Atlantic salmon, Salmo salar, is the most farmed marine fish worldwide, with an annual production of 2,248 million metric tons in 2016. Salmon hatcheries are increasingly changing from flowthrough toward recirculating aquaculture system (RAS) design to accommodate more control over production along with improved environmental sustainability due to lower impacts on water consumption. To date, microbiome studies of hatcheries have focused either on the fish mucosal microbiota or on the built environment microbiota but have not combined the two to understand their interactions. Our study evaluates how the water and tank biofilm microbiota influences the fish microbiota across three mucosal environments (gill, skin, and digesta). Results from this study highlight how the built environment is a unique source of microbes to colonize fish mucus and, furthermore, how this can influence fish health. Further studies can use this knowledge to engineer built environments to modulate fish microbiota for beneficial phenotypes.
KEYWORDS: 16S, aquaculture, built environment, environmental microbiology, microbial ecology, microbiome
ABSTRACT
Successful rearing of fish in hatcheries is critical for conservation, recreational fishing, commercial fishing through wild stock enhancements, and aquaculture production. Flowthrough (FT) hatcheries require more water than recirculating aquaculture systems (RAS), which enable up to 99% of their water to be recycled, thus significantly reducing environmental impacts. Here, we evaluated the biological and physical microbiome interactions of three Atlantic salmon hatcheries (RAS n = 2, FT n = 1). Gill, skin, and digesta from six juvenile fish along with tank biofilms and water were sampled from tanks in each of the hatcheries (60 fish across 10 tanks) to assess the built environment and mucosal microbiota using 16S rRNA gene sequencing. The water and tank biofilm had more microbial richness than fish mucus, while skin and digesta from RAS fish had 2 times the richness of FT fish. Body sites each had unique microbiomes (P < 0.001) and were influenced by hatchery system type (P < 0.001), with RAS being more similar. A strong association between the tank and fish microbiome was observed. Water and tank biofilm richness was positively correlated with skin and digesta richness. Strikingly, the gill, skin, and digesta communities were more similar to that in the origin tank biofilm than those in all other experimental tanks, suggesting that the tank biofilm has a direct influence on fish-associated microbial communities. Lastly, microbial diversity and mucous cell density were positively associated with fish growth and length. The results from this study provide evidence for a link between the tank microbiome and the fish microbiome, with the skin microbiome as an important intermediate.
IMPORTANCE Atlantic salmon, Salmo salar, is the most farmed marine fish worldwide, with an annual production of 2,248 million metric tons in 2016. Salmon hatcheries are increasingly changing from flowthrough toward recirculating aquaculture system (RAS) design to accommodate more control over production along with improved environmental sustainability due to lower impacts on water consumption. To date, microbiome studies of hatcheries have focused either on the fish mucosal microbiota or on the built environment microbiota but have not combined the two to understand their interactions. Our study evaluates how the water and tank biofilm microbiota influences the fish microbiota across three mucosal environments (gill, skin, and digesta). Results from this study highlight how the built environment is a unique source of microbes to colonize fish mucus and, furthermore, how this can influence fish health. Further studies can use this knowledge to engineer built environments to modulate fish microbiota for beneficial phenotypes.
INTRODUCTION
Aquaculture is the fastest-growing agricultural industry, now producing over 50% of seafood by volume globally (1). While freshwater systems currently outproduce marine systems (51.4 million metric tons versus 28.7 million metric tons in 2016), marine aquaculture has tremendous potential to expand, with estimates of theoretical production of 15 billion metric tons (522× increase) (2, 3). One of the primary challenges to scaling aquaculture production is improving seed quality by increasing survival rates and strengthening immune development of larvae and juveniles in the hatchery environment (4). This is challenging particularly as there are an estimated 369 different species of fish currently grown for commercial aquaculture, with additional species in experimental production (3). In terms of global aquaculture production, Atlantic salmon, Salmo salar, ranks first among marine fish and is the 9th largest aquaculture fish species overall (3). Global growth in Atlantic salmon production has primarily been driven by technological advancements in automated feeding machines, reduced reliance on fishmeal-based feeds, selective breeding to reduce growout time to market from 3 years to 1 year (at sea), and in the Northern hemisphere, disease control through commercial adoption of vaccine development along with biological control of parasite infection using cleaner fish (5). Note that neither Australia nor New Zealand has issues with sea lice but, like the Northern hemisphere, both have had issues with amoebic gill disease (6, 7). Improvements in land-based hatchery technology are further reducing the environmental footprint of aquaculture by decreasing water and land use (8, 9). Optimizing hatchery conditions is also important for mariculture, capture fisheries, recreational fisheries, and conservation, as many government programs rely on ocean enhancement efforts to replenish wild populations. For example, in 2018, 29 Alaska salmon hatcheries used for ocean enhancement contributed to 34% of commercial harvest worth $453 million (10). Understanding the factors for which hatchery-reared salmon exhibit altered performance compared to wild salmon, including higher growth rates, lower age to maturity, higher overall survival, lowered lifetime reproductive success, and increased aggression, including competitiveness, may be important for improved ocean enhancement (11–13).
Salmon are reared in two primary types of freshwater hatchery systems, flowthrough (FT), which requires continuous new water, and recirculating aquaculture systems (RAS), where up to 99% of the water is recycled. FT hatcheries, however, take in and release relatively large volumes of water, usually from natural surface waters, and require water treatment and settlement systems. The RAS have the potential to significantly reduce freshwater requirements and thereby lower environmental impacts. Some producers are concerned about switching to RAS over traditional flowthrough (FT) systems due to potential negative effects on fish health. Since RAS utilize microbes for biofiltration, this overall enrichment in microbes in a system may influence fish growout (14–16). Atlantic salmon hatcheries are primarily built near freshwater inputs such as streams or rivers whereby water is filtered and either continually flowed through the tanks at approximately 300% daily or used to replenish the RAS tanks at 2 to 7% daily. During the freshwater stage in both hatchery systems, juvenile salmon, parr, are reared in circular tanks ranging from 3 m to 10 m in diameter and 2 to 5 m in depth made from fiberglass, concrete, or other materials and equipped with oxygen injectors (aerators). This period is crucial for salmon survival, as disease outbreaks can cause costly die-offs in the system. Compared to FT systems, enclosed RAS have the benefit of requiring 93% less water from the environment and a 26 to 38% reduction in eutrophication potential to the environment (17, 18) but can also be more costly in energy use (24 to 40% higher) (9). Because RAS are enclosed batch systems, biosecurity is theoretically improved, as conditions can be regulated and controlled much more easily than in FT systems. In addition, the feed conversion ratio (FCR) can be lower in an RAS due to the ability to control variables such as temperature and salinity (17). RAS may enable establishment of stable, slow-growing bacterial communities in the hatchery systems, which has been shown to improve survival rates in cod (19). Other studies, however, have suggested that water quality (higher recirculating microbial loads, accumulation of metabolites, or accumulation of heavy metals from feeds) in RAS was detrimental for larvae survival and/or growth of common carp (20), sea bass (21), and Nile tilapia (22). For postsmolt Atlantic salmon reared in an RAS, both salinity (12, 22, and 32 parts per thousand [ppt]) and time (3, 4.5, and 7 months) influenced microbial communities of the water column, while the tank biofilm (which differed from the water column) remained stable (23). Since microbial communities are indicated as an important factor in RAS water quality, and thereby fish health, it is important to understand how microbiomes of both the built environment along with the fish mucus are influenced in RAS versus FT systems.
The importance of the mucus microbiome, the collection of microbial eukaryotes, bacteria, archaea, and viruses inhabiting a mucosal body site, to animal health has been well documented and shown to reduce pathogen growth and colonization through mechanisms such as competitive exclusion (24, 25), production of antimicrobial compounds (25, 26), and microenvironment control to reduce pathogen growth and colonization (27, 28). Mucosal environments, including the gill (29), skin (30), and gut (31), serve as important physical barriers for disease and are critical components of the immune response. The skin and gut microbiomes of Atlantic salmon are unique, differ by life stage (parr, smolt, and adult), and differ depending upon rearing environment (wild versus hatchery) (32–34). Furthermore, water has been shown to primarily influence the skin community (34), which is further exemplified during migration from freshwater to saltwater (33). Atlantic salmon gut microbial assemblages are primarily driven by the life stage rather than the environment (35, 36) in the wild, which has been hypothesized to be due to changes in diet along with increased consumption of water during the marine stage (37). The hatchery built environment is a unique microbial habitat which has largely remained unexplored (38, 39). Understanding the relationship between the built environment of the hatchery along with the mucosal microbiome of the fish may be important for predicting and improving fish health.
The purpose of this study was to evaluate how hatchery type (FT versus RAS) influences the microbial community of fish mucus and, subsequently, fish health. This study comprehensively evaluates the gill, skin, and gut microbiome of Atlantic salmon. We further combined histological analyses of mucosal sites to connect microbial changes to mucus health.
RESULTS
Microbiome sample success.
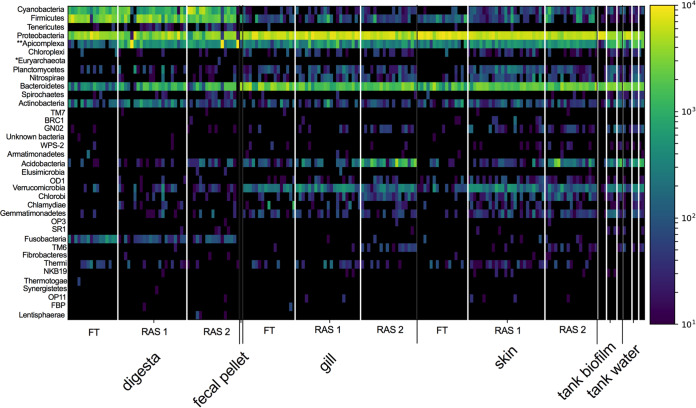
A total of 60 fish were sampled from three unique hatcheries, one flowthrough (FT) and two recirculating aquaculture systems (RAS). Within the hatcheries, a total of 6 fish were sampled from each of 10 unique tanks (3 tanks per hatchery). To evaluate their health status, fish were examined for histopathological measurements within the gill, skin, and gastrointestinal tract. In addition, the mucosal microbiome of three body sites (gill, skin, and digesta) was sampled across all 60 fish along with environmental controls, including the tank water and tank-associated biofilms. After calculating sample cutoff measures and rarefying to 1,000 reads, a total of 185 samples (out of 201) passed quality assurance/quality control (QA/QC), resulting in a total of 6,197 total unique suboperational taxonomic units (sOTUs) (see Fig. S1 in the supplemental material). This rarefaction depth maximized sample size while providing sufficient coverage to accurately describe microbial community structure. Failures were not associated with any particular hatchery system (success rate: 72/78 RAS 1, 56/60 RAS 2, 56/60 FT) or body site (success rate: 56/60 gill, 58/60 skin, 55/60 digesta). A total of 37 microbial phyla were represented in the data set, including one archaea (Euryarchaeota) and one eukaryote (Apicomplexa) (Fig. 1). Digesta samples generally had higher levels of Cyanobacteria, Firmicutes, Actinobacteria, and Fusobacteria, whereas the skin and gill were enriched with Bacteroidetes, Verrucomicrobia, and Acidobacteria. Across all body sites and built environment samples, Proteobacteria was most dominant.
