Acyl-CoA carboxylases play key roles in primary and secondary metabolism. However, the regulation of ACCase genes transcription in Streptomyces spp. remains unclear. Here, we demonstrated that AccR responded to intracellular acetyl-, propionyl-, and methylcrotonyl-CoA availability and mediated transcription of the genes related to production and assimilation of these compounds in S. avermitilis. When intracellular concentrations of these compounds are low, AccR binds to target genes and represses their transcription, resulting in low production of malonyl- and methylmalonyl-CoAs. When intracellular acetyl-, propionyl-, and methylcrotonyl-CoA concentrations are high, these compounds bind to AccR to dissociate AccR from target DNA, promoting the conversion of these compounds to malonyl- and methylmalonyl-CoAs. This investigation revealed how AccR coordinates short-chain acyl-CoA homeostasis in Streptomyces.
KEYWORDS: Streptomyces, TetR family transcriptional regulator, acyl-CoA carboxylase, polyketides, short-chain acyl-CoAs
ABSTRACT
Malonyl coenzyme A (malonyl-CoA) and methylmalonyl-CoA are the most common extender units for the biosynthesis of fatty acids and polyketides in Streptomyces, an industrially important producer of polyketides. Carboxylation of acetyl- and propionyl-CoAs is an essential source of malonyl- and methylmalonyl-CoAs; therefore, acyl-CoA carboxylases (ACCases) play key roles in primary and secondary metabolism. The regulation of the expression of ACCases in Streptomyces spp. has not been investigated previously. We characterized a TetR family transcriptional repressor, AccR, that mediates intracellular acetyl-, propionyl-, methylcrotonyl-, malonyl-, and methylmalonyl-CoA levels by controlling the transcription of genes that encode the main ACCase and enzymes associated with branched-chain amino acid metabolism in S. avermitilis. AccR bound to a 16-nucleotide palindromic binding motif (GTTAA-N6-TTAAC) in promoter regions and repressed the transcription of the accD1A1-hmgL-fadE4 operon, echA8, echA9, and fadE2, which are involved in the production and assimilation of acetyl- and propionyl-CoAs. Methylcrotonyl-, propionyl-, and acetyl-CoAs acted as effectors to release AccR from its target DNA, resulting in enhanced transcription of target genes by derepression. The affinity of methylcrotonyl- and propionyl-CoAs to AccR was stronger than that of acetyl-CoA. Deletion of accR resulted in increased concentrations of short-chain acyl-CoAs (acetyl-, propionyl-, malonyl-, and methylmalonyl-CoAs), leading to enhanced avermectin production. Avermectin production was increased by 14.5% in an accR deletion mutant of the industrial high-yield strain S. avermitilis A8. Our findings clarify the regulatory mechanisms that maintain the homeostasis of short-chain acyl-CoAs in Streptomyces.
IMPORTANCE Acyl-CoA carboxylases play key roles in primary and secondary metabolism. However, the regulation of ACCase genes transcription in Streptomyces spp. remains unclear. Here, we demonstrated that AccR responded to intracellular acetyl-, propionyl-, and methylcrotonyl-CoA availability and mediated transcription of the genes related to production and assimilation of these compounds in S. avermitilis. When intracellular concentrations of these compounds are low, AccR binds to target genes and represses their transcription, resulting in low production of malonyl- and methylmalonyl-CoAs. When intracellular acetyl-, propionyl-, and methylcrotonyl-CoA concentrations are high, these compounds bind to AccR to dissociate AccR from target DNA, promoting the conversion of these compounds to malonyl- and methylmalonyl-CoAs. This investigation revealed how AccR coordinates short-chain acyl-CoA homeostasis in Streptomyces.
INTRODUCTION
Polyketides are a large class of natural products with diverse structures and biological activities. Many of them (e.g., avermectin, erythromycin, lovastatin, rifamycin, and tetracycline) are widely used in medicine, agriculture, and animal husbandry (1). The formation of polyketides is initiated by polyketide synthases, which condense acyl coenzyme A (acyl-CoA) extender units onto the starter unit through repeated decarboxylative Claisen condensation reactions (2, 3). Malonyl- and methylmalonyl-CoAs are the most common extender units for polyketide biosynthesis (4). Malonyl-CoA is also a building block for fatty acid biosynthesis. Production of malonyl- and methylmalonyl-CoAs usually involves the carboxylation of acetyl- and propionyl-CoAs (5).
In actinobacteria, including Streptomyces, Mycobacterium, and Corynebacterium, the enzymes that carboxylate acetyl-CoA and propionyl-CoA are termed acyl-CoA carboxylases (ACCases) (6). Actinobacterial ACCases have broad substrate specificity and are able to simultaneously catalyze acetyl-CoA, propionyl-CoA, and butyryl-CoA to generate malonyl-CoA, methylmalonyl-CoA, and ethylmalonyl-CoA, respectively (7–9). The two major subunits of ACCase complex are a larger α subunit that contains biotin carboxylase (BC) and biotin carboxyl carrier protein (BCCP) domains and a smaller β subunit that contains the carboxyltransferase (CT) domain (10). In some cases, a noncatalytic ε subunit is required for holo-carboxylase activity (11). ACCases are categorized as acetyl-CoA carboxylase (ACC), propionyl-CoA carboxylase (PCC), or methylcrotonyl-CoA carboxylase (MCC) depending on substrate preference. Two ACCases have been characterized in Streptomyces coelicolor A3(2) (12–14). They have the same α subunit, AccA1 or AccA2 (encoded by identical genes accA1 or accA2). The β and ε components are, respectively, AccB and AccE for ACC and PccB and PccE for PCC (12–14). ACC is able to carboxylate acetyl-, propionyl-, and butyryl-CoAs with similar specificity, whereas PCC carboxylates only propionyl- and butyryl-CoAs. The major determinant of substrate specificity for ACCases is the β subunit (9). ACCases provide the building blocks for polyketide biosynthesis. Overexpression of ACC enhances intracellular malonyl-CoA levels, leading to the overproduction of actinorhodin in S. coelicolor A3(2) and of antitumor mithramycin in S. argillaceus (15, 16). PCC overexpression in S. hygroscopicus increased the rapamycin yield (17).
Despite the essential roles of ACCases in primary and secondary metabolism, the mechanisms that regulate their expression in actinobacteria are poorly understood. Several TetR family regulators (TFRs) have been identified to regulate the transcription of ACCase-encoding genes. TFRs are a ubiquitous group of transcriptional regulators, consisting of a highly conserved helix-turn-helix DNA binding motif at the N terminus and a diverse ligand binding domain at the C terminus (18). In Corynebacterium glutamicum, a highly conserved TFR FasR acts as a repressor for the gene accD1, which encodes the β subunit of the essential ACC. FasR also plays an important role in regulation of fatty acid synthesis by repressing the transcription of two fatty acid synthase encoding genes, fasA and fasB (19). In Saccharopolyspora erythraea, another TFR, PccD, represses PCC expression by binding to the upstream regions of the pccBC operon and pccA which encode the β, ε, and α subunits of PCC separately (20). Methylmalonic acid acts as a ligand of PccD, inhibiting the binding of PccD to its target DNA (20). In mycobacteria, the TetR family repressor BkaR (also known as Fad35R) binds to 16-nucleotide (nt) palindromic motifs (GTTANT-N4-ANTAAC) in upstream regions of target genes and represses the transcription of itself and of a divergently oriented operon, accD1A1-fadE19-Rv2499c-citE-bkdABC, involved in branched-chain keto-acid metabolism (21, 22). Regulation of ACCase expression in Streptomyces species has not been studied previously.
Avermectins, a series of 16-membered macrocyclic polyketides produced by S. avermitilis, are widely used in agricultural, veterinary, and medical fields because they display low host toxicity and broad-spectrum nematicidal, acaricidal, and insecticidal activities (23). Synthesis of avermectin aglycone is initiated by starter unit 2-methylbutyryl-CoA or isobutyryl-CoA and proceeds by the addition to the starter unit of seven acetate extender units from malonyl-CoA and five propionate extender units from methylmalonyl-CoA (23, 24). Here, we describe the identification of a TFR, AccR (SAV5279), that functions as a repressor for the control of intracellular short-chain acyl-CoA levels in S. avermitilis by inhibiting the transcription of ACCase coding genes and genes related to branched-chain amino acid metabolism. Methylcrotonyl-, propionyl-, and acetyl-CoAs act as ligands and interact with AccR to dissociate it from its target DNA. Deletion of accR resulted in an increase of short-chain acyl-CoA pools and consequent enhancement of avermectin production in S. avermitilis wild-type (WT) and industrial strains.
