Bacillus thuringiensis (Bt)-based biological insecticides are used extensively to control insect pests and vectors of human diseases. Bt-based products provide greater specificity and biosafety than broad-spectrum synthetic insecticides. The biological activity of this bacterium resides in spores and crystals comprising complex mixtures of toxic proteins. We developed and validated a fast, accurate, and reproducible method for quantitative determination of the crystal components of Bt-based products. This method will find clear applications in the improvement of various aspects of the industrial production process of Bt. An important aspect of the production of Bt-based insecticides is its quality control. By specifically quantifying the relative proportion of each of the toxins that make up the crystal, our method represents the most consistent and repeatable evaluation procedure in the quality control of different batches produced in successive fermentations. This method can also contribute to the design of specific culture media and fermentation conditions that optimize Bt crystal composition across a range of Bt strains that target different pestiferous insects. Quantitative information on crystal composition should also prove valuable to phytosanitary product registration authorities that oversee the safety and efficacy of crop protection products.
KEYWORDS: Bacillus thuringiensis, Bt-based commercial product, LC-MS/MS-MRM, crystal protein, protein quantification, proteomics, proteotypic peptides, relative protein abundance
ABSTRACT
Bacillus thuringiensis (Bt) is the most widely used active ingredient for biological insecticides. The composition of δ-endotoxins (Cry and Cyt proteins) in the parasporal crystal determines the toxicity profile of each Bt strain. However, a reliable method for their identification and quantification has not been available, due to the high sequence identity of the genes that encode the δ-endotoxins and the toxins themselves. Here, we have developed an accurate and reproducible mass spectrometry-based method (liquid chromatography-tandem mass spectrometry-multiple reaction monitoring [LC-MS/MS-MRM]) using isotopically labeled proteotypic peptides for each protein in a particular mixture to determine the relative proportion of each δ-endotoxin within the crystal. To validate the method, artificial mixtures containing Cry1Aa, Cry2Aa, and Cry6Aa were analyzed. Determination of the relative abundance of proteins (in molarity) with our method was in good agreement with the expected values. This method was then applied to the most common commercial Bt-based products, DiPel DF, XenTari GD, VectoBac 12S, and Novodor, in which between three and six δ-endotoxins were identified and quantified in each product. This novel approach is of great value for the characterization of Bt-based products, not only providing information on host range, but also for monitoring industrial crystal production and quality control and product registration for Bt-based insecticides.
IMPORTANCE Bacillus thuringiensis (Bt)-based biological insecticides are used extensively to control insect pests and vectors of human diseases. Bt-based products provide greater specificity and biosafety than broad-spectrum synthetic insecticides. The biological activity of this bacterium resides in spores and crystals comprising complex mixtures of toxic proteins. We developed and validated a fast, accurate, and reproducible method for quantitative determination of the crystal components of Bt-based products. This method will find clear applications in the improvement of various aspects of the industrial production process of Bt. An important aspect of the production of Bt-based insecticides is its quality control. By specifically quantifying the relative proportion of each of the toxins that make up the crystal, our method represents the most consistent and repeatable evaluation procedure in the quality control of different batches produced in successive fermentations. This method can also contribute to the design of specific culture media and fermentation conditions that optimize Bt crystal composition across a range of Bt strains that target different pestiferous insects. Quantitative information on crystal composition should also prove valuable to phytosanitary product registration authorities that oversee the safety and efficacy of crop protection products.
INTRODUCTION
For decades, the bacterium Bacillus thuringiensis (Berliner) has been the most important biological insecticide for crop protection. Its specificity and ecotoxicological profile have been key to its development as an alternative to synthetic pesticides. The world biopesticide market is currently valued at three billion dollars annually (1). Three-quarters of all biopesticides are Bacillus thuringiensis (Bt)-based products, which are among the safest and most environmentally benign insecticides available (2, 3).
Bt produces large parasporal crystals during sporulation. These crystals consist of various δ-endotoxins, mostly Cry proteins, and in some Bt strains, cytotoxic Cyt proteins (4). The Cry proteins are cleaved in the insect gut by host proteinases, which give rise to 65- to 70-kDa activated toxins that form pores in the columnar cell brush border membranes, leading to the disruption of ion and metabolite transport and subsequent insect death (5). Cyt proteins also undergo activation of protoxins (27 kDa) by proteolytic cleavage in the insect gut to produce an activated toxin of 25 kDa, which interacts directly with nonsaturated membrane lipids (6). In many Bt strains, including the well-known Bt serovar kurstaki strain HD1, the sequence identity and similarity among Cry proteins, and among Cyt proteins, are usually very high (86 to 90%). Nevertheless, small sequence differences in the critical regions of these proteins are responsible for pronounced differences in the insecticidal potency and effective range of target species (7). A large number of cry and cyt genes have been cloned, expressed, and shown to encode proteins with specific insecticidal activity against pests from the orders Lepidoptera, Diptera, Coleoptera, and Hymenoptera, as well as other invertebrates, such as nematodes and mites (8).
Historically, Bt strains have been classified based on the serological characteristics associated with the bacterial flagellar antigen (H) into more than 60 serotypes (H serotypes) and over 80 serological varieties (serovars, also known as subspecies), with broadly different cry gene profiles and insecticidal activity spectra (9). In the absence of any other classification method, the insecticidal spectrum of a Bt strain and its potential industrial applicability were frequently assumed based on the serological characteristics of the strain. In the past, this classification was useful. For example, many Bt strains toxic to lepidopterans belong to the serovars kurstaki or aizawai (4), strains toxic to mosquito larvae often belong to serovar israelensis (10), and strains active against coleopterans often belong to serovar morrisoni (11). However, many new Bt strains not belonging to these serovars also possess insecticidal activity against these orders of insects. Moreover, classification into a specific serovar does not guarantee the presence of a specific set of cry genes or their expression (12). This classification system is now considered largely obsolete.
Advances in DNA technologies allow Bt strains to be easily characterized according to their cry and cyt gene content (13). Characterization of the δ-endotoxin gene content is useful for strain classification but is of limited predictive value, as it is the expression of these genes that determines the spectrum of activity of a given strain. There are also many cry and cyt genes that are cryptic, or with insignificant levels of expression, that contribute little to the toxicity of a given strain (14, 15). Therefore, identification of the insecticidal proteins that make up the parasporal crystal is essential to infer the insecticidal activity of a particular strain or to understand why production batches of a Bt-based insecticide vary in their toxicity characteristics.
To date, attempts to quantify the δ-endotoxin content of Bt strains have relied on reverse-phase high-performance liquid chromatography (RP-HPLC) of urea-denatured and trypsinized proteins (16) or ion-exchange liquid chromatography at constant pH applied to alkaline-digested peptides (17). However, neither of these techniques can be used to reliably distinguish proteins with similar amino acid sequences, such as some Cry proteins, or those present at low concentrations. An alternative approach involves HPLC analysis of semitrypsinized crystal protein preparations (18). Nonetheless, the partial digestion used by these authors is not effective for differentiating among proteins with a high degree of similarity, which are commonly present in the crystals produced by many Bt strains. Liquid chromatography-stable isotope dilution-multiple reaction monitoring-tandem mass spectrometry (LC-SID-MRM-MS) has been used to quantify Cry1Ab protein in genetically modified maize leaves (19). Compared to the analysis of wild-type Bt strains, the problem faced by these authors was greatly simplified when analyzing a single protein, as the issue of high homology among Bt δ-endotoxins was avoided. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) has also been used to analyze samples of a Bt strain from the late sporulation phase by using a linear ion trap spectrometer (20), but in this case, none of the proteins detected were quantified. Thus, to date, no analytical method capable of identifying and quantifying the crystal proteins present in different strains of Bt has been published.
Due to the difficulties faced in the identification and quantification of the Cry and Cyt toxins present in mixtures of these proteins, commercial Bt-based bioinsecticides currently include no description of the composition of the active ingredients; their characterization is based entirely on their potency against a reference insect species in comparison with an international Bt standard (21). A major disadvantage of such bioassay-based characterization is that the potency of a Bt preparation depends on the insect species tested, notwithstanding the individual and population variation in susceptibility that affects the results of bioassays. Furthermore, the susceptibility of an insect species cannot be extrapolated to other insect species. For example, within the lepidopteran family Noctuidae, heliothine species are very susceptible to the Cry1Ac protein, whereas species of the genus Spodoptera are usually very tolerant (22).