FIG 1.
Microbial phyla (37) present in the study, including bacteria. *, archaea; **, one microbial eukaryote.
Microbiome driven by body site, hatchery, and then tank.
Statistical analyses of community composition (beta diversity) revealed that body site along with hatchery system and variation among tanks were all significant drivers of community composition, with body site being the strongest driver (P = 0.001, R2 = 0.127 unweighted UniFrac; P = 0.001, R2 = 0.340 weighted UniFrac; Adonis) (Table 1). Furthermore, when stratifying for each body site (gill, skin, and gut), microbial communities were significantly influenced by both hatchery location and across individual tanks using both unweighted and weighted UniFrac (Table 1).
TABLE 1.
Multivariate Adonis statistical testing of drivers of microbial beta diversityb
| Sample typea | Metadata name | Combined |
Gill |
Skin |
Gut |
||||
|---|---|---|---|---|---|---|---|---|---|
| R2 | P | R2 | P | R2 | P | R2 | P | ||
| Unw UniFrac | |||||||||
| Body site | Sample_type | 0.127 | 0.001 | Nan | Nan | Nan | Nan | Nan | Nan |
| Hatchery system | ylk_tank_system | 0.062 | 0.001 | 0.124 | 0.001 | 0.172 | 0.001 | 0.091 | 0.001 |
| Tank no. | ylk_sbt_tank_no. | 0.110 | 0.001 | 0.268 | 0.001 | 0.294 | 0.001 | 0.284 | 0.001 |
| W UniFrac | |||||||||
| Body site | Sample_type | 0.340 | 0.001 | Nan | Nan | Nan | Nan | Nan | Nan |
| Hatchery system | ylk_tank_system | 0.053 | 0.001 | 0.231 | 0.001 | 0.229 | 0.001 | 0.084 | 0.031 |
| Tank no. | ylk_sbt_tank_no. | 0.119 | 0.002 | 0.423 | 0.001 | 0.388 | 0.001 | 0.433 | 0.001 |
Unw UniFrac, unweighted UniFrac; W UniFrac, weighted normalized UniFrac.
Nan, not applicable.
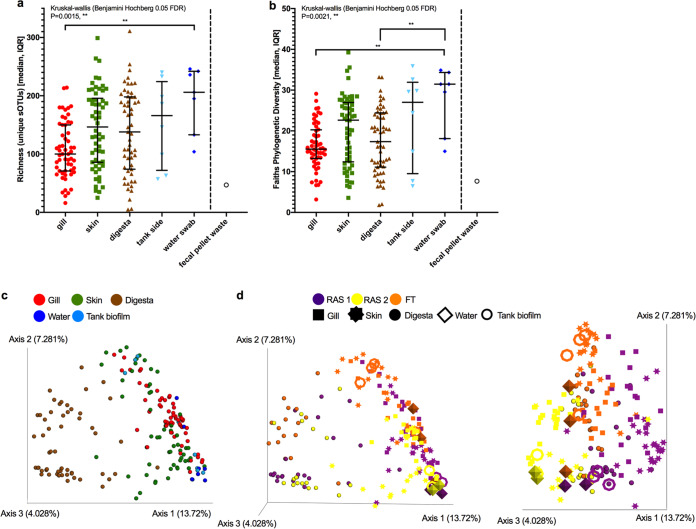
Microbial diversity differs according to sample type, with water samples having the highest richness (P = 0.0015, Kruskal-Wallis test [KW] = 17.52; Kruskal-Wallis, Benjamini Hochberg false-discovery rate [FDR] = 0.05) and phylogenetic diversity (P = 0.0021, KW = 16.79; Kruskal-Wallis, Benjamini Hochberg FDR = 0.05) (Fig. 2a and b). When only fish mucus samples were compared, the gill had less richness than the skin and digesta (P = 0.0056, KW = 10.37; Kruskal-Wallis, Benjamini Hochberg FDR = 0.05) and lower phylogenetic diversity than the skin (P = 0.0279, KW = 7.16; Kruskal-Wallis, Benjamini Hochberg FDR = 0.05). Microbial composition as assessed using unweighted UniFrac distances was primarily driven by sample type, followed by hatchery system, with samples from the RAS generally being more similar than the FT hatchery (Table 1, Fig. 2c and d). In addition, water and biofilm samples were highly distinguishable between the hatchery systems, particularly RAS versus FT, and clustered more closely to gill and skin samples, indicating that gill and skin microbiomes were more closely related to the built environment.
FIG 2.
Microbial ecology (16S rRNA) of three tanks each from three hatchery systems (water and tank biofilm) and Atlantic salmon gill mucus, skin mucus, and digesta. (a and b) Alpha diversity measures of (a) total richness and (b) Faith’s phylogenetic diversity evaluated with a nonparametric Kruskal-Wallis test with a Benjamini-Hochberg FDR of 0.05. (c and d) Beta diversity measures of unweighted UniFrac distances colored by (c) sample type and by (d) hatchery system. **, P < 0.01.
Impacts of hatchery type on fish and built environment microbiome.
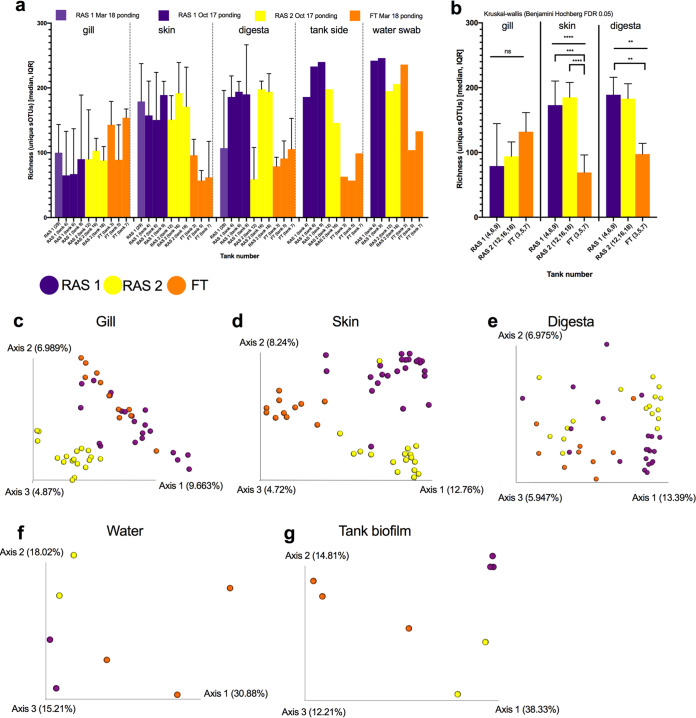
We next assessed how facility type influenced the microbiome of both the fish body sites and the built environment. The microbial richness of skin, digesta, tank biofilm, and tank water was generally higher in the RAS than in the FT system (Fig. 3a). When comparing only fish body sites, both skin (P < 0.0001, KW = 21.16; Kruskal-Wallis, Benjamini Hochberg FDR = 0.05) and digesta (P = 0.0058, KW = 10.29; Kruskal-Wallis, Benjamini Hochberg FDR = 0.05) richness was significantly different across hatcheries, with RAS having approximately 2 times more sOTUs associated with skin and digesta than FT systems (Fig. 3b). Post hoc multiple-comparison tests demonstrated that for skin, both RAS1 and RAS2 richnesses were greater than FT richness, whereas for digesta, only RAS1 was higher than FT (Fig. 3b). Compositionally, the microbial communities were significantly different across hatcheries for all samples combined (Table 1). When microbial communities of specific body sites, such as gill, skin, and digesta, were analyzed, a hatchery-specific microbiome was still observed (Fig. 3c to e). The hatchery-specific microbiome was also prevalent in the water column and tank biofilm (Fig. 3f and g). On closer observation, tank biofilm, tank water, and fish skin samples from the RAS were more similar compositionally than those from the FT system (Fig. 3, Fig. S2, Fig. S3), with RAS1 also having unique communities apart from RAS2.
FIG 3.
Interhatchery effects on microbial ecology of the built environment and fish body sites. (a) Richness (total observed sOTUs) distributions per each tank across body sites, tank biofilm, and water column from the three types of hatcheries, RAS1, RAS2, and FT. (b) Tank replicates are combined per hatchery to enable multiple-group statistical analysis of richness comparisons (Kruskal-Wallis). (c to g) Beta diversity distributions depicted through principal-coordinate analysis (PCoA) plots of unweighted UniFrac distances across tanks for each unique environment as follows: (c) salmon gill, (d) salmon skin, (e) salmon digesta, (f) tank water, and (g) tank biofilm. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Microenvironment “tank effect” drivers of the microbiome.