RESULTS
The expression of accD1 and accA1 was strongly induced by acetate, propionate, and l-leucine.
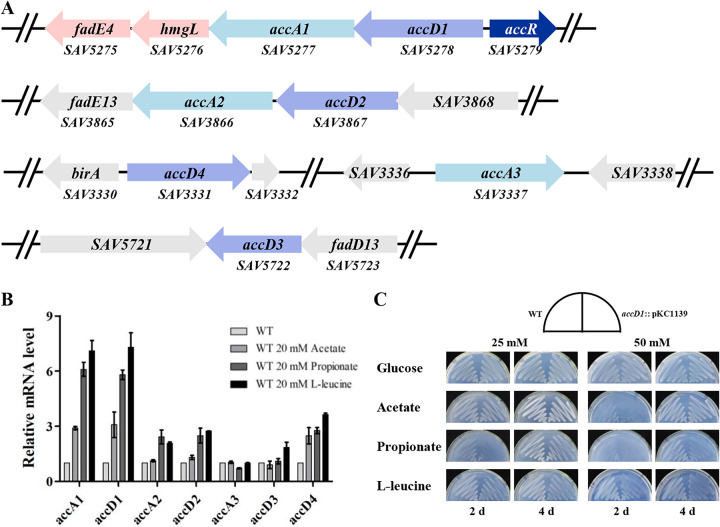
Seven ACCase coding genes were found in the S. avermitilis genome. Among these, it is predicted that accA1 (SAV5277), accA2 (SAV3866), and accA3 (SAV3337) encode α subunits of ACCases and that accD1 (SAV5278), accD2 (SAV3867), accD3 (SAV5722), and accD4 (SAV3331) encode β subunits of ACCases (Fig. 1A). S. avermitilis AccA1, AccA2, AccA3, AccD1, AccD2, and AccD3 share 78, 82, 93, 90, 92, and 78% sequence identities to SCO2777, SCO4381, AccA1/AccA2 (SCO6271/SCO4921, identical proteins), AccD1 (SCO2776), SCO4380, and SCO2445 in S. coelicolor A3(2), respectively. AccD4 has 91% identity with S. coelicolor A3(2) PCC β subunit (PccB, SCO4926) and presumably functions as a PCC in S. avermitilis. accD1, accA1, and their downstream genes (hmgL and fadE4) are oriented in the same direction, with very short intergenic regions, and were shown by reverse transcription-PCR analysis to be organized in the same operon (see Fig. S1 in the supplemental material). accD2 and adjacent accA2 are also oriented in the same direction. It is reasonable to presume that AccD1 and AccA1 form a heteromeric multisubunit ACCase, whereas AccD2 and AccA2 form another. The functions of these ACCases in S. avermitilis remain to be elucidated.
FIG 1.
Analysis of acyl-CoA carboxylase (ACCase)-encoding genes in S. avermitilis. (A) Organization of ACCase genes in S. avermitilis. accA and accD encode the α and β subunits of ACCase, respectively. accR encodes a TetR family regulator. hmgL and fadE4 were cotranscribed with accD1A1. Gray, adjacent genes of acc genes. (B) Induction of acc genes by acetate, propionate, or l-leucine in S. avermitilis. The transcription levels of acc genes were determined by RT-qPCR. RNAs were extracted from cells cultured in FM-I with supplemental acetate, propionate, or l-leucine for 24 h. (C) Phenotypic analyses of WT and accD1::pKC1139. The strains were cultured on MM supplemented with glucose, acetate, propionate, or l-leucine as the sole carbon source.
In bacteria, acetate and propionate are converted to acetyl-CoA and propionyl-CoA by acetyl/propionyl-CoA synthetase and then utilized by ACCases (25). To determine which acc gene encodes the main ACCase that carboxylates acetyl- and propionyl-CoAs in S. avermitilis, we analyzed the transcription levels of all acc genes in the WT strain ATCC 31267 after the addition of 20 mM acetate or propionate. In acetate-induced cells, the transcription levels of accA1 and accD1 were ∼3-fold higher than in cells without acetate induction, which was the highest fold increase of all acc genes under the same condition (Fig. 1B). The transcription levels of accA1 and accD1 were increased ∼6-fold under treatment with propionate. accD4 expression was enhanced (∼2.5-fold) by acetate and (∼2.8-fold) by propionate, indicating that the ACCase consisting of AccD4 functions mainly as a PCC. Other acc genes shown no notable induction by acetate, and only accA2 and accD2 were slightly induced by propionate. In Mycobacterium tuberculosis, the homolog of AccD1A1 was identified as an MCC involved in leucine metabolism, which catalyzes carboxylation of methylcrotonyl-CoA during leucine degradation (26). Therefore, we also detected the leucine induction of the acc genes. accD1A1 was significantly induced (∼7-fold) by l-leucine, which is much higher than the fold increases for other acc genes (Fig. 1B). These findings collectively indicate that acetate, propionate, and l-leucine induced transcription of accD1A1, and the induction of propionate and l-leucine was stronger than that of acetate. The fold increases of accA1 and accD1 were more pronounced than the others under all treatments. Thus, ACCase consisting of AccD1 and AccA1 appears to play a crucial role in the assimilation of acetyl-CoA, propionyl-CoA, and methylcrotonyl-CoA in S. avermitilis.
Disruption of accD1 affected growth on media with acetate, propionate, or l-leucine as the sole carbon source.
The β subunit of the ACCase determines substrate specificity. To identify the substrate specificity of AccD1A1, we disrupted the accD1 gene in the S. avermitilis genome by inserting plasmid pKC1139 into the accD1 open reading frame (ORF). The growth of WT and accD1::pKC1139 was investigated on minimal medium (MM) with glucose, acetate, propionate, or l-leucine as the sole carbon source. Both accD1::pKC1139 and WT grew well on MM supplemented with glucose. The growth of the WT was slightly inhibited by acetate and propionate but not by l-leucine. Compared to the WT, the growth of accD1::pKC1139 on MM with acetate, propionate, or l-leucine was significantly inhibited, and inhibition was more pronounced on the media with higher concentrations (50 mM) of these compounds (Fig. 1C). l-Leucine and propionate had more obvious inhibitory effects on accD1::pKC1139 than acetate, which was consistent with the higher fold inductions of accD1 and accA1 by l-leucine and propionate (Fig. 1B and C). These findings indicated that AccD1A1 functions as the main ACCase assimilating acetyl-CoA, propionyl-CoA, and methylcrotonyl-CoA in S. avermitilis.
AccR directly represses the expression of accD1A1 and its own gene.