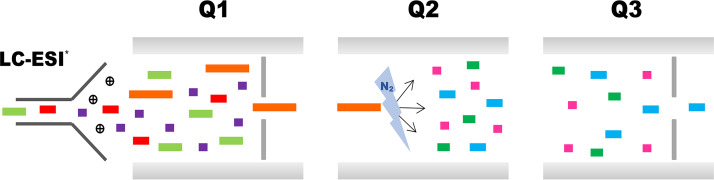
Advances in the field of proteomics now allow the quantitative characterization of proteins in a mixture. Specifically, the use of liquid chromatography coupled to mass spectrometry (LC-MS/MS) in combination with multiple reaction monitoring (MRM) allows the precise analysis of complex samples in which each component differs by one or more proteotypic peptides (peptides generated by protease treatment that are unique to a given protein), charge, and hydrophobicity (23). Indeed, the targeted nature of MRM, its high selectivity, and its wide dynamic range render this technique ideal for quantitative proteomics, especially when combined with known quantities of stable isotope-labeled (SIL) synthetic peptides (24, 25). The process involves a triple quadrupole mass spectrometer operating in an MRM assay to provide a highly sensitive, specific, and cost-effective analysis (Fig. 1).
FIG 1.
Diagram representing the operation of a triple quadrupole mass spectrometer in an MRM assay. First, peptides are filtered in the first quadrupole (Q1) according to the mass to charge ratio (m/z) of the precursor ion. In the collision cell (second quadrupole [Q2]), peptides are fragmented by collision-induced dissociation using nitrogen as the collision gas. Finally, predefined peptide-specific fragments are selected in the second mass filter (third quadrupole [Q3]), which is followed by measurement of the intensity of the transitions. Transitions are the precursor/product ion pairs, and several transitions are monitored over time for each peptide. LC-ESI, liquid chromatography coupled to electrospray ionization.
Here, we describe a novel application of LC-MS/MS-MRM that, for the first time, allows reliable quantitative determination of the components of insecticidal crystals of Bt strains. Following verification using artificial mixtures of Cry proteins, we applied the technique to four Bt-based products (DiPel DF, XenTari GD, VectoBac 12S, and Novodor) that are widely used for the control of lepidopteran, dipteran, and coleopteran pests. The method is rapid, accurate, and more reproducible than the previous analytical techniques developed for this purpose. This technique will also enable improved monitoring of Bt production processes, optimization of fermentation media and conditions, and accurate monitoring of batch variation for the production of high-potency Bt-based insecticides. It will also provide phytosanitary product registration authorities with precise information on the composition of Bt-based crop protection products.
RESULTS
Validation of the LC-MS/MS-MRM method by analysis of artificial Cry protein mixtures.
For initial validation of the LC-MS/MS-MRM technique, two protein mixtures containing Cry1Aa, Cry2Aa, and Cry6Aa were examined. All three proteins were identified in both Cry protein mixtures via proteotypic peptide detection (Fig. 2).
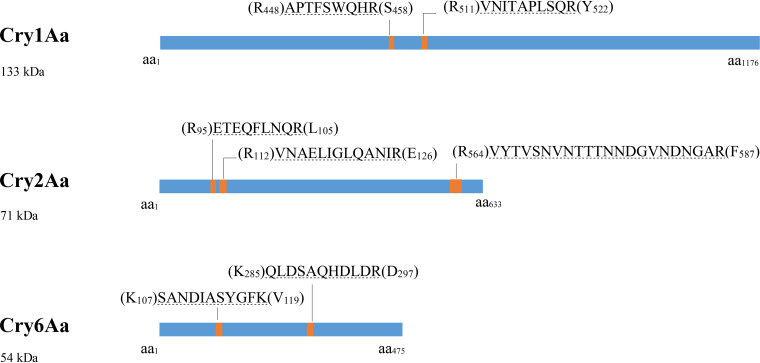
FIG 2.
Schematic overview of the targeted proteotypic peptide in three Cry proteins present in known amounts in artificial mixtures used for the LC-MS/MS-MRM method validation. These peptides were detected in the IDA analysis and checked against in silico digestion results of the artificial protein mixture. Two proteotypic peptides were used to identify Cry1Aa and Cry6Aa, and three were used to identify Cry2Aa. The locations of the proteotypic peptides (orange bands) within full-length proteins (blue bars) are shown, with different letters corresponding to different amino acids (aa) and a number indicating the position of the amino acid in the protein. The residues in parentheses are the previous and subsequent amino acids in the protein sequence.
The MS/MS spectra and extracted ion chromatograms (data not shown) and the transitions selected for each target peptide (data not shown) were determined. SIL peptides were synthesized and used for protein quantification of both artificial mixtures. In mixture 1, which contained equal amounts of each Cry protein, the relative abundance of Cry1Aa was determined at 32 to 35%, that of Cry2Aa at 24 to 26%, and that of Cry6Aa at 39 to 44% (Table 1). In mixture 2, containing Cry1Aa, Cry2Aa, and Cry6Aa in a molar ratio of 13:4:13, the measured relative abundances of the different proteins were also comparable with the expected relative abundances (in molarity) of 43, 13, and 43%, respectively, as follows: Cry1Aa, 43 to 46%; Cry2Aa, 6%; Cry6Aa, 48 to 51% (Table 1). The limits of detection were approximately 30 fmol for Cry1Aa, 300 fmol for Cry2Aa, and 500 fmol for Cry6Aa. The underestimation of Cry2Aa and the overestimation of Cry6Aa observed in both experimental mixtures are attributed to the low number of peptides and reduced number of replicates per peptide used in the quantification of these proteins.
TABLE 1.
Validation of the LC-MS/MS-MRM methoda
| Mixture and digestion Cry protein | Proteotypic peptide | Peptide mean ± SD (fmol) | Protein mean ± SD (fmol) | Relative molar composition (%) |
|---|---|---|---|---|
| Mixture 1 | ||||
| D1 | ||||
| Cry1Aa | APTFSWQHR | 2,511 ± 410 | 2,226 ± 405 | 32 |
| VNITAPLSQR | 1,942 ± 32 | |||
| Cry2Aa | ETEQFLNQR | 2,558 ± 254 | 1,666 ± 724 | 24 |
| VNAELIGLQANIR | 1,407 ± 155 | |||
| VYTVSNVNTTTNNDGVNDNGAR | 1,032 ± 28 | |||
| Cry6Aa | QLDSAQHDLDR | 3,092 ± 51 | 3,126 ± 128 | 44 |
| SANDIASYGFK | 3,159 ± 204 | |||
| D2 | ||||
| Cry1Aa | APTFSWQHR | 2,834 ± 103 | 2,402 ± 511 | 35 |
| VNITAPLSQR | 1,970 ± 157 | |||
| Cry2Aa | ETEQFLNQR | 2,614 ± 458 | 1,754 ± 772 | 26 |
| VNAELIGLQANIR | 1,567 ± 39 | |||
| VYTVSNVNTTTNNDGVNDNGAR | 1,081 ± 561 | |||
| Cry6Aa | QLDSAQHDLDR | 2,218 ± 283 | 2,654 ± 576 | 39 |
| SANDIASYGFK | 3,090 ± 392 | |||
| Mixture 2 | ||||
| D1 | ||||
| Cry1Aa | APTFSWQHR | 4,510 ± 854 | 3,630 ± 1,172 | 43 |
| VNITAPLSQR | 2,750 ± 543 | |||
| Cry2Aa | ETEQFLNQR | 714 ± 210 | 483 ± 211 | 6 |
| VNAELIGLQANIR | 430 ± 20 | |||
| VYTVSNVNTTTNNDGVNDNGAR | 304 ± 55 | |||
| Cry6Aa | QLDSAQHDLDR | 4,258 ± 416 | 4,380 ± 305 | 51 |
| SANDIASYGFK | 4,503 ± 213 | |||
| D2 | ||||
| Cry1Aa | APTFSWQHR | 5,906 ± 1,484 | 4,703 ± 1,648 | 46 |
| VNITAPLSQR | 3,501 ± 406 | |||
| Cry2Aa | ETEQFLNQR | 732 ± 72 | 591 ± 155 | 6 |
| VNAELIGLQANIR | 561 ± 84 | |||
| VYTVSNVNTTTNNDGVNDNGAR | 480 ± 203 | |||
| Cry6Aa | QLDSAQHDLDR | 4,507 ± 518 | 4,821 ± 486 | 48 |
| SANDIASYGFK | 5,134 ± 217 |
Proteins were identified and quantified in two artificial mixtures of three Cry proteins. Cry1Aa, Cry2Aa, and Cry6Aa were mixed in a molar ratio of 1:1:1 for mixture 1 (33.3% of each Cry protein) and a molar ratio of 13:4:13 for mixture 2 (equivalent to 43.3% of Cry1Aa, 13.3% of Cry2Aa, and 43.3% of Cry6Aa). The mixtures were digested with trypsin, and proteins were identified and quantified based on proteotypic peptide abundance and by comparing the signal intensities of the endogenous and corresponding SIL peptide. Mean values represent averages of two independent analyses for each tryptic digestion (D1 and D2).