Next, we directly evaluated the relationship between the environmental microbiome of the tank water and tank biofilm with the fish mucus. For each individual tank, the microbial richness of the biofilm (Fig. 4a) and that of the tank water (Fig. 4b) were compared to the richness of fish within that tank for the three body sites gill, skin, and digesta. Both skin and digesta richnesses were positively correlated with tank biofilm richness (skin P = 0.0001, R2 = 0.2835; digesta P = 0.002, R2 = 0.2042) and water richness (skin P = 0.0014, R2 = 0.2336; digesta P = 0.0264, R2 = 0.1296). This indicates that the skin and gut microbial communities interact with the built environment more than the gill microbial community does. In addition, just skin and just digesta were compared, the R2 values for the tank biofilm were higher than those for tank water, indicating that tank biofilms have a slightly stronger impact than tank water on fish mucus richness (or vice versa) (Fig. 4a and b). Since hatchery environmental microbes seemed to influence fish mucus microbes and unique microbial populations exist across hatcheries and within tank replicates within a hatchery, we hypothesized that within a tank, fish mucus microbial composition should be more similar to the biofilm and water of that tank than to those of tanks from other hatcheries. Here, we report that the gill (P = 0.0022, Mann-Whitney U test = 4,816), skin (P < 0.0001, Mann-Whitney U test = 3,318), and digesta (P = 0.02, Mann-Whitney U test = 5,051) of the fish are more similar to the tank biofilm of origin than to tanks from other hatcheries (Fig. 4c), whereas for tank water, this only is true for the gill (P = 0.0014, Mann-Whitney U test = 3,516) and skin (P = 0.0051, Mann-Whitney U test = 4,087) (Fig. 4d). Based on beta diversity distances, we tested whether certain body sites are more similar to the tank biofilm or water than others. Both gill and skin are more similar to the tank biofilm (P < 0.0001, KW = 66.72; Kruskal-Wallis, Benjamini Hochberg FDR = 0.05) and tank water (P < 0.0001, KW = 61.55; Kruskal-Wallis, Benjamini Hochberg FDR = 0.05) than digesta (Fig. 4e).
FIG 4.
Relationship between the built environment and the fish mucosal microbiome. (a and b) Correlation between (a) tank biofilm richness and fish mucus richness along with (b) tank water richness and fish mucus (linear regression). (c and d) Beta diversity measures to test similarity (unweighted UniFrac) of fish mucus to (c) tank biofilm and (d) tank water. Pairwise comparisons of similarities within a tank versus similarities to other tanks from across hatcheries with the Mann-Whitney test. (e) Overall fish mucosal similarities compared to tank biofilm and water indicate that gill and skin are more similar to the environment than digesta (Kruskal-Wallis). *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
To understand how microbial communities differ across hatchery types, we compared the beta diversity within sample types from the three hatcheries. We hypothesized that microbial communities from the RAS facilities would have more similar microbial communities compared to the FT and tested this across the various sample types. For unweighted UniFrac distances, tank biofilm (P = 0.0129, KW = 7.938; Kruskal-Wallis, Benjamini Hochberg FDR = 0.05; multiple comparisons of FT-RAS1 versus RAS1-RAS2 P = 0.0288, FT-RAS2 versus RAS1-RAS2 P = 0.0073) and skin communities (P < 0.0001, KW = 333.2; Kruskal-Wallis, Benjamini Hochberg FDR = 0.05; multiple comparisons of FT-RAS1 versus RAS1-RAS2 P < 0.0001, FT-RAS2 versus RAS1-RAS2 P < 0.0001) are more similar between RAS hatcheries compared to the FT hatchery (Fig. S3a). For weighted UniFrac distances (Fig. S3b), tank biofilm (P = 0.0002, KW = 12.86; Kruskal-Wallis, Benjamini Hochberg FDR = 0.05; multiple comparisons of FT-RAS1 versus RAS1-RAS2 P = 0.0037, FT-RAS2 versus RAS1-RAS2 P = 0.0008), skin (P < 0.0001, KW = 48.22; Kruskal-Wallis, Benjamini Hochberg FDR = 0.05), and gill (P < 0.0001, KW = 44.8; Kruskal-Wallis, Benjamini Hochberg FDR = 0.05) showed greater similarity between the RAS systems than between the FT system and either RAS system. Although water was not statistically significant, the trend is strong and similar to tank biofilm, suggesting that sample dropout and lower replication may be the reason it did not demonstrate significance. Since the skin microbiome was the most influenced body site on the fish, we calculated the differentially abundant sOTUs (n = 65) between the RAS and FT systems (Fig. S3c). Of the 65 differentially abundant skin sOTUs, 44 were present in the water or tank biofilm communities, while 17 were only found on the skin (Fig. S3d). Skin microbes that were associated with RAS included Saprospirales, Cytophagales, Sphingobacteriales, Verrucomicrobia, and Methylophilales (Methylotenera sp.), whereas the FT was enriched in Pseudomonas, Pseudomonadales, and Enterobacteriales (Fig. S3e). Additionally, Aeromonadales were highly enriched in the fecal detritus in the FT hatchery, while many of the FT-associated microbes were not found in the detritus, suggesting that they are indeed water or biofilm specific.
Microbiome associations with fish health.
Upon establishing a direct relationship between the microbiome of the hatchery environment, we next assessed how fish health is related to these changes. Broad mucosal histopathology was performed on eight endpoint measures across the gill (Fig. S4), skin (Fig. S5), and gastrointestinal (GI) tract (Fig. S6). Fish histology across the GI tract, gill, and skin mucosal surfaces differed across hatcheries (GI fold height: P = 0.0037, KW = 13.73; GI mucosa thickness: P = 0.001, KW = 13.73; GI fold width: not significant [NS]; GI muscularis thickness: P < 0.0001, KW = 27.9, GI goblet cells: P < 0.0001, KW = 24.83; gill mucous cell density: P < 0.0001, KW = 36.4; skin mucous cell density: P < 0.0001, KW = 33.51; Kruskal-Wallis, Benjamini Hochberg FDR = 0.05) (Fig. 5a). In all but one measure, a heightened score was demonstrated in RAS compared to the FT system for the fish sampled, with RAS1 being slightly higher than RAS2. Furthermore, we tested if the microbiome of the fish was driven by these histology scores and found that for unweighted UniFrac measures, where rare taxa are more heavily weighted in a phylogenetic context, the skin microbiome was significantly associated with mucous cell numbers in the gill (Adonis: P = 0.025) and skin communities (Adonis: P = 0.006 and P = 0.003), while the gut microbiome was also associated with mucous cell numbers in the skin (Adonis: P = 0.015) (Table 2). When weighted UniFrac, which looks primarily at relative abundances of sOTUs in a phylogenetic context, was analyzed, gill and skin microbial communities were associated with GI mucous cell numbers (Adonis: gill P = 0.026, skin P = 0.014), while the gut microbiome was associated with mucous cell numbers in the skin (Adonis: P = 0.002) (Table 2).
FIG 5.
Relationship between microbiome, mucosal cell density (immune cells), and size. (a) Histopathology analysis including gut morphometry and mucous cell counts from skin and gill of the fish from one flowthrough (FT) and two RAS hatcheries (RAS 1 and RAS 2 hatchery systems). Hatchery systems were compared using a nonparametric Kruskal-Wallis test. Skin mucous cell counts shown as both per length of epidermis section and per surface area or the epidermis. All fish sampled were clinically normal. (b and c) Comparison of RAS-reared fish (tanks 4, 6, 9, 12, 16, and 18) (b) skin, and (c) GI mucous cell counts to length and mass (positive correlation, linear regression). (d and e) Comparison of (d) skin and (e) digesta microbiome alpha diversity “Shannon evenness” to length and mass (positive correlation, linear regression). **, P < 0.01; ****, P < 0.0001.
TABLE 2.
Multivariate statistical testing of effects of microbiome on fish health metrics (continuous variables), Adonis 999 permutationsa
| Parameter | Metadata name | Unweighted UniFrac |
Weighted UniFrac |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gill |
Skin |
Gut |
Gill |
Skin |
Gut |
||||||||
| R2 | P | R2 | P | R2 | P | R2 | P | R2 | P | R2 | P | ||
| Fish length | host_height | x | 0.726 | x | 0.106 | 0.569 | x | 0.116 | x | 0.051 | x | 0.873 | |
| Fish mass | host_weight | x | 1.000 | x | 1.000 | 1.000 | x | 1.000 | x | 1.000 | x | 1.000 | |
| Fish K factor | sal_k_factor | x | 1.000 | x | 1.000 | 1.000 | x | 1.000 | x | 1.000 | x | 1.000 | |
| Gut histology (fold ht) | sal_histo_gi_fold_height | x | 1.000 | x | 1.000 | 1.000 | x | 1.000 | x | 1.000 | x | 1.000 | |
| Gut histology (mucosa thickness) | sal_histo_gi_mucosa_thickness | x | 1.000 | x | 1.000 | 1.000 | x | 1.000 | x | 1.000 | x | 1.000 | |
| Gut histology (fold width) | sal_histo_gi_fold_width | x | 1.000 | x | 1.000 | 1.000 | x | 1.000 | x | 1.000 | x | 1.000 | |
| Gut histology (muscularis thickness) | sal_histo_gi_muscularis_thickness | x | 1.000 | x | 1.000 | 1.000 | x | 1.000 | x | 1.000 | x | 1.000 | |
| Gut histology (no. of goblet cells) | sal_histo_gi_goblet_cells | x | 0.148 | x | 0.073 | 0.666 | 0.936 | 0.026 | 0.946 | 0.014 | x | 0.294 | |
| Gill histology (mucous cells per 0.2 mm2) | sal_histo_gill_mucous_cell_num_gill | x | 0.081 | 0.749 | 0.025 | 0.056 | x | 0.203 | x | 0.378 | x | 0.131 | |
| Skin histology (mucous cells per 0.2 mm2) | sal_histo_skin_mucous_cell_num_skin | x | 0.459 | 0.905 | 0.006 | 0.915 | 0.015 | x | 0.369 | x | 0.142 | 0.970 | 0.002 |
| Skin histology (mucous cells per mm2) | sal_histo_skin_mucous_cells_permm2_skin | x | 0.424 | 0.905 | 0.003 | 0.915 | 0.015 | x | 0.360 | x | 0.154 | 0.970 | 0.002 |
x, not reported.
