Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is an emerging coronavirus that belongs to the β‐genus, causing the outbreak of coronavirus disease 19 (COVID‐19). SARS‐CoV‐2 infection can stimulate a pronounced immune response in the host, which embodies in the decrease of lymphocytes and aberrant increase of cytokines in COVID‐19 patients. SARS‐CoV‐2 RNA and proteins interact with various pattern recognition receptors that switch on antiviral immune responses to regulate viral replication and spreading within the host in vivo. However, overactive and impaired immune responses also cause immune damage and subsequent tissue inflammation. This article focuses on the dual roles of immune system during SARS‐CoV‐2 infection, providing a theoretical basic for identifying therapeutic targets in a situation with an unfavourable immune reaction.
Keywords: coronavirus disease 19, cytokine storm, inflammation, severe acute respiratory syndrome coronavirus 2
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is a highly transmissible and pathogenic virus in humans. It currently has spread from China to other countries and become a global threat. The genome of SARS‐CoV‐2 contains 14 open reading frames (ORFs) and encodes 27 different proteins, including the spike (S) protein, envelope (E) protein, membrane (M) glycoproteins and nucleocapsid (N) protein. 1 It is currently believed that SARS‐CoV‐2 belongs to the species of SARS‐related coronavirus, with angiotensin‐converting enzyme 2 (ACE2) as the viral receptor, suggesting a similar tropism and entry route with SARS‐CoV. 2 , 3 SARS‐CoV‐2 infection can cause serious respiratory disease similar to SARS‐CoV, namely novel coronavirus disease 19 (COVID‐19). Common symptoms are fever, cough, shortness of breath and myalgia or fatigue. Some patients with severe disease could progress to acute respiratory distress syndrome (ARDS) and die of multiple organ failure. 4 , 5 Despite the identification of this virus, no specific antivirals or vaccines are currently developed for the treatment of COVID‐19, and the mechanism exacerbating the disease still remains largely undetermined.
Severe lung and systemic inflammation of COVID‐19 patients is currently believed to result from cytokine dysregulation. Recent research have indicated that COVID‐19 is associated with the induction of inflammatory cytokines including IL‐1β, IL‐6, IL‐8, IL‐12, IFN‐γ, GM‐CSF and TNF‐α, many of which were highly expressed in severe COVID‐19 patients. 6 In addition, laboratory investigation of infected patients showed lymphopenia as a universal feature for COVID‐19, and analysis of the lymphocyte subset showed a significant decline in the number of CD4+ and CD8+ T cells. 6 , 7 SARS‐COV‐2‐infected patients were observed to have massive accumulation of inflammatory cytokines and aberrant T cell responses compared to healthy individuals, providing evidence that COVID‐19 may be an immune interrelated disease. Therefore, it is crucial to assess the positive and negative roles of the immune system in SARS‐CoV‐2 infection for a more comprehensive and detailed understanding of the molecular mechanisms underlying the pathogenesis of SARS‐CoV‐2. Such dual functions need to be carefully evaluated when developing therapeutic intervention strategies targeting the immune system during SARS‐CoV‐2 infection. Even though the clinical symptoms exhibited by SARS‐CoV‐2 infection indicate that it can bring about immune responses, there is currently very little knowledge about how SARS‐CoV‐2 activates the immune system. SARS‐CoV‐2 has a high degree of sequence similarity to SARS‐CoV, with 76.47% identity on S proteins. 2 , 8 It has been reported that many B‐ and T cell epitopes are also highly conserved between SARS‐CoV and SARS‐CoV‐2, and antibodies against SARS‐CoV will cross‐neutralize SARS‐CoV‐2. 9 , 10 , 11 Therefore, SARS‐CoV‐2 may be similar to SARS‐CoV in antigenicity, and there exist cross‐reactive epitopes. In this article, the latest research about SARS‐CoV‐2 is combined with immunological studies of SARS‐CoV to analyse the possible roles of immune responses during SARS‐CoV‐2 infection.