Therefore, using known mixtures, three aspects of the procedure were validated: (i) all three types of Cry proteins were detected independently of their proportions in the mixture, (ii) the relative abundance determined by LC/MS-MS-based quantification was close to the true abundance of Cry proteins in both mixtures, and (iii) a high degree of reproducibility of the relative abundance values was observed in the analyses of different digestions of samples (Table 1). This indicated that the method was suitable for the quantification of Cry protein mixtures.
Gene content of the Bt strains in four commercial biopesticides and identification of the proteins present in the parasporal crystals.
Prior to the proteomic analysis, it is necessary to determine which proteins are likely to be present in the parasporal crystal based on the genes present in the genome of a given Bt strain. This information is necessary to perform the information-dependent acquisition (IDA) analysis and to select the proteotypic peptides that will be used to identify and quantify each crystal protein. Total genomic DNA sequencing revealed the different types of Cry and Cyt proteins encoded by the four strains of Bt present in the commercial products (Table 2). Once the potential composition of the crystals was established, IDA analyses were performed to identify which proteins encoded in the genome of each Bt strain were actually expressed and integrated into the parasporal crystal (Table 3). In the DiPel DF sample, peptides from five proteins were detected, namely, Cry1Aa, Cry1Ab, Cry1Ac, Cry2Aa, and Cry2Ab. Five proteins were detected in the parasporal crystal of XenTari GD—Cry1Aa, Cry1Ab, Cry1Ca, Cry1Da, and Cry2Ab. For VectoBac 12AS, seven different proteins were identified—five Cry proteins (Cry4Aa, Cry4Ba, Cry11Aa, Cry60Aa, and Cry60Ba) and two Cyt proteins (Cyt1Aa1 and Cyt1Da). In the Novodor sample, all the Cry proteins predicted by genome analysis were detected—Cry3Aa, Cry23Aa, and Cry37Aa. These results agreed with the detection of putative insecticidal proteins using SDS-PAGE of spore-crystal mixtures and solubilized proteins (data not shown).
TABLE 2.
Insecticidal protein genes identified in the genome of Bt strains isolated from four commercial insecticidesa
| Gene | DiPel (ABTS-351) | XenTari (ABTS-1857) | VectoBac (AM65-52) | Novodor (NB-176) |
|---|---|---|---|---|
| cry1Aa | + | + | – | – |
| cry1Ab | + | + | – | – |
| cry1Ac | + | – | – | – |
| cry1Ca | – | + | – | – |
| cry1Da | – | + | – | – |
| cry2Aa | + | – | – | – |
| cry2Ab | + | + | – | – |
| cry3Aa | – | – | – | + |
| cry4Aa | – | – | + | – |
| cry4Ba | – | – | + | – |
| cry9Ea | – | + | – | – |
| cry10Aa | – | – | + | – |
| cry11Aa | – | – | + | – |
| cry23Aa | – | – | – | + |
| cry37Aa | – | – | – | + |
| cry60Aa | – | – | + | – |
| cry60Ba | – | – | + | – |
| cyt1Aa | – | – | + | – |
| cyt1Da | – | – | + | – |
| cyt2Ba | – | – | + | – |
The Bt strains ABTS-351, ABTS-1857, AM65-52, and NB-176 were isolated from the commercial products DiPel DF, XenTari GD, VectoBac 12AS, and Novodor, respectively. Genomes of these strains were sequenced using an Illumina NextSeq 500 sequencer, and the resulting reads were assembled using the CLC Genomic Workbench version 10.1.1 with the de novo assembly tool. Contigs were then analyzed against a custom insecticidal toxin database constructed using insecticidal protein sequences. + indicates presence and – indicates absence.
TABLE 3.
Cry and Cyt proteins identified by IDA analysis of the Bt strains present in four insecticidal products
| Product (Bt strain) | Proteina | GenBank accession no. | Number of peptides used for protein identification |
|---|---|---|---|
| DiPel DF (ABTS-351) | Cry1Aa | MK184461 | 6 |
| DiPel DF (ABTS-351) | Cry1Ab | MK184462 | 7 |
| DiPel DF (ABTS-351) | Cry1Ac | MK184463 | 9 |
| DiPel DF (ABTS-351) | Cry2Aa | MK184464 | 11 |
| DiPel DF (ABTS-351) | (Cry2Ab) | MK184465 | (1) |
| XenTari GD (ABTS-1857) | Cry1Aa | MK184475 | 4 |
| XenTari GD (ABTS-1857) | Cry1Ab | MK184476 | 12 |
| XenTari GD (ABTS-1857) | Cry1Ca | MK184477 | 12 |
| XenTari GD (ABTS-1857) | Cry1Da | MK184478 | 4 |
| XenTari GD (ABTS-1857) | Cry2Ab | MK184479 | 5 |
| VectoBac 12AS (AM65-52) | Cry4Aa | MK184469 | 3 |
| VectoBac 12AS (AM65-52) | Cry4Ba | MK184470 | 17 |
| VectoBac 12AS (AM65-52) | Cry11Aa | MK184471 | 17 |
| VectoBac 12AS (AM65-52) | Cry60Aa | MK184472 | 5 |
| VectoBac 12AS (AM65-52) | Cry60Ba | MK184473 | 8 |
| VectoBac 12AS (AM65-52) | Cyt1Aa | MK184474 | 2 |
| VectoBac 12AS (AM65-52) | (Cyt1Da) | CAD30104 | (1) |
| Novodor (NB-176) | Cry3Aa | MK184466 | 16 |
| Novodor (NB-176) | Cry23Aa | MK184467 | 7 |
| Novodor (NB-176) | Cry37Aa | MK184468 | 4 |
Proteins in parentheses were detected on the basis of the presence of a single peptide. They were therefore considered to be potential false positives or present at a low concentration, close to the limit of detection of this technique.
In the IDA analyses, most proteins were identified by at least four peptides, whereas Cry2Ab in DiPel DF and Cyt1Da in VectoBac 12AS were each detected by a single peptide. Consequently, they were excluded from the analysis, although these proteins may have been present at concentrations that were at or below the detection limit. Similarly, the insecticidal proteins of three genes that had been identified by genome sequencing (Table 2) were not detected in the crystals from the commercial Bt insecticides. These were Cry9Ea in XenTari GD and Cry10Aa and Cyt2Ba in VectoBac 12AS. We assume either that the respective genes are not expressed or that these proteins are present at concentrations below the detection threshold of the current method.
Selection of proteotypic peptides and MRM parameters for improved protein quantification.
Next, we proceeded to select the proteotypic peptides that were unique to each protein within each mixture and would provide a clear response in the instrument. Proteotypic peptides were identified with in silico trypsin digestion of the amino acid sequences of the proteins identified by IDA analysis (data not shown). Cry proteins have a high degree of similarity, and therefore the selection of target proteotypic peptides depends on the specific composition of each Bt product. For example, the selected peptides for Cry1Aa in DiPel DF were not necessarily proteotypic for Cry1Aa in XenTari GD. Identifying proteins based on a specific list of Bt proteotypic peptides has the advantage of increasing the precision of ensuing identification and quantification.
