Mycoplasma haemocanis is enzootic in Darwin’s foxes. There is a higher M. haemocanis genetic diversity and prevalence in foxes than in sympatric dogs, although haplotypes are shared between the two carnivore species. There is an apparent tolerance of Darwin’s foxes to Mycoplasma haemocanis.
KEYWORDS: Canidae , Lycalopex, Mollicutes, risk factors, South America
ABSTRACT
Mycoplasma haemocanis is prevalent in the endangered Darwin’s fox (Lycalopex fulvipes) in its main stronghold, Chiloé Island (Chile). The origin of the infection, its dynamics, its presence in other fox populations and the potential consequences for fox health remain unexplored. For 8 years, hemoplasmal DNA was screened and characterized in blood from 82 foxes in Chiloé and two other fox populations and in 250 free-ranging dogs from Chiloé. The prevalence of M. haemocanis in foxes was constant during the study years, and coinfection with “Candidatus Mycoplasma haematoparvum” was confirmed in 30% of the foxes. Both hemoplasma species were detected in the two mainland fox populations and in Chiloé dogs. M. haemocanis was significantly more prevalent and more genetically diverse in foxes than in dogs. Two of the seven M. haemocanis haplotypes identified were shared between these species. Network analyses did not show genetic structure by species (foxes versus dogs), geographic (island versus mainland populations), or temporal (years of study) factors. The probability of infection with M. haemocanis increased with fox age but was not associated with sex, season, or degree of anthropization of individual fox habitats. Some foxes recaptured years apart were infected with the same haplotype in both events, and no hematological alterations were associated with hemoplasma infection, suggesting tolerance to the infection. Altogether, our results indicate that M. haemocanis is enzootic in the Darwin’s fox and that intraspecific transmission is predominant. Nevertheless, such a prevalent pathogen in a threatened species represents a concern that must be considered in conservation actions.
IMPORTANCE Mycoplasma haemocanis is enzootic in Darwin’s foxes. There is a higher M. haemocanis genetic diversity and prevalence in foxes than in sympatric dogs, although haplotypes are shared between the two carnivore species. There is an apparent tolerance of Darwin’s foxes to Mycoplasma haemocanis.
INTRODUCTION
The Darwin’s fox (Lycalopex fulvipes) is an endangered carnivore native to Chile. Its distribution range includes three metapopulations: one on Chiloé Island, with an estimated population of 412 mature individuals, and two isolated mainland populations (Nahuelbuta Park and Valdivian Coastal Range) with about 227 mature individuals in total (1). The presence of dogs in and around parks comprises one of the major threats to the conservation of this carnivore (2). The presence of free-ranging dogs in the entire Darwin’s fox distribution range has been documented (3), and the populations in the continent are completely surrounded by human-dominated lands. Rural dogs in Chile are usually allowed to range freely (4, 5) and often lack any kind of prophylactic treatment or veterinary care (6). The naive behavior of the Darwin’s fox can predispose it to interactions with dogs, ending in physical attacks and potential pathogen transmission (7).
Hemotropic mycoplasmas (also called hemoplasmas) are small bacteria that attach to the surface of red blood cells of mammals (8). These species are widely distributed and can infect humans (9), domestic animals (including dogs and cats) (10), and wildlife (including wild carnivores) (11, 12). Three species of mycoplasmas have been described in canids: Mycoplasma haemocanis, “Candidatus Mycoplasma haematoparvum,” and “Candidatus Mycoplasma turicensis” (10, 13–15). Hemoplasma infection in dogs can cause acute and chronic hemolytic anemia, with results ranging from asymptomatic infection or slight lethargy to death; M. haemocanis is the most pathogenic species (16). Thus far, no clinical signs have been reported in hemoplasma-infected wild canids, and the pathological and epidemiological relevance for wildlife remains unknown. The transmission route of hemoplasmas is still under debate. Some species infecting dogs and cats are believed to be vector borne, but direct and/or vertical transmission has been demonstrated for others (17, 18). M. haemocanis has recently been proved to be transmitted vertically (19) in a dog, although it was classically considered to be transmitted by the brown dog tick, Rhipicephalus sanguineus (20). Nevertheless, since there are no canine ticks on Chiloé Island, other ways of transmission (i.e., direct transmission) must be operating.
During a molecular disease survey, Cabello et al. (21) reported an unexpected high prevalence of hemoplasma DNA among 30 free-living Darwin’s foxes captured in Chiloé in the period from 2009 to 2012. Sequencing showed that 80% of the sequences obtained corresponded to M. haemocanis, another to M. haemofelis, and one to an as-yet-uncharacterized Mycoplasma sp., which was later found to be shared with domestic cats and the wild cat guigna (Leopardus guigna) in Chile (22). All the foxes studied by Cabello et al. (21) were apparently healthy and were negative for almost every other of the nine vector-borne pathogen groups for which these animals were tested. Pathogen transmission is hindered in small populations of solitary species such as the Darwin’s fox because of their low rate of interspecific contact (23). Therefore, the high prevalence of infection reported by Cabello et al. (21) would support the hypothesis that hemoplasmas more likely persist in foxes based on an interspecific (i.e., dog-to-fox) rather than an intraspecific (fox-to-fox) transmission pathway. Considering that poorly managed free-ranging dogs are abundant in rural parts of Chile (24), including those areas sustaining Darwin’s fox populations, a role of the domestic dog as a reservoir and source of infection for the fox is a likely scenario.
Different epidemiological questions regarding hemoplasma infection in the Darwin’s fox remain. Whether Darwin’s foxes in Chiloé and mainland populations are reservoir or spillover hosts for M. haemocanis is still unknown. It has been observed in wild felids that the domestic cat would act as a source host of hemoplasma (25, 26). However, proving a role as reservoir for a pathogen is complex. In order to disentangle the origin of hemoplasma infection in the Darwin’s fox, we took different approaches. If these bacteria are being transmitted from dogs to foxes, we predicted that shared sequences would be found in both hosts, with greater hemoplasma genetic diversity in dogs. We also expected a higher prevalence of infection in foxes living closer to human settlements. We also aimed to provide some insights into risk factors and effects of hemoplasma infection in the Darwin’s fox and to determine whether the pathogen is present in other Darwin’s fox populations. The ultimate goal of our study was to determine if hemoplasma infection should be considered a disease threat for the last Darwin’s foxes.
RESULTS
Molecular detection and characterization.