2. INNATE IMMUNE RESPONSES TO SARS‐COV‐2 INFECTION
2.1. Innate immune response‐mediated antiviral response
The innate immune signalling pathways usually begin with the recognition of specific pathogen‐associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs), mainly including the RIG‐I‐like receptors (RLRs) and Toll‐like receptors (TLRs). SARS‐related coronaviruses usually enter the host cells though binding to cellular receptors and receptor‐mediated endocytosis; the viral RNA is subsequently released to the cytosol when the S protein induces fusion between the envelope of virus and endosome. 11 , 12 So, viral RNA and S protein of SARS‐related coronaviruses may have evolved as major PAMPs which can mediate innate immune signalling cascades, initiating an antiviral state in infected cells.
2.1.1. RLRs‐dependent antiviral signalling pathway
RIG‐I and MDA5 are RNA helicases that precisely target viral RNA in the cytoplasm. RIG‐I directly recognizes and binds to viral 5′‐PPP RNA and short dsRNA through its helicase and repressor domain (RD), while MDA5 senses long dsRNAs. 13 , 14 As positive, single‐stranded RNA virus, SARS‐CoV‐2 is likely to have similar replication intermediates (putative RLR ligands) to other RNA viruses, which could be detected by the same sensors. After sensing virus, RIG‐I and MDA5 converge on mitochondrial adaptor protein, including mitochondrial antiviral signalling protein (MAVS), interferon‐β promoter stimulator 1 (IPS‐1) or virus‐induced signalling adaptor (VISA), which mediate the signalling cascade. 15 , 16 , 17 These adaptor proteins use various TRAFs to trigger TBK1/IKKE and IKKα/IKKβ that, respectively, mediate the activation of different transcription factors. 18 The RLRs signalling pathway eventually activates interferon regulatory factor (IRF)‐3 and IRF‐7 before they are translocated to the nucleus and stimulate expression of type I interferon (IFN‐I). 19 , 20 , 21 IFN‐I (IFN‐α and IFN‐β) utilizes autocrine and paracrine signalling to make sure cells express a myriad of interferon‐stimulated genes (ISGs), which establish an antiviral state. 21 , 22
2.1.2. TLR‐dependent antiviral signalling pathway
Toll‐like receptors are also important pattern recognition receptors of virus, recognizing viral components or replication intermediates. TLR3, TLR7, TLR8 and TLR9 detect viral nucleic acid in the intracellular compartments, while TLR2 and TLR4 recognize viral proteins on the cell surface. 23 It has been identified that TLR2 mRNA increased in PBMC among SARS‐CoV patients at acute phase, so TLR2 may recognize S protein of SARS‐CoV. 24 S protein of SARS‐CoV and SARS‐CoV‐2 has a high degree of sequence similarity and fuse with the same receptor ACE2 to enter host cells. 2 , 8 So TLR2 is also likely to detect SARS‐CoV‐2 S protein even though no TLRs have been confirmed to be related to the recognition of SARS‐CoV‐2. In hACE2 receptor‐positive lung epithelial and fibroblast cells, SARS‐CoV S1 protein induced IL‐8 through hACE2 signalling. 25 Then, activated TLRs combine with the adaptor molecule MyD88 and TRIF, leading to the activation of IRF3, IRF7 and NF‐kB; these transcription factors subsequently initiate transcription of IFN‐I and other cytokines, respectively. 26 , 27 , 28 In COVID‐19 patients, it has been observed that the activity of multiple IRFs is enhanced, which may assist the occurrence of IFN‐I‐related immune response to prevent viral spreading. 29
2.2. Innate immune response‐mediated inflammatory response
Although innate immune signalling pathways eventually caused the production of antiviral factor IFN‐I, innate immune signalling cascades also lead to the activation of NF‐κB that would lead to the production of inflammatory mediators, especially that of IL‐6 and IL‐8. 24 , 30 These innate immune effector molecules continue to mediate inflammation and cellular antiviral processes. It is worth noting that SARS‐CoV infection activates NF‐κB at 12‐hour post‐infection in vitro studies, while IFNs and ISGs are delayed in expression until 48‐hour post‐infection. 31 Early activation of NF‐κB and delayed production of IFN‐I could exacerbate host innate inflammatory responses by modulating, in part, the intrinsic functions of macrophages (MΦ) and dendritic cells (DC). 32 It has been confirmed that delayed IFN‐I response leads to a highly pathogenic IFN‐I‐dependent inflammatory response driven by inflammatory monocyte‐macrophages (IMMs) in susceptible mice. 33 Accumulation of pathogenic IMMs to the site of viral infection results in elevated lung cytokine levels and impaired virus‐specific T cell responses. 33
In fact, various inflammation‐related cytokines did increase due to the SARS‐CoV‐2 infection, which was correlated with the severity of the disease. 6 Such an intense cytokine response may also be attributed to hyper‐activation of IMM lineage cells; it has been reported that patients with severe disease have a larger accumulation of inflammatory macrophages in the lungs than patients with mild disease. 29 , 34 If SARS‐CoV‐2 is similar to SARS‐CoV, the speed and efficiency by which SARS‐CoV‐2 circumvents and delays the IFN‐I response may be a critical determinant of its pathogenicity.