To further evaluate how the microbiome may be related to fish health and growth, we subsampled our data set assessing only fish reared in the RAS facility from tanks 4, 6, and 9 (RAS1) and 12, 16, and 18 (RAS2). First, we compared three alpha diversity measures (richness, Shannon’s evenness, and Faith’s phylogenetic diversity [PD]) to mucosal cell density (in gill, skin, and digesta) along with fish growth phenotypes of condition factor, mass, and length using linear regression (Table S1). Note that all variables were normally distributed as verified using an Anderson-Darling test (40). Mucosal cell density in the skin (Fig. 5b) and GI tract (Fig. 5c) was positively associated with fork length and mass. Skin Shannon diversity was positively associated with fork length (P = 0.019, R2 = 0.155, m [slope] = 13.22) and mass (P = 0.031, R2 = 0.133, m = 23.6) (Fig. 5d). For digesta microbiome samples, both Shannon diversity (Fig. 5e) and Faith’s PD were positively associated with fork length (Shannon diversity: P = 0.016, R2 = 0.168, m = 4.42; Faith’s PD: P = 0.024, R2 = 0.149, m = 1.038) and mass (Shannon diversity: P = 0.024, R2 = 0.15, m = 7.993; Faith’s PD: P = 0.033, R2 = 0.134, m = 1.883) as well (Table S1).
In addition to alpha diversity measures, we tested if community composition at the genera level for gill, skin, and digesta could predict salmon fork length and mass. Testing via principal variance component analysis (PVCA) confirmed the presence of nontrivial (∼5% of total variance) tank-specific effects, regardless of relative abundance or compositionally coherent centered log-ratio (CLR) transformed data formats. Using Voom-SNM, a previously applied supervised method for removing technical variation in microbiome data sets, we then demonstrated sharp removal of tank-specific effects from the fish microbiomes while maintaining substantial biological variation between fish body sites (Fig. S7) (see Materials and Methods for details).
Downstream random forest machine learning using leave-one-out cross-validation revealed that Voom-SNM-corrected data accurately predicted fish length and mass solely using digesta (Fig. S7a and b), skin (Fig. S7c and d), or gill microbiomes (Fig. S7e and f) (regressing observed versus predicted values; Pearson’s R > 0 and P ≤ 0.05). Examining the Voom-SNM-based models further, we found that fish digesta microbiomes best predicted fish length and mass (Fig. S7). Taxa ranked by feature importance (see Materials and Methods) are available in the supplemental material (Table S2). Together, these data suggest the potential value of evaluating fish microbiomes to predict fish health and allude to the benefit of microbiome modulation as a way to possibly enhance fish growth, although interventional studies will be needed to confirm this.
Prevalence of RAS-associated microbes in hatchery tanks.
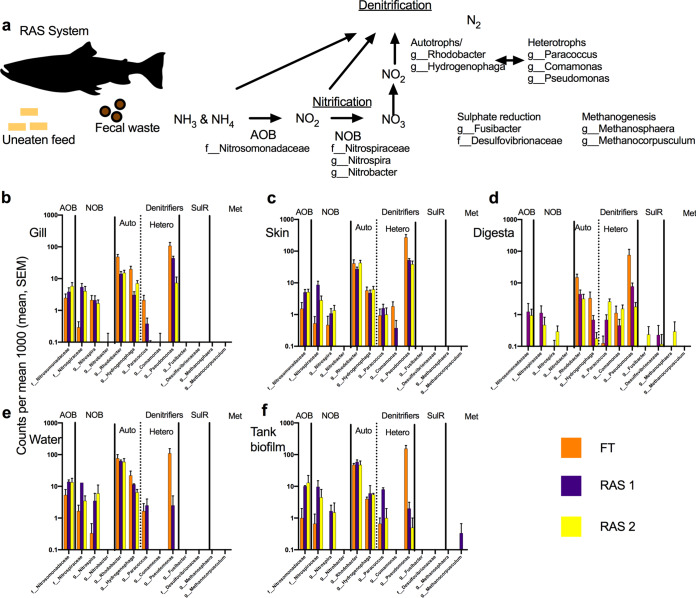
RAS utilize microbes to recycle and remove nitrogenous waste products generated from uneaten feed, fish feces, and other organic waste. We identified and quantified the types and relative abundances of these bacteria and archaea in this system to understand if known RAS-associated microbes were playing a role in colonization within fish mucus or the environment (Fig. 6a). The only known RAS-associated ammonia-oxidizing bacterium (AOB) found in the system was the family Nitrosomonadaceae, which was present in all of the hatcheries and sample types (Fig. 6b to e) and slightly enriched in the tank biofilm community (Fig. 6f). Nitrite-oxidizing bacteria (NOB), primarily the family Nitrospiraceae and Nitrospira spp., were generally in higher relative abundances in the RAS environmental components, including the water and biofilm (Fig. 6e and f), along with the skin, digesta, and gill (Nitrospiraceae only), indicating a possible transfer event (Fig. 6b to d). Note that for digesta samples, no NOB organisms were detected in any of the FT-reared fish. Among denitrifiers, Rhodobacter spp. and Hydrogenophaga spp. were enriched across all hatcheries and sample types, with Rhodobacter spp. showing higher relative abundances in some FT systems. For heterotrophic denitrifiers, Pseudomonas spp. were the most dominant and, specifically, were approximately 20 to 100 times higher in the FT water and tank biofilms than in the RAS (Fig. 6e and f). In addition to being enriched in the environment, Pseudomonas spp. were also consistently higher in the gill, skin, and digesta of fish reared in the FT system compared to RAS (Fig. 6b to d). Lastly, two methanogens were detected, albeit at very low frequencies and only in the tank biofilm (a Methanocorpusculum sp.) and the digesta (a Methanosphaera sp.) from the RAS.
FIG 6.
Distribution of RAS-associated microbes in Salmon hatcheries. (a) RAS are designed to recycle nitrogenous waste, primarily from uneaten feed and fish feces, using a series of nitrification and denitrification steps through microbial filters. The primary microbes involved in these processes and detected in the systems (AOB [ammonia-oxidizing bacteria], NOB [nitrite-oxidizing bacteria], and denitrification) are listed. (b to f) The distributions of these RAS-associated microbes are listed as mean relative counts per 1,000 according to each hatchery type (FT, orange; RAS 1, purple; RAS 2, yellow) across each particular sample type, i.e., (b) gill, (c) skin, (d) digesta, (e) tank water, and (f) tank biofilm.
DISCUSSION
The mucosal environment is paramount for fish health, as it is the first line of defense against pathogen invasion. Fish mucus contains various immune components, such as lysozymes, immunoglobins, lectins, crinotoxins, and antimicrobial peptides (41). Specifically, a healthy mucosal environment protects against infection through several endogenous mechanisms, including mucus production, immune components such as lysozymes, antimicrobial peptides, immunoglobulins, and exogenous mechanisms through establishment of a healthy microbiome. In this study, we investigated the means by which the mucosal environment of Atlantic salmon is influenced by the rearing environment. We evaluated three unique hatcheries utilizing two rearing methodologies, recirculating aquaculture systems (RAS) and flowthrough (FT) systems.
In both the biosecure RAS and FT hatchery environments, Atlantic salmon have unique microbial communities on their gill, skin, and digesta. Body sites were the strongest predictor of microbial community composition. In comparison to other taxa, the most enriched genus within the gill samples was Limnohabitans, while the skin harbored Pseudomonas, Acinetobacter, and Rhodobacter. Further research to identify the spatial structure of microbial communities within the gill and skin is needed (42, 43). Taxon coverage was more even in the gut, with Leuconostoc and Acinetobacter genera being slightly more enriched. In the Northern Atlantic salmon, Mycoplasma was enriched in wild adults but less prominent in juveniles. We observed very few Mycoplasma in our hatchery-reared juvenile fish digesta samples, suggesting that Mycoplasma exposure and colonization of the gut may occur during growout only (35). Aside from body sites, hatchery type was the next biggest driver of community composition.
These fish-associated mucosal microbiomes along with the tank and biofilm communities are further differentiated across hatchery systems by comparing RAS versus FT systems. RAS are known to harbor their own unique microbial communities in the biofilter but also within the hatchery system where fish are reared (16). Previous studies, however, have not looked at the built environment microbiomes simultaneously with the fish mucosal microbiomes.
For these hatchery systems, alpha diversity is higher in RAS compared to FT hatcheries for the following sample types: skin, digesta, tank water, and tank biofilm microbiomes. Fish skin and digesta richness is further positively associated with both tank biofilm and tank water richness, suggesting an influence of the environment microbiota on fish-associated microbiota, with the biofilm association being the strongest. Along with the association between alpha diversity, the built environment was linked to the fish mucous through beta diversity. Fish mucosal sites were more phylogenetically similar to both water and biofilms within their own tank compared to tanks from other hatcheries, indicating a microenvironmental effect. Skin microbiomes have been implicated as important for maintaining fish health, and thus, understanding any potential negative implications or drivers of dysbiosis is important for fish welfare (28, 44). One caveat of our study was that all sample types were obtained using swabs rather than biopsies. In trout, the epithelia of the skin can host up to 27% of the total microbial diversity (53 out of 199 genera) (42). An advantage of collecting microbiomes with a swab is that animal lives can be spared. Future work should be focused on describing impacts of swabbing versus biopsies for microbial community sampling. Tank biofilms can be challenging to monitor and control. Further research should focus on how manipulating tank surfaces through materials science and engineering could be used to promote fish health. Our study uncovers an important link between the built environment of the hatchery and the fish mucus. Hatchery types differ in their construction and operation. Thus, understanding the differences is important to realizing potential impacts to the microbiome.