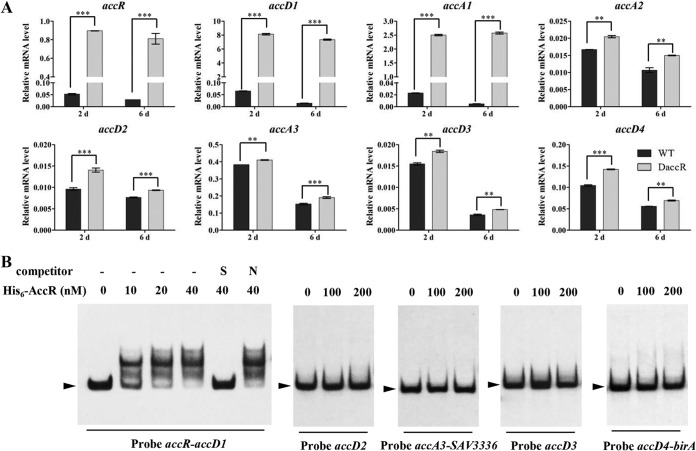
SAV5279 (designated accR) shares a bidirectional promoter region with the accD1A1-hmgL-fadE4 operon and was predicted to encode a TetR family regulator. AccR is highly conserved in actinobacteria, with 76 to 91% sequence identities to its homologs in Streptomyces, 53% identity to Nocardia farcinica Nfa50360, and 51% identity to M. tuberculosis BkaR (Fig. S2A). Its adjacent acc operon is also conserved in actinobacteria, except for the absence of the hmgL gene in N. farcinica and M. tuberculosis. To investigate possible regulation of adjacent acc operon by AccR, we constructed an accR deletion strain (termed DaccR) with an internal 421-bp deletion in the accR ORF by homologous recombination in the WT. The expression of accR and acc genes in WT and DaccR was analyzed by reverse transcription-quantitative PCR (RT-qPCR) during the exponential (day 2) and stationary (day 6) phases (Fig. 2A). accR expression in DaccR was, respectively, ∼17- and ∼27-fold higher than in the WT on days 2 and 6. The transcription levels of accA1 and accD1 in DaccR, relative to WT, were ∼120-fold higher on day 2 and >500-fold higher on day 6. The expression of other acc genes in DaccR increased to much smaller degrees. These findings indicated that AccR represses the expression of its own gene and of acc genes, particularly accD1 and accA1.
FIG 2.
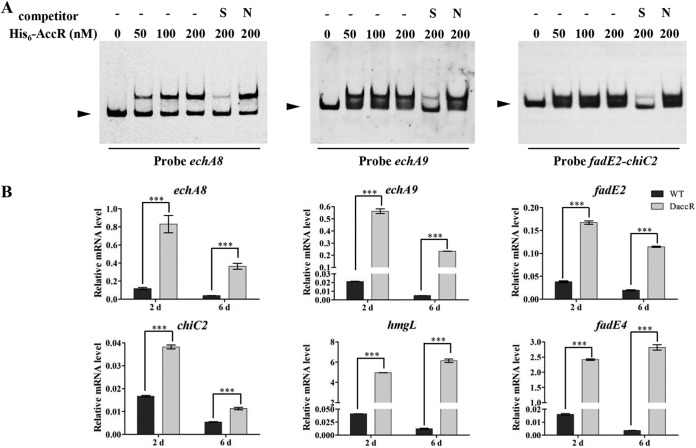
Interaction of AccR with acc genes in S. avermitilis. (A) Transcriptional levels of acc genes in WT and DaccR were determined by RT-qPCR. Values are means ± the standard deviations from three replicate experiments. **, P < 0.01; ***, P < 0.001 (Student t test). (B) EMSAs of AccR with promoter regions of acc genes. A 200-fold excess of specific (S) or nonspecific (N) unlabeled probes was used as a competitor of the labeled probe. Arrowheads indicate free DNA probes.
To determine whether AccR directly regulates acc genes, we performed electrophoretic mobility shift assays (EMSAs) using upstream regions of acc genes and His6-AccR purified from Escherichia coli (Fig. S3). His6-AccR bound to the intergenic region of accR-accD1, leading to clear shifting of the bands (Fig. 2B). Shifting of the bands was abolished by an unlabeled specific probe but not by a nonspecific probe (Fig. 2B), indicating that interaction of AccR with the accR-accD1 intergenic region is specific. No shifted bands were observed in upstream regions of accD2, accD3, or the intergenic regions of accA3-SAV3336 and accD4-birA with His6-AccR. These findings, taken together, indicate that AccR directly represses the expression of accR, accD1, and accA1 and indirectly inhibits the expression of other acc genes.
Determination of the AccR binding site in the accR-accD1 intergenic region.
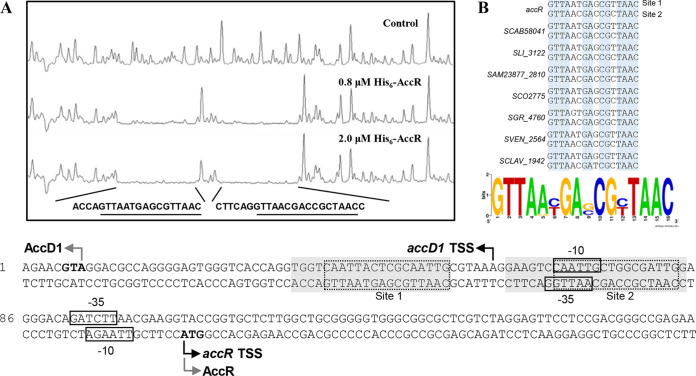
To clarify the regulatory mechanism of AccR on accD1A1 expression, we determined the binding sites of AccR by DNase I footprinting assays, using His6-AccR and a FAM-labeled DNA probe of the accR-accD1 intergenic region. Two DNA regions in the accR-accD1 intergenic region were clearly protected by increased concentrations of AccR (0.8 and 2.0 μM) (Fig. 3A). Analysis of the protected regions revealed two similar 16-nt palindromic sequences (GTTAA-N6-TTAAC), consistent with previous findings that TetR family regulators usually form homodimers and bind to palindromic sequences (27). Similar palindromic sequences were found in upstream regions of its homologous genes in other Streptomyces species (Fig. 3B; see also Fig. S2B in the supplemental material), indicating that the 16-nt GTTAA-N6-TTAAC palindromic sequence is the consensus sequence of AccR binding sites. Transcriptional start sites of accR and accD1 were determined by 5′ RACE and found to be located at nt –52 (A) for accD1 and nt +1 (A) for accR relative to their translation initiation codons. The AccR binding sites are located at the −35 region and upstream of the −35 region for accR and in the −10 region and downstream of the −10 region for accD1. Thus, AccR represses transcription of the accD1A1 operon and accR by blocking attachment of the RNA polymerase to their promoters, thereby preventing transcription initiation and extension.
FIG 3.
Identification of AccR binding sites in the intergenic region of accR-accD1. (A) DNase I footprinting assay of AccR with the accR-accD1 intergenic region. Gray-shaded boxes indicate DNA regions protected by AccR; dotted frames indicate palindromic binding sites. Dark and gray curved arrows indicate transcriptional start sites and translational initiation codons, respectively. Boxes indicate putative −10 and −35 regions. (B) Alignment of AccR binding sites of accR-accD1 intergenic regions in Streptomyces species. The consensus sequence logo was created using WebLogo.
Methylcrotonyl-CoA, propionyl-CoA, and acetyl-CoA are ligands of AccR.
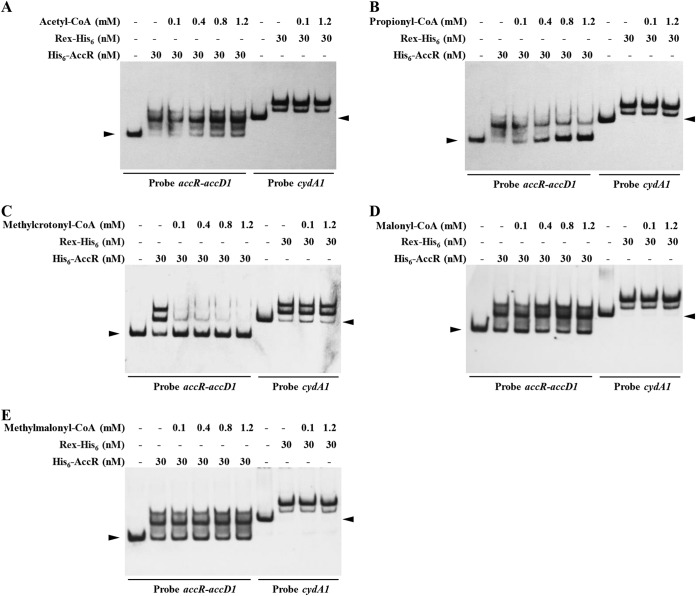
TetR family regulators bind and respond to one or more ligands, which change their DNA binding ability for DNA targeting, thus coordinating expression of target genes. AccR represses the transcription of accD1A1, which encodes the ACCase catalyzing assimilation of acetyl-, propionyl-, and methylcrotonyl-CoAs. Therefore, we performed EMSAs to determine which ACCase substrate (acetyl-, propionyl-, and methylcrotonyl-CoAs) or product (malonyl- and methylmalonyl-CoAs) affects the binding ability of AccR to the accR-accD1 intergenic region. Methylcrotonyl-, propionyl-, and acetyl-CoAs inhibited interaction of AccR with the accR-accD1 intergenic region (Fig. 4A to C), whereas malonyl- and methylmalonyl-CoAs had no effect on the DNA-binding activity of AccR (Fig. 4D and E). The effect of methylcrotonyl- and propionyl-CoAs on AccR activity was much more pronounced than that of acetyl-CoA. Methylcrotonyl-CoA at 5 μM or propionyl-CoA at 50 μM was sufficient to attenuate the interaction of AccR with accR-accD1 intergenic region, whereas a much higher concentration of acetyl-CoA was required to achieve a similar effect (Fig. 4A; see also Fig. S4 in the supplemental material). The redox-sensing regulator Rex of S. avermitilis, which recognizes NADH as a ligand (28), was used as a negative control. Taken together, these findings indicate that the ACCase substrates methylcrotonyl-CoA, propionyl-CoA, and acetyl-CoA are the true effectors of AccR to release AccR from its target genes. When intracellular methylcrotonyl-, propionyl-, and acetyl-CoA pools are abundant, these compounds bind to AccR to dissociate AccR from DNA, resulting in the enhanced expression of accD1A1 by derepression, which then promotes the utilization of these compounds.