3. T CELL IMMUNE RESPONSES TO SARS‐COV‐2 INFECTION
3.1. T cell‐mediated antiviral immune response
T cell immune responses are specific and can memorize the pathogens, playing an important role in fighting the virus. During the course of SARS‐CoV‐2 infection, activated CD4+ and CD8+ T cells are recruited to the lung of the COVID‐19 patients, and the levels of these T cells may be related to the outcome of the disease. 6 , 7 The expansion levels in both total T and CD8+ T cells are significantly higher in patients with mild disease, mediating a robust adaptive immune response. 29 But in multiple patients with severe disease, T cells have experienced a severe decline, leading to virus transmission, cytokine storm and high mortality. 4 , 5 , 6 , 7 It provides clinical evidence indicating that T cells are necessary for virus clearance during SARS‐CoV‐2 infection.
3.1.1. CD8+ T cell‐mediated immune responses
It is reported that a high number of activated CD8+ T cells were detected in blood of a patient with mild‐to‐moderate COVID‐19, suggesting a role of CD8+ T cells against SARS‐CoV‐2 infection. 35 It has been confirmed that CD8+ T cell responses are critical for virus clearance and protection from clinical disease in mice or human infected with other coronaviruses, such as SARS‐CoV, Middle East Respiratory Syndrome Coronavirus (MERS‐CoV) and mouse hepatitis virus (MHV). 36 , 37 , 38 Activated virus‐specific CD8+ T cells produce antiviral cytokines (IFN‐γ, TNF‐α and IL‐2), cytotoxic molecules (perforin and granzyme B), which mediate the clearance of virus and virus‐infected cells. 38 , 39 , 40 According to report of Liao et al, 29 CD8+ T cells of mild COVID‐19 patients expressed high levels of cytotoxic molecules, including granzyme A, granzyme K and FASL, which may kill virus‐infected cells by two contact‐dependent mechanisms. In the granzyme pathway, T cell receptor activation and release of lytic granule containing serine proteases lead to lysis of target cells; while in the FAS/ FASL pathway, target cell cytotoxicity is triggered when FASL expresses predominantly on activated T cells and binds FAS on the virus‐infected cells. 41 But in CD8+ T cells of the patients with severe disease, display reduced amount of cytotoxic molecules, which leads to lower proportion of cytotoxic T lymphocyte (CTL) compared with patients with mild disease, thereby failing to provide a robust response against SARS‐CoV‐2 infection. 29 So, in the lung microenvironment of COVID‐19 patients, highly expanded and functionally competent tissue resident clonal CD8+ T cells and timely CTL responses may connect with a better control of infection.