MRM-initiated detection and sequencing (MIDAS) analyses (see “Protein Quantification by Multiple Reaction Monitoring,” below) were performed to select the best MRM transition, in terms of sensitivity and selectivity, for each peptide. To increase sensitivity, the most intense m/z (mass/charge) ratio of each targeted peptide was selected at the first quadrupole, and in order to maximize selectivity, fragment ions with m/z ratios higher than those of the precursor ion were monitored. During the design of the MRM assays, several transitions from each peptide were tested, and those with the highest signal intensity and lowest level of interference signals were chosen (data not shown).
We selected the most suitable proteotypic peptides and MRM transitions for each crystal to enable highly specific quantification of the Cry and Cyt proteins of interest. We attempted to select at least two proteotypic peptides for each crystal protein in DiPel DF, XenTari GD, VectoBac 12AS, and Novodor (data not shown). However, this was not always possible because of different limitations, especially those involving the high similarity between some of these proteins and the difficulty encountered when synthesizing some peptides (see Discussion for more details).
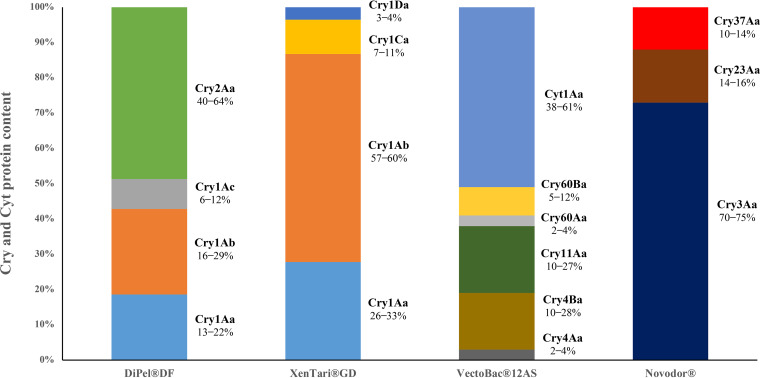
Having established the proteins present in each sample, we proceeded to determine the relative composition of the Bt crystals. The relative molar abundance of each crystal protein in the four commercial Bt-based insecticides is shown in Fig. 3. In all cases, the crystals contained a major protein that accounted for approximately 50% of all identified Cry and Cyt proteins. Parasporal crystals from DiPel DF were found to be mainly composed of Cry2Aa, followed by Cry1Ab and Cry1Aa and by Cry1Ac as a minor component. Crystals from XenTari GD mainly contained Cry1Ab and Cry1Aa, with Cry1Ca and Cry1Da as minor components. The major component of VectoBac 12AS crystals was Cyt1Aa, followed by Cry11Aa, Cry4Ba, and Cry60Ba, with a very low abundance of Cry60Aa and Cry4Aa. Finally, Cry3Aa was the major protein in Novodor crystals, followed by the Cry23Aa and Cry37Aa proteins, which function as binary toxins (Fig. 3). These results demonstrate that the LC-MS/MS-MRM method is suitable for the determination of the crystal protein content of Bt strains with very distinct protein compositions and toxicity spectra.
FIG 3.
Relative molar composition of proteins in parasporal crystals of Bt-based insecticides. The composition is expressed as a range from two independent tryptic digestions with two technical replicates each.
DISCUSSION
In the current study, we developed an LC-MS/MS-MRM method to evaluate the protein composition of parasporal crystals purified directly from Bt-based commercial insecticides or from any Bt product or culture. The mass spectrometry-based method can be used not only to identify the components of the crystal, but also to determine their relative abundance, thus providing a method for qualitative and quantitative characterization of the toxic protein crystals in Bt-based insecticidal preparations. This proteomic approach provides a tool that will improve some essential aspects of the industrial production of Bt, such as (i) quality control of batches produced during different fermentations, (ii) selection of culture media and fermentation conditions that optimize the protein composition of the crystal, and (iii) definition of the qualitative and quantitative composition of the different active substances (crystal proteins) that make up the active ingredient of each commercial Bt formulation.
The setup of an MRM experiment for the analysis of Cry and Cyt proteins is complex and requires several intermediate steps. The major experimental hurdle to overcome is the complexity of the mixtures of these proteins, which complicates the selection of suitable proteotypic peptides for each of the crystal proteins in the mixture. This selection can be challenging for the following reasons: (i) the small size of some proteins reduces the number of candidate peptides; (ii) there is a lack of spectral libraries for Bt; and (iii) certain proteins show high amino acid sequence similarity (26), which limits the number of candidate proteotypic peptides. Moreover, once the candidate proteotypic peptides have been identified using in silico analysis, they have to be filtered according to certain analytical criteria. These include sequence length (ideally 6 to 16 amino acids), chemical stability throughout the analytical process, and good detectability and MS response characteristics (27–29). Finally, SIL peptides that are too long or have strong hydrophobic characteristics can be difficult to synthesize and should be avoided (30). For some Bt proteins, these factors make it difficult to select a minimum of two proteotypic peptides for every crystal protein detected in order to provide highly selective and sensitive assays. Specifically, Cry1Ab and Cry2Aa in the DiPel DF sample were quantified based on a single proteotypic peptide that was specific for each protein. In these cases, the most efficient way to obtain a more accurate estimate would be to increase the number of replicates performed for each single proteotypic peptide. However, given the novelty of the method developed in this study and the high reproducibility observed in the results obtained from different digestions of the samples, we are confident that the use of a single proteotypic peptide for the quantification of some proteins can provide a valid estimate of protein relative abundance.
Careful selection of the MRM transitions is critical for the specificity of the assay and the ability to detect and quantify target peptides (31). Several transitions for each peptide were monitored in the current study (data not shown), which resulted in improved discrimination among the different crystal toxins. Typically, monitoring three or four transitions per peptide provides a suitable balance between selectivity and throughput (32, 33). For this method, we monitored between 4 and 6 transitions per peptide, and in some cases, we were even able to select transitions for the double- and triple-charged precursor ion. Overall, double-charged precursor ions are favored by electrospray ionization, but in some cases, such as when the peptide sequence contains a histidine residue, triple-charged ions are also favored (34). Because of the lack of spectral libraries for Bt Cry proteins, we had to determine the precursor ion charge state as well as the product ions to be monitored based on the experimental data obtained. In addition, to maximize selectivity and minimize background interference, the following features were considered: (i) product ions with m/z values higher than those of the precursor ion were preferentially selected (23); (ii) for a given MRM method, there were no peptides with the same m/z (even for those that differed in amino acid sequence) or retention time; (iii) runs without a spike of SIL peptides were performed previously to confirm the lack of interference in SIL transitions; (iv) the reference intensity dot product, which is the relative intensities of the transitions for endogenous and SIL peptides, had to be similar for each type of peptide. In this way, we ensured that maximum sensitivity and peptide discrimination were achieved.
Common Bt-based products for controlling pests of different orders (DiPel DF, Xentari GD, Vectobac 12S, and Novodor) were characterized in the current study using the novel validated method. The relative abundances of different Cry and Cyt proteins within the parasporal crystal of the Bt strain used in each product have been revealed for the first time.
DiPel DF and XenTari GD products are widely used in the control of lepidopteran pests. Although the former is effective against many leaf-feeding lepidopteran pests, it is less active against species from the genus Spodoptera (35). According to the manufacturer, the active ingredient of DiPel DF, Bt strain ABTS-351, harbors the cry1Aa, cry1Ab, cry1Ac, and cry2 genes (the manufacturer did not specify precisely which cry2 genes), and XenTari GD Bt strain ABTS-1857 contains cry1Aa, cry1Ab, cry1C, and cry1D. The differences in the gene content explain the activity spectra of these products; however, the level of expression of these genes in each product is not given by the manufacturer (Kenogard).