FIG 4.
Effects of acetyl-CoA (A), propionyl-CoA (B), methylcrotonyl-CoA (C), malonyl-CoA (D), and methylmalonyl-CoA (E) on in vitro binding of AccR to the accR-accD1 intergenic region. The concentrations of the five acyl-CoAs are shown. Binding of Rex-His6 to cydA1 promoter region was used as a negative control. Arrowheads indicate free probes.
Identification of other target genes regulated by AccR.
To further clarify the mechanisms for regulation of AccR in S. avermitilis, a consensus sequence was created (29) by aligning AccR binding sites of accR-accD1 intergenic regions from eight Streptomyces species (including S. avermitilis) (Fig. 3B) and used to predict AccR target genes in the S. avermitilis genome by PREDetector (30). With a score set to 10, a total of 13 genes were revealed (see Table S1 in the supplemental material), including accR and accD1. Interestingly, 8 of these 13 genes are involved in lipid and carbohydrate metabolism (chiC2 and udgA in carbohydrate metabolism; echA8, echA9, fadE1, fadE2, and icmB in lipid metabolism; and SAV1133 in lipid transport). The upstream regions of these 8 genes were used for EMSAs. AccR bound to promoter regions of echA8 and echA9 and the fadE2-chiC2 intergenic region but not to SAV1133 promoter region or intergenic regions of fadE1-udgA and icmB-SAV3461 (Fig. 5A; see also Fig. S5 in the supplemental material).
FIG 5.
Verification of putative AccR target genes. (A) EMSAs of AccR with promoter regions of putative target genes. Arrowheads indicate free DNA probes. (B) RT-qPCR analysis of the expression of putative target genes in WT and DaccR strains. ***, P < 0.001 (Student t test).
The transcription levels of echA8, echA9, fadE2, and chiC2 in DaccR were much higher than in the WT (Fig. 5B). Because hmgL and fadE4 are in the same operon as accD1A1, we analyzed the transcriptional levels of hmgL and fadE4. hmgL and fadE4 levels in DaccR were more than 100-fold higher than in the WT strain, similarly to accD1A1 (Fig. 2A and 5B). Thus, AccR also represses the expression of chiC2, echA8, echA9, fadE2, fadE4, and hmgL. chiC2 encodes a chitinase that degrades chitin. echA8 and echA9 encode enoyl-CoA hydratase, and fadE2 and fadE4 encode acyl-CoA dehydrogenase. These enzymes are involved in the β-oxidation process and branched-chain amino acid metabolism, an important source of acetyl- and propionyl-CoAs. hmgL encodes a hydroxymethylglutaryl-CoA cleavage enzyme that cleaves HMG-CoA (downstream product of carboxylation of methylcrotonyl-CoA in leucine degradation) to produce acetyl-CoA and acetoacetate. Thus, in addition to the repression of ACCase genes, AccR also represses the expression of genes related to acetyl- and propionyl-CoA production.
AccR mediates homeostasis of intracellular short-chain acyl-CoAs.
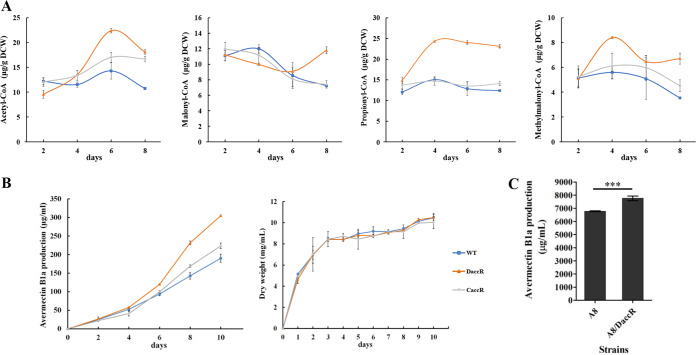
As described above, AccR represses the transcription of acyl-CoA metabolism genes involved in the production and assimilation of acetyl- and propionyl-CoAs. To elucidate the regulatory role of AccR in the metabolism of short-chain acyl-CoAs related to avermectin biosynthesis, we assessed the intracellular availability of malonyl- and methylmalonyl-CoAs and of their substrates, acetyl- and propionyl-CoAs, in WT, DaccR, and complemented CaccR strains. Titers of acetyl- and propionyl-CoAs were maintained at relatively constant levels in the WT but increased in DaccR in the early and middle stages of fermentation and then were maintained at much higher levels than in the WT strain (Fig. 6A). Malonyl- and methylmalonyl-CoA titers in WT increased in the early stage and reached maximal values on day 4 but, in DaccR, declined in the middle stage of fermentation, reached minimal values on day 6, and increased thereafter. In particular, malonyl- and methylmalonyl-CoA levels in DaccR were much higher than those in the WT in late-stage fermentation when secondary metabolites were rapidly biosynthesized (Fig. 6A). The titer curves of CaccR were similar to those of WT, indicating that altered levels of short-chain acyl-CoAs in DaccR were due to the loss of accR. Overall, the levels of short-chain acyl-CoAs in DaccR were strongly increased, an observation consistent with the notably increased expression of accD1A1, echA8, echA9, fadE2, fadE4, and hmgL in DaccR, which promoted the accumulation and conversion of acetyl- and propionyl-CoAs to malonyl- and methylmalonyl-CoAs (Fig. 2A and 5B), suggesting that AccR plays an important role in maintaining the homeostasis of intracellular short-chain acyl-CoAs.
FIG 6.
Determination of intracellular short-chain acyl-CoA contents in WT, DaccR, and CaccR strains. (A) Intracellular contents of acetyl-, malonyl-, propionyl-, and methylmalonyl-CoAs at the indicated times. (B) Avermectin production by WT, DaccR, and CaccR strains in FM-I and growth curves in FM-II. Error bars represent the standard deviations of three replicates. (C) Effect on avermectin production of accR deletion in the industrial high-yield strain S. avermitilis A8. ***, P < 0.001 (Student t test).
Deletion of accR increased avermectin production.
DaccR showed increased pools of intracellular malonyl- and methylmalonyl-CoAs, the precursors of avermectin biosynthesis. We evaluated avermectin production in related mutants to determine whether acyl-CoA metabolism affects secondary metabolism. Avermectin B1a production in DaccR (304 μg ml−1) was higher than in WT (190 μg ml−1) and in CaccR it was close to the WT level. accR deletion had no effect on growth rate and biomass (Fig. 6B). The findings indicated that the enhanced avermectin yield was attributed to the deletion of accR. Overexpression of accD1A1 led to enhanced avermectin production in the WT strain but not in DaccR, and production in the WT/accD1A1-overexpressing strain was less than in DaccR (Fig. S6), indicating that enhanced expression of other target genes besides accD1A1 contributed to the increase of avermectin production in DaccR. Although the transcription levels of the avermectin pathway-specific regulatory gene aveR and the biosynthetic genes aveA1, aveA4, and aveD were increased in DaccR, AccR did not bind to the upstream regions of these genes. Taken together, these findings indicate that AccR indirectly regulates avermectin production (see Fig. S7A and B in the supplemental material), mainly by affecting intracellular metabolic flux.