3.1.2. CD4+ T cell‐mediated immune responses
CD8+ CTLs alone may be not sufficient to control SARS‐CoV‐2 infection. CD4+T cells are also essential for viral clearance, which is likely associated with the production of specific antibodies and antiviral cytokines. At present, as appraised by Ramaiah et al, 42 eight high‐binding affinity CD4+ T cell epitopes are present in the S, E, M and N proteins of SARS‐CoV‐2, which can be commonly recognized by human leucocyte antigen‐DR (HLA‐DR) alleles of populations of the Asia and Asia‐Pacific regions. These antigenic epitopes may provide the basic for initiating the CD4+T cell‐mediated immune response that leads to the development of specific antibodies by activating T‐dependent B cells in vivo. According to the report, the level of follicular helper T cells (Tfh cells) and antibody‐secreting cells (ASCs) increased in a COVID‐19 patient, suggesting that CD4+T cells may bring a strong humoral immunity during SARS‐CoV‐2 infection. 35 High CXCR5 expression in Tfh cells facilitates their homing to B cell follicles and subsequently provides selection signals to germinal centre B cells. 43 In germinal centres, Tfh cells promote B cell differentiation to memory B cells and long‐lived plasma cells, which is essential for long‐lived antibody responses. 43 , 44 Accompanying the ascension of Tfh cells, there existed high levels of IgM and IgG SARS‐CoV‐2‐binding antibodies in the blood of COVID‐19 patients, which may contribute to the viral clearance via neutralizing effect and promoting phagocytosis of phagocytes. 35
3.2. T cell‐mediated aberrant immune response
Multiple reports indicate that COVID‐19 patients have experienced a severe decline in T cell numbers, and the expression of IFN‐γ in CD4+ T cells decreases in the late stage, indicating that Th1 cells or their secretory capacity may be restricted. 45 , 46 Different from SARS‐CoV infection, SARS‐CoV‐2 infection has a bias towards Th2 system dominance, which may lead to increased influx of activated macrophages in the lung microenvironment. 6 , 47 , 48 And under this circumstance, pathogenic microorganisms that had previously coexisted with the host may be no longer suppressed by the immune system, despite a paucity of evidence for bacterial co‐infection. 48 According to reports, 8% patients have experienced bacterial/fungal co‐infection during SARS‐CoV‐2 infection, and 16% of COVID‐19 deaths occurred in patients with secondary infection. 49 , 50 Therefore, whether T cell exhaustion causes secondary infections is a question that needs further investigation in the context of SARS‐CoV‐2 infection. However, although there are fewer lymphocytes, the proportion of activated T cells was increased, as evidenced by the higher double‐positive ratio of HLA‐DR to CD38 in COVID‐19 patients. 35 , 51 , 52 Highly cytotoxic CD8+T cells express high concentrations of cytotoxic particles (granulysin and perforin), causing immune damage to the tissue while clearing the infected cells. 51 In addition, high levels of pro‐inflammatory Th17 cells have been detected by testing patients who have died of COVID‐19. 51 The accumulation of Th17 cells leads to the release of a large number of pro‐inflammatory factors, such as IL‐17 and GM‐CSF that may recruit inflammatory monocytes and neutrophils to the site of inflammation and infection, increasing damage to tissues and organs. 34 , 53 Therefore, aberrant immune response may play an important role in the formation of cytokine storm and the development of macrophages and neutrophils that meditate a profibrotic environment within the lung.
4. CONCLUSION AND PROSPECTS
Returning to the question of the title, it seems that a rapid and coordinated innate and T cell immune response may rapidly control the spread of the virus, while a delayed and aberrant immune response leads to severe lung or systemic inflammation and high mortality. The immune defence triggered by SARS‐CoV‐2 may include initiation of IFN response, and the occurrence of CTL killing activity and neutralizing antibodies; the immunopathogenesis of SARS‐CoV‐2‐induced respiratory distress syndrome may involve deranged innate immune effector molecule production, abnormal elevation of inflammatory immune cells and cytokine storms (Figure 1).
FIGURE 1.