RAS are becoming popular for growing salmon smolts and offer many benefits, including minimized water use and waste generation along with improving survival rates of fish during transfer to net pens (45–48). Waste water is purified by processing through one of two main types of biofilters (fixed film or single sludge) which utilize a variety of bacteria and archaea (49, 50). The biofilters primarily comprise heterotrophs and chemoautotrophs that transform and detoxify ammonia and nitrate species (51). The common ammonia-oxidizing archaea and bacteria found in these systems include Nitrosopumilus (archaeon), Nitrosomonas, Nitrosococcus, and Nitrosospira (16). In our study, only ammonia-oxidizing bacteria within the family Nitrosomonadaceae were present, and they were highest in the RAS tank and water systems as well as in RAS-reared fish gill, skin, and digesta. Following ammonia oxidation, Nitrospira and Nitrobacter are the primary bacteria responsible for nitrite oxidation in RAS biofilters (16). Bacterial sOTUs from Nitrospira spp. and unclassified sOTUs within the family Nitrospiraceae were in higher relative abundance in the RAS hatcheries for tank water, tank biofilm, skin, and digesta and moderately abuntant in the gill. In the final step of nitrogen recycling, denitrification is carried out by both autotrophs and heterotrophs. The primary autotrophic bacteria associated with denitrification in RAS include Thiomicrospira, Thiothrix, Rhodobacter, and Hydrogenophaga (16). Both Rhodobacter and Hydrogenophaga were found in the hatcheries although in similar relative abundances across the FT and RAS hatcheries in both the tank environment and fish mucus. The primary heterotrophic microbes associated with denitrification in RAS include Pseudomonas, Paracoccus, and Comamonas spp. (16). All three were abundant in the hatcheries, with Pseudomonas being the highest of the three and generally higher in the flowthrough hatchery than in the RAS. In conclusion, various RAS-associated microbes which are responsible for nitrogen cycling, particularly nitrification, in the biofilters were present in our study and higher in the RAS built environment along with RAS-reared fish mucus, suggesting that these microbes are not being solely sequestered in the biofilter but are also are circulated through the fish tanks and may be colonizing fish mucus.
When excess organic matter, including fish feed and fish feces, accumulates in an RAS tank, the heterotrophs can quickly bloom and outcompete nitrifying microbes (16). This overgrowth and imbalance may contribute negatively to flesh flavor. Future studies are needed to understand which microbes and what metabolic pathways may be responsible for these effects (52). While most hatcheries are used for producing seed to then transfer to ocean growout cages, complete salmon production cycles in land-based RAS are becoming more common. Furthermore, both FT systems and RAS may be colonized by various microbial inputs from the air, water, fish feed, fish flesh, technicians, and biofilter type. Thus, understanding the contributions of each in a system will be important for both future experimental designs and for fish health (14).
The built environment microbiome may originally be colonized by animal excrement, including mucus, along with environmental sources such as water. The sustained built environment microbiome is a result of the new animal host deposition of cellular material but can also propagate based on host-associated animal matter. Furthermore, the built environment community can then influence the microbial communities of animal hosts residing there. Understanding the extent to which the animal’s microbiome can be influenced by its surroundings and then associated with a phenotype such as fish health or development will be important for experimental design where microbiome readout is a standard measure. This is commonly referred to as the “cage effect” and has primarily been demonstrated in mouse studies where animals which share the same cage have more similar fecal samples, likely due to coprophagy (53, 54). The cage effect can explain up to 31% of the variation in mouse feces compared to only 19% resolved by host genetics (55). For this reason, our experimental design included three separate tanks per treatment group (hatchery) along with multiple fish biological replicates per tank. Our experiment offers an explanation for an observed tank effect in fish. The tank effect, which has been observed in at least one other species of fish (56), is driven partially by the water and tank biofilm and influences primarily the fish skin and digesta. Since aquariums use a variety of material types to culture fish, it will be important for future studies to evaluate how biofilm formation changes with respect to tank material type (e.g., concrete, polyvinyl chloride [PVC], high-density polyethylene [HDPE], fiberglass, etc.).
Quantifying fish health can be a challenging and expensive endeavor which does not often easily scale for large hatchery operations. Here, we used histology as a measure of mucosal health across the gastrointestinal tract, gill, and skin. Elevated skin mucous cell numbers are generally reflective of healthy fish, whereas depleted mucosal cells may indicate a recent mucosal discharge due to stress or disease (57, 58). Mucosal cell numbers can also be influenced by the sampling location on the fish body, sex, diet, and age or developmental stage; thus, care must be taken when interpreting results (59–61). In our study, skin and gill mucosal cell numbers were elevated in both RAS compared to the FT system, suggesting that RAS fish may have been healthier or less stressed. Furthermore, these elevated skin mucous cell numbers positively correlated with microbial richness and phylogenetic diversity on the skin and were associated with changes in microbial composition. For the gastrointestinal tract, the fold height, mucosa thickness, muscularis thickness, and mucous cell number were higher in fish reared in the RAS compared to the FT hatchery. Overall, the RAS-reared fish had a more complex GI tract compared to fish reared in flowthrough systems. Vertebrate gut microbiomes are often driven by diet, habitat, and age (62). Since diet and age were controlled for in this study, we hypothesize that differences in microbial communities in the tank water column and biofilm are driving gut microbiome differences by drinking or grazing. The microbiome is essential for development and differentiation of mucous cells. For example, a germfree model of zebrafish showed reduced numbers of mucous cells in their intestines (63). For each unique mucosal site, distinct microbial communities were present and differentiated between the RAS and FT systems. Differences in GI communities between RAS and open water systems are likely reflective of variable microbial exposure in their environment (64). Atlantic salmon reared in an RAS which were infected by Aeromonas salmonicida also had differentiated gut microbiomes compared to healthy fish (65). By demonstrating how the environmental microbiome is influenced by hatchery design, which in turn influences the fish mucosal microbiome and subsequent health, our study demonstrates the utility of developing environmental and/or fish microbiome sampling as a potential fish tank health indicator. Future studies should evaluate more tanks and include metrics such as survival rate, growth rate, and body composition analysis to determine how the environmental microbiome may drive fish performance in the hatchery setting.
MATERIALS AND METHODS
A total of 60 clinically normal, juvenile Atlantic salmon were sampled for this study. Six fish were randomly sampled from each of ten tanks across three freshwater hatcheries within a 4-day span (14 August 2018 to 17 August 2018). All hatcheries were operated by the same company, Tassal, and fish were fed Skretting Nutra at a 1.3 to 1.5 “specific feeding rate.” As a control for comparing the ponding effect of RAS for FT, one RAS hatchery had an additional tank sampled (which had the same ponding date as FT), while others each had three tanks sampled. “Ponding” refers to when fish are first transferred from incubation trays to a tank. Fish were collected and euthanized with AQUI-S anesthetic by husbandry technicians according to label instructions. Biometrics, including total length, mass, and condition factor, were measured. Tank conditions, such as water temperature, salinity, and diameter, were recorded and can be found in the Qiita metadata (66). For each of the 60 fish, microbiome samples of the gill, skin, and gut were collected for a total of 180 fish microbiome samples. For each of the 10 tanks, a water microbiome sample and tank side microbiome sample were collected for a total of 20 samples. All fish were collected as part of standard welfare practices by the company and according to the University of Tasmania Animal Ethics Committee (A0017497).
Histology.
Samples of gills, skin, and intestine, fixed in 10% buffered formalin, were trimmed, processed using standard protocols for histology, and embedded in paraffin. Sections of 4 μm were cut, and one section was used for each fish. Briefly, the sections were stained with alcian blue-periodic acid-Schiff stain (AB/PAS) at pH 2.5 to quantify mucous cells in the gills per interlamellar unit (ILU) under a bright-field light microscope (DM1000; Leica, Hamburg, Germany) (67, 68). Mucous cells were counted using established methods, and the results were normalized per area (59). For the gastrointestinal tract, morphometric measurements of the fold height, mucosa thickness, fold width, and muscularis thickness, were done as previously described (69). Intestine sections stained with hematoxylin and eosin (H&E) were analyzed using Image-Pro Premier software. Ten intestinal folds from one section from each region were included in the analysis.
Fish and hatchery microbiome processing.
For each of the ten tanks, six fish were collected individually using hand nets and placed directly into a sterile sampling bucket and anesthetized using AQUI-S. The mucosal microbiome was sampled as follows: gill by swabbing the second gill arch on the left lateral side, skin by swabbing a 2 cm by 2 cm area posterior of the operculum on the left lateral side under the dorsal fin, and digesta by massaging the GI until a fecal pellet emerged. Swabs were then placed directly into a 2-ml PowerSoil tube and frozen at –20°C. In addition to biological samples, two environmental samples were taken per tank—a 2 cm by 2 cm swab of the inside of the tank just below the water line (biofilm) and a 400-μl bulk water sample from the tank. In total, three body sites across 60 fish (180 samples) and two environmental samples across 10 tanks (20 samples) were collected and processed for microbiome analysis. In addition, 21 technical controls were included.
DNA extraction was performed at the University of Tasmania, Hobart, Australia, using the Earth Microbiome Project protocols (earthmicrobiome.org), specifically using the manual single-tube MoBio PowerSoil kit to reduce well-to-well contamination (70). A total of 21 positive controls of a microbial isolate (replicates of 10-fold serial dilutions) were processed alongside the samples and then used to determine sample success rate by calculating the sample exclusion criteria based on read counts described in the Katharoseq method (38). Samples were processed in triplicate 5-μl PCRs (71) using the 16S v4 515/806 primers (72, 73) and then pooled at equal volume according to the Katharoseq method (38). The final amplicon pool was processed using the Qiagen PCR cleanup kit following EMP protocols and sequenced on a MiSeq 2 × 250-bp run (74). Sequencing runs were processed in Qiita (66) using Qiime2 commands (75) and can be found on Qiita study ID 12227, analysis 22399 (ENA accession number ERP120036). Samples were trimmed to 150 bp and then processed through the Deblur (76) pipeline, which generates unique, single sOTUs. To determine which samples had been sequenced successfully, the Katharoseq method (38), developed for low-biomass sequencing, was applied. The cutoff value for composition of a sample aligning to the target within the positive controls was 90%. In this case, the cutoff value was 405 reads, but we rarified it to 1,000 reads to have a higher depth of sequencing. This coincides with other studies demonstrating sufficient rarefaction depths of 1,250 reads per sample for the American Gut Project and 600 to 1,600 sequences per sample for trout mucus (42, 77). Within Qiita, samples which did not have histology metadata were excluded.
Statistical analysis.