To experimentally increase avermectin production in an industrial context, we deleted accR in the industrial high-yield strain S. avermitilis A8. The avermectin B1a yield of the A8/DaccR strain (7,757.23 μg ml−1) was 14.5% higher than that of the parental strain (6,770.46 μg ml−1) (Fig. 6C), indicating that industrial avermectin production can be effectively enhanced by genetic manipulation of accR.
DISCUSSION
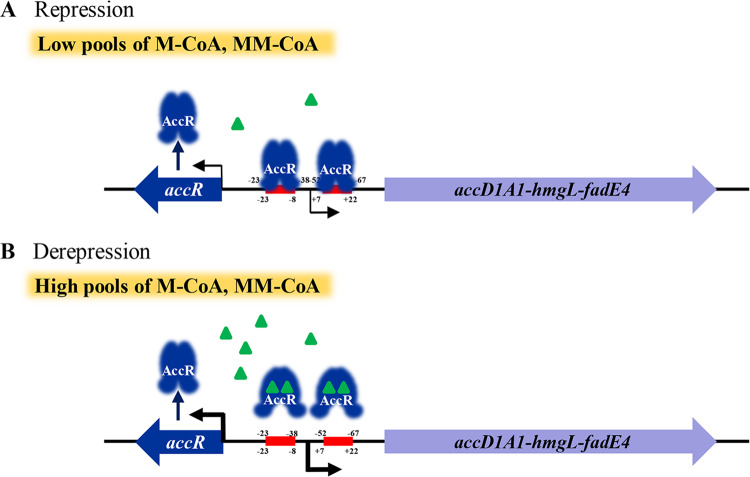
Malonyl- and methylmalonyl-CoAs are the building blocks for biosynthesis of polyketides and fatty acids. We demonstrated in this study that intracellular pools of malonyl- and methylmalonyl-CoAs are regulated by a TetR family regulator, AccR. Through binding to a 16-nt palindromic sequence (GTTAA-N6-TTAAC) in upstream regions of target genes, AccR represses the expression of branched-chain amino acid metabolism genes and the ACCase coding gene accD1A1, which is involved in the production and assimilation of acetyl- and propionyl-CoAs (see Fig. S8 in the supplemental material). AccR represses accD1A1 expression but does not directly control the expression of other acc genes. Methylcrotonyl-CoA, propionyl-CoA, and acetyl-CoA act as the ligands of AccR, inhibiting the interaction of AccR with its target DNA. When the intracellular concentrations of methylcrotonyl-, propionyl-, and acetyl-CoAs are low, AccR binds to target genes and represses the expression of these genes and also its own gene, resulting in low production of malonyl- and methylmalonyl-CoAs. Conversely, when intracellular methylcrotonyl-, propionyl-, and acetyl-CoA concentrations are high, these compounds form complexes with AccR, resulting in the dissociation of AccR from target DNA. The expression of target genes is thereby enhanced through derepression, promoting the conversion of methylcrotonyl-, propionyl-, and acetyl-CoAs to malonyl- and methylmalonyl-CoAs (Fig. 7; see also Fig. S8 in the supplemental material). In summary, AccR responds to intracellular methylcrotonyl-, propionyl-, and acetyl-CoA availability and plays a key role in regulating short-chain acyl-CoA pools in S. avermitilis.
FIG 7.
Proposed model of AccR-mediated regulation of ACCase (AccD1A1) in S. avermitilis. (A) When the intracellular concentrations of methylcrotonyl-, propionyl-, or acetyl-CoA are low, AccR binds to the intergenic region of accR-accD1 and represses the expression of the accD1A1-hmgL-fadE4 operon and of its own gene, resulting in low production of malonyl- and methylmalonyl-CoAs. (B) When the methylcrotonyl-, propionyl-, or acetyl-CoA levels are sufficient, these compounds form complexes with AccR, leading to dissociation of AccR from DNA, and the expression of target genes is enhanced by derepression. Red bars indicate AccR binding sites. Numbers indicate the position relative to the transcriptional start site. The thickness of the arrows indicates the transcriptional levels of accR and accD1A1-hmgL-fadE4. Green triangles indicate methylcrotonyl-, propionyl- or acetyl-CoA.
Several TetR family repressors have been shown to play essential roles in controlling expression of ACCase in bacteria; examples include CcrR in Methylobacterium extorquens, FasR in Corynebacterium glutamicum, PccD in S. erythraea, and BkaR (Fad35R) in mycobacteria (19–21, 31). In this study, we demonstrated that the TetR family regulator AccR in S. avermitilis represses the expression of echA8, echA9, fadE2, and the accD1A1-hmgL-fadE4 operon, which are involved in the generation and assimilation of acetyl- and propionyl-CoAs. The diversity of C-terminal structures found in TetR family regulators allows them to interact with a wide variety of small molecules, including signaling molecules, metabolites, and antibiotics. Such interaction will cause a structural change in the C-terminal helixes and release the regulator from the operator, allowing the transcription of target genes (27, 32). Fatty acids and activated fatty acids (acyl-CoAs) have been identified as effector molecules for several TetR family regulators involved in regulation of acyl-CoA metabolism. Long-chain acyl-CoA (C14–20) acts as a ligand of YsiA (a functional homolog of E. coli FadR) and inhibits the binding of YsiA to promoter regions of genes involved in fatty acid degradation in Bacillus subtilis (33). Methylmalonic acid inhibits the interaction of the propionyl-CoA assimilation repressor PccD with the upstream regions of pccBC operon and pccA in S. erythraea (20). The present study shows that AccR in Streptomyces utilizes methylcrotonyl-, propionyl-, and acetyl-CoAs, the substrates of target genes, as ligands. Our findings expand the known ligand classes of TetR family regulators. AccR acts as a repressor in response to intracellular methylcrotonyl-, propionyl-, and acetyl-CoA levels and coordinates the gene expression of ACCase, which is responsible for methylcrotonyl-, propionyl-, and acetyl-CoA assimilation, in a substrate-dependent manner.
AccR is highly conserved in Streptomyces species, sharing 75 to 91% amino acid sequence identities and exhibiting high identities for the DNA binding domain (>97%) and the ligand binding domain (>78%) (Fig. S2A). AccR is an ortholog of BkaR from Mycobacterium (50% identity) that also plays a vital role in regulating catabolism of branched-chain amino acids (21, 22), although the genomic context of accR in Streptomyces is quite different from that of bkaR in Mycobacterium. In Mycobacterium spp., the neighboring genes fadD35, scoAB, accD1A1, fadE19, Rv2499c, citE, and bkdABC (fadD35 and scoAB are absent from some species), divergently oriented from bkaR, are also under the negative control of BkaR (21, 22). accD1A1 encodes an MCC which catalyzes the carboxylation of methylcrotonyl-CoA (26). In S. avermitilis, transcription of accD1A1 was strongly induced by acetate, propionate, or l-leucine (Fig. 1B), indicating that the ACCase encoded by accD1A1 has MCC, PCC, and ACC activities, which is consistent with the broad substrate specificity of actinobacterial ACCases. The 16-nt palindromic binding motif of AccR (GTTAA-N6-TTAAC) is similar to the motif (GTTANT-N4-ANTAAC) recognized by BkaR, consistent with the high N-terminal sequence identity of the two regulators (Fig. S2A and B). Two conserved motifs are also present in the intergenic regions of accR and bkaR, suggesting a similar regulatory mechanism of AccR (BkaR) in actinobacteria. S. avermitilis harbors the homologous genes fadD35, scoAB, Rv2499c, citE, and bkdABC, which are apparently not under the direct control of AccR, because neither conserved AccR motif was found in their promoter regions, and AccR did not bind to them (see Fig. S9 in the supplemental material). bkdABC expression is under the negative control of the AsnC family regulator BkdR in Streptomyces (34, 35). Thus, it appears that only accD1A1 and fadE19 (the homolog of fadE4 in S. avermitilis) are conserved genes of the AccR/BkaR operon in actinobacteria.