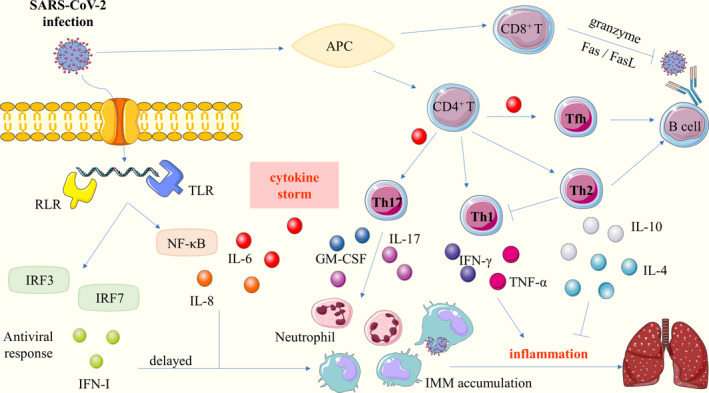
SARS‐CoV‐2 infection‐induced immune responses. SARS‐CoV‐2 enters the host cells though binding to cellular receptors ACE2. After SARS‐CoV‐2 infects cells, the innate immune response begins with the recognition of pathogen‐associated molecular patterns by the RLR and TLR. It activates IRF3, IRF7 and NF‐κB that lead to antiviral response and the production of inflammatory cytokines, respectively (left panel). With the assistance of APC, activated T cells exert different immune effects by distinct immune effector molecule (right panel). The innate and T cell immune responses eventually lead to the recruitment of IMM and neutrophil in the lung, and release high amounts of cytokines that mediate inflammatory damage
Hence, not only should attention be paid to direct virus‐induced cytopathic effects, carrying out antiviral treatment, but also to monitor the patient's immune status to prevent secondary damage caused by SARS‐CoV‐2 infection‐induced exuberant immune response. But it is not completely understood why some patients manifest aberrant immune responses and develop severe disease, but others suffer from mild or even asymptomatic diseases from infection with the same. Therefore, before using immunotherapy, it should be noted that it is necessary to fully understand the patient's current immune status to provide specific treatment, including the degree of T cell activation, the secretion of cytokines and the level of SARS‐CoV‐2‐specific antibodies. Cytokine blockers can be used to treat COVID‐19 patients with cytokine storms, such as IL‐6 receptor antagonist (tocilizumab), IL‐17 inhibitor (secukinumab) or anti‐GM‐CSF monoclonal antibodies (lenzilumab). The plasma of convalescent patients can be injected for emergency immunotherapy to the patients with humoral immunity immunodeficiency.
However, most of the current research were generated from studies about SARS‐CoV or other respiratory viruses, not directly from SARS‐CoV‐2. So, more investigations using SARS‐CoV‐2‐infected animal models and COVID‐19 patient samples are needed to explore the relevant immune protection or pathogenic mechanism, thereby facilitating the treatment and vaccine development.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGMENT
We thank Xiangyan Shan of Sanquan College of Xinxiang Medical University for the linguistic assistance during the preparation of this manuscript.
Li K, Hao Z, Zhao X, Du J, Zhou Y. SARS‐CoV‐2 infection‐induced immune responses: Friends or foes?. Scand J Immunol. 2020;92:e12895. 10.1111/sji.12895
Funding information
This work was funded by Key Science and Technology Project of Henan Province (No. 192102310333); Medical Science and Technology Project Jointly Built of Henan Province (No. 2018020385); Key scientific research projects of Henan Colleges and Universities (No. 19B320015); and Undergraduate Innovation and Entrepreneurship Training Program (No. S201913505004).
REFERENCES
- 1. Wu A, Peng Y, Huang B, et al. Genome composition and divergence of the novel coronavirus (2019‐nCoV) originating in China. Cell Host Microbe. 2020;27:325‐328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xu XW, Wu XX, Jiang XG, et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS‐Cov‐2) outside of Wuhan, China: retrospective case series. BMJ. 2020;368:m606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang JJ, Dong X, Cao YY, et al. Clinical characteristics of 140 patients infected with SARS‐CoV‐2 in Wuhan, China. Allergy. 2020;1–12. [DOI] [PubMed] [Google Scholar]
- 6. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of 2019 novel coronavirus infection in China. medRxiv. 2002;2020.2002.2006.20020974. [Google Scholar]
- 8. Xu X, Chen P, Wang J, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63:457‐460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grifoni A, Sidney J, Zhang Y, et al. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS‐CoV‐2. Cell Host Microbe. 2020;27:671‐680.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qiu T, Mao T, Wang Y, et al. Identification of potential cross‐protective epitope between a new type of coronavirus (2019‐nCoV) and severe acute respiratory syndrome virus. J Genet Genomics. 2020;47:115‐117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hoffmann M, Kleine‐Weber H, Krüger N, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and Is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271‐280.e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qinfen Z, Jinming C, Xiaojun H, et al. The life cycle of SARS coronavirus in Vero E6 cells. J Med Virol. 2004;73:332‐337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kowalinski E, Lunardi T, McCarthy Andrew A, et al. Structural basis for the activation of innate immune pattern‐recognition receptor RIG‐I by viral RNA. Cell. 2011;147:423‐435. [DOI] [PubMed] [Google Scholar]