In the current study, we initially corroborated that the genes expressed in DiPel DF were indeed cry1Aa, cry1Ab, cry1Ac, and cry2Aa. Next, we determined that the protein content in the crystal is distributed approximately evenly between Cry1A and Cry2A proteins (Fig. 3). The broad spectrum of toxicity against lepidopterans attributed to Cry2A proteins, along with a balanced contribution of Cry1 proteins, explains the efficacy of this Bt-based product against the recommended target species. In XenTari GD, we confirmed the presence of Cry1Aa, Cry1Ab, Cry1C, and Cry1D and determined that crystals comprised up to 15% of Cry1C and Cry1D (Fig. 3). Despite this low percentage, the occurrence of these two toxins is responsible for the unique toxicity spectrum of this product, which has high activity against species in the genus Spodoptera that are major pests of horticultural and other field crops (36, 37).
VectoBac 12AS, which is based on Bt serovar israelensis, is used for controlling medically important vectors, such as mosquitoes (38). In the present study, we showed that Cry4Aa, Cry4Ba, Cry11Aa, Cry60Aa, Cry60Ba, Cyt1Aa, and Cyt1Da were expressed, but we found no evidence for the presence of Cry10Aa or Cyt2Ba in the commercial product. In contrast, other studies using different techniques have reported small amounts of these proteins in the parasporal crystals of different strains of Bt serovar israelensis (39). Approximately 90% of the protein content of the crystals in VectoBac 12AS was found to comprise Cry4Aa, Cry4Ba, Cry11Aa, and Cyt1Aa (Fig. 3), which are toxins that, when present in mixtures, display high larvicidal activity in dipterans (40, 41). The low concentration of Cry60Aa and Cry60Ba detected in the current study may indicate that minor crystal components could contribute to the toxicity of the parasporal crystal by synergizing the potency of other Cry proteins (42) and may also contribute to the lack of reports of resistance to Bt serovar israelensis in dipteran populations subjected to long-term treatment with this type of insecticide (43, 44).
Novodor is widely used for the control of coleopteran pests. We confirmed the expression of three Cry proteins in this product (Fig. 3). The major insecticidal protein in Novodor crystals was Cry3Aa, which is toxic to insects in the orders Coleoptera, Hemiptera, and Hymenoptera (8). Another identified component, Cry23A, is active against certain species of Coleoptera only in the presence of Cry37 (45), which is believed to facilitate binding of the channel-forming toxins to midgut epithelial cells (46). The observation that the relative abundance of Cry37Aa in Novodor crystals was similar to that of Cry23Aa supports the notion of a positive interaction between these two proteins.
Although the applicability of the LC-MS/MS-MRM method may be limited by the selection of proteotypic peptides and by the availability of the corresponding SIL peptides, when proteins with a high homology are present in the same sample, we successfully analyzed commercial strains that contained several isomorphic proteins in their crystals. Also, the need for genome sequencing of the bacterium no longer poses a limitation to the use of this method, as genome sequencing technology is now routine in most laboratories across the world.
Accurate determination of the composition of Bt parasporal crystals can provide direct economic and environmental benefits associated with the production of effective biological insecticides with minimal environmental impact (47, 48). Application of the present method to quantify the effect of growth media and fermentation conditions on decisive aspects of the efficacy of Bt-based insecticides, such as the size and precise composition of insecticidal crystals, or production processes that influence batch variation will allow unprecedented control over key characteristics of these products. Such control would likely contribute to increasing the efficacy and the commercial competitiveness of Bt-based products. Precise information on crystal composition could also prove informative to phytosanitary registration authorities responsible for the safety and efficacy of crop protection products based on this uniquely valuable pathogen. In conclusion, the method developed in the current study allows, for the first time, accurate characterization of the insecticidal crystals of Bt isolates, as well as strains recovered from commercial pesticides targeted at a diversity of insect pests and vectors of medical importance.
MATERIALS AND METHODS
Bt recombinant strains expressing a single Cry protein.
Three recombinant Bt BMB171 strains, each producing Cry1Aa, Cry2Aa, or Cry6Aa, were obtained from Colin Berry, University of Cardiff, UK. Luria-Bertani (LB) medium was used to grow the samples that were incubated at 28°C overnight. A single colony of each strain was isolated and grown in 50 ml of casein hydrolysate and yeast extract (CCY) medium (49), enriched with mineral salts and pH adjusted to 7.5. Erlenmeyer flasks were used for the 96 h of incubation in a shaker-incubator (New Brunswick Innova 42R) at 28 ± 1°C and 200 rpm. NaCl-EDTA was added in order to obtain a 10-mM suspension before centrifugation at 15,000 × g for 10 min. The resulting pellet was washed twice in cold 1 M NaCl and six times in cold Milli-Q water and was finally resuspended in 1.5 ml of 10 mM KCl.
Purification of parasporal crystals.
Crystals were purified from the mixture by ultracentrifugation in a discontinuous sucrose gradient as previously described (50). Briefly, the spore-crystal mixture was sonicated for 20 s using a Soniprep 150 MSE apparatus (Curtin Matheson Scientific, Houston, TX) and immediately loaded onto a two-layer sucrose gradient composed of 16 ml of 67% (wt/vol) sucrose solution and 16 ml of 79% (wt/vol) sucrose solution. After centrifugation at 70,000 × g for 16 h (Optima L-100 XP; Beckman Coulter; SW 32 Ti rotor), the interphase containing the crystals was recovered using a Pasteur pipette, mixed with sterile Milli-Q water to a final volume of 50 ml, and centrifuged again (15,000 × g, 15 min). This step was repeated twice, and the crystal pellet was finally resuspended in 1 ml of sterile Milli-Q water. Crystal purity was assessed using phase-contrast microscopy (Zeiss AX-10) under 1,000× magnification using immersion oil (data not shown).
Polyacrylamide gel electrophoresis of Bt crystal proteins.
A 100 μl volume of purified crystals was solubilized in 500 μl of a solution of 50 mM Na2CO3 (pH 11.3) and 10 mM dithiothreitol (DTT) by gentle agitation for 2 h at 28°C. Unsolubilized crystals were removed by centrifugation at 9,000 × g for 10 min at 4°C. Aliquots (10 μl) of the solubilized proteins in the supernatant and of the spore and crystal mixture were then resolved using 10% SDS-PAGE (100:1 acrylamide/bis-acrylamide ratio) at 50 mA for 1 h using a mini-Protean III apparatus (Bio-Rad, Hercules, CA). The gels were stained with 50% (vol/vol) ethanol, 10% (vol/vol) acetic acid, and 0.1% (wt/vol) Coomassie brilliant blue R250 for 40 min and then destained in a solution of 6.75% (vol/vol) glacial acetic acid and 9.45% (vol/vol) ethanol. Protein mass-band patterns were determined by comparison with a broad-range protein marker (Precision Plus Protein dual color standards; Bio-Rad).
Protein quantification and tryptic digestion of Cry proteins.
Prior to tryptic digestion of the Cry proteins in each sample, the concentration of the solubilized proteins in the supernatant was determined using the Bradford assay (developed by Bradford in 1976 [52]) using bovine serum albumin as a standard. Preliminary tests were performed to establish the appropriate digestion protocol for crystal proteins, with urea or RapiGest SF (Waters Corp., Milford, MA) as the denaturant, followed by one or two steps of trypsin digestion (data not shown). The following optimal protocol was established. For protein denaturation, 10 μl of denaturing buffer (6 M urea and 100 mM Tris, pH 7.8) was added to previously evaporated crystal samples (protein content of approximately 30 μg). Cysteines were then reduced with 25 mM (final concentration) DTT for 30 min at 37°C and alkylated with 70 mM (final concentration) iodoacetamide for 30 min in the dark. Unreacted iodoacetamide was neutralized by the addition of 6 μl of a stock solution (200 mM) of DTT and incubated for 30 min at room temperature. The samples were then diluted with 75 μl of 50 mM ammonium bicarbonate so that the final concentration of urea was below 1 M. Digestion was then performed at 37°C overnight with trypsin (Gold Trypsin; Promega, Madison, WI), at an enzyme:protein weight ratio of 1:20. The reaction was stopped by adding 1 μl of concentrated formic acid (Sigma-Aldrich, Saint Louis, MO). Two independent tryptic digestions were performed for each sample.
Preparation of artificial mixtures of Cry proteins.