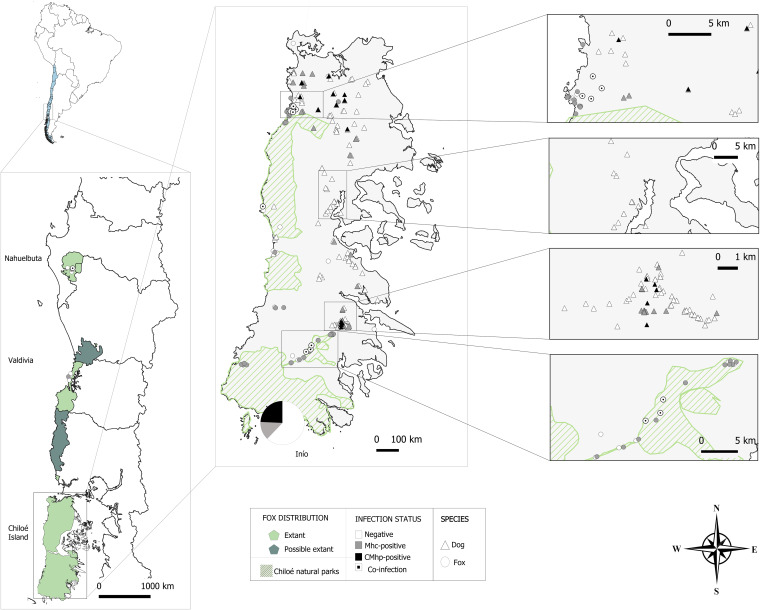
The overall observed prevalence of Mycoplasma DNA for the whole study period was 56.6% (95% confidence interval [CI] = 46.0% to 67.3%; 47 foxes positive). All the newly obtained 16S rRNA sequences showed between 99.7 and 100% identity with published sequences of M. haemocanis/M. haemofelis (Table 1). Sequencing of a portion of the RNase P gene (accession no. CP003199) indicated 100% identity with M. haemocanis sequences in all cases. Species-specific primers confirmed coinfection with “Ca. Mycoplasma haematoparvum” in nine of the 30 M. haemocanis-positive foxes for the period from 2013 to 2017 (30.0%, 95% CI = 13.6% to 46.4%). In the foxes from the mainland, M. haemocanis DNA was found in two individuals from Nahuelbuta, one of which was coinfected with “Ca. Mycoplasma haematoparvum,” and in one from Valdivia (Fig. 1). Six of the recaptured foxes were positive for M. haemocanis (two of which were coinfected with “Ca. Mycoplasma haematoparvum”), and one was negative at both capture events. Another fox, which was positive for M. haemocanis at both capture events, did not present coinfection with “Ca. Mycoplasma haematoparvum” at the first sampling but 2 years later was coinfected with this species (Table 2).
TABLE 1.
Nucleotide sequence types detected in hemoplasmas in blood samples from Darwin’s foxes and rural sympatric dogs and their closest GenBank sequences
| ntST | Animal(s) in which detected (n) | Closest sequence | Species | Identity (%) | Host | Country |
|---|---|---|---|---|---|---|
| 1 | Dog from Chiloé (10), fox from Chiloé (17), fox from Nahuelbuta (2) | EF416566 | Mycoplasma haemocanis | 100 | Canis lupus familiaris | Switzerland |
| 2 | Fox from Chiloé (1) | AF197337 | Mycoplasma haemocanis | 99.92 | Canis lupus familiaris | USA |
| 3 | Fox from Chiloé (1) | GQ129116 | Mycoplasma haemocanis | 99.92 | Canis lupus familiaris | Italy |
| 4 | Fox from Chiloé (1), dog from Chiloé (1) | GQ129116 | Mycoplasma haemocanis | 99.84 | Canis lupus familiaris | Italy |
| 5 | Fox from Chiloé (2) | DQ825458 | Mycoplasma haemofelis | 99.92 | Lynx lynx | Switzerland |
| 6 | Fox from Chiloé (1) | MK064162 | Mycoplasma haemocanis | 99.77 | Pulex irritans on Lycalopex culpaeus | Argentina |
| 7 | Dog from Chiloé (1) | KY117659 | Mycoplasma haemocanis | 99.73 | Canis lupus familiaris | Chile |
FIG 1.
Map of the study areas, showing hemoplasma infection status in the surveyed Darwin’s foxes and rural dogs. Dogs from Inio, a small fishing village in the south of Chiloé, are shown pooled in a pie chart.
TABLE 2.
Hemotropic mycoplasma infection status in Darwin’s foxes recaptured during the study period and Mycoplasma haemocanis nucleotide sequence type detected in each capture event
| Fox | Yr | Site | Season | Age | M. haemocanis status | M. haemocanis ntST | “Candidatus M. haematoparvum” status |
|---|---|---|---|---|---|---|---|
| 1 | 2014 | Chiloé | Spring | Adult | + | 4 | − |
| 2016 | Chiloé | Winter | Adult | + | 1 | + | |
| 2 | 2013 | Nahuelbuta | Spring | Adult | − | − | − |
| 2016 | Nahuelbuta | Summer | Adult | − | − | − | |
| 3 | 2014 | Chiloé | Spring | Juvenile | + | 1 | − |
| 2016 | Chiloé | Winter | Adult | + | 1 | − | |
| 4 | 2015 | Chiloé | Fall | Adult | + | 1 | + |
| 2017 | Chiloé | Fall | Adult | + | 1 | + | |
| 5 | 2016 | Chiloé | Fall | Adult | + | 3 | − |
| 2017 | Chiloé | Fall | Adult | + | 1 | − | |
| 6 | 2014 | Chiloé | Winter | Adult | + | 1 | − |
| 2015 | Chiloé | Fall | Adult | + | 1 | − | |
| 7 | 2013 | Chiloé | Fall | Adult | + | 1 | + |
| 2014 | Chiloé | Spring | Adult | + | 1 | + | |
| 8 | 2014 | Chiloé | Winter | Adult | + | 1 | − |
| 2015 | Chiloé | Fall | Adult | + | 1 | − |
Sixty dogs were positive for Mycoplasma sp. DNA (24.0%; 95% CI = 18.7% to 29.3%) across Chiloé Island (Fig. 1). Of the fifty-three readable sequences, 53.2% corresponded to M. haemocanis/M. haemofelis and 46.8% to “Ca. Mycoplasma haematoparvum.” M. haemocanis prevalence in foxes was significantly higher than in dogs (χ2 = 15.908, P < 0.001), whereas “Ca. Mycoplasma haematoparvum” prevalence was not significantly different between species (χ2 = 2.641, P > 0.1). We did not analyze dogs for potential M. haemocanis/“Ca. Mycoplasma haematoparvum” coinfections, but even if all dogs positive for “Ca. Mycoplasma haematoparvum” were coinfected with M. haemocanis, prevalence of M. haemocanis in foxes would still be significantly higher than in dogs (χ2 = 28.9, P < 0.001).
Only one of the foxes surveyed in the period from 2009 to 2017 period was parasitized by a tick (a larvae of Ixodes sigelos), a typical species of pudu (Pudu puda), an endemic deer from Chile, and none hosted fleas. No ticks were recovered from dogs, and 57 individuals (25.2%; 95% CI = 19.6% to 30.8%) hosted fleas.
Genetic analysis.