Alpha diversity was calculated using richness (total observed unique sOTUs) and Faith’s phylogenetic diversity. Differences between body sites and environmental variables were tested using the nonparametric Kruskal-Wallis test with a Benjamini Hochberg FDR of 0.05 (78, 79). Correlations between the richness of environmental variables (tank and biofilm) and salmon body site were calculated using linear regression. For beta diversity, we used weighted and unweighted UniFrac (80, 81). Multivariate statistical testing of both continuous and categorical variables was performed using Adonis within Qiime (82). Pairwise statistical comparisons of beta diversity measures were calculated using the Mann-Whitney test, while multiple comparisons were conducted using the Kruskal-Wallis test. To identify correlations between histological measures and specific microbes, a nonparametric Spearman correlation was calculated for the entire data set using the Calour analysis tool (83).
Analysis and correction of tank “batch” effects for downstream machine learning.
With the possibility of fish microbiome differences being influenced by experimental technical variation, specifically due to tank-driven effects, we estimated the amount of “signal” in the data set across fish body site and tank number using principal variance-component analysis (PVCA) (84, 85). The mathematical basis for PVCA was previously described by the National Institute of Environmental Health Sciences (NIEHS) (86), and we set the one tunable parameter to 75%, based on their recommendation of 60 to 90%. Briefly, this procedure reduces the dimensionality of the data by principal-component analysis and then partitions the amount of variance attributed to variables in the data set via variance-component analysis. Input data were either relative abundances of genera-level taxonomies or centered log-ratio transformed (CLR) data of the raw genera-level counts after adding a small pseudocount of 0.5 (87).
After observing a nontrivial amount of signal in the data set (∼5% of variance) attributed to tank-specific effects among fish microbiomes, we employed a pipeline called Voom-SNM, which was previously utilized by our lab to correct for technical batch effects in microbiome data while maintaining signal from biological variables (88). Briefly, the Voom algorithm (89) transformed genera-level counts to approximately normally distributed, log-count per million (log-cpm) data by modeling the data’s heteroskedasticity, accounting for the sample library size, and applying quantile normalization; then, supervised normalization via the SNM algorithm (90) removed tank-specific effects (i.e., tank number) while preserving expected biological effects (i.e., swabbed body site). The Voom and SNM model matrices were equivalent and built using “body site” as the target biological variable (n = 3; i.e., digesta, gill, skin) due to expected biological differences between them, for which signal should be preserved during the SNM; conversely, the tank number was modeled as the technical covariate to be mitigated during SNM. PVCA was run after Voom-SNM correction to ensure removal of tank number-driven effects.
Machine learning, performance estimation, and feature extraction.
We employed machine learning as a way to predict fish health, specifically measured as fish length (in millimeters) and fish mass (in grams). Samples from each body site were divided into subsets (e.g., digesta-only samples) and then utilized for leave-one-out cross-validation (LOOCV) modeling. Random forest (RF) machine learning models were trained, automatically tuned, and evaluated using the R programming language (https://www.r-project.org/), randomForest package (91), and Caret package (92). No further preprocessing was performed prior to machine learning; moreover, RF models used 500 trees and were tuned to minimize the root mean squared error (RMSE) by varying the number of features randomly sampled at each node in a tree (“mtry”; available options by default in Caret package: mtry = 2, 33, 555). After iteratively leaving out one sample, building a model on remaining samples, and then testing on the left-out sample, model regression performance was estimated by evaluating Pearson’s R, R2, and the mean absolute error (MAE) on the predictions versus the actual values. Additionally, a linear regression and two-sided hypothesis test of the slope between the observed (y axis) and predicted (x axis) values (see reference 93 for choice of axes) were also calculated (inset on plots). Machine learning models that led to positive (Pearson’s R > 0), strong (P ≤ 0.05) correlations between the observed and predicted values were deemed to be good predictors of fish health. Models and underlying data that failed to fit the data well (i.e., Pearson’s R < 0 or P > 0.05) were deemed to be poor predictors of fish health. Lastly, taxonomy features were extracted from the tuned models using the importance() function in the randomForest package and ranked by the percent increase in mean squared error when each taxon was omitted from the model. These were saved as CSV files for Voom-SNM-based models, which were solely significant at predicting fish health, and are provided in Table S2.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tassal for providing the opportunity to conduct this project and the University of Tasmania for research support.
This work was supported by an Australia Academy of Sciences Australia-America Ph.D. Research Internship Program award to J.J.M. and National Science Foundation grant OCE-1837116 to E.E.A.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.FAO. 2016. The state of world fisheries and aquaculture 2016 (SOFIA): contributing to food security and nutrition for all. FAO, Rome, Italy. [Google Scholar]
- 2.Gentry RR, Froehlich HE, Grimm D, Kareiva P, Parke M, Rust M, Gaines SD, Halpern BS. 2017. Mapping the global potential for marine aquaculture. Nat Ecol Evol 1:1317–1324. doi: 10.1038/s41559-017-0257-9. [DOI] [PubMed] [Google Scholar]
- 3.FAO. 2018. The state of world fisheries and aquaculture 2018—meeting the sustainable development goals. FAO, Rome, Italy. [Google Scholar]
- 4.Tave D, Hutson AM. 2019. Is good fish culture management harming recovery efforts in aquaculture-assisted fisheries? North Am J Aquac 81:333–339. doi: 10.1002/naaq.10107. [DOI] [Google Scholar]
- 5.Kumar G, Engle CR. 2016. Technological advances that led to growth of shrimp, salmon, and tilapia farming. Rev Fish Sci Aquac 24:136–152. doi: 10.1080/23308249.2015.1112357. [DOI] [Google Scholar]
- 6.Clark A, Nowak BF. 1999. Field investigations of amoebic gill disease in Atlantic salmon, Salmo salar L., in Tasmania. J Fish Dis 22:433–443. doi: 10.1046/j.1365-2761.1999.00175.x. [DOI] [Google Scholar]
- 7.Munday BL, Foster CK, Roubal FR, Lester R. 1990. Paramoebic gill infection and associated pathology of Atlantic salmon, Salmo salar and rainbow trout, Salmo gairdneri in Tasmania, p 215–222. In Pathology in marine science. Proceedings of the Third International Colloquium on Pathology in Marine Aquaculture, held in Gloucester Point, Virginia, USA, October 2-6, 1988. Academic Press, San Diego, CA. [Google Scholar]
- 8.Tal Y, Schreier HJ, Sowers KR, Stubblefield JD, Place AR, Zohar Y. 2009. Environmentally sustainable land-based marine aquaculture. Aquaculture 286:28–35. doi: 10.1016/j.aquaculture.2008.08.043. [DOI] [Google Scholar]
- 9.Badiola M, Basurko OC, Piedrahita R, Hundley P, Mendiola D. 2018. Energy use in recirculating aquaculture systems (RAS): a review. Aquac Eng 81:57–70. doi: 10.1016/j.aquaeng.2018.03.003. [DOI] [Google Scholar]
- 10.Stopha M. 2018. Alaska salmon fisheries enhancement annual report. Alaska Department of Fish and Game, Juneau, AK: http://www.adfg.alaska.gov/FedAidPDFs/RIR.5J.2019.01.pdf. [Google Scholar]
- 11.Jonsson B, Jonsson N. 2006. Cultured Atlantic salmon in nature: a review of their ecology and interaction with wild fish. ICES J Mar Sci 63:1162–1181. doi: 10.1016/j.icesjms.2006.03.004. [DOI] [Google Scholar]
- 12.Jonsson N, Jonsson B, Hansen LP. 2003. The marine survival and growth of wild and hatchery-reared Atlantic salmon. J Appl Ecol 40:900–911. doi: 10.1046/j.1365-2664.2003.00851.x. [DOI] [Google Scholar]
- 13.Jensen AJ, Berg M, Bremset G, Finstad B, Hvidsten NA, Jensås JG, Johnsen BO, Lund E. 2016. Passing a seawater challenge test is not indicative of hatchery-reared Atlantic salmon Salmo salar smolts performing as well at sea as their naturally produced conspecifics. J Fish Biol 88:2219–2235. doi: 10.1111/jfb.12984. [DOI] [PubMed] [Google Scholar]
- 14.Blancheton JP, Attramadal KJK, Michaud L, d’Orbcastel ER, Vadstein O. 2013. Insight into bacterial population in aquaculture systems and its implication. Aquac Eng 53:30–39. doi: 10.1016/j.aquaeng.2012.11.009. [DOI] [Google Scholar]
- 15.Liu D, Straus DL, Pedersen L-F, Meinelt T. 2018. Periodic bacterial control with peracetic acid in a recirculating aquaculture system and its long-term beneficial effect on fish health. Aquaculture 485:154–159. doi: 10.1016/j.aquaculture.2017.11.050. [DOI] [Google Scholar]
- 16.Rurangwa E, Verdegem MCJ. 2015. Microorganisms in recirculating aquaculture systems and their management. Rev Aquac 7:117–130. doi: 10.1111/raq.12057. [DOI] [Google Scholar]
- 17.d’Orbcastel ER, Blancheton J-P, Aubin J. 2009. Towards environmentally sustainable aquaculture: comparison between two trout farming systems using Life Cycle Assessment. Aquac Eng 40:113–119. doi: 10.1016/j.aquaeng.2008.12.002. [DOI] [Google Scholar]
- 18.Eding EH, Kamstra A, Verreth JAJ, Huisman EA, Klapwijk A. 2006. Design and operation of nitrifying trickling filters in recirculating aquaculture: a review. Aquac Eng 34:234–260. doi: 10.1016/j.aquaeng.2005.09.007. [DOI] [Google Scholar]
- 19.Attramadal KJK, Truong TMH, Bakke I, Skjermo J, Olsen Y, Vadstein O. 2014. RAS and microbial maturation as tools for K-selection of microbial communities improve survival in cod larvae. Aquaculture 432:483–490. doi: 10.1016/j.aquaculture.2014.05.052. [DOI] [Google Scholar]
- 20.Martins CIM, Pistrin MG, Ende SSW, Eding EH, Verreth AJ. 2009. The accumulation of substances in recirculating aquaculture systems (RAS) affects embryonic and larval development in common carp Cyprinus carpio. Aquaculture 291:65–73. doi: 10.1016/j.aquaculture.2009.03.001. [DOI] [Google Scholar]
- 21.Deviller G, Palluel O, Aliaume C, Asanthi H, Sanchez W, Franco Nava MA, Blancheton J-P, Casellas C. 2005. Impact assessment of various rearing systems on fish health using multibiomarker response and metal accumulation. Ecotoxicol Environ Saf 61:89–97. doi: 10.1016/j.ecoenv.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 22.Martins CI, Ende SSW, Ochola D, Eding EH, Verreth JAJ, 2007. Growth retardation in Nile tilapia (Oreochromis niloticus) cultured in recirculation aquaculture systems, p 162–163. In Proceedings of the International Conference and Industry Forum, Aquaculture Europe 2007: Competing Claims, Istanbul, Turkey, 24 to 27 October 2007.