Malonyl- and methylmalonyl-CoAs are extender units for avermectin production. AccR modulates avermectin production by controlling intracellular pools of malonyl- and methylmalonyl-CoAs. The intracellular pools of acetyl-, propionyl-, malonyl-, and methylmalonyl-CoAs were increased in DaccR because of the derepression of target genes, and the enhanced supply of precursors led to avermectin overproduction. accR and target genes (accD1A1, echA, fadE, and hmgL) are highly conserved in Streptomyces species; we therefore expect that future studies will provide improved, effective strategies for increasing the yields of polyketides for which malonyl- and methylmalonyl-CoAs are precursors, through the genetic manipulation of accR in natural or recombinant Streptomyces strains.
MATERIALS AND METHODS
Strains and culture conditions.
Plasmids and strains of E. coli and S. avermitilis used in this study are listed in Table 1. E. coli was cultured in Luria-Bertani (LB) medium at 37°C. S. avermitilis was grown on solid YMS (yeast extract-malt extract-soluble starch) medium at 28°C for sporulation. Minimal medium (MM) supplemented with different carbon sources was used for phenotype observation of S. avermitilis. Modified YEME liquid medium and RM14 solid medium were used to culture mycelia for protoplast preparation and regeneration, respectively (36). Insoluble fermentation medium FM-I and soluble fermentation medium FM-II were used for avermectin production and biomass measurement, respectively (37). Apramycin at 100 μg ml−1 or kanamycin at 50 μg ml−1 was used to culture E. coli, and apramycin at 10 μg ml−1 was used to culture S. avermitilis.
TABLE 1.
Plasmids and strains used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| S. avermitilis | ||
| ATCC 31267 | Wild-type strain (WT) | Laboratory stock |
| accD1::pKC1139 | accD1 disruption strain by insertion of pKC1139 | This study |
| DaccR | accR deletion mutant of strain ATCC 31267 | This study |
| CaccR | DaccR with insertion of plasmid pSET-CaccR | This study |
| A8 | Industrial high-yield avermectin-producing strain | Qilu Pharmaceutical |
| A8/DaccR | accR deletion mutant of strain A8 | This study |
| WT(pKC-ermE-accD1A1) | accD1A1 overexpression strain based on WT | This study |
| DaccR(pKC-ermE-accD1A1) | accD1A1 overexpression strain based on DaccR | This study |
| E. coli | ||
| JM109 | Host for DNA cloning | Laboratory stock |
| BL21(DE3) | Host for protein overexpression | Laboratory stock |
| Plasmids | ||
| pKC1139 | Temperature-sensitive shuttle vector for E. coli-Streptomyces | 41 |
| pSET152 | Integrated shuttle vector in Streptomyces | 41 |
| pET-28a(+) | Expression vector for His6-tagged protein purification in E. coli | Novagen |
| pKC-DaccD1 | accD1 disruption plasmid based on pKC1139 | This study |
| pKC-DaccR | accR deletion plasmid based on pKC1139 | This study |
| pSET-CaccR | accR complementation vector based on pSET152 | This study |
| pET-AccR | accR overexpression vector based on pET-28a(+) | This study |
| pKC-ermE-accD1A1 | accD1A1 overexpression vector based on pKC1139 carrying promoter ermE*p | This study |
Construction of S. avermitilis derived strains.
For the disruption of accD1, a 594-bp internal fragment of accD1 was amplified with the primer pair accD1-Fw/accD1-Rev (Table 2). The fragment was cloned into pKC1139 to produce disruption plasmid pKC-DaccD1. Disruption plasmid was transformed into protoplasts of WT strain, and the disruption strain (accD1::pKC1139) was generated by single crossover. For the deletion of accR, a 534-bp upstream fragment (from positions −479 to +55 relative to the accR translation start codon) and a 541-bp downstream fragment (from positions +477 to +1017) were amplified by PCR using WT genomic DNA as the template and the primer pairs DaccR-up-Fw/DaccR-up-Rev and DaccR-down-Fw/DaccR-down-Rev, respectively. Fragments were digested with EcoRI/KpnI and KpnI/BamHI, respectively, and then ligated into EcoRI/BamHI-cleaved pKC1139 to construct the accR deletion vector (termed pKC-DaccR). The deletion vector was transformed into protoplasts of the WT strain and industrial strain A8, and the deletion strain (DaccR) was screened as described previously (38). Putative accR deletion strains were verified using the primer pairs DaccR-ex-Fw/DaccR-ex-Rev and DaccR-Int-Fw/DaccR-Int-Rev, which were flanking and located within the exchange region. For complementation, an 839-bp fragment, including the accR coding region and its promoter, was amplified from the WT genomic DNA with the primer pair DaccR-c-Fw/DaccR-c-Rev. The fragment was digested with EcoRI/BamHI and cloned into pSET152 to generate the complement vector pSET-CaccR, which was then transferred into protoplast of DaccR to obtain a complemented strain (termed CaccR). For the overexpression of accD1A1 in S. avermitilis, a 3,746-bp DNA fragment containing the full ORF of accD1A1 was amplified from WT genomic DNA with the primer pair EaccD1A1-Fw/EaccD1A1-Rev. The fragment was cloned into pKC1139 under the strong constitutive promoter ermE*p to generate the accD1A1 overexpression plasmid pKC-ermE-accD1A1. The resulting plasmid was transformed into WT and DaccR to obtain the WT(pKC-ermE-accD1A1) and DaccR(pKC-ermE-accD1A1) overexpression strains.
TABLE 2.
Primers used in this study
| Purpose | Primer | Sequence (5′–3′)a |
|---|---|---|
| Construction of accD1::pKC1139 | accD1-Fw | CCG GAATTC TCTACAACCAGGCACGGATGTC (EcoRI) |
| accD1-Rev | CCCAAGCTT GGTCGCACAGCTCGATGAAGT (HindIII) | |
| Construction and verification of DaccR | DaccR-up-Fw | CGGAATTC GCTTCTTCACGGTCATCGGGTA (EcoRI) |
| DaccR-up-Rev | GGGGTACC CAGCCTCCTTGAGGATCTGCTC (KpnI) | |
| DaccR-down-Fw | GGGGTACC CCTGCTGAACTCCACGCCACAC (KpnI) | |
| DaccR-down-Rev | CGGGATCCCGCTTCTTCTGCTCGTCGGT (BamHI) | |
| DaccR-ex-Fw | CGACATCCGTGCCTGGTTGTAG | |
| DaccR-ex-Rev | GCCACCCGTGATGAAGGTCTTG | |
| DaccR-Int-Fw | GGTCTGTACCGGCACTTC | |
| DaccR-Int-Rev | CACCACCTCCACCCACAG | |
| Complementation of DaccR mutant | DaccR-c-Fw | CGGAATTC CTCCGGTGCCTCTTGCATCCT (EcoRI) |
| DaccR-c-Rev | CGGGATCC CGCCTCGTGATCCTCGTTGAAC (BamHI) | |
| Construction of pKC-ermE-accD1A1 | EaccD1A1-Fw | GTGCCGGTTGGTAGGATCCAGCGGTGAGCATGCAAGAGGCACCGGAG |
| EaccD1A1-Rev | GACGTTGTAAAACGACGGCCAGTGCCAAGCTTGCTCATGCCGTCTCCTCCT | |
| Construction of His6-AccR | His6-AccR-Fw | CGGGATCC ATGGCCACGAGAACCGAC (BamHI) |
| His6-AccR-Rev | CGGAATTC ACAGTCGTCCAGGGCAACC (EcoRI) | |
| DNase I footprinting | accR-FAM-Fw | GTCAGCTCCGGTGCCTCTT |
| accR-Rev | TTGAGGATCTGCTCGCGG | |
| EMSAs | accR-accD1-Fw | GTCAGCTCCGGTGCCTCTT |
| accR-accD1-Rev | TTGAGGATCTGCTCGCGG | |
| accD2-Fw | CCTGGCGGTGGCTCGTCC | |
| accD2-Rev | TGCTCGGCGGTCAGTTGG | |
| accA3-SAV3336-Fw | AGACCTTGACCACATCGG | |
| accA3-SAV3336-Rev | ATGACTCCCTCTCCTTGAAAC | |
| accD3-Fw | CCGAACCTCTTCACCGAGTACC | |
| accD3-Rev | GTCAGCCATGCGTCAGTG | |
| accD4-birA-Fw | GCACCTCGGACGGCTTCAG | |
| accD4-birA-Rev | CGGCTCGGACATCGGGATG | |
| echA8-Fw | CGGCTACGGCGGCGAGTA | |
| echA8-Rev | GACGGTCGAGGCGGATGGT | |
| echA9-Fw | ATCCCGCCCGCCCGTCAG | |
| echA9-Rev | GTCCGCTCGCTCCGCCATG | |
| fadE2-chiC2-Fw | GCGGACTGAGCATCGTGTCTC | |
| fadE2-chiC2-Rev | GGCATGGCGACGCTCTCCT |
Underlining is used to indicate restriction enzyme sites.