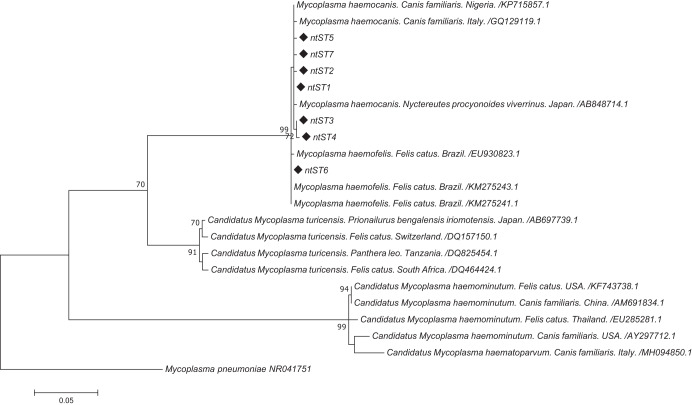
Sequencing of nearly 900 bp the 16S rRNA gene from 25 foxes and 12 dogs revealed the presence of seven different nucleotide sequence types (ntST), with 99.9% identity among them and between 99.7% and 99.9% identity with other M. haemocanis sequences (Fig. 2). Two of the seven ntST were shared between foxes and dogs (Table 1). ntST-1 was the most frequent; it was detected in 29 individuals (19 foxes and 10 dogs). The other ntST shared between species (ntST-4) was detected in one dog and one fox. ntST-5 was detected in two foxes, whereas the other four ntST were found in only a single individual, either a fox (n = 3) or a dog (n = 1). Five of the recaptured foxes presented ntST-1 in both capture events, while the other two presented a different ntST (Table 2). All these sequences were placed in the M. haemocanis/M. haemofelis clade in the phylogenetic tree (Fig. 2).
FIG 2.
Maximum-likelihood tree of the 16S rRNA gene (893 bp) of Mycoplasma haemocanis for Darwin’s foxes and domestic dogs. A Mycoplasma pneumoniae sequence was used as an outgroup. Bootstrap values of ≥70 are given at the nodes of the tree. Diamonds mark the nucleotide sequence types (ntST) from our study.
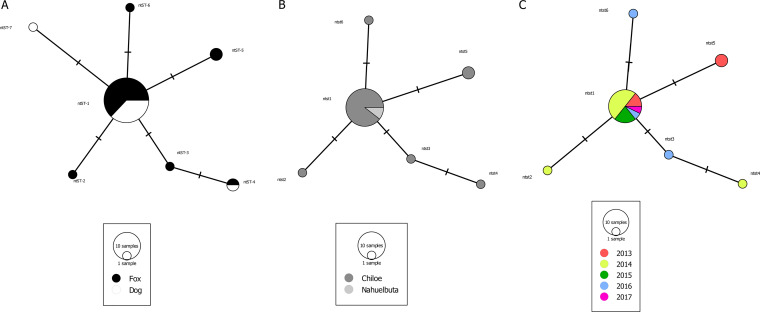
The haplotype diversity (Hd) in foxes was 0.427 (standard deviation [SD] = 0.122), the nucleotide diversity (Pi) was 0.00116 (SD = 0.00040), and the average number of nucleotide differences (k) was 0.547. In dogs, Hd was 0.345 (SD = 0.172), Pi was 0.00108 (SD = 0.00057), and k was 0.727. The network analysis showed no genetic structure between dogs and foxes (FST = 0.02919, P > 0.05; nearest-neighbor statistic [Snn] = 0.51243, P > 0.05) (Fig. 3) and no geographic structure between the island and mainland populations (FST = 0.030079, P > 0.5; Snn = 0.83647, P > 0.5) (Fig. 3). The most prevalent ntST was present through the entire sampling period, and in concordance, no genetic structure was detected among years (FST = 0.03809, P > 0.05; Snn = 0.0790, P > 0.05) (Fig. 3).
FIG 3.
Median joining network of the 16S gene (893 bp) of Mycoplasma haemocanis in rural dogs and Darwin’s foxes. Each circle in the network corresponds to a different nucleotide sequence type (ntST), the sizes of the circles correspond to ntST frequencies, the colors of the circles correspond to the two host species (Darwin’s foxes and rural dogs) (A), two geographic sampling sites (Chiloé Island and Nahuelbuta) (B), and years of sampling (C). The networks in panels B and C were performed with fox samples only.
Risk factor analysis.
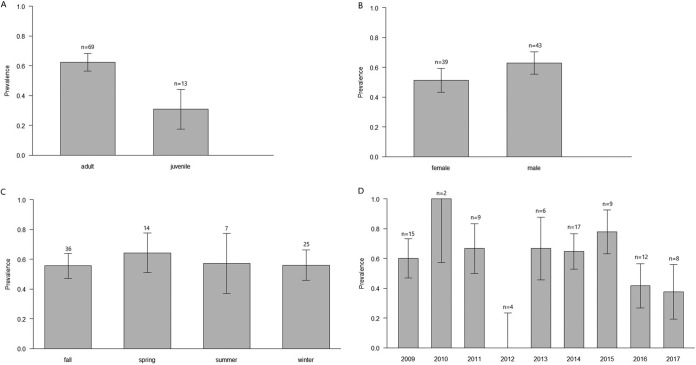
Models indicated that prevalence of infection of M. haemocanis was significantly higher in adult foxes than in juveniles (62.3% versus 20.0%; Z value = −2.247, P < 0.05) (Table 3). No other risk factor was related to the probability of M. haemocanis infection (Fig. 4). Prevalence of M. haemocanis-“Ca. Mycoplasma haematoparvum” coinfections did not differ depending on fox age (χ2 = 0.099, P > 0.5).
TABLE 3.
Best model representing multivariate relationships between predictor variables and detection of Mycoplasma haemocanis in Darwin’s foxes, using logistic regression analysis
| Variable | Estimate ± SE | Z valuea | AIC | Deviance | df | Hosmer-Lemeshow test P value |
|---|---|---|---|---|---|---|
| Intercept | 0.32 ± 0.58 | 0.56 | 76.134 | 68.134 | 59 | |
| Age juvenile | −2.07 ± 0.92 | −2.24* | 0.8 | |||
| No. of houses | −0.17 ± 0.07 | −2.30* | ||||
| Distance to nearest house | 0.02 ± 0.02 | 1.41 |
*, P = 0.01.
FIG 4.
Prevalence of Mycoplasma haemocanis depending on different intrinsic and extrinsic variables in Darwin’s foxes. (A) Age groups; (B) sex groups; (C) seasons; (D) study years.
Prevalence was also higher in adult dogs than in juveniles (χ2 = 63.2, P < 0.01), and no sex-related differences were found (χ2 = 1.7, P > 0.05).
Hematological analysis.
No significant differences were found in the hematological and biochemical variables evaluated depending on the sex or age of the animals, so the samples were pooled for further comparisons. None of the studied variables differed between the infection pattern groups that were compared.
DISCUSSION
Our survey revealed that M. haemocanis was present in the Darwin’s fox samples through the entire study period, indicating a constant exposure of the species to M. haemocanis. Moreover, the prevalence was higher in the endangered Darwin’s fox than in dogs. Although the observed prevalence in dogs is in the range of that found in a previous study in Chile (15) and even higher than the rates of infection detected elsewhere (20, 27), the higher prevalence in foxes may be explained by a greater risk of exposure and/or different susceptibility to the infection.
