 1 (2017): 285–90. [Google Scholar]
- 53.Amić Dragan, Beslo Drago, Lucić Bono, Nikolić Sonja, and Trinajstić Nenad, “The Vertex-Connectivity Index Revisited,” Journal of Chemical Information and Computer Sciences 38, no. 5 (1998): 819–22. [Google Scholar]
- 54.Ranjini P. S., Lokesha V., and Usha A., “Relation between Phenylene and Hexagonal Squeeze Using Harmonic Index,” International Journal of Graph Theory 1, no. 4 (2013): 116–21. [Google Scholar]
- 55.Vukicevic Damir. “Bond Additive Modeling 2 Mathematical Properties of Max-Min Rodeg Index,” Croatica Chemica Acta 54, no. 3 (2010): 261–73. [Google Scholar]
- 56.Fajtlowicz Siemion. “On Conjectures of Graffiti II,” Congr. Numer 60 (1987): 189–97. [Google Scholar]
- 57.Balaban Alexandru T. “Highly Discriminating Distance-Based Topological Index,” Chemical Physics Letters. 89, no. 5 (1982): 399–404. [Google Scholar]
- 58.Furtula Boris, Graovac Ante, and Vukićević Damir, “Augmented Zagreb Index,” Journal of Mathematical Chemistry 48, no. 2 (2010): 370–80. [Google Scholar]
- 59.Ghorbani Modjtaba and Hosseinzadeh Mohammad A., “The Third Version of Zagreb Index,” Discrete Mathematics, Algorithms and Applications 05, no. 04 (2013): 1350039. [Google Scholar]
- 60.Hosamani Sunilkumar M. “Computing Sanskruti Index of Certain Nanostructures,” Journal of Applied Mathematics and Computing 54, no. 1–2 (2017): 425–33. [Google Scholar]
- 61.Gutman Ivan, Ruscić Branko, Trinajstić Nenad, and Wilcox Charles F., “Graph Theory and Molecular Orbitals, XII. Acyclic Polyenes,” The Journal of Chemical Physics 62, no. 9 (1975): 3399–405. [Google Scholar]
- 62.Lokesha V., and Deepika T., “Symmetric Division Deg Index of Tricyclic and Tetracyclic Graphs,” International Journal of Scientific and Engineering Research 7, no. 5 (2016): 53–5. [Google Scholar]