Overexpression and purification of His6-AccR.
The accR coding region was amplified from WT genomic DNA using the primer pair His6-AccR-Fw/His6-AccR-Rev (Table 2) and inserted into pET-28a(+) to construct pET-AccR. pET-AccR was transformed into E. coli BL21(DE3) for overexpression of His6-AccR. The cells were cultured in LB medium with kanamycin at 50 μg ml−1 at 37°C until the optical density at 600 nm reached 0.6. His6-AccR was then induced by 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h. The cells were collected, resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole [pH 8.0]), sonicated on ice, and centrifuged. His6-AccR in the supernatant was purified by Ni2+-NTA agarose chromatography (Novagen) according to the manufacturer’s protocol.
Electrophoretic mobility shift assays.
EMSAs were performed using a DIG gel shift kit (2nd Generation; Roche) as described previously (39). DNA probes were amplified by PCR with primers (Table 2) and then labeled with digoxigenin (DIG) using terminal transferase. Reaction mixtures contained 0.15 nM DIG-labeled probe, 0.5 μl of poly(dI-dC) (2 μg μl−1), and various amounts of His6-AccR. For the ligand binding assays, His6-AccR and various amounts of acyl-CoAs were preincubated for 15 min and then added to the reaction mixtures.
RNA extraction and RT-qPCR analysis.
Total RNAs were extracted from mycelia grown in FM-I for various durations using TRIzol reagent (Tiangen; China) as described previously (39). To examine the induction of acc genes by acetate, propionate, or l-leucine, S. avermitilis cells were cultured in FM-I for 36 h. Then, 20 mM acetate, propionate, or l-leucine was added, and the sample was cultured for 24 h. The transcription levels of the tested genes were detected by RT-qPCR, using the hrdB transcription level as an internal control.
DNase I footprinting assays.
These assays were performed using a fluorescence labeling procedure. A 167-bp fragment containing the accR-accD1 intergenic region was amplified using the primers accR-FAM-Fw and accR-Rev (Table 2). A FAM-labeled DNA fragment and various amounts of His6-AccR were mixed, incubated at 25°C for 30 min, and digested by DNase I (0.016 U) at 37°C for 50 s. The reaction was terminated by the addition of EDTA (final concentration, 50 mM), and the reaction mixture was incubated for 10 min at 80°C. DNA samples were purified by chloroform extraction and ethanol precipitation and then sequenced by using a 3730XL DNA analyzer (Applied Biosystems).
HPLC analysis of avermectin.
Fermentation of S. avermitilis strains and high-pressure liquid chromatography (HPLC) analysis of avermectin were performed as described previously (37).
Detection of intracellular acyl-CoAs.
Extraction and HPLC analysis of acyl-CoAs were performed as described by Dayem et al. (40). In brief, S. avermitilis mycelia cultured in FM-I were collected at various times, frozen rapidly in liquid nitrogen, and ground to fine powder. Then, 0.1 g of the power was extracted with 135 μl of 10% trichloroacetic acid at 4°C, followed by centrifugation. Supernatants were analyzed by HPLC using a C18 reverse-phase column (5 μm; 4.6 × 250 mm) and a multiple-gradient elution procedure. HPLC buffer A contained 75 mM sodium acetate and 100 mM NaH2PO4 (pH 4.6), and buffer B contained 70% buffer A and 30% methanol. The column was equilibrated with 90% buffer A and 10% buffer B at a flow rate of 1 ml min−1. Samples (20 μl) were injected, and the percentage of buffer B was increased linearly to 40% over 35 min, increased linearly to 90% over 20 min, and then reduced linearly to 10% over 10 min. Acyl-CoAs were detected at a wavelength of 260 nm. Malonyl-, methylmalonyl-, acetyl-, and propionyl-CoAs (Sigma-Aldrich) were used as standards.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (grants 31470190 and 31861143004).
We are grateful to S. Anderson for English editing of the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Newman DJ, Cragg GM. 2016. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod 79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 2.Williams GJ. 2013. Engineering polyketide synthases and nonribosomal peptide synthetases. Curr Opin Struct Biol 23:603–612. doi: 10.1016/j.sbi.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hertweck C. 2009. The biosynthetic logic of polyketide diversity. Angew Chem Int Ed Engl 48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 4.Fischbach MA, Walsh CT. 2006. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem Rev 106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 5.Chan YA, Podevels AM, Kevany BM, Thomas MG. 2009. Biosynthesis of polyketide synthase extender units. Nat Prod Rep 26:90–114. doi: 10.1039/b801658p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Livieri AL, Navone L, Marcellin E, Gramajo H, Rodriguez E. 2019. A novel multidomain acyl-CoA carboxylase in Saccharopolyspora erythraea provides malonyl-CoA for de novo fatty acid biosynthesis. Sci Rep 9:6725. doi: 10.1038/s41598-019-43223-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demirev AV, Khanal A, Nguyen PKH, Nam KT, Nam DH. 2011. Biochemical characterization of propionyl-coenzyme A carboxylase complex of Streptomyces toxytricini. J Microbiol 49:407–412. doi: 10.1007/s12275-011-1122-1. [DOI] [PubMed] [Google Scholar]
- 8.Lin TW, Melgar MM, Kurth D, Swamidass SJ, Purdon J, Tseng T, Gago G, Baldi P, Gramajo H, Tsai SC. 2006. Structure-based inhibitor design of AccD5, an essential acyl-CoA carboxylase carboxyltransferase domain of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 103:3072–3077. doi: 10.1073/pnas.0510580103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arabolaza A, Shillito ME, Lin TW, Diacovich L, Melgar M, Pham H, Amick D, Gramajo H, Tsai SC. 2010. Crystal structures and mutational analyses of acyl-CoA carboxylase beta subunit of Streptomyces coelicolor. Biochemistry 49:7367–7376. doi: 10.1021/bi1005305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tong L. 2013. Structure and function of biotin-dependent carboxylases. Cell Mol Life Sci 70:863–891. doi: 10.1007/s00018-012-1096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh TJ, Daniel J, Kim HJ, Sirakova TD, Kolattukudy PE. 2006. Identification and characterization of Rv3281 as a novel subunit of a biotin-dependent acyl-CoA carboxylase in Mycobacterium tuberculosis H37Rv. J Biol Chem 281:3899–3908. doi: 10.1074/jbc.M511761200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodríguez E, Gramajo H. 1999. Genetic and biochemical characterization of the alpha and beta components of a propionyl-CoA carboxylase complex of Streptomyces coelicolor A3(2). Microbiology 145:3109–3119. doi: 10.1099/00221287-145-11-3109. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez E, Banchio C, Diacovich L, Bibb MJ, Gramajo H. 2001. Role of an essential acyl coenzyme A carboxylase in the primary and secondary metabolism of Streptomyces coelicolor A3(2). Appl Environ Microbiol 67:4166–4176. doi: 10.1128/aem.67.9.4166-4176.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diacovich L, Peiru S, Kurth D, Rodriguez E, Podesta F, Khosla C, Gramajo H. 2002. Kinetic and structural analysis of a new group of acyl-CoA carboxylases found in Streptomyces coelicolor A3(2). J Biol Chem 277:31228–31236. doi: 10.1074/jbc.M203263200. [DOI] [PubMed] [Google Scholar]