The sharing of the most frequent ntST of the 16S rRNA gene of M. haemocanis in dogs and foxes suggests cross-infection between these species. Whether dogs are the main source of infection for Darwin’s foxes cannot be proved with our data. However, considering that dogs markedly outnumber foxes and that dogs are moved large distances by their owners (24), it could be assumed that dogs were the origin of M. haemocanis infection for the fox. This is also supported by the fact that ntST-1 was found in foxes from both Chiloé Island and Nahuelbuta, which can be explained only by the movement of dogs between the continent and Chiloé. However, since this pathogen was introduced in the population, fox-to-fox transmission seems to be frequent now. This is supported by the higher prevalence in foxes than in dogs, the higher haplotype diversity detected in foxes, and the fact that the most prevalent ntST was found in foxes during all the studied years. This last observation would be better explained by intraspecific transmission than by periodic spillovers from dogs. Persistent intraspecific transmission of hemoplasmas following spillover from domestic animals has been also observed in felids (22, 26).
The higher prevalence observed in adult foxes compared to juveniles coincides with studies of free-ranging dogs (9) and cats (28) and with the data of this study in rural dogs of Chiloé. This indicates that both foxes and dogs face an increasing possibility of exposure to the pathogen with age. This, together with the apparent lack of seasonality and interannual differences in the prevalence indicates that M. haemocanis infection behaves enzootically in the Darwin’s fox. The data from the recaptured individuals further confirmed the widespread nature of the infection in foxes and that foxes can be infected more than once, as proven by the finding of foxes reinfected by a different haplotype. Host sex does not appear to be a risk factor for infection with M. haemocanis in foxes, which concurs with previous studies in domestic dogs (27, 29).
The observed prevalence of “Ca. Mycoplasma haematoparvum” in dogs in our study is in the range of that reported in Argentina (30) and higher than that reported in the above-metioned city dogs in Chile (15). Unfortunately, we were unable to characterize “Ca. Mycoplasma haematoparvum” further to confirm whether this species is also shared by both hosts.
The lack of effect of landscape features on hemoplasma prevalence in the Darwin’s fox is in disagreement with a previous study on feline hemoplasmas in a threatened sympatric felid, the guigna (22). Our results likely reflect a heterogeneity of the risk of transmission of a multihost pathogen that may be using more than a single method of transmission (horizontal, vertical, and/or vector borne) and/or more than a single arthropod vector.
Infections with hemoplasmas appear to occur as chronic conditions in domestic species (27, 29). This seems to be the case in the Darwin’s fox, as indicated by the high prevalence observed in the absence of clinical signs and the high proportion of foxes that were found to be infected by the same ntST in both capture events, which had up to 3 years of difference between them (although the possibility of periodic reinfections with the same ntST cannot be ruled out). The chronicity could explain the absence of hematological alterations associated with the presence of the pathogen. The possible tolerance of the Darwin’s fox to M. haemocanis is supported by the genetic variability of the pathogen in the fox population, with up to six different ntST detected in 25 individuals, which according to Kutzer and Armitage (31) may reflect some degree of tolerance to the infection. Kutzer and Armitage (31) also proposed that tolerance to a pathogen will confer a fitness advantage to the host, which could fluctuate by pathogen load and intrinsic factors of the host, reasons that may explain why we did not detect any risk factor associated with the infection other than age. Nevertheless, it is worth noting that when the apparent tolerance is lost, subclinical infections can lead to a reduction in host fitness, reproduction, survival, or dispersal, any of which can be detrimental for the population of an endangered species such as the Darwin’s fox (32).
Previous studies identified coinfection with other agents as a risk factor for becoming infected with hemoplasmas (27, 33), and it is known that the presence of other agents can aggravate the pathogenicity of a primary agent. For example, distemper outbreaks in African lions were exacerbated by concomitant Babesia spp. (34). We did not investigate coinfections with other vector-borne pathogens because our preliminary survey indicated absence of all the other main canine vector-borne pathogens (21). Moreover, a recent study showed that the same foxes studied here were not exposed to canine distemper virus (CDV) (35). Therefore, it appears that coinfection is not an important driver of hemoplasma infection for this species. Rynkiewicz et al. (36) proposed that a host that has a tolerance response to a pathogen will have higher fitness due to the avoidance of the energetic cost of clearing an infection, which seems to be the case for the fox according to the hematological analyses. Nevertheless, asymptomatic infection could revert, triggered by other concomitant infectious or noninfectious causes.
Despite the small sample size, we confirmed the presence of both canine hemotropic mycoplasmas in foxes from both mainland metapopulations. This could be relevant during health assessment protocols before potential fox translocations (37). In these two mainland populations, two other sympatric anthropophilic and more-abundant fox species, the Andean fox (Lycalopex culpaeus) and the gray fox (L. griseus), could add a potential bridge between dogs and Darwin’s foxes, increasing the complexity of interspecific transmission of this multihost pathogen.
Infectious diseases can pose a threat to the survival of endangered species, especially those such as the Darwin’s fox, which are losing their natural habitat and encountering dogs more frequently, increasing the opportunities for pathogen transmission (23, 38). Therefore, information about the health of their populations and the evaluation of risk factors for infection are imperative.
Conclusion.
We showed that the Darwin’s fox can sustain a constant prevalence of infection with a relatively high genetic diversity of M. haemocanis and with apparent lack of associated pathology. Nevertheless, although hemotropic mycoplasmas may seem asymptomatic, the constant circulation of this disease agent in the reduced Darwin’s fox population is a concern. The apparent tolerance of foxes to hemoplasma infection could be disrupted due to other factors that could be favored by poorly managed sympatric dogs. This disease risk is enhanced by the rapid habitat loss and degradation that the Darwin’s fox is currently suffering (39).
MATERIALS AND METHODS
Study areas and field techniques.
The samples included in this study were obtained between 2009 and 2017. The samples from 2009 to 2012 corresponded to the 30 fox blood samples from Chiloé Island included in the previous survey by Cabello et al. (21). From 2013 to 2017, we collected 52 additional fox samples in the three populations of Darwin’s fox: the Nahuelbuta area (n = 5) (37°45′S, 73°00′W), comprising Nahuelbuta National Park and surrounding native forests; the Valdivian Coastal Range (n = 5) (40°07′S, 73°33′W); and Chiloé Island (n = 42) (42°S, 74°W) (Fig. 1). Throughout the years of our study, seven foxes in Chiloé and one in Nahuelbuta were recaptured once. One individual was juvenile at the initial sampling and adult when recaptured, while all the others were adults in both capture events.
Foxes were captured using Tomahawk traps baited with chicken or canned fish. Traps were activated at dusk and checked the next morning at dawn. Foxes were anesthetized with a combination of 1 mg/kg xylazine (xylazine 2%; Centrovet, Chile) plus 10 mg/kg ketamine (Ketostop; DragPharma, Chile) or with 0.04 mg/kg dexmedetomidine (Dexdormitor; Zoetis, Chile) plus 5 mg/kg ketamine. The latter was reversed with 0.4 mg/kg atipamezole (Antisedan; Zoetis, Chile). A veterinarian performed an external clinical evaluation of the anesthetized animals. Blood was obtained from the cephalic, saphenous, or jugular vein. Foxes were classified as juveniles (less than 1 year) or adults (older than 1 year) based on tooth eruption.