Two mixtures of Cry proteins were prepared in order to validate the MS-based Cry protein quantification method. In mixture 1, equal molar amounts of each Cry protein were combined (equivalent to a percentage composition of 33.3% each of Cry1Aa, Cry2Aa, and Cry6Aa). In mixture 2, Cry1Aa, Cry2Aa, and Cry6Aa were combined in a molar ratio of 13:4:13, respectively (equivalent to a percentage composition of 43.3% Cry1Aa, 13.3% Cry2Aa, and 43.3% Cry6Aa). Each mixture was trypsinized in duplicate, as described in “Protein Quantification and Tryptic Digestion of Cry Proteins,” above, and then analyzed using LC-MS/MS-MRM.
Protein identification using LC-MS/MS and in silico digestion.
Proteins were identified using a nano-LC system (Tempo MDLC; AB Sciex, Framingham, MA) coupled to a hybrid triple quadrupole/linear ion trap mass spectrometer (4000 QTRAP; AB Sciex). After precolumn desalting, the tryptic digests were separated on a C18 column (Thermo Scientific, Waltham, MA) at a flow rate of 300 nl/min with a 90-min linear gradient from 5 to 35% acetonitrile in 0.1% formic acid. The mass spectrometer was interfaced with a nanospray source equipped with an uncoated fused silica emitter tip (20 μm inner diameter, 10 μm tip; New Objective, Woburn, MA) and was operated in the positive ion mode. The MS source parameters were as follows: capillary voltage, 2,800 V; source temperature, 150°C; declustering potential, 110 V; curtain and ion source gas (nitrogen), 20 lb/in2; and collision gas (nitrogen) set to high. Analyses were performed using an information-dependent acquisition (IDA) method as follows: single enhanced mass spectra (400 to 1,400 m/z) were acquired, and the eight most intense peaks were automatically chosen by the mass spectrometer and subjected to an enhanced product ion scan. Proteins were identified using the search engine Mascot version 2.3 (Matrix Science, Boston, MA) to examine the deduced amino acid sequences of the Cry proteins predicted from the genome sequence. The database used for Mascot searches was built in-house. The following search parameters were used: one missed cleavage; carbamidomethylation of cysteines as a fixed modification; 0.5 Da peptide mass tolerance; and 0.3 Da fragment mass tolerance. Two separate LC-MS/MS analyses were performed for each digestion.
An in silico trypsin digestion of the predicted Cry protein sequences was performed using MS-Digest, a bioinformatics tool in the software package Protein Prospector of the University of California San Francisco (http://prospector.ucsf.edu/prospector/mshome.htm).
Synthesis of SIL peptides for protein quantification.
SIL peptides, synthesized by stepwise solid-phase peptide synthesis on an automated peptide synthesizer (Multipep; Intavis Bioanalytical Instruments, Cologne, Germany), were obtained from the Proteomics Facility of the National Biotechnology Center (CNB, Madrid, Spain). Amino acid polymerization was performed using standard N-(9-fluorenyl) methoxycarbonyl (Fmoc) chemistry and PyBOP/N-methylmorpholine as coupling activation reagents. Fmoc-derivatized amino acid monomers were obtained from Merck (St. Louis, MO). l-lysine (13C6, 15N2)- and l-arginine (13C6, 15N4)-preloaded 2-chlorotrityl resins (Cambridge Isotope Laboratories, Tewksbury, MA) were used as a solid support. Once synthesized, the peptides were cleaved from the resin using a scavenger-containing trifluoroacetic acid-water cleavage solution and precipitated by the addition of cold ether. Crude peptides were purified using HPLC (Jasco PU-2089) equipped with a semipreparative Kromasil C18 column (Teknokroma, Barcelona, Spain). Purity and labeling efficiency were analyzed with mass spectrometry using a MALDI-TOF/TOF 4800 instrument (AB Sciex) and freeze-dried.
Prior to quantification, SIL peptides, isotopically labeled with either (13C6,15N2) lysine (+8 Da) or (13C6, 15N4) arginine (+10 Da), were reconstituted in 30% acetonitrile and 1% formic acid to produce a 5 nmol/μl stock solution. The stock solution was further diluted with 2% acetonitrile and 0.1% formic acid as required.
Protein quantification by multiple reaction monitoring.
The development of multiple reaction monitoring (MRM) methods and protein quantification were performed using Skyline version 4.2.0.19009 (MacCoss Lab Software, Seattle, WA). In MRM assays, the first quadrupole (Q1) acts as an m/z filter of the precursor ion of the different peptides, and after the fragmentation in the collision cell (Q2), predefined product ions are selected in the second mass filter (Q3). The precursor/product ion pair is called a transition (Fig. 1). Five transitions were selected for most peptides based on the intensity of y- or b-fragment ions in the MS/MS spectra obtained in the LC-MS/MS analysis described in “Protein Identification Using LC-MS/MS and In Silico Digestion,” above. MRM analyses were conducted using the nano-LC system (AB Sciex) coupled to the 4000QTRAP mass spectrometer (AB Sciex) with the chromatographic and source parameter settings described in “Protein Identification Using LC-MS/MS and In Silico Digestion,” above. MRM transitions for each peptide were recorded with a dwell time of 20 ms. Collision energies were automatically computed using the embedded rolling collision energy equations of the Skyline software. To confirm the identity of peptides, an MRM-initiated detection and sequencing (MIDAS) experiment was performed for each peptide. The mass spectrometer was instructed to switch from MRM to enhanced product ion scanning mode when an individual MRM transition signal exceeded 2,000 counts. MS/MS data were analyzed using an in-house Mascot server version 2.3. The data were compared against data deposited in the corresponding in-house database.
For stable isotope dilution, 20 to 2,000 fmol of SIL peptides (depending on the signal intensity of the endogenous peptide) were spiked into trypsin-digested Cry protein samples. To quantify the tryptic peptides in the Cry samples, the sum of the transition signal intensities of the endogenous peptides was calculated in reference to the sum of the transition intensities of the SIL peptides.
Purification of Bt crystals from Bt-based insecticides.
The crystals of Bt serovar kurstaki ABTS-351 and Bt serovar aizawai ABTS-1857 were recovered directly from water-dispersible granules of the commercial products DiPel DF (Kenogard, Valent BioScience Corp., Barcelona, Spain; manufacturing batch 261-355-PG; manufactured in January 2016) and XenTari GD (Kenogard; manufacturing batch 264-637-PG; manufactured in April 2016), respectively. Similarly, the crystals of Bt serovar israelensis AM65-52 and Bt serovar tenebrionis NB-176 were recovered directly from suspension concentrate formulations of the commercial products VectoBac 12AS (Kenogard; manufacturing batch 276-006; manufactured in April 2017) and Novodor (Kenogard; manufacturing batch 272-803-PG; manufactured in December 2016), respectively. In all cases, a sample of 1 g (solid formulations) or 1 ml (liquid formulations) was taken and washed six times in cold Milli-Q water by centrifugation at 15,000 × g for 10 min. The final pellet was resuspended in 1 ml of 10 mM KCl, and parasporal crystals were then purified as described in “Purification of Parasporal Crystals,” above.
Total DNA extraction, genome sequencing, and computational analysis.
Bt strains ABTS-351, ABTS-1857, AM65-52, and NB-176 were directly isolated from the commercial products DiPel DF, XenTari GD, VectoBac 12AS, and Novodor, respectively. Each bacterial strain was grown at 28°C for 12 h in 5 ml of sterile CCY medium (49). Total DNA (chromosomal and plasmid DNA) was extracted from vegetative cells following the protocol for DNA extraction from Gram-positive bacteria supplied in the Wizard genomic DNA purification kit (Promega, Madison, WI). Purified DNA samples were used for the preparation of DNA libraries and were sequenced using an Illumina NextSeq 500 Sequencer (Genomics Research Hub Laboratory, Cardiff University, UK). The resulting reads were assembled using CLC Genomic Workbench version 10.1.1 (Qiagen, Hilden, Germany) with the de novo assembly tool. Contigs were then analyzed using BLASTp (51) against a custom insecticidal toxin database constructed using insecticidal protein sequences obtained from GenBank and the BtToxin-scanner (13).
Determination of the relative proportion of Cry and Cyt proteins in Bt commercial products.