- 14. Nikonov A, Mölder T, Sikut R, et al. RIG‐I and MDA‐5 detection of viral RNA‐dependent RNA polymerase activity restricts positive‐strand RNA virus replication. PLoS Pathog. 2013;9:e1003610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu T, Chu Q, Cui J, Bi D. Inducible microRNA‐3570 feedback inhibits the RIG‐I‐dependent innate immune response to rhabdovirus in teleost fish by targeting MAVS/IPS‐1. J Virol. 2018;92:e01594‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee NR, Ban J, Lee NJ, et al. Activation of RIG‐I‐mediated antiviral signaling triggers autophagy through the MAVS‐TRAF6‐Beclin‐1 signaling axis. Front Immunol. 2018;9:2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xie T, Chen T, Li C, et al. RACK1 attenuates RLR antiviral signaling by targeting VISA‐TRAF complexes. Biochem Biophys Res Commun. 2019;508:667‐674. [DOI] [PubMed] [Google Scholar]
- 18. Clément JFO, Meloche S, Servant MJ. The IKK‐related kinases: from innate immunity to oncogenesis. Cell Res. 2008;18:889‐899. [DOI] [PubMed] [Google Scholar]
- 19. Parisien JP, Lenoir JJ, Mandhana R, et al. RNA sensor LGP2 inhibits TRAF ubiquitin ligase to negatively regulate innate immune signaling. EMBO Rep. 2018;19:e45176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cheng Y, Zhu W, Ding C, et al. IRF7 is involved in both STING and MAVS mediating IFN‐beta signaling in IRF3‐lacking chickens. J Immunol. 2019;203:1930‐1942. [DOI] [PubMed] [Google Scholar]
- 21. Channappanavar R, Fehr AR, Zheng J, et al. IFN‐I response timing relative to virus replication determines MERS coronavirus infection outcomes. J Clin Invest. 2019;130:3625‐3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Totura AL, Baric RS. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr Opin Virol. 2012;2:264‐275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arpaia N, Barton GM. Toll‐like receptors: key players in antiviral immunity. Curr Opin Virol. 2011;1:447‐454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dosch SF, Mahajan SD, Collins AR. SARS coronavirus spike protein‐induced innate immune response occurs via activation of the NF‐κB pathway in human monocyte macrophages in vitro. Virus Res. 2009;142:19‐27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chang YJ, Liu YY, Chiang BL, et al. Induction of IL‐8 release in lung cells via activator protein‐1 by recombinant baculovirus displaying severe acute respiratory syndrome‐coronavirus spike proteins: identification of two functional regions. J Immunol. 2004;173:7602‐7614. [DOI] [PubMed] [Google Scholar]
- 26. Barton GM, Medzhitov R. Toll‐like receptor signaling pathways. Science. 2003;6:712‐721. [DOI] [PubMed] [Google Scholar]
- 27. Spiegel M, Pichlmair A, Martinez‐Sobrido L, et al. Inhibition of Beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two‐step model for activation of interferon regulatory factor 3. J Virol. 2005;79:2079‐2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kostoula C, Shaker T, Cerovic M, et al. TLR3 preconditioning induces anti‐inflammatory and anti‐ictogenic effects in mice mediated by the IRF3/IFN‐beta axis. Brain Behav Immun. 2019;81:598‐607. [DOI] [PubMed] [Google Scholar]
- 29. Liao M, Liu Y, Yuan J, et al. Single‐cell landscape of bronchoalveolar immune cells in patients with COVID‐19. Nat Med. 2020. 10.1038/s41591-020-0901-9 [DOI] [PubMed] [Google Scholar]
- 30. Mulero MC, Huxford T, Ghosh G. NF‐κB, IκB, and IKK: integral components of immune system signaling. Struct Immunol. 2019;1172:207‐226. [DOI] [PubMed] [Google Scholar]