Between the years 2015 and 2018, 250 rural dogs were sampled across Chiloé (Fig. 1). Blood was collected individually after the consent of the owner. Whole blood from dogs and foxes was placed in EDTA tubes. When possible, whole blood was sent by courier to our laboratory for hematological analyses. Otherwise, it was stored at –20°C. For serum biochemistry analysis, sera of foxes were extracted and stored at –20°C when possible. Dogs were classified as juveniles (less than 1 year) or adults (older than 1 year) based on tooth eruption.
All captures were made with the permission of the Servicio Agricola y Ganadero of the Chilean Government under permits 1262/2009, 2263/2010, 206/2012, 3155/2013, 1492/2014, 3363/2015, 3035/2016, 2288/2016, and 5029/2017. The study was approved by the authorities on bioethics of Universidad Andres Bello (permit no. 08/2016).
Molecular detection and characterization.
DNA was isolated from the 302 fox and dog blood samples using a DNeasy blood and tissue kit (Qiagen) according to the manufacturer’s instructions. All the newly obtained samples from foxes and dogs were initially screened for Mycoplasma spp. by a conventional PCR targeting a 391-bp fragment of the 16S rRNA gene (40). Amplicons were selected and assigned to a nucleotide sequence type (ntST). Since M. haemocanis and M. haemofelis are undistinguishable based on the characterization of the 16S rRNA gene alone, we confirmed that all positive samples corresponded to M. haemocanis by a conventional PCR targeting a 175-bp fragment of the RNase P gene. All these PCR protocols were as described by Millán et al. (40). The complete 16S rRNA gene (∼1,400 bp) was then characterized in all the M. haemocanis-positive foxes and in two positive dogs per hemoplasma ntST (Table 4). In order to detect if foxes had so-far-undetected coinfections with “Ca. Mycoplasma haematoparvum,” all the sequences corresponding to M. haemocanis/M. haemofelis from 2013 to 2017 were screened through a “Ca. Mycoplasma haematoparvum”-specific protocol targeting a 112-bp fragment of the 16S rRNA gene as described by Martínez-Díaz et al. (41). Positive controls were obtained from clinical samples of M. haemocanis and “Ca. Mycoplasma haematoparvum” from previously sequenced dog blood samples, and ultrapure water was used as a negative PCR control. Two percent agarose gel electrophoresis was performed, and PCR products were visualized under an UV transilluminator. All positive samples were sequenced by Macrogen, and the sequences obtained were compared with sequences deposited in the GenBank database (NCBI).
TABLE 4.
Genes targeted and primers used for PCR screening and characterization of hemotropic mycoplasmas in Darwin’s foxes and rural dogs in Chile
| Target | Primer sequences | Primer names | Fragment length (bp) | Purpose | Reference |
|---|---|---|---|---|---|
| Mycoplasma sp. 16S rRNA | 5′-ATGTTGCTTAATTCGATAATACACGAAA-3′ (forward), 5′-ACRGGATTACTAGTGATTCCAACTTCAA-3′ (reverse) | Mycop16S rRNA-F, Mycop16S rRNA-R | 384 | Screening | 40 |
| Mycoplasma sp. 16S rRNA (seminested) | Characterization | This study | |||
| Template | 5′-AGAGTTTGATCCTGGCTCAG-3′ (forward), 5′-TACCTTGTTACGACTTAACT-3′ (reverse) | HemoF1, HemoR2 | 1,428 | ||
| First | 5′-ATATTCCTACGGGAAGCAGC-3′ (forward), 5′-TACCTTGTTACGACTTAACT-3′ (reverse) | HemoF2, HemoR2 | 1,107 | ||
| Second | 5′-GCCCATATTCCTACGGGAAGCAGCAGT-3′ (forward), 5′-GTTTGACGGGCGGTGTGTACAAGACC-3′ (reverse) | HemMycop16S-322s, HemMycop16S-1420as | 1,029 | ||
| Third | 5′-GYATGCMTAAYACATGCAAGTCGARCG-3′ (forward), 5′-CTCCACCACTTGTTCAGGTCCCCGTC-3′ (reverse) | HemMycop16S-41s, HemMyco16S-938as | 870 | ||
| M. haemocanis/M. haemofelis RNase P | 5′-CCTGCGATGGTCGTAATGTTG-3′ (forward), 5′-GAGGRGTTTACCGCGTTTCAC-3′ (reverse) | RNAseP-F, RNAseP-R | 175 | Characterization | 40 |
| “Ca. Mycoplasma haematoparvum” 16S rRNA | 5′-GGAATCACTAGTAATCCYGTGTCAGCTATAT-3′ (forward), 5′-AATTAAATACGGTTTCAACTAGTACGTTTCTTT-3′ (reverse) | Mycoplasma Species-F, Candidatus Mycoplasma haematoparvum-R | 112 | Characterization | 41 |
The sequences of the 16S rRNA genes obtained from foxes and dogs were aligned using ClustalW executed in Geneious Prime 2019.2.1 (Biomatters Ltd.). To determine genetic relationships between the sequences from wild and domestic canids, we constructed a maximum-likelihood phylogenetic tree using MEGA 7.0.26 (42) and median joining networks using PopART (43). A network containing the sequences from dogs and Darwin’s foxes from this study was used to infer genetic relationships among hemoplasma species. A second network was developed to infer relationships according to the sampling site (Chiloé versus Nahuelbuta only, since the sequence from Valdivia was not readable) using only the fox sequences. Finally, a network considering the year of fox sampling was used to infer the dynamics of infection in the Darwin’s fox. The genetic structure was estimated using the pairwise PhiST test implemented in Arlequin (44) (level of significance assessed with 1,000 permutations) and the nearest-neighbor statistic Snn (45) executed in DnaSP.5 (46). An analysis of the nucleotide polymorphisms of 16S rRNA sequences obtained was performed using the software DnaSP.5 (46) in order to determine the genetic differentiation of hemoplasmas among hosts.
Hematology and serum chemistry analyses.
The harsh climatic conditions in southern Chile and distant locations of many study sites prevented the proper preservation of some of the blood and serum samples until laboratory analyses. Therefore, hematological and biochemical variables were obtained for 31 of the foxes during the study period. Seven hematological variables (hematocrit, red blood cell, platelet and total leukocyte counts, hemoglobin concentration, mean corpuscular volume, and mean corpuscular hemoglobin concentration) were calculated using a HumaCount 80 cell counter (Human GmbH, Germany). Relative leukocyte differentiation was estimated by microscopic observation after Diff-Quick staining. Fourteen biochemical variables were analyzed using a BA400 Analyzer (BioSystem SA, Barcelona, Spain). Measurement units and variables included are listed in Table 5.
TABLE 5.