- 23.Bakke I, Åm AL, Kolarevic J, Ytrestøyl T, Vadstein O, Attramadal KJK, Terjesen BF. 2017. Microbial community dynamics in semi-commercial RAS for production of Atlantic salmon post-smolts at different salinities. Aquac Eng 78:42–49. doi: 10.1016/j.aquaeng.2016.10.002. [DOI] [Google Scholar]
- 24.Balcázar JL, de Blas I, Ruiz-Zarzuela I, Cunningham D, Vendrell D, Múzquiz JL. 2006. The role of probiotics in aquaculture. Vet Microbiol 114:173–186. doi: 10.1016/j.vetmic.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 25.Balcázar JL, Vendrell D, de Blas I, Ruiz-Zarzuela I, Gironés O, Múzquiz JL. 2007. In vitro competitive adhesion and production of antagonistic compounds by lactic acid bacteria against fish pathogens. Vet Microbiol 122:373–380. doi: 10.1016/j.vetmic.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 26.Pérez-Sánchez T, Balcázar JL, García Y, Halaihel N, Vendrell D, de Blas I, Merrifield DL, Ruiz-Zarzuela I. 2011. Identification and characterization of lactic acid bacteria isolated from rainbow trout, Oncorhynchus mykiss (Walbaum), with inhibitory activity against Lactococcus garvieae. J Fish Dis 34:499–507. doi: 10.1111/j.1365-2761.2011.01260.x. [DOI] [PubMed] [Google Scholar]
- 27.Beck BH, Peatman E. 2015. Mucosal health in aquaculture. Academic Press, San Diego, CA. [Google Scholar]
- 28.Merrifield DL, Rodiles A. 2015. The fish microbiome and its interactions with mucosal tissues, p 273–295. In Beck BH, Peatman E (ed), Mucosal health in aquaculture. Academic Press, San Diego, CA. [Google Scholar]
- 29.Adams MB, Nowak BF. 2003. Amoebic gill disease: sequential pathology in cultured Atlantic salmon, Salmo salar L. J Fish Dis 26:601–614. doi: 10.1046/j.1365-2761.2003.00496.x. [DOI] [PubMed] [Google Scholar]
- 30.Llewellyn MS, Leadbeater S, Garcia C, Sylvain F-E, Custodio M, Ang KP, Powell F, Carvalho GR, Creer S, Elliot J, Derome N. 2017. Parasitism perturbs the mucosal microbiome of Atlantic salmon. Sci Rep 7:43465. doi: 10.1038/srep43465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bakke-McKellep AM, Press CM, Baeverfjord G, Krogdahl Å, Landsverk T. 2000. Changes in immune and enzyme histochemical phenotypes of cells in the intestinal mucosa of Atlantic salmon, Salmo salar L., with soybean meal-induced enteritis. J Fish Dis 23:115–127. doi: 10.1046/j.1365-2761.2000.00218.x. [DOI] [Google Scholar]
- 32.Lavoie C, Courcelle M, Redivo B, Derome N. 2018. Structural and compositional mismatch between captive and wild Atlantic salmon (Salmo salar) parrs’ gut microbiota highlights the relevance of integrating molecular ecology for management and conservation methods. Evol Appl 11:1671–1685. doi: 10.1111/eva.12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lokesh J, Kiron V. 2016. Transition from freshwater to seawater reshapes the skin-associated microbiota of Atlantic salmon. Sci Rep 6:19707. doi: 10.1038/srep19707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webster TMU, Consuegra S, Hitchings M, De Leaniz CG. 2018. Interpopulation variation in the Atlantic salmon microbiome reflects environmental and genetic diversity. Appl Environ Microbiol 84:e00691-18. doi: 10.1128/AEM.00691-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Llewellyn MS, McGinnity P, Dionne M, Letourneau J, Thonier F, Carvalho GR, Creer S, Derome N. 2016. The biogeography of the Atlantic salmon (Salmo salar) gut microbiome. ISME J 10:1280–1284. doi: 10.1038/ismej.2015.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hovda MB, Fontanillas R, McGurk C, Obach A, Rosnes JT. 2012. Seasonal variations in the intestinal microbiota of farmed Atlantic salmon (Salmo salar L.). Aquac Res 43:154–159. doi: 10.1111/j.1365-2109.2011.02805.x. [DOI] [Google Scholar]
- 37.Dehler CE, Secombes CJ, Martin SAM. 2017. Seawater transfer alters the intestinal microbiota profiles of Atlantic salmon (Salmo salar L.). Sci Rep 7:13877. doi: 10.1038/s41598-017-13249-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minich JJ, Zhu Q, Janssen S, Hendrickson R, Amir A, Vetter R, Hyde J, Doty MM, Stillwell K, Benardini J, Kim JH, Allen EE, Venkateswaran K, Knight R. 2018. KatharoSeq enables high-throughput microbiome analysis from low-biomass samples. mSystems 3:e00218-17. doi: 10.1128/mSystems.00218-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilbert JA, Stephens B. 2018. Microbiology of the built environment. Nat Rev Microbiol 16:661–670. doi: 10.1038/s41579-018-0065-5. [DOI] [PubMed] [Google Scholar]
- 40.Stephens MA. 1974. EDF statistics for goodness of fit and some comparisons. J Am Stat Assoc 69:730–737. doi: 10.1080/01621459.1974.10480196. [DOI] [Google Scholar]
- 41.Shephard KL. 1994. Functions for fish mucus. Rev Fish Biol Fisheries 4:401–429. doi: 10.1007/BF00042888. [DOI] [Google Scholar]
- 42.Lowrey L, Woodhams DC, Tacchi L, Salinas I. 2015. Topographical mapping of the rainbow trout (Oncorhynchus mykiss) microbiome reveals a diverse bacterial community with antifungal properties in the skin. Appl Environ Microbiol 81:6915–6925. doi: 10.1128/AEM.01826-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quinn RA, Melnik AV, Vrbanac A, Fu T, Patras KA, Christy MP, Bodai Z, Belda-Ferre P, Tripathi A, Chung LK, Downes M, Welch RD, Quinn M, Humphrey G, Panitchpakdi M, Weldon KC, Aksenov A, da Silva R, Avila-Pacheco J, Clish C, Bae S, Mallick H, Franzosa EA, Lloyd-Price J, Bussell R, Thron T, Nelson AT, Wang M, Leszczynski E, Vargas F, Gauglitz JM, Meehan MJ, Gentry E, Arthur TD, Komor AC, Poulsen O, Boland BS, Chang JT, Sandborn WJ, Lim M, Garg N, Lumeng JC, Xavier RJ, Kazmierczak BI, Jain R, Egan M, Rhee KE, Ferguson D, Raffatellu M, Vlamakis H, et al. 2020. Global chemical effects of the microbiome include new bile-acid conjugations. Nature 579:123–129. doi: 10.1038/s41586-020-2047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Legrand TPRA, Catalano SR, Wos-Oxley ML, Stephens F, Landos M, Bansemer MS, Stone DAJ, Qin JG, Oxley APA. 2018. The inner workings of the outer surface: skin and gill microbiota as indicators of changing gut health in yellowtail kingfish. Front Microbiol 8:2664. doi: 10.3389/fmicb.2017.02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terjesen B, Ulgenes Y, Fjæra S, Summerfelt S, Brunsvik P, Baeverfjord G, Nerland S, Takle H, Norvik O, Kittelsen A. 2008. RAS research facility dimensioning and design: a special case compared to planning production systems. Nofima Marine, Sunndalsøra, Norway: https://www.nofima.no/filearchive/terjesen-et-al-aes-issues-forum-paper_1.pdf. [Google Scholar]
- 46.Bergheim A, Drengstig A, Ulgenes Y, Fivelstad S. 2009. Production of Atlantic salmon smolts in Europe: current characteristics and future trends. Aquac Eng 41:46–52. doi: 10.1016/j.aquaeng.2009.04.004. [DOI] [Google Scholar]
- 47.Drengstig A, Ulgenes Y, Liltved H, Bergheim A. 2011. Recent RAS trends of commercial salmon smolt farming in Norway. In Aquaculture America, New Orleans, LA, 28 February to 3 March 2011. [Google Scholar]
- 48.Martins C, Eding EH, Verdegem MC, Heinsbroek LT, Schneider O, Blancheton J-P, d’Orbcastel ER, Verreth J. 2010. New developments in recirculating aquaculture systems in Europe: a perspective on environmental sustainability. Aquac Eng 43:83–93. doi: 10.1016/j.aquaeng.2010.09.002. [DOI] [Google Scholar]
- 49.Leonard N, Blancheton J, Guiraud J. 2000. Populations of heterotrophic bacteria in an experimental recirculating aquaculture system. Aquac Eng 22:109–120. doi: 10.1016/S0144-8609(00)00035-2. [DOI] [Google Scholar]
- 50.Schreier HJ, Mirzoyan N, Saito K. 2010. Microbial diversity of biological filters in recirculating aquaculture systems. Curr Opin Biotechnol 21:318–325. doi: 10.1016/j.copbio.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 51.Itoi S, Niki A, Sugita H. 2006. Changes in microbial communities associated with the conditioning of filter material in recirculating aquaculture systems of the pufferfish Takifugu rubripes. Aquaculture 256:287–295. doi: 10.1016/j.aquaculture.2006.02.037. [DOI] [Google Scholar]
- 52.Burr GS, Wolters WR, Schrader KK, Summerfelt ST. 2012. Impact of depuration of earthy-musty off-flavors on fillet quality of Atlantic salmon, Salmo salar, cultured in a recirculating aquaculture system. Aquac Eng 50:28–36. doi: 10.1016/j.aquaeng.2012.03.002. [DOI] [Google Scholar]