Protein quality and band patterns were analyzed as described in “Polyacrylamide Gel Electrophoresis of Bt Crystal Proteins,” above, and quantification and tryptic digestion of the purified crystals were performed (see “Protein Quantification and Tryptic Digestion of Cry Proteins,” above). Protein identification was achieved with LC-MS/MS analyses, and in silico trypsin digestions were then performed using the amino acid sequences of the proteins identified with IDA analysis (see “Protein Identification Using LC-MS/MS and In Silico Digestion,” above) of Bt strains ABTS-351, ABTS-1857, AM65-52, and NB-176; SIL peptides were synthesized and prepared prior to quantification (see “Synthesis of SIL Peptides for Protein Quantification,” above). Finally, MRM analyses were performed as described in “Protein Quantification by Multiple Reaction Monitoring,” above, to quantify the protein content of each commercial product.
Data availability.
The GenBank accession numbers for the proteins discussed in this article can be found in Table 3.
ACKNOWLEDGMENT
This study received financial support from the Programa Nacional Español de I + D + I (projects AGL2015-70584-C2-2-R and RTI2018-095204-B-C22).
REFERENCES
- 1.Olson S. 2015. An analysis of the biopesticide market now and where it is going. Outlook Pest Man 26:203–206. doi: 10.1564/v26_oct_04. [DOI] [Google Scholar]
- 2.Raymond B, Federici BA. 2017. In defence of Bacillus thuringiensis, the safest and most successful microbial insecticide available to humanity—a response to EFSA. FEMS Microbiol Ecol 93:fix084. doi: 10.1093/femsec/fix084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel JP. 2001. The mammalian safety of Bacillus thuringiensis-based insecticides. J Invertebr Pathol 77:13–21. doi: 10.1006/jipa.2000.5000. [DOI] [PubMed] [Google Scholar]
- 4.Palma L, Muñoz D, Berry C, Murillo J, Caballero P. 2014. Bacillus thuringiensis toxins: an overview of their biocidal activity. Toxins (Basel) 6:3296–3325. doi: 10.3390/toxins6123296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bravo A, Gill SS, Soberón M. 2007. Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon 49:423–435. doi: 10.1016/j.toxicon.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Koni PA, Ellar DJ. 1996. Structure of the mosquitocidal δ-endotoxin CytB from Bacillus thuringiensis sp. kyushuensis and implications for membrane pore formation. J Mol Biol 257:129–152. doi: 10.1006/jmbi.1996.0152. [DOI] [PubMed] [Google Scholar]
- 7.Bravo A. 1997. Phylogenetic relationships of Bacillus thuringiensis δ-endotoxin family proteins and their functional domains. J Bacteriol 179:2793–2801. doi: 10.1128/jb.179.9.2793-2801.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Frankenhuyzen K. 2009. Insecticidal activity of Bacillus thuringiensis crystal proteins. J Invertebr Pathol 101:1–16. doi: 10.1016/j.jip.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 9.Ibrahim MA, Griko N, Junker M, Bulla LA. 2010. Bacillus thuringiensis: a genomics and proteomics perspective. Bioeng Bugs 1:31–50. doi: 10.4161/bbug.1.1.10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Floore TG. 2006. Mosquito larval control practices: past and present. J Am Mosq Control Assoc 22:527–533. doi: 10.2987/8756-971X(2006)22[527:MLCPPA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 11.Kati H, Sezen K, Demirbağ Z. 2007. Characterization of a highly pathogenic Bacillus thuringiensis strain isolated from common cockchafer, Melolontha melolontha. Folia Microbiol (Praha) 52:146–152. doi: 10.1007/BF02932153. [DOI] [PubMed] [Google Scholar]
- 12.Porcar M, Juárez-Pérez V. 2003. PCR-based identification of Bacillus thuringiensis pesticidal crystal genes. FEMS Microbiol Rev 26:419–432. doi: 10.1111/j.1574-6976.2003.tb00624.x. [DOI] [PubMed] [Google Scholar]
- 13.Ye W, Zhu L, Liu Y, Crickmore N, Peng D, Ruan L, Sun M. 2012. Mining new crystal protein genes from Bacillus thuringiensis on the basis of mixed plasmid-enriched genome sequencing and a computational pipeline. Appl Environ Microbiol 78:4795–4801. doi: 10.1128/AEM.00340-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dankocsik C, Donovan WP, Jany CS. 1990. Activation of a cryptic crystal protein gene of Bacillus thuringiensis subspecies kurstaki by gene fusion and determination of the crystal protein insecticidal specificity. Mol Microbiol 4:2087–2094. doi: 10.1111/j.1365-2958.1990.tb00569.x. [DOI] [PubMed] [Google Scholar]
- 15.Hodgman TC, Ziniu Y, Ming S, Sawyer T, Nicholls CM, Ellar DJ. 1993. Characterization of a Bacillus thuringiensis strain which is toxic to the housefly Musca domestica. FEMS Microbiol Lett 114:17–22. doi: 10.1016/0378-1097(93)90135-o. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto T. 1983. Identification of entomocidal toxins of Bacillus thuringiensis by high performance liquid chromatography. J Gen Microbiol 129:2595–2603. doi: 10.1099/00221287-129-8-2595. [DOI] [Google Scholar]
- 17.Pusztai-Carey M, Carey PR, Lessard T, Yaguchi M. October 1994. Isolation, quantitation and purification of insecticidal proteins from Bacillus thuringiensis. US patent no. 5,356,788.
- 18.Masson L, Erlandson M, Puzstai-Carey M, Brousseau R, Juárez-Pérez V, Frutos R. 1998. A holistic approach for determining the entomopathogenic potential of Bacillus thuringiensis strains. Appl Environ Microbiol 64:4782–4788. doi: 10.1128/AEM.64.12.4782-4788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Lai C, Su R, Zhang M, Xiong Y, Qing H, Deng Y. 2012. Quantification of Cry1Ab in genetically modified maize leaves by liquid chromatography multiple reaction monitoring tandem mass spectrometry using 18O stable isotope dilution. Analyst 137:2699–2705. doi: 10.1039/c2an35383k. [DOI] [PubMed] [Google Scholar]
- 20.Yang Q, Tang S, Rang J, Zuo M, Ding X, Sun Y, Feng P, Xia L. 2015. Detection of toxin proteins from Bacillus thuringiensis strain 4.0718 by strategy of 2D-LC–MS/MS. Curr Microbiol 70:457–463. doi: 10.1007/s00284-014-0747-9. [DOI] [PubMed] [Google Scholar]
- 21.Dulmage HT, Boening OP, Rehnborg CS, Hansen GD. 1971. A proposed standardized bioassay for formulations of Bacillus thuringiensis based on the international unit. J Invertebr Pathol 18:240–245. doi: 10.1016/0022-2011(71)90151-0. [DOI] [PubMed] [Google Scholar]
- 22.Bernardi O, Sorgatto RJ, Barbosa AD, Domingues FA, Dourado PM, Carvalho RA, Martinelli S, Head GP, Omoto C. 2014. Low susceptibility of Spodoptera cosmioides, Spodoptera eridania and Spodoptera frugiperda (Lepidoptera: Noctuidae) to genetically-modified soybean expressing Cry1Ac protein. Crop Prot 58:33–40. doi: 10.1016/j.cropro.2014.01.001. [DOI] [Google Scholar]
- 23.Gallien S, Duriez E, Domon B. 2011. Selected reaction monitoring applied to proteomics. J Mass Spectrom 46:298–312. doi: 10.1002/jms.1895. [DOI] [PubMed] [Google Scholar]
- 24.Lange V, Picotti P, Domon B, Aebersold R. 2008. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol 4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holman SW, Sims PFG, Eyers CE. 2012. The use of selected reaction monitoring in quantitative proteomics. Bioanalysis 4:1763–1786. doi: 10.4155/bio.12.126. [DOI] [PubMed] [Google Scholar]