Hematological and serum chemistry values in Darwin’s foxes from Chiloé Island
| Variable (unit)a | Sample size | Median value | SD | Range |
|---|---|---|---|---|
| WBC (μl) | 31 | 15,850 | 5,020 | 5,960–29,600 |
| RBC (μl) | 31 | 5,870,000 | 957,455 | 3,310,000–7,880,000 |
| HB (g/liter) | 31 | 13.9 | 2.14 | 7.3–18.7 |
| HTO (%) | 31 | 42.3 | 7 | 23.9–59.0 |
| MCV (fl) | 25 | 72 | 23.31 | 16.4–90.0 |
| MCH (fl) | 31 | 23.6 | 0.97 | 21.8–26.3 |
| MCHC (g/liter) | 31 | 32.3 | 1.7 | 27–35.5 |
| PLT (μl) | 31 | 304,000 | 109,250 | 141,000–580,000 |
| N (μl) | 25 | 12,648 | 3,845 | 4,410–18,480 |
| L (μl) | 30 | 1,738 | 1,717 | 752–9,400 |
| M (μl) | 30 | 510 | 1,167 | 138–6,150 |
| E (μl) | 26 | 150 | 681 | 0–3,071 |
| Ca (mmol/liter) | 26 | 2.46 | 0.39 | 1.35–2.94 |
| P (mmol/liter) | 28 | 1.95 | 3.50 | 0.1–11.3 |
| BUN (mmol/liter) | 28 | 9.11 | 5.02 | 4.82–27.4 |
| Crea (mmol/liter) | 28 | 0.07 | 0.3 | 0.04–0.18 |
| Bil (μmol/liter) | 21 | 3.93 | 2.57 | 1.71–13.68 |
| Gluc (mmol/liter) | 28 | 2.6 | 1.69 | 0.06–6.16 |
| Chol (mmol/liter) | 28 | 5 | 1.11 | 2.92–8.02 |
| ALP (IU/liter) | 22 | 40 | 76.59 | 3–227 |
| ALT (IU/liter) | 26 | 77 | 33.40 | 30–152 |
| AST (IU/liter) | 26 | 83 | 53.46 | 21–225 |
| GGT (IU/liter) | 24 | 2 | 4 | 1–21 |
| Prot (g/liter) | 25 | 76 | 11 | 45–96 |
| Glob (g/liter) | 23 | 40 | 73 | 32–58 |
| Alb (g/liter) | 28 | 30 | 66 | 17.2–47 |
WBC, white blood cells, RBC, red blood cells; HB, hemoglobin; HTO, hematocrit; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; PLT, platelet count; N, neutrophils; L, lymphocytes; M, monocytes; E, eosinophils; Ca, calcium; P, phosphorus; BUN, blood urea nitrogen; Crea, creatinine; Bil, bilirubin; Gluc, glucose; Chol, cholesterol; ALP, alkakine phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; GGT, gamma-glutamyl transpeptidase; Prot, total proteins; Glob, globulins; Alb, albumin.
Data analysis.
M. haemocanis presence/absence was binary coded and examined with a set of models of possible intrinsic and extrinsic variables affecting animal exposure to hemoplasmas. We included generalized linear models considering age, sex, and their interaction in relation to M. haemocanis infection. A chi-square test was used to determine differences in infection depending on these factors in dogs and to determine differences in prevalence between foxes and dogs.
Spatial analyses were performed only in Chiloé to test the effect of extrinsic variables on the M. haemocanis presence/absence, due to insufficient sample sizes in the other study areas. In order to analyze if foxes’ exposure to hemoplasmas depends on spillover events from dogs more than on intraspecific transmission (47), we used a model considering landscape anthropization in the fox home range area. We created a buffer zone around the fox capture site based on individual fox locations and the home range size described for the species (3.06 km2 for males and 2.72 km2 for females) (48). Independent variables were as follows: presence/absence of houses in the buffer area, total number of houses in the buffer area, distance of the capture site to the nearest house, land use, and proportion of vegetation cover in the buffer area (49). To determine if pathogen exposure showed temporal variation (e.g., epizootic or seasonal), a third model, including the variables season and year, was developed (50). Finally, a full model considering all the mentioned variables and a null model were developed. All variables were compared with presence/absence of M. haemocanis using generalized linear models. Variables were initially analyzed with univariate models, and then the variables with significant P values were included in multivariate models. The Akaike information criterion (AIC) was used for model selection, and its fit was assessed using the Hosmer-Lemeshow goodness-of-fit test (51).
Hematological and biochemical variables were compared for each of the following fox infection statuses: hemoplasma-positive versus -negative foxes, M. haemocanis-infected foxes versus M. haemocanis- and “Ca. Mycoplasma haematoparvum”-coinfected foxes, and M. haemocanis- and “Ca. Mycoplasma haematoparvum”-coinfected foxes versus negative foxes. We used the Shapiro-Wilk normality test to determine if the data were distributed normally. The two-sample Mann-Whitney U test was performed to determine differences among the variables. Following the “Guidelines for the Determination of Reference Intervals in Veterinary Species” of the American Society for Veterinary Clinical Pathology (available at https://cdn.ymaws.com/www.asvcp.org/resource/resmgr/QALS/Other_Publications/RI_Guidelines_For_ASVCP_webs.pdf), the median, standard deviation, and range values were provided for each parameter. All data analyses were performed using R software version 3.4.1 (52).
Data availability.
The new sequences obtained in the present study were submitted to GenBank with accession numbers MN164349 to MN164353.
ACKNOWLEDGMENTS
This study was funded by Morris Animal Foundation grant D16Z0-825, FONDECYT-Regular 1161593, CONICYT-FONDECYT Iniciación 11150934 (C.N.), CONICYT-FONDECYT Iniciación 11181180 (D.M.-A.), Morris Animal Foundation (MAF) Fellowship Training Award D15ZO-413 (C.N.), National Geographic Society Conservation Trust C309-15 (C.N.), Mohamed bin Zayed Species Conservation Fund 152510351 (C.N.), and CONICYT PAI Convocatoria Nacional Subvención a Instalación en la Academia Convocatoria Año 2019 Folio 77190064 (C.N.).
We thank Catherine Chirgwin, Alan Bannister, and the Tantauco Foundation, Cooperativa de Pescadores Mar Adentro, Corporación Nacional Forestal (CONAF), Parque Ahuenco, Parque Tablaruca, Forestal Arauco. We thank Carla Barría for laboratory assistance and Daniel González for identifying the tick.