- 31. Yoshikawa T, Hill TE, Yoshikawa N, et al. Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome‐associated coronavirus infection. PLoS One. 2010;5:e8729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yoshikawa T, Hill T, Li K, Peters CJ, Tseng CT. Severe acute respiratory syndrome (SARS) coronavirus‐induced lung epithelial cytokines exacerbate SARS pathogenesis by modulating intrinsic functions of monocyte‐derived macrophages and dendritic cells. J Virol. 2009;83:3039‐3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Channappanavar R, Fehr AR, Vijay R, et al. Dysregulated type I interferon and inflammatory monocyte‐macrophage responses cause lethal pneumonia in SARS‐CoV‐infected mice. Cell Host Microbe. 2016;19:181‐193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhou Y, Fu B, Zheng X, et al. Aberrant pathogenic GM‐CSF+ T cells and inflammatory CD14+CD16+ monocytes in severe pulmonary syndrome patients of a new coronavirus. bioRxiv. 2002;2020.2002.2012.945576. [Google Scholar]
- 35. Thevarajan I, Nguyen THO, Koutsakos M, Druce J, Kedzierska K. Breadth of concomitant immune responses prior to patient recovery: a case report of non‐severe COVID‐19. Nat Med. 2020;26:453‐455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hyoung‐Shik S, Yeonjae K, Gayeon K, et al. Immune responses to middle east respiratory syndrome coronavirus during the acute and convalescent phases of human infection. Clin Infect Dis. 2018;6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhao J, Zhao J, Perlman S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus‐infected mice▿. J Virol. 2010;84:9318‐9325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Channappanavar R, Fett C, Zhao J, Meyerholz DK, Perlman S. Virus‐specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J Virol. 2014;88:11034‐11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen H, Hou J, Jiang X, et al. Response of memory CD8+ T cells to severe acute respiratory syndrome (SARS) coronavirus in recovered SARS patients and healthy individuals. J Immunol. 2005;175:591‐598. [DOI] [PubMed] [Google Scholar]
- 40. van den Brand JM, Haagmans BL, van Riel D, Osterhaus AD, Kuiken T. The pathology and pathogenesis of experimental severe acute respiratory syndrome and influenza in animal models. J Comp Pathol. 2014;151:83‐112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ma G, Zhu LP, Zhang W. Cytotoxic T cell. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2002;24(4):439‐442. [PubMed] [Google Scholar]
- 42. Ramaiah A, Arumugaswami V. Insights into cross‐species evolution of novel human coronavirus 2019‐nCoV and defining immune determinants for vaccine development. bioRxiv. 2020;2020.01.29.925867. [Google Scholar]
- 43. Yu D, Rao S, Tsai LM, et al. The transcriptional repressor Bcl‐6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457‐468. [DOI] [PubMed] [Google Scholar]
- 44. Zhang Y, Garcia‐Ibanez L, Toellner KM. Regulation of germinal center B‐cell differentiation. Immunol Rev. 2016;270:8‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu B, Fan CY, Wang AL, et al. Suppressed T cell‐mediated immunity in patients with COVID‐ 19: a clinical retrospective study in Wuhan, China. J Infect. 2020. 10.1016/j.jinf.2020.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li G, Fan Y, Lai Y, et al. Coronavirus infections and immune responses. J Med Virol. 2020;92:424‐432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zornetzer GA, Frieman MB, Rosenzweig E, et al. Transcriptomic analysis reveals a mechanism for a prefibrotic phenotype in STAT1 knockout mice during severe acute respiratory syndrome coronavirus infection. J Virol. 2010;84:11297‐11309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dashraath P, Jeslyn WJL, Karen LMX, et al. Coronavirus disease 2019 (COVID‐19) pandemic and pregnancy. Am J Obstet Gynecol. 2020;222:521‐531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rawson TM, Moore LSP, Zhu N, et al. Bacterial and fungal co‐infection in individuals with coronavirus: a rapid review to support COVID‐19 antimicrobial prescribing. Clin Infect Dis. 2020;ciaa530. 10.1093/cid/ciaa530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID‐19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020.46:846‐848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Russell CD, Millar JE, Baillie JK. Clinical evidence does not support corticosteroid treatment for 2019‐nCoV lung injury. Lancet. 2020;395:473‐475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID‐19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420‐422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chuammitri P, Wongsawan K, Pringproa K, Thanawongnuwech R. Interleukin 17 (IL‐17) manipulates mouse bone marrow‐ derived neutrophils in response to acute lung inflammation. Comp Immunol Microbiol Infect Dis. 2019;67:101356. [DOI] [PubMed] [Google Scholar]