- 26.Brun V, Masselon C, Garin J, Dupuis A. 2009. Isotope dilution strategies for absolute quantitative proteomics. J Proteomics 72:740–749. doi: 10.1016/j.jprot.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 27.Kamiie J, Ohtsuki S, Iwase R, Ohmine K, Katsukura Y, Yanai K, Sekine Y, Uchida Y, Ito S, Terasaki T. 2008. Quantitative atlas of membrane transporter proteins: development and application of a highly sensitive simultaneous LC/MS/MS method combined with novel in-silico peptide selection criteria. Pharm Res 25:1469–1483. doi: 10.1007/s11095-008-9532-4. [DOI] [PubMed] [Google Scholar]
- 28.Alves G, Ogurtsov AY, Yu YK. 2011. Assigning statistical significance to proteotypic peptides via database searches. J Proteomics 74:199–211. doi: 10.1016/j.jprot.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuster B, Schirle M, Mallick P, Aebersold R. 2005. Scoring proteomes with proteotypic peptide probes. Nat Rev Mol Cell Biol 6:577–583. doi: 10.1038/nrm1683. [DOI] [PubMed] [Google Scholar]
- 30.Hoofnagle AN, Whiteaker JR, Carr SA, Kuhn E, Liu T, Massoni SA, Thomas SN, Townsend R, Zimmerman LJ, Boja E, Chen J, Crimmins DL, Davies SR, Gao Y, Hiltke TR, Ketchum KA, Kinsinger CR, Mesri M, Meyer MR, Qian WJ, Schoenherr RM, Scott MG, Shi T, Whiteley GR, Wrobel JA, Wu C, Ackermann BL, Aebersold R, Barnidge DR, Bunk DM, Clarke N, Fishman JB, Grant RP, Kusebauch U, Kushnir MM, Lowenthal MS, Moritz RL, Neubert H, Patterson SD, Rockwood AL, Rogers J, Singh RJ, Van Eyk JE, Wong SH, Zhang S, Chan DW, Chen X, Ellis MJ, Liebler DC, Rodland KD, et al. 2016. Recommendations for the generation, quantification, storage, and handling of peptides used for mass spectrometry-based assays. Clin Chem 62:48–69. doi: 10.1373/clinchem.2015.250563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Picotti P, Aebersold R. 2012. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods 9:555–566. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 32.Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham AJL, Keshishian H, Hall SC, Allen S, Blackman RK, Borchers CH, Buck C, Cardasis HL, Cusack MP, Dodder NG, Gibson BW, Held JM, Hiltke T, Jackson A, Johansen EB, Kinsinger CR, Li J, Mesri M, Neubert TA, Niles RK, Pulsipher TC, Ransohoff D, Rodriguez H, Rudnick PA, Smith D, Tabb DL, Tegeler TJ, Variyath AM, Vega-Montoto LJ, Wahlander Å, Waldemarson S, Wang M, Whiteaker JR, Zhao L, Anderson NL, Fisher SJ, Liebler DC, Paulovich AG, Regnier FE, Tempst P, Carr SA. 2009. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol 27:633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reiter L, Rinner O, Picotti P, Hüttenhain R, Beck M, Brusniak MY, Hengartner MO, Aebersold R. 2011. MProphet: automated data processing and statistical validation for large-scale SRM experiments. Nat Methods 8:430–435. doi: 10.1038/nmeth.1584. [DOI] [PubMed] [Google Scholar]
- 34.Willard BB, Kinter M. 2001. Effects of the position of internal histidine residues on the collision-induced fragmentation of triply protonated tryptic peptides. J Am Soc Mass Spectrom 12:1262–1271. doi: 10.1016/S1044-0305(01)00312-9. [DOI] [PubMed] [Google Scholar]
- 35.Bravo A, Likitvivatanavong S, Gill SS, Soberón M. 2011. Bacillus thuringiensis: a story of a successful bioinsecticide. Insect Biochem Mol Biol 41:423–431. doi: 10.1016/j.ibmb.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hernández-Martínez P, Ferré J, Escriche B. 2008. Susceptibility of Spodoptera exigua to 9 toxins from Bacillus thuringiensis. J Invertebr Pathol 97:245–250. doi: 10.1016/j.jip.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 37.Porcar M, Martinez C, Caballero P. 2000. Host range and gene contents of Bacillus thuringiensis strains toxic towards Spodoptera exigua. Entomol Exp Appl 97:339–346. doi: 10.1046/j.1570-7458.2000.00748.x. [DOI] [Google Scholar]
- 38.Dominic Amalraj D, Sahu SS, Jambulingam P, Boopathi Doss PS, Kalyanasundaram M, Das PK. 2000. Efficacy of aqueous suspension and granular formulations of Bacillus thuringiensis (Vectobac) against mosquito vectors. Acta Trop 75:243–246. doi: 10.1016/s0001-706x(00)00054-1. [DOI] [PubMed] [Google Scholar]
- 39.Ben-Dov E. 2014. Bacillus thuringiensis subsp. israelensis and its dipteran-specific toxins. Toxins (Basel) 6:1222–1243. doi: 10.3390/toxins6041222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crickmore N, Bone EJ, Williams JA, Ellar DJ. 1995. Contribution of the individual components of the δ-endotoxin crystal to the mosquitocidal activity of Bacillus thuringiensis subsp. israelensis. FEMS Microbiol Lett 131:249–254. [Google Scholar]
- 41.Monnerat R, Pereira E, Teles B, Martins E, Praça L, Queiroz P, Soberon M, Bravo A, Ramos F, Soares CM. 2014. Synergistic activity of Bacillus thuringiensis toxins against Simulium spp. larvae. J Invertebr Pathol 121:70–73. doi: 10.1016/j.jip.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 42.Berry C, O’Neil S, Ben-Dov E, Jones AF, Murphy L, Quail MA, Holden MTG, Harris D, Zaritsky A, Parkhill J. 2002. Complete sequence and organization of pBtoxis, the toxin-coding plasmid of Bacillus thuringiensis subsp. israelensis. Appl Environ Microbiol 68:5082–5095. doi: 10.1128/AEM.68.10.5082-5095.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Becker N, Ludwig M, Su T. 2018. Lack of resistance in Aedes vexans field populations after 36 years of Bacillus thuringiensis subsp. israelensis applications in the upper Rhine Valley, Germany. J Am Mosq Control Assoc 34:154–157. doi: 10.2987/17-6694.1. [DOI] [PubMed] [Google Scholar]
- 44.Ferré J, Van Rie J. 2002. Biochemistry and genetics of insect resistance to Bacillus thuringiensis. Annu Rev Entomol 47:501–533. doi: 10.1146/annurev.ento.47.091201.145234. [DOI] [PubMed] [Google Scholar]
- 45.Donovan WP, Donovan JC, Slaney AC. 2000. Bacillus thuringiensis CryET33 and CryET34 compositions and uses therefor. U.S. patent 6063756A.
- 46.de Maagd RA, Bravo A, Berry C, Crickmore N, Schnepf HE. 2003. Structure, diversity, and evolution of protein toxins from spore-forming entomopathogenic bacteria. Annu Rev Genet 37:409–433. doi: 10.1146/annurev.genet.37.110801.143042. [DOI] [PubMed] [Google Scholar]
- 47.Hokkanen HMT, Hajek A. 2003. Environmental impacts of microbial insecticides: need and methods for risk assessment. Kluwer Academic, Dordrecht, The Netherlands. [Google Scholar]
- 48.Jouzani GS, Valijanian E, Sharafi R. 2017. Bacillus thuringiensis: a successful insecticide with new environmental features and tidings. Appl Microbiol Biotechnol 101:2691–2711. doi: 10.1007/s00253-017-8175-y. [DOI] [PubMed] [Google Scholar]
- 49.Stewart GS, Johnstone K, Hagelberg E, Ellar DJ. 1981. Commitment of bacterial spores to germinate: a measure of the trigger reaction. Biochem J 198:101–106. doi: 10.1042/bj1980101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas WE, Ellar DJ. 1983. Bacillus thuringiensis var israelensis crystal delta-endotoxin: effects on insect and mammalian cells in vitro and in vivo. J Cell Sci 60:181–197. [DOI] [PubMed] [Google Scholar]
- 51.Altschul S, Gish W, Miller W, Myers E. 1990. Basic Local Alignment Search Tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 52.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The GenBank accession numbers for the proteins discussed in this article can be found in Table 3.