REFERENCES
- 1.Silva-Rodriguez E, Farias A, Moreira-Arce D, Cabello J, Hidalgo-Hermoso E, Lucherini M, Jiménez J. 2016. Lycalopex fulvipes, Darwin’s fox In The IUCN red list of threatened species. IUCN, Gland, Switzerland. [Google Scholar]
- 2.Moreira-Arce D, Vergara PM, Boutin S. 2015. Diurnal human activity and introduced species affect occurrence of carnivores in a human-dominated landscape. PLoS One 10:e0137854-19. doi: 10.1371/journal.pone.0137854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silva-Rodríguez EA, Ovando E, González D, Zambrano B, Sepúlveda MA, Svensson GL, Cárdenas R, Contreras P, Farías AA. 2018. Large-scale assessment of the presence of Darwin’s fox across its newly discovered range. Mamm Biol 92:45–53. doi: 10.1016/j.mambio.2018.04.003. [DOI] [Google Scholar]
- 4.Villatoro F, Naughton-Treves L, Sepulveda M, Stowhas P, Mardones FO, Silva-Rodríguez EA. 2019. When free-ranging dogs threaten wildlife: public attitudes toward management strategies in southern Chile. J Environ Manage 229:67–75. doi: 10.1016/j.jenvman.2018.06.035. [DOI] [PubMed] [Google Scholar]
- 5.Zorondo-Rodríguez F, Moreira-Arce D, Boutin S. 2019. Underlying social attitudes towards conservation of threatened carnivores in human-dominated landscapes. Oryx doi: 10.1017/S0030605318000832. [DOI] [Google Scholar]
- 6.Acosta-Jamett G, Surot D, Cortés M, Marambio V, Valenzuela C, Vallverdu A, Ward MP. 2015. Epidemiology of canine distemper and canine parvovirus in domestic dogs in urban and rural areas of the Araucanía region in Chile. Vet Microbiol 178:260–264. doi: 10.1016/j.vetmic.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 7.Lessa I, Corrêa T, Guimarães S, De Godoy H, Cunha A, Vieira EM. 2016. Domestic dogs in protected areas: a threat to Brazilian mammals? Nat Conserv 14:46–56. doi: 10.1016/j.ncon.2016.05.001. [DOI] [Google Scholar]
- 8.Messick JB. 2004. Hemotrophic mycoplasmas (hemoplasmas): a review and new insights into pathogenic potential. Vet Clin Pathol 33:2–13. doi: 10.1111/j.1939-165x.2004.tb00342.x. [DOI] [PubMed] [Google Scholar]
- 9.Vieira RE, Vidotto O, Vieira T, Guimaraes AMS, dos Santos AP, Nascimento NC, dos Santos NJR, Martins TF, Labruna MB, Marcondes M, Biondo AW, Messick JB. 2015. Molecular investigation of hemotropic mycoplasmas in human beings, dogs and horses in a rural settlement in southern Brazil. Rev Inst Med Trop Sao Paulo 57:353–357. doi: 10.1590/S0036-46652015000400014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sykes JE. 2010. Feline hemotropic mycoplasmas. Vet Clin North Am Small Anim Pract 40:1157–1170. doi: 10.1016/j.cvsm.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Millán J, Velarde R, Delicado V, Negre N, Ribas A, Oleaga Á, Llaneza L, Esperón F. 2018. High diversity of hemotropic mycoplasmas in Iberian wild carnivores. Comp Immunol Microbiol Infect Dis 60:11–16. doi: 10.1016/j.cimid.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 12.Volokhov D, Hwang J, Chizhikov V, Danaceau H, Gottdenker N. 2017. Prevalence, genotype richness, and coinfection patterns of hemotropic mycoplasmas in raccoons (Procyon lotor) on environmentally protected and urbanized barrier islands. Appl Environ Microbiol 83:e00211-17. doi: 10.1128/AEM.00211-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.André MR, Adania CH, Allegretti SM, Machado RZ. 2011. Hemoplasmas in wild canids and felids in Brazil. J Zoo Wildl Med 42:342–347. doi: 10.1638/2010-0198.1. [DOI] [PubMed] [Google Scholar]
- 14.Koneval M, Miterpáková M, Hurníková Z, Blaňarová L, Víchová B. 2017. Neglected intravascular pathogens, Babesia vulpes and haemotropic Mycoplasma spp. in European red fox (Vulpes vulpes) population. Vet Parasitol 243:176–182. doi: 10.1016/j.vetpar.2017.06.029. [DOI] [PubMed] [Google Scholar]
- 15.Soto F, Walker R, Sepulveda M, Bittencourt P, Acosta-Jamett G, Müller A. 2017. Occurrence of canine hemotropic mycoplasmas in domestic dogs from urban and rural areas of the Valdivia Province, southern Chile. Comp Immunol Microbiol Infect Dis 50:70–77. doi: 10.1016/j.cimid.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 16.Greene CE. 2012. Infectious diseases of the dog and cat, 4th ed Elsevier, Philadelphia, PA. [Google Scholar]
- 17.Museux K, Boretti FS, Willi B, Riond B, Hoelzle K, Hoelzle LE, Wittenbrink MM, Tasker S, Wengi N, Reusch CE, Lutz H, Hofmann-Lehmann R. 2009. In vivo transmission studies of “Candidatus Mycoplasma turicensis” in the domestic cat. Vet Res 40:45. doi: 10.1051/vetres/2009028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornok S, Micsutka A, Meli ML, Lutz H, Hofmann-Lehmann R. 2011. Molecular investigation of transplacental and vector-borne transmission of bovine haemoplasmas. Vet Microbiol 152:411–414. doi: 10.1016/j.vetmic.2011.04.031. [DOI] [PubMed] [Google Scholar]
- 19.Lashnits E, Grant S, Thomas B, Qurollo B, Breitschwerdt EB, Breitschwerdt CB. 2019. Evidence for vertical transmission of Mycoplasma haemocanis, but not Ehrlichia ewingii, in a dog. J Vet Intern Med 33:1747–1746. doi: 10.1111/jvim.15517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willi B, Novacco M, Meli ML, Wolf-Jäckel GA, Boretti FS, Wengi N, Lutz H, Hofmann-Lehmann R. 2010. Haemotropic mycoplasmas of cats and dogs: transmission, diagnosis, prevalence and importance in Europe. Schweiz Arch Tierheilkd 152:237–244. doi: 10.1024/0036-7281/a000055. [DOI] [PubMed] [Google Scholar]
- 21.Cabello J, Altet L, Napolitano C, Sastre N, Hidalgo E, Dávila JA, Millán J. 2013. Survey of infectious agents in the endangered Darwin’s fox (Lycalopex fulvipes): high prevalence and diversity of hemotrophic mycoplasmas. Vet Microbiol 167:448–454. doi: 10.1016/j.vetmic.2013.09.034. [DOI] [PubMed] [Google Scholar]
- 22.Sacristán I, Acuña F, Aguilar E, García S, López MJ, Cevidanes A, Cabello J, Hidalgo-Hermoso E, Johnson WE, Poulin E, Millán J, Napolitano C. 2019. Assessing cross-species transmission of hemoplasmas at the wild-domestic felid interface in Chile using genetic and landscape variables analysis. Sci Rep 9. doi: 10.1038/s41598-019-53184-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Millán J, Candela MG, Palomares F, Cubero MJ, Rodríguez A, Barral M, de la Fuente J, Almería S, León-Vizcaíno L. 2009. Disease threats to the endangered Iberian lynx (Lynx pardinus). Vet J 182:114–124. doi: 10.1016/j.tvjl.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villatoro F, Sepúlveda MA, Stowhas P, Silva-Rodríguez EA. 2016. Urban dogs in rural areas: human-mediated movement defines dog populations in southern Chile. Prev Vet Med 135:59–66. doi: 10.1016/j.prevetmed.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Hirata H, Tateno M, Sakuma M, Nakanishi N, Izawa M, Asari Y, Okamura M, Shimokawa Miyama T, Setoguchi A, Endo Y. 2012. An epidemiological survey of hemoplasma infection in Iriomote cats (Prionailurus bengalensis iriomotensis). J Vet Med Sci 74:1531–1537. doi: 10.1292/jvms.12-0094. [DOI] [PubMed] [Google Scholar]