- 53.Kim D, Hofstaedter CE, Zhao C, Mattei L, Tanes C, Clarke E, Lauder A, Sherrill-Mix S, Chehoud C, Kelsen J, Conrad M, Collman RG, Baldassano R, Bushman FD, Bittinger K. 2017. Optimizing methods and dodging pitfalls in microbiome research. Microbiome 5:52. doi: 10.1186/s40168-017-0267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Campbell JH, Foster CM, Vishnivetskaya T, Campbell AG, Yang ZK, Wymore A, Palumbo AV, Chesler EJ, Podar M. 2012. Host genetic and environmental effects on mouse intestinal microbiota. ISME J 6:2033–2044. doi: 10.1038/ismej.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hildebrand F, Nguyen TLA, Brinkman B, Yunta RG, Cauwe B, Vandenabeele P, Liston A, Raes J. 2013. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol 14:R4. doi: 10.1186/gb-2013-14-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown RM, Wiens GD, Salinas I. 2019. Analysis of the gut and gill microbiome of resistant and susceptible lines of rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol 86:497–506. doi: 10.1016/j.fsi.2018.11.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ledy K, Giambérini L, Pihan JC. 2003. Mucous cell responses in gill and skin of brown trout Salmo trutta fario in acidic, aluminium-containing stream water. Dis Aquat Organ 56:235–240. doi: 10.3354/dao056235. [DOI] [PubMed] [Google Scholar]
- 58.Vatsos IN, Kotzamanis Y, Henry M, Angelidis P, Alexis MN. 2010. Monitoring stress in fish by applying image analysis to their skin mucous cells. Eur J Histochem 54:e22. doi: 10.4081/ejh.2010.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pittman K, Sourd P, Ravnøy B, Espeland Ø, Fiksdal IU, Oen T, Pittman A, Redmond K, Sweetman J. 2011. Novel method for quantifying salmonid mucous cells: quantification of salmonid mucous cells. J Fish Dis 34:931–936. doi: 10.1111/j.1365-2761.2011.01308.x. [DOI] [PubMed] [Google Scholar]
- 60.Karlsen C, Ytteborg E, Timmerhaus G, Høst V, Handeland S, Jørgensen SM, Krasnov A. 2018. Atlantic salmon skin barrier functions gradually enhance after seawater transfer. Sci Rep 8:9510. doi: 10.1038/s41598-018-27818-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pittman K, Pittman A, Karlson S, Cieplinska T, Sourd P, Redmond K, Ravnøy B, Sweetman E. 2013. Body site matters: an evaluation and application of a novel histological methodology on the quantification of mucous cells in the skin of Atlantic salmon, Salmo salar L. J Fish Dis 36:115–127. doi: 10.1111/jfd.12002. [DOI] [PubMed] [Google Scholar]
- 62.Tarnecki AM, Burgos FA, Ray CL, Arias CR. 2017. Fish intestinal microbiome: diversity and symbiosis unravelled by metagenomics. J Appl Microbiol 123:2–17. doi: 10.1111/jam.13415. [DOI] [PubMed] [Google Scholar]
- 63.Bates JM, Mittge E, Kuhlman J, Baden KN, Cheesman SE, Guillemin K. 2006. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev Biol 297:374–386. doi: 10.1016/j.ydbio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 64.Dehler CE, Secombes CJ, Martin SAM. 2017. Environmental and physiological factors shape the gut microbiota of Atlantic salmon parr (Salmo salar L.). Aquaculture 467:149–157. doi: 10.1016/j.aquaculture.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang C, Sun G, Li S, Li X, Liu Y. 2018. Intestinal microbiota of healthy and unhealthy Atlantic salmon Salmo salar L. in a recirculating aquaculture system. J Ocean Limnol 36:414–426. doi: 10.1007/s00343-017-6203-5. [DOI] [Google Scholar]
- 66.Gonzalez A, Navas-Molina JA, Kosciolek T, McDonald D, Vázquez-Baeza Y, Ackermann G, DeReus J, Janssen S, Swafford AD, Orchanian SB, Sanders JG, Shorenstein J, Holste H, Petrus S, Robbins-Pianka A, Brislawn CJ, Wang M, Rideout JR, Bolyen E, Dillon M, Caporaso JG, Dorrestein PC, Knight R. 2018. Qiita: rapid, Web-enabled microbiome meta-analysis. Nat Methods 15:796–798. doi: 10.1038/s41592-018-0141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dang M, Nørregaard R, Bach L, Sonne C, Søndergaard J, Gustavson K, Aastrup P, Nowak B. 2017. Metal residues, histopathology and presence of parasites in the liver and gills of fourhorn sculpin (Myoxocephalus quadricornis) and shorthorn sculpin (Myoxocephalus scorpius) near a former lead-zinc mine in East Greenland. Environ Res 153:171–180. doi: 10.1016/j.envres.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 68.Roberts SD, Powell MD. 2003. Comparative ionic flux and gill mucous cell histochemistry: effects of salinity and disease status in Atlantic salmon (Salmo salar L.). Comp Biochem Physiol A Mol Integr Physiol 134:525–537. doi: 10.1016/s1095-6433(02)00327-6. [DOI] [PubMed] [Google Scholar]
- 69.Løkka G, Austbø L, Falk K, Bjerkås I, Koppang EO. 2013. Intestinal morphology of the wild Atlantic salmon (Salmo salar). J Morphol 274:859–876. doi: 10.1002/jmor.20142. [DOI] [PubMed] [Google Scholar]
- 70.Minich JJ, Sanders JG, Amir A, Humphrey G, Gilbert JA, Knight R. 2019. Quantifying and understanding well-to-well contamination in microbiome research. mSystems 4:e00186-19. doi: 10.1128/mSystems.00186-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Minich JJ, Humphrey G, Benitez RAS, Sanders J, Swafford A, Allen EE, Knight R. 2018. High-throughput miniaturized 16S rRNA amplicon library preparation reduces costs while preserving microbiome integrity. mSystems 3:e00166-18. doi: 10.1128/mSystems.00166-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Parada AE, Needham DM, Fuhrman JA. 2016. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 73.Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, Gilbert JA, Jansson JK, Caporaso JG, Fuhrman JA, Apprill A, Knight R. 2016. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e00009-15. doi: 10.1128/mSystems.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Zech Xu Z, Kightley EP, Thompson LR, Hyde ER, Gonzalez A, Knight R. 2017. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2:e00191-16. doi: 10.1128/mSystems.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McDonald D, Hyde E, Debelius JW, Morton JT, Gonzalez A, Ackermann G, Aksenov AA, Behsaz B, Brennan C, Chen Y, DeRight Goldasich L, Dorrestein PC, Dunn RR, Fahimipour AK, Gaffney J, Gilbert JA, Gogul G, Green JL, Hugenholtz P, Humphrey G, Huttenhower C, Jackson MA, Janssen S, Jeste DV, Jiang L, Kelley ST, Knights D, Kosciolek T, Ladau J, Leach J, Marotz C, Meleshko D, Melnik AV, Metcalf JL, Mohimani H, Montassier E, Navas-Molina J, Nguyen TT, Peddada S, Pevzner P, Pollard KS, Rahnavard G, Robbins-Pianka A, Sangwan N, Shorenstein J, Smarr L, Song SJ, Spector T, Swafford AD, Thackray VG, et al. 2018. American Gut: an open platform for citizen science microbiome research. mSystems 3:e00031-18. doi: 10.1128/mSystems.00031-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kruskal WH, Wallis WA. 1952. Use of ranks in one-criterion variance analysis. J Am Stat Assoc 47:583–621. doi: 10.1080/01621459.1952.10483441. [DOI] [Google Scholar]
- 79.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 80.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chang Q, Luan Y, Sun F. 2011. Variance adjusted weighted UniFrac: a powerful beta diversity measure for comparing communities based on phylogeny. BMC Bioinformatics 12:118. doi: 10.1186/1471-2105-12-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46. [Google Scholar]
- 83.Xu ZZ, Amir A, Sanders J, Zhu Q, Morton JT, Bletz MC, Tripathi A, Huang S, McDonald D, Jiang L, Knight R. 2019. Calour: an interactive, microbe-centric analysis tool. mSystems 4:e00269-18. doi: 10.1128/mSystems.00269-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Scherer A. 2009. Batch effects and noise in microarray experiments: sources and solutions. John Wiley & Sons, Chichester, UK. [Google Scholar]
- 85.Boedigheimer MJ, Wolfinger RD, Bass MB, Bushel PR, Chou JW, Cooper M, Corton JC, Fostel J, Hester S, Lee JS, Liu F, Liu J, Qian H-R, Quackenbush J, Pettit S, Thompson KL. 2008. Sources of variation in baseline gene expression levels from toxicogenomics study control animals across multiple laboratories. BMC Genomics 9:285. doi: 10.1186/1471-2164-9-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bushel PR. 2019. Principal variance component analysis. National Institute of Environmental Health Sciences, Research Triangle Park, NC: https://www.niehs.nih.gov/research/resources/software/biostatistics/pvca/index.cfm. [Google Scholar]
- 87.Zhou Y-H, Gallins P. 2019. A review and tutorial of machine learning methods for microbiome host trait prediction. Front Genet 10:579. doi: 10.3389/fgene.2019.00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, Kosciolek T, Janssen S, Metcalf J, Song SJ, Kanbar J, Miller-Montgomery S, Heaton R, Mckay R, Patel SP, Swafford AD, Knight R. 2020. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 579:567–574. doi: 10.1038/s41586-020-2095-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 89.Law CW, Chen Y, Shi W, Smyth GK. 2014. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mecham BH, Nelson PS, Storey JD. 2010. Supervised normalization of microarrays. Bioinformatics 26:1308–1315. doi: 10.1093/bioinformatics/btq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liaw A, Wiener M. 2002. Classification and egression by randomForest 2:5.
- 92.Smola AJ. 2007. An introduction to machine learning—L1: basics and probability theory 271. http://alex.smola.org/teaching/pune2007/pune_1.pdf.
- 93.Piñeiro G, Perelman S, Guerschman JP, Paruelo JM. 2008. How to evaluate models: observed vs. predicted or predicted vs. observed? Ecol Model 216:316–322. doi: 10.1016/j.ecolmodel.2008.05.006. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.