- 15.Zabala D, Brana AF, Florez AB, Salas JA, Mendez C. 2013. Engineering precursor metabolite pools for increasing production of antitumor mithramycins in Streptomyces argillaceus. Metab Eng 20:187–197. doi: 10.1016/j.ymben.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Ryu YG, Butler MJ, Chater KF, Lee KJ. 2006. Engineering of primary carbohydrate metabolism for increased production of actinorhodin in Streptomyces coelicolor. Appl Environ Microbiol 72:7132–7139. doi: 10.1128/AEM.01308-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung WS, Yoo YJ, Park JW, Park SR, Han AR, Ban YH, Kim EJ, Kim E, Yoon YJ. 2011. A combined approach of classical mutagenesis and rational metabolic engineering improves rapamycin biosynthesis and provides insights into methylmalonyl-CoA precursor supply pathway in Streptomyces hygroscopicus ATCC 29253. Appl Microbiol Biotechnol 91:1389–1397. doi: 10.1007/s00253-011-3348-6. [DOI] [PubMed] [Google Scholar]
- 18.Deng W, Li C, Xie J. 2013. The underling mechanism of bacterial TetR/AcrR family transcriptional repressors. Cell Signal 25:1608–1613. doi: 10.1016/j.cellsig.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Nickel J, Irzik K, van Ooyen J, Eggeling L. 2010. The TetR-type transcriptional regulator FasR of Corynebacterium glutamicum controls genes of lipid synthesis during growth on acetate. Mol Microbiol 78:253–265. doi: 10.1111/j.1365-2958.2010.07337.x. [DOI] [PubMed] [Google Scholar]
- 20.Xu Z, Wang M, Ye BC. 2017. TetR family transcriptional regulator PccD negatively controls propionyl coenzyme A assimilation in Saccharopolyspora erythraea. J Bacteriol 199:e00281-17. doi: 10.1128/JB.00281-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balhana RJ, Swanston SN, Coade S, Withers M, Sikder MH, Stoker NG, Kendall SL. 2013. bkaR is a TetR-type repressor that controls an operon associated with branched-chain keto-acid metabolism in mycobacteria. FEMS Microbiol Lett 345:132–140. doi: 10.1111/1574-6968.12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anand S, Singh V, Singh AK, Mittal M, Datt M, Subramani B, Kumaran S. 2012. Equilibrium binding and kinetic characterization of putative tetracycline repressor family transcription regulator Fad35R from Mycobacterium tuberculosis. FEBS J 279:3214–3228. doi: 10.1111/j.1742-4658.2012.08707.x. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda H, Omura S. 1997. Avermectin biosynthesis. Chem Rev 97:2591–2610. doi: 10.1021/cr960023p. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda H, Nonomiya T, Omura S. 2001. Organization of biosynthetic gene cluster for avermectin in Streptomyces avermitilis: analysis of enzymatic domains in four polyketide synthases. J Ind Microbiol Biotechnol 27:170–176. doi: 10.1038/sj.jim.7000092. [DOI] [PubMed] [Google Scholar]
- 25.You D, Wang MM, Ye BC. 2017. Acetyl-CoA synthetases of Saccharopolyspora erythraea are regulated by the nitrogen response regulator GlnR at both transcriptional and posttranslational levels. Mol Microbiol 103:845–859. doi: 10.1111/mmi.13595. [DOI] [PubMed] [Google Scholar]
- 26.Ehebauer MT, Zimmermann M, Jakobi AJ, Noens EE, Laubitz D, Cichocki B, Marrakchi H, Laneelle MA, Daffe M, Sachse C, Dziembowski A, Sauer U, Wilmanns M. 2015. Characterization of the mycobacterial acyl-CoA carboxylase holo complexes reveals their functional expansion into amino acid catabolism. PLoS Pathog 11:e1004623. doi: 10.1371/journal.ppat.1004623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuthbertson L, Nodwell JR. 2013. The TetR family of regulators. Microbiol Mol Biol Rev 77:440–475. doi: 10.1128/MMBR.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Cheng Y, Lyu M, Wen Y, Song Y, Chen Z, Li J. 2017. Redox-sensing regulator Rex regulates aerobic metabolism, morphological differentiation, and avermectin production in Streptomyces avermitilis. Sci Rep 7:44567. doi: 10.1038/srep44567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hiard S, Maree R, Colson S, Hoskisson PA, Titgemeyer F, van Wezel GP, Joris B, Wehenkel L, Rigali S. 2007. PREDetector: a new tool to identify regulatory elements in bacterial genomes. Biochem Biophys Res Commun 357:861–864. doi: 10.1016/j.bbrc.2007.03.180. [DOI] [PubMed] [Google Scholar]
- 31.Hu B, Lidstrom M. 2012. CcrR, a TetR family transcriptional regulator, activates the transcription of a gene of the ethylmalonyl coenzyme A pathway in Methylobacterium extorquens AM1. J Bacteriol 194:2802–2808. doi: 10.1128/JB.00061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Issa KHB, Phan G, Broutin I. 2018. Functional mechanism of the efflux pumps transcription regulators from Pseudomonas aeruginosa based on 3D structures. Front Mol Biosci 5:57. doi: 10.3389/fmolb.2018.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuoka H, Hirooka K, Fujita Y. 2007. Organization and function of the YsiA regulon of Bacillus subtilis involved in fatty acid degradation. J Biol Chem 282:5180–5194. doi: 10.1074/jbc.M606831200. [DOI] [PubMed] [Google Scholar]
- 34.Sprusansky O, Stirrett K, Skinner D, Denoya C, Westpheling J. 2005. The bkdR gene of Streptomyces coelicolor is required for morphogenesis and antibiotic production and encodes a transcriptional regulator of a branched-chain amino acid dehydrogenase complex. J Bacteriol 187:664–671. doi: 10.1128/JB.187.2.664-671.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skinner DD, Morgenstern MR, Fedechko RW, Denoya CD. 1995. Cloning and sequencing of a cluster of genes encoding branched-chain alpha-keto acid dehydrogenase from Streptomyces avermitilis and the production of a functional E1 [alpha beta] component in Escherichia coli. J Bacteriol 177:183–190. doi: 10.1128/jb.177.1.183-190.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macneil DJ, Klapko LM. 1987. Transformation of Streptomyces-avermitilis by plasmid DNA. J Ind Microbiol 2:209–218. doi: 10.1007/BF01569542. [DOI] [Google Scholar]
- 37.Jiang L, Liu Y, Wang P, Wen Y, Song Y, Chen Z, Li J. 2011. Inactivation of the extracytoplasmic function sigma factor Sig6 stimulates avermectin production in Streptomyces avermitilis. Biotechnol Lett 33:1955–1961. doi: 10.1007/s10529-011-0673-x. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Yan T, Jiang L, Wen Y, Song Y, Chen Z, Li J. 2013. Characterization of SAV7471, a TetR family transcriptional regulator involved in the regulation of coenzyme A metabolism in Streptomyces avermitilis. J Bacteriol 195:4365–4372. doi: 10.1128/JB.00716-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo J, Zhang X, Chen Z, Wen Y, Li J. 2014. Two adjacent and similar TetR family transcriptional regulator genes, SAV577 and SAV576, coregulate avermectin production in Streptomyces avermitilis. PLoS One 9:e99224. doi: 10.1371/journal.pone.0099224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dayem LC, Carney JR, Santi DV, Pfeifer BA, Khosla C, Kealey JT. 2002. Metabolic engineering of a methylmalonyl-CoA mutase-epimerase pathway for complex polyketide biosynthesis in Escherichia coli. Biochemistry 41:5193–5201. doi: 10.1021/bi015593k. [DOI] [PubMed] [Google Scholar]
- 41.Bierman M, Logan R, O’Brien K, Seno ET, Rao RN, Schoner BE. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.