- 26.Kellner A, Carver S, Scorza V, McKee CD, Lappin M, Crooks KR, VandeWoude S, Antolin MF. 2018. Transmission pathways and spillover of an erythrocytic bacterial pathogen from domestic cats to wild felids. Ecol Evol 8:9779–9792. doi: 10.1002/ece3.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roura X, Peters IR, Altet L, Tabar MD, Barker EN, Planellas M, Helps CR, Francino O, Shaw SE, Tasker S. 2010. Prevalence of hemotropic mycoplasmas in healthy and unhealthy cats and dogs in Spain. J Vet Diagn Invest 22:270–274. doi: 10.1177/104063871002200219. [DOI] [PubMed] [Google Scholar]
- 28.Walker Vergara R, Morera Galleguillos F, Gómez Jaramillo M, Pereira Almosny NR, Arauna Martínez P, Grob Behne P, Acosta-Jamett G, Müller A. 2016. Prevalence, risk factor analysis, and hematological findings of hemoplasma infection in domestic cats from Valdivia, southern Chile. Comp Immunol Microbiol Infect Dis 46:20–26. doi: 10.1016/j.cimid.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Wengi N, Willi B, Boretti FS, Cattori V, Riond B, Meli ML, Reusch CE, Lutz H, Hofmann-Lehmann R. 2008. Real-time PCR-based prevalence study, infection follow-up and molecular characterization of canine hemotropic mycoplasmas. Vet Microbiol 126:132–141. doi: 10.1016/j.vetmic.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 30.Mascarelli PE, Tartara GP, Pereyra NB, Maggi RG. 2016. Detection of Mycoplasma haemocanis, Mycoplasma haematoparvum, Mycoplasma suis and other vector-borne pathogens in dogs from Córdoba and Santa Fé, Argentina. Parasit Vectors 9:642. doi: 10.1186/s13071-016-1920-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kutzer MAM, Armitage S. 2016. Maximising fitness in the face of parasites: a review of host tolerance. Zoology (Jena) 119:281–289. doi: 10.1016/j.zool.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 32.Scott ME. 1988. The impact of infection and disease on animal populations: implications for conservation biology. Conserv Biol 2:40–56. doi: 10.1111/j.1523-1739.1988.tb00334.x. [DOI] [Google Scholar]
- 33.Bergmann M, Englert T, Stuetzer B, Hawley JR, Lappin MR, Hartmann K. 2017. Risk factors of different hemoplasma species infections in cats. BMC Vet Res 13:52–52. doi: 10.1186/s12917-017-0953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munson L, Terio KA, Kock R, Mlengeya T, Roelke ME, Dubovi E, Summers B, Sinclair ARE, Packer C. 2008. Climate extremes promote fatal co-infections during canine distemper epidemics in African lions. PLoS One 3:e2545-10. doi: 10.1371/journal.pone.0002545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hidalgo-Hermoso E, Cabello J, Vega C, Kroeger-Gómez H, Moreira-Arce D, Napolitano C, Navarro C, Sacristán I, Cevidanes A, Di Cataldo S, Dubovi EJ, Mathieu-Benson C, Millán J. 2020. An eight-year survey for canine distemper virus indicates lack of exposure in the endangered Darwin’s fox (Lycalopex fulvipes). J Wildl Dis 56:482–485. doi: 10.7589/2019-08-195. [DOI] [PubMed] [Google Scholar]
- 36.Rynkiewicz EC, Pedersen AB, Fenton A. 2015. An ecosystem approach to understanding and managing within-host parasite community dynamics. Trends Parasitol 31:212–221. doi: 10.1016/j.pt.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Lewis J, Tomlinson A, Gilbert M, Alshinetski M, Arzhanova T, Goncharuk M, Goodrich J, Kerley L, Korotkova I, Miquelle D, Naidenko S, Sulikhan N, Uphyrkina O. 2019. Assessing the health risks of reintroduction: the example of the Amur leopard, Panthera pardus orientalis. Transbound Emerg Dis doi: 10.1111/tbed.13449. [DOI] [PubMed] [Google Scholar]
- 38.Kapil S, Yeary TJ. 2011. Canine distemper spillover in domestic dogs from urban wildlife. Vet Clin North Am Small Anim Pract 41:1069–1086. doi: 10.1016/j.cvsm.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delahay RJ, Smith GC, Hutchings MR. 2009. Management of disease in wild mammals, p 1–8. Springer, Tokyo, Japan. [Google Scholar]
- 40.Millán J, Travaini A, Cevidanes A, Sacristán I, Rodríguez A. 2019. Assessing the natural circulation of canine vector-borne pathogens in foxes, ticks and fleas in protected areas of Argentine Patagonia with negligible dog participation. Int J Parasitol Parasites Wildl 8:63–70. doi: 10.1016/j.ijppaw.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martínez-Díaz VL, Silvestre-Ferreira AC, Vilhena H, Pastor J, Francino O, Altet L. 2013. Prevalence and co-infection of haemotropic mycoplasmas in Portuguese cats by real-time polymerase chain reaction. J Feline Med Surg 15:879–885. doi: 10.1177/1098612X13480985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar S, Stecher G, Tamura K, Dudley J. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bandelt H, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 44.Excoffier L, Lischer HE. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 45.Hudson RR. 2000. A new statistic for detecting genetic differentiation. Genet Soc Am 155:2011–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Librado P, Rozas J. 2009. DnaSP v5:a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 47.McFarlane R, Sleigh A, McMichael T. 2012. Synanthropy of wild mammals as a determinant of emerging infectious diseases in the Asian-Australasian region. Ecohealth 9:24–35. doi: 10.1007/s10393-012-0763-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiménez JE. 2007. Ecology of a coastal population of the critically endangered Darwin’s fox (Pseudalopex fulvipes) on Chiloé Island, southern Chile. J Zool 271:63–77. doi: 10.1111/j.1469-7998.2006.00218.x. [DOI] [Google Scholar]
- 49.Hansen M, Potapov P, Moore R, Hancher M, Turubanova S, Tyukavina A, Thau D, Stehman S, Goetz S, Loveland T, Kommareddy A, Egorov A, Chini L, Justice C, Townshend J. 2013. High-resolution global maps of 21st-century forest cover change. Science 342:850–853. doi: 10.1126/science.1244693. [DOI] [PubMed] [Google Scholar]
- 50.Millán J, López-Bao JV, Garcıá EJ, Oleaga Á, Llaneza L, Palacios V, De La Torre A, Rodríguez A, Dubovi EJ, Esperón F. 2016. Patterns of exposure of Iberian wolves (Canis lupus) to canine viruses in human-dominated landscapes. Ecohealth 13:123–134. doi: 10.1007/s10393-015-1074-8. [DOI] [PubMed] [Google Scholar]
- 51.Hosmer DW, Hosmer T, Le Cessie S, Lemeshow S. 1997. A comparison of the goodness-of-fit test for the logistic regression model. Statist Med 16:965–980. doi:. [DOI] [PubMed] [Google Scholar]
- 52.R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The new sequences obtained in the present study were submitted to GenBank with accession numbers MN164349 to MN164353.